Abstract
The SARS-CoV-2 pandemic has continued for about three years since emerging in late December 2019, resulting in millions of deaths. Therefore, there is an urgent need to develop a safe and effective vaccine to control SARS-CoV-2. In this study, we developed a bacterium-like particle vaccine that displays the SARS-CoV-2 receptor binding domain (RBD) (named Trim-RBD-GEM) using the GEM-PA system. We evaluated the immunogenicity and protective efficacy of the Trim-RBD-GEM vaccine with the oil-in-water adjuvant AddaVax in C57BL/6 N mice intramuscularly. We found that Trim-RBD-GEM&AddaVax induced high levels of humoral immunity in C57BL/6 N mice. Additionally, the lung virus loads in the immunized group were significantly decreased compared to the adjuvant control and mock groups. Therefore, this vaccine provides protection against lethal infection in a C57BL/6 N mouse model. Our Trim-RBD-GEM&AddaVax vaccine is potentially a promising, rapid, and safe subunit vaccine for preventing and controlling SARS-CoV-2.
Keywords: SARS-CoV-2 pandemic, Bacterium-like particle, AddaVax adjuvant, Al(OH)3 adjuvant
1. Introduction
The SARS-CoV-2 pandemic has resulted in more than 759 million confirmed human cases and over 6 million deaths as of March 2023(). Notably, vaccinations were one of the primary measures against the SARS-CoV-2 pandemic (Cattel et al., 2022). As a result, several kinds of vaccines have been approved worldwide, including protein subunit vaccines, inactivated vaccines, viral vector vaccines, and nucleic acid vaccines (Chi et al., 2022; Hadj Hassine, 2022). These vaccines provided an effective measure to control the pandemic.
According to reports published by the World Health Organization, as of March 2023, 183 SARS-CoV-2 vaccines are in clinical studies, and 59 subunit vaccine candidates are currently in the clinical phase. The subunit vaccine has its advantages, including easy production and high safety, and has been used to prevent diseases such as hepatitis B and tetanus (Khalaj-Hedayati et al., 2020; Liu et al., 2020; Wang et al., 2020b). On the other hand, it has some noted disadvantages, such as lower immunogenicity and difficulty in inducing a strong and long-term immune response when administered alone. Adjuvants can stimulate and enhance the strength and durability of the immune response in the subunit vaccine (Kaur and Gupta, 2020). Therefore, some adjuvants have been approved for commercial vaccines. Notably, a novel adjuvant named AddaVax, an oil-in-water adjuvant containing squalene, can induce type 1 T-helper (Th1) and type 2 T-helper (Th2) immune responses (Li et al., 2012; Ten Brinke et al., 2007).
SARS-CoV-2 contains various proteins such as nucleocapsid, spike, membrane, envelope, and other proteins. The target antigen for SARS-CoV-2 vaccines is the spike protein, which contains S1 and S2 structures, with S1 functioning to bind to the receptor of the host cells and S2 functioning to be responsible for the fusion of the cell membrane with the viral membrane (Wang et al., 2020a). Importantly, the S1 subunit contains the N-terminal domain (NTD) and the receptor binding domain (RBD). The RBD region contains about 220 amino acids. In the variants of SARS-CoV-2, some mutations have developed in this protein and possibly influenced the antigenicity of the virus (Tao et al., 2021). Notably, the RBD is the key neutralizing antibody production region, allowing focus on the immune response to interfering with receptor binding, which can effectively block viral infection (Dai et al., 2020; Yang et al., 2021). Therefore, the RBD is often selected as the target antigen in SARS-CoV-2 vaccines.
In recent years, Lactococcus lactis expression systems have become widely used in the vaccine and biopharmaceutical industries, consisting of Gram-positive enhanced matrix (GEM) and a protein anchor (PA) (Lee et al., 2003; Zhang et al., 2022b). After trichloroacetic acid (TCA) treatment, the Gram-positive GEM particles obtained from Lactococcus lactis retain the peptidoglycan structure with removal of the protein and DNA (Li et al., 2019). PA is produced by the lysin motif (LysM) of the C-terminal peptidoglycan-binding structural domain of AcmA, an autolysin of Lactobacillus, and consists of three LysMs (Buist et al., 2008). The GEM-PA system could display exogenous proteins on the surface of Gram-positive bacteria, in which PA is anchored to the surface of the cell wall in a non-covalent form, and binding of the exogenous protein to the cell wall of the bacteria enhances the immunogenicity in vaccines (Michon et al., 2016). The GEM-PA system has several advantages when used as a vaccine. First, this system has a high safety profile, and Lactobacillus is a food-grade bacterium with a long usage history (Li et al., 2018; Raha et al., 2005). Second, it has a high expression level of the exogenous protein, and although it is non-covalent binding, the affinity of the binding is robust (Bosma et al., 2006; Steen et al., 2003). Third, it can be used as a mucosal adjuvant to improve the ability of a mucosal system to induce an immune response (Audouy et al., 2007; Li et al., 2019).
In this study, we designed a protein subunit vaccine based on the GEM-PA system with RBD proteins named Trim-RBD-GEM. This subunit vaccine induced high antigen-specific immune responses in the C57BL/6 N mouse model and provided protective efficacy against the SARS-CoV-2 challenge.
2. Materials and methods
2.1. Cells, bacteria, and viruses
Vero E6 cells were cultured in Dulbecco's modified Eagle medium (DMEM) containing sodium pyruvate, 2 mM l-glutamine, 1% penicillin-streptomycin (Gibco) and 10% Fetal Bovine Serum (Gibco). Adherent Spodoptera frugiperda 9 (Sf9) cells from Life Technologies (USA) were cultured in Grace's Insect Cell Culture medium (Gibco) with 10% Fetal Bovine Serum and 1% penicillin-streptomycin at 27 °C; Sf9 suspension cells were cultured in SIM SF Expression Medium (Sino Biological) with 1% penicillin-streptomycin at 27 °C. Lactobacillus MG1363 was cultured in M17 medium (Cat: HB0391, Qingdao Hope Biotechnology Company) with 1% glucose at 30 °C and was stored in our laboratory.
The isolated SARS-CoV-2 virus (BetaCoV/Beijing/IME-BJ05-2020, abbreviated as BJ05, GenBank accession No. MT291835) and a SARS-CoV-2 mouse-adapted strain (C57MA14) were grown in Vero E6 cells for 3 days at 37 °C and 5% CO2. The C57MA14 strain was passaged 14 times in aged C57BL/6 N mice. Furthermore, BJ05 and C57MA14 were cultured as described previously (Yan et al., 2022).
2.2. Recombinant RBD proteins
The Trim-RBD was constructed as follows: the RBD region was selected for codon optimization based on the S gene of the SARS-CoV-2 WH01 strain published in GenBank (GenBank: MN908947.3), ligated with the T4F motif to form a trimer-based highly aggregated state, and then linked to the protein anchor by a flexible linker (GGGGS) (Walsh et al., 2020). Importantly, PA can bind to GEM particles in a non-covalent manner, thus binding the target protein to the GEM particle surface (Bosma et al., 2006). The recombinant plasmid was synthesized by Sangon Biotech (Shanghai) Co., Ltd, and then linked to the PA gene through the flexible linker (GGGGS) and attached to the pFastbac1 vector. The recombinant plasmid was transformed into DH10Bac competent cells and blue-white spot screened, and recombinant baculovirus was rescued. Additionally, the recombinant baculovirus was transfected into Sf9 adherent cells with Lipofectamine 3000 according to the Bac-to-Bac expression system standard protocol to obtain the recombinant baculovirus. Furthermore, we continued inoculation into Sf9 cells at a 5% volume ratio to obtain the P3 generation virus. Then we inoculated into Sf9 suspension cells to obtain the P4 generation virus, and the P4 generation virus was used for subsequent experiments.
2.3. GEM preparation
In short, Lactococcus lactis MG1363 was grown in GM17 for 16 h at 30 °C and shaken at 195 rpm on a temperate orbital shaker. The Lactococcus lactis MG1363 was collected by centrifugation at 6000 rpm for 20 min, and the precipitate was resuspended with phosphate-buffered saline (PBS). After a thoroughly cleaning, the precipitate was resuspended in 10% trichloroacetic acid (Sigma-Aldrich, USA) and boiled for 60 min at 98 °C. Subsequently, the precipitate was resuspended in PBS after being washed three times. Approximately 2.5 × 109 particles were considered one unit (1U) of GEM particles.
To determine the maximum amount of virus supernatant and GEM particles bound, 0 mL, 2 mL, 4 mL, 6 mL, 8 mL, and 10 mL of virus supernatant were bound to 1 U GEM particles and subjected to SDS-PAGE experiments. Additionally, different concentrations of BSA (0.5 mg/mL, 0.25 mg/mL, 0.125 mg/mL, 0.06245 mg/mL, 0.03125 mg/mL, 0.015625 mg/mL) and Trim-RBD-GEM were subjected to SDS-PAGE after reduction treatment. The bound pellets were then reduced and electrophoresed using 8–16% SDS-polyacrylamide gels. Additionally, the SDS-PAGE gel was incubated at room temperature for 1 h using Komas Brilliant Blue staining solution (Beyotime) and decolorized for 2 h using decolorizing solution (Beyotime).
The 8 ml supernatant containing Trim-RBD protein was mixed gently at 37 °C and 60 min with 1U of GEM particles. The particles that had bonded to the Trim-RBD protein were removed by centrifugation them at 4500 rpm for 5 min before being washed them with PBS and resuspended. SDS-PAGE and Western blot methods were used to analyze and identify the samples. Notably, GEM particles were named Trim-RBD-GEM when banded with the Trim-RBD protein.
2.4. Immunofluorescence and Western blot
Indirect immunofluorescence was used to identify the Trim-RBD protein. Sf9 adherent cells were cultured in 6-well plates at a density of 5 × 104 cells per well, infected with recombinant baculovirus at 0.5 MOI, and then incubated at 27 °C for 48 h. Noninfected cells served as a negative control. After infection, cells were fixed with 4% paraformaldehyde (Solarbio) for 10 min at room temperature, washed with PBS, and then permeabilized with 0.2% Triton X-100 for 15 min at room temperature, followed by incubation with SARS-CoV-2 Spike RBD antibody for 1 h at 37 °C. The cells were then washed four times with PBS and incubated in a 1:1000 dilution of goat anti-rabbit IgG/FITC antibody (Cat: ab6717, Abcam) for 1 h at 37 °C. The plates were washed four times with PBS and photographed using a ZEISS fluorescence microscope (Zeiss, Germany).
Western blot assay was performed on both the cell lysate and supernatant from Sf9 cells infected with recombinant baculovirus to verify the expression of the Trim-RBD protein. After passaging at a density of 1.8 × 106 cells in Erlenmeyer shake flasks, Sf9 suspension cells were infected with a 3% volume ratio of recombinant baculovirus and incubated at 27 °C and shaken at 120 rpm for 60 h. Proteins from cell lysates and supernatants were boiled at 100 °C for 10 min, electrophoresed on 8–16% SDS-polyacrylamide gels, and then transferred to 0.45 μm polyvinylidene difluoride membranes (Biorad) using the wet transfer method. Membranes were then blocked with TBST solution (Beyotime) containing 3% bovine serum albumin (Sigma) for 1.5 h at room temperature. After blocking, the membrane was incubated with SARS-CoV-2 Spike Antibody, Rabbit PAb (Cat: GTX635692, GeneTex), diluted 1:2000 in TBST solution containing 10% bovine serum albumin (Sigma) overnight at 4 °C. The membrane was washed three times with TBST (0.5% Tween-20). Following washing, the membrane was incubated with goat anti-rabbit IgG/HRP secondary antibody (Cat: BS13278, Bioworld) diluted 1:25,000 in TBST containing 5% bovine serum albumin for 1 h at room temperature on the next day. After TBST (0.5% Tween-20) washes, the membrane was photographed using Gel Image System version 4.2 (Tanon).
2.5. Thin layer chromatography (TLC) and transmission electron microscopy (TEM)
The gels obtained by SDS-PAGE were subjected to the TLC method for precise quantification of proteins. The mobile thin layer plate was placed in the appropriate position, the scanning wavelength was set to 595 nm, the scan mode was set to Zigzag, Lambda was selected as Single according to the requirements, and the scanning was started. Furthermore, the data were processed after the scanning was completed.
In preparation for TEM, the Trim-RBD-GEM vaccines were fixed with 2.5% glutaraldehyde for 24 h at 4 °C. The GEM particles were also post-fixed with 1% osmium tetroxide. The samples were dehydrated using a succession of ethanol gradients and acetone solutions before being embedded in an Epon 812 resin mixture. Using a JEM 1200EXII electron microscope for imaging (JEOL, Japan), ultrathin slices (70 nm) were stained with 2% uranyl acetate in 70% ethanol and Reynold's lead solution.
2.6. Vaccination and virus challenge experiments
Specific Pathogen Free (SPF) 6-8-week-old female C57BL/6 N mice were purchased from the Charles River Experimental Animal Center. We used C57BL/6 N mice to evaluate the immunogenicity and protective efficacy of the Trim-RBD-GEM subunit vaccine. All female C57BL/6 N mice were randomly divided into seven groups (n = 5). Notably, the immunized mouse group contained the Trim-RBD group, the Trim-RBD-GEM group, the Trim-RBD-GEM&AddaVax group, the Trim-RBD-GEM&Al(OH)3 group, the GEM&AddaVax group, the GEM&Al(OH)3 group, and the mock group. Mice in the Trim-RBD group, Trim-RBD-GEM group, Trim-RBD-GEM&AddaVax group, and Trim-RBD-GEM&Al(OH)3 group were injected intramuscularly (IM) with a 10 μg dose of vaccine. The adjuvant group used 10 μg of GEM mixed with adjuvant, and mice in the mock group were injected with 100 μL PBS. Furthermore, AddaVax adjuvant was purchased from InvivoGen, France, and Al(OH)3 adjuvant was purchased from Thermo Fisher Scientific. When adjuvants were used, the antigen and adjuvant were mixed in a 1:1 ratio before immunization. Mice were immunized three times on days 0, 14, and 28. Furthermore, blood samples were collected from the retroorbital venous plexus 10 days after immunization, and the serum was used to measure humoral antibodies.
According to the immunogenicity of these groups, we chose the Trim-RBD-GEM&AddaVax group, the GEM&AddaVax group, and the mock group for the challenge. Mice were anesthetized with isoflurane and challenged with the intranasally mouse-adapted SARS-CoV-2 strain (C57MA14) with 50 LD50 (5 × 105 TCID50) in a volume of 50 μL (Yan et al., 2022). The body weights and survival were recorded for 14 days. Nasal turbinates and lungs were collected on day 3 after the challenge. Moreover, tissues were used to measure the virus loads via reverse transcription-quantitative PCR and virus titers by 50% tissue culture infectious dose (TCID50) assays. Furthermore, the lungs were fixed in 4% paraformaldehyde for 72 h. Sections were then stained with hematoxylin and eosin (H&E), and the remaining mice were humanely euthanized when the experiment was finished.
2.7. Enzyme-linked immunosorbent assay (ELISA)
ELISA was used to assess the humoral antibody titers in serum samples. The coated antigen protein was purchased from Sino Biological (Cat: 40592-V08H). RBD protein (2 μg/mL, 100 μL/well) was coated on 96-well plates (Corning) at 4 °C overnight and then blocked with 1% bovine serum albumin (Sigma) at 37 °C for 2 h. We then serially diluted the serum 2-fold, starting at 1:200, and added it to the plates. The plates were first incubated for 1 h at 37 °C, then washed three times with PBST (PBS containing 0.05% Tween-20), and then incubated for 1 h at 37 °C with goat anti-mouse IgG-HRP at a dilution of 1:25,000 (Cat: BS12478, Bioworld). We also tested for IgG subtypes, including IgG1, IgG2a, IgG2b, IgG2c, and IgG3 at a dilution of 1:5000 (Cat: 1071–05, 1081–05, 1091–05, 1078–05, 1101–05, Southern Biotech) by serially diluting the sera 2-fold from 1:200 and adding them to the plates the same as IgG. Then the plates were washed four times with PBST, and TMB substrate was used to produce the signals (Sigma). The colorimetric reaction was halted by applying a stop buffer, and the optical density (OD) was determined at 450 nm using an ELISA plate reader (Bio-Rad).
2.8. Neutralization assay
The neutralization assay measured the neutralizing antibody titers in the serum. In this study, the BJ05 strain was utilized. 100 TCID50 of SARS-CoV-2 was incubated with serially diluted serum at 37 °C for 1 h and incubated with 2 × 104 Vero E6 cells in 96-well plates to observe the cytopathic effects. The final results were recorded by observing the cytopathic effects at 72 h. The final neutralizing antibody titer was defined as the maximum dilution that could prevent the virus from infecting the cells.
2.9. ELISpot assay
The secretion levels of IL-4 and IFN-γ cytokines in mouse splenocytes were measured using the ELISpot kit (Mabtech). The splenocytes from each group on day 35 after the first immunization were isolated, and the density was adjusted to 2.5 × 106 cells/mL and stimulated with 10 μg/mL of RBD protein for 36 h. After the cells were washed, antibodies against IL-4 and IFN-γ were added. Following an additional washing, antibodies against Streptavidin-HRP were added and subsequently washed with water as spots appeared. Finally, we used the AID enzyme-linked spot analyzer to calculate the number of spots.
2.10. Intracellular cytokine staining (ICS) assay
The secretions of IFN-γ, IL-4, and IL-2 in the splenocytes were measured using flow cytometry. The density of splenocytes was adjusted to 2.5 × 106 cells/mL, stimulated with 10 μg/mL of SARS-CoV-2 RBD protein for 20 h, and incubation was done Protein Transport Inhibitor Cocktail for 4 h. The plates were washed three times with a wash buffer. CD3-BV510, CD4-FITC, and CD8-BV785 antibodies were used to stain the splenocytes. The cells were gently mixed with these antibodies and incubated at 4 °C for 30 min in the dark. Then we added Fixation/Permeabilization solution in the dark, and incubation proceeded for 20 min at 4 °C. Furthermore, IL-2-APC, TNF-BV421, and IFN-γ-PE antibodies (BD Biosciences) were used to stain the splenocytes. Finally, the fluorescent signal was detected by using the ACEA system.
2.11. Virus titers and virus loads in lungs and nasal turbinate
Nasal turbinate and lungs were collected and ground for 400 s with a Tissue Lyser (Qiagen, Germany), then centrifuged at 4 °C at 17,000 rpm for 10 min, and the supernatants were collected for subsequent experiments. To detect the virus titers in the supernatants by the TCID50 method, Vero E6 cells were inoculated into 96-well plates at 5 × 104/mL per well starting from 1:10 at 10 times dilution of the supernatants to be tested. When the confluence reached 85%, the supernatants to be tested were added and incubated for 1 h at 37 °C in an incubator containing 5% CO2. Subsequently, DMEM containing 2% FBS and 1% penicillin were added, and the cytopathic effects were observed under a light microscope after 72 h incubation.
Quantitative real-time PCR was performed using the QIAamp Viral RNA Mini kit (Qiagen) to extract RNA, and then the 2019-nCoV nucleic acid test kit was used to detect the N gene, with the following primers and probes: NF (5′GGGGAACTTCTCCTGCTAGAAT-3′); NR(5′-CAGACATTTTGCTCTCAAGCTG-3′); and NP (5′-FAMTTGCTGCTGCTTGACAGATT-TAMRA-3′).
2.12. Statistical analysis
GraphPad Prism 9.0 software (GraphPad Software, San Diego, CA, USA) was used for statistical analysis. One-way ANOVA tests or two-way ANOVA tests were used and analyzed when comparing the data from more than two groups. P values less than 0.05 were regarded as significant.
2.13. Ethics statement
All mice were in good health and were housed and had access to water, food, and periodic light in good health, according to the National Standards of Laboratory Animal Requirements (GB 14925–2010). All C57BL/6 N mouse experiments were approved by the Animal Welfare and Ethics Committee of the Changchun Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (approval number: IACUC of AMMS-11-2020-020).
3. Results
3.1. Construction and identification of RBD proteins
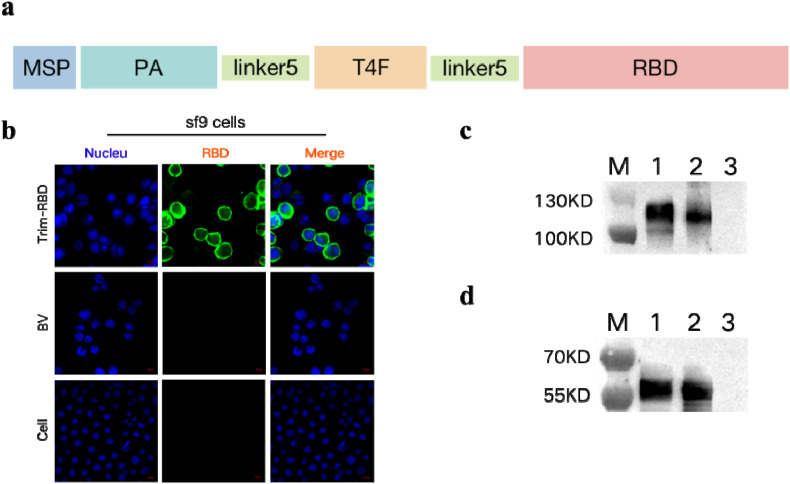
The design strategy of the Trim-RBD protein is shown in Fig. 1 a. The recombinant baculovirus, which contains the MSP signal peptide, PA, T4F, and RBD proteins, was linked with the linker (GGGGS). We employed indirect immunofluorescence and Western blot assays to identify the Trim-RBD protein. Indirect immunofluorescence experiments demonstrated a clear green fluorescent signal for Sf9 cells infected with recombinant baculovirus but not for uninfected cells (Fig. 1b). Protein expression of the Trim-RBD protein was analyzed by Western blot. Sf9 cells were infected with the virus at a 3% volume ratio and collected as supernatant and precipitate after non-reduction and reduction treatments, respectively. The results showed bands between 100 and 130 KDa and 55 and 70 KDa. This was consistent with the protein size, and the results indicate that Trim-RBD protein was expressed in the supernatants (Fig. 1c and d).
Fig. 1.
Construction and identification of Trim-RBD protein.
(a) Design of Trim-RBD structures, MSP: mastoparan; PA: protein anchor; linker5: (GGGGS)5 linker; T4F: T4 fibritin foldon domain. (b) Indirect immunofluorescence identification of recombinant baculovirus, infected with Sf9 cells at 3% volume ratio, incubated with rabbit anti-SARS-CoV-2 RBD protein antibody. (c) Western blot identification of recombinant baculovirus with non-reduction treatment; lane 1 is recombinant baculovirus supernatant, and lane 2 is recombinant baculovirus precipitate, and lane 3 is baculovirus (BV) culture suspensions. (d) Western blot identification of recombinant baculovirus with reduction treatment; lane 1 is recombinant baculovirus supernatant, lane 2 is recombinant baculovirus precipitate, and lane 3 is BV culture cell suspension.
3.2. The construction and characterization of Trim-RBD-GEM
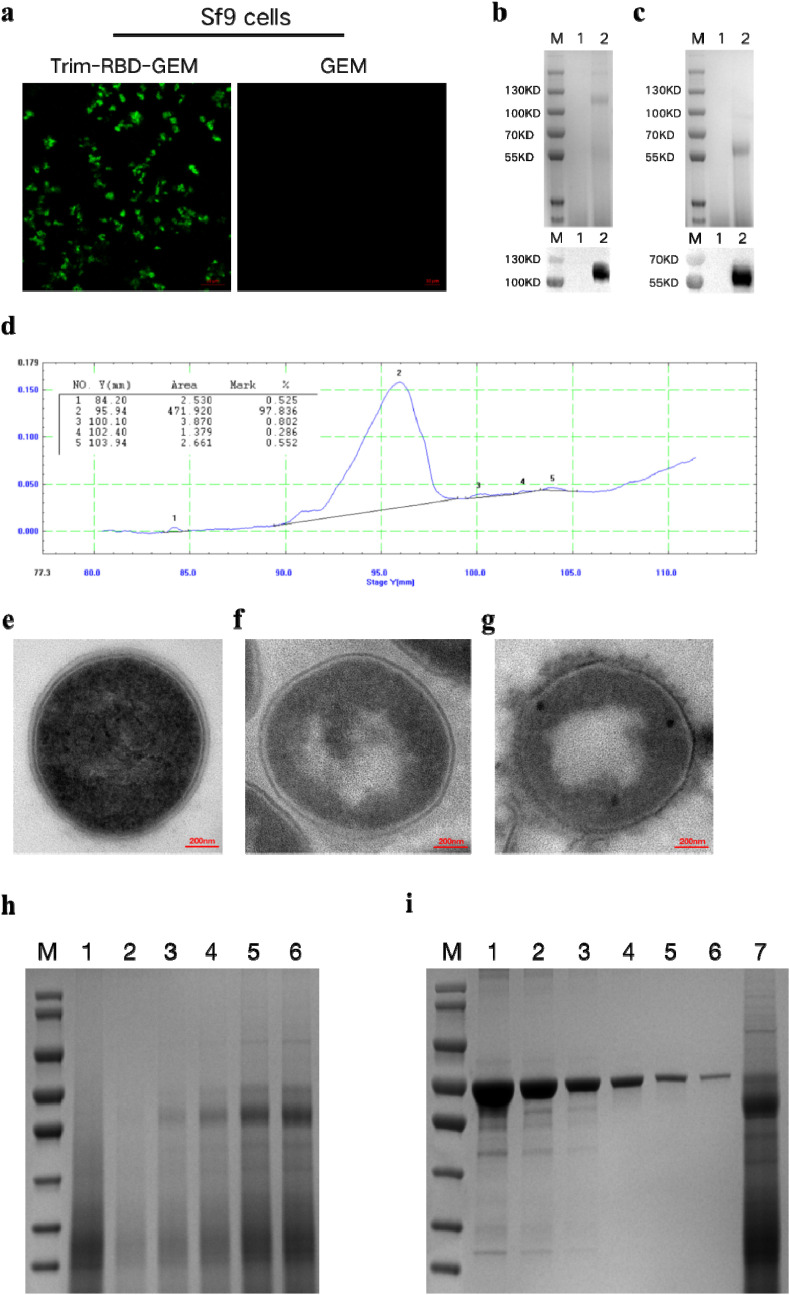
Indirect immunofluorescence experiments were performed to determine whether the Trim-RBD protein was successfully bound to GEM particles. The Trim-RBD-GEM had a significant green fluorescent signal, while no fluorescence was seen for GEM particles as a negative control (Fig. 2 a). In SDS-PAGE and Western blot, bands could be seen between 100 and 130 KDa for the non-reducing Trim-RBD-GEM and between 55 and 70 KDa for the reducing treatment, consistent with the Trim-RBD results (Fig. 2b and c). Analysis of Trim-RBD-GEM after reduction treatment by TLC experiments showed a purity of over 97% (Fig. 2d). To determine whether the Trim-RBD protein was successfully bound to the GEM particles, we analyzed the morphological differences between untreated L. lactis MG1363 and L. lactis MG1363 heat-treated with 10% TCA and GEM incorporating exogenous proteins by TEM. The internal structure of untreated L. lactis MG1363 was homogeneous, and the cell wall and contents of the bacteria could be observed. In contrast, we could see the peptidoglycan skeleton in the GEM particles, but the internal structure was changed. The results of Trim-RBD-GEM demonstrate that the protein is attached to the surface of the GEM particles (Fig. 2e–g). Following grayscale analysis, it was found that the binding amount reached saturation when 8 mL of using BSA was used as the standard protein, a standard curve was made with Y = 0.0000002X-0.2074 (Y is the protein concentration and X is the volume), and the binding amount of Trim-RBD-GEM was calculated as 68.62 μg/U (Fig. 2h–i).
Fig. 2.
Identification of Trim-RBD-GEM.
(a) Indirect immunofluorescence identification of Trim-RBD after binding to GEM particles, with GEM as a negative control. (b) SDS-PAGE and Western blot results of Trim-RBD-GEM non-reduction treatment; lane 1 is GEM particles, and lane 2 is the non-reduction treatment of Trim-RBD-GEM. (c) SDS-PAGE and Western blot results of Trim-RBD-GEM reduction treatment; lane 1 is the GEM particles, and lane 2 is the Trim-RBD-GEM reduction treatment. (d) Thin slice scanning results after Trim-RBD-GEM reduction treatment. (e) TEM images of MG1363. (f) TEM images of GEM particles. (g) TEM images of Trim-RBD-GEM. (h) 1 U GEM particles were mixed with 0 mL, 2 mL, 4 mL, 6 mL, 8 mL, and 10 mL of virus culture supernatants. (i) Protein amounts of fusion proteins were determined for lanes 1–6 for different concentrations of BSA protein and lane 7 for Trim-RBD-GEM.
3.3. The humoral immune responses induced by Trim-RBD-GEM in mice
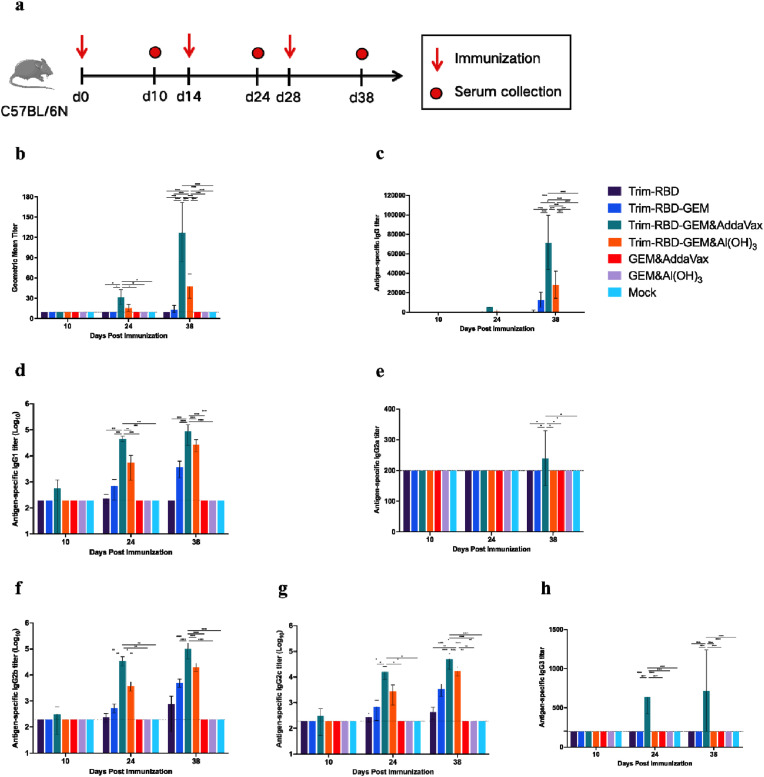
The immunogenicity of Trim-RBD-GEM was evaluated in C57BL/6 N mice, which were injected intramuscularly for a total of three doses (days 0, 14, and 28) (Fig. 3 a). Blood was collected and isolated from the serum 10 days after each immunization, and ELISA antibody and neutralizing antibody levels were measured. The neutralizing antibody levels in the serum were measured using the BJ05 strain, and the results showed that the Trim-RBD-GEM&AddaVax group had significantly higher neutralizing antibody levels than the other groups (Fig. 3b). We observed that the adjuvant significantly increased the antibody titers. The ELISA antibody titers were significantly higher in the Trim-RBD-GEM&AddaVax group and Trim-RBD-GEM&Al(OH)3 group than in the no-adjuvant group after the first, second, and third immunizations under the same conditions (Fig. 3c). Additionally, the level of ELISA antibody in the Trim-RBD-GEM group was significantly higher than that in the Trim-RBD group, indicating that the GEM particles also enhanced the immune response of mice. The same results were found for the IgG subtypes, with the Trim-RBD-GEM&AddaVax and Trim-RBD-GEM&Al(OH)3 groups having significantly higher antibody levels in IgG1, IgG2a, IgG2b, IgG2c, and IgG3 than in the no-adjuvant group (Fig. 3d–h). Therefore, our experiment results showed that the Trim-RBD-GEM vaccine group produced humoral immune responses in the immunization mice.
Fig. 3.
Immunization schedule and immunogenicity of Trim-RBD-GEM in mice. C57BL/6 N mice (n = 5) were immunized 3 times at days 0, 14, and 28. (a) Immunization schedule. (b) SARS-CoV-2 Neutralization Titer. (c) SARS-CoV-2 RBD-specific IgG titer. (d) SARS-CoV-2 RBD-specific IgG1 titer. (e) SARS-CoV-2 RBD-specific IgG2a titer. (f) SARS-CoV-2 RBD-specific IgG2b titer. (g) SARS-CoV-2 RBD-specific IgG2c titer. (h) SARS-CoV-2 RBD-specific IgG3 titer. Data are shown as the mean ± SD. All the data were analyzed using a two-way ANOVA combined with a Šídák's multiple comparisons test over three groups. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
3.4. Vaccination with Trim-RBD-GEM protects against the SARS-CoV-2 challenge
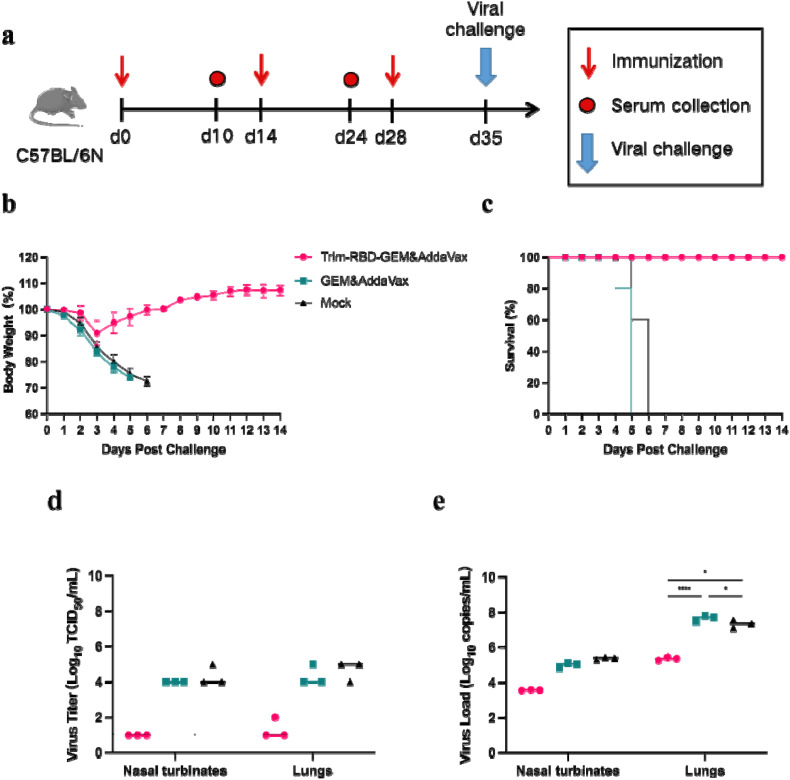
The protective efficacy of the Trim-RBD-GEM&AddaVax vaccines was evaluated using C57BL/6 N mice. Mice were divided into Trim-RBD-GEM&AddaVax, GEM&AddaVax, and mock groups and immunized intramuscularly in three doses on days 0, 14, and 28. On day 35, after the first immunization, mice were challenged with 50 LD50 C57MA14 (Fig. 4 a). After the SARS-CoV-2 C57MA14 challenge, mice in the Trim-RBD-GEM&AddaVax group had little weight loss, whereas mice in the GEM&AddaVax and mock groups had severe weight loss (Fig. 4b). In addition, the Trim-RBD-GEM&AddaVax group mice had a survival rate of 100%, while the adjuvant control and mock group mice lost less than 75% of their body weight on days 5 and 6 after the challenge and had a survival rate of 0%. Mice in the Trim-RBD-GEM&AddaVax group provided lethal protection (Fig. 4c).
Fig. 4.
Immune protection of Trim-RBD-GEM against the SARS-CoV-2 challenge in mice. C57BL/6 N mice (n = 5) were immunized intramuscularly with 10 μg Trim-RBD-GEM&AddaVax, GEM&AddaVax, and mock 3 times, on days 0, 14, and 28. The mice were then challenged on day 35 with 50 LD50 C57MA14 intranasally. (a) Immunization schedule. (b) Body weights. (c) Survivals. (d) Virus titers (TCID50/mL). (e) Virus loads.
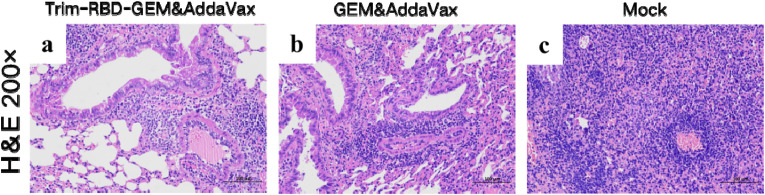
Lung tissue and nasal turbinates were collected by dissection at 3dpi for reverse transcription-quantitative PCR assay and virus titer determination. Viral RNA was barely detectable in lung tissue and nasal turbinates in the Trim-RBD-GEM&AddaVax group, whereas high levels of viral RNA copies could be detected in the GEM&AddaVax and mock groups (Fig. 4d and e). Given the location of the viral challenge, detection of viral RNA in nasal swabs is possible and does not necessarily indicate viral replication in vivo. Notably, histopathological analysis of lungs confirmed no significant changes after infection with SARS-CoV-2 C57MA14 in mice inoculated with Trim-RBD-GEM&AddaVax (Fig. 5 ). Therefore, these results indicate that our Trim-RBD-GEM with AddaVax adjuvant could protect mice from the SARS-CoV-2 challenge.
Fig. 5.
Pathological changes in lungs after SARS-CoV-2 challenge in C57BL/6N mice. (a) HE results of Trim-RBD-GEM&AddaVax group. (b) HE results of GEM&AddaVax group. (c) HE results of mock group.
4. Discussion
The SARS-CoV-2 pandemic has been ongoing for three years, and safe and effective vaccines are essential to prevent and reduce the spread of the disease (Zhang et al., 2022a). Currently, several SARS-CoV-2 vaccines have primarily been used worldwide. Notably, these include the mRNA vaccines, the virus-vector vaccines, the inactivated vaccines, and the subunit vaccines (Cui et al., 2022). Accounting for 32%, according to the data published by WHO, exploratory research on SARS-CoV-2 vaccines is one of the current priorities, and recombinant subunit vaccines play a significant role in developing SARS-CoV-2 vaccines.
In this study, the RBD region was selected for codon optimization based on the S gene and linked to the T4F motif to form a trimeric-dominated highly aggregated state, which was then linked to the PA gene via a flexible linker (GGGGS). The flexible linker is generally composed of polar or non-polar amino acids, which provide functional and structural flexibility for linkage between proteins, and the hydrogen bonds formed by serine and threonine with water molecules allow the fusion protein to maintain structural and functional stability even in aqueous solution (Chen et al., 2013). The flexible linker added in this study should ensure that the GEM-PA can bind in the correct position and ensure the stability of the SARS-CoV-2 RBD protein structure to not affect immunogenicity. Finally, the (GGGGS)*5 sequence was chosen for linkage. In mice, Trim-RBD-GEM was immunized with AddaVax adjuvant and Al(OH)3 adjuvant. The results showed that it induced cellular and humoral immunity in mice and provided better immune protection (Fig. S1 and Fig. S2).
The S protein contains non-neutralizing epitopes that increase the risk of antibody-dependent enhancement (ADE). In contrast, the RBD protein is smaller, reducing exposure to non-neutralizing epitopes, thereby reducing the risk of pathology and focusing the immune response on interfering viruses, thereby reducing the risk of ADE (Dai and Gao, 2021). Furthermore, the combination of RBD and neutralizing antibody reduces the interaction between ACE-2 and RBD, thus preventing the virus from entering the cells, making the RBD protein an attractive target for antigen design (Dai et al., 2020; Robbiani et al., 2020; Shi et al., 2020; Walls et al., 2020; Zhu et al., 2020).
The GEM-PA surface display system has been applied to animal models, and the results show that it can block viruses, bacteria, parasites, etc. (Heine et al., 2015; Li et al., 2016, 2022; Nganou-Makamdop et al., 2012; Rigter et al., 2013). Non-transgenic, non-living GEM has little recombinant DNA, reducing the risk of transmission to the environment (Li et al., 2019). Additionally, GEM particles bind non-covalently to PA with high affinity, with approximately 1 million PA molecules binding to one particle with high binding efficiency (Ramirez et al., 2010). Lactococcus lactis is very safe, frequently used for cheese making, and can be an alternative to using attenuated vaccines. Furthermore, the GEM-PA display system allows multiple PA fusion proteins to be anchored to a single particle, enabling the production and development of multivalent vaccines and providing a new strategy for vaccine study (van Roosmalen et al., 2006).
We purified the protein using preliminary purification by one-step centrifugation to obtain a recombinant protein with more than 97% purity. Notably, this dramatically saved time and antigen usage compared to S-trimer (Liang et al., 2021). In addition, Trim-RBD-GEM can be stored for a long time at 2–8 °C, which makes transportation to remote areas more accessible and convenient.
However, our experiments have some limitations. The immunogenicity induced by the subunit vaccine was weak, and mice in the Trim-RBD-GEM&AddaVax group only reached a neutralizing antibody titer of 1:128 after the third immunization. However, ZF2001, a vaccine also designed to target SARS-CoV-2 RBD as an antigen, induced a neutralizing antibody level of 1:256 on day 35 after immunization of mice and induced a specific IgG antibody titer of 47051 (An et al., 2022). In contrast, mice in the Trim-RBD-GEM&AddaVax group induced a mean specific IgG antibody titer of 71680 on day 38 after the initial immunization. The specific IgG antibody titers induced on day 35 in rodents immunized with the 9 μg dose of S-Trimer combined with AS03 adjuvant were 23207 (Liang et al., 2021), and the IgG antibody titers induced on day 35 in mice immunized with the 10 μg antigen dose of NVX-Co2373 were 139000 (Tian et al., 2021). These results demonstrate that the level of immunity induced by the subunit vaccine was low. However, the data after the challenge in mice can show that our vaccine Trim-RBD-GEM&AddaVax is effective, providing lethal protection to mice. Based on the humoral immunity, cellular immunity levels, and protective efficacy, we next want to study the antibody levels of Trim-RBD-GEM&AddaVax against the Omicron strain. The Omicron variant with the risk of immune escape poses a new challenge to achieving population-wide herd immunity, being more insidious and infectious; it spreads faster, and makes outbreak prevention and control more complex than with other variant strains (Wang et al., 2023). As we know from the literature, infection with Omicron is ineffective in boosting immunity, and we still need vaccinations (Guo et al., 2022; Khandia et al., 2022; Vitiello et al., 2022). Therefore, we need to develop highly immunogenic and safe vaccines against Omicron variants as essential to control the global SARS-CoV-2 pandemic and prevent further disease and death (Zhang et al., 2022c).
According to previous studies, with vaccination with two doses of the mRNA vaccine, compared to the original strain or the Delta strain, the neutralizing antibodies against the Omicron mutant strain were barely detectable in the serum after five months. However, in individuals who received the third booster shot, the neutralizing effect of the serum against the three mutant strains was improved after seven months, and the neutralizing effect for Omicron was still significantly low (Planas et al., 2022). Although the effectiveness of current vaccines against Omicron is reduced substantially, booster vaccination can increase the level of neutralizing antibodies. Therefore, vaccination still has a positive effect on preventing SARS-CoV-2 pandemics.
5. Conclusions
In summary, the Trim-RBD-GEM vaccine was successfully constructed and induced high levels of antibody titers after immunization with AddaVax adjuvant, significantly higher than with other groups. The Trim-RBD-GEM&AddaVax group provided lethal protection to the mice and significantly reduced the virus loads and virus titers of the lungs. Our vaccine provides a new choice for preventing and controlling SARS-CoV-2 pandemics.
Consent for publication
All authors approved the submission of the manuscript for publication.
Formatting of funding sources
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Rina Su: Methodology, Validation, Investigation, Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. Zhuangzhuang Shi: Methodology, Validation, Investigation, Formal analysis, Validation, Visualization, Writing – original draft, Writing – review & editing. Entao Li: Conceptualization, Methodology. Menghan Zhu: Investigation. Dongxu Li: Investigation. Xiawei Liu: Investigation. Yue Sun: Investigation. Na Feng: Investigation, Data curation. Jianzhong Wang: Investigation, Data curation. Tiecheng Wang: Investigation, Data curation. Xianzhu Xia: Supervision, Project administration. Weiyang Sun: Resources, Data curation, Validation, Writing – review & editing. Yuwei Gao: Conceptualization, Methodology, Validation, Resources, Data curation, Writing – review & editing, Supervision, Project administration.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Handling Editor: Alexander E. Gorbalenya
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.virol.2023.06.005.
Abbreviations
- GEM
Gram-positive enhancer matrix
- PA
protein anchor
- RBD
receptor binding domain
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- TCA
trichloroacetic acid
- BLP
bacterium-like particle
- NTD
N-terminal domain
- LysM
lysin motif
- Sf9
Spodoptera frugiperda 9
- MOI
multiplicity of infection
- PBS
phosphate-buffered saline
- TBST
Tris buffer solution and Tween 20
- BSA
bovine serum albumin
- TLC
thin layer chromatography
- TEM
transmission electron microscopy
- IM
intramuscularly
- BV
baculovirus
- H&E
hematoxylin and eosin
- OD
optical density
- TCID50
50% tissue culture infective dose
- FBS
Fetal Bovine Serum
- DMEM
Dulbeccos's Modified Eagle Medium
- SPF
Specific Pathogen Free
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- LD50
50% lethal dose
- PCR
Polymerase Chain Reaction
- WHO
World Health Organization
- LPS
lipopolysaccharide
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- An Y., Li S., Jin X., Han J.B., Xu K., Xu S., Han Y., Liu C., Zheng T., Liu M., Yang M., Song T.Z., Huang B., Zhao L., Wang W., A R., Cheng Y., Wu C., Huang E., Yang S., Wong G., Bi Y., Ke C., Tan W., Yan J., Zheng Y.T., Dai L., Gao G.F. A tandem-repeat dimeric RBD protein-based covid-19 vaccine zf2001 protects mice and nonhuman primates. Emerg. Microb. Infect. 2022;11:1058–1071. doi: 10.1080/22221751.2022.2056524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audouy S.A., van Selm S., van Roosmalen M.L., Post E., Kanninga R., Neef J., Estevao S., Nieuwenhuis E.E., Adrian P.V., Leenhouts K., Hermans P.W. Development of lactococcal GEM-based pneumococcal vaccines. Vaccine. 2007;25:2497–2506. doi: 10.1016/j.vaccine.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Bosma T., Kanninga R., Neef J., Audouy S.A., van Roosmalen M.L., Steen A., Buist G., Kok J., Kuipers O.P., Robillard G., Leenhouts K. Novel surface display system for proteins on non-genetically modified gram-positive bacteria. Appl. Environ. Microbiol. 2006;72:880–889. doi: 10.1128/AEM.72.1.880-889.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buist G., Steen A., Kok J., Kuipers O.P. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- Cattel L., Giordano S., Traina S., Lupia T., Corcione S., Angelone L., La Valle G., De Rosa F.G., Cattel F. Vaccine development and technology for SARS-CoV-2: current insight. J. Med. Virol. 2022;94:878–896. doi: 10.1002/jmv.27425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zaro J.L., Shen W.C. Fusion protein linkers: property, design and functionality. Adv. Drug Deliv. Rev. 2013;65:1357–1369. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi W.Y., Li Y.D., Huang H.C., Chan T.E.H., Chow S.Y., Su J.H., Ferrall L., Hung C.F., Wu T.C. COVID-19 vaccine update: vaccine effectiveness, SARS-CoV-2 variants, boosters, adverse effects, and immune correlates of protection. J. Biomed. Sci. 2022;29:82. doi: 10.1186/s12929-022-00853-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Zhao K., Zhang C., Lin J., Sun S., Li Q., Du L., Zhang C., Liu J., Gao F., He W., Gao Y., Guo Z., Guan J. Parapoxvirus-based therapy eliminates SARS-CoV-2-loaded fine aerosol and blocks viral transmission in hamster models. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.1086627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat. Rev. Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., An Y., Cheng Y., Li S., Liu M., Yang M., Li Y., Cheng H., Yuan Y., Zhang W., Ke C., Wong G., Qi J., Qin C., Yan J., Gao G.F. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182:722–733 e711. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z., Zhang C., Zhang C., Cui H., Chen Z., Jiang X., Wang T., Li Y., Liu J., Wan Z., Meng K., Li J., Tong Y., Gao Y. SARS-CoV-2-related pangolin coronavirus exhibits similar infection characteristics to SARS-CoV-2 and direct contact transmissibility in hamsters. iScience. 2022;25 doi: 10.1016/j.isci.2022.104350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadj Hassine I. Covid-19 vaccines and variants of concern: a review. Rev. Med. Virol. 2022;32 doi: 10.1002/rmv.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine S.J., Franco-Mahecha O.L., Chen X., Choudhari S., Blackwelder W.C., van Roosmalen M.L., Leenhouts K., Picking W.L., Pasetti M.F. Shigella IpaB and IpaD displayed on L. lactis bacterium-like particles induce protective immunity in adult and infant mice. Immunol. Cell Biol. 2015;93:641–652. doi: 10.1038/icb.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaur S.P., Gupta V. COVID-19 Vaccine: a comprehensive status report. Virus Res. 2020;288 doi: 10.1016/j.virusres.2020.198114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaj-Hedayati A., Chua C.L.L., Smooker P., Lee K.W. Nanoparticles in influenza subunit vaccine development: immunogenicity enhancement. Influenza Other Respir. Viruses. 2020;14:92–101. doi: 10.1111/irv.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandia R., Singhal S., Alqahtani T., Kamal M.A., El-Shall N.A., Nainu F., Desingu P.A., Dhama K. Emergence of SARS-CoV-2 Omicron (B.1.1.529) variant, salient features, high global health concerns and strategies to counter it amid ongoing COVID-19 pandemic. Environ. Res. 2022;209 doi: 10.1016/j.envres.2022.112816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Choi J.H., Xu Z. Microbial cell-surface display. Trends Biotechnol. 2003;21:45–52. doi: 10.1016/s0167-7799(02)00006-9. [DOI] [PubMed] [Google Scholar]
- Li E., Chi H., Huang P., Yan F., Zhang Y., Liu C., Wang Z., Li G., Zhang S., Mo R., Jin H., Wang H., Feng N., Wang J., Bi Y., Wang T., Sun W., Gao Y., Zhao Y., Yang S., Xia X. A novel bacterium-like particle vaccine displaying the MERS-CoV receptor-binding domain induces specific mucosal and systemic immune responses in mice. Viruses. 2019;11 doi: 10.3390/v11090799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H.B., Zhang J.Y., He Y.F., Chen L., Li B., Liu K.Y., Yang W.C., Zhao Z., Zou Q.M., Wu C. Systemic immunization with an epitope-based vaccine elicits a Th1-biased response and provides protection against Helicobacter pylori in mice. Vaccine. 2012;31:120–126. doi: 10.1016/j.vaccine.2012.10.091. [DOI] [PubMed] [Google Scholar]
- Li L., Qiao X., Chen J., Zhang Y., Zheng Q., Hou J. Surface-displayed porcine reproductive and respiratory syndrome virus from cell culture onto gram-positive enhancer matrix particles. J. Ind. Microbiol. Biotechnol. 2018;45:889–898. doi: 10.1007/s10295-018-2061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P.C., Qiao X.W., Zheng Q.S., Hou J.B. Immunogenicity and immunoprotection of porcine circovirus type 2 (PCV2) Cap protein displayed by Lactococcus lactis. Vaccine. 2016;34:696–702. doi: 10.1016/j.vaccine.2015.09.007. [DOI] [PubMed] [Google Scholar]
- Li W., Li J., Dai X., Liu M., Khalique A., Wang Z., Zeng Y., Zhang D., Ni X., Zeng D., Jing B., Pan K. Surface Display of porcine circovirus type 2 antigen protein cap on the spores of bacillus subtilis 168: an effective mucosal vaccine candidate. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1007202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J.G., Su D., Song T.Z., Zeng Y., Huang W., Wu J., Xu R., Luo P., Yang X., Zhang X., Luo S., Liang Y., Li X., Huang J., Wang Q., Huang X., Xu Q., Luo M., Huang A., Luo D., Zhao C., Yang F., Han J.B., Zheng Y.T., Liang P. S-Trimer, a COVID-19 subunit vaccine candidate, induces protective immunity in nonhuman primates. Nat. Commun. 2021;12:1346. doi: 10.1038/s41467-021-21634-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F.J., Shi D.Y., Li Z.Y., Lu J.S., Wang R., Pang X.B., Yang Z.X., Yu Y.Z. Evaluation of a recombinant tetanus toxin subunit vaccine. Toxicon. 2020;187:75–81. doi: 10.1016/j.toxicon.2020.08.001. [DOI] [PubMed] [Google Scholar]
- Michon C., Langella P., Eijsink V.G., Mathiesen G., Chatel J.M. Display of recombinant proteins at the surface of lactic acid bacteria: strategies and applications. Microb. Cell Factories. 2016;15:70. doi: 10.1186/s12934-016-0468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nganou-Makamdop K., van Roosmalen M.L., Audouy S.A.L., van Gemert G.-J., Leenhouts K., Hermsen C.C., Sauerwein R.W. Bacterium-like particles as multi-epitope delivery platform for Plasmodium berghei circumsporozoite protein induce complete protection against malaria in mice. Malar. J. 2012;11:50. doi: 10.1186/1475-2875-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Saunders N., Maes P., Guivel-Benhassine F., Planchais C., Buchrieser J., Bolland W.H., Porrot F., Staropoli I., Lemoine F., Pere H., Veyer D., Puech J., Rodary J., Baele G., Dellicour S., Raymenants J., Gorissen S., Geenen C., Vanmechelen B., Wawina-Bokalanga T., Marti-Carreras J., Cuypers L., Seve A., Hocqueloux L., Prazuck T., Rey F.A., Simon-Loriere E., Bruel T., Mouquet H., Andre E., Schwartz O. Considerable escape of SARS-CoV-2 Omicron to antibody neutralization. Nature. 2022;602:671–675. doi: 10.1038/s41586-021-04389-z. [DOI] [PubMed] [Google Scholar]
- Raha A.R., Varma N.R., Yusoff K., Ross E., Foo H.L. Cell surface display system for Lactococcus lactis: a novel development for oral vaccine. Appl. Microbiol. Biotechnol. 2005;68:75–81. doi: 10.1007/s00253-004-1851-8. [DOI] [PubMed] [Google Scholar]
- Ramirez K., Ditamo Y., Rodriguez L., Picking W.L., van Roosmalen M.L., Leenhouts K., Pasetti M.F. Neonatal mucosal immunization with a non-living, non-genetically modified Lactococcus lactis vaccine carrier induces systemic and local Th1-type immunity and protects against lethal bacterial infection. Mucosal Immunol. 2010;3:159–171. doi: 10.1038/mi.2009.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigter A., Widjaja I., Versantvoort H., Coenjaerts F.E., van Roosmalen M., Leenhouts K., Rottier P.J., Haijema B.J., de Haan C.A. A protective and safe intranasal RSV vaccine based on a recombinant prefusion-like form of the F protein bound to bacterium-like particles. PLoS One. 2013;8 doi: 10.1371/journal.pone.0071072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S., Hagglof T., Oliveira T.Y., Viant C., Hurley A., Hoffmann H.H., Millard K.G., Kost R.G., Cipolla M., Gordon K., Bianchini F., Chen S.T., Ramos V., Patel R., Dizon J., Shimeliovich I., Mendoza P., Hartweger H., Nogueira L., Pack M., Horowitz J., Schmidt F., Weisblum Y., Michailidis E., Ashbrook A.W., Waltari E., Pak J.E., Huey-Tubman K.E., Koranda N., Hoffman P.R., West A.P., Jr., Rice C.M., Hatziioannou T., Bjorkman P.J., Bieniasz P.D., Caskey M., Nussenzweig M.C. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Shan C., Duan X., Chen Z., Liu P., Song J., Song T., Bi X., Han C., Wu L., Gao G., Hu X., Zhang Y., Tong Z., Huang W., Liu W.J., Wu G., Zhang B., Wang L., Qi J., Feng H., Wang F.S., Wang Q., Gao G.F., Yuan Z., Yan J. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature. 2020;584:120–124. doi: 10.1038/s41586-020-2381-y. [DOI] [PubMed] [Google Scholar]
- Steen A., Buist G., Leenhouts K.J., El Khattabi M., Grijpstra F., Zomer A.L., Venema G., Kuipers O.P., Kok J. Cell wall attachment of a widely distributed peptidoglycan binding domain is hindered by cell wall constituents. J. Biol. Chem. 2003;278:23874–23881. doi: 10.1074/jbc.M211055200. [DOI] [PubMed] [Google Scholar]
- Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22:757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ten Brinke A., Karsten M.L., Dieker M.C., Zwaginga J.J., van Ham S.M. The clinical grade maturation cocktail monophosphoryl lipid A plus IFNgamma generates monocyte-derived dendritic cells with the capacity to migrate and induce Th1 polarization. Vaccine. 2007;25:7145–7152. doi: 10.1016/j.vaccine.2007.07.031. [DOI] [PubMed] [Google Scholar]
- Tian J.H., Patel N., Haupt R., Zhou H., Weston S., Hammond H., Logue J., Portnoff A.D., Norton J., Guebre-Xabier M., Zhou B., Jacobson K., Maciejewski S., Khatoon R., Wisniewska M., Moffitt W., Kluepfel-Stahl S., Ekechukwu B., Papin J., Boddapati S., Jason Wong C., Piedra P.A., Frieman M.B., Massare M.J., Fries L., Bengtsson K.L., Stertman L., Ellingsworth L., Glenn G., Smith G. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat. Commun. 2021;12:372. doi: 10.1038/s41467-020-20653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Roosmalen M.L., Kanninga R., El Khattabi M., Neef J., Audouy S., Bosma T., Kuipers A., Post E., Steen A., Kok J., Buist G., Kuipers O.P., Robillard G., Leenhouts K. Mucosal vaccine delivery of antigens tightly bound to an adjuvant particle made from food-grade bacteria. Methods. 2006;38:144–149. doi: 10.1016/j.ymeth.2005.09.015. [DOI] [PubMed] [Google Scholar]
- Vitiello A., Ferrara F., Auti A.M., Di Domenico M., Boccellino M. Advances in the Omicron variant development. J. Intern. Med. 2022;292:81–90. doi: 10.1111/joim.13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Fiala B., Schafer A., Wrenn S., Pham M.N., Murphy M., Tse L.V., Shehata L., O'Connor M.A., Chen C., Navarro M.J., Miranda M.C., Pettie D., Ravichandran R., Kraft J.C., Ogohara C., Palser A., Chalk S., Lee E.C., Guerriero K., Kepl E., Chow C.M., Sydeman C., Hodge E.A., Brown B., Fuller J.T., Dinnon K.H., 3rd, Gralinski L.E., Leist S.R., Gully K.L., Lewis T.B., Guttman M., Chu H.Y., Lee K.K., Fuller D.H., Baric R.S., Kellam P., Carter L., Pepper M., Sheahan T.P., Veesler D., King N.P. Elicitation of potent neutralizing antibody responses by designed protein nanoparticle vaccines for SARS-CoV-2. Cell. 2020;183:1367–1382 e1317. doi: 10.1016/j.cell.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R., Swanson K.A., Li P., Koury K., Kalina W., Cooper D., Fontes-Garfias C., Shi P.Y., Tureci O., Tompkins K.R., Lyke K.E., Raabe V., Dormitzer P.R., Jansen K.U., Sahin U., Gruber W.C. Safety and immunogenicity of two RNA-based covid-19 vaccine candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.Y., Zhao R., Gao L.J., Gao X.F., Wang D.P., Cao J.M. SARS-CoV-2: structure, biology, and structure-based therapeutics development. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.587269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Iketani S., Li Z., Liu L., Guo Y., Huang Y., Bowen A.D., Liu M., Wang M., Yu J., Valdez R., Lauring A.S., Sheng Z., Wang H.H., Gordon A., Liu L., Ho D.D. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186:279–286 e278. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Zhou X., Bian Y., Wang S., Chai Q., Guo Z., Wang Z., Zhu P., Peng H., Yan X., Li W., Fu Y.X., Zhu M. Dual-targeting nanoparticle vaccine elicits a therapeutic antibody response against chronic hepatitis B. Nat. Nanotechnol. 2020;15:406–416. doi: 10.1038/s41565-020-0648-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Li E., Wang T., Li Y., Liu J., Wang W., Qin T., Su R., Pei H., Wang S., Feng N., Zhao Y., Yang S., Xia X., Gao Y. Characterization of two heterogeneous lethal mouse-adapted SARS-CoV-2 variants recapitulating representative aspects of human COVID-19. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.821664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Li Y., Dai L., Wang J., He P., Li C., Fang X., Wang C., Zhao X., Huang E., Wu C., Zhong Z., Wang F., Duan X., Tian S., Wu L., Liu Y., Luo Y., Chen Z., Li F., Li J., Yu X., Ren H., Liu L., Meng S., Yan J., Hu Z., Gao L., Gao G.F. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect. Dis. 2021;21:1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Cui H., Guo Z., Chen Z., Yan F., Li Y., Liu J., Gao Y., Zhang C. SARS-CoV-2 virus culture, genomic and subgenomic RNA load, and rapid antigen test in experimentally infected Syrian hamsters. J. Virol. 2022;96 doi: 10.1128/jvi.01034-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Yan F., Liu D., Li E., Feng N., Xu S., Wang H., Gao Y., Yang S., Zhao Y., Xia X. Bacterium-like particles displaying the rift valley fever virus Gn head protein induces efficacious immune responses in immunized mice. Front. Microbiol. 2022;13 doi: 10.3389/fmicb.2022.799942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Tan W., Lou Z., Zhao Y., Zhang J., Liang H., Li N., Zhu X., Ding L., Huang B., Zhou W., Guo Y., Yang Z., Qiao Y., He Z., Ma B., He Y., Zhu D., Wang Z., Chang Z., Zhao X., Wang W., Xu Y., Zhu H., Zheng X., Wang C., Xu G., Wu G., Wang H., Yang X. Vaccination with Omicron inactivated vaccine in pre-vaccinated mice protects against SARS-CoV-2 prototype and Omicron variants. Vaccines (Basel) 2022;10 doi: 10.3390/vaccines10071149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., Zhao X., Huang B., Shi W., Lu R., Niu P., Zhan F., Ma X., Wang D., Xu W., Wu G., Gao G.F., Tan W., China Novel Coronavirus I., Research T. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.