Abstract
Helix–hairpin–helix (HhH) is a widespread motif involved in non-sequence-specific DNA binding. The majority of HhH motifs function as DNA-binding modules, however, some of them are used to mediate protein–protein interactions or have acquired enzymatic activity by incorporating catalytic residues (DNA glycosylases). From sequence and structural analysis of HhH-containing proteins we conclude that most HhH motifs are integrated as a part of a five-helical domain, termed (HhH)2 domain here. It typically consists of two consecutive HhH motifs that are linked by a connector helix and displays pseudo-2-fold symmetry. (HhH)2 domains show clear structural integrity and a conserved hydrophobic core composed of seven residues, one residue from each α-helix and each hairpin, and deserves recognition as a distinct protein fold. In addition to known HhH in the structures of RuvA, RadA, MutY and DNA-polymerases, we have detected new HhH motifs in sterile alpha motif and barrier-to-autointegration factor domains, the α-subunit of Escherichia coli RNA-polymerase, DNA-helicase PcrA and DNA glycosylases. Statistically significant sequence similarity of HhH motifs and pronounced structural conservation argue for homology between (HhH)2 domains in different protein families. Our analysis helps to clarify how non-symmetric protein motifs bind to the double helix of DNA through the formation of a pseudo-2-fold symmetric (HhH)2 functional unit.
INTRODUCTION
The vast growth of sequence and structural data necessitates the classification of proteins to understand the relationship between sequence, structure and function. Such classification should bring together our knowledge on protein structure with theoretical views of evolutionary processes in protein molecules. Evolution-based structural classification is particularly difficult for small domains, since structure-based similarity statistics become marginal due to the small number of secondary structural elements and superimposable residues. Furthermore, with decreasing domain size it is increasingly difficult to separate similarities originating from common evolutionary history (homology) from those resulting from the general rules of molecular packing in the absence of evolutionary connection (analogy). With the development of powerful profile-based tools such as HMMer (1–3) and PSI-BLAST (4,5) for detection of very weak but significant sequence similarity, it became traditional to use them in structure-classification studies (6–11). It is believed that sequence similarity detected with the profile-based methods indicates homology (7). Thus combination of sequence-based and structure-based methods has been proven most efficient for recognition of remote evolutionary connections between protein domains to aid their classification (6). Here we apply this combined approach to the analysis of helix–hairpin–helix (HhH) proteins.
The HhH motif plays a role in non-sequence-specific DNA binding and has been detected in a variety of protein families exemplified by DNA-polymerases, NAD+-dependent DNA ligases, S13 ribosomal proteins and DNA glycosylases (12,13). Structurally, the motif forms into a pair of anti-parallel α-helices connected by a hairpin-like loop. This loop is involved in interactions with DNA (14–16) and usually contains a consensus glycine-hydrophobic amino acid-glycine sequence pattern (GhG), where h is a hydrophobic residue, most commonly Ile, Val or Leu. The last G of the consensus serves as an N-terminal cap of the second α-helix, and the hydrophobic residue h contributes to the interactions between the two α-helices of the motif. The two α-helices are packed at an acute angle of ~25–50° that dictates the characteristic pattern of hydrophobicity in the sequences (Fig. 1) (13). This packing of the two α-helices is different from the ones found in other α-helical DNA-binding proteins. For example, the helix–turn–helix (HTH) motif, which is also formed by a pair of helices, can be easily distinguished by the packing of the helices at an almost right angle.
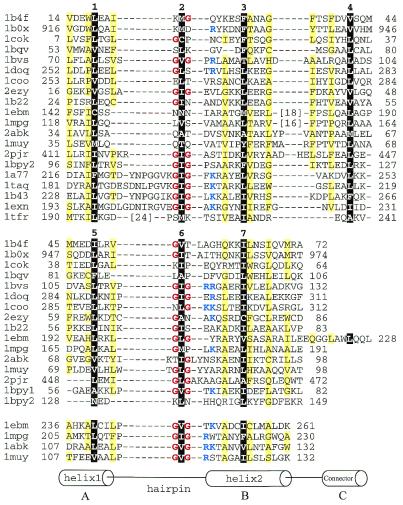
Figure 1.
Structure-based sequence alignment of HhH proteins. For each sequence, PDB entry name and starting and ending residue numbers are given. Protein name, chain name (if any) and gene identifier (gi) number for each entry are: 1b4f, human EphB2 receptor, chain A, 4558093; 1b0x, mouse EphA4 receptor tyrosine kinase, chain A, 4929864; 1cok, human p73 C-terminal domain, chain A, 5822025; 1bqv, mouse Ets-a transcription factor pointed domain, 3891925; 1bvs, Mycobacterium leprae DNA-helicase RuvA middle domain, chain A, 3660156; 1doq, T.thermophilus RNA-polymerase α-subunit C-terminal domain, chain A, 6730428; 1coo, E.coli RNA-polymerase α-subunit C-terminal domain, 1421046; 2ezy, human BAF, chain A, 4389121; 1b22, human DNA repair protein Rad51 N-terminal domain, chain A, 6730074; 1ebm, human 8-oxoguanine glycosylase central domain, chain A, 2078294; 1mpg, E.coli 3-methyladenine DNA glycosylase II 2 C-terminal domains, chain A, 2914353; 2abk, E.coli endonuclease III, 1311214; 1muy, E.coli MutY catalytic domain, chain A, 5822134; 2pjr, Bacillus stearothermophilus DNA-helicase PcrA insertion domain, chain A, 4930184; 1bpy1 and 1bpy2, human DNA-polymerase beta N-terminal (8 kDa) domain and ‘fingers’ domain, respectively, chain A, 2392200; 1a77, Methanococcus jannaschii Flap endonuclease-1, 5821778; 1taq, Thermus aquaticus DNA-polymerase Taq 5′ to 3′ exonuclease domain, 1942938; 1b43, Pyrococcus furiosus Fen-1 nuclease, chain A, 6980604; 1exn, bacteriophage T5 5′-exonuclease, chain A, 2392326; 1tfr, bacteriophage T4 RNase H, 1943457. The first HhH motifs are aligned in the top panel. For proteins with more than one HhH motif, the second and the third motifs are aligned in the middle and the bottom panels, respectively. All three panels are also aligned with each other. Positions of hydrophobic core residues are highlighted in black and are labeled with numbers corresponding to those in Figure 2c. Residues in the third HhH motif are not numbered. Glycines in the signature sequence GhG (h is a hydrophobic residue) of the hairpin regions are in red and positively charged residues following the signature are in blue. Positions with mostly uncharged residues are shaded in yellow. Numbers in brackets indicate the number of omitted residues in the sequence.
The structure of an individual HhH motif in several proteins has been analyzed in detail by Doherty et al. (13), who concluded that HhH-containing proteins do not share a common fold and HhH motifs in them exist as rather separate units. Here we show that HhH motifs are typically involved in formation of a larger structure of five α-helices that we term (HhH)2 domain. (HhH)2 is a pseudo-2-fold unit composed of two HhH motifs linked by a connector α-helix (Fig. 2). We demonstrate that (HhH)2 domain is a structurally compact entity with a well-defined hydrophobic core and thus deserves recognition as a distinct common protein fold. Strong sequence similarity in HhH motif regions suggests common evolutionary origin of (HhH)2 domains in different proteins. Additionally, we detect the presence of the HhH-like structures in sterile alpha motif (SAM) and barrier-to-autointegration factor (BAF) domains, RNA-polymerase α-subunit, and in DNA-helicase PcrA, and find new HhH motifs in DNA glycosylases.
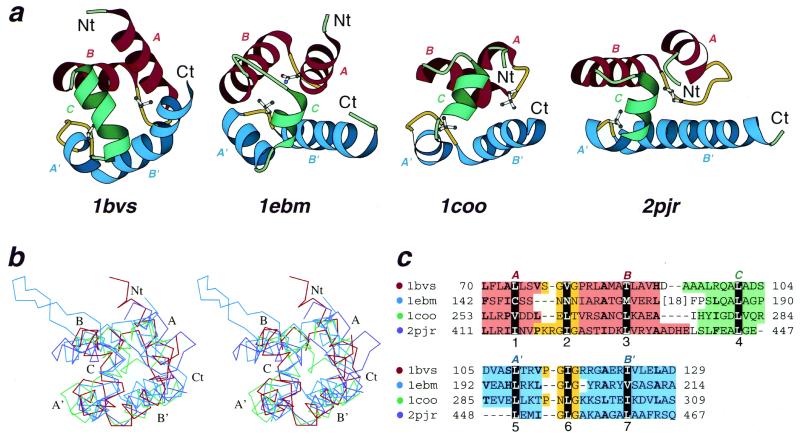
Figure 2.
Structural comparisons of divergent (HhH)2 domains. (a) Structural diagrams of DNA-helicase RuvA middle domain (1bvs, chain A, residues 63–134), 8-oxoguanine glycosylase central domain (1ebm, chain A, 135–221), C-terminal domain of RNA-polymerase α-subunit (1coo, 253–296) and DNA-helicase PcrA insertion domain (2pjr, chain A, 405–478), showing the HhH motifs from each protein. For each protein, N- and C-termini are labeled with Nt and Ct, respectively. The helices in the first HhH motif are labeled with A and B, and are in red. The corresponding helices in the second HhH motif are labeled with A′ and B′ and are in blue. The hairpin regions of both motifs are in yellow. Side-chains of central hydrophobic residues in hairpins are shown using ball-and-stick representation. The helices connecting the two HhH motifs are labeled with C and are in green. The ribbon diagrams were rendered by Bobscript (28), a modified version of Molscript (29). (b) Stereo diagram of superimposed Cα traces of DNA-helicase RuvA subunit (red), 8-oxoguanine glycosylase (blue), C-terminal domain of RNA-polymerase α-subunit (green) and DNA-helicase PcrA (purple). Superpositions were made using InsightII package (Molecular Simulations Inc.). Labels match those described in (a). (c) Structure-based sequence alignment of HhH motif regions of the four illustrated protein domains. For each sequence the PDB entry name and starting and ending residue numbers are given. The dot color scheme (in front of each PDB entry) matches those in (b). Color shading and helix labels correspond to those in (a). The two HhH motifs (upper and lower panels) are aligned with each other. Sites of conserved core hydrophobic residues are highlighted in black, and are labeled 1, 2 and 3 in the first HhH motif, 5, 6 and 7 in the second motif, and 4 in the connector helix C (also see Fig. 1). Additional conserved hydrophobic residues are shown in bold.
MATERIALS AND METHODS
Sequence similarity searches
Sequence similarity searches against the non-redundant protein database (nr) maintained at the National Center for Biotechnology Information (Bethesda, MD) were performed using the PSI-BLAST program (4,5) with the default parameters run to convergence as described previously (17). The non-redundant database (March 14, 2000 release, 440 253 sequences, 135 345 493 letters) sequences were filtered for low-complexity regions using the SEG program (18,19) with parameters: window 40, trigger 2.7 and extension 3.2. The query sequences were not subjected to SEG filtering. The BLOSUM62 matrix (20) was used for scoring, and 0.01 was used as an E-value threshold (4,5,17) for inclusion in the profile calculation. All sequences of HhH domains in proteins with known structure were used as initial PSI-BLAST queries. Since the results of searches strongly depend on the query sequence used (17), the database hits identified in the initial searches were used as queries for additional PSI-BLAST searches. All significantly different (<1.0 bits per site or ~50% identical) sequences found in the course of iterations were used as queries to extend the searches. If a protein of determined structure was detected, PSI-BLAST alignment was verified by the structure-based alignment to ensure the validity of the match. Sequence analysis protocols were carried out using SEALS (21).
Structure analysis
Structure similarity searches against the protein data bank (PDB) (22) maintained at the Research Collaboratory for Structural Bioinformatics (RCSB) were performed using DALI (23,24), VAST (25) and CE (26) programs with default parameters. The classical proteins with HhH motifs, such as RuvA (1bvs) (PDB entry is given for each structure in parenthesis) (15) and DNA-polymerase β (1bpy) (14,27) were used to initiate the searches that were continued using each detected protein as a query. The structure-based alignments generated by the above programs were used as starting points for the multiple structure-based alignment shown in Figure 1. SCOP (Structural Classification Of Proteins) database (November 1, 1999 release 1.48, 9912 PDB entries) (6,8,9,26) was used as a source of protein classification. Protein structures were visualized and superimposed using InsightII package (Molecular Simulations Inc., San Diego, CA) and the multiple structure-based alignment was built on the basis of the superpositions made in InsightII. The detection of the (HhH)2 hydrophobic core residues (one in each helix and each hairpin, seven in total) aided in the multiple alignment construction. Depending on the number of HhH motif(s) present in each protein, between 18 and 65 residues were used for superpositions. Residues used for superpositions cover regions (–4 1 +2), (–1 2 +1), (–5 3 +4), (–5 4 +3), (–4 5 +3), (–1 6 +1) and (–5 7 +6), where the middle number in parentheses indicates the consensus core residues (see numbers 1–7 in Fig. 1) and the – and + numbers indicate offsets from those core residues. The region covering the third motif was also included in superposition when present in both proteins. For each pair of proteins, the maximum possible number of residues within the above-defined region were used for superposition. Structure diagrams were rendered using Bobscript (28), a modified version of Molscript (29).
RESULTS AND DISCUSSION
Structural description of (HhH)2 domain
Sequence and structure searches for HhH motifs (see Materials and Methods) revealed the presence of the motif in the following proteins and domains of known structure: RuvA (15), Rad51 (30), DNA-polymerase β (14,27), SAM domain (31–34), RNA-polymerase α-subunit (35), BAF domain (36), DNA glycosylases (12,16,37,38), DNA-helicase PcrA (39) and 5′ to 3′ exonucleases (40–42). In all analyzed protein structures the hydrophobic core of each HhH motif is completed by a pair of additional α-helices. In most cases this additional helical pair appears to be a second HhH motif, and the two HhH motifs are linked by a connector helix (Fig. 2a). However, in several proteins, such as 5′ to 3′ exonucleases and the 8 kDa N-terminal domain of DNA-polymerase β, the second helical pair might not be evolutionarily related to HhH motif as indicated by its significant sequence and structural differences. This helical pair was not included in the alignment (Fig. 1). Available structures of HhH protein–DNA complexes (16,43,44) show that hairpins of both helical pairs are involved in symmetric contacts with the DNA-backbone. Thus the functional and structural unit containing HhH motifs is a five-helical domain that we term (HhH)2. This domain consists of two HhH motifs related by pseudo-2-fold symmetry and packed onto each other (Fig. 2a). The connector helix links the two HhH motifs and completes the hydrophobic core of the molecule. Due to the small size of the (HhH)2 domain and its significant sequence variability, superposition of different (HhH)2 structures has proved to be difficult. To aid structural comparisons, we outlined the positions of the hydrophobic core as the sites, one from each secondary structural element, that contain the least exposed residues. Five helices and two hairpins of the (HhH)2 domain yielded seven sites for hydrophobic core residues (Fig. 3a), which were superimposed using InsightII package (Molecular Simulations Inc.). Since the two HhH motifs of the domain are homologous and can be treated as repeats, they were subsequently superimposed with each other to generate the alignment presented in Figures 1 and 2.
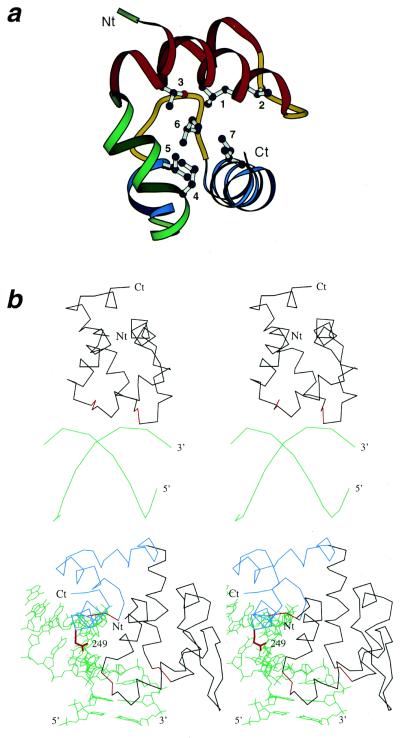
Figure 3.
Conserved hydrophobic core residues in the (HhH)2 proteins and DNA binding by HhH proteins. (a) Ribbon diagram of DNA-helicase RuvA middle domain (1bvs, chain A, residues 63–134) showing its hydrophobic core. The coloring and termini labeling scheme follows that in Figure 1a. Conserved core hydrophobic residues L74, V80, T88, L109, L115 and I123 are shown using ball-and-stick representation, and are labeled with numbers 1–7 corresponding to those in Figures 1c and 2. (b) Stereo diagrams showing interaction of (HhH)2 domains and DNA. Upper panel: E.coli DNA-helicase RuvA middle domain (1bdx, chain A, residues 65–142) with bound DNA at a Holliday junction (alpha carbons and phosphate atoms only). Lower panel: human 8-oxoguanine glycosylase (1ebm, 135–262) with bound DNA. DNA chains are in green and termini are labeled with 5′ and 3′. Protein Cα traces are in black, except that the third (catalytic) HhH motif in human 8-oxoguanine glycosylase is in blue, and the signature sequences GhG of the hairpin regions are in red. N- and C-termini are labeled with Nt and Ct, respectively. Side chain of residue 249 (Lys, mutated to Gln in the structure) in human 8-oxoguanine glycosylase, which is involved in the lyase activity of the protein, is shown in red.
The alignment clearly shows the conservation of the GhG pattern in the hairpin region, where h is a hydrophobic residue (conserved sites 2 and 6, Fig. 2), and the conserved hydrophobicity pattern in i, i+3, i+4 and i+7 positions of the ‘helical wheel’. This alignment represents the comparison of available HhH structures and illustrates several features that have been difficult to appreciate using purely sequence-based alignments in the absence of structural superpositions. Most significantly, insertions/deletions can be present before and after the conserved GhG element in the hairpin. Reliable detection of these is possible only on the basis of structural comparisons. The longest insertions are present in the 5′ to 3′ exonuclease family proteins before the GhG element in the first HhH motif (Fig. 1, pdb entries 1tfr, 1taq, 1exn, 1a77, 1b43). These insertions are 7–24 residues long and have been termed 3T regions, while the term H3TH has been coined for this deviant HhH motif (45). Notably, the positively charged residues (Lys, Arg) are concentrated near the N-terminus of the second α-helix in the HhH motif (Fig. 1). These regions, just after the GhG pattern, are close to the DNA-backbone in the protein–DNA-complexes (Fig. 3b). Sequence conservation of the HhH structure is significantly less than usually expected from purely sequence-based comparisons. Indeed, the HhH consensus given in Rafferty et al. (15) is only rarely matched, and even the most conserved GhG element in the hairpin is frequently replaced by other residues. Additionally, the first α-helix of the HhH motif shows a tendency to deteriorate. Helix-disrupting prolines are frequently present in the middle of this α-helix (Fig. 1) and it is reduced to a single turn in such structures as the C-terminal domain of RNA-polymerase α-subunit (first HhH motif) and DNA-helicase PcrA insertion domain (second HhH motif) (Fig. 2a and b).
In some proteins, for example in 5′ to 3′ exonucleases, the first helix of what should be a second HhH motif is not present at all. In these proteins, the hydrophobic core of the (HhH-connector) motif is completed with a single α-helix. It is not clear if this is a reflection of a deteriorated second HhH motif or an alternative way to complete the hydrophobic core of the first. Using caution, we stick to the last option and do not include this region in the alignment and superpositions (Fig. 1 and Table 1). However, the domain containing a single HhH motif in H3TH proteins appears to be a four-helical bundle displaying pseudo-2-fold symmetry. Finally in DNA glycosylases we detect three HhH motifs which form into the structure more appropriately described as (HhH)3 (Fig. 3b). The discussion of this structure and its relation to the typical (HhH)2 domain follows.
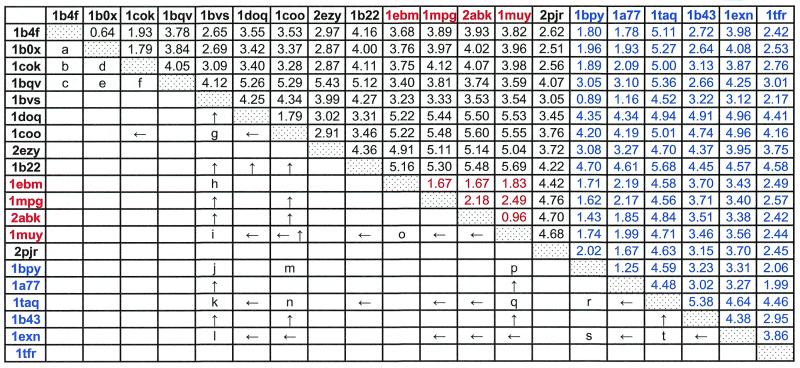
Table 1. Structural comparison among HhH motif-containing proteins and their sequence relations.
PDB entries of 20 representative HhH motif-containing proteins are listed in the top row and left column. The HhH domain proteins, containing two HhH motifs with a connector helix in between, are in black; those containing one additional HhH motif are in red; those that have only one HhH domain are in blue. Numbers in the upper right half of the table are r.m.s.d. values between each pair of proteins. Each pair of proteins was superimposed by Cα traces with InsightII package (Molecular Simulations Inc.). Numbers in black were obtained by superimposing the two HhH motifs and the connector helix; those in red by superimposing the same region plus the additional HhH motif; those in blue by superimposing only one HhH motif. See Materials and Methods for the definition of superimposed regions. The lower left half of the table shows PSI-BLAST search results. A letter indicates a single starting query sequence that can detect the proteins in the row and in the column in PSI-BLAST iterations with 0.01 as an E-value threshold before convergence (see Materials and Methods). The gi numbers:residue ranges of them are: a, 4929864:23_98; b, 4929864:23_98; c, 3386625:220_356; d, 4929864:23_98; e, 3386625:220_356; f, 3386625:220_356; g, 3560537:1_124; h, 3182983:477_593; i, 3182983:415_589; j, 2127858:1113_1192; k, 2127858:1113_1192; l, 586902:220_348; m, 42144:404_494; n, 6458154:80_175; o, 2982908:48_146; p, 4981594:11_181; q, 1651660:446_601; r, 549012:123_252; s, 4980590:436_598; t, 2983968:158_289. An arrow indicates that a sequence of one protein can find the other protein sequence through multiple rounds of PSI-BLAST iterations using all significantly different detected homologs as queries (see Materials and Methods) in subsequence iterations. An ‘up’ arrow shows that the protein in the row was used as a starting query to detect the protein in the column, and a ‘left’ arrow indicates the opposite situation.
RuvA, Rad51 and DNA-polymerase β: classical HhH-containing proteins
The middle domain of Escherichia coli RuvA protein (1bvs) (15) represents a clear duplication of HhH motifs with both motifs showing significant sequence and structural similarity to each other (Figs 1 and 2; Table 1). RuvA structure is the most symmetric version of (HhH)2 domain in the current structure database. The recently solved structure of the N-terminal domain of Rad51 protein (1b22) (30) that participates in DNA repair and recombination confirmed the presence of two HhH motifs.
We also detect two (HhH)2 domains in the structure of DNA-polymerase β (1bpy). The first one corresponds to the 8 kDa N-terminal domain, and the second one has been termed ‘fingers’ in the literature (46,47). Both domains are composed of four to five α-helices with the hairpins placed in the DNA proximity in the compexed structures. However, only a single HhH motif in each of the two (HhH)2 domains has been detected previously and discussed in detail (14,48). Our analysis indicates that the ‘fingers’ domains is likely to contain a second, highly divergent HhH motif (Fig. 1), in which the first α-helix is reduced to a single turn. The first pair of helices in the 8 kDa domain is rather long, does not superimpose well with other HhH structures, and was not included in this analysis.
SAM/pointed-domain
The SAM/pointed-domain is found in different signaling molecules, such as protein kinases and GTPases, and in Ets transcription factors (31–34). It has been proposed to mediate protein–protein interactions through homo/hetero-oligomerization (49) rather than to bind DNA. The four available structures of this domain are very similar (r.m.s.d. 0.64–4.12 Å) and show significant sequence similarity to each other (Table 1). Indeed, for each pair of structures from this family we were able to find a query sequence that retrieves sequences of both structures in PSI-BLAST iterations with 0.01 as an E-value threshold (see Materials and Methods; Table 1).
SCOP attributes classical HhH proteins such as RuvA middle domain and DNA-polymerase β N-terminal domain to the same fold with the SAM domain. Indeed, the structural similarity between the SAM domain and other (HhH)2 domains is pronounced. For example, DALI aligns EphB2 receptor SAM domain (1b4f_A) (32) with the 65 residues of HhH motifs in endonuclease III (2abk) giving a Z-score of 5.0 and r.m.s.d. of 2.8 Å, with 60 residues of RuvA (1hjp) (50) giving a Z-score of 3.5 and r.m.s.d. of 2.5 Å, and with 89 residues of BAF (1qck_A) (51) giving a Z-score of 4.5 and r.m.s.d. of 2.3 Å. VAST finds significant structural similarity between Eph receptor SAM domain (1b0x) (33) and HhH containing the N-terminal domain of DNA-polymerase β (1bno) (52) resulting in a P-value of 0.0154, a r.m.s.d. of 2.0 Å and a sequence identity of 13.3% between 30 superimposed residues. However, despite the significant structural similarity of the SAM domain to the (HhH)2 domains, especially to the (HhH)2 domain of RuvA as shown in our analysis (Table 1), the presence of HhH motif was not reported before for any member of the SAM family.
We were able to find a sequence similarity link between the SAM domain of Eph receptors (1b4f) (32) and HhH motif in the C-terminal domain of RNA-polymerase α-subunit (1doq and 1coo) (35) using PSI-BLAST. When the sequence of mouse EphA2 receptor SAM domain (gi│1706570, residues 877–975) was taken as a query, the sequence of RNA-polymerase from Deinococcus radiodurans (gi│6459926, residues 243–337) was detected on the third iteration with a score of 37.7 bits and an E-value of 0.026 and appears above the 0.01 threshold with a score of 39.4 bits and an E-value of 0.008 on the sixth iteration. The resulting PSI-BLAST alignment shows 17% identity and is in agreement with the alignment inferred from the superposition of EphB2 receptor SAM domain (1b4f) (32) and the C-terminal domain of Thermus thermophilus RNA-polymerase α-subunit (1doq), which validates the match. Therefore the presence of detectable sequence and pronounced structural similarity of SAM/pointed-domain family with other HhH proteins argues for their common evolutionary origin.
RNA-polymerase α-subunit and BAF-domain
SCOP (release 1.48) places these structurally similar domains into a separate fold in all-alpha proteins class (BAF-like, fold number 1.38). Most of other HhH-containing proteins are attributed to the SCOP fold termed SAM domain-like (fold number 1.62). However, we were able to obtain many links of statistically significant sequence similarity between the C-terminal domain of RNA-polymerase α-subunit (1coo) (35) and classical HhH domain proteins such as RuvA and DNA-polymerase β (Table 1). SMART (53) detects the first HhH motif in the sequence of T.thermophilus RNA-polymerase α-subunit (gi│4519423) with an E-value of 6.66e-01. PSI-BLAST finds E.coli RNA-polymerase α-subunit (1coo) on the first iteration with the sequence gi│4519423 (residues 237–314) as a query with a score of 89 bits and an E-value of 4e-18. CE (26) finds structural similarity between 1coo and Rad51 N-terminal HhH containing domain (pdb entry 1b22) with a Z-score of 4.1 (r.m.s.d. 4.0 Å and 12.9 sequence identity). This extensive sequence and structural similarity firmly establishes the presence of the two HhH motifs in E.coli RNA-polymerase α-subunit structure (1coo), despite the fact that the two GhG patterns in the hairpins are replaced with ELT and NLG, respectively, and strongly indicates the homology of 1coo domain with other (HhH)2 domains. Given that according to SCOP definitions homologous proteins should be grouped in the same superfamily within the same fold, unification of BAF-like and SAM-like SCOP folds seems reasonable.
BAF is a DNA-binding domain that functions as an inhibitor of retroviral DNA autointegration (36). BAF domain structure shows pronounced similarity to the C-terminal domain of RNA-polymerase α-subunit (pdb entry 1coo) and SAM-domains (Table 1). DALI finds 60 residues between 1coo and BAF (1qck_A) similar with a Z-score of 3.6, r.m.s.d. of 2.8 and 14% of sequence identity. Moreover, structure-based sequence alignment shows the strong conservation of the first GhG pattern (GIG in pdb entry 2ezy). It is therefore surprising that we were not able to support sequence similarity between 2ezy and other HhH motif proteins by PSI-BLAST statistics (Table 1). However, when PSI-BLAST searches are initiated with BAF domain (2ezy) several HhH proteins are found below the 0.01 threshold with the structurally correct alignments, for example E.coli impB (gi│6009442), Pyrobaculum islandicum RadA (gi│6683006) and DNA repair protein from Pyrococcus horikoshii (gi│7521024).
DNA glycosylases (MutY family)
HhH motif has been originally named and described from the structure of endonuclease III (2abk) (12). Only one copy of HhH motif has been widely discussed in the literature on DNA glycosylases so far (12,16,37,38). Notably, this HhH copy contains a catalytic Lys residue in 8-oxoguanine DNA glycosylase structure (1emb) (16). This Lys249 has been proposed to displace the oxoguanine base and to assist elimination of the 3′-phosphodiester through Schiff base chemistry (54,55). Thus HhH motif traditionally used for pure DNA binding acquires enzymatic function in some DNA glycosylases.
Our analysis detects two additional HhH motifs in DNA glycosylases that are placed N-terminal to the classical HhH motif (Figs 1–3). The second of these has been previously referred to as a ‘pseudo-HhH’ and the first one has been called ‘DNA-minor groove reading motif’ (38). These two motifs form a clear (HhH)2 domain (Fig. 2), which interacts with DNA in a manner similar to the one found in RuvA (HhH)2 domain (Fig. 3b). DALI detects a highly significant structural match between the (HhH)2 domain of EphB SAM (1b4f) and those two additional HhH motifs in endonuclease III (2abk) (Z-score of 5.0, r.m.s.d. 2.8 Å for 65 residues). Classical HhH in the RuvA middle domain (1hjp) is also found in this search (Z-score of 3.1, r.m.s.d. 4.0 Å for 79 residues). All three HhH motifs in DNA glycosylases form a compact seven- or eight-helical domain, which represents a (HhH)3 fold rather than (HhH)2. This domain secondary structure pattern can be described as H11h1H12C1H21h2H22C2H31h3H32, where H is a motif helix, h is a hairpin and C is a connector helix (second connector helix may be lacking). The first index refers to the motif and the second index shows the helix number within the motif. The classical (HhH)2 domain is embedded into such (HhH)3 structures and this can be rationalized in two ways. First, H11h1H12C1H21h2H22 can be considered an (HhH)2 unit and the third HhH motif, C2H31h3H32, is therefore a C-terminal addition (Fig. 3b). Alternatively, H11h2H22C2H31h3H32 might represent a (HhH)2 domain, while h1H12C1H21 is an insertion into the hairpin region (Fig. 3b). Thus the DNA glycosylase structure reveals how the HhH motifs might ‘nest’ into each other and form structures containing more than two motifs in one compact domain. However, due to the common evolutionary origin of the motifs, we do not think that these seven/eight-helical structures should be classified as a separate fold.
SCOP attributes DNA glycosylases to a separate fold (fold number 1.92). Given the presence of three HhH motifs that comprise the core of the structure, it might be reasonable to classify the N-terminal domain of DNA glycosylases together with other HhH proteins and consider regions outside the HhH motifs as insertions and additions to the common fold.
DNA-helicase PcrA
A VAST search initiated with the E.coli RuvA (HhH)2 domain (1bdx) produced a match with the 42 residues of the insertion domain of DNA-helicase PcrA (Fig. 2) (39). DALI searches initiated with PcrA (2pjr) yielded EphB2 SAM domain (1b4f, Z-score 3.1, r.m.s.d. 3.2 for 54 residues). DNA-helicase PcrA is a P-loop ATPase composed of two homologous α/β domains, each of which bears long all α-helical insertions (39). The role in DNA binding is suggested for these insertions (39). One of the domains in the long α-helical insertion displays similarity with HhH proteins. Significant structural similarity combined with the functional similarity of this helicase domain with (HhH)2 proteins suggests common evolutionary origin. Indeed, hairpin regions of the two HhH motifs have been proposed to be in close proximity with DNA as indicated by the crystal structure of the PcrA–DNA complex (39). To our knowledge the presence of HhH motifs in DNA-helicase PcrA has not been reported previously. In SCOP, all-helical insertion domains of DNA helicases are not split from the P-loop-containing core and are not classified separately. Thus we suggest that the insertion domain (residues 411–467 of 2pjr) comprise a (HhH)2 domain and should be classified accordingly. Despite the presence of both GhG patterns in the two HhH motifs (GIG in the first motif and GLG in the second) as shown in structure-based alignment (Fig. 1), we were not able to detect statistically supported sequence similarity between the HhH domain of this helicase and any other proteins using the PSI-BLAST program.
5′ to 3′ exonucleases (H3TH proteins)
Spatial structures are available for the five close homologs of this family: nuclease domain of Taq polymerase (1taq) (40), T5 5′ exonuclease (1exn) (41), T4 RNase H (1tfr) (42), Flap endonuclease-1 (1a77) (56) and Fen-1 nuclease (1b43) (45). PSI-BLAST searches identify a single copy of HhH motif placed in the C-terminal domain in the proteins of this family (Table 1). Clearly detectable HhH motif contains a long insertion in the hairpin before the GhG pattern (7–24 residues) and was termed ‘H3TH motif’ due to the presence of two additional turns in the insertion region (45,57). We find this terminology confusing, since H3TH motif shows homology to HhH motif (Table 1 and Fig. 1) and should not be confused with HTH motif. Single HhH motif in 5′ to 3′ exonucleases is expected to bind dsDNA in a manner typical for classical HhH motifs (57). Although the hydrophobic core of this single HhH motif is completed by a pair of α-helices to form a structure similar to the (HhH)2 domain, the orientation of these helices is rather distinct from the one found in HhH motif. Therefore we did not include these α-helices in our analysis and consider only a single copy of HhH motif in a (HhH)2 domain of 5′ to 3′ exonucleases.
(HhH)2 and DNA binding: functional implications
DNA functions as a double helix, which is a pseudo-2-fold structure. DNA-binding proteins are intrinsically asymmetric and they should solve the problem of fitting into the symmetric DNA molecules. The simplest solution is to bind a single α-helix in a DNA major groove, which is used by HTH proteins (58,59). In this case, an α-helix itself could be considered as an approximation of a 2-fold structure (N-to-C versus C-to-N main-chain orientations). The second solution involves symmetrization by a domain duplication. Such examples include HTH proteins and various Zn-fingers (60,61), where more than one domain of the same class is used for DNA binding. A more complex idea of symmetrization is utilized by HLH (13) or leucine zipper proteins (62,63), which are obligatory functional dimers, many with true 2-fold symmetry. This mode is similar to duplication; however, the two DNA-binding units do not constitute separate structural domains but rather form a hydrophobic core on the dimer interface with both subunits contributing to it. In this case the HLH dimer itself (and not separate monomers) forms a structural domain. The same mode of symmetrization is used also by Arc transcription regulators (64).
More complex symmetrization is found in HhH proteins. Most HhH motifs are arranged in pairs and each pair forms into a five-helical (HhH)2 domain, which is a pseudo-2-fold object with two HhH motifs connected by an α-helix. The formation of (HhH)2 domains serves at least two functions. First, a short helical hairpin of a single HhH motif is unlikely to have the stability of an average protein due to the incomplete hydrophobic core. Such structure needs to be stabilized and its hydrophobic core completed. Second, symmetrization of the DNA-binding unit that mirrors the symmetry of the DNA-double helix facilitates stronger DNA-binding properties. HhH proteins are rather unique given that DNA recognition is non-sequence-specific. In accord with this protein–DNA contacts are not built around DNA bases but rather on a sugar–phosphate backbone. HhH motif binds DNA via hydrogen bonds between nitrogens of protein backbone and oxygens of DNA phosphate groups (13,16,43,44). The presence of two motifs in a (HhH)2 domain provides symmetric binding to both DNA chains in a duplex and facilitates stronger interactions. The DNA-binding modes of different (HhH)2 proteins with known structure in a DNA complex are rather similar (Fig. 3b) and in all of them the bound DNA appears to be bent (13,16,43,44). These include the classical HhH proteins such as RuvA and DNA-polymerase β, and two newly detected additional HhH motifs in DNA glycosylases (Fig. 3b).
As is often the case, some proteins in this diversified superfamily have changed functions. Indeed, SAM domain has been proposed to mediate protein–protein interactions and not DNA binding (49). The third (classical) HhH motif in some DNA glycosylases acquired enzymatic function in addition to DNA binding by incorporating a catalytic lysine residue (16).
Acknowledgments
ACKNOWLEDGEMENTS
The authors are grateful to Hong Zhang and Monica Horvath for critical reading of the manuscript and helpful comments.
REFERENCES
- 1.Bateman A., Birney,E., Durbin,R., Eddy,S.R., Finn,R.D. and Sonnhammer,E.L. (1999) Nucleic Acids Res., 27, 260–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McClure M.A., Smith,C. and Elton,P. (1996) ISMB, 4, 155–164. [PubMed] [Google Scholar]
- 3.Hughey R. and Krogh,A. (1996) Comput. Appl. Biosci., 12, 95–107. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altschul S.F. and Koonin,E.V. (1998) Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- 6.Murzin A.G., Brenner,S.E., Hubbard,T. and Chothia,C. (1995) J. Mol. Biol., 247, 536–540. [DOI] [PubMed] [Google Scholar]
- 7.Murzin A.G. (1998) Curr. Opin. Struct. Biol., 8, 380–387. [DOI] [PubMed] [Google Scholar]
- 8.Hubbard T.J.P., Murzin,A.G., Brenner,S.E. and Chothia,C. (1997) Nucleic Acids Res., 25, 236–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hubbard T.J., Ailey,B., Brenner,S.E., Murzin,A.G. and Chothia,C. (1999) Nucleic Acids Res., 27, 254–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H. and Grishin,N.V. (1999) Protein Sci., 8, 1658–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova K.S. and Grishin,N.V. (1999) J. Mol. Biol., 292, 11–17. [DOI] [PubMed] [Google Scholar]
- 12.Thayer M.M., Ahern,H., Xing,D., Cunningham,R.P. and Tainer,J.A. (1995) EMBO J., 14, 4108–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doherty A.J., Serpell,L.C. and Ponting,C.P. (1996) Nucleic Acids Res., 24, 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawaya M.R., Prasad,R., Wilson,S.H., Kraut,J. and Pelletier,H. (1997) Biochemistry, 36, 11205–11215. [DOI] [PubMed] [Google Scholar]
- 15.Rafferty J.B., Ingleston,S.M., Hargreaves,D., Artymiuk,P.J., Sharples,G.J., Lloyd,R.G. and Rice,D.W. (1998) J. Mol. Biol., 278, 105–116. [DOI] [PubMed] [Google Scholar]
- 16.Bruner S.D., Norman,D.P. and Verdine,G.L. (2000) Nature, 403, 859–866. [DOI] [PubMed] [Google Scholar]
- 17.Aravind L. and Koonin,E.V. (1999) J. Mol. Biol., 287, 1023–1040. [DOI] [PubMed] [Google Scholar]
- 18.Wootton J.C. (1994) Comput. Chem., 18, 269–285. [DOI] [PubMed] [Google Scholar]
- 19.Wootton J.C. and Federhen,S. (1996) Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 20.Henikoff S. and Henikoff,J.G. (1992) Proc. Natl Acad. Sci. USA, 89, 10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker D.R. and Koonin,E.V. (1997) ISMB, 5, 333–339. [PubMed] [Google Scholar]
- 22.Abola E.E., Sussman,J.L., Prilusky,J. and Manning,N.O. (1997) Methods Enzymol., 277, 556–571. [DOI] [PubMed] [Google Scholar]
- 23.Holm L. and Sander,C. (1996) Nucleic Acids Res., 24, 206–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holm L. and Sander,C. (1997) Nucleic Acids Res., 25, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibrat J.F., Madej,T. and Bryant,S.H. (1996) Curr. Opin. Struct. Biol., 6, 377–385. [DOI] [PubMed] [Google Scholar]
- 26.Shindyalov I.N. and Bourne,P.E. (1998) Protein Eng., 11, 739–747. [DOI] [PubMed] [Google Scholar]
- 27.Pelletier H., Sawaya,M.R., Wolfle,W., Wilson,S.H. and Kraut,J. (1996) Biochemistry, 35, 12742–12761. [DOI] [PubMed] [Google Scholar]
- 28.Esnouf R.M. (1997) J. Mol. Graph. Model., 15, 133–138. [DOI] [PubMed] [Google Scholar]
- 29.Kraulis P.J. (1991) J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- 30.Aihara H., Ito,Y., Kurumizaka,H., Yokoyama,S. and Shibata,T. (1999) J. Mol. Biol., 290, 495–504. [DOI] [PubMed] [Google Scholar]
- 31.Slupsky C.M., Gentile,L.N., Donaldson,L.W., Mackereth,C.D., Seidel,J.J., Graves,B.J. and McIntosh,L.P. (1998) Proc. Natl Acad. Sci. USA, 95, 12129–12134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thanos C.D., Goodwill,K.E. and Bowie,J.U. (1999) Science, 283, 833–836. [DOI] [PubMed] [Google Scholar]
- 33.Stapleton D., Balan,I., Pawson,T. and Sicheri,F. (1999) Nat. Struct. Biol., 6, 44–49. [DOI] [PubMed] [Google Scholar]
- 34.Chi S.W., Ayed,A. and Arrowsmith,C.H. (1999) EMBO J., 18, 4438–4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeon Y.H., Negishi,T., Shirakawa,M., Yamazaki,T., Fujita,N., Ishihama,A. and Kyogoku,Y. (1995) Science, 270, 1495–1497. [DOI] [PubMed] [Google Scholar]
- 36.Cai M., Huang,Y., Zheng,R., Wei,S.Q., Ghirlando,R., Lee,M.S., Craigie,R., Gronenborn,A.M. and Clore,G.M. (1998) Nat. Struct. Biol., 5, 903–909. [DOI] [PubMed] [Google Scholar]
- 37.Labahn J., Scharer,O.D., Long,A., Ezaz-Nikpay,K., Verdine,G.L. and Ellenberger,T.E. (1996) Cell, 86, 321–329. [DOI] [PubMed] [Google Scholar]
- 38.Guan Y., Manuel,R.C., Arvai,A.S., Parikh,S.S., Mol,C.D., Miller,J.H., Lloyd,S. and Tainer,J.A. (1998) Nat. Struct. Biol., 5, 1058–1064. [DOI] [PubMed] [Google Scholar]
- 39.Velankar S.S., Soultanas,P., Dillingham,M.S., Subramanya,H.S. and Wigley,D.B. (1999) Cell, 97, 75–84. [DOI] [PubMed] [Google Scholar]
- 40.Kim Y., Eom,S.H., Wang,J., Lee,D.S., Suh,S.W. and Steitz,T.A. (1995) Nature, 376, 612–616. [DOI] [PubMed] [Google Scholar]
- 41.Ceska T.A., Sayers,J.R., Stier,G. and Suck,D. (1996) Nature, 382, 90–93. [DOI] [PubMed] [Google Scholar]
- 42.Mueser T.C., Nossal,N.G. and Hyde,C.C. (1996) Cell, 85, 1101–1112. [DOI] [PubMed] [Google Scholar]
- 43.Hargreaves D., Rice,D.W., Sedelnikova,S.E., Artymiuk,P.J., Lloyd,R.G. and Rafferty,J.B. (1998) Nat. Struct. Biol., 5, 441–446. [DOI] [PubMed] [Google Scholar]
- 44.Hollis T., Ichikawa,Y. and Ellenberger,T. (2000) EMBO J., 19, 758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hosfield D.J., Mol,C.D., Shen,B. and Tainer,J.A. (1998) Cell, 95, 135–146. [DOI] [PubMed] [Google Scholar]
- 46.Davies J.F.,II, Almassy,R.J., Hostomska,Z., Ferre,R.A. and Hostomsky,Z. (1994) Cell, 76, 1123–1133. [DOI] [PubMed] [Google Scholar]
- 47.Sawaya M.R., Pelletier,H., Kumar,A., Wilson,S.H. and Kraut,J. (1994) Science, 264, 1930–1935. [DOI] [PubMed] [Google Scholar]
- 48.Pelletier H. and Sawaya,M.R. (1996) Biochemistry, 35, 12778–12787. [DOI] [PubMed] [Google Scholar]
- 49.Schultz J., Ponting,C.P., Hofmann,K. and Bork,P. (1997) Protein Sci., 6, 249–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishino T., Ariyoshi,M., Iwasaki,H., Shinagawa,H. and Morikawa,K. (1998) Structure, 6, 11–21. [DOI] [PubMed] [Google Scholar]
- 51.Kuszewski J., Gronenborn,A.M. and Clore,G.M. (1996) Protein Sci., 5, 1067–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu D., Prasad,R., Wilson,S.H., DeRose,E.F. and Mullen,G.P. (1996) Biochemistry, 35, 6188–6200. [DOI] [PubMed] [Google Scholar]
- 53.Schultz J., Milpetz,F., Bork,P. and Ponting,C.P. (1998) Proc. Natl Acad. Sci. USA, 95, 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dodson M.L., Michaels,M.L. and Lloyd,R.S. (1994) J. Biol. Chem., 269, 32709–32712. [PubMed] [Google Scholar]
- 55.Nash H.M., Bruner,S.D., Scharer,O.D., Kawate,T., Addona,T.A., Spooner,E., Lane,W.S. and Verdine,G.L. (1996) Curr. Biol., 6, 968–980. [DOI] [PubMed] [Google Scholar]
- 56.Hwang K.Y., Baek,K., Kim,H.Y. and Cho,Y. (1998) Nat. Struct. Biol., 5, 707–713. [DOI] [PubMed] [Google Scholar]
- 57.Parikh S.S., Mol,C.D., Hosfield,D.J. and Tainer,J.A. (1999) Curr. Opin. Struct. Biol., 9, 37–47. [DOI] [PubMed] [Google Scholar]
- 58.Suzuki M., Yagi,N. and Gerstein,M. (1995) Protein Eng., 8, 329–338. [DOI] [PubMed] [Google Scholar]
- 59.Aravind L. and Koonin,E.V. (1999) Nucleic Acids Res., 27, 4658–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Takatsuji H. (1999) Plant Mol. Biol., 39, 1073–1078. [DOI] [PubMed] [Google Scholar]
- 61.Krempler A. and Brenig,B. (1999) Mol. Gen. Genet., 261, 209–215. [DOI] [PubMed] [Google Scholar]
- 62.Hagerman P.J. (1996) Proc. Natl Acad. Sci. USA, 93, 9993–9996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Luscher B. and Larsson,L.G. (1999) Oncogene, 18, 2955–2966. [DOI] [PubMed] [Google Scholar]
- 64.Gallegos M.T., Schleif,R., Bairoch,A., Hofmann,K. and Ramos,J.L. (1997) Microbiol. Mol. Biol. Rev., 61, 393–410. [DOI] [PMC free article] [PubMed] [Google Scholar]