Abstract
We have developed a method for simultaneous deposition and covalent cross-linking of oligonucleotide or PCR products on unmodified glass surfaces. By covalently conjugating an active silyl moiety onto oligonucleotides or cDNA in solutions we have generated a new class of modified nucleic acids, namely silanized nucleic acids. Such silanized molecules can be immobilized instantly onto glass surfaces after manual or automated deposition. This method provides a simple and rapid, yet very efficient, solution to the immobilization of prefabricated oligonucleotides and DNA for chip production.
INTRODUCTION
Solid phase nucleic acid hybridization has been used in a wide variety of applications including monitoring gene expression (1–3), polymorphism analysis (4,5), disease screening and diagnostics (6,7), nucleic acid sequencing (8–10) and genome analysis (11,12). A number of different substances have been tested as the solid support for nucleic acid immobilization (13–16), but glass slides are generally favored for DNA and oligonucleotide microarrays (17–21).
There are two ways to build DNA or oligonucleotide arrays on the glass surface: direct on-surface synthesis (20–23) and immobilization of prefabricated DNA or oligonucleotides (the deposition method) (13–19,24,25). On-chip synthesis, using photolithography or ink-jet methods, is by far the most efficient method of generating high-density oligonucleotide chips on a glass surface, but has practical limitations in terms of flexibility and affordability. Immobilization of pre-fabricated nucleic acid, on the other hand, offers excellent flexibility that can accommodate most research and clinical applications. For making chips of medium or low complexity, the deposition method can afford a much higher production speed than on-surface synthesis. Such technologies are becoming widely available and importantly, much more affordable for most researchers and clinicians.
Central to the deposition technologies is the development of efficient chemistries for covalent attachment of nucleic acids on glass and silicon surfaces. A great number of attachment methods have been published, which vary widely in chemical mechanisms, ease of use, probe density and stability (17–19,26–28). In all these methods, glass and silicon surfaces have been modified to different extents in order to achieve reactivity against corresponding modified (or unmodified) nucleic acids. Most of these procedures are laborious and time-consuming.
Here we report a new nucleic acid modification method that allows the modified molecules to be attached covalently to unmodified glass surface directly. We have demonstrated different procedures to covalently conjugate an active silyl moiety on the oligonucleotides or cDNA in solutions to form a new class of modified nucleic acid, namely silanized nucleic acids. The silanized oligonucleotides and cDNA were shown to immobilize readily to glass slides upon deposition. The immobilization is fast and chips produced with this method gave strong hybridization signals with negligible background.
MATERIALS AND METHODS
Chemicals and oligonucleotides
Chemicals were purchased from Sigma-Aldrich Sweden, Stockholm, Sweden unless otherwise indicated. Acrylic-oligonucleotides were from Eurogentec, Seraing, Belgium and all other oligonucleotides were from Interactiva, Ulm, Germany. Unmodified glass slides were purchased from Kebo-lab, Stockholm, Sweden. Polylysine-coated slides PolyPrep™ and aminopropylsilane-coated slides AminoPrep™ were from Sigma. Aminopropylsilane-coated slides CMT-GAPS™ were from Corning, Corning, USA. Nylon- and nitrocellulose-coated slides were from Schleicher and Schuell, Dassel, Germany. Cy3-labeled strepavidin was from Amersham Pharmacia Biotech, Uppsala, Sweden.
Conjugation of silane with oligonucleotides and nucleic acids
Mercaptosilane [(3-Mercaptopropyl)-trimethoxysilane] was diluted to 5 mM stock solution with one of the two reaction buffers: NaOAc (sodium acetate) buffer (30 mM, pH 4.3) or sodium citrate (30 mM, pH 4). For conjugation of 5′-thiol-labeled oligonucleotides (Lac-thio, TCA TGG TCA TAG CTG TTT CC; Lac-thio-sen, GGA AAC AGC TAT GAC CAT GA) with mercaptosilane, 1 nmol oligonucleotides were reacted with 5 nmol mercaptosilane in 20 µl of the same buffer. The reaction was allowed to proceed for 10 min to 2 h at room temperature. Then the reaction mixture was used directly or diluted with the reaction buffer to desired concentration for immobilization on glass surface. 5′-acrylic-labeled oligonucleotides (Lac-acrylic, TCA TGG TCA TAG CTG TTT CC; Lac-acrylic-sen, GGA AAC AGC TAT GAC CAT GA) were also conjugated to mercaptosilane using an identical procedure.
Thiol-labeled 1 kb LacZ DNA fragment was generated by PCR using one 5′-thiol-labeled primer (Lac-thio-sen) with an unlabeled reverse primer (LacR1, GCA GGC TTC TGC TTC AAT CA). The PCR product was either directly used for subsequent conjugation reaction or concentrated 5-fold before proceeding to the conjugation step. An aliquot of 8 µl of the cDNA solution was reacted with 2.5 nmol mercaptosilane in 30 mM NaOAc, pH 4.3 for 1 h (total volume 10 µl). Similarly, 21 cDNA random clones were PCR amplified from a subtracted mouse cDNA library with a pair of thiol-labeled T3 and T7 primers. The PCR products were concentrated 10-fold and conjugated with mercaptosilane.
Lac-thio and Lac-thio-sen were conjugated with aminosilane [(3-aminopropyl)-trimethoxysilane] in dimethylsulfoxide (DMSO) in the presence of heterobifunctional linkers N-succinimidyl-3-(2-pyridyldithiol)-propionate (SPDP) or succinimidyl-6-(iodoacetyl-amino)-hexanoate (SIAX). Different amounts of oligonucleotides (final concentration 5–50 µM) were combined with 2.5 nmol aminosilane (added from 5 mM solution in ethanol) and 2.5 nmol bifunctional reagents (added from 5 mM stock solution in DMSO) in 10 µl DMSO. The reaction was allowed to proceed for 1–2 h at room temperature.
Acrylic-labeled oligonucleotides (Lac-acrylic and Lac-acrylic-sen) (50–500 pmol) were combined with 2.5 nmol acrylicsilane (γ-methacryloxy-propyl-trimethoxysilane) in 10 µl of 30 mM NaOAc, pH 4.3. Ammonium persulfate (10% in H2O) and N,N,N′,N′-tetramethylethylenediamine (TEMED) were added to final concentration of 0.5 and 2%, respectively, and the mixture was allowed to react for 30 min at room temperature.
After the conjugation reactions, the reaction mixture was referred to as silanized nucleic acid and directly used for spotting.
Washing of glass slides and nucleic acid deposition
Glass slides were washed by ultrasonication in water for 30 min, soaked in 10% NaOH for 30 min and then rinsed with tap water, distilled water and dried in an 80°C oven for 10 min or air-dried overnight.
Silanized nucleic acids were spotted on the glass slides either manually (120 nl/spot) or with an automated arrayer (Genetic Microsystem, Woburn, USA) (1 nl/spot). For nucleic acids in aqueous solutions, the glass slides were kept in a humidified chamber for 15 min at room temperature after spotting and then dried at 50°C for 5 min. The slides were then dipped into boiling water for 30 s to remove non-covalently bound nucleic acids and dried with nitrogen before proceeding to the hybridization step. For oligonucleotides in DMSO, the slides were left at room temperature for 15 min after spotting and then dried at 50°C for 10 min. These slides were sequentially washed with DMSO (3 × 2 min), ethanol (3 × 2 min) and boiling water (2 min) and then dried with nitrogen for further use.
Hybridization
5′-Cy3-labeled oligonucleotide probes (Lac-Cy3, GGA AAC AGC TAT GAC CAT GA; LacR1-Cy3, GCA GGC TTC TGC TTC AAT CA) were diluted to between 20 nM and 1 µM in 5× SSC (750 mM NaCl, 125 mM sodium citrate, pH 7) with 0.1% Tween-20. Hybridization was done under coverslips in a humidifier at 37°C for 30 min to overnight. Un-hybridized and non-specific probes were removed by washing with 5× SSC containing 0.1% Tween-20 (3 × 1 min) followed by 1× SSC containing 0.1% Tween-20 (2 × 15 min).
For the 21 cDNA chip, 1 µl of the PCR products of each of 96 random mouse cDNA clones (including the 21 clones that have been immobilized on chip, see above) were pooled. An aliquot of 5 µl of this mixture was then transcribed with T7 RNA polymerase in the presence of 0.1 mM biotin-labeled UTP. The RNA product was then purified by precipitation and used as probe. Hybridization was carried out at 65°C for 4 h in 3 × SSC with 0.1% SDS and 1 µg/µl yeast tRNA. The slides were then washed with 1× SSC containing 0.1% SDS (3× 2 min) and 0.1× SSC containing 0.1% SDS (3 × 5 min) at room temperature. Cy3-conjugated streptavidin (0.1 µg/µl in 5× SSC with 0.1% Tween-20) was added on the slides and incubated for 30 min. The slides were rinsed three times with 5× SSC and 1× SSC sequentially.
After washing, the slides were dried with nitrogen gas and scanned on GMS418 fluorescent scanner (Genetic Microsystem) to visualize hybridization signals. For repeated hybridization on the same chips, the chips were boiled in water for 1 min and then dried with nitrogen gas before proceeding to the next hybridization reaction.
Comparison of background fluorescence of differently coated slides
Unmodified glass slides and differently coated slides, without any prior treatment, were scanned with green (532 nm) and red (635 nm) lasers separately using GMS418 scanner. All background readings were scanned at 100% laser power and 100% photo multiplier tube (PMT) gain, except for nylon and nitrocellulose coated surfaces. For nylon and nitrocellulose coated surfaces the readings in the green channel were recorded at 50% laser power and 50% PMT gain (because of their intensive background signals) and then multiplexed by 4 to reconstitute the background levels corresponding to readings at 100% laser power and 100% PMT gain. Using the ImageQuantTM software (Molecular Dynamics, Sunnyvale, USA), an area of 4.25 cm2 was selected from each of the slide types and divided into 625 segments of 6764 pixels each. The reading (in units of photons/pixel) for each area was then used to calculate the average background level and the background variation for each type of slides.
Quantification of oligonucleotide immobilization
Lac-Cy3 was diluted with 30 mM NaOAc pH 4.3 to a 2-fold dilution series ranging from 2 µM to 30 nM and four spots, each 120 nl in volume, were made on unmodified glass slide for each dilution. The fluorescent intensity of the spots was examined on a scanner and quantified with ImageQuantTM to obtain a standard curve. In a separate experiment, silanized Lac-thio and Lac-acrylic were spotted at final concentrations of 50, 25, 12.5, 6.25 and 3.2 µM on unmodified glass and then treated for hybridization with Lac-Cy3. The fluorescence intensities of the spots were quantified and the amount of immobilized oligonucleotides available for hybridization was then deduced from the standard curve.
RESULTS AND DISCUSSION
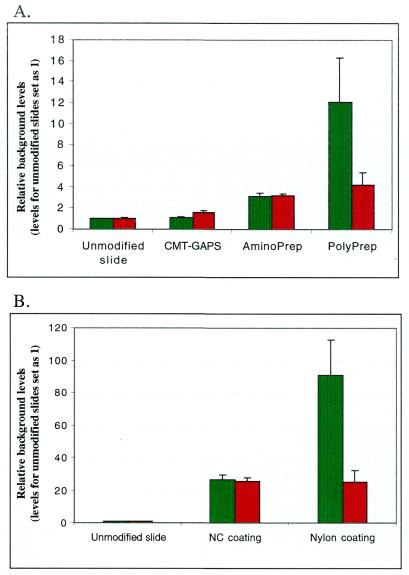
To determine whether unmodified glass surface has advantages over modified glass surfaces for DNA chip production, we tested background levels of five differently coated glass slides available from the commercial suppliers and compared them with unmodified glass surface. As depicted in Figure 1A, the background fluorescence of polylysine coated slides was 12-fold higher than the unmodified glass when observed under the green laser. Under the red laser, however, the difference was 4-fold. Additionally, the polylysine coated slides also displayed a much greater degree of background variation. Aminosilane coated slides AminoPrep™ showed a 3-fold higher background as compared to unmodified slides in both laser channels, but for aminosilane coated slides CMT-GAPS the background fluorescence was just slightly higher than unmodified chips. We found that the background reading of nylon membrane was 20 (red channel) to 90 times (green channel) higher than unmodified glass slides. Nitrocellulose coated slides had slightly lower background than nylon but the reading was still 20 times (red channel) to 30 times (green channel) higher than unmodified glass (Figure 1B). Thus, the unmodified glass slides showed the lowest background fluorescence and the best uniformity as compared to all surface modifications examined here.
Figure 1.
Background comparison of different solid supports with unmodified glass slides. (A) Aminosilane coated slides CMT-GAPS, AminoPrep and polylysine coated slides PolyPrep. (B) Nylon and nitrocellulose coated slides. Each slide was scanned under green laser (532 nm, green columns) and red laser (635 nm, red columns). Readings for unmodified glass slides were assigned a value of 1, and all other readings were normalized to this value.
We further explored whether a silane-oligonucleotide conjugate molecule can be constructed and reacted with unmodified glass surface. If so, different silylating reagents and correspondingly modified oligonucleotides can then be conjugated with each other in their respective optimized solutions and the conjugated molecules can be deposited pointedly on the glass chips. Such an approach could provide a general platform of DNA immobilization that can reconcile most of the existing chemical pathways that have been developed for immobilizing prefabricated nucleic acids.
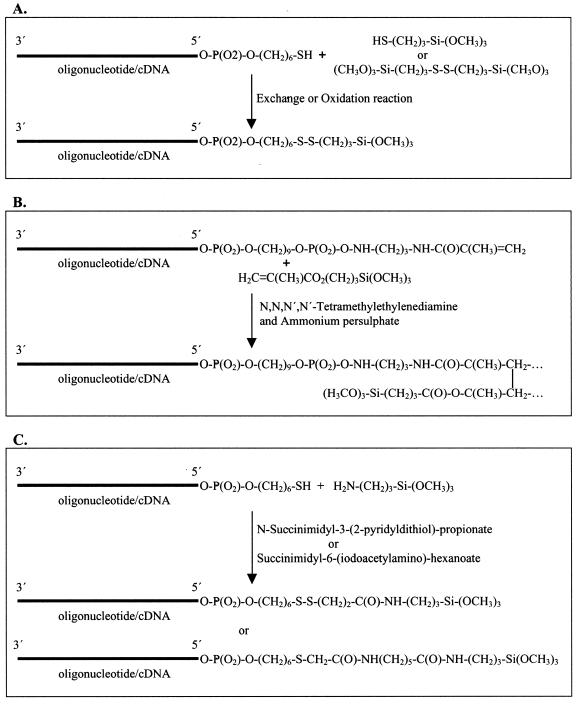
We first chose to conjugate thiol-labeled oligonucleotides (Lac-thio and Lac-thio-sen) with mercaptosilane because of the known specificity of the reaction (17) (Fig. 2A). A 5- fold excess of mercaptosilane dissolved in a pH 4.3 buffer was used in this experiment. Silanes are unstable at high pH in aqueous solutions and their degradation is minimized under the relatively low pH buffers used in our system. The conjugated oligonucleotide (referred to as silanized oligonucleotide) was used directly for spotting on the pre-cleaned glass slides without additional purification. Then the slides were examined by hybridization to Lac-Cy3. As shown in Figure 3A and B, the conjugated molecules were immobilized on glass surfaces within a few minutes and were available for hybridization. Quantification of the hybridization signals showed that with a starting concentration of 20 µM in the spotting buffer, oligonucleotides can routinely be immobilized at 2 × 105 molecules/µm2 densities on unmodified glass slides.
Figure 2.
Diagram showing three chemical pathways used to generate silanized nucleic acids. (A) Conjugating thiol-labeled nucleic acids to mercapto- or disulfide silanes. (B) Conjugating acrylic-labeled nucleic acids to acrylic-silane by polymerization. (C) Conjugating thiol-labeled nucleic acids to amino silane using a heterobifunctional-crosslinker.
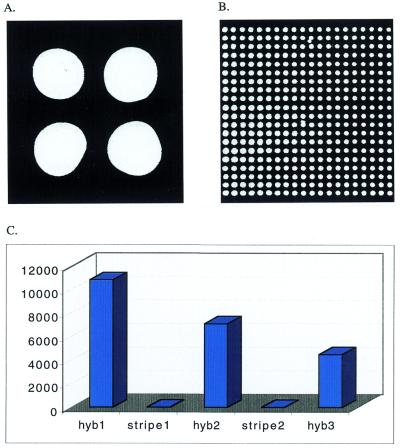
Figure 3.
Silanized Lac-Thio arrayed on unmodified glass slides. Chips spotted manually (A) or using an automated arrayer (B) were detected by hybridizing to a Cy3-labeled complementary probe. Hand-spotted chips were used in re-probing assays and the signals (photons/pixel) from the three successive probing and stripping cycles were illustrated (C). The hybridization in (C) was done for 30 min at 2 µM probe concentration and stripping was done by boiling the chips in a microwave oven for 1 min in water. Even at the third round of hybridization, the signal was thousands of fold higher than background.
The oligonucleotide chips were stripped and re-hybridized to examine the durability of the chips. We observed significant signal loss after each round of hybridization and stripping presumably due to the harsh stripping condition (100°C for 1 min) (Fig. 3C). However, even after three to four rounds of hybridization/stripping, the signal levels were still more than 1000 times higher than background signal. This confirms that the immobilization process employed here results in very durable covalent bonds between the nucleic acid moiety and glass surface.
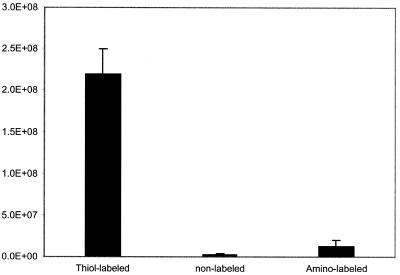
To demonstrate the specificity of the immobilization chemistry, equal molar amount of oligonucleotides containing either thiol groups, amino groups or no label groups was reacted with mercaptosilane separately in solution. After spotting the samples on glass slides, the chips were hybridized with LacCy3 to determine the correlation between immobilization and the modification of oligonucleotides. It was observed that oligonucleotides without any modification or with amino groups were not immobilized significantly (2–5% of the signal as compared to that obtained from thiol modified oligonucleotides) (Fig. 4). These results suggest that immobilization of thiol-labeled oligonucleotides is thiol group specific and the nucleic acid backbone does not contribute to the attachment.
Figure 4.
The specificity of the silanization reaction between thiol-labeled oligonucleotides and mercaptosilane. After reacting thiol-labeled, unlabeled and amino-labeled oligonucleotides with the mercaptosilane, the oligonucleotides were hand-spotted on unmodified glass slides and immobilization was analyzed by hybridization. The signal (photons/spot) from unlabeled and amino-labeled oligonucleotides was only 2 and 5%, respectively, of that of thiol-labeled oligonucleotides.
We also tested other methods of conjugating oligonucleotide and silane. Acrylic-labeled oligonucleotides (Lac-acrylic and Lac-acrylic-sen) with acrylicsilane (γ-methacryloxy-propyl-trimethoxysilane) were conjugated by polymerization (Fig. 2B) and the conjugated molecules were spotted manually and also with an automated arrayer on glass slides. It was estimated that 20% of the input oligonucleotides were immobilized. After stripping with boiling water, the chips were re-hybridized with the same probe and a comparable level of signals was observed. Similarly, the conjugation of thiol-labeled oligonucleotides (Lac-thio and Lac-thio-sen) with aminosilane (3-aminopropyl-trimethoxysilane) was carried out in DMSO using bifunctional cross-linking reagents SPDP or SIAX. Since oligonucleotides cannot be directly dissolved in DMSO, a concentrated oligonucleotide solution was made in water and then the appropriate amount of this stock solution was added to the reaction mixture in DMSO. The conjugated oligonucleotides were spotted onto glass chips directly in DMSO. Detection by hybridization illustrated that this procedure results in a good level of oligonucleotide immobilization and DMSO is compatible with the automated arrayer (data not shown).
The silanized oligonucleotides in DMSO, because of their slow evaporation rate, were exploited to form DNA monolayers on glass slides. Such monolayers will be very useful for fabricating DNA-based biosensors. Aliquots of 10 µl of 20 µM silanized Lac-thio and Lac-thio-sen in DMSO were used to form an oligonucleotide monolayer under a coverslip. The oligonucleotide monolayers formed were evaluated by hybridization with LacCy3. The uniformity of monolayers was further analyzed using ImageQuant™ by sampling 1600 spots from the Lac-thio coated area. Statistics data showed that the standard deviation of the intensity across the 1600 spots was only 16% of the signals. The signal from the control oligonucleotide (Lac-thio-sen) was >80-fold lower.
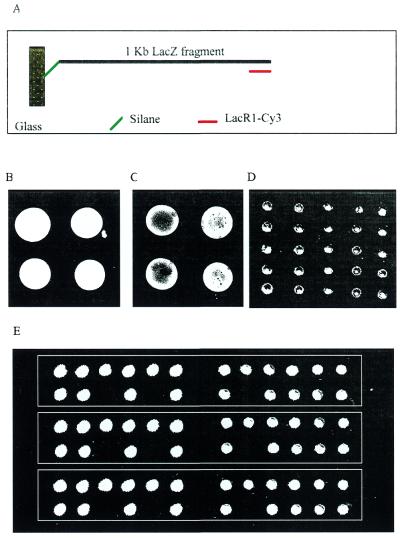
Immobilization of PCR amplified cDNA was attempted. A 1 kb LacZ cDNA fragment was PCR amplified using Lac-Thio-sen and LacR1 so that one strand of the cDNA was thiol-labeled. The PCR products were conjugated with mercaptosilane and the silanized cDNA was spotted on glass slides directly. After routine processing, the chips were hybridized to an oligonucleotide probe complementary to the distal end of the attached cDNA strand (Fig. 5A). The results demonstrate that the method results in rapid immobilization of cDNA also (Fig. 5B–D). The cDNA chips were also stripped and re-hybridized with the same probe, and the result suggested that even for cDNA chips, where hydrolysis of long cDNA chains during prolonged incubation could be substantial, re-use is possible (data not shown).
Figure 5.
Immobilization of thiol-labeled cDNA. (A) Diagram of the immobilization and detection of a LacZ cDNA. The 1 kb LacZ fragment was PCR amplified with one thiol-labeled primer and one unlabeled primer and then conjugated to mercaptosilane. After removing the unlabeled strand of the cDNA, the chips were hybridized to a probe complementary to the distal end of the attached strand. (B) PCR product concentrated 5-fold and spotted manually. (C) and (D) PCR product without concentration spotted manually or with an arrayer, respectively. (E) Twenty-one random mouse cDNAs were PCR amplified and spotted in triplicate after 10-fold concentration and silane conjugation. Immobilization was detected by biotin-labeled RNA probe in combination with Cy3-labeled streptavidin.
For immobilization of random cDNA that has no directional information, we labeled both strands of the cDNA by using a pair of 5′-thiol-labeled primers in PCR amplification. Twenty-one random cDNAs were thus amplified and concentrated 10-fold by precipitation and spotted in triplicate onto unmodified slides. As shown in Figure 5E, the hybridization signals were very strong and uniform. Three slots were left open during arraying to monitor possible carryover of samples. It was demonstrated here that no sample-to-sample carryover can be detected on these chips.
CONCLUSIONS
The emergence of DNA chip (microarray) technologies has revolutionized many aspects of biological research. Access to these important research tools, however, has been limited largely due to the poor availability and affordability of the technologies. The two barriers are related to the low speed and high complexity of the current chip-manufacture methods. A novel maskless method of photolithographic synthesis has been developed recently with the promise of reducing the cost and time of chip production by on-chip synthesis (22). In an effort to simplify the production of microarrays by deposition methods, we introduced the concept of silanized nucleic acids as a potentially universal platform for DNA immobilization. We hope that application of these new methods in chip production will improve the availability and affordability of the DNA chips in the near future.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Prof. Claes Wahlestedt for support, Dr Harold Swerdlow for discussions and Dr Liam Good for help with the manuscript. The research was supported by an intramural grant to Z.L. from Center for Genomics Research, Karolinska Institutet and Pharmacia Corporation.
REFERENCES
- 1.Johnston M. (1998) Curr. Biol., 8, 171–174. [DOI] [PubMed] [Google Scholar]
- 2.de Saizieu A., Certa,U., Warrington,J., Gray,C., Keck,W. and Mous,J. (1998) Nature Biotechnol., 16, 45–48. [DOI] [PubMed] [Google Scholar]
- 3.Pollack J.R., Perou,C.M., Alizadeh,A.A., Eisen,M.B., Pergamenschikov,A., Williams,C.F., Jeffrey,S.S., Botstein,D. and Brown,P.O. (1999) Nature Genet., 23, 41–46. [DOI] [PubMed] [Google Scholar]
- 4.Hacia J.G., Fan,J.B., Ryder,O., Jin,L., Edgemon,K., Ghandour,G., Mayer,R.A., Sun,B., Hsie,L., Robbins,C.M., Brody,L.C., Wang,D., Lander,E.S., Lipshutz,R., Fodor,S.P. and Collins,F.S. (1999) Nature Genet., 22, 164–167. [DOI] [PubMed] [Google Scholar]
- 5.Gilles P.N., Wu,D.J., Foster,C.B., Dillon,P.J. and Chanock,S.J (1999) Nature Biotechnol., 17, 365–370. [DOI] [PubMed] [Google Scholar]
- 6.Dill K., Stanker,L.H. and Young,C.R. (1999) J. Biochem. Biophys. Methods, 41, 61–67. [DOI] [PubMed] [Google Scholar]
- 7.Struelens M.J., De Gheldre,Y. and Deplano,A. (1998) Infect. Control Hosp. Epidemiol., 19, 565–569. [DOI] [PubMed] [Google Scholar]
- 8.O’Donnell-Maloney M.J. and Little,D.P. (1996) Genet. Anal., 13, 151–157. [DOI] [PubMed] [Google Scholar]
- 9.Kurian K.M., Watson,C.J. and Wyllie,A.H. (1999) J. Pathol., 187, 267–271. [DOI] [PubMed] [Google Scholar]
- 10.Drmanac S., Kita,D., Labat,I., Hauser,B., Schmidt,C., Burczak,J.D. and Drmanac,R. (1998) Nature Biotechnol., 16, 54–58. [DOI] [PubMed] [Google Scholar]
- 11.Hacia J.G., Brody,L.C. and Collins,F.S. (1998) Mol. Psychiatry, 3, 483–492. [DOI] [PubMed] [Google Scholar]
- 12.Milosavljevic A., Savkovic,S., Crkvenjakov,R., Salbego,D., Serrato,H., Kreuzer,H., Gemmell,A., Batus,S., Grujic,D., Carnahan,S., Paunesku,T. and Tepavcevic,J. (1996) Genomics, 37, 77–86. [DOI] [PubMed] [Google Scholar]
- 13.Proudnikov D., Timofeev,E. and Mirzabekov,A. (1998) Anal. Biochem., 259, 34–41. [DOI] [PubMed] [Google Scholar]
- 14.Chrisey L.A., Lee,G.U. and O’Ferrall,C.E. (1996) Nucleic Acids Res., 24, 3031–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasmussen S.R., Larsen,M.R. and Rasmussen,S.E (1991) Anal. Biochem., 198, 138–142. [DOI] [PubMed] [Google Scholar]
- 16.Salo H., Virta,P., Hakala,H., Prakash,T.P., Kawasaki,A.M., Manoharan,M. and Lonnberg,H. (1999) Bioconjug. Chem., 10, 815–823. [DOI] [PubMed] [Google Scholar]
- 17.Rogers Y.H., Jiang-Baucom,P., Huang,Z.J., Bogdanov,V., Anderson,S. and Boyce-Jacino,M.T. (1999) Anal. Biochem., 266, 23–30. [DOI] [PubMed] [Google Scholar]
- 18.Cohen G., Deutsch J., Fineberg J. and Levine A. (1997) Nucleic Acids Res., 25, 911–912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beier M. and Hoheisel,J.D. (1999) Nucleic Acids Res., 27, 1970–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipshutz R.J., Fodor,S.P., Gingeras,T.R. and Lockhart,D.J. (1999) Nature Genet., 21, 20–24. [DOI] [PubMed] [Google Scholar]
- 21.McGall G., Labadie,J., Brock,P., Wallraff,G., Nguyen,T. and Hinsberg,W. (1996) Proc. Natl Acad. Sci. USA, 93, 13555–13560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh-Gasson S., Green,R.D., Yue,Y., Nelson,C., Blattner,F., Sussman,M.R. and Cerrina,F. (1999) Nature Biotechnol., 17, 974–978 [DOI] [PubMed] [Google Scholar]
- 23.Milner N., Mir,K.U. and Southern,E.M. (1997) Nature Biotechnol., 15, 537–541. [DOI] [PubMed] [Google Scholar]
- 24.Cheung V.G., Morley,M., Aguilar,F., Massimi,A., Kucherlapati,R. and Childs,G. (1999) Nature Genet., 21, 15–19. [DOI] [PubMed] [Google Scholar]
- 25.Morozov V.N. and Morozova,T.Y.A. (1999) Anal. Chem., 71, 3110–3117. [DOI] [PubMed] [Google Scholar]
- 26.Herne T.M. and Tarlov,M.J. (1997) J. Am. Chem. Soc., 119, 8916–8920. [Google Scholar]
- 27.Ramsay G. (1998) Nature Biotechnol., 16, 40–44. [DOI] [PubMed] [Google Scholar]
- 28.Henke L., Piunno,P.A.E., McClure,A.C., and Krull,U. (1997) Anal. Chim. Acta, 344, 20–30. [Google Scholar]