Abstract
A U3 snoRNA gene isolated from a Chlamydomonas reinhardtii (Cre) genomic library contains putative pol III-specific transcription signals similar to those of RNA polymerase III-specific small nuclear (sn)RNA genes of higher plants. The 222 nt long Cre U3 snoRNA was immunoprecipitated by anti-γ-mpppN antisera, but not by anti-m2,2,7G antibodies, supporting the notion that it is a RNA polymerase III transcript. Tagged Cre U3 snoRNA gene constructs were expressed in Cre cells. Results of chemical and enzymatic structure probing of Cre U3 snoRNA in solution and of DMS modification of Cre U3 snoRNA under in vivo conditions revealed that the two-hairpin structure of the 5′-domain that is found in solution is no longer detected under in vivo conditions. The observed differences can be explained by the formation of several base pair interactions with the 18S and 5′-ETS parts of the pre-rRNA. A model that involves five intermolecular helices is proposed.
INTRODUCTION
The small nucleolar RNAs (snoRNAs) play essential roles in post-transcriptional maturation of RNA polymerase I (pol I) transcripts (pre-rRNA) (reviewed in 1–3). Only a few of the snoRNAs are required for cleavages of this long transcript, which contains the 17/18S, 5.8S and 25/28S rRNAs. The others serve as guide RNAs in nucleotide modification reactions (for reviews see 4,5). Among the snoRNAs involved in the ordered series of cleavages leading to mature 17/18S, 5.8S and 25/28S rRNAs, U3 snoRNA plays an essential role. Abolition of U3 snoRNA gene expression in Saccharomyces cerevisiae blocks 18S rRNA synthesis, whereas the 5.8S and 26S rRNAs are produced in normal amounts (6). U3 snoRNA is involved in the very early cleavage (A0), which takes place in the 5′-external transcribed spacer (5′-ETS) of the pre-rRNA, as demonstrated in mouse cell extracts, in Xenopus oocyte extracts and in yeast (7–10). U3 snoRNA is also involved in cleavages occurring on both sides of the 18S rRNA coding region as shown in yeast and in Xenopus oocytes (6,11) and at a site near the boundary between internal transcribed spacer 1 (ITS1) and 5.8S rRNA, as found in Xenopus oocytes (11). Based on in vivo crosslinking experiments, a base pair interaction (helix V) was first discovered between U3 snoRNA and the 5′-ETS region (12). The biological significance of this interaction was then largely documented by genetic experiments, phylogenetic evidence and direct in vivo analysis of the U3 snoRNA structure in yeast (10,13,14). More recently, based on a phylogenetic study, three intermolecular base pair interactions (helices I, II and III) between U3 snoRNA and pre-rRNA were proposed (15). The U3 snoRNA segments involved are contained in the phylogenetically conserved boxes A and A′ and the pre-rRNA segments concerned are located in the 18S rRNA region. These segments form a pseudoknot structure in mature 18S rRNA (15). Later on, based on an in vivo analysis of the S.cerevisiae U3 snoRNA structure, the model proposed by Hughes was revised and a fourth bimolecular interaction (helix IV) that involves the fourth segment of the 18S rRNA pseudoknot structure was proposed (14). Mutation in the U3 snoRNA region involved in the formation of helices I–III (14–16) demonstrated the functional importance of these U3 snoRNA sequences. More recently, the functional importance of helix II was demonstrated by generation of compensatory mutations in U3 snoRNA and the 18S part of pre-rRNA (17). When the same kind of experiment was performed for helix I, no compensatory effect was observed (17). However, this may reflect an importance of the primary structure, together with the secondary structure. Hence, complementary experiments are needed to confirm the proposed model of interaction between U3 and the pre-rRNA. One way to obtain additional support for this model is to probe the in vivo structure of U3 snoRNA in a phylogenetically distant species. Chlamydomonas reinhardtii (Cre) is an ideal model for such a study, since DMS can be used as a probe of RNA structure in this unicellular alga.
Another point of interest in using Cre U3 snoRNA as a model concerns the enzyme specificity of gene transcription. The U3 snoRNA gene and the telomerase RNA gene are the only known examples of genes that are transcribed by different RNA polymerases in different organisms (for the telomerase see 18,19). In vertebrates and yeasts U3 snoRNA is transcribed by RNA polymerase II (pol II) (20,21) and in higher plants by RNA polymerase III (pol III) (22,23). This difference is also evident in the presence of two different cap structures at the 5′-end of vertebrate and yeast U3 snoRNAs (m2,2,7Gppp) and plant U3 snoRNAs (γmppp) (20,24,25). It was thus of interest to determine at which stage of evolution the change in RNA polymerase took place. The study of a U3 gene of a unicellular alga, which is probably a remote ancestor of higher plants, was expected to shed some light on this problem. Combining direct U3 snoRNA 3′-terminal sequence analysis with cDNA synthesis and sequence analysis, the complete sequence of Cre U3 snoRNA was established. The cDNA generated using U3 snoRNA as the template was used to isolate a U3 gene from a Cre genomic library. Cre cell transformation experiments indicated that the isolated gene was an expressed bona fide U3 gene. The results obtained strongly support the idea that the Cre U3 gene is a pol III transcript.
In this paper we describe the nucleotide sequence of Cre U3 snoRNA and its gene, we characterize the cap structure of Cre U3 snoRNA and we analyze under in vivo conditions the structure of the Cre U3 snoRNA 5′-terminal domain. Finally, we propose a model for the interaction between Cre U3 snoRNA and the 18S part of the Cre pre-rRNA and identify hinge 2 of Cre U3 as a potential target for an interaction with the 5′-ETS region of pre-rRNA.
MATERIALS AND METHODS
Isolation of U3 snoRNA from Cre cells and nucleotide sequence determination
U3 snoRNA was extracted from strain CW15 of Chlamydomonas reinhardtii (a kind gift of Dr C.F. Beck) grown under conditions described previously (26). First, total RNA from Cre cells was phenol extracted at 65°C as described by Steele et al. (27) and separated on 10% polyacrylamide–8 M urea gels. The band corresponding to U3 snoRNA, detected by staining with ethidium bromide, was cut from the gel. Elution from the gel slices, 3′-end-labeling and chemical sequencing of the 3′-portion of the RNA were done as reported earlier (28). For the synthesis of a complete cDNA corresponding to the Cre U3 snoRNA, the first strand was obtained with a synthetic oligonucleotide primer (P/3′Eco) complementary to the 3′-end of the RNA, 5′-GGAATTCACCCATCTGAATGCCC-3′. Elongation with M-MLV reverse transcriptase (Gibco BRL, UK) was done according to the manufacturer’s protocol, with addition of 10 µCi [α-32P]dCTP. After completion of the reaction, a 1/20 vol of 3 M NaOH was added to the mixture, followed by boiling for 5 min. The labeled first strand cDNA was repurified from a 10% polyacrylamide–8 M urea gel. The recovered single-stranded cDNA was dG tailed using terminal deoxynucleotidyl transferase (US Biochemical, USA) according to Eschenfeldt et al. (29) and PCR amplified by Vent DNA polymerase (Biolabs, UK) using a synthetic oligonucleotide [oligo(dC)12/EcoRI] as the second primer. After purification on a 2% agarose gel, the amplified DNA fragment was cloned into the SmaI site of the pBS(–) vector pCRU3C (Stratagene, USA). For double-stranded DNA sequencing Sequenase (US Biochemical) was used according to the supplier. Identification of the cap structure carried by the Cre U3 snoRNA was achieved by immunoaffinity chromatography using anti-m2,2,7G IgG and anti-γmpppN IgG, as previously described (26,30).
Secondary structure alignment of Cre U3 snoRNA with those of different organisms
Different U3 snoRNA sequences proposed to be folded into a two stem–loop structure were in a first step treated with the program described by Lück et al. (31), then alignment was refined manually in order to align the phylogenetically conserved boxes and for a better representation of compensatory mutations.
Expression of Cre U3 snoRNA gene constructs in a homologous system
The gene coding for Cre U3 snoRNA (the Cre U3 gene) was isolated from a Chlamydomonas reinhardtii wild-type 2137 A(+) genomic library constructed in the λ-EMBL3 vector (kindly provided by M. Goldschmidt-Clermont, Geneva, Switzerland). The genomic library was screened with an [α-32P]-labeled DNA fragment generated by PCR amplification of the Cre U3 snoRNA cDNA (pCRU3C) with M13 forward and reverse sequencing primers (32). Hybridization was performed for 20 h at 42°C in 6× SSC, 5× Denhardt’s solution, 0.5% SDS, 100 mg/ml denatured chicken blood DNA and 50% formamide. Filters were washed twice in 2× SSC, 0.1% SDS at room temperature and twice in 0.1× SSC, 0.1% SDS at 55°C for 30 min. Five positive phage were isolated after screening of 2 × 105 plaques. One λ-EMBL3 recombinant phage encoding a Cre U3 gene was mapped with restriction enzymes. Upon hybridization with the pCRU3C probe a 3 kb SalI–PstI fragment from the insert was found to contain the Cre U3 gene. This SalI–PstI fragment was subcloned and sequenced and a 524 bp fragment was found to carry the entire 222 nt long U3 snoRNA coding region with 171 and 131 nt long upstream and downstream flanking sequences, respectively. This 524 bp fragment was amplified by PCR using primers 5′-GCGCCCCCCGCCGCTTTTC-3′ (5′primer) and 5′-CCACGCACTTCATGCACC-3′ (3′primer) and Pfu DNA polymerase (Stratagene) and ligated into the EcoRV site of plasmid pBKSII(+) to give the recombinant plasmid pCRU3G. For the production of variant U3 genes, plasmid pCRU3G served as the template for PCR-directed mutagenesis using the ‘add-on primer’ strategy (33). Construct pCRU3GM1 produced a Cre U3 snoRNA (225 nt) with a substitution of the loop Ib sequence CUUC (positions 53–56) by the sequence GAAUUCU and construct pCRU3GM2 produced a Cre U3 snoRNA (223 nt) where the sequence UCCUC (positions 70–74) was replaced by GAAUUC. RNase A/T1 mapping was carried out as described by Goodall et al. (34). Sequence-specific RNA probes were synthesized in vitro using T3 or T7 RNA polymerase in the presence of [α-32P]UTP (sp. act. 30–40 Ci/mmol) on HindIII-linearized pCRU3G, pCRU3GM1 and pCRU3GM2 plasmids.
Chemical and enzymatic probing of in vitro produced Cre U3 snoRNA
To obtain the DNA matrix for in vitro transcription of Cre U3 snoRNA, the Cre U3 snoRNA coding sequence of plasmid pCRU3G was PCR amplified. One of the primers, 5′-CGGGATCCTAATACGACTCACTATAGGATGACCGTACTTGAA-3′ (P/Bam/T7/5′), generated a BamHI restriction site (underlined), a T7 RNA polymerase promoter (bold) and contained the 5′-sequence of Cre U3 snoRNA (italic). The second primer was oligonucleotide P/3′Eco. The amplification product was cloned into the BamHI and EcoRI sites of plasmid pBSKII(+) (pCRU3WT). Prior to transcription by T7 RNA polymerase, plasmid pCRU3WT was linearized with EcoRI. Owing to the location of the EcoRI restriction site, the U3 snoRNA transcript contained five additional nucleotides at its 3′-end as compared to the authentic RNA. In vitro transcription was carried out as described earlier (35). In vitro structure probing with chemical reagents (DMS and CMCT) was performed on unlabeled Cre U3 snoRNA transcript under the conditions described previously (35). In addition, methylations at N-7 G positions were detected by further treatment of DMS-modified samples with sodium borohydride and aniline, respectively, according to Peattie (36). Positions of modifications were identified by primer extension analysis, using the primers P/Internal Loop (5′-GCATGTCTTCGTCTGGGTTGAGGA-3′) and P/BoxB (5′-CCACAGTGGTCATACG-3′) complementary to sequences between positions 70–87 and 125–115, respectively, and AMV reverse transcriptase (Life Sciences, USA). The conditions used for primer extension were as previously described (35). Structure probing with RNase V1 (cobra venom; US Biochemical) was carried out on 5′-end-labeled RNA transcripts. Between 10 and 15 pmol gel-purified, dephosphorylated RNA transcript was 5′-end-labeled in the presence of 100 µCi [γ-32P]ATP (Dupont/NEN, USA) and 10 U T4 polynucleotide kinase (Biolabs) (37). Limited digestion of 5000 c.p.m. RNA was performed in the presence of 2.5 µg tRNA, with 0.01 or 0.03 U/µg RNase V1. The digestion products were separated on a 10% sequencing gel. As a control, limited alkaline hydrolysis and RNase T1 sequencing were performed according to Kiss et al. (38).
Structure probing in vivo
The procedure described by Zaug and Cech (39) was used for structure probing analysis of Cre U3 snoRNA in vivo. Cre cells were grown up to a concentration of 105 cells/ml (exponential phase), harvested and resuspended at 108 cells/ml concentration, in 10 ml TMS buffer (10 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 3 mM CaCl2, 250 mM sucrose). Two milliliters of this culture were treated with 20, 40 or 80 µl of DMS for 2 min at room temperature with gentle shaking, then 200 µl of 14.3 M β-mercaptoethanol (0.7 M final concentration) were added to quench the DMS reaction. As a control, an aliquot was treated in the same way except that no DMS was added. RNA was isolated from Cre cells as described by Jakab et al. (26). The RNA pellet was dissolved at 10 mg/ml concentration and 1 µl was used for primer extension analysis, according to Méreau et al. (14).
RESULTS
Identification and nucleotide sequence analysis of Cre U3 snoRNA
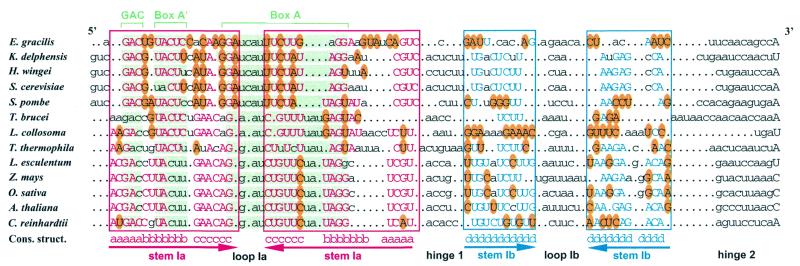
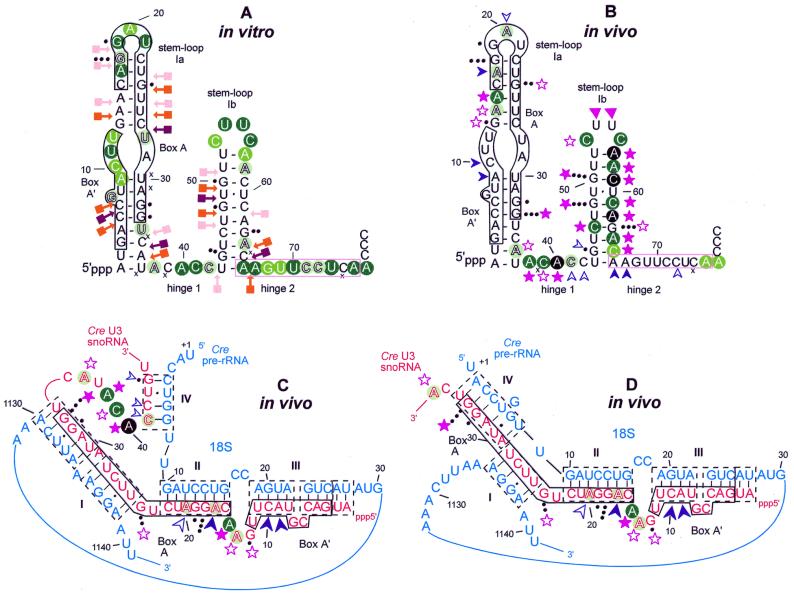
When low molecular weight RNAs isolated from Cre cells were fractionated on 10% polyacrylamide gels under denaturing conditions, a band with a mobility similar to that of U3 snoRNA from other organisms appeared on the gel (not shown). The 3′-part of this RNA was chemically sequenced and shown to contain the characteristic box D sequence of U3 snoRNAs. An oligonucleotide primer complementary to the very 3′-end of this molecule was used for first strand cDNA synthesis. After G tailing, a dC oligonucleotide was used for synthesis of the second strand. The resulting cDNA was PCR amplified, cloned (pCRU3C) and the entire sequence established (EMBL accession no. AJ001179). The 222 nt long sequence of Cre U3 snoRNA was compared to those of U3 snoRNAs from various species. An alignment of the 5′-portion of U3 snoRNAs according to secondary structure is shown in Figure 1. The 75 nt long 5′-domain of the Cre U3 snoRNA which contains the conserved boxes A′ and A is assumed to be involved in base pair interaction with phylogenetically conserved regions of the 18S moiety of pre-rRNA (12,15). The 147 nt long 3′-domain of U3 snoRNAs encompasses the evolutionarily conserved boxes B, C, C′ and D, supposed to be associated with proteins (14,16,40–45). The high degree of sequence similarity observed between the 5′-domain of Cre U3 snoRNA and its counterparts in U3 snoRNAs from other species is in line with the above proposal. It should be noted that, whereas the GC content of most spliceosomal UsnRNAs from Cre is ∼65%, that of the 5′-domain of U3 snoRNA is as low as 42.7%. This supports a functional difference between the two domains of Cre U3 snoRNA. With respect to both sequence and proposed secondary structure, among the U3 snoRNAs that we compared [including yeasts, trypanosomes, unicellular algae, plants (Fig. 1) and vertebrates (not shown)], Cre U3 snoRNA shows the strongest similarity with the higher plant U3 snoRNAs. Whereas the 5′-domain of vertebrate U3 snoRNAs is proposed to be folded into a single stem–loop structure, the 5′-domain of plant U3 snoRNAs is proposed to be folded into a two stem–loop structure (46), and a similar two stem–loop structure has been experimentally found in yeast U3 snoRNAs (14,44,47). The comparison shown in Figure 1 predicts a two stem–loop structure for the Cre U3 snoRNA 5′-domain and, as illustrated in Figure 1, numerous base-compensatory mutations have preserved the possibility to form a two stem–loop structure in the 5′-domain of U3 snoRNA in a large number of species, including C.reinhardtii. In the present paper, we provide experimental evidence for the presence of this structure in the free Cre U3 snoRNA and demonstrate its complete opening in vivo, due to base pair interactions with the pre-rRNA.
Figure 1.
Compensatory base mutations conserved the possibility of forming a two stem–loop structure in yeast, ciliate, unicellular algal and plant U3 snoRNAs. The sequences of the 5′-terminal domain from Euglena gracilis (Eg27297), Kluyveromyces delphensis (Kdz78433), Hansenula wingei (Hwu3sno), Saccharomyces cerevisiae (Scsnr17a), Schizosaccharomyces pombe (Spsnru3), Trypanosoma brucei (Tburb), Leptomonas collosoma (Lctgv3snr), Tetrahymena thermophila (Ttsnru31), Lycopersicon esculentum (Leu3snr), Zea mays (Zmu3snrng), Oryza sativa (Osu3snrn), Arabidopsis thaliana (Atu3csnr) and Chlamydomonas reinhardtii (Crj001179) were first subjected to the program developed by Lück et al. (31). Then alignment was manually refined for a better alignment of the phylogenetically conserved boxes GAC, A′ and A (highlighted in green) and for a better representation of co-variations. The nucleotide sequences involved in the stem of stem–loop Ia are boxed in red. Stems are marked below by inverted arrows. The base paired residues are in capital letters. Red capital letters correspond to residues fitting the consensus sequence established from the alignment. Nucleotide variations from this consensus sequence that were found to preserve base pair interactions are circled in yellow. Dots indicate missing nucleotides as referred to the alignment. The same representation is used for stem–loop Ib, except that blue is used instead of red. The sequences can be accessed via the EMBL-ID at http://srs.ebi.ac.uk:5000/site
Cre U3 snoRNA has a γmppp cap and its gene is likely to be transcribed by RNA polymerase III
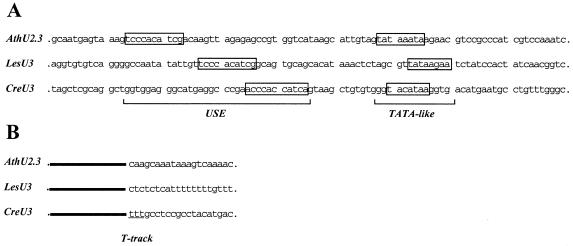
Figure 2 shows that Cre U3 snoRNA is precipitated by anti-γmpppN antibodies, as found for U3 snoRNA from broad bean, but was not precipitated by anti-m2,2,7GpppN antibodies, as was broad bean U2 snRNA. This indicates that its cap structure is γmppp, like that of the higher plant U3 snoRNAs (25). The presence of a γmppp cap at the 5′-extremity of Cre U3 snoRNA strongly suggests that the Cre U3 snoRNA gene is transcribed by pol III. To obtain more information on this point, a Cre genomic library was first screened with labeled U3 snoRNA as probe. The isolated gene candidate was cloned (pCRU3G) and sequenced as described in Materials and Methods. The sequence of the coding region of the gene was identical to that of Cre U3 snoRNA established above by cDNA sequence analysis. The 5′-flanking region contains a TATA-like box (TACATAA) located between positions –31 and –25 and an ACCCACCATCA sequence positioned between –56 and –46 (Fig. 3). This sequence is highly similar to that of the upstream sequence element (USE) characterized in plant U3 snoRNA genes (Fig. 3; 48). The distance observed between these two putative promoter elements, as well as the presence of a U track abutting the 3′-end of the coding region, are strong indications that the isolated gene is transcribed by pol III.
Figure 2.
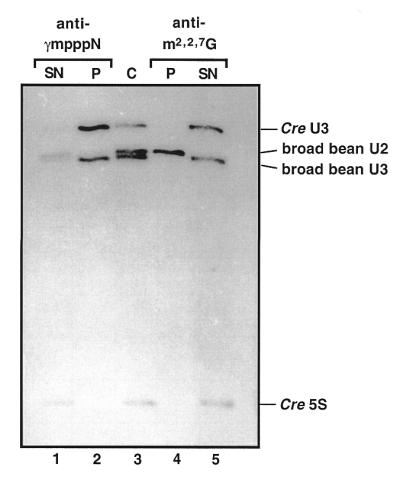
Cap structure of Cre U3 snoRNA. U3 snoRNA from Cre and from broad bean, U2 snRNA from broad bean and 5S RNA from Cre were 3-end-labeled, mixed and run on a 10% polyacrylamide gel under semi-denaturing conditions either directly (C, lane 3) or after immunoprecipitation with anti-γmpppN (lanes 1 and 2) or anti-m2,2,7G (lanes 4 and 5) antibodies. RNAs present in the precipitates (P) and in the supernatants (SN) are shown in lanes 2 and 4 and lanes 1 and 5, respectively. Their identities are indicated on the right side of the figure.
Figure 3.
Comparative analysis of the Cre U3 gene. The upstream (A) and downstream (B) non-coding regions of the Cre U3 gene that we isolated were aligned with those of the A.thaliana (AthU2.3) (55) and L.esculentum (LesU3) (48) U3 genes. In (A) the upstream (USE) and TATA-like promoter elements identified for the AthU2.3 and LesU3 genes are boxed. A sequence of the Cre U3 gene showing 72.7% identity with the AthU2.3 and LesU3 USE elements is boxed as well as a putative TATA box. In (B) the track of T residues following the Cre U3 coding sequence is underlined.
To see whether this gene candidate is a bona fide U3 snoRNA gene, we checked its expression in transformed Cre cells. To differentiate its expression from that of the endogenous Cre U3 gene, the cloned Cre gene (pCRU3G) was tagged. On the basis of the secondary structure prediction in Figure 1, tag sequences were introduced into segments known to form stem–loop structure Ib and hinge 2 (see Materials and Methods), resulting in constructs pCRU3GM1 and pCRU3GM2, respectively. Cre cells were transformed with plasmids encoding either of the two mutant Cre U3 genes. The mutant genes can express 225 and 223 nt long transcripts, respectively. Expression of each of the two mutant genes was tested by RNase A/T1 protection assay, using 527 and 525 nt single-stranded RNA probes, complementary to the U3 snoRNAs expressed by plasmids pCRU3GM1 and pCRU3GM2, respectively. In the control experiment, the probe was complementary to the wild-type (222 nt long) RNA. As shown in Figure 4, the expression of both mutants could be detected. However, only a low level of protected probe was observed for a construct derived from pCRU3GM2, which is an indication of a low level of RNA corresponding to this mutant. Altogether, the results show that the isolated Cre U3 snoRNA gene is a bona fide gene.
Figure 4.
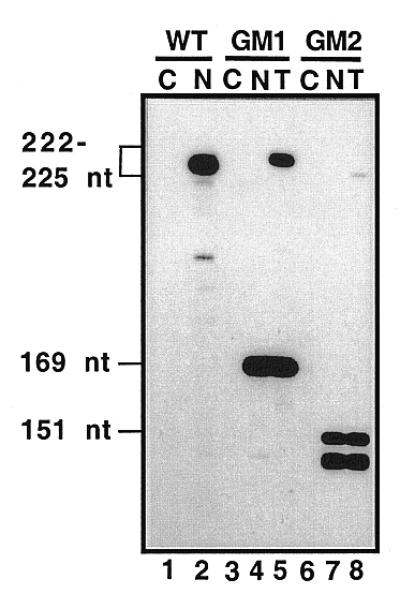
Expression of Cre U3 snoRNA genes in Cre cells analyzed by RNase A/T1 mapping. Cre cells were transformed with plasmid pCRU3GM1 or pCRU3GM2, coding for a Cre U3 snoRNA (225 nt) with a substitution in loop Ib (positions 53–56) and a Cre U3 snoRNA (223 nt) with a substitution in sequence 70–74, respectively (see Materials and Methods). RNAs extracted from non-transformed cells (N, lanes 2, 4 and 7) and transformed cells (T, lanes 5 and 8), as well as carrier tRNA from E.coli used as a control (C, lanes 1, 3 and 6), were hybridized with uniformly labeled probes prepared by transcription of plasmid pCRU3G (WT, lanes 1 and 2), pCRU3GM1 (GM1, lanes 3–5) or pCRU3GM2 (GM2, lanes 6–8). After RNase A/T1 digestion, the resistant fragments from the RNA probes were analyzed by electrophoresis on a 7% polyacrylamide gel. In lane 2, the wild-type probe (222 nt in length) is protected by the endogenous U3. In lanes 4 and 5, a 165 nt long fragment resulted from partial protection of the mutated probe by endogenous U3 and in lane 5, a full-length probe resulted from protection by the expressed GM1 U3 mutant RNA (225 nt). In lanes 7 and 8, the 150 nt long fragment corresponds to partial protection of the GM2 probe by endogenous U3. In lane 8, the full-length probe was only protected in tiny amounts, indicating a low level of GM2 mutant U3 snoRNA (223 nt) in the transformed cells.
Structure of the Cre U3 snoRNA 5′-terminal domain in vivo
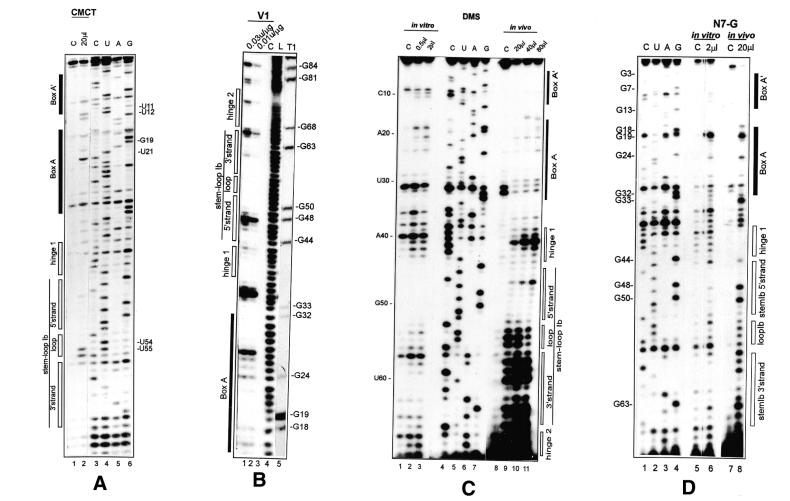
Recent literature data show that owing to its interaction with pre-rRNA, the 5′-terminal structure of U3 snoRNA is different in semi-purified U3 snoRNP and in vivo (14,41,43). To obtain information on the pre-rRNA–U3 snoRNA interaction in Cre, the structure of the 5′-terminal domain of Cre U3 snoRNA was probed: (i) in solution using chemical reagents and enzymes; (ii) in vivo using DMS. The in vitro study of the Cre U3 snoRNA 5′-domain confirmed the prediction of a two stem–loop structure (Figs 5 and 6A). Note that since neither U49 nor U60 residues were modified by CMCT and that the bond G48–U49 was cleaved by RNase V1, these two residues may be hydrogen bonded in stem Ib. To test for U3 snoRNA structural changes in vivo, Cre cells were exposed to DMS and the methylation pattern of U3 snoRNA was analyzed by primer extension as described in Materials and Methods (Fig. 5). The results are summarized in Figure 6B and models of interaction with the 18S part of the pre-rRNA are proposed in Figure 6C and D.
Figure 5.
Secondary structure analysis of the Cre U3 snoRNA 5′-domain in solution (A–E, lanes marked in vitro) and in vivo (C–E, lanes marked in vivo). Chemical reagents and enzymes were as described in Materials and Methods. Positions of cleavages and modifications were identified by extension of primer P/Internal Loop (A–D) or P/BoxB (E) with reverse transcriptase. The cDNAs were fractionated on a 7% sequencing gel. The amounts of chemical reagents (DMS and CMCT) and enzymes (RNases V1 and T1) used are indicated at the top of the Figure. Control lanes (C) correspond to primer extensions made on RNA incubated in the absence of chemical reagent or enzyme. Lanes C, U, A and G correspond to the sequencing ladder obtained with the same primer. Nucleotide positions in Cre U3 and secondary structural elements are marked on the left and right sides of the pictures.
Figure 6.
Schematic representation of the results of chemical and enzymatic probing on the secondary structure proposed for the Cre U3 snoRNA 5′-domain. (A) In vitro probing. The phylogenically conserved boxes A′ and A are framed in black. Nucleotides modified by DMS or CMCT at Watson–Crick positions are circled. N7-G methylations are shown by black dots; the number of dots reflects the intensity of the modification. Positions of RNase V1 cleavages are indicated by arrows linked to squares. Intensity of the colors (green for chemicals, orange for RNase V1) indicates the intensity of the modification or yield of cleavage. Crosses (×) indicate pauses of the reverse transcriptase. (B) In vivo probing. The code for DMS modification is as in (A). In addition, extremely strong modifications are shown in black. Decreased sensitivity to DMS in vivo as compared to in vitro is indicated by a blue arrow: moderate protection by an open arrow, strong protection by a full arrow. Increased sensitivity is indicated by a purple star: a moderate increase by an open star, a strong increase by a full star. Unusual modifications of U residues are indicated by purple triangles (see text for explanation). The hinge 2 sequence proposed to be complementary to the 5′-ETS rRNA is framed in pink. (C and D) Two alternative models of the Cre U3 snoRNA–pre-rRNA interaction. Bimolecular helices I–IV between the Cre U3 snoRNA (sequence in red) and the Cre 18S rRNA (sequence in blue) according to Hughes (15) and Méreau et al. (14) are shown. The phylogenetically conserved boxes A and A′ are boxed in black. Variation of reactivity of nucleotides to DMS in vivo, as compared to in vitro, is indicated by the same code as in (B).
Residues A9 and C10, which are constituents of box A′, and residue A20, a constituent of box A, were protected in vivo but not in vitro. This is in agreement with the involvement of residues A9 and C10, and A20 in bimolecular helices III and II, respectively (Fig. 6C). In contrast to their behavior in vitro, residues A14 and, particularly, A15, which are both located between box A′ and box A, were highly available in vivo (Fig. 6B). This fits with their location between two heterologous helices in vivo (Fig. 6C). Interestingly, in the region from position 1 to 28 only residues G13 and G24 were found to have an increased sensitivity to DMS at the N-7 position in vivo. They are both located in a single-stranded segment joining two intermolecular helices. Residue A29 was not reactive to DMS in vivo. As discussed below, there are different possible explanations for this protection.
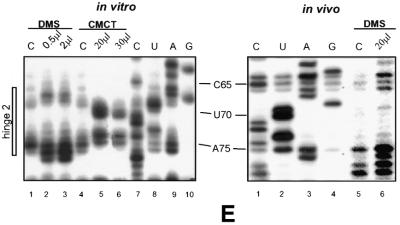
The data also reflected strong structural changes in the hinge 1, helix Ib and hinge 2 regions in vivo. Indeed, an increased reactivity of residue A36 at the border of hinge 1 was observed in vivo, as well as a strong reactivity of residues A38 and C39 in hinge 1. Even the reactivity of residue 40, which is strong in vitro, was reinforced in vivo. In contrast, residues C41 and C42 of hinge 1 were highly protected under in vivo, as compared to in vitro, conditions (Fig. 6B). In the region corresponding to the 3′-strand of stem Ib (Fig. 6B), all the A and C residues, located between positions A57 and A64, showed a very strong increase in reactivity in vivo. This strongly suggests that stem Ib is not present in vivo and that the portion of the Cre U3 snoRNA between positions 53 and 67 is single-stranded. Owing to the limited number of A and C residues in the segment corresponding to the 5′-strand of stem Ib, in vivo DMS modification gave limited information on this region. The only C residue of this strand (C46) was much more reactive in vivo than in vitro. These results suggest that in vivo stem Ib is open and the segment corresponding to nucleotides 41–45 may be base paired with a pre-rRNA sequence. In the hinge 2 sequence, protection against DMS was observed at positions A66, A67, C71, C72 and A76. Altogether, the experimental data suggest that the entire 5′-terminal domain from position 1 to 75 is subjected to conformational changes in vivo. Implication of base pair interactions with the pre-rRNA in these conformational changes is discussed below.
DISCUSSION
Cre U3 snoRNA is a RNA polymerase III transcript
We have isolated a Cre U3 snoRNA gene that was expressed in C.reinhardtii under the control of its own promoter. As the coding sequence of this gene was identical to that of the expressed U3 snoRNA, we assume that the isolated gene is a bona fide Cre U3 snoRNA gene.
U3 snoRNA genes are either transcribed by RNA pol II (vertebrates and yeasts) (21,24,49) or by RNA pol III (higher plants) (23,50; for reviews see 46,51). Our data strongly indicate that U3 snoRNA from the unicellular alga C.reinhardtii is transcribed by RNA pol III. As C.reinhardtii is considered to derive from an early ancestor of higher plants (52), transcription of U3 snoRNA genes by pol III is a very ancient property of photosynthetic eukaryotes. It is difficult to confirm pol III transcription of the Cre U3 snoRNA gene by simple inspection of its promoter sequence elements. Indeed, in plants, there are two upstream promoter elements of basically identical sequence in the pol II and the pol III transcribed UsnRNA and U3 snoRNA genes. Only the distance between these two elements (USE and TATA elements) discriminates pol II from pol III transcripts (about four helical turns in the pol II-transcribed U1, U2, U4 and U5 genes, as against three helical turns in the pol III-transcribed U6 and U3 genes; 53,54). In the Cre U3 snoRNA gene, the distance between the putative USE and TATA elements is of 2.25 helical turns, as against 4 helical turns in the Arabidopsis thaliana U2-3 snRNA gene (55; Fig. 3), which fits well with pol III transcription of the Cre U3 snoRNA. The putative USE element of the Cre U3 snoRNA gene shares stronger homology with the plant USE element (ACCCACCATCA in Cre, TCCCACATCG in plants) than with the DSE element found in the genes of vertebrate UsnRNAs [(Y)ATGYARAT; for reviews see 56,57]. This is in accord with our previous results on the U1 snRNA gene from a slime mold, Physarum polycephalum, which derives from an ancestor at the divergence point between plants and animals (58,59). Altogether, results in P.polycephalum and C.reinhardtii strongly suggest that the characteristic sequence elements found in the promoters of UsnRNA and U3 snoRNA genes of higher plants appeared concomitantly or very soon after the divergence of plants and animals.
The 5′-terminal region of Cre U3 snoRNA forms two stem–loop structures in the free snoRNA
The 5′-domain of U3 snoRNA plays an essential role in U3 snoRNA function. Indeed, base pair interactions with the pre-rRNA is required for pre-rRNA cleavage at sites A0, A1 and A2 in yeast (10,15,17) and at sites 1 and 2 in Xenopus laevis (11,60). Whereas the 5′-domain of vertebrate U3 snoRNA can be folded into a single stem–loop structure denoted I (40,41), the 5′-terminal domain of plant U3 snoRNA and of U3 snoRNAs from several lower eukaryotes (yeasts, ciliates, etc.) can be folded into a two stem–loop structure (Fig. 1; 14,44,48,61,62; for a review see 46). Experimental demonstration of this two stem–loop structure has only been obtained for yeasts (14,44,47). However, Figure 1 clearly shows that several base compensatory mutations contribute to the conservation of this structure in yeasts, ciliates, unicellular algae and higher plants. Trypanosoma brucei is a special case, as only a very short stem–loop structure Ib can be formed (Fig. 1) and this structure may be unstable (43). Here we provide an experimental demonstration of the occurrence of a two stem–loop structure of the Cre U3 snoRNA 5′-terminal domain. Stem–loop structure Ia contains the three phylogenetically highly conserved boxes GAC, A′ and A (16,24,63). As shown in Figure 1, the precise position of these conserved boxes as referred to the helical and bulged parts of helix Ia is not strictly conserved in U3 snoRNAs. This is in agreement with the idea that stem–loop Ia does not correspond to the functional conformation of U3 snoRNA. Interestingly, the helical part of stem–loop Ib shows a high level of sequence conservation in yeasts, some of the ciliates and plants (Fig. 1). However, very strong sequence divergences are observed in the ciliate Leptomonas collosoma and the unicellular alga Euglena gracilis (Fig. 1). Comparison of the 5′-domains of yeast, ciliate, unicellular alga and plant U3 snoRNAs reveals a high degree of variability of the terminal loop of stem–loop Ib, hinge 1 and hinge 2.
The 5′-terminal domain of Cre U3 snoRNA forms at least three heterologous helices with the 18S part of the pre-rRNA
As previously found for S.cerevisiae, the DMS modification pattern of the Cre U3 snoRNA 5′-domain is completely different under in vitro and in vivo conditions. The in vivo data are in accord with formation of the bimolecular helices I–III, involving boxes A′ and A of U3 snoRNA and the 18S part of pre-rRNA, as previously proposed by Hughes (15) and revised by Méreau et al. (14). The Cre U3 nucleotides expected to be involved in helices I and II formation showed DMS protection in vivo. Nucleotides in U3 segments linking these helices showed an increased reactivity in vivo. Based on S.cerevisiae U3 snoRNA analysis in vivo, helix II was proposed to contain 7 bp, as against 8 bp in the Hughes model (14). The present data on Cre confirm the yeast data. Experimental data on yeast (14) also led to the proposal of an extended helix III. Based on sequence comparisons, such an extension is possible in several lower eukaryotes and in plants, but not in vertebrates (14). Here we demonstrate the occurrence of this extension in C.reinhardtii. This is evident from the absence of DMS reactivity of the 5 nt sequence at the 5′-terminus of Cre U3 snoRNA in vivo, while part of its partner sequence in the structure of the free Cre U3 snoRNA becomes available to DMS in vivo (increased reactivity of the N-1 position of residue A36; Fig. 6A and B).
Formation of a fourth helix (IV) between U3 snoRNA and the 18S part of pre-rRNA was also proposed in S.cerevisiae (14). The S.cerevisiae U3 snoRNA sequence involved in this base pair interaction is located downstream of the segment that base pairs with the 5′-ETS region. The corresponding region in Cre U3 snoRNA may be the in vivo protected segment from position 41 to 44. This segment can form a 4 bp interaction, including three G-C pairs and a non-canonical U.U pair, with the 5′-terminal sequence of the 18S rRNA (Fig. 6C). The additional protection of the 3′-terminal part of box A observed in C.reinhardtii, but not in S.cerevisiae, may be explained by extended base pairing with the 18S region from position 1126 to 1130. Another possibility for the formation of helix IV in the Cre system is represented in Figure 6D: in this alternative model, the 5′-terminal part of the 18S rRNA region is base paired with the 3′-terminal part of box A. From the present data, it is difficult to choose between these two alternatives.
As already found in yeast, the single-stranded Cre U3 snoRNA residues located between the bimolecular helices I and II, on the one hand, and II and III, on the other, are accessible to DMS in vivo. This suggests the absence of protection by proteins. In S.cerevisiae, the Mpp10p protein associated with the recently identified proteins Imp3p and Imp4p was proposed to interact with the U3 snoRNA 5′-terminal domain (64,65) and to play an important role for U3 snoRNA function (for a review see 66). As is evident from the DMS reactivity of U3 snoRNA in S.cerevisiae (14) and in C.reinhardtii in vivo, either Mpp10p and its possible counterpart in C.reihardtii exhibit a transitory interaction with the 5′-terminal domain of U3 snoRNA, which does not result in protection of the single-stranded regions involved, or these proteins interact with the bimolecular helices.
The Cre U3 snoRNA stem–loop Ib is disrupted in vivo
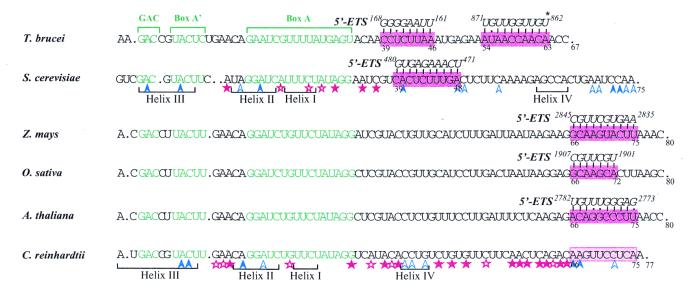
In vivo probing of Cre U3 snoRNA with DMS revealed strong conformational changes in hinges 1 and 2 and stem–loop structure Ib of Cre U3 snoRNA as compared to the structure in vitro. Clearly, stem–loop structure Ib is not formed in vivo. Based on its very high sensitivity to DMS, the segment from position 57 to 65 is single-stranded and devoid of protein interaction in vivo. However, protection of part of hinge 1 and of the N-7 position of residue G44 is observed. As already mentioned, hinge 1 of yeast U3 snoRNA was found to be involved in a base pair interaction with the 5′-ETS region (12,13) and this interaction was found to be necessary for cleavage at sites A0, A1 and A2 of pre-rRNA (10). Similarly, the hinge region of T.brucei U3 snoRNA was proposed to interact with a segment of the 5′-ETS region, adjacent to a recently discovered cleavage site, which shows similarity with the yeast A0 site (67). Hence, in addition to the possible formation of a helix IV with the 5′-end of 18S rRNA, a possible explanation for the observed protection of the Cre U3 segment 41–45 and for the absence of stem–loop Ib formation in vivo may be the formation of a base pair interaction with the 5′-ETS region of the pre-rRNA. Unfortunately, no 5′-ETS sequence from C.reinhardtii is available to confirm this hypothesis. The sequence of the 5′-ETS region has been established for several plants, namely rice (Oryza sativa, X54194), maize (Zea mays, X03989) and Arabidopsis (Arabidopsis thaliana, X15550). As shown in Figure 7, sequence complementarities are observed between the plant 5′-ETS regions and the hinge 1 region of plant U3 snoRNAs. However, the corresponding base pair interactions are of low stability. On the basis of cross-linking data, Hartshorne and Toyofuku (68) proposed several base pair interactions between U3 snoRNA and the 5′-ETS region in T.brucei (Fig. 7). Thus, we made a more extensive analysis of the complementarities between the rice, maize and Arabidopsis 5′-ETS regions and the U3 snoRNA 5′-domains. As shown in Figure 7, a stable interaction may be formed in plants between the hinge 2 region of U3 snoRNAs and the 5′-ETS regions (Fig. 7). A similar base pair interaction in C.reinhardtii would explain the protection observed in hinge 2 from the Cre U3 snoRNA in vivo. However, we cannot exclude the possibility that some of the observed protections are due to RNA–protein interactions. Interestingly, an unusual reactivity to DMS of the two U residues U54 and U55 was detected in vivo. This may be explained by some protein interactions altering the electronegativity of the base ring, as recently observed at the N-7 position of an adenosine residue of the UsnRNA Sm binding sites (69).
Figure 7.
Alignment of the 5′-portion of U3 snoRNAs from T.brucei, S.cerevisiae, Z.mays, O.sativa, A.thaliana and C.reinhardtii showing sequence complementarities with the 5′-ETS regions of pre-rRNAs. Database accession numbers of the U3 snoRNA sequences are indicated in the legend to Figure 1. The phylogenetically conserved boxes GAC, A′ and A are shown in green. The complementary 5′-ETS sequences are in italic and positions of their extremities as referred to the pre-rRNA transcription start site are indicated. The two complementary 5′-ETS sequences given for T.brucei were proposed by Hartshorne and Tokoyufu (68), U862 from the 5′-ETS shown to be cross-linked to U3 snoRNA is marked by an asterisk. The S.cerevisiae 5′-ETS complementarity corresponds to the well-documented helix V (12). Positions of helices I–IV, formed with the 18S part of the pre-rRNA, are shown, as well as increased (red stars) or decreased (blue arrows) sensitivity of A and C residues to DMS in vivo (14). The U3–5′-ETS complementarities proposed for Z.mays, O.sativa and A.thaliana were obtained by a search performed with pattern(n) software (Infobiogen). For C.reinhardtii, the variations of DMS reactivities observed in the present paper are represented by the same code as for S.cerevisiae. Sequences that may form bimolecular helices I–IV with the 18S part of pre-rRNA are indicated, as well as the potential site of interaction with the 5′-ETS region (highlighted in pink).
Altogether, the present data reflect a conformational change of the entire 5′-domain of Cre U3 snoRNA in vivo, due to the formation of several base pair interactions with the 5′-ETS and 18S parts of the pre-rRNA.
Acknowledgments
ACKNOWLEDGEMENTS
This work was supported by laboratory funds from the Ministère de la Recherche et de l’Enseignement Supérieur, the Hungarian OTKA-T016373, the French Centre National de la Recherche Scientifique and EC contract no. CMRX-CT93-0191.
DDBJ/EMBL/GenBank accession no. AJ001179
REFERENCES
- 1.Eichler D.C. and Craig,N. (1994) Prog. Nucleic Acid Res. Mol. Biol., 49, 197–239. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell E.S. and Fournier,M.J. (1995) Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- 3.Raué H.A. and Planta,R.J. (1995) Gene Expr., 5, 71–77. [PMC free article] [PubMed] [Google Scholar]
- 4.Bachellerie J.P. and Cavaillé,J. (1998) In Grosjean,H. and Benne,R. (eds), The Modification and Editing of RNA. ASM Press, Washington, DC, pp. 255–272.
- 5.Ofengand J. and Fournier,M.J. (1998) In Grosjean,H. and Benne,R. (eds), The Modification and Editing of RNA. ASM Press, Washington, DC, pp. 229–253.
- 6.Hughes J.M. and Ares,M.,Jr (1991) EMBO J., 10, 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Craig N., Kass,S. and Sollner-Webb,B. (1987) Proc. Natl Acad. Sci. USA, 84, 629–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kass S., Tyc,K., Steitz,J.A. and Sollner-Webb,B. (1990) Cell, 60, 897–908. [DOI] [PubMed] [Google Scholar]
- 9.Mougey E.B., Pape,L.K. and Sollner-Webb,B. (1993) Mol. Cell. Biol., 13, 5990–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beltrame M. and Tollervey,D. (1995) EMBO J., 14, 4350–4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Savino R. and Gerbi,S.A. (1990) EMBO J., 9, 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beltrame M. and Tollervey,D. (1992) EMBO J., 11, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brulé F., Venema,J., Ségault,V., Tollervey,D. and Branlant,C. (1996) RNA, 2, 183–197. [PMC free article] [PubMed] [Google Scholar]
- 14.Méreau A., Fournier,R., Grégoire,A., Mougin,A., Fabrizio,P., Lührmann,R. and Branlant,C. (1997) J. Mol. Biol., 273, 552–571. [DOI] [PubMed] [Google Scholar]
- 15.Hughes J.M. (1996) J. Mol. Biol., 259, 645–654. [DOI] [PubMed] [Google Scholar]
- 16.Samarsky D.A. and Fournier,M.J. (1998) Mol. Cell. Biol., 18, 3431–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharma K. and Tollervey,D. (1999) Mol. Cell. Biol., 19, 6012–6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greider C.W. (1996) Annu. Rev. Biochem., 65, 337–365. [DOI] [PubMed] [Google Scholar]
- 19.Chapon C., Cech,T.R. and Zaug,A.J. (1997) RNA, 3, 1337–1351. [PMC free article] [PubMed] [Google Scholar]
- 20.Busch H., Reddy,R., Rothblum,L. and Choi,Y.C. (1982) Annu. Rev. Biochem., 51, 617–654. [DOI] [PubMed] [Google Scholar]
- 21.Hughes J.M., Konings,D.A. and Cesareni,G. (1987) EMBO J., 6, 2145–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipowicz W., Kiss,T., Marshallsay,C. and Waibel,F. (1990) Mol. Biol. Rep., 14, 125–129. [DOI] [PubMed] [Google Scholar]
- 23.Kiss T., Marshallsay,C. and Filipowicz,W. (1991) Cell, 65, 517–526. [DOI] [PubMed] [Google Scholar]
- 24.Myslinski E., Ségault,V. and Branlant,C. (1990) Science, 247, 1213–1216. [DOI] [PubMed] [Google Scholar]
- 25.Shimba S., Buckley,B., Reddy,R., Kiss,T. and Filipowicz,W. (1992) J. Biol. Chem., 267, 13772–13777. [PubMed] [Google Scholar]
- 26.Jakab G., Mougin,A., Kis,M., Pollak,T., Antal,M., Branlant,C. and Solymosy,F. (1997) Biochimie, 79, 387–395. [DOI] [PubMed] [Google Scholar]
- 27.Steele W.J., Okamura,N. and Busch,H. (1965) J. Biol. Chem., 240, 1742–1749. [PubMed] [Google Scholar]
- 28.Kiss T., Toth,M. and Solymosy,F. (1985) Eur. J. Biochem., 152, 259–266. [DOI] [PubMed] [Google Scholar]
- 29.Eschenfeldt W.H., Puskas,R.S. and Berger,S.L. (1987) Methods Enzymol., 152, 304–305. [Google Scholar]
- 30.Liu M.H., Busch,R.K., Buckley,B. and Reddy,R. (1992) Nucleic Acids Res., 20, 4299–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lück R., Graf,S. and Steger,G. (1999) Nucleic Acids Res., 27, 4208–4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Balakrishnan R., Frohlich,M., Rahaim,P.T., Backman,K. and Yocum,R.R. (1993) J. Biol. Chem., 268, 24792–24795. [PubMed] [Google Scholar]
- 33.Scharf S.J., Horn,G.T. and Erlich,H.A. (1986) Science, 233, 1076–1078. [DOI] [PubMed] [Google Scholar]
- 34.Goodall G.J., Wiebauer,K. and Filipowicz,W. (1990) Methods Enzymol., 181, 148–161. [DOI] [PubMed] [Google Scholar]
- 35.Mougin A., Grégoire,A., Banroques,J., Ségault,V., Fournier,R., Brulé,F., Chevrier-Miller,M. and Branlant,C. (1996) RNA, 2, 1079–1093. [PMC free article] [PubMed] [Google Scholar]
- 36.Peattie D.A. (1979) Proc. Natl Acad. Sci. USA, 76, 1760–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Donis-Keller H. (1979) Nucleic Acids Res., 7, 179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kiss T., Antal,M. and Solymosy,F. (1987) Nucleic Acids Res., 15, 543–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zaug A.J. and Cech,T.R. (1995) RNA, 1, 363–374. [PMC free article] [PubMed] [Google Scholar]
- 40.Parker K.A. and Steitz,J.A. (1987) Mol. Cell. Biol., 7, 2899–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeppesen C., Stebbins-Boaz,B. and Gerbi,S.A. (1988) Nucleic Acids Res., 16, 2127–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lübben B., Marshallsay,C., Rottmann,N. and Lührmann,R. (1993) Nucleic Acids Res., 21, 5377–5385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hartshorne T. and Agabian,N. (1994) Nucleic Acids Res., 22, 3354–3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fournier R., Brulé,F., Ségault,V., Mougin,A. and Branlant,C. (1998) RNA, 4, 285–302. [PMC free article] [PubMed] [Google Scholar]
- 45.Lafontaine D.L. and Tollervey,D. (1999) RNA, 5, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Solymosy F. and Pollak,T. (1993) Crit. Rev. Plant Sci., 12, 275–369. [Google Scholar]
- 47.Ségault V., Mougin,A., Grégoire,A., Banroques,J. and Branlant,C. (1992) Nucleic Acids Res., 20, 3443–3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kiss T. and Solymosy,F. (1990) Nucleic Acids Res., 18, 1941–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazan S. and Bachellerie,J.P. (1990) Gene, 94, 263–272. [DOI] [PubMed] [Google Scholar]
- 50.Marshallsay C., Connelly,S. and Filipowicz,W. (1992) Plant Mol. Biol., 19, 973–983. [DOI] [PubMed] [Google Scholar]
- 51.Reddy R., Singh,R. and Shimba,S. (1992) Pharmacol. Ther., 54, 249–267. [DOI] [PubMed] [Google Scholar]
- 52.Harris E.H. (1989) The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. Academic Press, San Diego, CA. [DOI] [PubMed]
- 53.Waibel F. and Filipowicz,W. (1990) Nature, 346, 199–202. [DOI] [PubMed] [Google Scholar]
- 54.Waibel F. and Filipowicz,W. (1990) Nucleic Acids Res., 18, 3451–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vankan P. and Filipowicz,W. (1988) EMBO J., 7, 791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dahlberg J.E. and Lund,E. (1988) In Birnstiel,M.L. (ed.), Small Nuclear RNAs. Springer Verlag, Heidelberg, Germany, pp. 38–70.
- 57.Parry H.D., Scherly,D. and Mattaj,I.W. (1989) Trends Biochem. Sci., 14, 15–19. [Google Scholar]
- 58.Myslinski E. and Branlant,C. (1989) Nucleic Acids Res., 17, 9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szkukalek A., Mougin,A., Grégoire,A., Solymosy,F. and Branlant,C. (1996) Biochimie, 78, 425–435. [DOI] [PubMed] [Google Scholar]
- 60.Borovjagin A.V. and Gerbi,S.A. (1999) J. Mol. Biol., 286, 1347–1363. [DOI] [PubMed] [Google Scholar]
- 61.Porter G.L., Brennwald,P.J., Holm,K.A. and Wise,J.A. (1988) Nucleic Acids Res., 16, 10131–10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marshallsay C., Kiss,T. and Filipowicz,W. (1990) Nucleic Acids Res., 18, 3459–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wise J.A. and Weiner,A.M. (1980) Cell, 22, 109–118. [DOI] [PubMed] [Google Scholar]
- 64.Lee S.J. and Baserga,S.J. (1997) Proc. Natl Acad. Sci. USA, 94, 13536–13541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lee S.J. and Baserga,S.J. (1999) Mol. Cell. Biol., 19, 5441–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Baserga S.J., Agentis,T.M., Wormsley,S., Dunbar,D.A. and Lee,S. (1997) Nucleic Acids Symp. Ser., 36, 64–67 [PubMed] [Google Scholar]
- 67.Hartshorne T. (1998) Nucleic Acids Res., 26, 2541–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hartshorne T. and Toyofuku,W. (1999) Nucleic Acids Res., 27, 3300–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hartmuth K., Raker,V.A., Huber,J., Branlant,C. and Lührmann,R. (1999) J. Mol. Biol., 285, 133–147. [DOI] [PubMed] [Google Scholar]