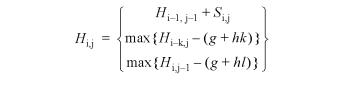Abstract
DbClustal addresses the important problem of the automatic multiple alignment of the top scoring full-length sequences detected by a database homology search. By combining the advantages of both local and global alignment algorithms into a single system, DbClustal is able to provide accurate global alignments of highly divergent, complex sequence sets. Local alignment information is incorporated into a ClustalW global alignment in the form of a list of anchor points between pairs of sequences. The method is demonstrated using anchors supplied by the Blast post-processing program, Ballast. The rapidity and reliability of DbClustal have been demonstrated using the recently annotated Pyrococcus abyssi proteome where the number of alignments with totally misaligned sequences was reduced from 20% to <2%. A web site has been implemented proposing BlastP database searches with automatic alignment of the top hits by DbClustal.
INTRODUCTION
Multiple sequence alignment is one of the most widely used tools in computational biology. It gives a synthetic overview of the homologous regions in a family of proteins in their entirety. This is essential for reliable phylogenetic studies, domain identification and potential splice site recognition and facilitates start codon verification. One important application of multiple alignments is the alignment of a set of sequences detected by a database homology search. Much research has been concentrated on developing efficient and sensitive database search programs such as Psi-Blast (1), SAM-T98 (2) and Fasta3 (3), resulting in the detection of distant, subtle relationships between sequences. These sequences often contain large extensions and insertions, multiple domains, repeats, circular permutations, etc., posing special problems for multiple alignment methods (4). Also, with exponential growth of the sequence databases, the time requirement has become a major limiting factor for automatic database search analysis methods. In particular, the comparative analysis of complete genomes necessitates high throughput, automatic processing of thousands of database homology searches.
A number of systems have already been developed that construct a local multiple alignment of the top scoring sequences identified in a database search. The MaxHom program (5) aligns database search hits to a query sequence, excising unaligned regions from the hits. Taylor (6) used a pattern matching search method to identify sequence segments that are then multiply aligned using Multal (7). Koretke (8) used Macaw (9) to align sequences detected by Psi-blast. The Probe program (10) searches for homologues of a query sequence in a database and locates and aligns conserved patterns in the sequences. However, these alignments only include the most conserved segments, ignoring the more variable regions in between. In contrast, several (semi-)automatic systems incorporate global multiple alignment programs for sequences detected by database searching, but these systems require a certain amount of manual intervention by an expert. Gracy and Argos (11) used a progressive multiple alignment method with manual correction for automatic protein sequence database classification. Jiang and Jacob in EbEST (12) constructed alignments of expressed sequence tags (EST). Baxevanis and Landsman (13) constructed manual alignments of histone fold motifs, then used ClustalW (14) to align the remainder of the sequences. Srinivasarao et al. (15) also used ClustalW to generate multiple sequence alignments in the PIR-ALN database, followed by manual correction.
Some recent developments (16,17) in multiple alignment algorithms have been directed towards combining alignments from various sources, including both local (or motif-finding) and global algorithms, in order to deal with the complex patterns induced by highly variable, modular proteins. Although these methods are capable of producing accurate alignments for many difficult alignment problems, the computer resources required are too expensive for large alignments of numerous proteins.
Here, we present a new program called DbClustal, which is capable of producing high quality global alignments of the top scoring sequences found in a Blast database search, automatically and within the time limits essential to large scale genome analysis projects. DbClustal combines the advantages of both local and global alignment algorithms in a traditional tree-based progressive alignment. The widely used global alignment program ClustalW (14) has been modified to incorporate local alignment data in the form of a list of anchor points between pairs of sequences in the dataset. Here, we use the Blast post-processing program, Ballast (18), to create the anchor points, although other sources of local conservation information could be used. Ballast identifies conserved segments in the sequences detected by Blast and creates a file containing a list of self-consistent anchors between the query sequence and the database hits. By weighting the DbClustal global alignment towards the anchor points, an accurate multiple alignment can be constructed incorporating very long gaps for the terminal extensions and internal insertions. The weighting scheme implemented in DbClustal means that the global alignment is encouraged towards, but not constrained to, the conserved motifs.
A number of examples are presented from a recent genome annotation project (http://www.genoscope.cns.fr/Pab/ ), demonstrating the accuracy and reliability of the DbClustal global alignments even in the case of non-collinear sequences. DbClustal has been incorporated into a web site proposing BlastP database searches with post-processing by Ballast and multiple alignment of the top scoring database sequences with DbClustal.
MATERIALS AND METHODS
The DbClustal program is a modified version of the ClustalW multiple alignment program. It is written in the ANSI C programming language and has been tested on various UNIX (Digital UNIX, SGI IRIX and SUN Solaris), PC (MS-DOS and Windows), Macintosh 68000 and PowerMac systems. The Ballast program is also written in ANSI C and is available by ftp from ftp://ftp-igbmc.u-strasbg.fr/pub/Ballast . A web site (http://igbmc.u-strasbg.fr:8080/ballast.html ) has been implemented which provides searches in protein databases (SwissProt and SPTrEMBL) using the BlastP program. Post-processing the database search results by Ballast takes <2 s on a DEC Alpha Server 8200 with a query sequence of 1000 amino acids. The 50 top scoring sequences detected by BlastP with an E-value less than 10–3 are aligned with DbClustal. The source code for DbClustal is freely available by ftp from ftp://ftp-igbmc.u-strasbg.fr/pub/DbClustal
Modularisation of the ClustalW program
To facilitate the addition of new modules in ClustalW, some modularisation of the program was necessary. The modified program retains the same functionality as ClustalW v.1.81 and exactly the same alignments are produced by the two versions of the program. The modular version of the ClustalW source code is available at the DbClustal ftp address. Future versions will be made available separately when new modules are incorporated into the program.
Creation of anchors by Ballast
Ballast stacks ungapped segment pairs deduced from a BlastP search (gapped alignments or HSPs) to build a profile of conservation along the query sequence. This profile, which uses only those HSPs detected with an E-value less than 0.1, is then used to predict local maximum segments (LMS) in the query sequence, i.e. sequence segments conserved relative to their flanking regions. The length of an LMS is equal to the width of the corresponding peak in the conservation profile, measured between the points of inflection. Local maximum segment pairs (LMSPs) are then defined as the segments of the Blast alignment overlapping the LMSs. Each LMSP is assigned a score depending on the profile height and the similarity between the query and the database sequence, as described in Plewniak et al. (18). LMSPs can thus define anchors between the query and database sequences with a weight equal to the LMSP score divided by the maximum possible score in the current search. One of the outputs from the Ballast program is a file containing a consistent set of anchors between the query sequence and the top scoring database sequences. The first line of the anchor file contains the word ‘Ballast’ followed by the maximum possible score for a motif. Each anchor is then entered on a single line with the following format:
seq: NAME1 NAME2 pos: P beg: R1 R2 len: L weight: W
where NAME1 and NAME2 are the names of two sequences, P is the position in sequence 1 of the corresponding local peak of the Ballast conservation profile (this is not currently used by DbClustal), R1 and R2 are the first residues in the motif for sequences 1 and 2, respectively, L is the length of the anchor and W is the Ballast weight for the anchor.
Incorporating the anchors in the global alignment
ClustalW uses the global dynamic programming algorithm first developed by Needleman and Wunsch (19) to construct a multiple alignment by aligning the most closely related sequences first and progressively aligning larger and larger groups of sequences until all sequences are aligned. The dynamic programming algorithm for two sequences X and Y of length N and M requires a score for aligning any two residues Xi and Yj, plus penalties for opening and extending gaps in the alignment. For details of the ClustalW alignment algorithm see Thompson et al. (14). The recursive algorithm may be summarised as follows:
where Si,j is the score for aligning residues Xi and Yj, g is the penalty for opening a gap and h is the penalty for extending a gap by one residue. For ClustalW, the score Si,j is simply equal to the residue comparison matrix score Ci,j for the two residues. The alignment of two groups of sequences (or profiles) is a simple extension of the algorithm where the score for aligning two residues is replaced by the score for aligning two columns in the respective profiles.
The score for aligning two residues (or profile columns) has been further modified in DbClustal to incorporate local conservation information. DbClustal reads a list of anchors from a Ballast format anchor file. During the progressive multiple alignment, an M × N position-specific anchor matrix is calculated for each pair (or group) of sequences to be aligned. For column i in the first group of sequences and column j in the second group, the anchor matrix score ANCHORij is:
ANCHORi,j = maxk = 1,L(0,Wk)
for all anchors containing any pair of residues in columns i,j, where Wk is the weight defined in the Ballast format file.
For a pair of sequences, the score for aligning residues Ai and Bj is now defined as:
Ci,j + ANCHORi,j
where Ci,j is the residue comparison matrix score for Ai and Bj.
Similarly, the score for aligning two groups of sequences is defined as:
Pi,j + ANCHORi,j
where Pi,j is the profile-to-profile score for Ai and Bj.
The penalties for opening and extending gaps remain the same.
Propagation of the anchors
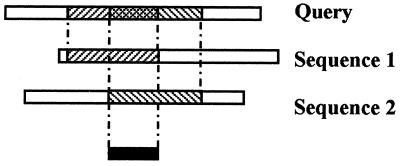
The Ballast program determines anchors for each database sequence relative to the query sequence only. No anchors are provided between pairs of database sequences. Therefore, DbClustal provides an option to propagate these anchors between all the sequences. If two anchors in two different database sequences overlap on the query sequence, a new anchor is created between the two database sequences. The method is demonstrated in Figure 1.
Figure 1.
Propagation of the anchors between the query and each database sequence (shaded regions). The overlapping segments in the database sequences are used as an anchor between the two sequences (shown in black).
Implementation
As DbClustal is intended for use in automatic systems, all options to the program are provided through a command line interface similar to that used by ClustalW. All the alignment options provided by ClustalW are also available in DbClustal. Thus, to perform a multiple alignment using default parameters, the command line would be:
dbclustal seq_file
where seq_file is the name of the file containing the sequences to be aligned.
The minimum input to DbClustal is a file containing the unaligned sequences and the Ballast format anchor file. Two new options have been implemented in order to incorporate the anchors:
motifs=anchor_file where anchor_file is the name of
the file containing the anchors.
propagate propagate the anchors between all
sequences.
Evaluating the quality of a multiple alignment
In order to compare the alignments produced by DbClustal with other methods, an estimate of the quality of a multiple alignment is required. The most popular way to determine whether an alignment program has successfully aligned a group of sequences has been to compare the program alignment with a ‘standard of truth’, generally an alignment that has been generated by superposing the secondary or tertiary structures of the proteins (4,20–22). In the absence of such a ‘reference’ alignment, three measures of alignment quality are generally used: the (weighted) sum of pairs, the minimum entropy and the mean distance (MD) score first introduced in the ClustalX program (23). In ClustalX the conservation of each column in the alignment is estimated using a geometric analysis based on an N-dimensional sequence space, where N is the number of residues. The method is fully described by Thompson et al. (23); however, a brief description is presented here. For a specified column in the alignment, each sequence i is assigned a point Si in the space and a consensus value X is calculated depending on the observed frequencies of the N residues in the column. The score for the column is defined as the mean of the distances between each sequence point Si and the consensus position X. Finally the column scores are normalized by multiplying by the percentage of sequences which have residues (and not gaps) at this position. We have used the MD method to estimate the quality of the full global alignment by summing the conservation scores over all columns in the alignment. As the absolute value of all three quality measures depends on the similarity of the sequences to be aligned, none of the methods is capable of defining the biologically ‘correct’ alignment. Nevertheless, in tests performed using the BAliBASE benchmark alignment database (24), the MD score has proved to be better correlated with validated structural alignments. Notably, the MD score is better able to identify significant improvements in alignments containing sequences sharing low per cent residue identity, those containing a family of sequences aligned with a single, more divergent family member or sequences with large N/C-terminal extensions or internal insertions. In the tests, we used the ratio of the MD scores to compare two alignments obtained by different methods. An MD score ratio greater than 1.05 generally indicated significant differences in the two alignments. Below this threshold, the differences between the alignments were generally observed to be negligible.
RESULTS AND DISCUSSION
Modularisation of the ClustalW code facilitates the introduction of new functions into the existing program. We have included a new module to incorporate local alignment information in the global alignment. Here, we demonstrate the method using locally conserved segments recuperated automatically from the protein sequence databases, via post-processing of the BlastP search. However, it would be possible to use other sources of local alignment information.
It has previously been shown that neither global nor local alignment-based algorithms are capable of providing reliably accurate alignments under all conditions (4). It is therefore crucial to incorporate the two aspects of the multiple alignment in a single algorithm. One possibility would be to use a system of ‘hard’ constraints, where the multiple alignment is fixed at the anchor points and only the regions between the anchors are aligned. However, this method is only possible when the anchor points are sure. For highly variable, complex proteins the automatic detection of locally conserved segments in a set of sequences becomes more difficult. DbClustal therefore uses a system of ‘soft’ constraints, where the global alignment is weighted towards, but not constrained to, the locally conserved segments. The weight assigned to each anchor depends on the residue conservation and the frequency of occurrence of the corresponding conserved segment in the sequence database. Thus, well-conserved motifs that are found in most of the homologous sequences detected during the database search are more strongly weighted and in fact act as ‘hard’ constraints. Motifs that are less well conserved are assigned less weight and may be overridden by the residue comparison matrix scores and gap penalties in DbClustal. In addition, DbClustal performs a progressive multiple alignment following the branching order of a guide tree and it is therefore possible that a number of database sequences may be aligned before the query sequence is included in the alignment. Therefore the anchors provided by Ballast are propagated between all pairs of sequences in the dataset. One alternative would be to construct a multiple alignment centred on the query sequence, by aligning each database sequence in turn to the query.
Figure 2 illustrates the advantages of both the tree-based progressive alignment method and the soft constraints implemented in DbClustal. Both the query-based and the tree-based methods were used to construct an alignment of six sequences. The Thermotoga maritima transcription anti-termination protein (accession no. P29397) contains an insertion of 148 residues. Using the query-based method, the N-terminal region of P29397 is misaligned, partly due to the insertion and partly because of an incorrect anchor between P29397 and the query sequence (shown boxed in Fig. 2). Using DbClustal, the sequence P29397 is aligned with its nearest neighbours first and the higher residue comparison scores thus obtained override the weight of the incorrect anchor.
Figure 2.
Alignment of a set of six proteins by (A) a query-based alignment method and (B) DbClustal using a progressive tree-based method. The anchors supplied by Ballast are shown in red. The boxed segment in P29397 indicates an incorrect anchor.
Evaluation of DbClustal in a genome project
In order to evaluate the performance of DbClustal under realistic conditions, we used the set of proteins coded by a complete genome as test cases. We chose Pyrococcus abyssi, as annotation of the genome had recently been completed (http://www.genoscope.cns.fr/Pab/ ). Using all the proteins coded by the genome enabled us to test DbClustal with a wide variety of sequences with extensions and/or insertions, multi-domain proteins, sequences with repeats or circular permutations and transmembrane proteins. By searching for homologous sequences of each genome protein in the Swissprot and SPTrEMBL protein databases, we were able to evaluate, firstly, the ability of Ballast to define reliable anchor points and, secondly, the accuracy of the DbClustal global alignments.
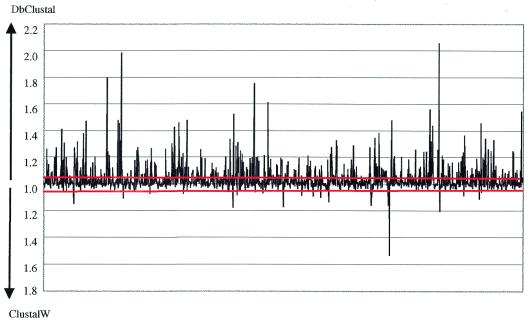
For each of the 1765 ORFs coded by the genome, a BlastP database search was performed to identify possible sequence homologues. For the 1683 searches which detected at least one other homologous protein, the top 50 hits found by Blast with an E-value less than 0.1 were aligned using both DbClustal and ClustalW (v.1.81). The alignments obtained by the two methods were compared using the MD score described in Materials and Methods. The MD score ratios for the 1683 alignments are shown in Figure 3. Score ratios above 1.05, indicating a significant difference between the alignments of DbClustal and ClustalW, were observed for 366 (22%) of the 1683 alignments. DbClustal scored significantly higher in 341 (20.3%) of the total alignments and ClustalW scored higher in 24 (1.4%) alignments.
Figure 3.
A plot of the ratio of the MD scores for 1683 alignments obtained by ClustalW and DbClustal. The red lines denote the score ratio threshold of 1.05, above which the two alignments are considered to be significantly different.
To further verify the MD significance threshold, we calculated the percentage of identical columns between the two alignments using the bali_score program (4). Of the 366 alignments with a score ratio above 1.05, only 6.8% contained >80% identical columns, while 62% shared <50% column identity. In contrast, of the 1317 alignments with a non-significant MD score ratio, 55% shared >80% column identity, while only 16% shared <50% column identity.
To determine the nature of the sequence sets and to identify possible causes of misalignments, all of the 366 alignments with a score ratio above 1.05 were analysed visually. All the alignments presenting significant differences between ClustalW and DbClustal may be viewed on the World Wide Web at http://www-igbmc.u-strasbg.fr/BioInfo/DbClustal . A summary of the analysis is shown in Table 1. A number of typical problems encountered by multiple alignment programs have previously been identified (4), including small numbers of sequences, highly divergent sequences and large extensions and insertions. The problems of large extensions and insertions are mainly resolved by the anchors incorporated in DbClustal. Even in the presence of ‘orphan’ sequences or fragments, DbClustal successfully aligns the homologous regions. In these tests using the P.abyssi proteome, a relatively high number of ‘orphan’ sequences were observed, i.e. sequences sharing <25% overall residue identity with all other sequences. This was to be expected when aligning sequences detected by BlastP with E-values as high as 0.1. Nevertheless, local regions of homology were still observed in most of the sequences. Of the alignments where ClustalW achieved significantly higher scores, four contained repeated domains and 17 contained sequences with low complexity regions, including transmembrane helices. Proteins containing non-linear elements such as repeats, inversions or circular permutations remain a problem for all global multiple alignment programs. Work is now in progress to develop a system that will pre-process a set of sequences, alerting the user to the presence of these regions.
Table 1. A summary of the manual analysis of the 366 alignments aligned differently by ClustalW and DbClustal.
| Alignment problem | Number of alignment sets |
|---|---|
| ‘Orphan’ sequences | 297 |
| Large extensions | 219 |
| Large insertions | 25 |
| Sequence fragments | 39 |
| Small number of sequences (<7) | 44 |
| Repeats | 51 |
| Transmembrane regions | 48 |
The main core of the alignment was defined as the conserved regions present in most of the sequences. Large extensions and insertions consisted of at least 100 residues outside the main core of the alignment. Fragments were either annotated in Swissprot or were defined as sequences lacking a large part of the main core regions. Very divergent or ‘orphan’ sequences were detected automatically and are defined as sharing <25% residue identity with any other sequence in the alignment set. Repeats were detected automatically and consist of regions at least 20 residues long with at least 30% residue identity.
The alignments provided by DbClustal often furnish additional essential information complementary to the database search results. Figure 4 shows two alignments of the sequences detected by the P.abyssi protein ABY0882 (accession no. CAB50242), obtained with ClustalW (Fig. 4A) and DbClustal (Fig. 4B). The sequence BAA86961, which is detected by BlastP with an E-value of 10–3, is a human adenocarcinoma antigen. In the DbClustal alignment, it can be seen that the human antigen contains an insertion of 129 residues and a C-terminal extension of 100 residues. The yeast proteins with accession nos Q08444 and CAB62419 also contain large insertions in the same region of the protein. More importantly, the cysteine-rich region present in the C-terminus of all sequences except Q9YD10 from the Aeropyrum pernix genome is correctly aligned by DbClustal. A global multiple alignment also facilitates identification of the domain structures of both the query sequence and the proteins detected in the database search. Figure 5 shows alignments of the sequences detected with P.abyssi protein ABY1118 (accession no. CAB50613) using both ClustalW and DbClustal. The Mycobacterium leprae poly(A) polymerase protein (accession no. Q59534), which was detected with an E-value of 0.02 by BlastP, has two structural domains as annotated in Swissprot: a polymerase domain at residues 102–259 and a HD domain at residues 269–411. In the DbClustal alignment, the P.abyssi query sequence is well aligned on the C-terminal HD domain and the homologous regions shared by all the proteins in the alignment are easily distinguished. In the ClustalW alignment, the HD domain in the two sub-families is misaligned on the polymerase domain of Q59534.
Figure 4.
Alignment of a set of 11 proteins by (A) ClustalW and (B) DbClustal. The N- and C-terminal extensions in both alignments are not shown. The corresponding segments in the ClustalW alignments are shown for comparison purposes. The regions denoted by the blue arrows below the DbClustal alignment correspond to the blocks detected by the Probe program. Ten homologues were detected by Probe, with four sequences being included in the final model alignment. These are indicated by the underlined names.
Figure 5.
Alignment of a set of 15 proteins by (A) ClustalW and (B) DbClustal. The N- and C-terminal extensions in both alignments are not shown. Anchors are shown in red in the DbClustal alignment. The corresponding segments in the ClustalW alignments are shown for comparison purposes. The regions denoted by the blue arrows below the DbClustal alignment correspond to the blocks detected by the Probe program. Of 131 homologues detected by Probe, 46 sequences were included in the final model alignment. Those that were also included in the DbClustal alignment are indicated by the underlined names.
Comparison of DbClustal with a local alignment program
The above study represents a comparison between DbClustal and one of the best global alignment algorithms currently available (4). To compare the performance of DbClustal with a local alignment program, we chose Dialign (25), as it has been proved to be one of the most effective programs at finding locally conserved regions in a variety of sequence alignment problems. We re-aligned the 366 alignment test cases identified above using the Dialign program. No significant differences were observed between DbClustal and Dialign for 49% of the alignments. Of the 188 remaining alignments, Dialign scored better in 112 cases (30%). However, the computer resources required to perform the alignments were inhibitory. For the 366 alignment test cases, Dialign required 53 h 35 min, compared to DbClustal which required 3 h 7 min.
Finally, we compared the performance of DbClustal with the multiple alignment produced by the Probe (10) program which implements a Gibbs sampling strategy (26). Probe (v.1.0) uses a single query sequence to first perform a transitive pairwise search of the sequence databases. An alignment model of the protein family is then constructed and progressively refined using a combination of Gibbs sampling, a genetic algorithm and iterative database searches. Only a representative set of sequences are currently included in the Probe alignment, although a new version implementing a sequence weighting scheme is under development. This leads to frequent discrepancies between the sequences included in the DbClustal and Probe alignments. In addition, the multiple alignment produced by Probe is a local alignment consisting of a number of ungapped conserved regions (or blocks) corresponding to those positions in the alignment that have been identified as having some trace of conservation. In contrast, the DbClustal alignment includes the whole protein sequences. Thus, it is impossible to directly compare the MD scores obtained by DbClustal and Probe. Nevertheless, we used the Probe program to search the Swissprot and SPTrEMBL protein databases with various P.abyssi proteins, and the results for the two proteins ABY0882 and ABY1118 are presented in Figures 4 and 5, respectively. While the conserved regions identified by the two methods are comparable, the CPU time required by the iterative algorithm implemented in Probe is much greater than that required by DbClustal. For the two alignments in Figures 4 and 5, the Probe algorithm (excluding the first iteration in which the Blast search is performed) requires 726 and 10 506 s, respectively, compared to DbClustal which requires 16 and 65 s, respectively. In addition, the more variable regions of the sequences and N- and C-terminal extensions and internal insertions present in only a sub-set of the sequences are not included in the local Probe alignment.
CONCLUSION
We have here presented a new modular version of ClustalW which has permitted incorporation of a new module to combine local conservation information with a global alignment algorithm. We have illustrated the method using anchors provided by the Ballast program. However, anchors could be constructed from any source of local alignment information, providing that the anchor file is compatible with the Ballast format. Work is now in progress to investigate alternative methods of detecting conserved motifs in a predefined set of unaligned sequences, without reference to a query sequence. It is also foreseeable that other information could now be included in the multiple alignment, such as tertiary structure information or secondary structure predictions. A Web-based user interface is now being developed for the Clustal family of programs, based on the modular version of the ClustalW source code. This will allow automatic integration of information from a variety of sources, such as protein sequence and structure databases, into the global multiple alignment. The results shown highlight the ability of DbClustal to provide automatic multiple alignment of the full-length sequences detected during database searches. Sequences detected with E-values as low as 0.1 can now be investigated in the light of the overall global family alignment. In addition, the rapidity of DbClustal should make it suitable for use in automatic, high throughput systems such as automatic genome annotation and analysis projects.
Acknowledgments
ACKNOWLEDGEMENTS
We would like to thank R. Ripp and O. Lecompte for helpful discussions and D. Moras for his continuous support. This work was supported by institute funds from the Institut National de la Santé et de la Recherche Médicale, the Centre National de la Recherche Scientifique, the Hôpital Universitaire de Strasbourg and the Fond de Recherche Hoechst Marion Roussel.
REFERENCES
- 1.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karplus K., Barrett,C. and Hughey,R. (1998) Bioinformatics, 14, 846–856. [DOI] [PubMed] [Google Scholar]
- 3.Pearson W.R. (2000) Methods Mol. Biol., 132, 185–219. [DOI] [PubMed] [Google Scholar]
- 4.Thompson J.D., Plewniak,F. and Poch,O. (1999) Nucleic Acids Res., 27, 2682–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sander C. and Schneider R. (1991) Proteins, 9, 56–68. [DOI] [PubMed] [Google Scholar]
- 6.Taylor W.R. (1998) J. Mol. Biol., 280, 375–406. [DOI] [PubMed] [Google Scholar]
- 7.Taylor W.R. (1988) J. Mol. Evol., 28, 161–169. [DOI] [PubMed] [Google Scholar]
- 8.Koretke K.K., Russell,R.B., Copley,R.R. and Lupas,A.N. (1999) Proteins, 37, 141–148. [Google Scholar]
- 9.Schuler G.D., Altschul,S.F. and Lipman,D.J. (1991) Proteins, 9, 180–190. [DOI] [PubMed] [Google Scholar]
- 10.Neuwald A.F., Liu,J.S., Lipman,D.J. and Lawrence,C.E. (1997) Nucleic Acids Res., 27, 1665–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gracy J. and Argos,P. (1998) Bioinformatics, 14, 164–173. [DOI] [PubMed] [Google Scholar]
- 12.Jiang J. and Jacob,H.J. (1998) Genome Res., 8, 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baxevanis A.D. and Landsman,D. (1998) Nucleic Acids Res., 26, 372–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srinivasarao G.Y, Yeh,L.S., Marzec,C.R., Orcutt,B.C. and Barker,W.C. (1999) Bioinformatics, 5, 382–390. [DOI] [PubMed] [Google Scholar]
- 16.Brocchieri L. and Karlin,S. (1998) J. Mol. Biol., 276, 249–264. [DOI] [PubMed] [Google Scholar]
- 17.Bucka-Lassen K., Caprani,O. and Hein,J. (1999) Bioinformatics, 15, 122–130. [DOI] [PubMed] [Google Scholar]
- 18.Plewniak F., Thompson,J.D. and Poch,O. (2000) Bioinformatics, 16, in press. [DOI] [PubMed] [Google Scholar]
- 19.Needleman S.B. and Wunsch,C.D. (1970) J. Mol. Biol., 48, 443–453. [DOI] [PubMed] [Google Scholar]
- 20.Vogt G., Etzold,T. and Argos,P. (1995) J. Mol. Biol., 249, 816–831 [DOI] [PubMed] [Google Scholar]
- 21.Gotoh O. (1996) J. Mol. Biol., 264, 823–838. [DOI] [PubMed] [Google Scholar]
- 22.Notredame C., Holm,L. and Higgins,D.G. (1998) Bioinformatics, 14, 407–422. [DOI] [PubMed] [Google Scholar]
- 23.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J.D., Plewniak,F. and Poch,O. (1999) Bioinformatics, 15, 87–88. [DOI] [PubMed] [Google Scholar]
- 25.Morgenstein B., Dress,A. and Werner,T. (1996) Proc. Natl Acad. Sci. USA, 93, 12098–12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wootton,J.C. (1993) Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]