Abstract
Introduction
Atezolizumab plus bevacizumab treatment is highly effective in patients with unresectable hepatocellular carcinoma (HCC). However, progressive disease (PD) occurs in approximately 20% of HCC patients treated with atezolizumab plus bevacizumab, resulting in a poor prognosis. Thus, the prediction and early detection of HCC is crucial.
Methods
Patients with unresectable HCC treated with atezolizumab plus bevacizumab and had baseline preserved serum (n = 68) were screened and classified according to their PD, 6 weeks after treatment initiation (early PD; n = 13). Of these, 4 patients each with and without early PD were selected for cytokine array and genetic analyses. The identified factors were validated in the validated cohort (n = 60) and evaluated in patients treated with lenvatinib.
Results
No significant differences were observed in the genetic alterations in circulating tumor DNA. Cytokine array data revealed that baseline MIG (CXCL9), ENA-78, and RANTES differed substantially between patients with and without early PD. Subsequent analysis in the validation cohort revealed that baseline CXCL9 was significantly lower in patients with early PD than that in patients without early PD, and the best cut-off value of serum CXCL9 to predict early PD was 333 pg/mL (sensitivity: 0.600, specificity: 0.923, AUC = 0.75). In patients with lower serum CXCL9 (<333 pg/mL), 35.3% (12/34) experienced early PD with atezolizumab plus bevacizumab, while progression-free survival (PFS) was significantly shorter relative to that in patients without (median PFS, 126 days vs. 227 days; HR: 2.41, 95% CI: 1.22–4.80, p = 0.0084). While patients with objective response to lenvatinib had significantly lower CXCL9 levels compared with those of patients without.
Conclusion
Baseline low serum CXCL9 (<333 pg/mL) levels may predict early PD in patients with unresectable HCC treated with atezolizumab plus bevacizumab.
Keywords: Atezolizumab, Bevacizumab, Early PD, Hepatocellular carcinoma, CXCL9, Baseline, Prognosis
Introduction
Hepatocellular carcinoma (HCC) is a major cause of cancer-related deaths, and its incidence has been increasing globally [1]. In the recent years, there have been drastic advancements in the therapeutic options of systematic chemotherapy for patients with unresectable HCC. In addition to sorafenib [2], lenvatinib [3], regorafenib [4], cabozantinib [5], ramucirumab [6], and the vascular endothelial growth factor (VEGF) inhibitor bevacizumab plus the programmed death ligand-1 inhibitor atezolizumab have been approved for systemic therapy in patients with unresectable HCC [7]; of which, atezolizumab plus bevacizumab is considered to be the first-line therapy [8, 9], while lenvatinib and sorafenib are considered alternative first-line therapies or as second-line therapies in cases where atezolizumab and bevacizumab prove to be ineffective [8].
The IMbrave150 trial, which was a phase 3 clinical trial of atezolizumab and bevacizumab for patients with unresectable HCC, revealed that atezolizumab and bevacizumab could achieve longer overall survival (OS) and progression-free survival (PFS) compared to those of sorafenib [7]. However, progressive disease (PD) occurred in 19.6% (71/326) of the patients treated with atezolizumab plus bevacizumab [7]. Additionally, early PD (PD at 6 weeks after atezolizumab plus bevacizumab initiation) occurred in 17.2% (10/58) of the patients in real-world settings [10].
Exploratory analysis of IMbrave150 on OS stratified by the treatment response (treatment response as defined by Response Evaluation Criteria in Solid Tumors 1.1 (RECIST 1.1)) revealed that the median OS of patients with the best response to PD treated with atezolizumab plus bevacizumab was much shorter (6.8 months) than that of the responders [11]. Therefore, clarifying the factors associated with early PD in patients treated with atezolizumab and bevacizumab plus development of alternative therapies for treatment of unresectable HCC is an urgent and clinically important issue.
Poor prognosis has been observed due to CTNNB-1 gene alteration in patients with unresectable HCC who undergo immune checkpoint inhibitor (ICI) monotherapy [12, 13, 14]. In addition, some cytokines and chemokines, including CCR5 and CXCL13, have been reported to be associated with the ICI treatment response [15, 16]. In the present study, we aimed to clarify the factors associated with early PD in patients with unresectable HCC − who are treated with atezolizumab and bevacizumab − by a comprehensive analysis, including assessment of clinical factors, genetic alterations in circulating tumor DNA, and cytokine arrays.
Materials and Methods
Patients
We retrospectively screened patients with unresectable HCC − who were treated with atezolizumab and bevacizumab between October 2020 and January 2022 and had complete clinical data and baseline preserved serum samples for biomarker analysis − in the NORTE study groups [17, 18, 19, 20, 21], while those patients who did not meet the above criteria or declined to participate in the study were excluded. Subsequently, we evaluated their treatment response by dynamic computed tomography (CT) or magnetic resonance imaging (MRI) at 6 weeks after treatment initiation.
We collected cell-free DNA samples from 29 patients. Of those, 4 patients each with and without early PD were selected according to the rate of change in tumor size after atezolizumab and bevacizumab treatment. Four patients each with the highest increment and shrinking rate of HCC tumor diameter after atezolizumab and bevacizumab treatment were selected from among the 29 patients (discovery cohort). The patients' clinical factors were compared, and the preserved serum samples and cell-free DNA samples were analyzed for cytokine array and genetic alterations. Subsequently, we evaluated these results in the validation cohort (n = 60), excluding 8 patients in the discovery cohort from all analyses.
Finally, we analyzed the treatment response to lenvatinib in patients with unresectable HCC − stratified by the factors that were identified in the above analysis − after screening these patients at Hokkaido University Hospital and JCHO Hokkaido Hospital between April 2018 and May 2022. Only patients with complete clinical information were evaluated for their treatment response every 2–3 months following treatment and had baseline preserved serum samples were included here.
The present study protocols conformed to the ethical guidelines of the Declaration of Helsinki and were approved by the Ethics Committee of Hokkaido University Hospital (approval no. 020-0267 and 017–0521) and the participating institutes. Written informed consent was obtained from all the included patients.
Treatment Protocols
Patients with unresectable HCC were treated with atezolizumab (1,200 mg) plus bevacizumab (15 mg/kg) every 3 weeks. Their treatment response was evaluated 6 weeks after treatment initiation. Treatment was discontinued if unacceptable adverse events or PD were observed, as previously described [10]. Patients who weighed <60 kg received 8 mg lenvatinib once a day orally, and those who weighed ≥ 60 kg received 12 mg once a day, as described previously [22].
Cytokine Array Analysis
Serum chemokines and cytokines, including angiogenin, chemokine ligand-1 (CCL1), CCL15, CCL17, CCL2, CCL22, CCL5, CCL7, CCL8, colony stimulating factor-1 (CSF1), CSF2, CSF3, C-X-C motif chemokine ligand-1 (CXCL1), CXCL12, CXCL2, CXCL3, CXCL5, CXCL8, CXCL9, epidermal growth factor, interferon-γ, insulin-like growth factor-1, IL-10, IL-12A, IL-13, IL-15, IL-1A, IL-1B, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, KIT ligand, leptin, lymphotoxin-α, oncostatin M, platelet-derived growth factor subunit B, transforming growth factor-β1, thrombopoietin, tumor necrosis factor, thyroperoxidase, and VEGFA, were analyzed using the RayBio Human Cytokine Antibody Array G Series 3 (RayBiotech Life, GA, USA), according to the manufacturer's protocols. All patient samples were analyzed in duplicates, and the signals were scanned using GenePix 4400A (Molecular Devices, San Jose, CA, USA) and analyzed using Microarray Data Analysis Tool v3.2 (Filgen, Nagoya, Aichi, Japan) and Array-Pro Analyzer v4.5 (Media Cybernetics, Rockville, MD, USA). To normalize the Array Data (signal intensity data), one sub-array was defined as the “reference,” against which all the other arrays were normalized. We calculated the normalized values as follows: X(Ny) = X(y) × P1/P(y), where P1 = mean signal intensity of POS pots on reference array, P(y) = mean signal intensity of POS spots on Array “y,” X(y) = mean signal intensity for spot “X” on Array “y,” and X(Ny) = normalized signal intensity for spot “X” on Array “y.” The normalized signal intensities were compared between patients with and without early PD (n = 4, each).
Analysis of Changes in Serum CXCL9, CCL5, ENA-78, M-CSF, and PDGF-BB
Serum CXCL9, CCL5, ENA-78, M-CSF, and PDGF-BB levels were evaluated using commercial ELISA kits (CXCL9, CCL5, ENA-78, M-CSF, and PDGF-BB; R&D Systems, Minneapolis, MN, USA) according to the manufacturer's protocols [23].
Evaluation of the Treatment Response
Dynamic computed tomography or MRI was performed on patients at baseline and at 6 weeks after the initiation of atezolizumab plus bevacizumab treatment, and the treatment responses were classified according to RECIST 1.1 and modified RECIST [24].
In patients treated with lenvatinib, the treatment response was evaluated every 2–3 months using modified RECIST [24]. In addition to the treatment response, PFS and OS were evaluated according to the factors associated with early PD.
Analysis of Genetic Alterations in Circulating Tumor DNA
Of the patients with and without PD treated with atezolizumab plus bevacizumab, cell-free DNA samples from 4 patients were analyzed.
Sample Preparation
Cell-free DNA was extracted from 4 mL of plasma using the AVENIO ctDNA Analysis Kit (Roche Diagnostics, Tokyo, Japan) according to the manufacturer's protocols. For sample quantity and quality check, the concentration of cell-free DNA was determined using the Qubit dsDNA HS Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA), and the fragment size was examined using the Agilent 2200 TapeStation System with High Sensitivity D5000 ScreenTape Assay (Agilent Technologies, Santa Clara, CA, USA).
Target Selection and Sequencing
Library preparation and sequencing were conducted according to the manufacturer's instructions (Thermo Fisher Scientific). PCR amplification of target regions and library preparation for sequencing was performed using the Oncomine Pan-Cancer Cell-Free Assay and Tag Sequencing Barcode Set (Thermo Fisher Scientific). Library concentration was estimated using the Ion Library TaqMan Quantitation Kit (Thermo Fisher Scientific). Emulsion PCR was conducted using the Ion 540 Kit-Chef and Ion Chef (Thermo Fisher Scientific). Sequencing was performed using Ion 540 Chip and Ion 540 Kit-Chef on an Ion S5 XL System (Thermo Fisher Scientific) at Riken Genesis Co. (Tokyo, Japan).
Mapping and Variant Calling
Quality control and mapping of sequencing reads were conducted using Torrent Suite Software 5.12.2 (Thermo Fisher Scientific). Variant calls were performed using Oncomine TagSeq Pan-Cancer Liquid Biopsy w2.1–Single Sample workflow in Ion Reporter Software 5.10 (Thermo Fisher Scientific). Annotations of the called variants were based on dbSNP151, CCDS (National Center for Biotechnology Information, Release 15), RefSeq (UCSC Genome Browser, November 2018), Gencode (UCSC Genome Browser, v19), and 1,000 Genomes (phase 3; release v5). These analyses were conducted at the Riken Genesis Co.
Statistical Analysis
Continuous variables were analyzed using the Mann-Whitney U test. Categorical variables were analyzed using the χ2 test and Fisher's exact test. The best cut-off value was determined based on the receiver operating characteristic (ROC) curve by maximizing the Youden's index. Survival curves of PFS and OS were calculated using Kaplan-Meier analysis and compared using the log-rank test.
In all analyses, p < 0.05 was defined as statistically significant. All statistical analyses were performed using SPSS Statistics (v22.0; IBM, Armonk, NY, USA).
Results
Patient Characteristics
A total of 68 patients who received atezolizumab plus bevacizumab between October 2020 and January 2022 in the NORTE study group, had preserved serum at baseline, completed clinical information, and had their evaluated treatment response at 6 weeks after atezolizumab plus bevacizumab treatment were included. As listed in Table 1, PD was observed in 19.1% (13/68) of the patients who received atezolizumab plus bevacizumab at 6 weeks after treatment. Comparison between patients with and without early PD did not show any significant differences, including NLR.
Table 1.
Comparison of baseline patient characteristics between those with or without early PD to atezolizumab and bevacizumab
| All patients (N = 68) | PD after 6 weeks (N = 13) | Non-PD after 6 weeks (N = 55) | p value | |
|---|---|---|---|---|
| Age (range), years | 72 (31–89) | 75 (37–83) | 70 (31–89) | 0.85 |
| Sex | ||||
| Male | 55 | 10 | 45 | 0.70 |
| Female | 13 | 3 | 10 | |
| Etiology | ||||
| Viral hepatitis | 30 | 4 | 26 | 0.36 |
| NBNC | 38 | 9 | 29 | |
| CH | 33 | 9 | 24 | 0.13 |
| LC | 35 | 4 | 31 | |
| ECOG PS | ||||
| 0 | 54 | 10 | 44 | >0.99 |
| 1 | 14 | 3 | 11 | |
| White blood cell count, mm3 | 5,040 (1,970–11800) | 4,800 (2700–11800) | 5,200 (1,970–11800) | 0.91 |
| Neutrophil count, mm3 | 3,285 (1,185–9,971) | 2760 (1,809–9,971) | 3,440 (1,185–9,204) | 0.84 |
| Lymphocyte count, mm3 | 1,120 (140–2881) | 1,296 (540–2366) | 1,088 (140–2881) | 0.41 |
| NLR | 2.83 (0.83–16.69) | 2.20 (0.83–7.52) | 2.98 (0.98–16.69) | 0.32 |
| Platelet, ×104/µL | 16.7 (3.6–46.4) | 16.8 (8.5–32.6) | 16.2 (3.6–46.4) | 0.79 |
| Prothrombin time, % | 95.9 (35.3–128.0) | 103.3 (81.8–113.8) | 95.0 (35.3–128.0) | 0.10 |
| NH3, µg/dL | 42 (8–136) | 30 (12–100) | 43 (8–136) | 0.24 |
| Albumin, g/dL | 3.7 (2.7–4.8) | 3.9 (3.1–4.8) | 3.7 (2.7–4.8) | 0.48 |
| ALBI grade | ||||
| 1 | 25 | 5 | 20 | 0.85 |
| 2a | 23 | 5 | 18 | |
| 2b | 20 | 3 | 17 | |
| AST, IU/L | 45 (14–672) | 44 (20–134) | 46 (14–672) | 0.72 |
| ALT, IU/L | 28 (7–278) | 35 (12–122) | 27 (7–278) | 0.53 |
| Child-Pugh grade | ||||
| A | 65 | 13 | 52 | >0.99 |
| B | 3 | 0 | 3 | |
| Child-Pugh score | ||||
| 5 | 38 | 9 | 29 | 0.67 |
| 6 | 27 | 4 | 23 | |
| 7 | 1 | 0 | 1 | |
| 8 | 2 | 0 | 2 | |
| AFP, ng/mL | 111.8 (0.8–200000.0) | 156.0 (2.4–15009.5) | 80.1 (0.8–200000.0) | 0.67 |
| DCP, mAU/mL | 887 (19–245000) | 839 (25–110159) | 1,116 (19–245000) | 0.59 |
| Maximum intrahepatic tumor size, mm | 32.0 (0–166.6) | 21.6 (0–119) | 36 (0–166.6) | 0.39 |
| More than 50% liver involvement | 11 | 2 | 9 | >0.99 |
| Diffuse type | 10 | 1 | 9 | 0.67 |
| Hepatic tumors | ||||
| 0 | 10 | 2 | 8 | 0.94 |
| 1 | 7 | 1 | 6 | |
| Multiple | 51 | 10 | 41 | |
| BCLC stage | ||||
| B | 23 | 6 | 17 | 0.34 |
| C | 45 | 7 | 38 | |
| Up to seven | ||||
| In | 23 | 4 | 19 | >0.99 |
| Out | 45 | 9 | 36 | |
| Positive for Vp | 13 | 3 | 10 | 0.70 |
| Positive for Vv | 1 | 0 | 1 | >0.99 |
| Positive for bile duct invasion | 1 | 0 | 1 | >0.99 |
| Positive for LN metastasis | 13 | 1 | 12 | 0.44 |
| Positive for EHM | 31 | 4 | 27 | 0.35 |
| Naive | 16 | 3 | 13 | >0.99 |
| Recurrence | 52 | 10 | 42 | |
| History of operation | 33 | 8 | 25 | 0.36 |
| History of RFA | 19 | 3 | 16 | >0.99 |
| History of TACE | 29 | 7 | 22 | 0.53 |
| History of HAIC | 1 | 0 | 1 | >0.99 |
| History of RT | 10 | 1 | 9 | 0.67 |
| History of TKI | 39 | 9 | 30 | 0.37 |
NBNC, non-HBV non-HCV; CH, chronic hepatitis; LC, liver cirrhosis; ECOG PS, Eastern Cooperative Oncology Group Performance Status; BMI, body mass index; NLR, neutrophil to lymphocyte ratio; ALBI, albumin-bilirubin; AST, aspartate aminotransferase; ALT alanine aminotransferase; AFP, alpha-fetoprotein; DCP, des-gamma-carboxy prothrombin; BCLC, Barcelona Clinic Liver Cancer; LN, lymph node; EHM, extrahepatic metastasis; RFA, radiofrequency ablation; TACE, transcatheter arterial chemoembolization; HAIC, hepatic arterial infusion chemotherapy; RT, radiation therapy; TKI, tyrosine kinase inhibitor
Analysis of Biomarkers Associated with Prediction of Early PD in HCC Patients Who Were Treated with Atezolizumab Plus Bevacizumab
We collected cell-free DNA from 29 patients at baseline. Of these, to investigate the factors associated with early PD, we selected 4 patients each with and without early PD, according to the rate of change in HCC at 6 weeks after treatment initiation (online supp. Table S1; see www.karger.com/doi/10.1159/000527759 for all online suppl. material). As shown in online supplementary Figure S1, no significant differences were observed in the genetic alterations in circulating tumor DNA between patients with and without early PD (median amplicon coverage in 8 patients was 41,198.33–62,859.96).
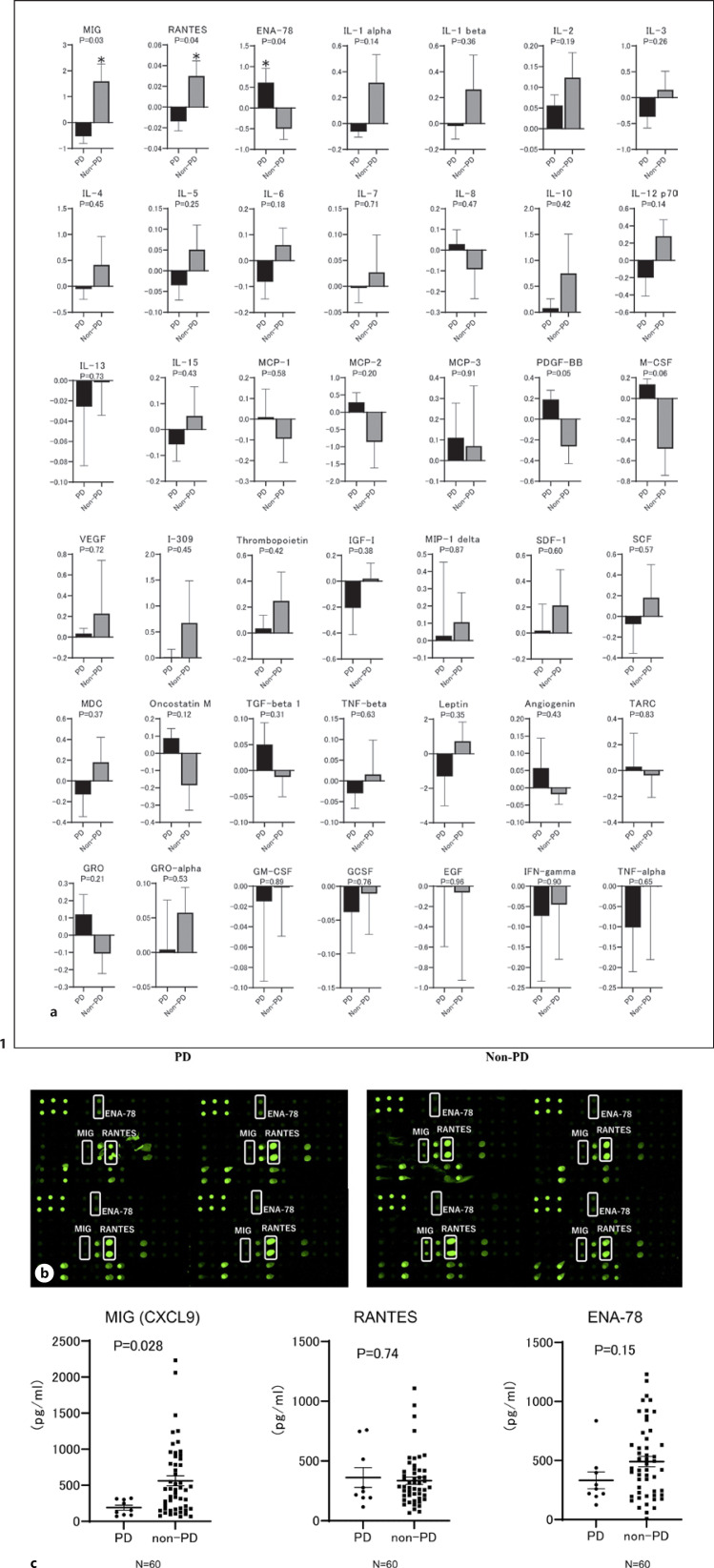
However, the signal levels of baseline cytokines and chemokines analyzed using the RayBio Human Cytokine Antibody Array G Series 3 revealed that MIG (CXCL9), ENA-78, and RANTES (CCL5) were significantly different between patients with early PD and those without early PD (Fig. 1a, b). Subsequently, to validate these results, we investigated the baseline serum MIG (CXCL9), ENA-78, and RANTES (CCL5) levels in the validation cohort consisting of 60 patients, excluding 8 from the discovery cohort, using ELISA and compared these values between patients with and without early PD who were treated with atezolizumab plus bevacizumab. As shown in Figure 1c, CXCL9 levels were significantly lower in patients with early PD relative to those in patients without early PD even in the validation cohort. Similarly, we analyzed cytokines and chemokines of marginal significance in the discovery cohort and M-CSF (p = 0.06) and PDGF-BB (p = 0.05) in validation cohort. As shown in the online supplementary Figure S2, M-CSF and PDGF-BB were not significantly associated with early PD in the validation cohort.
Fig. 1.
Comparison of serum cytokines and chemokines using cytokine array analysis between patients with and without early PD treated with atezolizumab plus bevacizumab. a In the cytokine array, the signals were scanned using GenePix 4400A (Molecular Devices) and analyzed using MicroArray Data Analysis Tool v3.2 (Filgen) and Array-Pro Analyzer v4.5 (Media Cybernetics). Signals were normalized, and the intensities were compared between patients with early PD (n = 4) and those without (n = 4). Mean signal intensity in the early PD and non-early PD groups is shown as the log2 ratio and SD *p < 0.05. b Complete images of array membranes of samples from patients with early PD (n = 4) and those without early PD (n = 4) treated with atezolizumab plus bevacizumab. c Comparison of baseline serum MIG (CXCL9), ENA-78, and RANTES levels between patients with and without early PD after treatment with atezolizumab and bevacizumab in the validation cohort (n = 60), excluding 8 patients of the discovery cohort from the entire cohort.
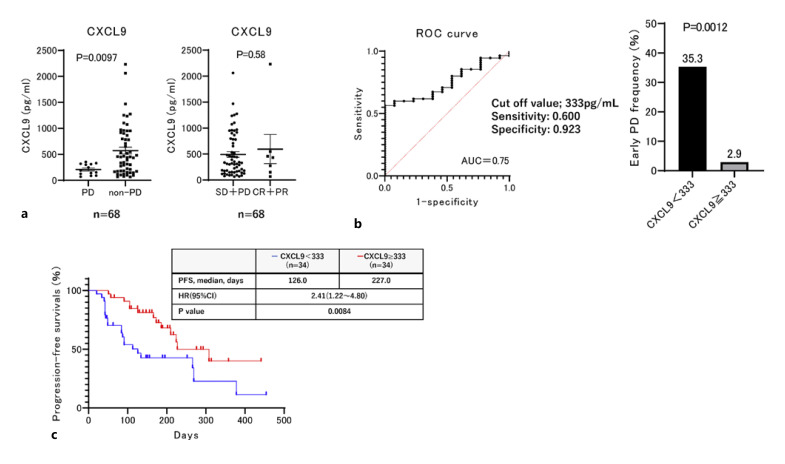
Subsequently, we analyzed the relationship between the baseline serum CXCL9 level and atezolizumab plus bevacizumab treatment response in the entire cohort. As shown in Figure 2a, CXCL9 levels were significantly lower in patients with early PD compared to those in patients without early PD; while CXCL9 levels were similar in patients with or without objective responses. We then analyzed and set the best cut-off value that predicted early PD after atezolizumab plus bevacizumab treatment initiation at 333 pg/mL based on the ROC curve that maximized the Youden index (sensitivity, 0.600; specificity, 0.923; AUC = 0.75), as shown in Figure 2b. Among these patients, 35.3% (12/34) of those with low baseline serum CXCL9 levels (<333 pg/mL) exhibited early PD. However, only 2.9% (1/34) of those with high baseline serum CXCL9 levels exhibited early PD (Fig. 2b). As shown in Figure 2c, PFS was significantly shorter in patients with low CXCL9 levels (<333 pg/mL) relative to that in patients with higher CXCL9 levels (median PFS, 126 days vs. 227 days; HR: 2.41, 95% CI: 1.22–4.80, p = 0.0084).
Fig. 2.
Association between baseline MIG (CXCL9) levels and early PD treated with atezolizumab plus bevacizumab. a Comparison of baseline serum CXCL9 levels between patients with and without early PD, or an objective response to atezolizumab and bevacizumab. b Best cut-off value of CXCL9 levels for predicting early PD following atezolizumab and bevacizumab treatment. The best cut-off value was determined based on the receiver operating characteristic curve obtained by maximizing the Youden index. The cut-off baseline CXCL9 level for predicting early PD after atezolizumab and bevacizumab was 333 pg/mL (sensitivity: 0.600, specificity: 0.923, AUC = 0.75). Rate of early PD in patients with ≥333 pg/mL and <333 pg/mL baseline CXCL9 following atezolizumab plus bevacizumab treatment. c Kaplan-Meier estimates of PFS stratified by <333 pg/mL, the baseline CXCL9. Median PFS in patients with ≥333 pg/mL and <333 pg/mL baseline CXCL9 is 126 days and 227 days, respectively (HR: 2.41, 95% CI: 1.22–4.80, p = 0.0084).
Characteristics of Patients with Unresectable HCC Who had Low Levels of Serum CXCL9
Table 2 lists a comparison between patients with unresectable HCC with low (<333 pg/mL) and high baseline CXCL9 (≥333 pg/mL) levels. Evidently, no significant differences were observed between patients with low and high baseline CXCL9 levels.
Table 2.
Comparison of baseline patient characteristics between those with and without low serum MIG levels upon treatment with atezolizumab and bevacizumab
| CXCL9 low (<333 pg/ml) (N = 34) | CXCL9 high (≥333 pg/ml) (N = 34) | p value | |
|---|---|---|---|
| Age (range), years | 71.5 (31–84) | 72.5 (47–89) | 0.29 |
| Sex | |||
| Male | 28 | 27 | >0.99 |
| Female | 6 | 7 | |
| Etiology | |||
| Viral hepatitis | 13 | 17 | 0.46 |
| NBNC | 21 | 17 | |
| CH | 17 | 16 | >0.99 |
| LC | 17 | 18 | |
| ECOG PS | |||
| 0 | 26 | 28 | 0.77 |
| 1 | 8 | 6 | |
| White blood cell count, mm3 | 4,900 (2700–11800) | 5,300 (1,970–9,300) | 0.39 |
| Neutrophil count, mm3 | 3,308 (1,185–9,971) | 3,270 (1,320–7,440) | 0.43 |
| Lymphocyte count, mm3 | 1,088 (480–2881) | 1,140 (140–2664) | 0.92 |
| NLR | 2.82 (0.84–16.69) | 2.84 (0.98–12.15) | 0.69 |
| Platelet, ×104/µL | 16.0 (6.1–46.4) | 17.0 (3.6–41.3) | 0.45 |
| Prothrombin time, % | 96.3 (35.3–118.4) | 95.9 (42.8–128.0) | 0.41 |
| NH3, µg/dL | 42 (12–136) | 42 (8–98) | 0.37 |
| Albumin, g/dL | 3.8 (2.7–4.8) | 3.7 (3.0–4.8) | 0.90 |
| ALBI grade | |||
| 1 | 12 | 13 | 0.87 |
| 2a | 11 | 12 | |
| 2b | 11 | 9 | |
| AST, IU/L | 39 (19–134) | 53 (14–672) | 0.10 |
| ALT, IU/L | 24 (12–122) | 32 (7–278) | 0.14 |
| Child-Pugh grade | |||
| A | 33 | 32 | >0.99 |
| B | 1 | 2 | |
| Child-Pugh score | |||
| 5 | 18 | 20 | 0.70 |
| 6 | 15 | 12 | |
| 7 | 0 | 1 | |
| 8 | 1 | 1 | |
| AFP, ng/mL | 63.2 (1.8–57125.2) | 182.8 (0.8–200000.0) | 0.37 |
| DCP, mAU/mL | 887 (19–213066) | 938 (21–245000) | 0.31 |
| Maximum intrahepatic tumor size, mm | 24.6 (0–160.0) | 48.5 (0–166.6) | 0.06 |
| More than 50% liver involvement | 4 | 7 | 0.51 |
| Diffuse type | 2 | 8 | 0.08 |
| Hepatic tumors | |||
| 0 | 6 | 4 | 0.43 |
| 1 | 2 | 5 | |
| Multiple | 26 | 25 | |
| BCLC stage | |||
| B | 13 | 10 | 0.61 |
| C | 21 | 24 | |
| Up to seven | |||
| In | 13 | 10 | 0.61 |
| Out | 21 | 24 | |
| Positive for Vp | 8 | 5 | 0.54 |
| Positive for Vv | 0 | 1 | >0.99 |
| Positive for bile duct invasion | 1 | 0 | >0.99 |
| Positive for LN metastasis | 3 | 10 | 0.06 |
| Positive for EHM | 12 | 19 | 0.14 |
| Naive | 9/25 | 7/27 | 0.78 |
| Recurrence | |||
| History of operation | 18 | 15 | 0.63 |
| History of RFA | 9 | 10 | >0.99 |
| History of TACE | 18 | 11 | 0.14 |
| History of HAIC | 0 | 1 | >0.99 |
| History of RT | 5 | 5 | >0.99 |
| History of TKI | 20 | 19 | >0.99 |
Treatment Response to Lenvatinib in Patients with Low Levels of Baseline Serum CXCL9
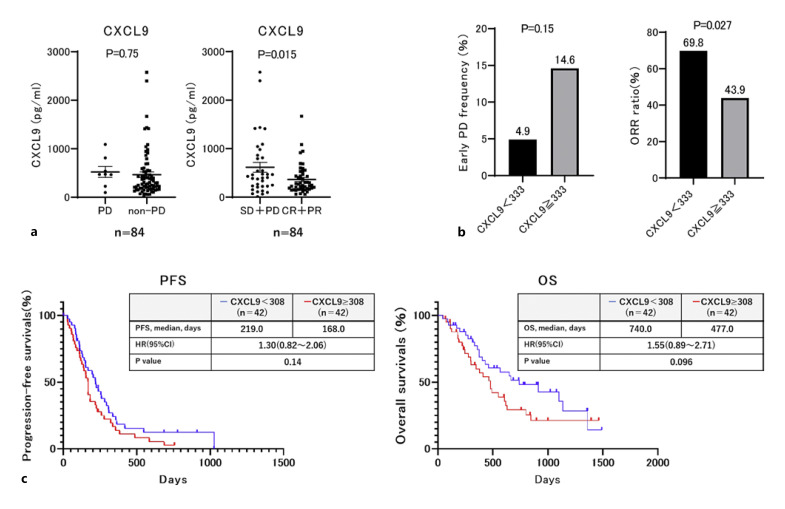
Subsequently, we analyzed the relationship between low baseline serum CXCL9 levels and lenvatinib treatment response in 84 patients with unresectable HCC. The baseline patient characteristics are shown in online supplementary Table S2. Varying results were obtained for atezolizumab plus bevacizumab treatment, whereas the baseline serum CXCL9 levels in patients with objective response (complete response + partial response; objective response rate [ORR]) were significantly lower compared with those in patients without objective response (p = 0.015; Fig. 3a). Furthermore, the ORR was significantly higher in patients with low baseline CXCL9 level (<333 pg/mL) compared with that in patients with high CXCL9 level (≥333 pg/mL) (p = 0.027; Fig. 3b). Subsequently, we set the best cut-off values of CXCL9 to 308 pg/mL to predict OR in patients treated with lenvatinib using ROC analysis. OS was marginally significantly longer in patients with low baseline CXCL9 (<308 pg/mL) compared with those with high baseline CXCL9 levels (≥308 pg/mL) (p = 0.096; Fig. 3c).
Fig. 3.
Comparison of treatment responses and PFS between unresectable HCC patients with or without low baseline MIG (CXCL9) treated with lenvatinib. a Comparison of baseline serum CXCL9 levels between patients with or without PD or objective responses to lenvatinib. b Comparison of rate of PD and objective response (OR) to lenvatinib between patients with ≥333 pg/mL and <333 pg/mL at baseline CXCL9. c PFS and overall survival (OS) stratified by <308 pg/mL the baseline MIG. Median PFS in patients with ≥308 pg/mL and <308 pg/mL baseline MIG was 168 and 219 days, respectively (HR: 1.30, 95% CI: 0.82–2.06, p = 0.14). Median OS in patients with ≥308 pg/mL and <308 pg/mL baseline MIG was 477 and 740 days, respectively (HR: 1.55, 95% CI: 0.89–2.71, p = 0.096).
Discussion
In the present study, we investigated the candidate predictive factors such as clinical factors, serum cytokines and chemokines, and genetic alterations in circulating tumor DNA associated with early PD after atezolizumab and bevacizumab treatment. Although no remarkable differences were observed in the clinical factors and genetic alterations in circulating cell-free DNA, cytokine array data showed that baseline MIG (CXCL9), ENA-78, and RANTES (CCL5) remarkably differed between the 4 patients with early PD and four without. Subsequent validation analysis of the data of 60 patients treated with atezolizumab and bevacizumab revealed that baseline CXCL9 levels were substantially lower in patients with early PD than in those patients without early PD. In contrast, lenvatinib exhibited the opposite effect in patients with low baseline CXCL9 levels. Patients with an objective response to lenvatinib had significantly lower baseline serum CXCL9 levels compared with those in patients without objective response. OS was marginally significantly longer in patients with low baseline CXCL9 compared with those with high baseline CXCL9 levels (p = 0.096).
We identified serum CXCL9 as a potential predictive factor associated with early PD after bevacizumab and atezolizumab treatment. In this study, the best cut-off value of serum CXCL9 for predicting early PD was set at 333 pg/mL. Among the 34 patients, 12 (35.3%) of them with low baseline serum CXCL9 levels (<333 pg/mL) exhibited early PD, while only 1 (2.9%) of them with high baseline serum CXCL9 (≥333 pg/mL) exhibited early PD. Thus, in patients with low baseline CXCL9 levels, evaluation of the treatment response in the early phase of atezolizumab and bevacizumab treatment might be required.
Recently, Litchfield et al. [16] analyzed whole-exome and transcriptome data of more than 1,000 patients who were treated with ICIs and revealed that in addition to clonal tumor mutational burden, high CXCL9 expression level is one of the strongest predictors of a favorable response to ICI treatment. CXCL9 is a critical chemokine that recruits cytotoxic CD8+ T cells into the tumor [25]; thus, high CXCL9 expression is a strong predictive marker of immune cell infiltration [26]. Similarly, in an inflamed class of HCC, higher levels of cytokines are expressed, which are involved in lymphocyte chemotaxis including CCL5, CXCL10, CXCL11, and CXCL9. Thus, HCC with low levels of CCL5, CXCL10, CXCL11, and CXCL9 in the tissue is classified as noninflamed tumor subtype with low level of CD8+ T and dendric cells infiltrating the tumor [27, 28]; therefore, this HCC is insufficient response to ICI. However, the relationship between serum levels of those cytokines and ICI response in HCC has not been well clarified. In this study, we firstly reported that serum CXCL9 levels are associated with early PD of atezolizumab and bevacizumab for unresectable HCC.
Whether serum CXCL9 can represent the status of an HCC immune microenvironment remains unclear. Therefore, we additionally analyzed the relationship between the serum CXCL9 level and CXCL9 expression in HCC samples. In this study, we analyzed 2 patients with HCC and simultaneously collected their biopsy and blood samples, one of which revealed a high baseline CXCL9 level (1,087.4 pg/mL), while the other had a relatively lower baseline serum CXCL9 level (379.7 pg/mL). As shown in online supplementary Figure S3, the expression of CXCL9 in HCC was higher in patients with a high serum CXCL9 level compared with that in patients with a relatively lower serum CXCL9 level. Additionally, to clarify the immune status of HCC with low CXCL9 expression levels, we analyzed the expression levels of CD8. CD8 expression levels in HCC were also significantly higher in patients with high serum CXCL9 levels than those in patients with relatively low serum CXCL9 levels. Therefore, in the two cases, CXCL9 and CD8 expression in HCC and serum CXCL9 levels were correlated. In addition, a recent study suggested that CXCL9 expression in ovarian tumors is significantly correlated with the serum CXCL9 level [29]. However, as we used the data of only 2 patients for the analysis of the relationship between the expression of CXCL9 in HCC and serum CXCL9 level, further analysis with a larger dataset is required.
Recently, it has been reported that anti-VEGF antibody in tumor increases CXCL9, CXCL10, and CXCL11 levels, thereby increasing CD8+T-cell infiltration into the tumor and the response to the combination immunotherapy with bevacizumab for HCC even in tumors with low expression levels of CXCL9, CXCL10, and CXCL11 [30, 31, 32]. Previous reports [30, 31] speculated the possible mechanisms, whereby anti-VEGF could increase CXCL9; anti-VEGF can induce hypoxic conditions which might contribute to an increased IFN-γ, thereby inducing the production of CXCL9. However, in this study, the reason as to why some patients with low serum CXCL9 level did not show good responses to combination therapy of anti-VEGF antibody and ICIs (atezolizumab plus bevacizumab) remains unclear.
We reasonably speculate that in some patients with low serum CXCL9 levels, tumor angiogenesis not only depends on VEGF but also on other tumor angiogenesis factors such as FGF, HGF, EGF, or placental-derived growth factor. Thus, the inducing effects of anti-VEGF on CXCL9 production via hypoxia-induced increased IFN-γ may be insufficient. Further analyses are required to investigate this hypothesis. Meanwhile, as shown in Figure 2a, even in patients with low serum CXCL9 baseline, those with objective response by atezolizumab and bevacizumab might achieve durable response and longer OS.
One of the potential causes of the shorter OS in patients with PD who were treated with atezolizumab and bevacizumab might be hyperprogressive disease. Hyperprogressive disease is sometimes observed in patients treated with ICIs and exhibits a paradoxical acceleration of tumor growth after ICI initiation. A study conducted on patients with HCC who were treated with programmed cell death protein-1 (PD-1) inhibitors revealed that 12.7% of those patients experienced hyperprogressive disease and had poor prognosis [33]. It was difficult to identify hyperprogressive disease in this study owing to the lack of data regarding HCC growth rate before treatment initiation. However, a total of 22.2% patients with early PD exhibited more than 50% tumor diameter expansion at 6 weeks after treatment initiation. Thus, predicting the outcome of such patients is clinically important, and baseline CXCL9 level is a potential predictive factor.
However, patients with low baseline serum CXCL9 levels exhibited a favorable response to lenvatinib treatment. Lenvatinib is a potent multi-kinase inhibitor that mainly targets fibroblast growth factor receptors-1–4 and VEGF receptor-1–3 [34, 35]. The efficacy and safety of lenvatinib for patients with unresectable HCC have been confirmed in clinical trials and real-world data [3, 22, 23, 36, 37, 38]. Thus, early response evaluation is necessary in atezolizumab plus bevacizumab treatment for unresectable HCC to determine the time of switching to the next line of treatment, especially in patients with low serum CXCL9 baseline level, which is a possible predictor of early PD. Although the cause of the favorable response to lenvatinib by patients with unresectable HCC and low CXCL9 expression could not be clarified, we hypothesized that CXCL9 might be associated with a signaling pathway, which lenvatinib could suppress. We obtained the “Human HCC” gene expression data (total of 405 records) from The Cancer Genome Atlas consortium expression database (https://portal.gdc.cancer.gov/) and converted them into TPM values for further analysis.
We classified patients into either high CXCL9 expression group (TPM ≥ 60) or low expression group (TPM < 60) and analyzed the association between CXCL9 and FGFR4 expression levels. As shown in the online supplementary Figure S4, expressions of FGFR4 − which are reportedly associated with lenvatinib treatment responses [39, 40, 41] − were significantly higher in patients with low CXCL9 expression levels. Thus, we hypothesize that the high FGFR4 expression levels in patients with low CXCL9 expression levels are attributable to a favorable response to lenvatinib treatment in patients with unresectable HCC and low CXCL9 expression. Subsequently, we confirmed the correlation between CXCL9 and FGFR4 expression levels in human hepatoma cell lines of HepG2 cells, Huh1 cells, Huh7 cells, JHH2 cells, JHH4 cells, and JHH6 cells. As shown in online supplementary Figure S5, the expression levels of FGFR4 and CXCL9 were significantly negatively correlated (R2 = 0.8041, p = 0.0155). Additionally, consistent with the results of TCGA analysis, and in vitro analysis of hepatoma cell lines, as shown in online supplementary Figure 3, HCC of patients with high serum CXCL9 levels had lower expression levels of FGFR4 and higher expression levels of CXCL9 in HCC. Further, to clarify the relationship between the expression levels of FGFR4 and CXCL9 in HCC, we investigated the effect of lenvatinib, which could suppress FGFR4-mediated signaling strongly [35], on intracellular CXCL9 expression levels in Huh7 cells and supernatant CXCL9 concentrations in Huh7 cells. As shown in online supplementary Figure S6, treatment of Huh7 cells with lenvatinib resulted in increased intracellular CXCL9 expression levels and increased supernatant CXCL9 concentrations. Thus, we speculated that FGFR4-mediated signaling is associated with the downregulation of CXCL9 expression. Subsequently, we knocked down FGFR4 in Huh7 cells by siRNA and analyzed the changes in intracellular CXCL9 expression levels. As shown in online supplementary Figure S7, the knock down of FGFR4 increased the CXCL9 expression level. The results indicated that FGFR4 signaling suppressed CXCL9 gene expression. In addition, the results may be consistent with the previous reports indicating that FGFR4-mediated signaling negatively regulates NF-kappa B signaling [42], which may activate the transcription of CXCL9 [43]. Finally, we analyzed the changes in serum CXCL9 levels at 7 days after lenvatinib initiation for unresectable HCC. As shown in online supplementary Figure S8, similar to in vitro analysis, the median serum CXCL9 increased in patients with objective response to lenvatinib, and the median changes in serum CXCL9 were significantly higher in patients with objective response to lenvatinib than in those without objective response to lenvatinib.
Recently, Harding et al. [12]revealed that ICI monotherapy had a poor response in patients with unresectable HCC carrying WNT/β-catenin mutations, including CTNNB-1 mutation, resulting in poor prognosis. Therefore, we analyzed the genetic alterations in circulating tumor DNA; however, in this small size analysis, no significant genetic alterations in circulating tumor DNA were observed. Using the combination of anti-VEGF antibody therapy with ICI might affect the results, although further analysis of larger patient cohorts is required.
CCL5 was selected as a predictive factor associated with early PD in exploratory analysis. In HCC, high CCL5 expression has been reported to induce the infiltration of CD8+ T cells [44]. In addition, low baseline CXCL5 levels are associated with poor PD-1 inhibitor response in advanced melanoma [45]. Although these factors did not show significant differences between patients with and without early PD in the validation analysis of 60 patients, further analysis is required.
Quite recently, Zhu et al. [46] reported detailed molecular analyses of clinical response to atezolizumab plus bevacizumab. The authors showed that the T effector signature, including CXCL9, was significantly correlated with response to atezolizumab plus bevacizumab treatment. The results are consistent with our data; however, to the best of our knowledge, our study is the first to show the potential predictive factor (baseline serum CXCL9 levels) associated with early PD post-atezolizumab plus bevacizumab treatment. In addition, we present an alternative potent therapy for patients at high risk of early PD development during atezolizumab plus bevacizumab treatment.
This study had a few limitations. This was a retrospective study; the number of included patients was relatively small, especially those analyzed for genetic alterations in circulating tumor DNA. The predictive utility of baseline serum CXCL9 levels for early PD was not high (AUC of 0.75 in ROC analysis), thus, this should be taken into consideration while interpreting these findings. In addition, the backgrounds of the 8 patients in the discovery cohort were not uniform. Thus, a larger prospective study is necessary to validate the results of this study. In conclusion, low serum CXCL9 (<333 pg/mL) levels may predict early PD in patients with unresectable HCC treated with atezolizumab plus bevacizumab.
Statement of Ethics
The present study protocols conformed to the ethical guidelines of the Declaration of Helsinki and were approved by the Ethics Committee of Hokkaido University Hospital (approval no. 020-0267 and 017–0521) and the participating institutes. Written informed consent was obtained from all the included patients.
Conflict of Interest Statement
Professor Naoya Sakamoto received lecture fees from Bristol Myers Squibb and Janssen Pharmaceutical K.K.; grants and endowments from MSD, Pharmaceutical K. K., and Chugai Pharmaceutical Co. Ltd.; and research grants from Gilead Sciences Inc. and AbbVie GK. Dr. Goki Suda received research grants from Merck & Co., Gilead Sciences Inc., and Bristol Myers Squibb. The remaining of the authors has no conflicts of interest to disclose.
Funding Sources
This study was supported in part by grants from the Japan Agency for Medical Research and Development (AMED; Grant No. JP21fk0210072, JP21fk0310101, JP21fk0210047, JP21fk0210064, JP21fk0210056, JP21fk0210048, JP21fk0210058, JP21fk0210066, and JP21fk0210067) and SPS KAKENHI (Grant No. 19K08458).
Author Contributions
Shunichi Hosoda and Goki Suda contributed equally to this work. Goki Suda designed the study, performed the experiments, and wrote the manuscript. Shunichi Hosoda conducted examinations. Takuya Sho, Koji Ogawa, Megumi Kimura, Zijian Yang, Sonoe Yoshida, Akinori Kubo, Yoshimasa Tokuchi, Takashi Kitagataya, Osamu Maehara, Shunsuke Ohnishi, Akihisa Nakamura, Ren Yamada, Masatsugu Ohara, Naoki Kawagishi, Mitsuteru Natsuizaka, Masato Nakai, Kenichi Morikawa, Ken Furuya, Masaru Baba, Yoshiya Yamamoto, Kazuharu Suzuki, Takaaki Izumi, Takashi Meguro, Katsumi Terashita, Jun Ito, and Takuto Miyagishima collected the data. Takuya Sho, Mitsuteru Natsuizaka, Koji Ogawa, and Naoya Sakamoto provided hepatological advice and edited the manuscript. Naoya Sakamoto revised the manuscript for important intellectual content.
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.
NBNC, non-HBV non-HCV; CH, chronic hepatitis; LC, liver cirrhosis; ECOG PS, Eastern Cooperative Oncology Group Performance Status; BMI, body mass index; NLR, neutrophil to lymphocyte ratio; ALBI, albumin-bilirubin; AST, aspartate aminotransferase; ALT alanine aminotransferase; AFP, alpha-fetoprotein; DCP, des-gamma-carboxy prothrombin; BCLC, Barcelona Clinic Liver Cancer; LN, lymph node; EHM, extrahepatic metastasis; RFA, radiofrequency ablation; TACE, transcatheter arterial chemoembolization; HAIC, hepatic arterial infusion chemotherapy; RT, radiation therapy; TKI, tyrosine kinase inhibitor.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Acknowledgments
The authors would like to thank all patients and their families as well as Yurina Ohta and the investigators and staff of the participating institutions, NORTE study group. The principal investigators of the NORTE study sites are listed below: Junichi Yoshida (JCHO Sapporo Hokushin Hospital), Atsushi Nagasaka (Sapporo City General Hospital), Akira Fuzinaga (Abashiri-Kosei General Hospital), Hideaki Kikuchi, Tomofumi Atarashi (Obihiro-Kosei General Hospital), Ken Furuya (JCHO Hokkaido Hospital), Shuichi Muto (National Hospital Organization Hokkaido Medical Center), Takashi Meguro (Hokkaido Gastroenterology Hospital), Akiyoshi Saga (Kaisei Hospital), Munenori Okamoto (Aiiku Hospital), Masaki Katagiri (Sapporo Hokuyu Hospital), Takuto Miyagishima (Kushiro Rosai Hospital), Jun Konno (Hakodate Central General Hospital), Kenichi Kumagai (Mori City National Health Insurance Hospital), Manabu Onodera (NTT EAST Sapporo Hospital), Tomoe Kobayashi (Tomakomai City Hospital), Minoru Uebayashi (Japanese Red Cross Kitami Hospital), Kanji Katou (Iwamizawa Municipal General Hospital), Yasuyuki Kunieda (Wakkanai City Hospital), Miki Tateyama (Tomakomai Nissho Hospital), Atsuhiko Kawakami (Sapporo Century Hospital), Izumi Tsunematsu (Touei Hospital), Keisuke Shinada (Keiwakai Ebetsu Hospital), and Yoshiya Yamamoto (Hakodate City General Hospital).
Funding Statement
This study was supported in part by grants from the Japan Agency for Medical Research and Development (AMED; Grant No. JP21fk0210072, JP21fk0310101, JP21fk0210047, JP21fk0210064, JP21fk0210056, JP21fk0210048, JP21fk0210058, JP21fk0210066, and JP21fk0210067) and SPS KAKENHI (Grant No. 19K08458).
References
- 1.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362((9399)):1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008 Jul 24;359((4)):378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 3.Kudo M, Finn RS, Qin S, Han KH, Ikeda K, Piscaglia F, et al. Lenvatinib versus sorafenib in first-line treatment of patients with unresectable hepatocellular carcinoma a randomised phase 3 non-inferiority trial. Lancet. 2018 Mar 24;391((10126)):1163–1173. doi: 10.1016/S0140-6736(18)30207-1. [DOI] [PubMed] [Google Scholar]
- 4.Bruix J, Qin S, Merle P, Granito A, Huang YH, Bodoky G, et al. Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE) a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017 Jan 7;389((10064)):56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 5.Kudo M, Tsuchiya K, Kato N, Hagihara A, Numata K, Aikata H, et al. Cabozantinib in Japanese patients with advanced hepatocellular carcinoma a phase 2 multicenter study. J Gastroenterol. 2021 Feb;56((2)):181–190. doi: 10.1007/s00535-020-01753-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu AX, Kang YK, Yen CJ, Finn RS, Galle PR, Llovet JM, et al. Ramucirumab after sorafenib in patients with advanced hepatocellular carcinoma and increased alpha-fetoprotein concentrations (REACH-2) a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2019 Feb;20((2)):282–296. doi: 10.1016/S1470-2045(18)30937-9. [DOI] [PubMed] [Google Scholar]
- 7.Finn RS, Qin S, Ikeda M, Galle PR, Ducreux M, Kim TY, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020 May 14;382((20)):1894–1905. doi: 10.1056/NEJMoa1915745. [DOI] [PubMed] [Google Scholar]
- 8.Gordan JD, Kennedy EB, Abou-Alfa GK, Beg MS, Brower ST, Gade TP, et al. Systemic therapy for advanced hepatocellular carcinoma ASCO guideline. J Clin Oncol. 2020 Dec 20;38((36)):4317–4345. doi: 10.1200/JCO.20.02672. [DOI] [PubMed] [Google Scholar]
- 9.Kudo M, Kawamura Y, Hasegawa K, Tateishi R, Kariyama K, Shiina S, et al. Management of hepatocellular carcinoma in Japan JSH consensus statements and recommendations 2021 update. Liver Cancer. 2021 Jun;10((3)):181–223. doi: 10.1159/000514174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sho T, Suda G, Ogawa K, Kimura M, Kubo A, Tokuchi Y, et al. Early response and safety of atezolizumab plus bevacizumab for unresectable hepatocellular carcinoma in patients who do not meet IMbrave150 eligibility criteria. Hepatol Res. 2021 Sep;51((9)):979–989. doi: 10.1111/hepr.13693. [DOI] [PubMed] [Google Scholar]
- 11.Ducreux M, Zhu AX, Cheng A-L, Galle PR, Ikeda M, Nicholas A, et al. IMbrave150 exploratory analysis to examine the association between treatment response and overall survival (OS) in patients (pts) with unresectable hepatocellular carcinoma (HCC) treated with atezolizumab (atezo) + bevacizumab (bev) versus sorafenib (sor) J Clin Oncol. 2021;39((15_Suppl l)):4071–4071. [Google Scholar]
- 12.Harding JJ, Nandakumar S, Armenia J, Khalil DN, Albano M, Ly M, et al. Prospective genotyping of hepatocellular carcinoma clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res. 2019 Apr 1;25((7)):2116–2126. doi: 10.1158/1078-0432.CCR-18-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kudo M. Gd-EOB-DTPA-MRI could predict WNT/β-Catenin mutation and resistance to immune checkpoint inhibitor therapy in hepatocellular carcinoma. Liver Cancer. 2020 Sep;9((5)):479–490. doi: 10.1159/000509554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubo A, Suda G, Kimura M, Maehara O, Tokuchi Y, Kitagataya T, et al. Characteristics and lenvatinib treatment response of unresectable hepatocellular carcinoma with iso-high intensity in the hepatobiliary phase of EOB-MRI. Cancers. 2021 Jul 20;13((14)):3633. doi: 10.3390/cancers13143633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura Y. Biomarkers for immune checkpoint inhibitor-mediated tumor response and adverse events. Front Med. 2019;6:119. doi: 10.3389/fmed.2019.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Litchfield K, Reading JL, Puttick C, Thakkar K, Abbosh C, Bentham R, et al. Meta-analysis of tumor- and T cell-intrinsic mechanisms of sensitization to checkpoint inhibition. Cell. 2021 Feb 4;184((3)):596.e14–614.e14. doi: 10.1016/j.cell.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suda G, Furusyo N, Toyoda H, Kawakami Y, Ikeda H, Suzuki M, et al. Daclatasvir and asunaprevir in hemodialysis patients with hepatitis C virus infection a nationwide retrospective study in Japan. J Gastroenterol. 2018 Jan;53((1)):119–128. doi: 10.1007/s00535-017-1353-y. [DOI] [PubMed] [Google Scholar]
- 18.Suzuki K, Suda G, Yamamoto Y, Furuya K, Baba M, Kimura M, et al. Entecavir treatment of hepatitis B virus-infected patients with severe renal impairment and those on hemodialysis. Hepatol Res. 2019 Nov;49((11)):1294–1304. doi: 10.1111/hepr.13399. [DOI] [PubMed] [Google Scholar]
- 19.Ogawa K, Kobayashi T, Furukawa JI, Hanamatsu H, Nakamura A, Suzuki K, et al. Tri-antennary tri-sialylated mono-fucosylated glycan of alpha-1 antitrypsin as a non-invasive biomarker for non-alcoholic steatohepatitis a novel glycobiomarker for non-alcoholic steatohepatitis. Sci Rep. 2020 Jan 15;10((1)):321. doi: 10.1038/s41598-019-56947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohara M, Suda G, Kimura M, Maehara O, Shimazaki T, Shigesawa T, et al. Analysis of the optimal psoas muscle mass index cut-off values as measured by computed tomography for the diagnosis of loss of skeletal muscle mass in Japanese people. Hepatol Res. 2020 Jun;50((6)):715–725. doi: 10.1111/hepr.13499. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki K, Suda G, Yamamoto Y, Furuya K, Baba M, Nakamura A, et al. Tenofovir-disoproxil-fumarate modulates lipid metabolism via hepatic CD36/PPAR-alpha activation in hepatitis B virus infection. J Gastroenterol. 2021 Feb;56((2)):168–180. doi: 10.1007/s00535-020-01750-3. [DOI] [PubMed] [Google Scholar]
- 22.Sho T, Suda G, Ogawa K, Shigesawa T, Suzuki K, Nakamura A, et al. Lenvatinib in patients with unresectable hepatocellular carcinoma who do not meet the REFLECT trial eligibility criteria. Hepatol Res. 2020 Aug;50((8)):966–977. doi: 10.1111/hepr.13511. [DOI] [PubMed] [Google Scholar]
- 23.Shigesawa T, Suda G, Kimura M, Shimazaki T, Maehara O, Yamada R, et al. Baseline angiopoietin-2 and FGF19 levels predict treatment response in patients receiving multikinase inhibitors for hepatocellular carcinoma. JGH Open. 2020;4((5)):880–888. doi: 10.1002/jgh3.12339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lencioni R, Llovet JM. Modified RECIST (mRECIST) assessment for hepatocellular carcinoma. Semin Liver Dis. 2010 Feb;30((01)):052–060. doi: 10.1055/s-0030-1247132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorbachev AV, Kobayashi H, Kudo D, Tannenbaum CS, Finke JH, Shu S, et al. CXC chemokine ligand 9/monokine induced by IFN-gamma production by tumor cells is critical for T cell-mediated suppression of cutaneous tumors. J Immunol. 2007 Feb 15;178((4)):2278–2286. doi: 10.4049/jimmunol.178.4.2278. [DOI] [PubMed] [Google Scholar]
- 26.House IG, Savas P, Lai J, Chen AXY, Oliver AJ, Teo ZL, et al. Macrophage-derived CXCL9 and CXCL10 are required for antitumor immune responses following immune checkpoint blockade. Clin Cancer Res. 2020 Jan 15;26((2)):487–504. doi: 10.1158/1078-0432.CCR-19-1868. [DOI] [PubMed] [Google Scholar]
- 27.Llovet JM, Castet F, Heikenwalder M, Maini MK, Mazzaferro V, Pinato DJ, et al. Immunotherapies for hepatocellular carcinoma. Nat Rev Clin Oncol. 2022 Mar;19((3)):151–172. doi: 10.1038/s41571-021-00573-2. [DOI] [PubMed] [Google Scholar]
- 28.Montironi C, Castet F, Haber PK, Pinyol R, Torres-Martin M, Torrens L, et al. Inflamed and non-inflamed classes of HCC a revised immunogenomic classification. Gut. 2022 Feb 23; doi: 10.1136/gutjnl-2021-325918. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mlynska A, Salciuniene G, Zilionyte K, Garberyte S, Strioga M, Intaite B, et al. Chemokine profiling in serum from patients with ovarian cancer reveals candidate biomarkers for recurrence and immune infiltration. Oncol Rep. 2019 Feb;41((2)):1238–1252. doi: 10.3892/or.2018.6886. [DOI] [PubMed] [Google Scholar]
- 30.Iwai T, Sugimoto M, Patil NS, Bower D, Suzuki M, Kato C, et al. Both T cell priming in lymph node and CXCR3-dependent migration are the key events for predicting the response of atezolizumab. Sci Rep. 2021 Jul 6;11((1)):13912. doi: 10.1038/s41598-021-93113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikura N, Sugimoto M, Yorozu K, Kurasawa M, Kondoh O. AntiVEGF antibody triggers the effect of antiPDL1 antibody in PDL1(low) and immune desertlike mouse tumors. Oncol Rep. 2021;47((2)):36. doi: 10.3892/or.2021.8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kudo M. Combination immunotherapy with anti-PD-1/PD-L1 antibody plus anti-VEGF antibody may promote cytotoxic T lymphocyte infiltration in hepatocellular carcinoma including in the noninflamed subclass. Liver Cancer. 2022;11((3)):185–191. doi: 10.1159/000524977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo S, Lee D, Shim JH, Kim KM, Lim YS, Lee HC, et al. Risk of hepatitis B virus reactivation in patients treated with immunotherapy for anti-cancer treatment. Clin Gastroenterol Hepatol. 2022;20((4)):898–907. doi: 10.1016/j.cgh.2021.06.019. [DOI] [PubMed] [Google Scholar]
- 34.Hiraoka A, Kumada T, Kariyama K, Takaguchi K, Itobayashi E, Shimada N, et al. Therapeutic potential of lenvatinib for unresectable hepatocellular carcinoma in clinical practice multicenter analysis. Hepatol Res. 2019 Jan;49((1)):111–117. doi: 10.1111/hepr.13243. [DOI] [PubMed] [Google Scholar]
- 35.Shigesawa T, Maehara O, Suda G, Natsuizaka M, Kimura M, Shimazaki T, et al. Lenvatinib suppresses cancer stem-like cells in HCC by inhibiting FGFR 1-3 signaling but not FGFR4 signaling. Carcinogenesis. 2020 May 25;42((1)):58–69. doi: 10.1093/carcin/bgaa049. [DOI] [PubMed] [Google Scholar]
- 36.Kawamura Y, Kobayashi M, Shindoh J, Kobayashi Y, Kasuya K, Sano T, et al. F-Fluorodeoxyglucose uptake in hepatocellular carcinoma as a useful predictor of an extremely rapid response to lenvatinib. Liver Cancer. 182020 Jan;9((1)):84–92. doi: 10.1159/000503577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maruta S, Ogasawara S, Ooka Y, Obu M, Inoue M, Itokawa N, et al. Potential of lenvatinib for an expanded indication from the REFLECT trial in patients with advanced hepatocellular carcinoma. Liver Cancer. 2020 Aug;9((4)):382–396. doi: 10.1159/000507022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sho T, Suda G, Ogawa K, Kimura M, Shimazaki T, Maehara O, et al. Early response and safety of lenvatinib for patients with advanced hepatocellular carcinoma in a real-world setting. JGH Open. 2020 Feb;4((1)):54–60. doi: 10.1002/jgh3.12209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamauchi M, Ono A, Ishikawa A, Kodama K, Uchikawa S, Hatooka H, et al. Tumor fibroblast growth factor receptor 4 level predicts the efficacy of lenvatinib in patients with advanced hepatocellular carcinoma. Clin Transl Gastroenterol. 2020 May;11((5)):e00179. doi: 10.14309/ctg.0000000000000179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myojin Y, Kodama T, Maesaka K, Motooka D, Sato Y, Tanaka S, et al. ST6GAL1 is a novel serum biomarker for lenvatinib-susceptible FGF19-driven hepatocellular carcinoma. Clin Cancer Res. 2021 Feb 15;27((4)):1150–1161. doi: 10.1158/1078-0432.CCR-20-3382. [DOI] [PubMed] [Google Scholar]
- 41.Shigesawa T, Suda G, Kimura M, Maehara O, Tokuchi Y, Kubo A, et al. Baseline serum angiopoietin-2 and VEGF levels predict the deterioration of the liver functional reserve during lenvatinib treatment for hepatocellular carcinoma. PLoS One. 2021;16((3)):e0247728. doi: 10.1371/journal.pone.0247728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drafahl KA, McAndrew CW, Meyer AN, Haas M, Donoghue DJ. The receptor tyrosine kinase FGFR4 negatively regulates NF-kappaB signaling. PLoS One. 2010 Dec 22;5((12)):e14412. doi: 10.1371/journal.pone.0014412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiroi M, Ohmori Y. The transcriptional coactivator CREB-binding protein cooperates with STAT1 and NF-kappa B for synergistic transcriptional activation of the CXC ligand 9/monokine induced by interferon-gamma gene. J Biol Chem. 2003 Jan 3;278((1)):651–660. doi: 10.1074/jbc.M204544200. [DOI] [PubMed] [Google Scholar]
- 44.Ruiz de Galarreta M, Bresnahan E, Molina-Sanchez P, Lindblad KE, Maier B, Sia D, et al. β-Catenin activation promotes immune escape and resistance to anti-PD-1 therapy in hepatocellular carcinoma. Cancer Discov. 2019 Aug;9((8)):1124–1141. doi: 10.1158/2159-8290.CD-19-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fujimura T, Sato Y, Tanita K, Lyu C, Kambayashi Y, Amagai R, et al. Association of baseline serum levels of CXCL5 with the efficacy of nivolumab in advanced melanoma. Front Med. 2019;6:86. doi: 10.3389/fmed.2019.00086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu AX, Abbas AR, de Galarreta MR, Guan Y, Lu S, Koeppen H, et al. Molecular correlates of clinical response and resistance to atezolizumab in combination with bevacizumab in advanced hepatocellular carcinoma. Nat Med. 2022 Aug;28((8)):1599–1611. doi: 10.1038/s41591-022-01868-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Data Availability Statement
All data generated or analyzed during this study are included in this article. Further inquiries can be directed to the corresponding author.
NBNC, non-HBV non-HCV; CH, chronic hepatitis; LC, liver cirrhosis; ECOG PS, Eastern Cooperative Oncology Group Performance Status; BMI, body mass index; NLR, neutrophil to lymphocyte ratio; ALBI, albumin-bilirubin; AST, aspartate aminotransferase; ALT alanine aminotransferase; AFP, alpha-fetoprotein; DCP, des-gamma-carboxy prothrombin; BCLC, Barcelona Clinic Liver Cancer; LN, lymph node; EHM, extrahepatic metastasis; RFA, radiofrequency ablation; TACE, transcatheter arterial chemoembolization; HAIC, hepatic arterial infusion chemotherapy; RT, radiation therapy; TKI, tyrosine kinase inhibitor.