Abstract
Bone development starts with condensations of undifferentiated mesenchymal cells that set a framework for future bones within the primordium. In the endochondral pathway, mesenchymal cells inside the condensation differentiate into chondrocytes and perichondrial cells in a SOX9-dependent mechanism. However, the identity of mesenchymal cells outside the condensation and how they participate in developing bones remain undefined. Here we show that mesenchymal cells surrounding the condensation contribute to both cartilage and perichondrium, robustly generating chondrocytes, osteoblasts, and marrow stromal cells in developing bones. Single-cell RNA-seq analysis of Prrx1-cre-marked limb bud mesenchymal cells at E11.5 reveals that Notch effector Hes1 is expressed in a mutually exclusive manner with Sox9 that is expressed in pre-cartilaginous condensations. Analysis of a Notch signaling reporter CBF1:H2B-Venus reveals that peri-condensation mesenchymal cells are active for Notch signaling. In vivo lineage-tracing analysis using Hes1-creER identifies that Hes1+ early mesenchymal cells surrounding the SOX9+ condensation at E10.5 contribute to both cartilage and perichondrium at E13.5, subsequently becoming growth plate chondrocytes, osteoblasts of trabecular and cortical bones, and marrow stromal cells in postnatal bones. In contrast, Hes1+ cells in the perichondrium at E12.5 or E14.5 do not generate chondrocytes within cartilage, contributing to osteoblasts and marrow stromal cells only through the perichondrial route. Therefore, Hes1+ peri-condensation mesenchymal cells give rise to cells of the skeletal lineage through cartilage-dependent and independent pathways, supporting the theory that early mesenchymal cells outside the condensation also play important roles in early bone development.
Keywords: Hes1, Notch signaling, mesenchymal condensation, perichondrium, cartilage, perichondrial cells, chondrocytes, osteoblasts, in vivo lineage-tracing, single-cell RNA-sequencing, endochondral bone development
Bone development involves highly sequential steps of mesenchymal cell proliferation and differentiation, thus represents a prime example of organogenesis deliberately executed through heterotypic cellular interactions. Condensation of undifferentiated mesenchymal cells is the first step of bone development, which sets a framework for future bones within the primordium (1, 2). In this process, a previously dispersed population of undifferentiated mesenchymal cells migrate to the site of the future skeleton in the limb bud and aggregate to form a vasculature-free element termed mesenchymal condensation (3). Mesenchymal condensations serve as an important modality to generate skeletal precursor cells on a large scale in a highly organized environment. Particularly in the endochondral pathway by which most of mammalian bones are formed, mesenchymal condensations give rise to both cartilage template and perichondrium in the subsequent phase. Chondrocytes within the cartilage template and perichondrial cells provide distinct cellular sources of growth plate chondrocytes, trabecular and cortical osteoblasts, and marrow stromal cells (4, 5, 6, 7). Therefore, mesenchymal condensations provide the foundation of developing bones.
The current concept holds that SOX9, a master transcription factor for chondrogenesis, is essential for condensing mesenchymal cells to differentiate into chondrocytes and perichondrial cells (1, 8, 9, 10). Indeed, SOX9 is absolutely required for these mesenchymal cells to stay organized within the condensation (11). Fate-mapping studies using Sox9-cre/creER demonstrate that Sox9+ cells within the condensation function as osteo-chondro-progenitor cells as they give rise to chondrocytes and perichondrial cells in the subsequent stage (8, 9, 12). In the limb, Prrx1-cre is used to mark the entire spectrum of cells of the skeletal lineage including “skeletal stem cells” and their derivatives, as Prrx1 is expressed not only by condensing mesenchymal cells but also by other cells that originate from the lateral plate mesoderm (13, 14). Interestingly, Prrx1 is predominantly expressed in the perichondrium at E13.5 when the cartilage template is established (4, 15, 16), suggesting that cells surrounding the condensation may also robustly contribute to the generation of skeletal lineage cells. However, the identity of early mesenchymal cells outside the condensation and how they participate in developing bones remain undefined.
We recently demonstrated that Dlx5+ early perichondrial cells located in the outer layer of the perichondrium at E12.5 sustainably contribute to cortical bone and marrow stromal compartments in developing bones (4). These Dlx5+ perichondrial cells remain in the perichondrium in subsequent stages, unlike osterix (Osx+) osteogenic perichodrial cells that are in the inner layer of the perichondrium (17). Osx+ cell descendants contribute to skeletogenesis only transiently and eventually disappear from the perichondrium and the skeletal element (8, 18). Interestingly, however, Dlx5+ early perichondrial cells do not contribute to cartilage. It has not been determined if peri-condensation mesenchymal cells preceding Dlx5+ perichondrial cells can generate chondrocytes within the cartilage template.
In this study, we hypothesize that early mesenchymal cells surrounding the condensation can contribute to chondrocytes of the cartilage template while also remaining outside the condensation and contributing to perichondrial cells. To test this hypothesis, we utilized in vivo lineage-tracing approaches to define the cell fates of peri-condensation mesenchymal cells. Using single-cell RNA-seq analyses, we identified Notch effector Hes1 as a potential marker of peri-condensation mesenchymal cells that are located outside the SOX9+ condensation and subsequently revealed their cell fates using a tamoxifen-inducible Hes1-creER allele. Our findings demonstrate that Hes1+ peri-condensation mesenchymal cells give rise to cells of the skeletal cell lineage through cartilage-dependent and independent pathways in growing bones and critically support early bone development.
Results
Single-cell RNA-seq identifies Hes1 as a potential marker of peri-condensation mesenchymal cells
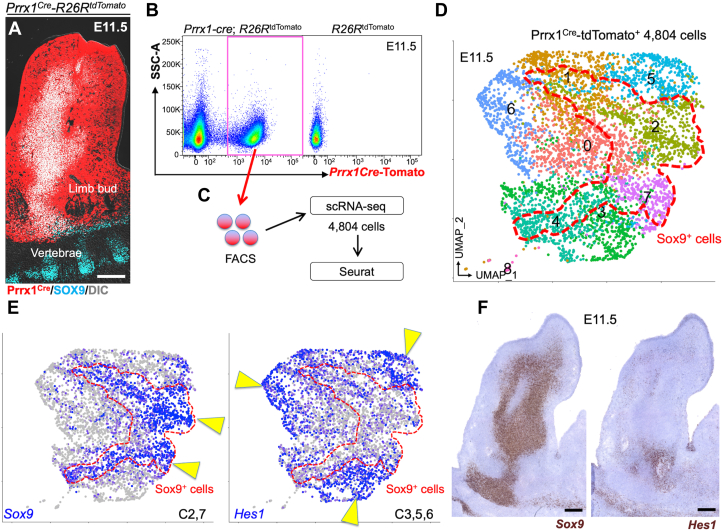
In the mouse limb bud, undifferentiated mesenchymal cells make condensations around embryonic day (E) 10.5. These condensing mesenchymal cells express SOX9 to activate the transcription machinery for chondrocyte differentiation (19). We first performed single-cell RNA-seq analyses to define cellular heterogeneity of the limb bud mesenchyme. Limb bud mesenchymal cells contributing to the skeletal element, including SOX9+ condensing mesenchymal cells are ubiquitously marked by a cre recombinase driven by a 2.4kb Prrx1 promoter/enhancer (13) (Fig. 1A). We dissociated limb mesenchymal cells of Prrx1-cre, R26RtdTomato mice at embryonic day 11.5 (E11.5), and isolated tdTomato+ cells by fluorescence-activated cell sorting (FACS) (Fig. 1B). We profiled 4804 cells Prrx1cre-tdTomato+ cells using the 10X Chromium Single-Cell Gene Expression Solution platform (Fig. 1C). A graph-based clustering analysis using Seurat (20) revealed 9 clusters (Fig. 1D), including two clusters of cells abundant in Sox9 (Cluster 2,7, arrowheads of Fig. 1E left) and six clusters of their surrounding cells (Cluster 0,1,3–6). Prrx1 was expressed in the surrounding cluster (Cluster 5) while another chondrogenic marker Col2a1 expression overlapped with Sox9 expression (cluster 2,7) (Fig. S1A). Notably, Sox9 and Col2a1 were also expressed in a part of clusters 1, 3, and 4 (red dotted contour of Fig. 1, D and E). The surrounding clusters were composed of mesenchymal cells expressing unique homeobox proteins, such as Msx1 (Cluster 5,6), Lhx9 (Cluster 5), Meox2 (Cluster 0), Emx2 (Cluster 4) and Irx3/5 (Cluster 3,7) (Fig. S1A). In addition, Shh was expressed in Cluster 6, indicating that this cluster corresponds to cells on the posterior margin of the limb bud (21). Therefore, the limb bud skeletal element is constituted by distinct groups of mesenchymal cells constituting the SOX9+ condensation and its surrounding structures.
Figure 1.
Single cell RNA-seq reveals the heterogeneity of early mesenchymal cells in the limb bud.A–E, Single cell RNA-seq analysis of Prrx1-cre-marked limb bud mesenchymal cells at E11.5. A, Prrx1-cre; R26RtdTomato femur stained for SOX9. Scale bar: 200 μm. n = 3 mice. B and C, FACS (B) and scRNA-seq (C) strategy for Prrx1cre-tdTomato+ cells (red box). Shown are cells isolated from Prrx1-cre; R26RtdTomato limb buds (left) or R26RtdTomato control limb buds (right). D, UMAP-based visualization of major classes of Prrx1cre-tdTomato+ cells (Cluster 0–8). E, feature plots. Blue: high expression. Arrowheads: clusters in which a given gene is identified as a cell type-specific marker. Cluster 2,7: Sox9+, Cluster 3,5,6: Hes1+. Dotted contour: Sox9+ cells. n = 4804 cells. Pooled from n = 5 mice. F, RNAscope in situ hybridization analysis of E11.5 limb bud for Hes1 and Sox9 mRNA. Scale bar: 200 μm. n = 3 mice.
In search for a marker for peri-condensation mesenchymal cells, we noticed that a Notch effector gene Hes1 exhibited a pattern negatively correlated with Sox9, identified as cell-type specific markers for Cluster 3,5,6 (Fig. 1E). Notch signaling is essential for maintaining skeletal progenitor cells, and its target gene Hes1 is most abundantly expressed in the fetal perichondrium (22, 23). RNAscope analysis revealed that Hes1 was expressed in a manner surrounding the Sox9-expressing condensation (Fig. 1F). To further validate Notch-responsive status of peri-condensation mesenchymal cells in situ, we utilized a Notch signaling reporter strain expressing a histone 2B (H2B)-bound Venus protein under a C promoter binding factor 1 (CBF1) promoter (CBF1:H2B-Venus) (24). Interestingly, Notch-responsive Venusbright cells were mostly located outside the SOX9+ domain of the condensation (Fig. S1B). Therefore, Notch signaling is active in mesenchymal cells surrounding the SOX9+ condensation. In subsequent in vivo lineage-tracing studies, we focused on Notch effector Hes1 as a marker of peri-condensation mesenchymal cells.
Hes1-creER+ peri-condensation mesenchymal cells can generate chondrocytes
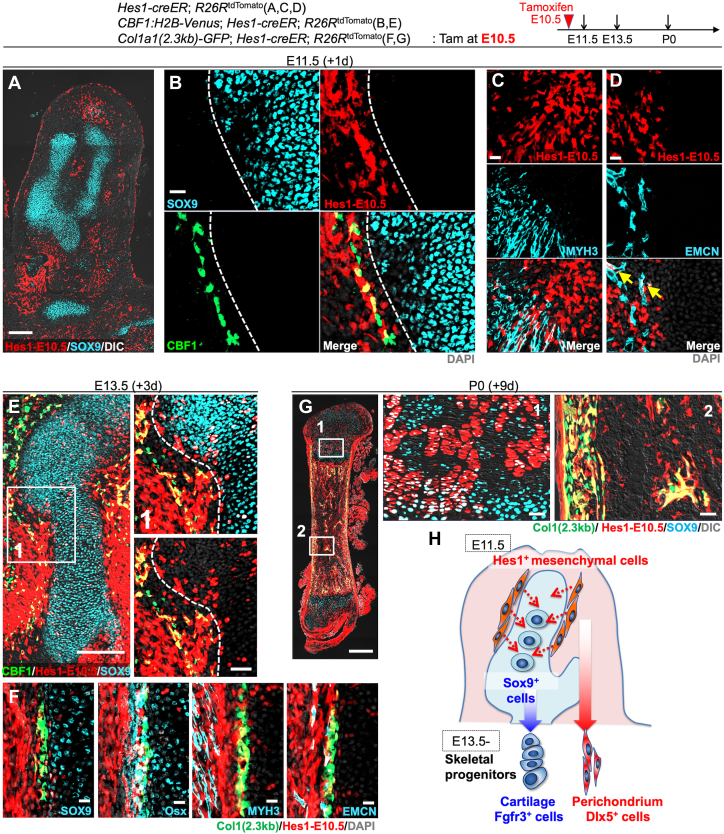
To study the cell fates of these Hes1+ cells surrounding Sox9+ condensation, we performed lineage-tracing experiments using a Hes1-creER knock-in allele (25) that activates an R26R-tdTomato reporter in a tamoxifen-dependent manner. First, we pulsed Hes1-creER; R26RtdTomato mice at E10.5 and analyzed these mice after 24 h at E11.5 to define the identities of Hes1-creER+ cells (Hes1CE-E10.5 cells). Hes1CE-E10.5 cells were located throughout the limb bud at E11.5 in a manner excluding the SOX9+ pre-cartilaginous condensation (Fig. 2A). Hes1CE-E10.5 cells were also located in the area immediately adjacent to SOX9+ cells associated with CBF1:H2B-Venus reporter activities (Fig. 2B). We also noticed that Hes1-creER simultaneously marked other types of cells outside the skeletal elements of the limb bud, including myosin heavy chain 3 (MYH3+) skeletal muscle cells and endomucin (EMCN+) endothelial cells (Fig. 2, C and D). Therefore, Hes1-creER can mark mesenchymal cells in the SOX9-negative domain of the condensation upon tamoxifen injection.
Figure 2.
Undifferentiated Hes1-creER+cells surrounding the mesenchymal condensation provide skeletal progenitor cells.A–G, Cell-fate analysis of Hes1-creER+ mesenchymal cells of the condensation stage, pulsed at E10.5. Hes1-creER; R26RtdTomato femurs carrying CBF1:Venus (B and E) or Col1a1(2.3kb)-GFP (F and G) reporters. A, Limb bud at E11.5. Scale bar: 200 μm. n = 4 mice. B, Magnified view of mesenchymal condensation at E11.5. Scale bar: 20 μm. n = 4 mice. C and D, Immunostaining for MYH3 (C) and endomucin (EMCN) (D). Arrow: Hes1-creER+Emcn+ cells. Scale bar: 20 μm. n = 4 mice. E, Cartilage template at E13.5. Right panel: magnified view of Area (1). Dotted line: cartilage-perichondrium border. Grey: DIC. Scale bar: 200 μm (left), 50 μm (right). n = 4 mice. F, Cartilage-perichondrium immunostaining for SOX9, osterix (OSX), MYH3 and EMCN. Scale bar: 20 μm. n = 4 mice. G, Neonatal femur at P0, after 9 days of chase. Right panels: magnified views of (1: growth plate, 2: bone marrow). Scale bar: 200 μm (left), 20 μm (right 2 panels). n = 4 mice. H, Diagram of Hes1+ mesenchymal cell fates. Hes1+So x 9neg cells surrounding the condensation are bona fide skeletal progenitor cells during endochondral bone development, contributing to both chondrocytes and perichondrial cells and subsequently to all limb skeletal cells. MYH3, myosin heavy chain 3.
Subsequently, we traced the fate of Hes1-creER+ cells in endochondral bone development (see Fig. S2, A and B for H&E and Alcian Blue staining of developing endochondral bones). After 3 days of chase at E13.5, Hes1CE-E10.5 cells contributed to both chondrocytes and perichondrial cells of the cartilage template at E13.5 (Fig. 2E). Hes1CE-E10.5 cells became SOX9+ chondrocytes within the cartilage template while contributing to a majority of perichondrial cells including those expressing CBF1:H2B-Venus (Fig. 2E, right panel). In fact, Hes1CE-E10.5 cells contributed to all layers of the perichondrium including OSX+ osteoblast precursors, in addition to MYH3+ skeletal muscle cells outside the perichondrium and EMCN+ endothelial cells (Figs. 2F and S3A). After 9 days of the chase at postnatal day (P) 0, Hes1CE-E10.5 cells contributed to many columnar chondrocytes of the growth plate, Col1a1-GFP+ osteoblasts on the cortical and trabecular bone, and stromal cells throughout the marrow space (Fig. 2G). We also examined the cell fate of Hes1+ cells at an earlier stage. For this purpose, we pulsed Hes1-creER; R26RtdTomato mice at E8.5 and analyzed these mice at E13.5 and E18.5. Hes1CE-E8.5 cells contributed to essentially all skeletal cells, robustly generating chondrocytes, perichondrial cells, osteoblasts, and marrow stromal cells (Fig. S3, C and D). Notably, Hes1CE-E8.5 cells contributed more substantially to the skeletal lineage than Hes1CE-E10.5 cells did, indicating that Hes1+ cells at a pre-condensation stage might possess a strong chondrogenic potential.
Importantly, skeletal muscle cells or endothelial cells that were initially marked by Hes1-creER at E10.5 did not contribute to these skeletal lineage cells, as cells marked by skeletal muscle-specific cre lines (Acta1-cre, Myl1-cre, and Mck-cre) or an endothelial cell-specific cre line (Tie2-cre) did not contribute to chondrocytes, osteoblasts, or marrow stromal cells at any time points (Fig. S4, A and B). This allows us to exclude the contribution of skeletal muscle cells and endothelial cells initially labelled by Hes1-creER at E10.5 to skeletal lineage cells.
Therefore, Hes1+ mesenchymal cells surrounding the condensation can differentiate not only into perichondrial cells but also into chondrocytes within the cartilage template, both of which robustly contribute to chondrocytes, osteoblasts, and marrow stromal cells in the later stages of endochondral bone development (Fig. 2H).
Hes1-creER+ perichondrial cells possess osteogenic but not chondrogenic potential
Subsequently, we sought to define the cell fate of Hes1+ cells in the later stages of endochondral bone development, at E12.5 when the perichondrium is established adjacent to the cartilage template, as well as at E14.5 when the formation of the marrow space starts due to vascular invasion into the cartilage template.
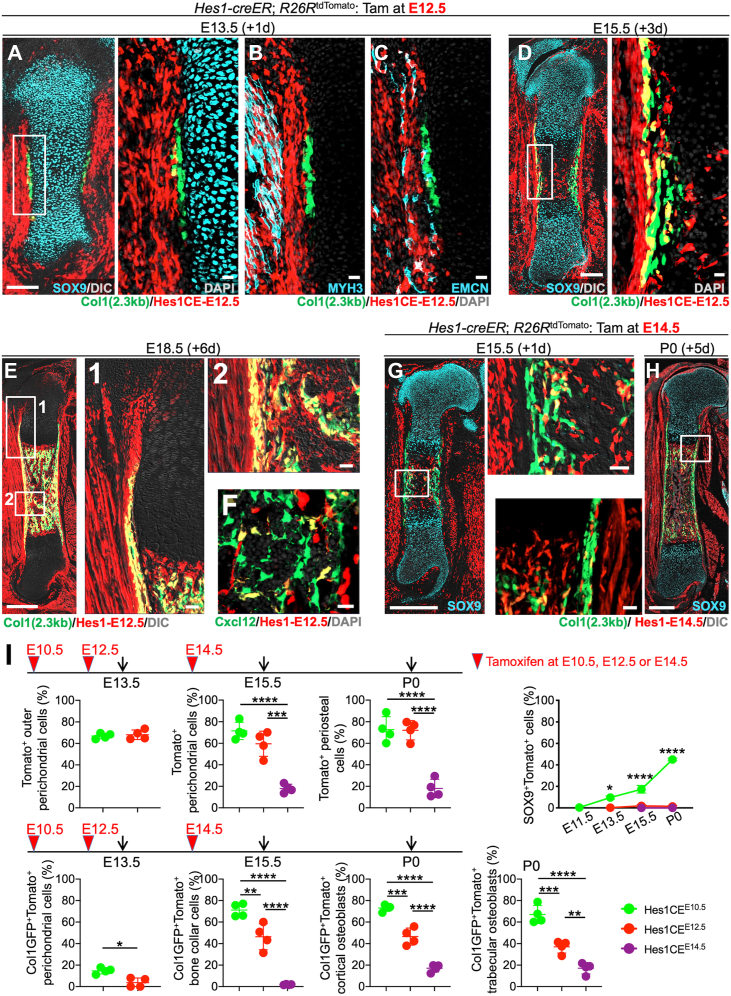
To this end, we first pulsed Col1a1(2.3kb)-GFP; Hes1-creER; R26RtdTomato mice at E12.5 and analyzed these mice after 24 h at E13.5 to define the characteristics of Hes1-creER+ cells in the perichondrium (Hes1CE-E12.5 cells). Hes1CE-E12.5 cells were located outside the SOX9+ domain of the cartilage template with minimal overlap with Col1a1-GFP+ osteoblasts in the perichondrium (Fig. 3A). Hes1CE-E12.5 cells were also found outside the skeletal element among MYH3+ skeletal muscle cells and EMCN+ endothelial cells (Fig. 3, B and C).
Figure 3.
Hes1-creER+cells provide fewer skeletal progenitor cells at becoming later embryonic stages.A–F, Cell-fate analysis of Hes1-creER+ cells, pulsed at E12.5, carrying Col1a1(2.3kb)-GFP reporters. A–C, Cartilage template at E13.5. Immunostaining for SOX9 (A), MYH3 (B), and EMCN (C). Right panel of (A): magnified view of the boxed area. Scale bar: 200 μm (A-left). 20 μm (A-right, B and C). n = 4 mice. D, Left panel: whole femur at E15.5. Scale bar: 200 μm. Right panel: magnified view of the boxed area. n = 4 mice. Scale bar: 20 μm. E, Whole femur at E18.5. Right panels: magnified view of the boxed area (1, 2). Perichondrium, bone collar and growth plate. Scale bar: 500 μm (left), 50 μm (right 2 panels). n = 4 mice. F, Bone marrow at E18.5. Scale bar: 20 μm. n = 3 mice. G and H, Cell-fate analysis of Hes1-creER+ cells, pulsed at E14.5, carrying Col1a1(2.3kb)-GFP reporters. G, Whole femur at E15.5. Right panel: magnified view of the boxed area. Scale bar: 500 μm (left), 50 μm (right). n = 4 mice. (H) Whole femur at E15.5. Left panel: magnified view of the boxed area. Scale bar: 500 μm (right), 50 μm (left). n = 4 mice. I, Quantitative analysis of Hes1-E10.5-, Hes1-E12.5- or Hes1-E14.5-tdTomato+ cells’ contribution to skeletons in embryonic and neonatal stages. Green: Hes1-E10.5, Red: Hes1-E12.5, Violet: Hes1-E14.5. Upper left three panels: Percentage of total tdTomato+ outer perichondrial cells among total outer perichondrial cells at E13.5 (leftmost) and E15.5 (second from the left), and tdTomato+ periosteal cells among total periosteal cells at P0 (third from the left). n = 4 mice per each group. Upper rightmost panel: Contribution of Hes1-creER+ (E10.5, E12.5 and E14.5) cells to SOX9+ chondrocytes within the cartilage template. n = 4 mice per each group. Lower panels: Percentage of Col1a1(2.3kb)-GFP+tdTomato+ cells among total Col1a1(2.3kb)-GFP+ cells at E13.5 (inner perichondrium, leftmost), E15.5 (bone collar, second from the left) and P0 (cortical bone, third from the left. Trabecular bone, rightmost). n = 4 mice per each group. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Two-tailed, Mann–Whitney U test (upper and lower leftmost panels). Two-tailed, One-way ANOVA followed by Tukey’s post hoc test (the others). Data are presented as mean ± SD. MYH3, myosin heavy chain 3.
We subsequently traced the fate of Hes1CE-E12.5 perichondrial cells during the formation of the primary ossification center. After 3 days of chase at E15.5, Hes1CE-E12.5 cells contributed to cells within the primary ossification center as well as Col1a1-GFP+ osteoblasts in the bone collar (Fig. 3D). After 6 days of chase at E18.5, Hes1CE-E12.5 cells contributed to Cxcl12-GFPhigh marrow stromal cells and Col1a1-GFP+ osteoblasts in the trabecular bone (Fig. 3, E and F). Importantly, unlike those pulsed at an earlier stage, Hes1CE-E12.5 cells did not contribute to SOX9+ chondrocytes in the growth plate but maintained themselves within the perichondrium, and robustly contributed to the bone collar and Col1a1-GFP+ osteoblasts in the cortical bone (Fig. 3E). Therefore, Hes1-creER marks perichondrial cells with robust capability to contribute to cortical and trabecular bone compartments as well as to marrow stromal compartments, akin to those marked by Dlx5-creER that we recently reported (4).
We also traced the fate of Hes1-creER+ cells at E14.5 (Hes1CE-E14.5 cells). At E15.5, Hes1CE-E14.5 cells were located among the perichondrium, the periosteum, and the primary ossification center, although most of these cells appeared to be non-skeletal with minimal overlap with Col1a1-GFP+ osteoblasts (Fig. 3G). After 5 days of chase at P0, Hes1CE-E14.5 cells did not robustly contribute to Col1a1-GFP+ osteoblasts in the cortical and trabecular bones (Fig. 3H), indicating that Hes1+ cells lose robust osteogenic potential at E14.5. Postnatally, Hes1-creER+ cells at P3 (Hes1CE-P3 cells) were localized in the groove of Ranvier, the metaphyseal primary spongiosa and the superficial layer of the articular cartilage at P5 (Fig. S5). Hes1CE-P3 cells did not overtly overlap with SOX9+ cells within the growth plate, whereas a few Hes1CE-P3 cells in a deeper layer of the articular cartilage expressed SOX9. Therefore, Hes1+ cells do not overlap with SOX9+ cells at a later stage of perichondrial development.
We further performed quantitative histological approaches to determine how Hes1+ cells at different stages (Hes1CE-E10.5, Hes1CE-E12.5, and Hes1CE-E14.5 cells) differentially contribute to endochondral bone development. First, we quantified the contribution of Hes1CE-E10.5 or Hes1CE-E12.5 cells to the E13.5 perichondrium among outer Col1a1-GFPneg perichondrial cells (top) or inner Col1a1-GFP+ osteogenic perichondrial cells (bottom). Hes1+ peri-condensation mesenchymal cells robustly contributed to osteogenic perichondrial cells, as Hes1CE-E10.5 cells contributed to a significantly higher fraction of Col1a1-GFP+ osteogenic perichondrial cells, while both Hes1CE-E10.5 and Hes1CE-E12.5 cells constituted an equivalent fraction of outer perichondrial cells (Fig. 3I, first panels from the left).
Second, we quantified the contribution of Hes1CE-E10.5, Hes1CE-E12.5 or Hes1CE-E14.5 cells to the E15.5 perichondrium and bone collar, among outer Col1a1-GFPneg perichondrial cells (top) or inner Col1a1-GFP+ osteogenic bone collar cells (bottom). Hes1-creER did not effectively mark perichondrial cells at E14.5, as Hes1CE-E14.5 cells contributed to a significantly fewer fraction of outer perichondrial cells than Hes1CE-E10.5 or Hes1CE-E12.5 cells did (Fig. 3I, second panel from the left in the upper panels). Hes1CE-E10.5 cells contributed to a majority of Col1a1-GFP+ osteogenic cells in the bone collar. However, the osteogenic potential of Hes1+ cells declined in later stages, as Hes1CE-E12.5 and Hes1CE-E14.5 cells contributed to progressively fewer fractions of inner Col1a1-GFP+ osteogenic cells in the bone collar (Fig. 3I, second panel from the left in the lower panels). These trends continued onto P0, as the contribution of Hes1+ cells to cortical and trabecular bone osteoblasts progressively declined in later stages (Fig. 3I, third panel from the left in the lower panels).
Third, we quantified the contribution of Hes1CE-E10.5, Hes1CE-E12.5, or Hes1CE-E14.5 cells to SOX9+ chondrocytes within the cartilage template. The chondrogenic potential was unique to Hes1CE-E10.5 cells. These cells contributed progressively to a larger fraction of SOX9+ chondrocytes within the growth plate during the chase (Fig. 3I, upper right panel).
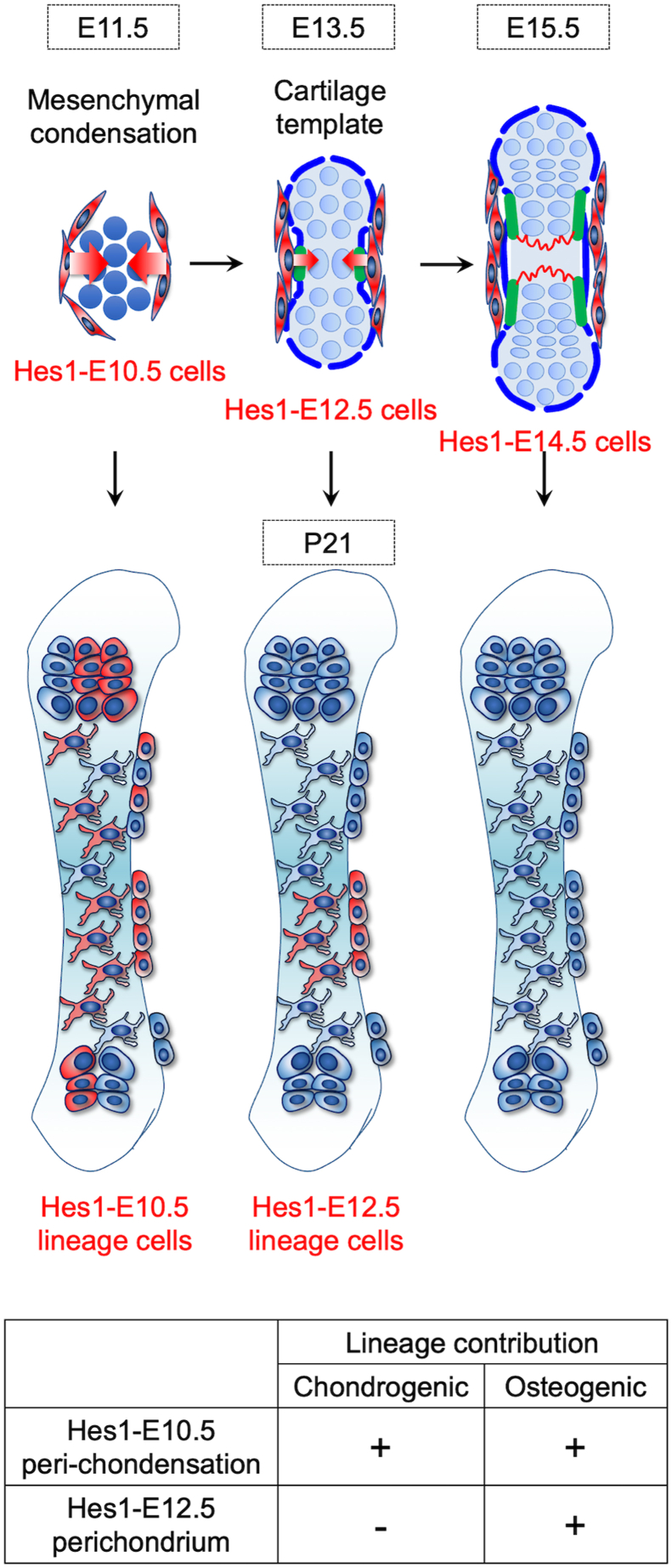
Therefore, Hes1CE-E10.5 peri-condensation mesenchymal cells have both chondrogenic and osteogenic potentials, while Hes1CE-E12.5 perichondrial cells have only osteogenic potentials. Importantly, Hes1+ descendants appeared to continue expressing Hes1 in the perichondrium, as the relative fraction of Hes1CE-E10.5 and Hes1CE-E12.5 cells in the perichondrium remained comparable at E13.5. Interestingly Hes1CE-E14.5 cells lose their robust potential to contribute neither chondrocyte or osteoblast, indicating that the ability of Hes1+ cells to generate chondrocytes and osteoblasts is restricted to an early stage of endochondral bone development.
Hes1-creER+ peri-condensation mesenchymal cells contribute to postnatal marrow stroma via cartilage-dependent and independent pathways
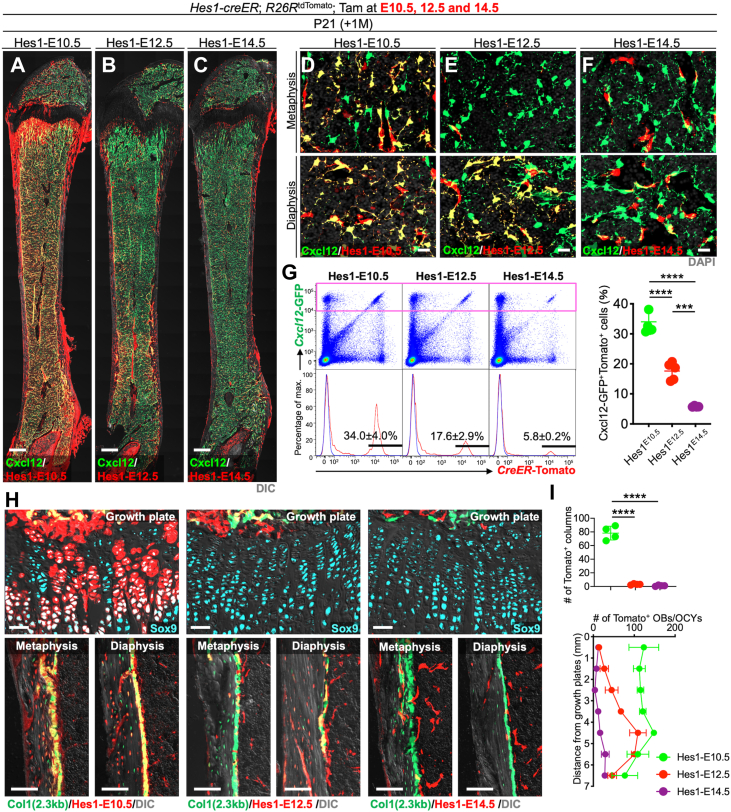
We further set out to define how Hes1+ cells contribute to the postnatal bone marrow stromal compartment. For this purpose, we utilized Hes1-creER; R26RtdTomato mice carrying Cxcl12-GFP reporter, and analyzed Hes1CE-E10.5, Hes1CE-E12.5, and Hes1CE-E14.5 cells at P21. Notably, Hes1CE-E10.5 cells contributed to the entire bone marrow compartment, generating Cxcl12-GFPhigh stromal cells both in the metaphyseal and diaphyseal marrow space (Fig. 4, A and D). In contrast, Hes1CE-E12.5 cells contributed only to Cxcl12-GFPhigh stromal cells in the diaphyseal bone marrow stromal compartment (Fig. 4, B and E). Interestingly, Hes1CE-E14.5 cells contributed to only a small number of Cxcl12-GFPhigh stromal cells; instead, these cells appeared to contribute to sinusoidal endothelial cells immediately adjacent to Cxcl12-GFPhigh stromal cells (Fig. 4, C and F).
Figure 4.
Early developmental Hes1-creER+cells outside the Sox9+cells contribute to the postnatal skeletal compartment in a time-dependent manner.A–G, Contribution of fetal Hes1-creER+ cells, pulsed at E10.5 (A and D), E12.5 (B and E), or E14.5 (C and F) to Cxcl12-GFP+ bone marrow stromal cells at P21. Cxcl12GFP/+; Hes1-creER; R26RtdTomato femurs with growth plates on top. A–C, Whole bone, upper panels in (D–F): metaphyseal bone marrow, lower panels in (D–F) diaphyseal bone marrow. Scale bar: 500 μm (A–C), 20 μm (D–F). n = 4 mice per group. G, Percentage of Cxcl12-GFPhightdTomato+ cells per total Cxcl12-GFPhigh cells. n = 4 mice per group. ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. Two-tailed, One-way ANOVA followed by Tukey’s post-hoc test (the others). Data are presented as mean ± s.d. H and I, Contribution of fetal Hes1-creER+ cells to growth plate chondrocytes and cortical osteoblasts and osteocytes at P21. H, Col1a1(2.3kb)-GFP; Hes1-creER; R26RtdTomato, pulsed at E10.5 (left), E12.5 (center) and E14.5 (right). Growth plate (upper), endocortical marrow space at metaphysis (lower left) and diaphysis (lower right). Scale bar: 50 μm (upper panels), 100 μm (lower panels). n = 4 mice per each group. I, upper, quantification of tdTomato+ columns in growth plate. Lower, Quantification of tdTomato+ osteoblasts/cytes, based on the distance from growth plate. Green: Hes1-E10.5, Red: Hes1-E12.5, Violet: Hes1-E14.5. n = 4 mice per each group. ∗∗∗∗p < 0.0001. Two-tailed, One-way ANOVA followed by Tukey’s post hoc test (the others). Data are presented as mean ± SD.
We quantitatively assessed how Hes1+ cells at different stages contribute differentially to Cxcl12-GFPhigh stromal cells using flow cytometry analysis, using bone marrow cells isolated from P21 femurs of Cxcl12GFP/+; Hes1-creER; R26RtdTomato triple transgenic mice pulsed at E10.5, E12.5 or E14.5 (Figs. 4G and S6A). Hes1+ peri-condensation mesenchymal cells at E10.5 demonstrated a robust potential to generate marrow stromal cells, as Hes1CE-E10.5 cells contributed to a substantial fraction of Cxcl12-GFPhigh stromal cells (34.0 ± 4.0%) (Fig. 4G, right panel). In contrast, Hes1+ perichondrial cells at E12.5 contributed to a sizable but significantly smaller fraction of Cxcl12-GFPhigh stromal cells (17.6 ± 2.9%). Hes1CE-E14.5 cells contributed to a much smaller fraction of Cxcl12-GFPhigh stromal cells than Hes1CE-E10.5 and Hes1CE-E12.5 cells did (5.8 ± 0.2%), indicating that Hes1+ cells progressively lose their potential to generate bone marrow stromal cells in later stages.
We further quantified the contribution of Hes1CE-E10.5, Hes1CE-E12.5, and Hes1CE-E14.5 cells to postnatal growth plate chondrocytes and osteoblasts using histological approaches (Fig. 4H). Notably, Hes1CE-E10.5 cells, but not Hes1CE-E12.5 or Hes1CE-E14.5 cells, generated postnatal growth plate chondrocytes (Fig. 4I). Further, Hes1CE-E10.5 cells contributed robustly to osteoblasts/cytes both in the metaphysis and the diaphysis, while Hes1CE-E12.5 cells predominantly contributed to osteoblasts/cytes in the diaphysis. Hes1CE-E14.5 cells made only a small contribution to osteoblasts/cytes in the diaphysis (Figs. 4, G and I and S6B). Taken together, these findings indicate that Hes1+ peri-condensation mesenchymal cells generate postnatal marrow stroma via cartilage-dependent and independent pathways.
Hes1-creER+ peri-condensation mesenchymal cells contribute to bone marrow colony forming fibroblasts
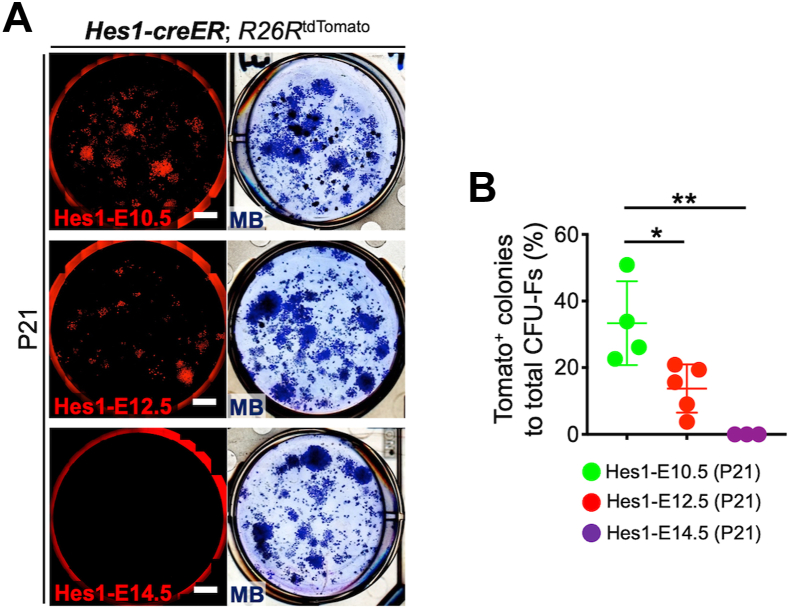
Lastly, we asked whether Hes1+ peri-condensation mesenchymal cells can contribute to a fraction of putative skeletal stem cells in the bone marrow. To this end, we performed colony-forming unit fibroblast (CFU-F) assays of P21 bone marrow cells isolated from Hes1-creER; R26RtdTomato mice pulsed at E10.5, E12.5 or E14.5, to discern the contribution of Hes1CE-E10.5, Hes1CE-E12.5 and Hes1CE-E14.5 cells to the clonogenic fraction. Hes1CE-E10.5 cells contributed to a substantial faction of CFU-Fs (Hes1CE-E10.5: 33.4 ± 12.6% of total CFU-Fs), while Hes1CE-E12.5 cells contributed to a significantly smaller fraction of CFU-Fs (Hes1CE-E12.5: 13.7 ± 7.2% of total CFU-Fs). In contrast, Hes1CE-E14.5 cells made no contribution to CFU-Fs (Hes1CE-E14.5: 0.0 ± 0.0% of total CFU-Fs, Fig. 5, A and B). Therefore, Hes1+ peri-condensation mesenchymal cells contribute to the clonogenic fraction of the postnatal bone marrow stroma, which may include a population of putative skeletal stem cells.
Figure 5.
Early developmental Hes1-creER+cell-derived marrow stromal cells behave as skeletal progenitor cells.A, Colony-forming unit fibroblast (CFU-F) assay of Hes1-creER; R26RtdTomato at P21, pulsed at E10.5 (top), E12.5 (middle), and E14.5 (bottom). Scale bar: 5 mm. B, Percentage of tdTomato+ colonies among total CFU-Fs. n = 4 (Hes1-E12.5), n = 5 (Hes1-E12.5), n = 3 (Hes1-E14.5) mice. ∗p < 0.05, ∗∗p < 0.01. Two-tailed, One-way ANOVA followed by Tukey’s post-hoc test. Data are presented as mean ± SD. MB, Methylene blue staining.
Discussion
Here, we demonstrate that a population of early mesenchymal cells defined by Notch effector Hes1 expression outside the SOX9+ condensation can provide an important source of chondrocytes and perichondrial cells in early endochondral bone development. These Hes1+ peri-condensation mesenchymal cells represent precursors of Dlx5+ early perichondrial cells that we reported recently (4) and robustly participate in chondrogenesis, osteogenesis, and subsequently the formation of the postnatal bone marrow stromal compartment through two routes of cartilage-dependent and independent pathways. Our Hes1-creER-based in vivo lineage-tracing findings establish the concept that SOX9-negative Hes1-expressing mesenchymal cells surrounding mesenchymal condensations play important roles in early endochondral bone development. Importantly, the robust chondrogenic and osteogenic potential of Hes1+ cells is unique to the mesenchymal condensation stage, whereas these cells progressively lose such potential in later stages.
We believe that the two lineages of chondrocytes and perichondrial cells diverge prior to E12.5, as Hes1CE-E10.5 cells, but not Hes1CE-E12.5 cells, contribute to chondrocytes within the cartilage template. Hes1+ cells at E10.5 and earlier time are likely to provide a common progenitor population for at least some of the chondrocytes and osteoblasts. We acknowledge, however, that our approach based on a single-color reporter cannot determine if an individual peri-condensation Hes1+ cell can contribute clonally both to chondrocytes and osteoblasts. An in vivo clonal analysis with a multi-color lineage reporter would facilitate the identification of a common progenitor population for chondrocytes and osteoblasts in the peri-condensation region.
Hes1 is one of the canonical Notch effector genes that is expressed by cells in which Notch signaling is activated. In fact, we observed that a Notch signaling reporter CBF1:H2B-Venus was specifically active in peri-condensation mesenchymal cells surrounding the SOX9+ pre-cartilaginous condensation. Notch signaling, which is primarily mediated by the CBF1-dependent target gene Hes1, regulates both Sox9 and Runx2 expression during skeletal cell differentiation (22). Hes1 inhibits Sox9 expression, and Hes1 inhibition promotes chondrogenesis by upregulating Sox9. Hes1 also inhibits osteoblast differentiation through the inhibition of Runx2 activities (23). Notch signaling regulates asymmetric cell division and lineage decision through lateral inhibition. It is possible that Hes1+ peri-condensation mesenchymal cells divide into Hes1-positive and negative cells through asymmetric cell division; the former Hes1-positive cells may stay in the perichondrium as undifferentiated skeletal progenitors, while the latter Hes1-negative cells enter into the cartilage template and differentiate into chondrocytes.
We show that Hes1-creER allows the marking of early mesenchymal cells in a SOX9-negative domain of the condensation. Historically, Prrx1-cre/creER has been widely utilized as a tool to mark “skeletal stem/progenitor cells” at the condensation stage (13, 26). However, the caveat is that Prrx1-cre simultaneously marks SOX9+ cells within the condensation, making it impossible to discern the contribution of SOX9-negative peri-condensation mesenchymal cells. Our findings from Hes1-creER-based lineage-tracing experiments revise the concept regarding the origin of osteo-chondroprogenitor cells within the limb bud, which is currently considered to rest solely upon SOX9+ (Sox9-cre+) cells (9). The emerging concept is that both SOX9+ cells within the condensation and SOX9-negative cells outside the condensation play equally important roles in early endochondral bone development. Identifying the unique role of each respective cell-of-origin of the limb bud mesenchyme remain as an important agenda for future studies.
In conclusion, we propose a new concept that peri-condensation mesenchymal cells surrounding the SOX9+ condensation provide an important source of skeletal progenitor cells in early endochondral bone development (see the proposed diagram in Fig. 6).
Figure 6.
Hes1-marked early mesenchymal cells surrounding the condensation behave as skeletal progenitors in endochondral bone development. Early perichondrial population of skeletal progenitor cells in endochondral bone development. Undifferentiated mesenchymal cells surrounding condensation or outer perichondrial cells outside the cartilage template provide an important source during fetal endochondral bone development. During the condensation stage, Hes1+ undifferentiated mesenchymal cells take two distinct routes to participate in endochondral bone development. First, these cells translocate into the cartilage template and directly differentiate into Sox9+ chondrocytes and then contribute to bone formation (chondrocyte-dependent pathway). Second, these cells contribute to the outer layer of the perichondrium and become Hes1+ perichondrial cells. These Hes1+ perichondrial cells translocate into the nascent marrow space and directly differentiate into marrow mesenchymal cells by bypassing a Sox9+ state (chondrocyte-independent pathway). These perichondrial skeletal progenitor cells keep providing osteoblasts and bone marrow stromal cells even during the postnatal stage. As a result, postnatal bone marrow is characterized by transitional mosaicism composed of both chondrocyte-derived and perichondrium-derived stromal cells.
Experimental procedures
Mouse strains
Hes1-creER (Hes1tm1(cre/ERT2)Lcm) (25) and Cxcl12GFP/+ (Cxcl12tm2Tng) (27) mice have been described previously. Prrx1-cre (JAX005584), Acta1-cre (JAX006139), Myl-cre (JAX024713), Mck-cre (JAX006475), Tie2-cre (JAX008863), Rosa26-CAG-loxP-stop-loxP-tdTomato (Ai14: R26R-tdTomato, JAX007914), Rosa26-SA-loxP-GFP-stop-loxP-DTA (JAX006331), Col1a1(2.3kb)-GFP (JAX013134), and CBF:H2B-Venus (JAX020942) mice were acquired from the Jackson laboratory. All procedures were conducted in compliance with the Guidelines for the Care and Use of Laboratory Animals approved by the University of Texas Health Science Center at Houston’s Animal Welfare Committee (AWC), protocol AWC-21-0070, and the University of Michigan’s Institutional Animal Care and Use Committee (IACUC), protocol 9496. All mice were housed in a specific pathogen-free condition and analyzed in a mixed background. Mice were housed in static microisolator cages (Allentown Caging). Access to water and food (irradiated LabDiet 5008) was ad libitum. Animal rooms were climate controlled to provide temperatures of 22 to 23 °C, 40 to 65% of humidity on a 12 h light/dark cycle (lights on at 0600). For all breeding experiments, creER transgenes were maintained in male breeders to avoid spontaneous germline recombination. Mice were identified by micro-tattooing or ear tags. Tail biopsies of mice were lysed by a HotShot protocol (incubating the tail sample at 95oC for 30 min in an alkaline lysis reagent followed by neutralization) and used for PCR-based genotyping (GoTaq Green Master Mix, Promega, and Nexus X2, Eppendorf). Perinatal mice were also genotyped fluorescently (BLS miner's lamp) whenever possible. Mice were euthanized by over-dosage of carbon dioxide or decapitation under inhalation anesthesia in a drop jar (Fluriso, Isoflurane USP, VetOne).
Tamoxifen and induction of cre-loxP recombination
Tamoxifen (Sigma T5648) was mixed with 100% ethanol until completely dissolved. Subsequently, a proper volume of sunflower seed oil (Sigma S5007) was added to the tamoxifen-ethanol mixture and rigorously mixed. The tamoxifen-ethanol-oil mixture was incubated at 60 °C in a chemical hood until the ethanol evaporated completely. The tamoxifen-oil mixture was stored at room temperature until use. Tamoxifen was injected at a dose of 3 mg into pregnant mice intraperitoneally using a 26 to 1/2-gauge needle (BD309597).
Histology and immunohistochemistry
Samples were dissected under a stereomicroscope (Nikon SMZ-800), and fixed in 4% paraformaldehyde for a proper period, typically ranging from 3 h to overnight at 4oC, then decalcified in 15% EDTA for a proper period, typically ranging from 3 h to 14 days. Embryonic samples were not decalcified. Subsequently, samples were cryoprotected in 30% sucrose/PBS solutions and then in 30% sucrose/PBS:OCT (1:1) solutions, each at least overnight at 4 °C. Samples were embedded in an OCT compound (Tissue-Tek, Sakura) under a stereomicroscope and transferred on a sheet of dry ice to solidify the compound. Embedded samples were cryosectioned at 14 μm using a cryostat (Leica CM1850) and adhered to positively charged glass slides (Fisherbrand ColorFrost Plus). Sections were postfixed in 4% paraformaldehyde for 15 min at room temperature. For immunostaining, sections were permeabilized with 0.25% TritonX/TBS for 30 min, blocked with 3% BSA/TBST for 30 min and incubated with rabbit anti-Sox9 polyclonal antibody (1:500, EMD-Millipore, AB5535), rat anti-endomucin (Emcn) monoclonal antibody (1:100, Santa Cruz Biotechnology, sc65495), rabbit anti-Myh3 polyclonal antibody (1:500, Abcam, ab124205),or rabbit anti-Osx polyclonal antibody (1:500, Abcam, ab22552) overnight at 4 °C, and subsequently with Alexa Fluor 647-conjugated donkey anti-rabbit IgG (A31573) or Alexa Fluor 633-conjugated goat anti-rat IgG (A21049) (1:400, Invitrogen) for 3 h at room temperature. Sections were further incubated with DAPI (4′,6-diamidino-2-phenylindole, 5 μg/ml, Invitrogen D1306) to stain nuclei prior to imaging.
RNAscope in situ hybridization
Samples were fixed in 4% paraformaldehyde overnight at 4 °C and then cryoprotected. Frozen sections at 14 μm were prepared on positively charged glass slides. In situ hybridization was performed with RNAscope 2.5 HD Reagent kit Brown (Advanced Cell Diagnostics 322,300) using the following probes: Hes1 (417701) and Sox9 (custom-designed) according to the manufacturer’s protocol.
Imaging and cell quantification
Images were captured by an automated inverted fluorescence microscope with a structured illumination system (Zeiss Axio Observer Z1 with ApoTome.2 system) and Zen 2 (blue edition) software. The filter settings used were: FL Filter Set 34 (Ex. 390/22, Em. 460/50 nm), Set 38 HE (Ex. 470/40, Em. 525/50 nm), Set 43 HE (Ex. 550/25, Em. 605/70 nm), Set 50 (Ex. 640/30, Em. 690/50 nm), and Set 63 HE (Ex. 572/25, Em. 629/62 nm). The objectives used were: Fluar 2.5x/0.12, EC Plan-Neofluar 5x/0.16, Plan-Apochromat 10x/0.45, EC Plan-Neofluar 20x/0.50, EC Plan-Neofluar 40x/0.75, Plan-Apochromat 63x/1.40. Images were typically tile-scanned with a motorized stage, Z-stacked, and reconstructed by a maximum intensity projection (MIP) function. Differential interference contrast (DIC) was used for objectives higher than 10×. Representative images of at least three independent biological samples are shown in the figures. Quantification of cells on sections was performed using NIH Image J software.
Cell preparation
For embryonic samples, hind limbs were harvested and incubated with 2 Wunsch units of Liberase TM (Sigma/Roche 5401127001) in 2 ml Ca2+, Mg2+-free Hank’s Balanced Salt Solution (HBSS, Sigma H6648) at 37 °C for 15 min on a shaking incubator (ThermomixerR, Eppendorf). For postnatal samples, soft tissues and epiphyses were carefully removed from dissected femurs. After removing distal epiphyseal growth plates and cutting off proximal ends, femurs were cut roughly and incubated with 2 Wunsch units of Liberase TM and 1 mg of Pronase (Sigma/Roche 10165921001) in 2 ml Ca2+, Mg2+-free HBSS at 37 °C for 60 min on a shaking incubator. After cell dissociation, cells were mechanically triturated using an 18-gauge needle with a 1 ml Luer-Lok syringe (BD) and a pestle with a mortar (Coors Tek), and subsequently filtered through a 70 μm cell strainer (BD) into a 50 ml tube on ice to prepare single cell suspension. These steps were repeated for five times, and dissociated cells were collected in the same tube. Cells were pelleted and resuspended in an appropriate medium for subsequent purposes. For cell culture experiments, cells were resuspended in 10 ml culture medium and counted on a hemocytometer.
Flow cytometry
Dissociated cells were stained by standard protocols with the following antibodies (1:500, eBioscience). Allophycocyanin (APC)-conjugated CD31 (390, endothelial/platelet), CD45 (30F-11, hematopoietic), and Ter119 (TER-119, erythrocytes). Flow cytometry analysis was performed using a four-laser BD LSR Fortessa (Ex. 405/488/561/640 nm) and FACSDiva software. Acquired raw data were further analyzed on FlowJo software (TreeStar). Representative plots of at least four independent biological samples are shown in the figures.
Single-cell RNA-seq analysis of fluorescence-activated cell sorting-isolated cells
Cell sorting was performed using a four-laser BD fluorescence-activated cell sorting (FACS) Aria III (E x .407/488/561/640 nm) high-speed cell sorter with a 100 μm nozzle. tdTomato+ cells were directly sorted into ice-cold DPBS/1% BSA, pelleted by centrifugation and resuspended in appropriate amount of DPBS/1% BSA (1000 cells/μl). Cell numbers were quantified by Countless II automated Cell Counter (ThermoFisher) before loading onto the Chromium Single Cell 3′ v2 microfluidics chip (10x Genomics Inc). cDNA libraries were sequenced by Illumina HiSeq 4000 using two lanes and 50 cycle paired-end read, generating a total of ∼770 million reads. The sequencing data was first pre-processed using the 10X Genomics software Cell Ranger. For alignment purposes, we generated and used a custom genome fasta and index file by including the sequences of tdTomato-WPRE to the mouse genome (mm10). Further downstream analysis steps were performed using the Seurat (20) R package. We filtered out cells with less than 500 genes per cell and with more than 20% mitochondrial read content. The downstream analysis steps include normalization, identification of highly variable genes across the single cells, scaling based on number of UMI, dimensionality reduction (PCA, CCA, and t-SNE), unsupervised clustering, and the discovery of differentially expressed cell-type specific markers. Differential gene expression to identify cell-type specific genes was performed using the nonparametric Wilcoxon rank sum test.
Colony-forming assay and subcloning
Nucleated bone marrow cells were plated into tissue culture 6-well plates (BD Falcon) at a density of <105 cells/cm2 and cultured in low-glucose DMEM with GlutaMAX supplement (Gibco 10567022) and 10% mesenchymal stem cell-qualified FBS (Gibco 12662029) containing penicillin-streptomycin (Sigma P0781) for 10∼14 days. Cell cultures were maintained at 37 °C in a 5% CO2 incubator. Representative images of at least three independent biological samples are shown in the figures. For CFU-Fs, cells were fixed with 70% Ethanol for 5 min and stained for 2% methylene blue.
Statistical analysis
Results are presented as mean values ± SD. Statistical evaluation was conducted using the Mann-Whitney's U-test or one-way ANOVA. A p value of < 0.05 was considered significant. No statistical method was used to predetermine sample size. Sample size was determined on the basis of previous literature and our previous experience to give sufficient standard deviations of the mean so as not to miss a biologically important difference between groups. The experiments were not randomized. All of the available mice of the desired genotypes were used for experiments. The investigators were not blinded during experiments and outcome assessment. One femur from each mouse was arbitrarily chosen for histological analysis. Genotypes were not particularly highlighted during quantification.
Data availability
The data generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. The single-cell RNA-seq data presented herein has been deposited in the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE144411 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144411).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank C. Murtaugh (University of Utah) for Hes1-creER mice, T. Nagasawa (Osaka University) for Cxcl12GFP/+ mice and S.Y. Wong (University of Michigan) for CBF1:Venus mice.
Author contributions
Y. M., N. O. conceptualization; Y. M., N. O. methodology; Y. M., H. M., T. O., S. N., and M. N. validation; Y. M., H. M., T. O., S. N., and M. N. formal analysis; Y. M., H. M., T. O., S. N., and M. N. investigation; Y. M., H. M., T. O., S. N., M. N., and N. O. data curation; Y. M. writing – original draft; Y. M., H. M., T. O., S. N., and M. N. visualization; Y. M., W. O., and N. O. supervision; Y. M. and N. O. project administration; Y. M., W. O., and N. O. funding acquisition; W. O. resources; N. O. software; N. O. writing – review and editing.
Funding and additional information
This research was supported by National Institute of Health grants R01DE030630, R01DE026666 (to N. O.), R01DE029181 (to W. O.), JSPS grant JP20KK0356, JP21H03124 and JST FOREST Program JPMJFR2111 Japan to Y. M. We thank M. Pihalja and K. Saiya-Cork of the University of Michigan Flow Cytometry Core, Yusuke Ono of Kumamoto University and T. Lau for supporting this study. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reviewed by members of the JBC Editorial Board. Edited by Robert Haltiwanger
Supporting information
References
- 1.Ono N., Balani D.H., Kronenberg H.M. Stem and progenitor cells in skeletal development. Curr. Top. Dev. Biol. 2019;133:1–24. doi: 10.1016/bs.ctdb.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizoguchi T., Ono N. The diverse origin of bone-forming osteoblasts. J. Bone Miner. Res. 2021;36:1432–1447. doi: 10.1002/jbmr.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall B.K., Miyake T. All for one and one for all: condensations and the initiation of skeletal development. Bioessays. 2000;22:138–147. doi: 10.1002/(SICI)1521-1878(200002)22:2<138::AID-BIES5>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 4.Matsushita Y., Chu A.K.Y., Tsutsumi-Arai C., Orikasa S., Nagata M., Wong S.Y., et al. The fate of early perichondrial cells in developing bones. Nat. Commun. 2022;13:7319. doi: 10.1038/s41467-022-34804-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colnot C., Zhang X., Knothe Tate M.L. Current insights on the regenerative potential of the periosteum: molecular, cellular, and endogenous engineering approaches. J. Orthop. Res. 2012;30:1869–1878. doi: 10.1002/jor.22181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eames B.F., de la Fuente L., Helms J.A. Molecular ontogeny of the skeleton. Birth Defects Res. C Embryo Today. 2003;69:93–101. doi: 10.1002/bdrc.10016. [DOI] [PubMed] [Google Scholar]
- 7.Long F., Chung U.I., Ohba S., McMahon J., Kronenberg H.M., McMahon A.P. Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development. 2004;131:1309–1318. doi: 10.1242/dev.01006. [DOI] [PubMed] [Google Scholar]
- 8.Ono N., Ono W., Nagasawa T., Kronenberg H.M. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat. Cell Biol. 2014;16:1157–1167. doi: 10.1038/ncb3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akiyama H., Kim J.E., Nakashima K., Balmes G., Iwai N., Deng J.M., et al. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc. Natl. Acad. Sci. U. S. A. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura E., Nguyen M.T., Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- 11.Bi W., Deng J.M., Zhang Z., Behringer R.R., de Crombrugghe B. Sox9 is required for cartilage formation. Nat. Genet. 1999;22:85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 12.Soeda T., Deng J.M., de Crombrugghe B., Behringer R.R., Nakamura T., Akiyama H. Sox9-expressing precursors are the cellular origin of the cruciate ligament of the knee joint and the limb tendons. Genesis. 2010;48:635–644. doi: 10.1002/dvg.20667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Logan M., Martin J.F., Nagy A., Lobe C., Olson E.N., Tabin C.J. Expression of Cre recombinase in the developing mouse limb bud driven by a Prxl enhancer. Genesis. 2002;33:77–80. doi: 10.1002/gene.10092. [DOI] [PubMed] [Google Scholar]
- 14.Martin J.F., Olson E.N. Identification of a prx1 limb enhancer. Genesis. 2000;26:225–229. [PubMed] [Google Scholar]
- 15.Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Currie J.D., Grosser L., Murawala P., Schuez M., Michel M., Tanaka E.M., et al. The Prrx1 limb enhancer marks an adult subpopulation of injury-responsive dermal fibroblasts. Biol. Open. 2019;8 doi: 10.1242/bio.043711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maes C., Kobayashi T., Selig M.K., Torrekens S., Roth S.I., Mackem S., et al. Osteoblast precursors, but not mature osteoblasts, move into developing and fractured bones along with invading blood vessels. Dev. Cell. 2010;19:329–344. doi: 10.1016/j.devcel.2010.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mizoguchi T., Pinho S., Ahmed J., Kunisaki Y., Hanoun M., Mendelson A., et al. Osterix marks distinct waves of primitive and definitive stromal progenitors during bone marrow development. Dev. Cell. 2014;29:340–349. doi: 10.1016/j.devcel.2014.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ohba S., He X., Hojo H., McMahon A.P. Distinct transcriptional programs underlie Sox9 regulation of the mammalian chondrocyte. Cell Rep. 2015;12:229–243. doi: 10.1016/j.celrep.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu J., Mackem S. John Saunders' ZPA, Sonic hedgehog and digit identity - how does it really all work? Dev. Biol. 2017;429:391–400. doi: 10.1016/j.ydbio.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong Y., Jesse A.M., Kohn A., Gunnell L.M., Honjo T., Zuscik M.J., et al. RBPjkappa-dependent Notch signaling regulates mesenchymal progenitor cell proliferation and differentiation during skeletal development. Development. 2010;137:1461–1471. doi: 10.1242/dev.042911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilton M.J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., et al. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 2008;14:306–314. doi: 10.1038/nm1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nowotschin S., Xenopoulos P., Schrode N., Hadjantonakis A.K. A bright single-cell resolution live imaging reporter of Notch signaling in the mouse. BMC Dev. Biol. 2013;13:15. doi: 10.1186/1471-213X-13-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopinke D., Brailsford M., Shea J.E., Leavitt R., Scaife C.L., Murtaugh L.C. Lineage tracing reveals the dynamic contribution of Hes1+ cells to the developing and adult pancreas. Development. 2011;138:431–441. doi: 10.1242/dev.053843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawanami A., Matsushita T., Chan Y.Y., Murakami S. Mice expressing GFP and CreER in osteochondro progenitor cells in the periosteum. Biochem. Biophys. Res. Commun. 2009;386:477–482. doi: 10.1016/j.bbrc.2009.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ara T., Tokoyoda K., Sugiyama T., Egawa T., Kawabata K., Nagasawa T. Long-term hematopoietic stem cells require stromal cell-derived factor-1 for colonizing bone marrow during ontogeny. Immunity. 2003;19:257–267. doi: 10.1016/s1074-7613(03)00201-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. The single-cell RNA-seq data presented herein has been deposited in the National Center for Biotechnology Information (NCBI)’s Gene Expression Omnibus (GEO) and are accessible through GEO Series accession number GSE144411 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE144411).