Abstract
As a proven source of potent and selective antimicrobials, Xenorhabdus bacteria are important to an age plagued with difficult-to-treat microbial infections. Yet, only 27 species have been described to date. In this study, a novel Xenorhabdus species was discovered through genomic studies on three isolates from Kenyan soils. Soils in Western Kenya were surveyed for steinernematids and Steinernema isolates VH1 and BG5 were recovered from red volcanic loam soils from cultivated land in Vihiga and clay soils from riverine land in Bungoma respectively. From the two nematode isolates, Xenorhabdus sp. BG5 and Xenorhabdus sp. VH1 were isolated. The genomes of these two, plus that of X. griffiniae XN45 – this was previously isolated from Steinernema sp. scarpo that also originated from Kenyan soils – were sequenced and assembled. Nascent genome assemblies of the three isolates were of good quality with over 70 % of their proteome having known functions. These three isolates formed the X. griffiniae clade in a phylogenomic reconstruction of the genus. Their species were delineated using three overall genome relatedness indices: an unnamed species of the genus, Xenorhabdus sp. BG5, X. griffiniae VH1 and X. griffiniae XN45. A pangenome analysis of this clade revealed that over 70 % of species-specific genes encoded unknown functions. Transposases were linked to genomic islands in Xenorhabdus sp. BG5. Thus, overall genome-related indices sufficiently delineated species of two new Xenorhabdus isolates from Kenya, both of which were closely related to X. griffiniae . The functions encoded by most species-specific genes in the X. griffiniae clade remain unknown.
Keywords: prokaryotic pangenomics, Steinernema endosymbionts, species delineation, Xenorhabdus bacteria
Data Summary
NCBI GenBank accession numbers of the three genome assemblies generated from this study are JACWFC000000000.1, JADEUF000000000.1 and JADEUG000000000.1. The metadata for soil samples collected in this study are listed in Table S2 (available in the online version of this article).
Accession numbers and strain names of publicly available genomes used in this study are listed in Table S3.
The supplementary workbook contains detailed raw data used for pangenome analyses. IS family transposases in the genome of strain BG5 have been deposited in the ISfinder Database under accession numbers ISXsp1, ISXsp2, ISXsp3, ISXsp4, ISXsp5, ISXsp6, ISXsp7, ISXsp8, ISXsp9, ISXsp10, ISXsp11, ISXsp12, ISXsp13, ISXsp14, ISXsp15, ISXsp16, ISXsp17, ISXsp18, ISXsp19 and ISXsp20.
Parts of the methods, results and discussion sections were previously reported in a doctoral thesis of the first author and are thus not considered a prior publication.
Impact Statement.
Xenorhabdus bacteria are important because they produce various antimicrobials. However, not many have been isolated from their natural habitat, which is the gut of soil-dwelling Steinernema nematodes. Two of these bacteria were thus isolated from nematodes of soils in Western Kenya. Their genome sequences were determined and used in genomic analyses, which revealed novel species. These genomes can be used to show the location of genes encoding antimicrobial production, thus making it easier for future isolation of antimicrobials from these new bacterial strains.
Introduction
Bacteria of the genus Xenorhabdus naturally produce specialized metabolites such as non-ribosomal peptides that have antiprotozoal, antifungal and antibacterial activities [1]. Yet, despite each Xenorhabdus species [2] – and sometimes even strain [3]– encoding a unique antimicrobial production profile, only 27 [4–13] Xenorhabdus bacteria have been isolated from their Steinernema nematode hosts and described as novel species. Thus, the aim of this study was to discover new Xenorhabdus species and strains.
Bacteria of the genus Xenorhabdus are naturally found in soil biota, specifically as autochthonous endosymbionts of insect-killing Steinernema nematodes. Once a steinernematid enters an insect prey via natural openings such as spiracles, it migrates to the haemocoel and defecates [14] its Xenorhabdus endosymbionts, which then secrete specialized metabolites such as insecticidal toxins [15] that result in quick death of the insect. Secreted antimicrobials function to deter soil microbial competitors from the nutrient-rich cadaver. Nematodes then utilize this nutrient-filled, cadaver enclosure to reproduce exponentially. Depletion of nutrients halts the reproductive cycle and triggers nematode bacterium re-association. This is succeeded by pre-infective juvenile steinernematids emigrating from the cadaver to the soil, where they lie in wait for insect prey. Thus, to isolate a Xenorhabdus bacterium one needs to first isolate its infective juvenile steinernematid host from soil, using a combination of insect larvae as bait [16] and White traps [17].
For species such as Xenorhabdus khoisanae (Table S1), X. bovienii , X. kozodoii , X. poinarii and X. hominckii, we see one Xenorhabdus species as the natural symbiont of numerous Steinernema species [18]. However, the reverse, one Steinernema species that naturally hosts, with equal fitness, two different Xenorhabdus species, has yet to be discovered. Thus, there is a high possibility of identifying new Xenorhabdus species from the over 50 described Steinernema species whose symbionts remain uncharacterized [19].
This research gap between steinernematid isolation and Xenorhabdus endosymbiont identification is also seen in Sub-Saharan Africa, where numerous steinernematids have been isolated (see Table S1 for a full list of species and location). In Kenya, apart from X. hominickii from Steinernema karii [13], and X. griffiniae XN45 that we isolated from Steinernema sp. scarpo [20], endosymbionts have yet to be isolated and described from strains that were isolated from Central and Coastal Regions including Steinernema sp. UH3 [21] and UH13 [22]. No steinernematid isolates have been documented from the Western region of Kenya.
Genome assemblies of >50× coverage of new isolates are not only required for the description of novel species/emendation of prokaryotic taxa [23] but also enable accurate species delineation via overall genome-related indices (OGRIs) such as orthologous average nucleotide identity (orthoANI) [24] and digital DNA–DNA hybridization (dDDH) [25], and phylogenomic reconstructions based on genome-genome distances [26]. Furthermore, comparative genome analyses of closely related strains are useful for the identification of not only genomic islands and mobile genetic elements but also the core and dispensable genes of a specific monophyletic group [27, 28].
In this study two Xenorhabdus strains, VH1 and BG5, were isolated from soil biota from Western Kenya. Their genomes, and that of X. griffiniae XN45 that we previously isolated from Steinernema sp. scarpo from Kenya, were sequenced and assembled. These three genomes were used for downstream species delineation and emendation, and comparative genome analyses.
Methods
Collection of field soil samples
Fieldwork was carried out from 16 October 2018 to 4 November 2018 in the Western and Rift Valley Regions of Kenya. No access permits were required as per the exceptions of section 3(d) of the Environmental Management and Coordination (Conservation of Biological Diversity and Resources, Access to Genetic Resources and Benefit Sharing) Regulations 2006 of the Environmental Management and Coordination Act, 1999 of the Laws of Kenya. Ten localities were selected for the collection of soil samples: Nandi Hills, Tinderet, Fort Tenan, Kakamega, Gisambai, Vihiga, Kisumu, Bungoma, Kaimosi and Mt. Elgon. Within each locality, collection points were selected from cultivated lands, fallow lands, forests, crop edges, shorelines, swamps and riverine areas. This resulted in a total of 76 soil collection points. To collect a soil sample, vegetation was first cleared from the topsoil. Then using a digging fork, soil was excavated to a depth of not more than 60 cm. Using a collection spade, the soil was scooped into a measuring cup to an amount of ca. 500 g. Twigs, branches and stones were removed before the soil sample was placed in labelled cotton bags. Dug-out soil was returned to the hole and soil samples were then transported at room temperature to the laboratories. Geographical coordinates, altitude and descriptions of soil collection points are provided in Table S2.
Isolation of nematodes from soils samples
To isolate entomopathogenic nematodes (EPNs) from soils, a soil sample was first spread out on a tray and crumbled. The soil was then redistributed into transparent polyethylene terephthalate (PET) plastic containers of 20 cm in diameter and 5 cm in depth. To bait EPNs from the soil, Galleria mellonella larvae were first obtained from a laboratory insect culture of KALRO-Horticulture Research Institute, where they had been reared as previously described [29]. From this culture, any healthy larvae were selected. Two/three larvae were then buried in the soil, in a hole of ca. 1 cm diameter and 5 cm depth. In total, about 15 G. mellonella larvae buried in five holes per container were used as bait. The container lids were replaced and set-ups were maintained at room temperature. After a maximum of 7 days, containers were checked. Dead larvae were assessed for the following characteristics that typify an EPN infection: limp cadaver, tan or red in colour, and minimal smell of putrefaction. Samples BG5 and VH1 had dead cadavers that were either light red or tan in colour. Sample BG5 was clay soil collected from fallow riverine land. Sample VH1 was collected from land cultivated with cabbages. To isolate putative EPNs from these cadavers, a modified White trap [17] was used. Briefly, clean PET containers of 20 cm diameter and 5 cm depth were filled with distilled water to a depth of ca. 4 mm. A clean Petri dish was placed upside down into the container such that the Petri dish surface was raised from the bottom of the PET container. Clean white cotton cloths of the same size as the Petri dish were placed on this raised surface. Selected cadavers were placed onto the cotton cloths. To allow putative EPNs to emigrate from the cadavers to the water, a part of the cloth was dipped in the distilled water. PET containers were covered and kept for 7 days. The distilled water was observed daily under a dissecting microscope for the presence of white motile, ca. 1 mm long nematodes. For positive samples, contaminants such as cadaver tissue debris were separated from nematodes by a series of sedimentation and decanting using clean distilled water. Nematodes were stored in contamination-free distilled water – to a depth of not more than 0.4 cm – in clear plastic containers in the dark. Stored EPN nematode cultures were named after their soil collection point.
Isolation of bacteria from nematodes
The indirect isolation of Xenorhabdus bacteria from haemolymph was based on a previously described method [30] with modifications [20]. First, nematode isolates from collection points BG5 and VH1 were selected. Cadavers with which BG5 and VH1 nematodes were baited were surface-sterilized and dissected under aseptic conditions. A light-yellow, viscous, heterogenous fluid was aseptically obtained and streaked onto nutrient agar supplemented with 0.0025 % (w/v) bromothymol blue and 0.004 % (w/v) 2,3,5 triphenyl tetrazolium chloride (NBTA). This was incubated at 30 °C for 96 h. Only colonies that had the following observed morphologies were selected for further pure culture techniques: blue/yellow pigmentation, irregular margins, umbonate shape and visible swarming patterns. On these pure cultures, a catalase test was performed, and the absence of bubble production indicated a catalase-negative isolate, and these were presumptively identified as Xenorhabdus species. They were named Xenorhabdus sp. strains BG5 and VH1.
Genome sequencing and assembly
Previously, we isolated X. griffiniae XN45 from Steinernema sp. scarpo, which was originally isolated from Muran’ga District in Kenya [20]. Thus, in addition to Xenorhabdus sp. strains VH1 and BG5, this strain was selected for genome sequencing and assembly. DNA from strain XN45 was extracted with FastDNA Spin Kit for Soil (Mp Bio) to yield a concentration of 20 ng µl–1 and UV absorbance ratio at 260nm/280nm (A260/280) >1.8. From this, only 0.1 ng of DNA was used to prepare a library using a Nextera XT kit (Illumina). Sequencing was done by CeGaT GmBH on a NovaSeq 6000 platform with the following parameters: short insert paired-end reads of 100 bp and targeted coverage of 100×. Output data were raw sequence reads in fastq.gz format (2.902 GB), which had Illumina standard Phred scores (offset +33) and adapter sequences already removed. In terms of quality, 91.32 % of reads had a Q30 value. Genome assembly was done with Spades 3.10.1 [31] with thresholds for minimum contig length and coverage set at 1000 bp and 5× respectively.
For VH1 and BG5, DNA was isolated with Gentra Puregene DNA extraction kit (Qiagen) to yield samples of 1 µg µl–1 concentration and A260/280 ratios of >1.8. At the Doherty Institute, University of Melbourne, Australia, DNA libraries were created using a Nextera XT DNA preparation kit (Illumina), and whole genome sequencing was performed on a NextSeq platform (Illumina) with paired-end reads of 150 bp and targeted coverage of >50×. Genome assembly was done with Spades 3.10.1 [31]. For assembled genomes of strains VH1, BG5 and X. griffiniae XN45 from this study and X. griffiniae strain BMMCB (doi: 10.1128/genomeA.00785–15) from Mothupi et al. [32], characteristics such as completeness, contamination, N50, L50, length and GC content were determined using the comprehensive genome analysis tool of the PATRIC platform [33].
Phylogenomic reconstruction and calculation of ANI values
For phylogenomic reconstruction of the genus Xenorhabdus , 27 fasta files (Table S3) were used as input data for a whole genome-based taxonomic analysis on the Type strain genome server platform (TYGS) [26]. On these, the MASH algorithm [34] was used to quickly calculate intergenomic relatedness and determine the strains with the smallest distances. All pairwise comparisons and inference of intergenomic distances among the set of genomes were conducted using the genome BLAST distance phylogeny (GBDP) 'trimming' algorithm and distance formula d5 [35]. One hundred distance replicates were calculated for each. The genome–genome distance calculator (GGDC) 2.1 formula was used to calculate dDDH values and confidence intervals [35]. Confidence intervals are given in workbook S1. Intergenomic distances were then used to infer a balanced minimum evolution tree with branch support via FASTME 2.1.4 including SPR post-processing [36]. Branch support was inferred from 100 pseudo-bootstrap replicates for each. Rooting of trees was done at the midpoint whereas visualization and graphics editing were done with iTOL [37] and Inkscape [38] respectively.
Using the same workflow, a phylogenomic reconstruction with only strains VH1, BG5, BMMCB and XN45 was made. Minimum thresholds for two strains to be classified as one species and sub-species were 70 % and 79 % dDDH respectively [26]. To calculate ANI values among species most closely related to strains XN45, VH1 and BG5, the orthoANI algorithm was used within the OAT software package, which was also used to obtain genome–genome distance (GGD) 2.1 values [24].
Creation of pangenomes
To determine whether the genus Xenorhabdus had an open pangenome, genomes of Xenorhabdus species were first used to construct a pangenome of the genus using the anvio v7.1 pangenome workflow [28, 39] with the following parameters: use ncbi-blast, MCL inflation=10, minbit=1, exclude-partial-gene-calls. A pangenome of strains VH1, BG5, XN45 and BMMCB only was also created using the same workflow and parameters. The strain names, accession numbers and total number of gene calls of each genome used are listed in Table S3. The mean α value was determined using the P-GAP platform running on the Panweb server [40]. Briefly, RAST-k [41] annotated genomes were used as data input on the Panweb server and the following parameters were selected for clustering genes into one gene cluster: minimum 80 % nucleotide similarity, and minimum 80 % coverage with gene family algorithm.
Estimation of the effect of draft genomes on the determination of the core genome
To estimate how the use of draft genomes affects the determination of the core genome, two additional pangenomes were created. The first contained six genomes, each of which was composed of fewer than two contigs: X. bovienii CS-03 (NZ_FO818637), X. hominickii ANU (NZ_CP016176), X. cabanillasii DSMZ 19705 (NZ_QTUB01000001), X. poinarii G6 (FO704551), X. nematophila AN6/1 (FN667742) and X. szentirmaii US123(NIUA01000001). The second contained draft genomes of similar species: X. bovienii T228 (JANAIF000000000.1), X. hominickii DSM 17903 (NJAI00000000.1), X. cabanillasii JM26 (NJGH00000000.1), X. poinarii SK (JADLIG000000000.1) and X. nematophila C2-3 (JRJV00000000.1). Pangenomes were created via the aforementioned anvio workflow and the sizes of their resultant core genomes were compared.
Analysis of gain and loss of gene clusters in the X. griffiniae clade
Using the gene clusters (GCs) of the VH1-BG5-XN45-BMMCB pangenome, a matrix of the presence and absence of GCs among the four strains was created (workbook S1). This matrix and the GBDP phylogeny of the four strains were then used as input data for gene gain and loss analysis in the COUNT program (downloaded 17 January 2023) using Wagner parsimony (penalty=1) [42].
Characterization of core, accessory, species and strain-specific genes
By using the ‘search’ and ‘bin’ functions of the anvi´o-interactive program, GCs that were present in all genomes under analysis as single copies were obtained and binned as single-copy core GCs (SCGs). Other binned GCs were: strain BG5 specific, strain BMMCB specific, strain XN45 specific, strain VH1 specific, XN45-VH1 accessory/ X. griffiniae species specific, XN45-VH1-BG5 accessory, XN45-VH1-BMMCB accessory and BMMCB-BG5 accessory. Using the anvi-get-sequences-for-gene-clusters program with the ‘report DNA sequences’ and ‘concatenate’ flags, sequences for the single-copy GCs of each of the bins were obtained. For those consisting of GCs that constituted genes from multiple genomes, sequences from a single genome were used to represent the GC. These sequences were annotated in PROKKA [43] to elucidate the functions encoded by predicted genes.
Clustering of gene clusters into functional groups
The functions of GCs were determined by manually querying the UniProt Knowledgebase [44] with each annotated gene symbol. Then, GC functions were assigned based on the described biological process the gene most clearly contributed to. This was supplemented by querying GCs with assigned cluster of orthologous groups (COG) functions against the respective database [45]. To aid the identification of genes that encode the biosynthesis of specialized metabolites, nucleotide sequences for each bin were annotated in antiSMASH [46]. Then, GC-function lists were compiled for each bin and manually curated. GCs were grouped according to similarity of function and visually represented in column graphs.
Elucidation of genomic islands in strain BG5
To highlight putative genomic islands flanked by transposase genes, an annotated record of the BG5 genome was concatenated and used as the reference genome in BRIG [47] and compared to genomes of VH1 and XN45 by the blast algorithm [48] utilizing an NCBI-blast 2.4.0+ bin library. Selected rings to be visualized were for BG5 genome guanine-cytosine (G+C) content (ring 1) and skew (ring 2), VH1 genome (ring 4), XN45 genome (ring 5) and loci of coding DNA sequences (CDS) annotated as transposases on the BG5 genome (ring 6). Output visualizations were obtained as .svg files and enhanced in Inkscape [38].
Results
Strains VH1, BG5, XN45 and BMMCB form a clade
Two putative Steinernema isolates, VH1 and BG5, were isolated from soils in Western Kenya. VH1 was isolated from red volcanic loam soils on cabbage cultivated land at a point with coordinates 0.06293, 34.72903 and altitude 1624 m, in Vihiga. BG5 was isolated from clay soils on riverine land at a crop edge at a point with coordinates 0.48044, 34.40836 and altitude 1239 m, in Bungoma. From these two, Xenorhabdus sp. strains VH1 and BG5 were respectively isolated. Soils from the Rift Valley Regions sampled did not yield any steinernematids. Previously, we isolated X. griffiniae XN45 from Steinernema sp. scarpo, which originated from soils in Muran’ga County, Kenya [20]. Xenorhabdus sp. strain BMMCB, which was designated as an X. griffiniae species, was previously isolated from Steinernema sp. BMMCB [32], whose natural habitat was red volcanic sandy-loam soils in Brits, North West Province, South Africa [49].
Thus, to investigate the phylogenomic relationships between these four strains, the quality of their genome assemblies was first determined. The draft genomes of XN45, VH1, BG5 and BMMCB were complete and consistent, with low contamination (Table 1), and of coverage >50×. They were thus of sufficient quality for species delineation via overall genome relatedness indices [23]. For the nascent BG5, XN45 and VH1 assemblies, 74.5, 72.2 and 71.5 % of proteins encoded in their respective genomes had known functions (Table 1), which were slightly below the 80 % average for genomes of Gammaproteobacteria [45].
Table 1.
Quality and characteristics of genome assemblies
Genomes of BG5, XN45 and VH1 were assembled in this study. BMMCB was obtained from NCBI GenBank (LDNM00000000.1). Annotation statistics were determined via the PATRIC platform.
|
Xenorhabdus sp. strain BG5 |
X. griffiniae XN45 |
Xenorhabdus sp. strain VH1 |
Xenorhabdus sp. strain BMMCB |
|
|---|---|---|---|---|
|
Contigs |
129 |
381 |
273 |
231 |
|
Guanine–cytosine content (%) |
43.80 |
43.57 |
43.65 |
44.68 |
|
Contig L50 |
12 |
29 |
43 |
21 |
|
Contig N50 (bp) |
102 633 |
45 29 |
29 298 |
57 901 |
|
Genome length (bp) |
3 933 551 |
4 215 754 |
4 224 998 |
4 183 760 |
|
Fine consistency (%) |
96.5 |
95.9 |
95.9 |
95.7 |
|
Coarse consistency (%) |
97.0 |
96.7 |
96.7 |
96.7 |
|
Contamination (%) |
0.7 |
0 |
0 |
0 |
|
Completeness (%) |
100 |
100 |
100 |
100 |
|
CDS |
3827 |
4232 |
4160 |
4318 |
|
Repeat regions |
127 |
66 |
69 |
70 |
|
Hypothetical proteins |
973 |
1175 |
1185 |
1193 |
|
Proteins with functional assignments |
2854 |
3057 |
2975 |
3125 |
Bp, base pairs; CDS, coding DNA sequences; Contig L50, the minimum number of contigs that contain 50% of the assembly; Contig N50, the shortest contig among that minimum number of contigs, which contain 50% of the assembly.
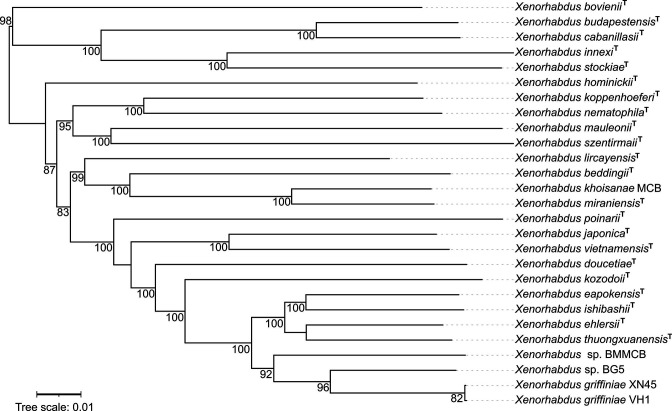
Strains VH1, BG5, BMMCB and XN45 formed an exclusive clade, as demonstrated in a phylogenomic reconstruction of 24/27 described species of the genus (Fig. 1). They were more closely related to each other than to other species, and this is demonstrated by the most closely related strains to VH1, BG5, XN45 and BMMCB being XN45, XN45, VH1 and BG5 (Fig. 2) at genome–genome distances (GGD) of 0.00, 0.059, 0.00 and 0.08 respectively (Table 2).
Fig. 1.
Phylogenomic reconstruction of Xenorhabdus species using genome blast distance phylogeny approach (GBDP) distances calculated from genome sequences using the d5 distance formula. Genome sequences of Xenorhabdus sp. strains VH1 and BG5 and X. griffiniae XN45 were obtained in this study. Xenorhabdus sp. strain VH1 and X. griffiniae XN45 clustered together. Xenorhabdus sp. strain BMMCB was previously classified as representing an X. griffiniae species. However, it did not cluster with X. griffiniae XN45. Xenorhabdus sp. strain BG5 did not form a clade with any extant species. GBDP pseudo-bootstrap values of above 40 % are shown. The scale bar represents substitutions per site.
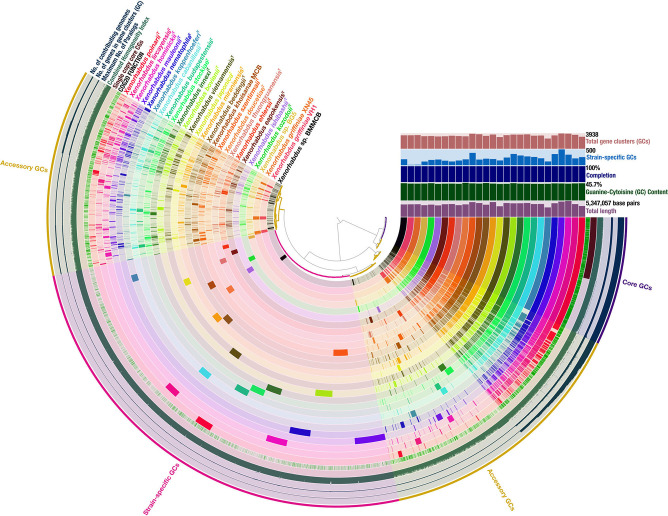
Fig. 2.
Graphical representation of the pangenome of 26 species of the genus Xenorhabdus . The largest genome was 5 347 057 bp and the highest guanine–cytosine content was 45.7 %. The pangenome was composed of a total of 13 469 gene clusters (GCs). Core GCs, those found in all 27 genomes, numbered 1654 in total. Accessory GCs, those found in two to 26 genomes, numbered 5992 in total. Strain-specific GCs, those found in one genome only, numbered 5820 GCs in total.
Table 2.
Orthologous average nucleotide identity (orthoANI), genome-to-genome distance (GGD) and digital DNA–DNA hybridization (%) (dDDH) values for type species most closely related to Xenorhabdus sp. strains VH1 and BG5, X. griffiniae XN45 and Xenorhabdus sp. strain BMMCB
OrthoANI values are in the top half of the matrix (top triangle), GGD in parentheses, and values for dDDH are in the bottom half of the matrix (bottom triangle). Values that are within the threshold for two strains to be classified as one species are shaded in grey. The thresholds for conspecific strains are orthoANI values above 95.1 %, dDDH values above 70 % and GGD values below 0.0361. Type strains are: DSM 2270, X. ishibashii; DL20, X. eapokensis; DSM 16337, X. ehlersii; 30TX1, X. thuongxuanensis.
|
DSM 22 670 |
XN45 |
DL20 |
DSM 16 337 |
VH1 |
BG5 |
30T×1 |
BM MCB |
|
|---|---|---|---|---|---|---|---|---|
|
BM MCB |
89.68 (0.103) |
91.41 (0.088) |
90.03 (0.010) |
91.95 (0.082) |
91.42 (0.088) |
92.25 (0.080) |
90.56 (0.094) |
– |
|
30T×1 |
92.55 (0.076) |
91.02 (0.091) |
93.42 (0.067) |
93.76 (0.064) |
91.01 (0.091) |
91.72 (0.085) |
– |
49.6 |
|
BG5 |
90.44 (0.097) |
94.24 (0.059) |
90.94 (0.092) |
93.08 (0.071) |
94.22 (0.059) |
– |
57.3 |
57.1 |
|
VH1 |
89.80 (0.102) |
99.99 (0.000) |
90.32 (0.097) |
91.95 (0.081) |
– |
67.4 |
51.1 |
50.5 |
|
DSM 16 337 |
92.11 (0.080) |
92.05 (0.081) |
92.55 (0.077) |
– |
59.9 |
66.7 |
53.50 |
56.4 |
|
DL20 |
93.23 (0.069) |
90.34 (0.097) |
– |
47.90 |
51.6 |
57.5 |
51.90 |
50.9 |
|
XN45 |
89.92 (0.102) |
– |
51.7 |
59.9 |
99.9 |
67.3 |
51.1 |
50.4 |
|
DSM 22 670 |
– |
50.3 |
51.2 |
46.4 |
50.3 |
55.0 |
48.3 |
48.9 |
Strains XN45 and VH1 were conspecific, as demonstrated by their GGD, dDDH and ANI values of 0.000, 99.9 % and 99.9 % respectively– these were all within the conspecific thresholds of <0.0361 for GGD, >70 % for dDDH and >95.1 % for orthoANI [24, 26, 35]. XN45 and BG5 were most closely related to each other but were not conspecific as their GGD, dDDH and ANI values of 0.059, 67.3 % and 94.24 % respectively were all outside conspecific thresholds. BG5 and BMMCB were most closely related to each other but were not conspecific, as seen from their GGD, dDDH and ANI values of 0.08, 57.1 % and 92.25 % respectively.
BMMCB and XN45 were both described as X. griffiniae species [20, 32]. We previously demonstrated XN45 and X. griffiniae ID10T as conspecific based on percentage nucleotide similarities for their 16S rRNA, recA and serC genes as 99.595, 98.571 and 97.686 % respectively. These were above the same species thresholds of 98.65, 97 and 97 % respectively [11, 50]. Conversely, strains BMMCB and XN45 were not conspecific, as demonstrated by their respective percentage nucleotide similarities values of 98.545, 93.67 and 92.066 % for 16S rRNA, recA and serC genes respectively [20]. Indeed, BMMCB was not an X. griffiniae species as its GGD, dDDH and ANI values with X. griffiniae XN45 were 0.08, 50.4 % and 91.41 % respectively – these were all outside conspecific thresholds. This was corroborated by a difference of 1.11 % in G+C content between the two genomes (Table 1), which was above the 1 % same species threshold [51]. Taken together, these results demonstrated that these four strains represented three species: X. griffiniae XN45, X. griffiniae VH1, and the two undescribed species Xenorhabdus sp. BG5, and Xenorhabdus sp. BMMCB.
The genus Xenorhabdus has an open pangenome
We hypothesized that the genus Xenorhabdus had a pangenome that included many strain-specific genes, due to the numerous Xenorhabdus strains and their respective genomes, which have not yet been isolated from under-investigated Steinernema species [19]. This would make it an open pangenome. The pangenome is the pool of genes from which all the taxon genomes are constituted. The core genes are those found in all genomes, accessory genes are those found in two or more genomes, and strain-specific genes are those found in one genome only [27].
The Xenorhabdus pangenome was composed of 27 genomes (from 24/27 described species) each of which had between 2771 ( X. koppenhoeferi ) and 4990 ( X. hominickii ) genes. It contained 101 832 genes, after the exclusion of 3668 partial gene calls from the analysis (Table S2). The pangenome contained a total of 13 469 GCs (Fig. 2). The core genome had 1654 GCs (12.3 %). However, this likely to be an underestimate since draft genomes were mostly used in the analysis, and the use of draft as opposed to complete genomes reduced the core genome size by 5 %, in our comparison of pangenomes of similar species (Fig. S3). The accessory genome had 5992 GCs (44.5 %) and the strain-specific genome had 5820 GCs (43 %). In total, 6834 GCs (50.7 %) encoded known functions as per the Clusters of Orthologous Genes (COG) database. The highest percentage of these were in the core genome whereas the strain-specific genome had the lowest. X. mauleonii had the largest number of strain-specific genes (500) whereas X. ehlersii had the fewest (118). X. griffiniae VH1 and XN45 had remarkably few strain-specific GCs, 13 and 17 respectively, as they were the only two strains from the same species (Fig. 2). Strain-specific genes from newly added genomes increase the pangenome, and the rate of new strain-specific genes per newly added genome decreases to zero. If this rate of decrease is high, the result is a closed pangenome [27] whose size does not change significantly with the addition of new genomes. Conversely, open pangenomes have a slow rate of decreasing number of new strain-specific genes per newly added genome. Moreover, they are typified by a small percentage of core genes [52]. Relatedly, the mean α exponent of Heaps' Law is used to estimate this rate of decrease [53], and pangenomes with values >1 are defined as closed whereas those <1 are defined as open [27]. Thus, Xenorhabdus has an open pangenome, as demonstrated by its mean α exponent of Heaps' Law of 0.2752 and a core genome of 12.2 % (1654/13 469). This corroborated previous global comparative genome analyses of the genus Xenorhabdus [2, 28, 54].
Most species-specific genes in a pangenome of the X. griffiniae clade encode unknown proteins
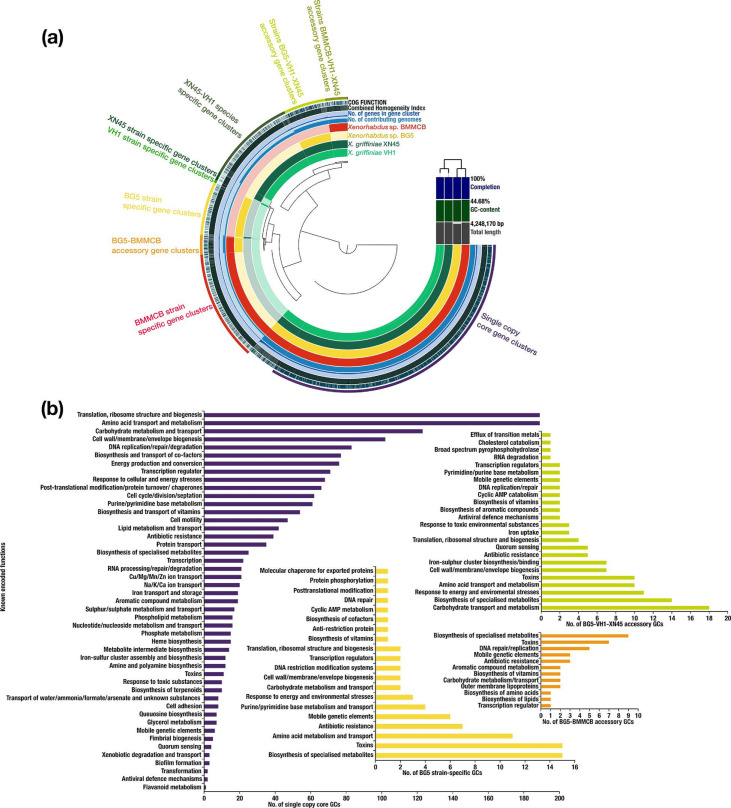
A pangenome analysis of the clade containing strains XN45, VH1, BG5 and BMMCB, ‘the X. griffiniae’ clade, was conducted. It had 15 411 genes that formed 4877 GCs. There were 2364 core GCs, of which 2231 were single-copy. Other groups included 766 strain BMMCB specific, 617 XN45-VH1 species specific, 377 strain BG5 specific, 297 BG5-VH1-XN45 accessory, 154 BMMCB-VH1-XN45 accessory, 150 BG5-BMMCB accessory, 54 strain XN45 specific, 26 strain VH1 specific, 14 XN45-BG5 accessory and five VH1-BMMCB accessory GCs (Fig. 3). From an analysis of gene gain and loss within this clade (Fig. S2), the species-specific genes, 79% of which encoded proteins with unknown functions, possibly resulted from a net acquisition of new genes [55].
Fig. 3.
(a) Graphical representation of a pangenome of a monophyletic group of three Xenorhabdus species. Core genes clusters (GCs) were those found in all four genomes, and numbered 2364. Accessory GCs were those found in two or three genomes. BG5-BMMCB, BG5-VH1-XN45 and BMMCB-VH1-XN45 accessory genomes had 150, 297 and 154 GCs respectively. Strain-specific GCs were those found in one genome only, and BMMCB, BG5, XN45 and VH1 had 766, 377, 54 and 26 strain-specific GCs respectively. (b) Bar charts of known functions encoded by Xenorhabdus sp. strain BG5 (strain BG5) GCs. Commensurate with their lifestyle as endosymbionts of entomopathogenic nematodes, these bacteria encoded the following non-canonical core functions: antibiotic resistance, and biosynthesis of specialized metabolites and toxins. For XN45-VH1-BG5 accessory GCs, those that encoded the metabolism and transport of carbohydrates such as apiose, fuculose, tagatose, galactose and sorbose, and those that encoded biosynthesis of specialized metabolites such as antibiotics, polyketides, non-ribosomal peptides and siderophores were enriched. For BG5-BMMCB accessory GCs, those that encoded the biosynthesis of specialized metabolites were enriched. For BG5 strain-specific GCs, those that encoded biosynthesis of specialized metabolites and toxins such as type II, III and IV secretion system toxins were enriched among those with known protein functions.
To determine which encoded functions were enriched in the core, accessory and strain-specific genomes, single-copy GCs of each were annotated in PROKKA [43]. The functions and biological processes that the GCs encoded were then determined from the UniProt Knowledgebase [44] and COG [45] descriptions. In total, 3352 (68.4 %) GCs had known functions. The core genome had the largest percentage of these while strain-specific genomes had the smallest. Specifically, 80 % (1735/2182) of the core, 46 % (117/252) of BG5-XN45-VH1 accessory, 36 % (38/107) of BG5-BMMCB accessory, 29 % (31/108) of BMMCB-XN45-VH1 accessory, 23 % (75/327) of strain BG5 specific, 23 % (154/655) of strain BMMCB specific and 18 % (102/561) of X. griffiniae species specific GCs had known functions (Figs 3 and S1).
The most enriched functions in the core were those of housekeeping, such as translation, ribosome structure and biogenesis, amino acid transport and metabolism, carbohydrate transport and metabolism, and cell wall/membrane/envelope (Fig. 3). For the last, the Gram-negative nature of the bacteria [30] was demonstrated by the presence of genes encoding lipopolysaccharide (LPS) biosynthesis such as lpt, rfa, lpx operons, lapA-B, msbA, waaA, galU, wbgU and yhjD. Xenorhabdus are characterized as motile, peritrichously flagellated rods [30] and this was demonstrated by the presence of genes encoding flagellum biogenesis and motility such as flgA, flgD, flgJ-L, flhA-D, fliC-T and fliZ. Strains XN45, VH1 and BG5 exhibited swarming motility, and this was supported by the presence of the following core genes: flgB-C, flgE-G and motA-B. Other core genes included those encoding antibiotic resistance such as acrABRZ, mdtABCK macAB fsr bsr and lmrA. The biosynthesis of a few specialized metabolites was also a core function (Fig. 3), and this corroborated a pangenome analysis of genes encoding specialized metabolites from both Xenorhabdus and Photorhabdus bacteria [28].
X. griffinae genomes (XN45 and VH1) were enriched with a wide selection of genes that encoded carbohydrate metabolism and transport. This was demonstrated by enrichment of genes encoding metabolism and transport of apiose, fuculose, tagatose, galactose and sorbose in the XN45-VH1-BG5 accessory GCs, ribose, galactose, myo-inositol, d-malate and galactonate in the XN45-VH1-BMMCB GCs, and glucoside, glycolate and glycerate in the X. griffiniae species-specific GCs (Figs 3 and S1). Biosynthesis of specialized metabolites – such as non-ribosomal peptides, polyketides siderophores and antibiotics – was highly species dependent, as demonstrated by its enrichment in species-specific GCs. This was similarly observed with the production of type II, III and IV secretion system toxins. However, these enrichments were from a small fraction (21 %) of species-specific GCs; most genes specific to these three Xenorhabdus species encoded proteins with unknown functions.
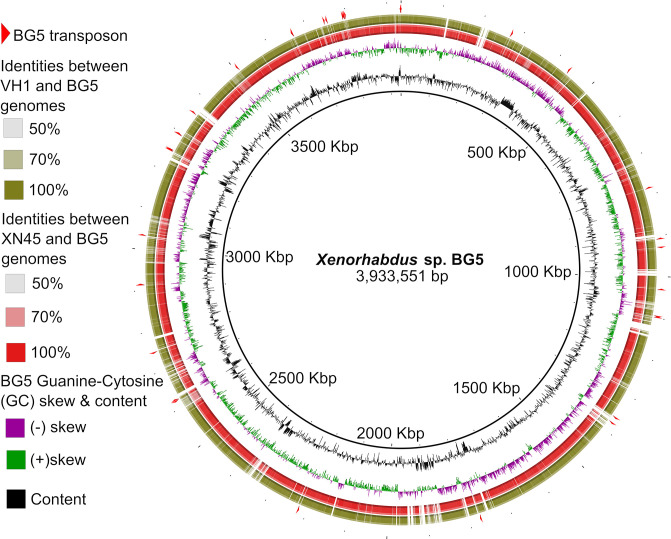
Xenorhabdus sp. BG5 genomic islands are flanked by transposase genes
To investigate whether genes specific to Xenorhabdus sp. BG5 formed genomic islands, its genome was compared to those of X. griffiniae XN45 and VH1. Regions of less than 50 % nucleotide similarity were determined and visualized in BRIG [47]. These were verified as genomic islands by genome alignments in Mauve. Most CDS in these genomic islands encoded hypothetical proteins. Genes encoding transposases in BG5 either flanked genomic islands/were the genomic island (Fig. 4). Insertion sequence (IS) elements predominated the type of transposases predicted to be encoded by these genes, as shown here: IS110 family transposase ISSfl8, IS3 family transposase ISKpn37, IS481 family transposase ISVvu4, IS5 family transposase ISSod6, IS630 family transposase ISPlu10, IS1 family transposase ISEhe5, IS1 family transposase ISPda1, IS110 family transposase ISPlu13, IS3 family transposase ISAlg, IS630 family transposase ISEc40 and IS982 family transposase ISNsp1 (workbook S1). IS elements contribute to genome reshuffling [56] and thus may be implicated in the creation of genomic islands in Xenorhabdus sp. BG5.
Fig. 4.
Genome visualizations of Xenorhabdus sp. BG5 when compared to X. griffiniae XN45 (red) and X. griffiniae VH1 (green). Genomic islands of Xenorhabdus sp. BG5 are denoted by white breaks in X. griffiniae genomes, and these represented cognate nucleotide sequences that were less than 50 % identical. Red triangles in the outermost ring denote loci of IS family transposase genes on the BG5 genome. All transposase genes that were not on contig edges flanked genomic islands. Genomic islands not associated with transposase genes are also shown. The visualization was created in Blast Ring Image Generator (BRIG).
Discussion
This study aimed to discover new Xenorhabdus strains because different species [2, 57] and even strains [3] from this genus have different antimicrobial production profiles. Soils of Western Kenya were selected as they had not hitherto been investigated for steinernematids, unlike those from Central Kenya [16, 21]. Soils were collected from Bungoma County and sites with the occurrence of steinernematids were clay soils on riverine land, corroborating previous studies on similar soils [58]. From Vihiga County, soils with the occurrence of steinernematids were red volcanic loam soils on cabbage cultivated land, corroborating previous studies on similar lands [21, 59]. From nematodes isolated from these soils, Xenorhabdus sp. strains VH1 and BG5 were isolated.
Previously we isolated X. griffiniae XN45 from Steinernema sp. scarpo which also originated from Kenyan soils. Using draft genome assemblies of strains XN45, VH1 and BG5 (Table 1) which were all of suitable quality for species delineation as per the standards of Chun et al. [23], the phylogenomic reconstruction of the genus (Fig. 1) demonstrated that these Kenyan strains formed a monophyletic group that could be enlarged to include strain BMMCB. This strain was previously designated as representing an X. griffiniae species. The draft assembly of XN45 was used for species delineation via analysis of orthoANI, DDH and G+C content thresholds for conspecific strains [24, 25, 51]. From these, strain BMMCB was identified as an undescribed species whereas strain VH1 was designated as X. griffiniae VH1. Strain BG5 was most closely related to XN45. However, ANI, dDDH and GGD values for the two were not consistent with those of conspecific strains. It was thus designated as an undescribed species of the genus Xenorhabdus . This demonstrated the importance of genome assemblies for accurate species delineation via overall genome relatedness indices, corroborating previous pivotal studies [23, 26, 60].
The open pangenome of the genus Xenorhabdus corroborated not only a larger pangenome analysis of 40 Xenorhabdus strains [54] but also the large number of Xenorhabdus strains –hosted by over 50 described Steinernema species [19] – that have yet to be isolated, identified and their genome sequences determined.
Using a pangenome analysis of the clade, the four closely related strains XN45, VH1, BG5 and BMMCB were further distinguished as representing three species based on the large numbers of species-specific genes – these were 377, 617 and 766 for Xenorhabdus sp. BG5, X. griffiniae and Xenorhabdus sp. BMMCB respectively. Conversely, strain VH1 was of the same species as XN45 as its genome only had 26 unique genes when compared to that of XN45. Similar pangenome analyses of other clades may elucidate a minimum number of strain-specific genes that delineate species in this genus. Notably, 79 % of the functions of proteins encoded by species-specific genes were unknown, compared to 20 % for proteins encoded by the core genome. It has long been established that most species-specific prokaryotic genes encode unknown functions [61]. Indeed, in 613 prokaryotic species, over 50 % of a subset of species-specific protein-coding genes encoded unknown functions [62]. These genes lead to speciation when they encode environmentally important traits [61]. For all three species, species-specific genes – only the subset that had known functions – were enriched for the biosynthesis of specialized metabolites. However, this enrichment was probably overestimated because genes encoding secondary metabolites of Xenorhabdus species are often long and clustered in genome loci that span thousands of base pairs, which leads to their frequent fragmentation in draft genomes, resulting in inflated counts [63]. Xenorhabdus sp. BG5 had genomic islands when its genome was compared to those of X. griffiniae strains. Some of these islands were flanked by genes encoding transposases, the vast majority of which were IS elements. IS elements are known to contribute to genome reshuffling [56] suggesting that IS transposases contributed to the creation of genomic islands in Xenorhabdus sp. BG5. However, the majority of the islands were not associated with genes encoding transposases, implicating other factors such as phages, as drivers of these differences. In conclusion, two Xenorhabdus bacteria isolated from steinernematids from soils in Western Kenya were identified as a novel species and strain. Within the X. griffiniae clade, most species-specific genes encoded unknown functions. These genomes, species delineations and genome analyses are useful for in silico-based discovery of antimicrobials from the genus Xenorhabdus .
Supplementary Data
Funding information
This study was supported by the Kenya National Research Fund grant NRF first CALL/MULTIDISCIPLINARY RESEARCH/127 ‘Drug Development of Antibiotics: Xenorhabdus bacteria from Kenya’ to N.O.A. This research was also supported by the German Academic Exchange Service (DAAD) under programme number 57 299 294 and reference number 91 653 288 and GRADE completion scholarship to R.M.A. Research in the Bode lab was supported by LOEWE Translational Biodiversity Genomics funded by the State of Hesse, Germany. Tim Stinear provided genome sequencing laboratory support for this study.
Author contributions
R.M.A. – conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, validation, visualization, writing-first draft, and writing-editing and reviewing. C.N.W. – methodology, resources, validation, and writing-editing and reviewing. S.J.P. – data curation, formal analysis, investigation, and writing-editing and reviewing. N.O.A. – funding acquisition, methodology, project administration, resources, supervision, and writing-editing and reviewing. H.B.B. – funding acquisition, methodology, project administration, resources, supervision, validation, and writing-editing and reviewing.
Conflicts of interest
The authors declare no competing financial interest. R.M.A. is a proprietor of Elakistos Biosciences.
Ethical statement
No humans or vertebrate organisms were investigated in this study.
Footnotes
Abbreviations: ANI, average nucleotide identity; BLAST, basic local alignment search tool; CDS, coding DNA sequence; COG, cluster of orthologous groups; dDDH, digital DNA–DNA hybridization; EPNs, entomopathogenic nematodes; GBDP, genome BLAST distance phylogeny; GC, gene clusters; GGD, genome–genome distance; IS, insertion sequence; LPS, lipopolysaccharide; MCL, Markov clustering; OGRIs, overall genome-related indices; orthoANI, orthologous average nucleotide identity; SPR, subtree pruning and regrafting.
Three supplementary figures and three supplementary tables are available with the online version of this article.
References
- 1.Booysen E, Dicks LMT. Does the future of antibiotics lie in secondary metabolites produced by Xenorhabdus spp.? A review. Probiotics Antimicrob Proteins. 2020;12:1310–1320. doi: 10.1007/s12602-020-09688-x. [DOI] [PubMed] [Google Scholar]
- 2.Tobias NJ, Wolff H, Djahanschiri B, Grundmann F, Kronenwerth M, et al. Natural product diversity associated with the nematode symbionts Photorhabdus and Xenorhabdus . Nat Microbiol. 2017;2:1676–1685. doi: 10.1038/s41564-017-0039-9. [DOI] [PubMed] [Google Scholar]
- 3.Muangpat P, Suwannaroj M, Yimthin T, Fukruksa C, Sitthisak S, et al. Antibacterial activity of Xenorhabdus and Photorhabdus isolated from entomopathogenic nematodes against antibiotic-resistant bacteria. PLoS One. 2020;15:e0234129. doi: 10.1371/journal.pone.0234129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akhurst RJ, Boemare NE. A numerical taxonomic study of the genus Xenorhabdus (Enterobacteriaceae) and proposed elevation of the subspecies of X. nematophilus to species. J Gen Microbiol. 1988;134:1835–1845. doi: 10.1099/00221287-134-7-1835. [DOI] [PubMed] [Google Scholar]
- 5.Castaneda-Alvarez C, Prodan S, Zamorano A, San-Blas E, Aballay E. Xenorhabdus lircayensis sp. nov., the symbiotic bacterium associated with the entomopathogenic nematode Steinernema unicornum . Int J Syst Evol Microbiol. 2021;71 doi: 10.1099/ijsem.0.005151. [DOI] [PubMed] [Google Scholar]
- 6.Ferreira T, van Reenen CA, Endo A, Spröer C, Malan AP, et al. Description of Xenorhabdus khoisanae sp. nov., the symbiont of the entomopathogenic nematode Steinernema khoisanae . Int J Syst Evol Microbiol. 2013;63:3220–3224. doi: 10.1099/ijs.0.049049-0. [DOI] [PubMed] [Google Scholar]
- 7.Kämpfer P, Tobias NJ, Ke LP, Bode HB, Glaeser SP. Xenorhabdus thuongxuanensis sp. nov. and Xenorhabdus eapokensis sp. nov., isolated from Steinernema species. Int J Syst Evol Microbiol. 2017;67:1107–1114. doi: 10.1099/ijsem.0.001770. [DOI] [PubMed] [Google Scholar]
- 8.Kuwata R, Qiu L-H, Wang W, Harada Y, Yoshida M, et al. Xenorhabdus ishibashii sp. nov., isolated from the entomopathogenic nematode Steinernema aciari . Int J Syst Evol Microbiol. 2013;63:1690–1695. doi: 10.1099/ijs.0.041145-0. [DOI] [PubMed] [Google Scholar]
- 9.Lengyel K, Lang E, Fodor A, Szállás E, Schumann P, et al. Description of four novel species of Xenorhabdus, family Enterobacteriaceae: Xenorhabdus budapestensis sp. nov., Xenorhabdus ehlersii sp. nov., Xenorhabdus innexi sp. nov., and Xenorhabdus szentirmaii sp. nov. Syst Appl Microbiol. 2005;28:115–122. doi: 10.1016/j.syapm.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 10.Nishimura Y, Hagiwara A, Suzuki T, Yamanaka S. Xenorhabdus japonicus sp. nov. associated with the nematode Steinernema kushidai . World J Microbiol Biotechnol. 1994;10:207–210. doi: 10.1007/BF00360889. [DOI] [PubMed] [Google Scholar]
- 11.Tailliez P, et al. Phylogeny of Photorhabdus and Xenorhabdus based on universally conserved protein-coding sequences and implications for the taxonomy of these two genera. proposal of new taxa: X. vietnamensis sp. nov., P. luminescens subsp. caribbeanensis subsp. nov., P.luminescens subsp. hainanensis subsp. nov., P. temperata subsp. khanii subsp. nov., P. temperata subsp. tasmaniensis subsp. nov., and the reclassification of P. luminescens subsp. thracensis as P. temperata subsp. thracensis comb. nov. Int J Syst Evol Microbiol. 2010;60:1921–1937. doi: 10.1099/ijs.0.014308-0. [DOI] [PubMed] [Google Scholar]
- 12.Tailliez P, Pagès S, Edgington S, Tymo LM, Buddie AG. Description of Xenorhabdus magdalenensis sp. nov., the symbiotic bacterium associated with Steinernema australe . Int J Syst Evol Microbiol. 2012;62:1761–1765. doi: 10.1099/ijs.0.034322-0. [DOI] [PubMed] [Google Scholar]
- 13.Tailliez P, Pagès S, Ginibre N, Boemare N. New insight into diversity in the genus Xenorhabdus, including the description of ten novel species. Int J Syst Evol Microbiol. 2006;56:2805–2818. doi: 10.1099/ijs.0.64287-0. [DOI] [PubMed] [Google Scholar]
- 14.Heryanto C, Eleftherianos I. Nematode endosymbiont competition: fortune favors the fittest. Mol Biochem Parasitol. 2020;238:111298. doi: 10.1016/j.molbiopara.2020.111298. [DOI] [PubMed] [Google Scholar]
- 15.Vigneux F, Zumbihl R, Jubelin G, Ribeiro C, Poncet J, et al. The xaxAB genes encoding a new apoptotic toxin from the insect pathogen Xenorhabdus nematophila are present in plant and human pathogens. J Biol Chem. 2007;282:9571–9580. doi: 10.1074/jbc.M604301200. [DOI] [PubMed] [Google Scholar]
- 16.Waturu CN, Hunt DJ, Reid AP. Steinernema karii sp. n.(Nematoda: Steinernematidae), a new entomopathogenic nematode from Kenya. Int J Nematol. 1997;7:68–75. [Google Scholar]
- 17.White GF. A method for obtaining infective nematode larvae from cultures. Science. 1927;66:302–303. doi: 10.1126/science.66.1709.302-a. [DOI] [PubMed] [Google Scholar]
- 18.Awori RM. Nematophilic bacteria associated with entomopathogenic nematodes and drug development of their biomolecules. Front Microbiol. 2022;13:993688. doi: 10.3389/fmicb.2022.993688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat AH, Chaubey AK, Askary TH. Global distribution of entomopathogenic nematodes, Steinernema and Heterorhabditis . Egypt J Biol Pest Control. 2020;30:31. doi: 10.1186/s41938-020-0212-y. [DOI] [Google Scholar]
- 20.Awori RM, Ng’ang’a PN, Nyongesa LN, Amugune NO, Masiga D. Mursamacin: a novel class of antibiotics from soil-dwelling roundworms of Central Kenya that inhibits methicillin-resistant Staphylococcus aureus . F1000Res. 2017;5:2431. doi: 10.12688/f1000research.9652.2. [DOI] [Google Scholar]
- 21.Mwaniki SW, Nderitu JH, Olubayo F, Kimenju JW, Nguyen K. Factors influencing the occurrence of entomopathogenic nematodes in the Central Rift Valley Region of Kenya. African J Ecol. 2008;46:79–84. doi: 10.1111/j.1365-2028.2008.00933.x. [DOI] [Google Scholar]
- 22.Spiridonov SE, Reid AP, Podrucka K, Subbotin SA, Moens M. Phylogenetic relationships within the genus Steinernema (Nematoda: Rhabditida) as inferred from analyses of sequences of the ITS1-5.8S-ITS2 region of rDNA and morphological features. Nematol. 2004;6:547–566. doi: 10.1163/1568541042665304. [DOI] [Google Scholar]
- 23.Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 24.Lee I, Ouk Kim Y, Park S-C, Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66:1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 25.Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 26.Meier-Kolthoff JP, Göker M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat Commun. 2019;10:2182. doi: 10.1038/s41467-019-10210-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Medini D, Donati C, Rappuoli R, Tettelin H. The Pangenome. Cham: Springer; 2020. The pangenome: a data-driven discovery in biology; pp. 3–20. [PubMed] [Google Scholar]
- 28.Shi Y-M, Hirschmann M, Shi Y-N, Ahmed S, Abebew D, et al. Global analysis of biosynthetic gene clusters reveals conserved and unique natural products in entomopathogenic nematode-symbiotic bacteria. Nat Chem. 2022;14:701–712. doi: 10.1038/s41557-022-00923-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngugi CN. Nairobi, Kenya: Faculty of Science and Technology, University of Nairobi; 2021. Characterization and Evaluation of Entomopathogenic Nematodes for the Management of Tomato Leafminer (Tuta absoluta Meyrick). PhD Dissertation. [Google Scholar]
- 30.Boemare N, Akhurst R, Stackebrandt E. In: The Prokaryotes: Volume 6: Proteobacteria: Gamma Subclass. Dworkin M, Falkow S, Rosenberg E, Schleifer KH, editors. New York, NY: Springer New York; 2006. The genera Photorhabdus and Xenorhabdus ; pp. 451–494. [DOI] [Google Scholar]
- 31.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mothupi B, Featherston J, Gray V. Draft whole-genome sequence and annotation of Xenorhabdus griffiniae strain BMMCB associated with the South African entomopathogenic nematode Steinernema khoisanae strain BMMCB. Genome Announc. 2015;3:e00785-15. doi: 10.1128/genomeA.00785-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wattam AR, Brettin T, Davis JJ, Gerdes S, Kenyon R, et al. Assembly, annotation, and comparative genomics in PATRIC, the all bacterial bioinformatics resource center. Methods Mol Biol. 2018;1704:79–101. doi: 10.1007/978-1-4939-7463-4_4. [DOI] [PubMed] [Google Scholar]
- 34.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, et al. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meier-Kolthoff JP, Auch AF, Klenk H-P, Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lefort V, Desper R, Gascuel O. FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol Biol Evol. 2015;32:2798–2800. doi: 10.1093/molbev/msv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bah T. Inkscape: Guide to a Vector Drawing Program. Prentice Hall Press; 2007. [Google Scholar]
- 39.Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, et al. Anvi’o: an advanced analysis and visualization platform for 'omics' data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pantoja Y, Pinheiro K, Veras A, Araújo F, Lopes de Sousa A, et al. PanWeb: a web interface for pan-genomic analysis. PLoS One. 2017;12:e0178154. doi: 10.1371/journal.pone.0178154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, et al. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep. 2015;5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Csurös M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics. 2010;26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 43.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 44.UniProt Consortium The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–74. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galperin MY, Kristensen DM, Makarova KS, Wolf YI, Koonin EV. Microbial genome analysis: the COG approach. Brief Bioinform. 2019;20:1063–1070. doi: 10.1093/bib/bbx117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, et al. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019;47:W81–W87. doi: 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alikhan N-F, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mothupi B. South Africa: MSc Dissertation, Faculty of Science, University of the Witwatersrand, Johannesburg; 2016. Genomics of entomopathogenic bacterial endosymbiont species associated with desiccation tolerant entomopathogenic nematode. [Google Scholar]
- 50.Kim M, Oh H-S, Park S-C, Chun J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int J Syst Evol Microbiol. 2014;64:346–351. doi: 10.1099/ijs.0.059774-0. [DOI] [PubMed] [Google Scholar]
- 51.Meier-Kolthoff JP, Klenk HP, Göker M. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol. 2014;64:352–356. doi: 10.1099/ijs.0.056994-0. [DOI] [PubMed] [Google Scholar]
- 52.Delmont TO, Eren AM. Linking pangenomes and metagenomes: the Prochlorococcus metapangenome. PeerJ. 2018;6:e4320. doi: 10.7717/peerj.4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heaps HS. Information Retrieval, Computational and Theoretical Aspects. Academic Press; 1978. [Google Scholar]
- 54.Rivera-Ramírez A, Salgado-Morales R, Jiménez-Pérez A, Pérez-Martínez R, García-Gómez BI, et al. Comparative genomics and pathogenicity analysis of two bacterial symbionts of entomopathogenic nematodes: the role of the GroEL protein in virulence. Microorganisms. 2022;10:486. doi: 10.3390/microorganisms10030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Iranzo J, Wolf YI, Koonin EV, Sela I. Gene gain and loss push prokaryotes beyond the homologous recombination barrier and accelerate genome sequence divergence. Nat Commun. 2019;10:5376. doi: 10.1038/s41467-019-13429-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fodor A, Fodor AM, Forst S. Comparative analysis of antibacterial activities of xenorhabdus species on related and non-related bacteria in vivo. J Microbiol Antimicrob. 2010;2:36–46. [Google Scholar]
- 58.Brodie G. Natural occurrence and distribution of entomopathogenic nematodes (Steinernematidae, Heterorhabditidae) in Viti Levu, Fiji Islands. J Nematol. 2020;52:1–17. doi: 10.21307/jofnem-2020-017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zepeda-Jazo I, Molina-Ochoa J, Lezama-Gutiérrez R, Skoda SR, Foster JE. Survey of entomopathogenic nematodes from the families Steinernematidae and Heterorhabditidae (Nematoda: Rhabditida) in Colima, México. Int J Trop Insect Sci. 2014;34:53–57. doi: 10.1017/S1742758413000416. [DOI] [Google Scholar]
- 60.Varghese NJ, Mukherjee S, Ivanova N, Konstantinidis KT, Mavrommatis K, et al. Microbial species delineation using whole genome sequences. Nucleic Acids Res. 2015;43:6761–6771. doi: 10.1093/nar/gkv657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci. 2005;102:2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koressaar T, Remm M. Characterization of species-specific repeats in 613 prokaryotic species. DNA Res. 2012;19:219–230. doi: 10.1093/dnares/dss006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Klassen JL, Currie CR. Gene fragmentation in bacterial draft genomes: extent, consequences and mitigation. BMC Genomics. 2012;13:14. doi: 10.1186/1471-2164-13-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.