Chi3l1 is a modulator of glioma stem cell states that can be targeted to promote differentiation and suppress growth of glioblastoma.
Abstract
Chitinase 3–like 1 (Chi3l1) is a secreted protein that is highly expressed in glioblastoma. Here, we show that Chi3l1 alters the state of glioma stem cells (GSC) to support tumor growth. Exposure of patient-derived GSCs to Chi3l1 reduced the frequency of CD133+SOX2+ cells and increased the CD44+Chi3l1+ cells. Chi3l1 bound to CD44 and induced phosphorylation and nuclear translocation of β-catenin, Akt, and STAT3. Single-cell RNA sequencing and RNA velocity following incubation of GSCs with Chi3l1 showed significant changes in GSC state dynamics driving GSCs towards a mesenchymal expression profile and reducing transition probabilities towards terminal cellular states. ATAC-seq revealed that Chi3l1 increases accessibility of promoters containing a Myc-associated zinc finger protein (MAZ) transcription factor footprint. Inhibition of MAZ downregulated a set of genes with high expression in cellular clusters that exhibit significant cell state transitions after treatment with Chi3l1, and MAZ deficiency rescued the Chi3L-induced increase of GSC self-renewal. Finally, targeting Chi3l1 in vivo with a blocking antibody inhibited tumor growth and increased the probability of survival. Overall, this work suggests that Chi3l1 interacts with CD44 on the surface of GSCs to induce Akt/β-catenin signaling and MAZ transcriptional activity, which in turn upregulates CD44 expression in a pro-mesenchymal feed-forward loop. The role of Chi3l1 in regulating cellular plasticity confers a targetable vulnerability to glioblastoma.
Significance:
Chi3l1 is a modulator of glioma stem cell states that can be targeted to promote differentiation and suppress growth of glioblastoma.
Introduction
Glioblastoma is the most prevalent and aggressive primary brain tumor. Standard treatment of glioblastoma includes surgical resection followed by radiation in the vicinity of the resection cavity and administration of temozolomide (1). Even with this therapeutic approach, tumor recurrence is inevitable (2) mainly due to the presence of glioma stem cells (GSC), which are characterized by high migratory potential (3), resistance to chemotherapy and radiation and the ability to form recurrent tumors (4). GSCs exhibit remarkable plasticity and dynamically transition between more differentiated and less differentiated states via mechanisms akin to cellular reprogramming (5). These mechanisms of GSC plasticity have led to examination of factors that might influence tumor propagating potential and tumor recurrence. One such concept relates to the role of tumor microenvironment to induce cellular reprogramming that results in generation of more stem-like cancer cells with the ability to propagate malignancy (6). Although each glioblastoma contains cells in multiple states, plasticity between states and the potential of a single cell to generate multiple transcriptomic and phenotypic states has been demonstrated (7). From all glioma cell phenotypes, the mesenchymal is associated with worst clinical outcomes and the main phenotypic markers expressed in mesenchymal GSCs are CD44 and chitinase 3–like protein 1 (Chi3l1; ref. 7).
Chi3l1, also known as YKL40, is a member of an evolutionary conserved 18-glycosyl-hydrolase protein family that was originally discovered in murine mammary adenocarcinoma cells as a secretory protein (8). Chi3l1 is secreted from a variety of cells including macrophages, neutrophils, epithelial, endothelial, synovial cells as well as cancer cells. Studies have demonstrated that circulating levels of Chi3l1 are increased in many malignancies, including prostate, colon, ovary, kidney, breast, glioblastoma, malignant melanoma, and lung cancer. High levels of Chi3l1 frequently associate with poor prognosis and mortality in these cancers (9–13). In glioblastoma, Chi3l1 is a key marker for identification of the mesenchymal subtype and has been implicated its increased expression in oncogenic pathways involving the NF-κB RelB and STAT-3/RTVP-1 signaling systems that are known to drive mesenchymal differentiation (14, 15).
Despite these findings, the role of Chi3l1 as paracrine modulator of GSC states and the transcriptional regulatory network induced by Chi3l1 in GSCs remain unknown.
Here, using single-cell RNA sequencing (scRNA-seq) and RNA velocity analysis, we show that incubation of patient-derived GSCs with recombinant Chi3l1 induces significant transitions of cells towards clusters with mesenchymal transcriptomic profiles. Chi3l1 significantly reduces the probability of GSCs to transition towards terminal transcriptomic states. This suggests that a function of Chi3l1 is to modulate cellular plasticity and to restrict cell commitment. Assay for transposase-accessible chromatin using sequencing (ATAC-seq) of Chi3l1-treated GSCs reveals targeted chromatin remodeling, increased accessibility on promoters enriched for the Myc-associated zinc finger protein (MAZ) motif and an increase in MAZ transcription factor (TF) footprinting. Knockdown of MAZ in GSCs results in inhibition of Chi3l1-induced genes that exhibit the highest expression in GSC cellular clusters undergoing cell state transitions. In vivo sustained local treatment of human glioblastoma xenografts with a blocking anti-Chi3l1 antibody results in inhibition of glioblastoma growth by more than 60%. Finally, human glioblastoma xenografts treated with anti-Chi3l1 show significant reduction of the mesenchymal transcriptomic signature. Our work implicates Chi3l1 as paracrine modulator of GSC cellular states and demonstrates preclinical efficacy of our anti-Chi3l1 antibody to reduce glioblastoma tumor burden.
Materials and Methods
Isolation and culture of patient-derived GSC cultures
The Institutional Review Board (IRB) of Rhode Island Hospital has approved the collection of patient-derived glioblastoma multiforme tissue. All collections were performed with written informed consent from patients in completely deidentified manner and the studies were performed in accordance to recognized ethical guidelines (Belmont Report). Primary GSC spheres were cultured from human glioma samples as previously described (16). GSCs used in this study were authenticated by ATCC using short tandem repeat analysis. GSCs used were between passages 5 and 30 and cultured either as spheres or as attached on fibronectin-coated plates (10 μg/mL; Millipore Sigma) in a medium of 1X neurobasal medium, B27 serum-free supplement, minus vitamin A, 100X Glutamax (Thermo Fisher Scientific), 1 mg/mL Heparin (STEMCELL Technologies), 20 ng/mL epidermal growth factor (Peprotech), 20 ng/mL bFGF (Peprotech). All GSC cultures are routinely tested for Mycoplasma contamination using the MycoSensor qPCR assay (Agilent).
Mouse studies
All animal experiments were approved by Rhode Island Hospital's Institutional Animal Care and Use Committee and conformed to the relevant regulatory standards and overseen by the IRB. Nine-week-old female and male NU/J homozygous mice were obtained from Jackson Laboratory (RRID:IMSR_JAX:002019) and used for the stereotactic injections and subcutaneous injections. Animals were housed together until the day before the surgery and then housed individually. Animals receiving treatment were randomly assigned.
Flow cytometry, Western blot, immunoprecipitations, NanoString expression analysis, bulk RNA sequencing (RNA-seq) analysis, anti-Chi3l1 antibody characterization, and Methods for stereotactic implantation and MRI imaging are described at Supplementary Data.
ATAC-seq
Sample preparation
ATAC-seq libraries were prepared according to the protocol established and optimized before (17). Specifically, cell lysis was performed using 50 μL of cold lysis buffer [resuspension buffer, 0.1% NP-40, 0.1% Tween-20, and 0.01% Digitonin (Promega)], and transposition of the resulting nuclei pellet from the lysate by Tn5 transposase was accomplished using the Nextera DNA Library Prep Kit (Illumina). DNA isolation from the transposition reaction was done using MinElute Reaction Cleanup Kit (Qiagen), and the library was generated from the isolated DNA through PCR amplification protocol. Purification of the library for optimal library preparation included removal of primer dimers and DNA fragments larger than 1,000 base pairs (bp) through double-sided bead purification with Agencourt AMPure XP beads (Beckman Coulter). Sequencing of the final library was performed by GENEWIZ using the Illumina HiSeq 2500 system.
ATAC-seq FASTQ alignment, BAM preprocessing, and peak calling
FASTQ files of sequencing reads for each sample were filtered, deduplicated, and aligned to the GRCh38/hg38 human reference genome using the paired-end alignment feature. BAM files from FASTQ alignment were sorted, indexed, and isolated for uniquely mapped reads using the SAMtools. Finally, peak calling was performed using Genrich with a FDR adjusted P value significance threshold of 0.05 (-q 0.05) and a minimum peak length of 200 bp (-l 200). Consensus peak set was derived for each group by performing individual peak regions based on Fisher combined probability test.
ATAC-seq analysis
ATAC-seq data analyses were performed using R. The processing of the narrowPeak files into GRanges object was done using readPeakFile. Peaks were annotated to “TxDb.Hsapiens.UCSC.hg38.knownGene” database and visualized with plotAvgProf2.
Accessibility changes in promoters following addition of Chi3l1
To identify differentially accessible chromatin regions, sequence read counts of each peak range in the consensus peakset for each sample were calculated with featureCounts. The resulting counts matrix was then converted into a DESeq data set and normalized via median-by-ratio normalization (MRN) for differential chromatin accessibility analysis by DESeq2 (18). Differentially accessible chromatin regions were defined as peak regions with a normalized count fold change greater than 1.5 and a P value smaller than 0.05. Differentially accessible chromatin regions were then annotated to promoters as followed: 1,000 bp upstream and downstream of a transcript start site; the flanking radius to search beyond the promoter was 3,000 bp upstream and downstream.
Motif enrichment and TF binding analysis
Motif enrichment within highly accessible promoter sites was performed using the HOMER motif analysis package with the following parameters: “hg38 -len 8,10,12 -mask -size given.” Both de novo and known motif enrichment results were considered.
Three platforms were used for consensus differential activity analysis: the Differential ATAC-seq Toolkit (DAStk), Hmm-based IdeNtification of Transcription factor footprints (HINT-ATAC) and Transcription factor Occupancy prediction by investigation of ATAC-seq signal (TOBIAS; refs. 19–21). Enriched motifs were probed for differential TF activity inference using DAStk differential MD score. MD scores were obtained by assessing the density of biologically active motifs active at enhancer sites (eRNA). Motif activity and hence TF binding probability was defined by the degree of motif colocalization with an enhancer RNA origin as previously described (22). To detect footprints, TOBIAS and HINT-ATAC were independently ran with, respectively, JASPAR CORE motif set and HOCOMOCO.
scRNA-seq
Sample preparation
GSCs were treated for a week with or without Chi3l1 (500 ng/mL). scRNA-seq libraries were generated using the Chromium Next GEM Single Cell 3′ GEM, Library & Gel Bead Kit v3.1, Chromium Single Cell 3′, Dual Index Kit TT Set A, Chromium Next GEM Chip G, and 10× Chromium Controller (10× Genomics) as per 10× Single Cell 3′ protocol. The sequencing was done by the Molecular Biology Core Facilities at Dana-Farber Cancer Institute using Illumina NovaSeq SP Flowcell (800M Reads) as well as the raw data processing. CellRanger count outputs were provided.
Analysis of scRNA-seq data
Data integration and clusters determination
Using 10x Genomics platform, GSC1/GSC2, 8000/7015 cells, and 7112/8961 cells passed the sequencing threshold quality control for the non-treated/control and Chi3l1 treated samples, respectively. Then, low complexity cells (<1,000 genes) and cells with >12% mitochondrial genomic content were excluded as previously described for glioblastoma (23), resulting in a final population of 6,102/5,892 and 4,060/7,558 cells for control and Chi3l1 samples for GSC1/GSC2, respectively.
Seurat (24) was used for downstream analysis. First each library was converted into a Seurat object using read10x and makeseuratobject. Data was normalized using SCTransform method then objects were integrated along identified anchors to identify cell types across conditions. To determine optimal parameters for dimensional reduction and evaluate cluster stability, Jaccard similarity index was applied involving a snakemake pipeline to scatter and gather Seurat object by subsampling the cells and repeat for multiple times as described in (25) with the following parameters: subsample rate: 0.8; number of subsample: 100; subsample ks: 8, 10, 12, 14, 16, 20, 30, 50, 80, 100; subsample resolutions 0.5, 0.6, 0.8, 1, 1.2, 1.5; subsample pcs: 20, 30, 40, 50.
Cells were then classified on the basis of top expressed genes and functional pathway enrichment. First, the cluster defining genes across conditions were identified using the FindConservedMarkers. Second, the most highly expressed genes per cluster were extracted from the Seurat top ranked “identity.” Third, MSigDB cancer hallmark geneset was used in enrichr platform for cluster specific functional enrichment analysis.
RNA velocity and trajectory inference
RNA velocity analysis was performed using both velocyto and scVelo. First, the cellranger count output was preprocessed using the velocyto command-line tool with default parameters specific for 10x Genomics data (26). The .loom files generated were filtered for complexity and mitochondrial content and were then loaded to scVelo (27) for downstream RNA velocity analysis. The data was preprocessed by selection the top 2,000 variable genes with minimum shared counts of at least 20 and log normalized for cell size. RNA velocity was estimated using the dynamical mode and projected unto anchored Seurat derived UMAP embeddings.
CellRank was used to predict overall cell state trajectories and cell–cell transition probabilities. Genes were ranked on the basis of likelihood scores and assigned a percentile rank. Differential ranking analysis was performed, cluster by cluster, by calculating the difference percentile rank between Chi3l1 versus control. EnrichR (MSigDB) was queried for pathways enrichment. Furthermore, a method was developed to characterize transition probabilities between cluster (Supplementary Data). Generalized Perron Cluster Analysis (28) was used to identify initial and terminal states as well as single-cell probabilities toward terminal states.
General statistical analysis methods
Our goal is to obtain results with greater than 95% confidence level. Assuming that data were normally distributed and that the standard deviation for measurements was no more than 3⁄4 of the mean, the t test of mean was used to estimate the number of required observations. For FACS, we used n = 4 biological replicates, for NanoString analysis using GSCs, we used n = 3 biological replicates and GSCs from two patients. For MRI quantification we used n = 8 animals per group, while for NanoString in vivo analysis we used n = 5 IgG control treated mice and n = 4 anti-Chi3l1 treated mice.
Age-matched female and male mice were randomly used for in vivo experiments and the quantification of tumor area was performed blindly in regard to treatment group assignment by two independent researchers.
To determine significance among the means of three or more independent groups, we used one-way ANOVA. The homogeneity of variances was confirmed with Brown and Forsythe test, and the significance between specific groups was calculated with a post hoc Dunnett test. To determine significance among the means of two independent groups, we performed an unpaired two-tailed t test. To verify Gaussian distribution of data before applying the t test, we performed the D'Agostino and Pearson and Shapiro-Wilk normality tests.
Data availability
Publicly available data generated by others were used by the authors. The data analyzed in this study were obtained from The Cancer Genome Atlas (TCGA) at
https://gdac.broadinstitute.org/runs/stddata__latest/samples_report/GBM.html
https://gdac.broadinstitute.org
The data discussed in this manuscript have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE154060 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154060).
High throughput microarray data (NanoString) and all other raw data are available upon request from the corresponding author.
Results
Chi3l1 induces expression of mesenchymal markers in proneural GSCs
GSCs from patients with IDH wild-type primary glioblastoma have been characterized by RNA-seq and shown to recapitulate the patient's tumor in orthotopic xenografts (16, 29).
To determine whether the presence of Chi3l1 in the microenvironment of GSCs induces phenotypic alterations, we cultured GSCs in the presence or absence of recombinant Chi3l1 for 7 days and performed Flow Cytometry analysis for the expression of markers that define the mesenchymal and proneural phenotype (30). We show that incubation of GSCs with Chi3l1 induces significant increase in CD44+/ Chi3l1+ and Chi3l1+ expression and concomitant significant decrease in the population of GSCs expressing SOX2+/CD133+ (Fig. 1A). The significant increase in CD44 expression following incubation of GSCs with Chi3l1 was also confirmed by Western blot (Fig. 1B). To evaluate bulk transcriptomic changes following exposure of GSCs to Chi3l1, we performed differential gene expression analysis using the NanoString PanCancer Progression Panel. This returned 53 genes with significantly increased expression and 12 genes with significantly decreased expression in the Chi3l1-treated set (Supplementary Fig. S1A). To determine whether these upregulated genes were correlated with a mesenchymal phenotype, we used the TCGA RNA-seq data for cross-validation and found significant correlation with mesenchymal gene enrichment (Supplementary Fig. S1B).
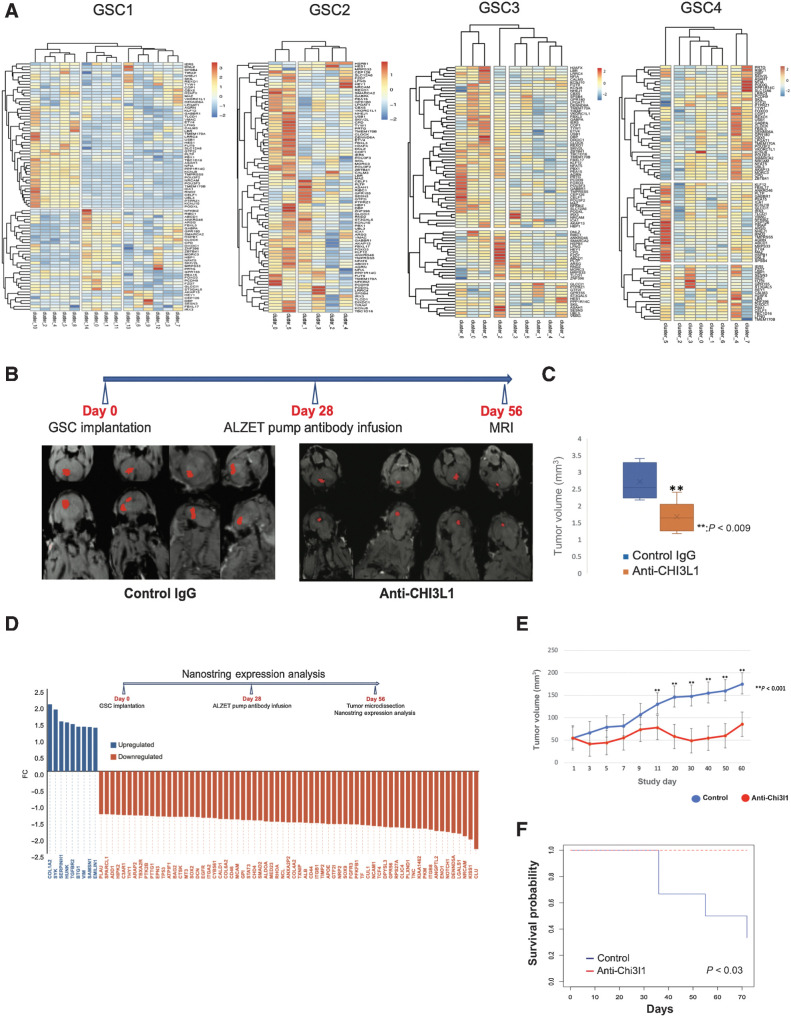
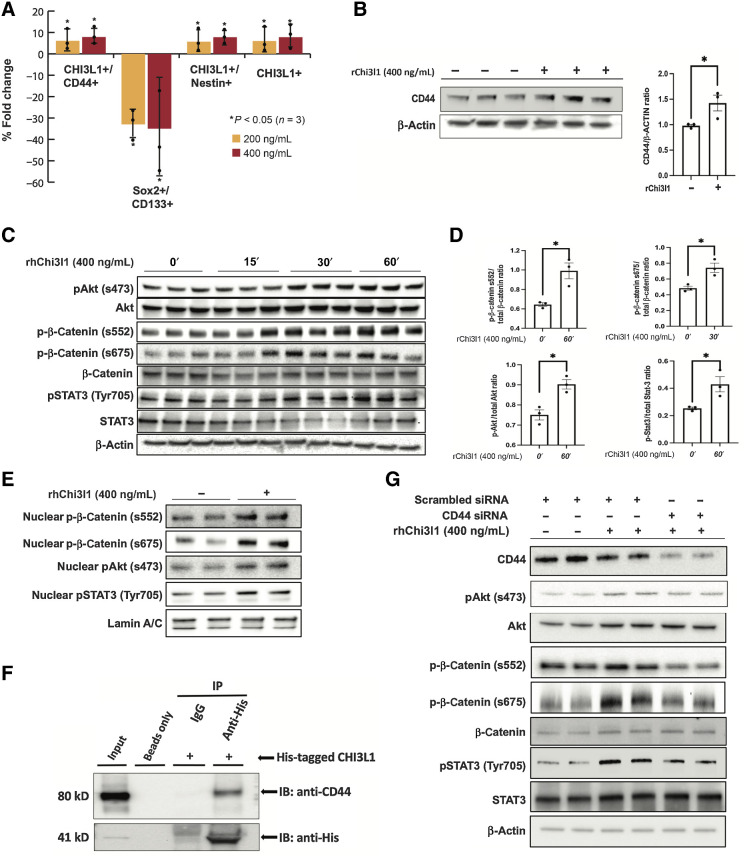
Figure 1.
Chi3l1 interacts with CD44 and induces phosphorylation and nuclear translocation of Akt, β-catenin, and STAT3. A, Percentage of fold change of mesenchymal and proneural markers in GSCs following Chi3l1 treatment quantified by flow cytometry. n = 4. B, Left, Western blot analysis of CD44 in GSCs treated with or without Chi3l1. Actin was used as a loading control. Right, densitometric analysis of the Western blot results showing that GSCs treated with ChiL3 exhibit significant higher expression of CD44. The results are normalized to the expression of actin and are presented as mean ± SD from three independent experiments. C and E, Western blot analysis showing that incubation of GSCs with Chi3l1 induces significant phosphorylation and nuclear translocation of Akt (s473), β-catenin (s552 & s675), and STAT3 (Tyr705). Actin (C) or lamin A/C (E) were used as a loading control. D, Densitometry quantification of phosphorylation of Akt (s473), β-catenin (s552 & s675), and STAT3 (Tyr705). Results are presented as mean ± SD from three independent experiments. F, Protein lysates of GSCs (Input) were immunoprecipitated with beads only, His-tagged antibody, and isotype-matched IgG. Western blot was performed using CD44 antibody, which showed that Chi3l1 interacts directly and coprecipitates with CD44. G, Western blot analysis of phosphorylation of Akt(s473), β-catenin (s552 & s675), and STAT3 (Tyr705) in GSCs with siRNA targeting negative control (scrambled siRNA) or CD44 and treated with or without Chi3l1 showing that knockdown of CD44 inhibits the Chi3l1-induced phosphorylation of Akt, β-catenin, and STAT3. Significance was calculated with Student t test. *, P < 0.05.
Chi3l1 interacts with CD44 and induces phosphorylation and nuclear translocation of akt, β-catenin, and STAT3
In glioblastoma cells, Chi3l1 has been shown to interact with IL13Ra2 and induce Erk1/2 and Akt phosphorylation (31). We show that incubation of GSCs with Chi3l1 induces significant phosphorylation and nuclear translocation of Akt (s473), β-catenin (s552 & s675), and STAT3 (Tyr705; Fig. 1C–E; Supplementary Fig. S1C). Chi3l1 interacts directly and coprecipitates with CD44 in GSCs (Fig. 1F). Knockdown of CD44 with small interfering RNAs (siRNA) inhibits the Chi3l1-induced phosphorylation of Akt, β-catenin, and STAT3, suggesting that CD44 is the receptor that mediates this Chi3l1 induced signaling cascade in GSCs (Fig. 1G; Supplementary Fig. S1C).
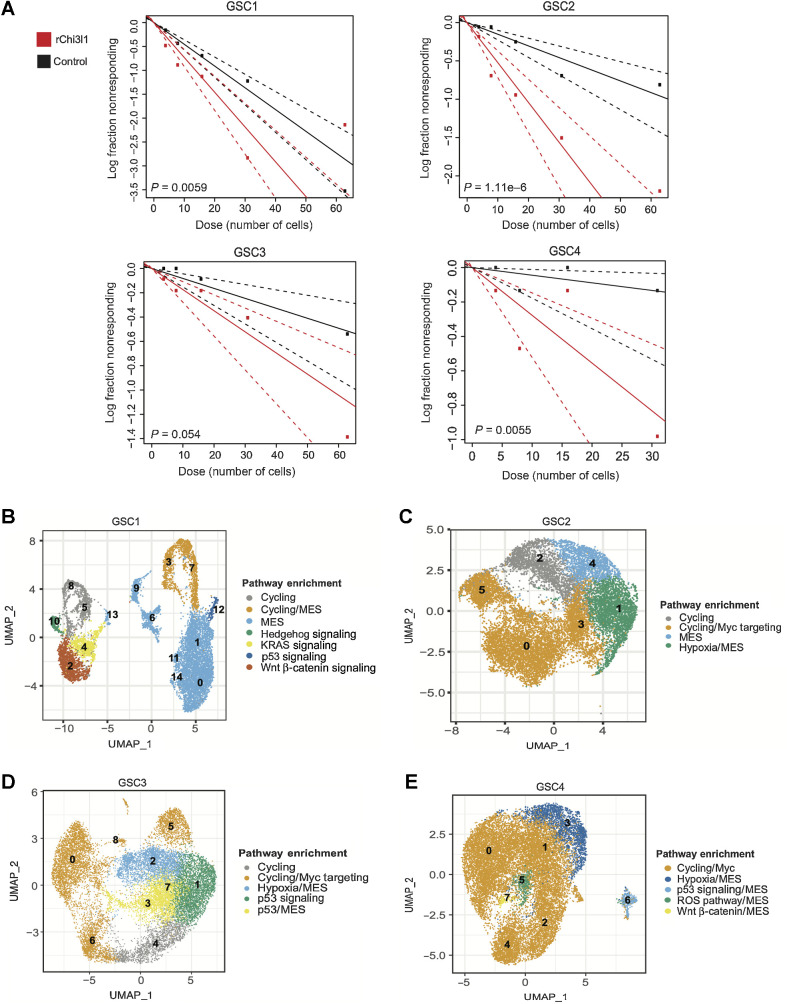
Because the mesenchymal phenotype has been linked to higher “stemness” potential, we examined whether culturing of GSCs in the presence of Chi3l1 affects the ability of GSCs to self-renew. We performed in vitro limiting dilution analysis (32) for 14 days with and without the addition of Chi3l1 using 4 patient-derived GSCs and found that Chi3l1 significantly increases their frequency of self-renewal (Fig. 2A).
Figure 2.
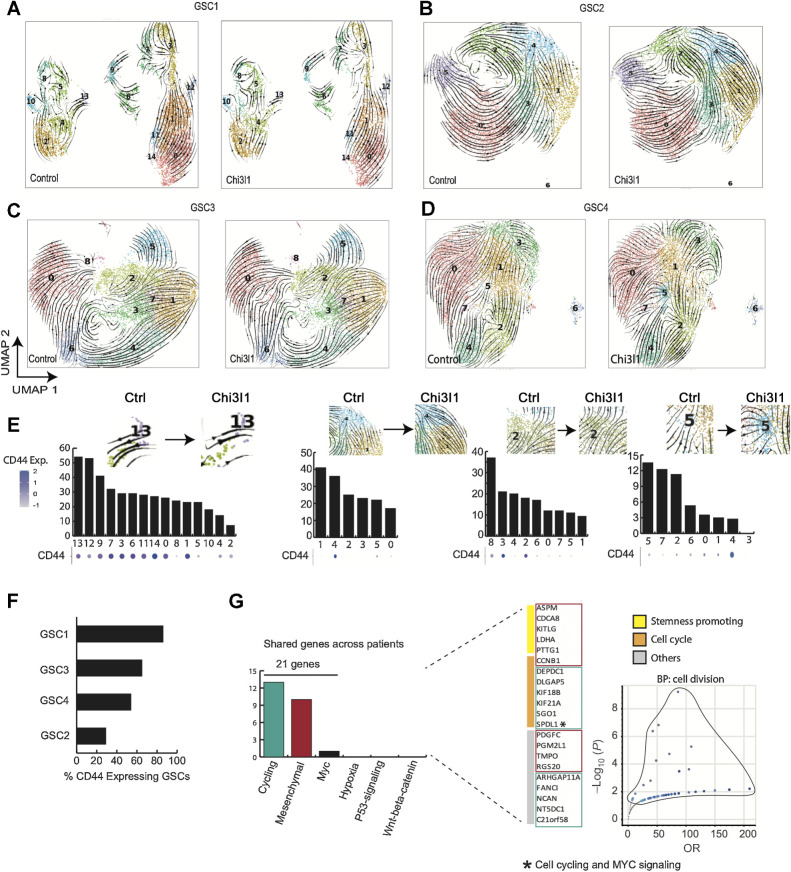
Functional role of Chi3l1 on self-renewal and corresponding single-cell characterization of GSCs subpopulations. A, Quantification of self-renewal changes across multiple GSCs following 14 days of incubation with recombinant Chi3l1. B–E, UMAP of GSCs colored by the most significantly enriched pathways for GSC1 through GSC4. These projections represented integrated control and Chi3l1-treated single-cell data per GSC. The clusters were anchored to facilitate cross condition comparison between cells of the same functional identity.
GSC single-cell characterization
To determine the effect of Chi3l1 on GCSs at the single-cell level, we applied scRNA-seq on 4 patient-derived GSCs treated with or without Chi3l1 for 7 days using the 10x Genomics Chromium platform. Following data integration and anchoring to allow for direct comparisons, we sought to identify stable clusters using the Jaccard similarity index pipeline (25). Jaccard indexing is a resampling method in which 80% of cells are resampled from the population and re-clustered. Clusters with a mean stability score greater than 0.85 are highly stable while clusters lower than 0.6 are considered unstable (25). For GSC1 using k.param of 12, resolution of 0.6 and PC of 50 provides 95% of all cells within 15 highly stable clusters. For GSC2, the optimal parameters were k.param of 50, resolution of 0.6 and PC of 50, yielding in 7 stable clusters. For GSC3 and GSC4, the parameters were k.param of 80, resolution of 0.6 and PC of 30; and k.param of 80, resolution of 0.6 and PC of 50, yielding 8 and 9 clusters, respectively (Supplementary Fig. S2A–S2H).
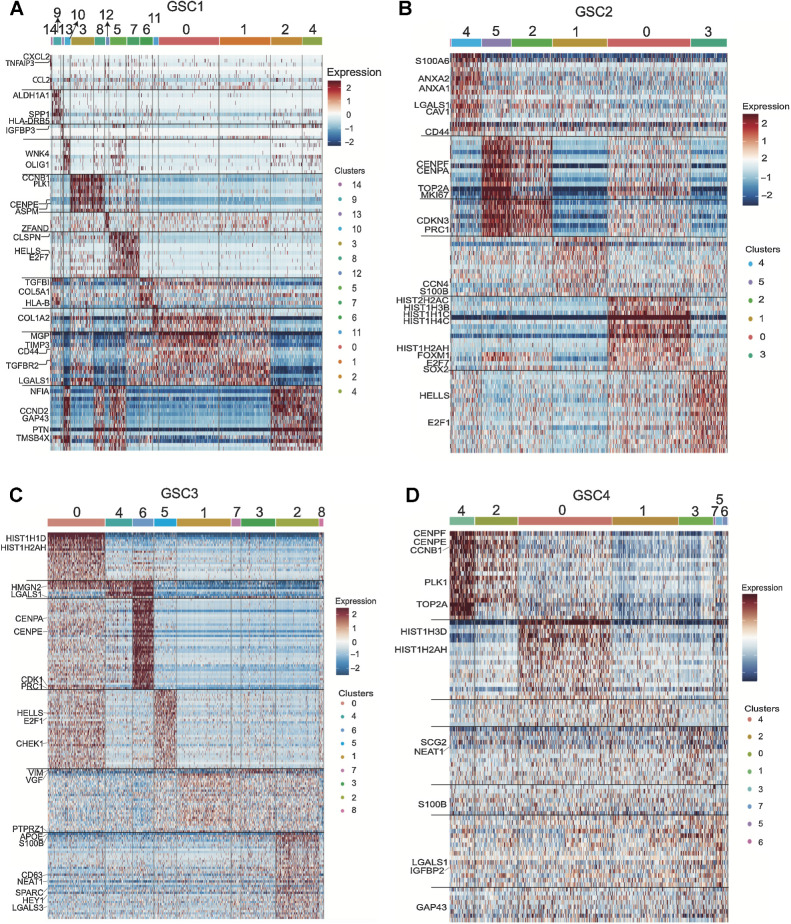
Next, we performed functional enrichment analysis to characterize each cluster. GSC1 shows enrichment for six major pathways including: epithelial–mesenchymal transition (EMT; clusters 0, 1, 6, 9, 11, 13, 14, 7, and 3), Hedgehog signaling (cluster 10), KRAS signaling (clusters 4), p53 pathway (cluster 12), cell cycling (clusters 3, 5, 7, 8) and WNT β-catenin signaling (cluster 2; Fig. 2B). Notably, clusters 10, 2 and 4 express OLIG1, CCND2, PTN, GAP43, and TMSB4X. Clusters 6, 14, 1, 0, and 11 show higher expression of CD44, TGFB1, COL1A2, TFGBR2, TIMP3, SERPINF1, MGP, LGALS1, and S100A6, which are considered mesenchymal markers. Clusters 3, 8, 5, and 7 display expressions of CCNB1, PLK1, CENPE, PRC1, ASPM, and CDKN3. Finally, cluster 13 exhibits a unique profile with expression of IGFBP3, SERPINE1, HLA-DRB5, SPP1, SCG2, and TGFBI (Fig. 3A). GSC2 is predominantly enriched for cell cycling (clusters 2, 5, 0, and 3) with high expression of genes such as FOXM1, followed by MYC targeting (clusters 0, 3, and 5), and EMT pathways (clusters 4 and 1; Fig. 2C). Interestingly, clusters 2, 3, 0, and 5 are also enriched for SOX2. Cluster 4 highly expresses genes such as ANXA1, CD44, LGALS1, and CAV1, while cluster 1 expresses WISP1 and the astro-mesenchymal marker S100B (Fig. 3B). GSC3 is enriched for cell cycling (cluster 4) and cycling cells that are also significantly enriched for MYC targeting (clusters 0, 5, 6, 8). In addition, GSC3 contains two EMT enriched subpopulations, one enriched for p53 signaling (clusters 3 and 7) and the other for hypoxic pathways (cluster 2; Fig. 2D). Clusters 3 and 7 exhibit distinctly high expression levels of VIM and VGF compared with any other clusters and cluster 2 comparatively expresses high levels of APOE and S100B (Fig. 3C). Finally, cluster 1 exhibits p53 signaling pathway enrichment (Fig. 2D). Finally, GSC4 is predominantly constituted of cycling cells that are enriched for MYC targets (cluster 0 to 4) and a subset of mesenchymal GSCs enriched for the EMT pathway. This minor subset of mesenchymal cells displays remarkable heterogeneity with specific enrichment for hypoxic pathway (cluster 3), p53 signaling (cluster 6), reactive oxygen species pathway (cluster 5), and WNT β-catenin pathway (cluster 7; Fig. 2E). We noted significant expression of SCG2 in cluster 3 and high expression of LGALS1 across this small subset of mesenchymal cells in GSC4, but not in cluster 6 (Fig. 3D).
Figure 3.
Single-cell characterization of the transcriptomic profile of GSCs displays the heterogeneity inherent to glioblastoma. A–D, Characterization heatmaps showing cluster-specific gene expression profiles across four patient-derived GSCs. The characterization was achieved by using integrated single-cell data across conditions per GSC and identifying cluster-specific genes/markers.
Finally, we confirmed that our classification of cell clusters with mesenchymal phenotype agrees with the Verhaak classification of mesenchymal GSCs and separates these cells from proneural cells (PN) in our GSC samples (Supplementary Fig. S5). In addition, we collected all cluster-defining genes across all GSCs to identify conserved identity genes. We found 76 genes that reproducibly define GSC subpopulation identity (Supplementary Table S1). To achieve this, we collected genes that were conserved in 3 or more of the GSCs. The 76 genes are involved in cell cycle, stemness regulation, epigenetic regulation, DNA repair, and ubiquitination. Amongst these genes, LGALS1 and S100B were remarkably conserved across all GSCs in mesenchymal subpopulations. (Supplementary Table S2). Interestingly, we also discover that the long noncoding RNA NEAT1 is conserved across GSCs and is highly expressed in mesenchymal GSCs (Supplementary Table S2).
RNA velocity predicts significant cell state transition changes following Chi3l1 exposure
To observe the effect of Chi3l1 on the transcriptomic state of GSCs, we employed RNA velocity analysis (the change in mRNA abundance) in order to predict cell state trajectories. To accomplish this, we used a likelihood-based dynamical model generalized to transient transcriptomic cell states. RNA velocity fields show velocity trends specific to each patient-derived GSC. In GSC1, we observe pronounced RNA velocity field changes following Chi3l1 exposure, suggesting treatment induced cell state transition changes. There are striking directionality changes in RNA velocities in the cycling cluster 5 and KRAS/EMT/IGFBP3 enriched cluster 13. Cluster 13 transitions away from KRAS enriched cluster 4 and towards EMT enriched populations. In addition, we notice transition changes in the KRAS enriched population (cluster 4), the WNT β-catenin (cluster 2) and EMT enriched population (cluster 9 and 6; Fig. 4A). In GSC2, prior to Chi3l1 exposure we observe a general state transition from the initial cluster 2 towards cluster 5. However, after exposure of GSC2 to Chi3l1, the velocity field becomes less linear/direct, suggesting a disruption of the GSC2 cell–cell state transition. We note a striking increase in transition towards mesenchymal populations, cluster 4 and cluster 1 (Fig. 4B). For GSC3, the state transitions experience visible vector changes at the center of the UMAP projection, at the mesenchymal enriched cluster 2 and 3 (Fig. 4C). For GSC4, we note significant changes in the mesenchymal clusters 7, 5, and 3 and major general increase in vector transition stochasticity. In addition, there is striking increase transition towards the hypoxia enriched mesenchymal cluster 3 (Fig. 4D).
Figure 4.
RNA velocity predicts significant cell state transition changes following Chi3l1 exposure. A–D, Velocity fields derived from the dynamical model (scVelo) are visualized as streamlines and projected unto UMAP embedding of anchored cells. Cells are colored by predefined clusters. E, Detection of driver genes. Genes were ranked on the basis of likelihood scores and assigned a percentile rank. Perturbation percent/fraction refers to the fraction of the top quartile driver genes that are displaced as top likelihood ranking genes following Chi3l1 exposure. F, Percentage of cells expressing CD44 per GSC. G, Driver genes that have reproducibly experienced significant changes across GSCs following incubation with Chi3l1. Genes are classified by pathway enrichment and biological function. Finally, biological process enrichment of the conserved, perturbed driver genes shows significant enrichment for cell division.
To further supplement these observations, we developed an analytical pipeline (Supplementary Methods) for unbiased statistical comparison of cell transition changes across conditions. The first approach is a rank-based method, while the second employs a cluster to cluster pairwise transition probability distribution comparison. These methods were developed to allow for: (i) the quantification of significant cell state transition changes across conditions, and (ii) to identify gene dynamics that drive these state changes.
Chi3l1-induced state transition dynamics are driven by changes in genes enriched for mesenchymal and stemness pathways
To evaluate our rank-based analysis, we assessed the concordance between observed velocity field changes and the percentage of top quartile genes that have been perturbed after Chi3l1 referred as percent driver gene perturbation. We established that this percentage was consistent with velocity map changes. For example, in GSC1, cluster 13 and 5, which experience the most significant vector changes, exhibit the highest percent perturbation. This is observed in mesenchymal clusters 1 and 4 for GSC2 as well (Fig. 4E). In GSC3, the same pattern is noted for mesenchymal clusters 2 and 3 (Fig. 4E). And finally, GSC4 follows the same pattern with mesenchymal clusters 5 and 7 experiencing the greatest perturbation in concordance with significant vectorized changes (Fig. 4E). We also note a general correlation of higher perturbation in state transition driver genes with the levels of CD44 expression (Fig. 4E and F), which could connect the proposed role of CD44 as the receptor for Chi3l1 functions (Fig. 1). Finally, to further probe Chi3l1 specific effects, we selected for driver genes that experienced a reproducible perturbation in at least 2 GSCs following treatment. We identify 21 such genes across the GSCs. We find that these genes belong to the mesenchymal and cycling transcriptomic states and play a key role in stemness and cell cycling processes (Fig. 4G).
Chi3l1 increases transition towards mesenchymal-enriched GSC states
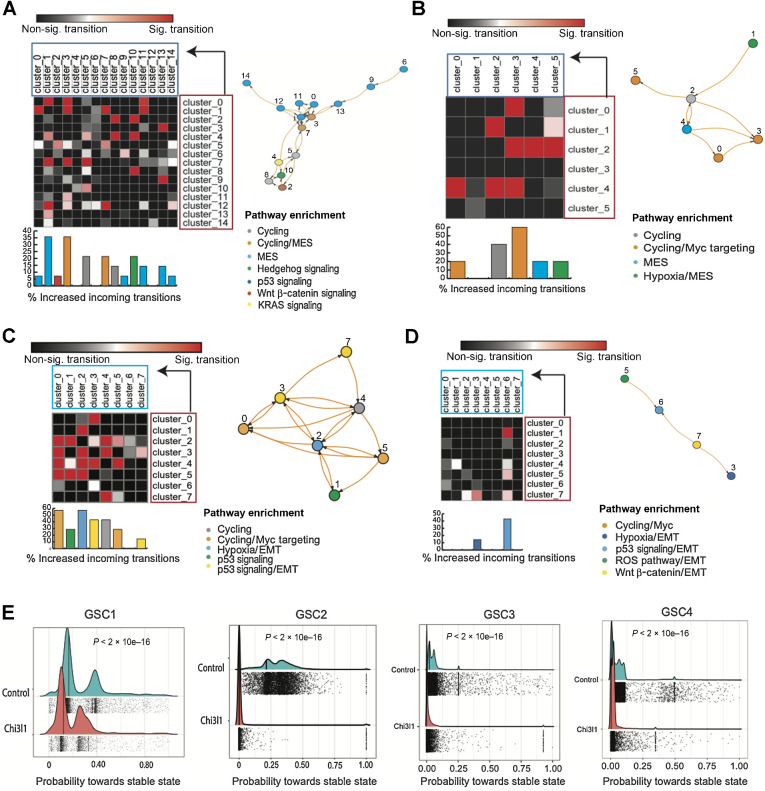
In GSC1, there is a notable increase in GSC state transition towards mesenchymal (MES) enriched clusters, particularly towards cluster 1 and cluster 3. These clusters exhibit the highest scores for significant increase in incoming transitions following Chi3l1 (about 40% of all transitions are significant). Together, CD44/MES enriched clusters 1 and 3 account for more than 1/3 of all significant increase in incoming transition (11 of 28 total significant increase in transitions in Chi3l1 relative to control; Fig. 5A). Furthermore, we also note an equal, marked increase in transition towards MES enriched cluster 7, cycling cluster 5 and hedgehog enriched cluster 10 (Fig. 5A). For GSC2, we report an increase in transition towards the MES enriched cluster 4. In addition, a notable transition towards cycling/MYC enriched cluster 3 is observed (Fig. 5B). In GSC3, we note the most significant increased transition towards the hypoxia enriched mesenchymal cluster 2 as well as the MYC targeting enriched cluster 0 (Fig. 5C). For GSC4, the most notable increase in transition is directed towards the mesenchymal clusters 6 and 3 (Fig. 5D). However, GSC4 interestingly had the least increase in transition changes in general given the simplicity of the network state transition model.
Figure 5.
Chi3l1 increases transition towards mesenchymal GSC states and decreases transition towards stable states. A–D, Matrices of pairwise cluster-to-cluster transition probability changes. Here is only shown cluster-to-cluster transitions that experience an increase in transition probability after exposure of GSCs to Chi3l1. Arrow, transition order from an initial cluster/state (crimson) to final state (turquoise). The increase in transition that was statistically significant is colored in the red/pink scale, while the nonsignificant ones are in the black/gray scale (P < 0.05, Mann–Whitney U Test). In addition, road map visualization of significant cluster-to-cluster increase in transitions following Chi3l1 treatment as a nodes and edges network. E, Plot shows significantly decreased probabilities towards terminal states following Chi3l1 exposure across all GSCs.
Chi3l1 decreases transition towards stable states
RNA velocity data suggest that Chi3l1 exposure induces significant changes in GSCs state transition. To better characterize the role of Chi3l1 in cell state dynamics and supplement our previous findings, we applied a Markov-based transition probability model (CellRank; https://www.biorxiv.org/content/10.1101/2020.10.19.345983v2.full) to our RNA velocity data. This trajectory inference model uses RNA velocity to identify “initial”/“terminal” transcriptomic states and assigns each cell a fate probability of assuming a “terminal” transcriptomic state (Fig. 5E). Initial states refer to cell states that are predicted to be the founding or precursor populations and therefore have the lowest incoming transition probabilities. Conversely, terminal states, which can be thought of as “attractor” states, refer to states with highest self-transition probabilities, in other words states that are most readily assumed by the cells (33). First, we identify initial and terminal transcriptomic states. Next, we examined transition probabilities towards terminal transcriptomic states. We found significantly lower probabilities towards terminal states following Chi3l1 exposure (Fig. 5E) for all patient-derived GSCs, suggesting a general loss of GSC commitment towards these attractor/terminal transcriptomic cellular states. In conclusion, these data suggest that Chi3l1 induces GSCs to acquire a poised, more plastic/transitional phenotype.
Effects of Chi3l1 on chromatin accessibility and TF footprinting of GSCs
To determine transcriptional regulatory mechanisms following exposure to Chi3l1, we performed ATAC-seq on Chi3l1-treated and control GSCs. Annotation of the peaks reveals that the peak densities at promoter regions increase in signal for the Chi3l1 treated group, indicating increased accessibility (Supplementary Fig. S3A). We confirmed that differential analysis was not biased towards genomic regions with intrinsically high counts by studying the differentially accessible sites relative to their average log normalized counts (Supplementary Fig. S3B). Analysis of chromatin accessibility identified 1,034 differentially accessible promoters, 525 of which with increased accessibility following Chi3l1 treatment (Fig. 6A). Motif enrichment analysis in promoters with increase in accessibility shows enrichment for SOX9, S0×10, MYB, ZNF281, WT1, CTCFL, and MAZ displaying the most pronounced enrichment (Fig. 6A).
Figure 6.
TF activity and RNA-seq integration identifies Chi3l1-activated MAZ as regulator of stemness in GSCs. A, Left, MA plot exhibiting differentially accessible sites following Chi3l1 exposure. Right, the corresponding motif enrichment at promoters with accessibility gain. B, Venn diagrams depicting TF activity consensus across the three TF activity platforms (DAStk, HINT-ATAC, and TOBIAS). C, Barcode plot encoding TF activity enrichment at MAZ motifs in Chi3l1-treated samples versus controls (P = 2.3E-05) using DAStk. D, HINT-ATAC aggregates footprint signal at MAZ motif in control versus treated samples (P = 0.06). E, Top plots show aggregate signals of TF binding across conditions (left, control; right, Chi3l1). Heatmaps depicting individual binding site signals show greater signal enrichment for bound sites compared with nonbound sites. Chi3l1 sample experienced an increase in bound site relative to control. F, Read count plot showing that MAZ is knocked down in siMAZ (siRNA) RNA-seq. G, Volcano plot displays differential gene expression following MAZ knockdown. NS, nonsignificant; up, upregulated in siMAZ; down, downregulated in siMAZ. H and I, Volcano plots depict differentially expressed genes following siMAZ [genes activated by MAZ (H) or repressed by MAZ (I)]. Green points emphasize differentially expressed genes with MAZ footprint/binding within their promoters. J, The activated or repressed genes by MAZ were queried for pathway enrichment. K and L, Examples depicting footprints within the promoters of MAZ-activated FOXO3 and MAZ-repressed ADAMS9. Footprints coincide with sites with (i) high binding scores and (ii) loss of signal within highly accessible sites that is characteristic of ATAC footprints. M, Knockdown of MAZ expression in GSCs rescues the activation of self-renewal induced by Chi3l1.
Then, TFs activity change was probed via DAStk, HINT-ATAC, and TOBIAS. Only 3 TFs are reproducibly identified across all platforms to experience a significant increase in activity following treatment: KLF15, VEZF1, and MAZ (Fig. 6B). In GSCs, KLF15 has very low expression, VEZF1 is moderately expressed and MAZ is highly expressed (Supplementary Fig. S3C). TF activity using change in motif displacement scores of 768 motifs from DAStk reveals that MAZ has the greatest significance in terms of expression and activity (Fig. 6C; Supplementary Fig. S3C and S3D). Furthermore, TF footprinting analysis using HINT-ATAC and TOBIAS independently validates MAZ as one of the most significantly activated TFs following Chi3l1 exposure (Fig. 6D and E). Consensus analysis across all three platforms shows that 25% of activated TFs are called by at least two of the programs, but 75% are non-reproducible/unique to each platform.
Chi3l1 induces a MAZ-regulated transcriptional network
To identify genes that are potentially directly regulated by MAZ following Chi3l1 treatment, we combined MAZ footprinting prediction from ATAC-seq data with RNA-seq data following MAZ knockdown. GSCs were treated with Chi3l1 and transfected with siCTR (n = 2) and siMAZ (n = 2). Following MAZ knockdown, 826 genes are downregulated and 1,062 are upregulated (Fig. 6F and G). To extricate potential direct MAZ regulated genes, the promoters of differentially expressed genes following MAZ perturbation were surveyed for the presence of a MAZ footprint. We identify 3,130 footprint sites or predicted binding sites. These footprints are predicted to be within the promoters of 1,836 genes. Of these 1,836 genes, 145 genes are found to be differentially expressed following MAZ knockdown, 86 of which downregulated so were deemed to be MAZ-activated genes and 58 of which upregulated, representing the potentially MAZ-repressed genes (Fig. 6H and I). Ultimately, pathway enrichment analysis reveals that potential MAZ activated gene network is enriched for stemness maintenance mechanisms (Notch1 and Hedgehog signaling) while MAZ repressed gene network is enriched for TNFa, hypoxia, and TGFβ signaling (Fig. 6J, representative MAZ peaks at Fig. 6K and L). Finally, to assess if MAZ participates in the Chi3l1 induced increase in self-renewal of GSCs (Fig. 2A), we transfected cells with two independent siRNA against MAZ, incubated with recombinant Chi3l1 for 7 days and conducted limited dilution assay and tumor sphere formation assay for self-renewal. We find that MAZ KD rescues the self-renewal phenotype of GSCs in the presence of Chi3l1 (Fig. 6M; Supplementary Fig. S4D and S4E). To determine if MAZ has the same pro-stemness role in GSCs independent of Chi3l1, we inhibited MAZ expression with siRNAs and performed self-renewal (LDA) and tumor sphere formation assay. This showed that MAZ knockdown significantly inhibits tumor sphere formation by GSCs, suggesting that MAZ regulates pro-stemness phenotypes of human GSCs independent of Chi3l1 (Supplementary Fig. S4F).
MAZ-regulated genes belong to clusters with cell transition changes following chi3l1 treatment
In GSC1, approximately 60% of MAZ activated genes are relatively more expressed in populations that showed significant cell state changes following Chi3l1 (clusters 10, 2, 4, 5, and 8; Fig. 7A). Interestingly clusters 5 and 10 display the highest expression of MAZ activated genes. Cluster 5 presents one of the most significant cell state changes, showing an increase in hedgehog and Notch signaling and increase in transition towards EMT and WNT β-catenin enriched cell states following Chi3l1 exposure. Cluster 10, which is hedgehog enriched, shows the highest expression of the MAZ regulated genes (Fig. 7A). In GSC2, we find the MAZ genes to be highly expressed in clusters 5, 0, and 1 (Fig. 7A), which are enriched EMT and cycling related genes (Fig. 2C). In GSC3, we find that MAZ regulated genes are enriched in cluster 8. A small subset of these genes is also relatively highly expressed in cluster 2. These two clusters experience significant transition changes, particularly cluster 2 (Figs. 7A and 4E). In GSC4, a subset of MAZ regulated genes shows strikingly significant relative enrichment in cluster 5. Cluster 5 experiences the most significant changes in GSC4, both numerically and in terms of transition (Figs. 7A and 4E). Cluster 7, which experiences major changes in transition, also shows high expression of MAZ regulated genes (Figs. 7A and 4D). However, important to note is that some clusters across all GSCs experience significant changes but show minimal MAZ regulated gene expression and vice versa.
Figure 7.
Chi3l1 induces a MAZ-regulated transcriptional network and anti-Chi3l1 blocking antibody inhibits glioblastoma growth in orthotopic xenografts. A, Hierarchically clustered heatmaps showing that MAZ-regulated genes are relatively highly expressed in clusters that have experienced significant cell transition changes following Chi3l1 exposure. B, MRI images of 16 mice treated with control IgG (8) or anti-Chi3l1 antibody (8), with segmented and reconstructed tumors pseudocolored in red. C, Volumetric surface quantification of control IgG (blue)- and anti-Chi3l1 (orange)–treated tumors (n = 8 animals per antibody group; t test: **, P < 0.009). D, Waterfall plot showing genes up- (blue) and downregulated (orange) following anti-Chi3l1 treatment. E, Quantification of subcutaneous flank tumor volume. Treatment with anti-Chi3l1 results in significant inhibition of flank tumor growth (P < 0.001). F, Kaplan–Meier survival curve of mice bearing subcutaneous flank glioblastomas treated with anti-Chi3l1 (n = 5) or control IgG (n = 6). Treatment with anti-Chi3l1 results in significant survival benefit (χ2 = 4.7, df = 1, P < 0.03).
Treatment with anti-Chi3l1 antibody results in significant reduction of human glioblastoma growth and significant survival benefit
To develop a potential therapeutic drug against Chi3l1, monoclonal blocking anti-Chi3l1 antibody isotype IgG2b, was generated and affinity purified against the peptide FRGQEDASPDRF corresponding to amino acids 223–234 of the full-length human Chi3l1 protein. Western blotting was performed to assess the specificity and sensitivity of the purified anti-Chi3l1 antibody against variable concentrations (250, 125, 61.25, 31.5, 15.6, 7.8 ng/mL) of recombinant human and mouse Chi3l1 protein (Supplementary Fig. S4A). Determination of the dose response curve of the antibody displays high sensitivity in detecting Chi3l1 (Kd = 1.1 × 10−8) and a limit of detection of 15 ng/mL (R2 = 0.991) as calculated by competitive ELISA (Supplementary Fig. S4B and S4C). To determine the in vivo efficacy of anti-Chi3l1 antibody as localized treatment for glioblastoma, we generated orthotopic xenograft glioblastoma in immunocompromised mice using patient-derived GSCs as shown before (16, 29). 28 days following the stereotactic implantation of GSCs, mice were implanted with an intracranial catheter connected to an osmotic pump (Alzet) for continuous infusion of anti-Chi3l1 antibody. After 28 days of continuous treatment, mice were subjected to MRI to determine the effect of anti-Chi3l1 on human glioblastoma growth. Volumetric measurements and quantification of tumor surface of mice treated with isotype matched IgG (n = 8) was compared with mice treated with anti-Chi3l1 antibody (n = 8; Fig. 7B) using 3D Slicer (v.4.10.2, Brigham & Women's Hospital). We show that continuous localized treatment with anti-Chi3l1 antibody results in more than 60% reduction of tumor volume compared with IgG treated control mice (Fig. 7C, Student t test: **, P value < 0.009).
Using GSCs from a different patient, we generated a subcutaneous flank tumor model in immunocompromised mice (Nu/J). Mice were randomized when tumors reached 50 mm3 and treated intraperitoneally with anti-Chi3l1 (n = 5) or control IgG (n = 6) every other day for a total of 21 days. Mice were euthanized when tumors reached 3 to 4 times the randomization size and Kaplan–Meier survival curve was plotted. Treatment with anti-Chi3l1 results in significant decrease in subcutaneous tumor growth (P < 0.001) and significant increase in survival (χ2 = 4.7, df = 1, P < 0.03; Fig. 7E and F).
In vivo treatment with anti-Chi3l1 inhibits the Chi3l1-regulated transcriptome of glioblastoma
To determine whether the effect of anti-Chi3l1 in vivo is the result of specific inhibition of the Chi3l1-induced expression of mesenchymal transcripts, we treated mice carrying orthotopic patient-derived glioblastoma xenografts with anti-Chi3l1 or isotype matched IgG as described above. At the end of the 28-day continuous treatment, we micro-dissected IgG-treated (n = 5) and anti-Chi3l1-treated (n = 4) tumors and performed NanoString expression analysis. We show that treatment with anti-Chi3l1 significantly downregulates the expression of transcripts with major roles in mesenchymal glioblastoma such as CD44, TNC, ANXA2, ITGB1, SMAD, and STAT3 as well as transcripts that have been linked to migration, invasion, and stemness of glioblastoma including SOX2, SOX9, EGFR, NOTCH1, TCF4, and MED23 (Fig. 7D).
Discussion
Functional heterogeneity in glioblastoma is defined not only by the genetic makeup of glioma cells but also through microenvironment-driven epigenetic influences that regulate glioma cell stemness. Recently, the hierarchical cancer stem cell model has been challenged by evidence suggesting that cancer stem cells may not constitute a defined cellular entity, but rather a cellular state adapting to autocrine and microenvironmental cues (6). The result of these epigenetic influences is activation of differentiation and dedifferentiation transcriptional programs, mechanisms of chromatin remodeling, noncoding RNA species and posttranscriptional RNA modification mechanisms that regulate phenotypic states of GSCs and confer tumor recurrence and therapeutic resistance.
Chi3l1, a member of the chitinase-like glycoprotein family, was first identified from the medium of a human osteosarcoma cell line MG-63 (34). Multiple studies have shown that high serum levels of Chi3l1 are correlated with metastasis and poor survival in a broad spectrum of human carcinomas including breast cancer (35), colorectal cancer (36), ovarian cancer (37), leukemia (38), lymphoma (39), and glioblastoma (40). Chi3l1, highly expressed in the microenvironment of human glioblastomas, is known to be associated with GSCs with a mesenchymal transcriptomic profile (7, 41). Phenotypic plasticity in human tumors can be driven by activation of EMT (42). This is the process by which cells acquire plasticity and gain the properties of stem cells (43, 44). EMT is linked with an undifferentiated cellular state, including the capacity for extended self-renewal and the acquisition of a stem-like gene expression program. Protein–protein interaction between Chi3l1 and CD44 at the surface of GSCs, activates a pAkt, p-β-catenin signaling cascade, which in turn induces CD44 expression in a pro-mesenchymal feed forward loop. Using RNA velocity at a single-cell resolution, we observe that the presence of Chi3l1 in the environment of GSCs induces marked transcriptomic cell state transitions towards mesenchymal phenotypes. Chi3l1 is a highly potent inducer of cellular plasticity and GSCs exposed to Chi3l1 express genes that define a “poised” uncommitted cancer stem cell state.
Reversible transitions between epigenetic states enable tumor cells to adapt to different microenvironment pressures, including therapy (45, 46). GSCs employ adaptive chromatin remodeling to drive cellular plasticity and therapy resistance (47). These types of chromatin alterations could also contribute to emergence of subclones in tumors with the least ability for reversion of stem-like properties or with the highest propensity to acquire stem-like characteristics. We show here that Chi3l1 as constituent of the GSC microenvironment, induces chromatin remodeling to facilitate accessibility of regulatory elements on promoters of genes with significant roles in the pathobiology of glioblastoma. We identify increase in MAZ TF footprint in accessible chromatin regulatory regions following exposure of GSCs to Chi3l1.
MAZ is a zing finger TF that is associated with Myc gene expression (48) and regulates a wide variety of genes. Recently, it was shown that MAZ interacts with CTCF and functions as a genome-wide insulator arresting cohesin movement and directly interacting with Rad21 (49). Interestingly, the second most significantly induced TF following Chi3l1 treatment of GSCs is Vezf1, which has been also shown capable of pausing RNA Pol II and affecting global RNA splicing (50). It is possible that Chi3l1 induces global architectural changes in the genome through alteration of MAZ transcriptional regulation and splicing deregulation through Vezf1. Indeed, MAZ knockdown in GSCs affects genes that belong to the clusters of cells that show the most significant changes in cell transition probabilities following exposure to Chi3l1. We posit that the Chi3l1 induction of the MAZ footprint in GSCs could result in stabilization of CTCF binding and in combination with increased Vezf1 binding could block RNA polymerase II advancement resulting in alternative RNA splicing patterns. This hypothesis could explain the significant cell state transition changes noted after Chi3l1 treatment of GSCs, which could represent global genomic effects of MAZ and Vezf1 because they reflect RNA splicing ratios inferred as RNA velocity patterns.
Elucidating state transition programs and mechanisms driving cellular plasticity will be essential to overcome current therapeutic deficiencies in glioblastoma. A limitation of the work described here, is that most of our observations are based on single-cell transcriptomic data. Application of additional biological assays to complement our scRNA-seq based observations, will be important to validate and extend our findings. It remains unclear whether the microenvironment selects for survival of specific GSCs or whether tumor cells adapt within new microenvironments. We favor the later and propose that Chi3l1 as a microenvironment factor could mediate cell state transitions in glioblastoma driving GSCs towards a poised uncommitted fate. We envision anti-Chi3l1 as a potent therapeutic agent in combination therapies targeting cellular adaptation in glioblastoma.
Supplementary Material
Supplementary Table 1
Supplementary Table 2
Supplementary Data and figure legends
Supplementary Figures and Legends
Acknowledgments
The authors would like to thank Owen Leary for analyzing the mouse MRI images with 3D Slicer. This work was supported by a Warren Alpert Foundation Grant to N. Tapinos and S.A. Toms and by funds of the Neurosurgery Department of Brown University to N. Tapinos.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
D. Karambizi reports grants from Warren Alpert Foundation during the conduct of the study. J.E. Fajardo reports grants from Warren Alpert Foundation during the conduct of the study. J.A. Elias reports other support from Ocean Biomedical outside the submitted work; in addition, J.A. Elias has a patent for 16/124,575 issued and a patent for 16/943,320 pending; and Dr. Elias is a cofounder of Elkurt Biologics and Ocean Biomedical. Both are working to develop cancer treatments based on the 18 glycosyl hydrolase gene family. N. Tapinos reports grants from Warren Alpert Foundation during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
C. Guetta-Terrier: Data curation, formal analysis, validation, investigation, methodology, writing–original draft. D. Karambizi: Data curation, formal analysis, investigation, visualization, writing–original draft, writing–review and editing. B. Akosman: Investigation. J.P. Zepecki: Investigation. J.-S. Chen: Investigation. S. Kamle: Investigation. J.E. Fajardo: Data curation, formal analysis. A. Fiser: Data curation, formal analysis. R. Singh: Formal analysis, methodology. S.A. Toms: Funding acquisition. C.G. Lee: Resources, supervision. J.A. Elias: Resources, supervision. N. Tapinos: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, methodology, writing–original draft, writing–review and editing.
References
- 1. Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med 2005;352:987–96. [DOI] [PubMed] [Google Scholar]
- 2. Stupp R, Weber DC. The role of radio- and chemotherapy in glioblastoma. Onkologie 2005;28:315–7. [DOI] [PubMed] [Google Scholar]
- 3. Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, et al. Identification of a cancer stem cell in human brain tumors. Cancer Res 2003;63:5821–8. [PubMed] [Google Scholar]
- 4. Soni P, Qayoom S, Husain N, Kumar P, Chandra A, Ojha BK, et al. CD24 and nanog expression in stem cells in glioblastoma: correlation with response to chemoradiation and overall survival. Asian Pac J Cancer Prev 2017;18:2215–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lopez-Bertoni H, Li Y, Laterra J. Cancer stem cells: dynamic entities in an ever-evolving paradigm. Biol Med 2015;7:001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dirkse A, Golebiewska A, Buder T, Nazarov PV, Muller A, Poovathingal S, et al. Stem cell–associated heterogeneity in glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun 2019;10:1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019;178:835–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Funkhouser JD, Aronson NN Jr. Chitinase family GH18: evolutionary insights from the genomic history of a diverse protein family. BMC Evol Biol 2007;7:96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Iwamoto FM, Hottinger AF, Karimi S, Riedel E, Dantis J, Jahdi M, et al. Serum YKL-40 is a marker of prognosis and disease status in high-grade gliomas. Neuro Oncol 2011;13:1244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt H, Johansen JS, Gehl J, Geertsen PF, Fode K, von der Maase H. Elevated serum level of YKL-40 is an independent prognostic factor for poor survival in patients with metastatic melanoma. Cancer 2006;106:1130–9. [DOI] [PubMed] [Google Scholar]
- 11. Vom Dorp F, Tschirdewahn S, Niedworok C, Reis H, Krause H, Kempkensteffen C, et al. Circulating and tissue expression levels of YKL-40 in renal cell cancer. J Urol 2016;195:1120–5. [DOI] [PubMed] [Google Scholar]
- 12. Bi J, Lau SH, Lv ZL, Xie D, Li W, Lai YR, et al. Overexpression of YKL-40 is an independent prognostic marker in gastric cancer. Hum Pathol 2009;40:1790–7. [DOI] [PubMed] [Google Scholar]
- 13. Johansen JS, Drivsholm L, Price PA, Christensen IJ. High serum YKL-40 level in patients with small cell lung cancer is related to early death. Lung Cancer 2004;46:333–40. [DOI] [PubMed] [Google Scholar]
- 14. Lee DW, Ramakrishnan D, Valenta J, Parney IF, Bayless KJ, Sitcheran R. The NF-kappaB RelB protein is an oncogenic driver of mesenchymal glioma. PLoS One 2013;8:e57489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giladi ND, Ziv-Av A, Lee HK, Finniss S, Cazacu S, Xiang C, et al. RTVP-1 promotes mesenchymal transformation of glioma via a STAT-3/IL6-dependent positive feedback loop. Oncotarget 2015;6:22680–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zepecki JP, Snyder KM, Moreno MM, Fajardo E, Fiser A, Ness J, et al. Regulation of human glioma cell migration, tumor growth, and stemness gene expression using a Lck targeted inhibitor. Oncogene 2019;38:1734–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 2013;10:1213–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bentsen M, Goymann P, Schultheis H, Klee K, Petrova A, Wiegandt R, et al. ATAC-seq footprinting unravels kinetics of transcription factor binding during zygotic genome activation. Nat Commun 2020;11:4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Z, Schulz MH, Look T, Begemann M, Zenke M, Costa IG. Identification of transcription factor binding sites using ATAC-seq. Genome Biol 2019;20:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tripodi IJ, Allen MA, Dowell RD. Detecting differential transcription factor activity from ATAC-seq data. Molecules 2018;23:1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Azofeifa JG, Allen MA, Hendrix JR, Read T, Rubin JD, Dowell RD. Enhancer RNA profiling predicts transcription factor activity. Genome Res 2018;28:334–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Couturier CP, Ayyadhury S, Le PU, Nadaf J, Monlong J, Riva G, et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat Commun 2020;11:3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tang M, Kaymaz Y, Logeman BL, Eichhorn S, Liang ZS, Dulac C, et al. Evaluating single-cell cluster stability using the Jaccard similarity index. Bioinformatics 2021;37:2212–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, et al. RNA velocity of single cells. Nature 2018;560:494–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 2020;38:1408–14. [DOI] [PubMed] [Google Scholar]
- 28. Reuter B, Weber M, Fackeldey K, Roblitz S, Garcia ME. Generalized Markov state modeling method for nonequilibrium biomolecular dynamics: exemplified on amyloid beta conformational dynamics driven by an oscillating electric field. J Chem Theory Comput 2018;14:3579–94. [DOI] [PubMed] [Google Scholar]
- 29. Zepecki JP, Karambizi D, Fajardo JE, Snyder KM, Guetta-Terrier C, Tang OY, et al. miRNA-mediated loss of m6A increases nascent translation in glioblastoma. PLos Genet 2021;17:e1009086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Behnan J, Finocchiaro G, Hanna G. The landscape of the mesenchymal signature in brain tumors. Brain 2019;142:847–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wurm J, Behringer SP, Ravi VM, Joseph K, Neidert N, Maier JP, et al. Astrogliosis releases pro-oncogenic chitinase 3–like 1 causing MAPK signaling in glioblastoma. Cancers 2019;11:1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods 2009;347:70–8. [DOI] [PubMed] [Google Scholar]
- 33. Prager BC, Bhargava S, Mahadev V, Hubert CG, Rich JN. Glioblastoma stem cells: driving resilience through chaos. Trends Cancer 2020;6:223–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Johansen JS, Williamson MK, Rice JS, Price PA. Identification of proteins secreted by human osteoblastic cells in culture. J Bone Miner Res 1992;7:501–12. [DOI] [PubMed] [Google Scholar]
- 35. Jensen BV, Johansen JS, Price PA. High levels of serum HER-2/neu and YKL-40 independently reflect aggressiveness of metastatic breast cancer. Clin Cancer Res 2003;9:4423–34. [PubMed] [Google Scholar]
- 36. Cintin C, Johansen JS, Christensen IJ, Price PA, Sorensen S, Nielsen HJ. Serum YKL-40 and colorectal cancer. Br J Cancer 1999;79:1494–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hogdall EV, Johansen JS, Kjaer SK, Price PA, Christensen L, Blaakaer J, et al. High plasma YKL-40 level in patients with ovarian cancer stage III is related to shorter survival. Oncol Rep 2003;10:1535–8. [PubMed] [Google Scholar]
- 38. Bergmann OJ, Johansen JS, Klausen TW, Mylin AK, Kristensen JS, Kjeldsen E, et al. High serum concentration of YKL-40 is associated with short survival in patients with acute myeloid leukemia. Clin Cancer Res 2005;11:8644–52. [DOI] [PubMed] [Google Scholar]
- 39. Hottinger AF, Iwamoto FM, Karimi S, Riedel E, Dantis J, Park J, et al. YKL-40 and MMP-9 as serum markers for patients with primary central nervous system lymphoma. Ann Neurol 2011;70:163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pelloski CE, Mahajan A, Maor M, Chang EL, Woo S, Gilbert M, et al. YKL-40 expression is associated with poorer response to radiation and shorter overall survival in glioblastoma. Clin Cancer Res 2005;11:3326–34. [DOI] [PubMed] [Google Scholar]
- 41. Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006;9:157–73. [DOI] [PubMed] [Google Scholar]
- 42. Gupta PB, Pastushenko I, Skibinski A, Blanpain C, Kuperwasser C. Phenotypic plasticity: driver of cancer initiation, progression, and therapy resistance. Cell Stem Cell 2019;24:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Thiery JP, Sleeman JP. Complex networks orchestrate epithelial–mesenchymal transitions. Nat Rev Mol Cell Biol 2006;7:131–42. [DOI] [PubMed] [Google Scholar]
- 44. Puisieux A, Pommier RM, Morel AP, Lavial F. Cellular pliancy and the multistep process of tumorigenesis. Cancer Cell 2018;33:164–72. [DOI] [PubMed] [Google Scholar]
- 45. Easwaran H, Tsai HC, Baylin SB. Cancer epigenetics: tumor heterogeneity, plasticity of stem-like states, and drug resistance. Mol Cell 2014;54:716–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 2010;141:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liau BB, Sievers C, Donohue LK, Gillespie SM, Flavahan WA, Miller TE, et al. Adaptive chromatin remodeling drives glioblastoma stem cell plasticity and drug tolerance. Cell Stem Cell 2017;20:233–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Marcu KB, Patel AJ, Yang Y. Differential regulation of the c-MYC P1 and P2 promoters in the absence of functional tumor suppressors: implications for mechanisms of deregulated MYC transcription. Curr Top Microbiol Immunol 1997;224:47–56. [DOI] [PubMed] [Google Scholar]
- 49. Xiao T, Li X, Felsenfeld G. The Myc-associated zinc finger protein (MAZ) works together with CTCF to control cohesin positioning and genome organization. Proc Natl Acad Sci USA 2021;118:e2023127118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gowher H, Brick K, Camerini-Otero RD, Felsenfeld G. Vezf1 protein binding sites genome-wide are associated with pausing of elongating RNA polymerase II. Proc Natl Acad Sci USA 2012;109:2370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1
Supplementary Table 2
Supplementary Data and figure legends
Supplementary Figures and Legends
Data Availability Statement
Publicly available data generated by others were used by the authors. The data analyzed in this study were obtained from The Cancer Genome Atlas (TCGA) at
https://gdac.broadinstitute.org/runs/stddata__latest/samples_report/GBM.html
https://gdac.broadinstitute.org
The data discussed in this manuscript have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE154060 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE154060).
High throughput microarray data (NanoString) and all other raw data are available upon request from the corresponding author.



![Figure 6. TF activity and RNA-seq integration identifies Chi3l1-activated MAZ as regulator of stemness in GSCs. A, Left, MA plot exhibiting differentially accessible sites following Chi3l1 exposure. Right, the corresponding motif enrichment at promoters with accessibility gain. B, Venn diagrams depicting TF activity consensus across the three TF activity platforms (DAStk, HINT-ATAC, and TOBIAS). C, Barcode plot encoding TF activity enrichment at MAZ motifs in Chi3l1-treated samples versus controls (P = 2.3E-05) using DAStk. D, HINT-ATAC aggregates footprint signal at MAZ motif in control versus treated samples (P = 0.06). E, Top plots show aggregate signals of TF binding across conditions (left, control; right, Chi3l1). Heatmaps depicting individual binding site signals show greater signal enrichment for bound sites compared with nonbound sites. Chi3l1 sample experienced an increase in bound site relative to control. F, Read count plot showing that MAZ is knocked down in siMAZ (siRNA) RNA-seq. G, Volcano plot displays differential gene expression following MAZ knockdown. NS, nonsignificant; up, upregulated in siMAZ; down, downregulated in siMAZ. H and I, Volcano plots depict differentially expressed genes following siMAZ [genes activated by MAZ (H) or repressed by MAZ (I)]. Green points emphasize differentially expressed genes with MAZ footprint/binding within their promoters. J, The activated or repressed genes by MAZ were queried for pathway enrichment. K and L, Examples depicting footprints within the promoters of MAZ-activated FOXO3 and MAZ-repressed ADAMS9. Footprints coincide with sites with (i) high binding scores and (ii) loss of signal within highly accessible sites that is characteristic of ATAC footprints. M, Knockdown of MAZ expression in GSCs rescues the activation of self-renewal induced by Chi3l1.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/8dbb/10267676/309fd76d2f99/1984fig6.jpg)