Abstract
Glypican-3 (GPC3) is a cell-surface glycoprotein that is frequently overexpressed in hepatocellular carcinoma (HCC). GPC3 undergoes extensive posttranslational modification (PTM) including cleavage and glycosylation. This review focuses on the structure and function of GPC3 in liver cancer, highlighting the PTM of the tertiary and quaternary structures of GPC3 as a potential oncogenic regulatory mechanism. We propose that the function of GPC3 in normal development can vary with extensive PTM and that dysregulation of these processes leads to disease. Defining the regulatory impact of these modifications can provide a deeper understanding of the role of GPC3 in oncogenesis, epithelial–mesenchymal transition, and drug development. Through review of current literature, this article provides a unique perspective on the role of GPC3 in liver cancer, focusing on potential regulatory mechanisms of PTM on GPC3 function at the molecular, cellular, and disease level.
Introduction
Glypican 3 (GPC3) has been implicated as a biomarker in many malignancies including liver cancer. Molecular overexpression occurs almost exclusively in the context of pathogenic processes after cessation of normal development. Furthermore, aberrant GPC3 expression is associated with poorer prognosis in cancer, making it a key candidate for molecular evaluation (1). GPC3 is one of six glypicans found in the mammalian genome that make up a family of heparan sulfate proteoglycans (HSPG; ref. 2). Glypicans are anchored to the cell membrane via a glycosyl-phosphatidyl-inositol (GPI) anchor that facilitates cell–extracellular matrix (ECM) and cell–cell interactions (3, 4). These proteins are structurally complex 60 to 70 kDa molecules that undergo endoproteolytic cleavage by Furin to form a 40 kDa amino (N) terminus and a 30 kDa carboxyl (C) terminus containing heparan sulfate (HS)-type glycosaminoglycan (GAG) side chains (2, 5, 6). The N and C terminus remain attached by disulfide bridges formed by 14 highly conserved cysteine residues (7, 8). Ultimately, glypicans can dissociate from the cell surface after cleavage of the GPI anchor by Notum (2, 3).
In normal human development, GPC3 is expressed in fetal tissue in a stage- and tissue-specific manner with higher mRNA levels found in lung, liver, kidney, and placenta while absent in most adult tissue (3, 9–11). Furthermore, studies in Drosophila, Xenopus, zebrafish, and mammals have shown that GPC3 modulates embryogenesis by interacting with cell signaling pathways including Wnt, Hedgehog (HH), bone morphogenetic proteins (BMP), and FGF (9, 12–16). Nonpathologic expression of GPC3 occurs in adult liver after partial hepatectomy to trigger parenchymal regeneration via the HH signaling pathway (17, 18).
Investigation of Simpson-Golabi-Behmel syndrome (SGBS) further clarified GPC3’s role in development and disease. SGBS is an X-linked overgrowth disorder caused by mutations in GPC3 (19–21). The mechanism leading to overgrowth remains unclear; however, there are more than 86 distinct GPC3 gene mutations that cause SGBS (22, 23). The majority are deletions or truncating mutations (frameshift, nonsense mutations) resulting in loss of function of GPC3 protein. Studies performed in constitutive GPC3 knockout (KO) mice have resulted in similar phenotypes and demonstrate increased cell number due to increased proliferation or reduced apoptosis (24–27). There are two described missense mutations resulting in functional GPC3, one within the furin cleavage site leading to convertase resistance and another in exon 8, resulting in truncated protein with loss of the signal sequence for HS attachment and GPI anchorage (22). Investigation of these mutations lead to functional, but altered, GPC3, and their phenotypic consequences will continue to advance our understanding of GPC3.
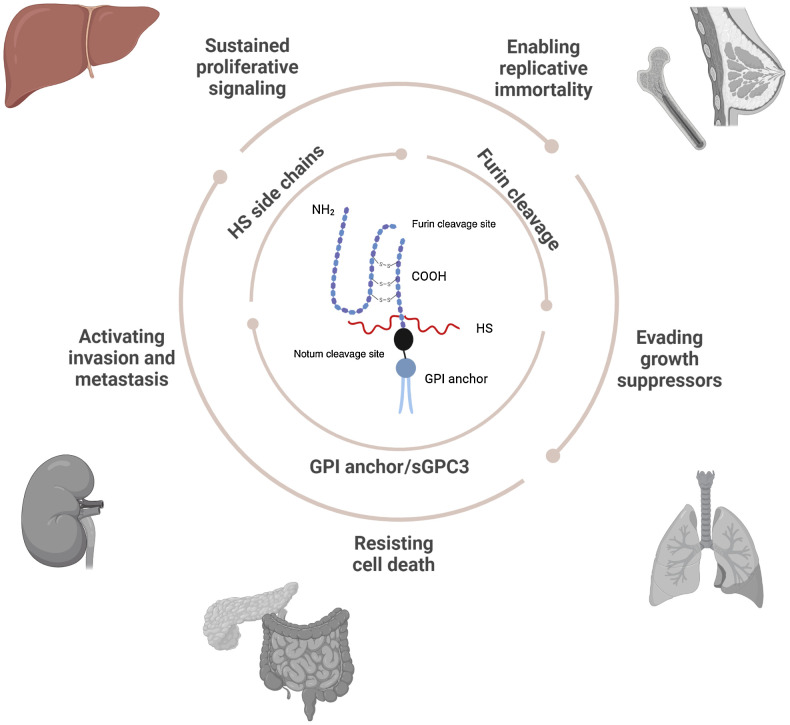
GPC3’s association with cancer was first discovered in pediatric embryonal tumors, including Wilms’ tumor, hepatoblastoma, and yolk sac tumor. Subsequently, aberrant expression of GPC3 has been implicated in the pathogenesis of additional cancers including hepatocellular carcinoma (HCC), ovarian carcinoma, breast carcinoma, squamous cell lung carcinoma, melanoma, and colorectal adenocarcinoma (Fig. 1; refs. 28–31).
Figure 1.
Schematic representing GPC3 structural and functional role in disease. (Created with BioRender.com.)
In this review, we highlight the importance of GPC3 structure on function in liver cancer as well as the diverse role of this protein in other cancers. We focus on specific structural modifications of GPC3 and proposed regulatory function of cleavage and glycosylation, including their impact on molecular signaling and phenotypic changes seen in GPC3-related malignancy. We hypothesize that GPC3’s role in normal development can vary with extensive posttranslational modification (PTM), and that dysregulation of these processes leads to disease. Therefore, mitigation of GPC3 modification in an organ- or cancer-specific manner could be an important molecular target for treatment. We explore cell signaling, cleavage, side chains, anchored, and soluble GPC3 to understand the numerous protein modifications that may impact oncogenesis.
GPC3 and Cell Signaling Pathways
GPC3 interacts with and modulates numerous cell signaling pathways. GPC3 interacts with canonical and noncanonical Wnt-signaling pathways in normal development and disease (3). Canonically, Wnt binds two coreceptors, frizzed (FZD) and low-density lipoprotein receptor-related protein 5/6 (LRP5/6) and transduces an intracellular signal to prevent β-catenin destruction (3). Ultimately, Wnt regulates cell differentiation, proliferation, and migration (32). β-catenin localizes within cytoplasm of hepatocytes and maintains adherens junctions (3, 33). β-catenin dysregulation results in cytoplasmic accumulation and eventual nuclear translocation that ultimately may lead to liver disease and cancer (34). In HCC, GPC3 promotes canonical Wnt signaling (1) via direct binding to Wnt3a and (2) indirectly via increased ligand binding to FZD receptor in HCC cell lines including Hep3B, ultimately leading to dysregulated cell proliferation (3, 35, 36). In hepatoblastoma, the most common pediatric liver malignancy, CTNNB1 is the most commonly mutated gene resulting in stabilized β-catenin (37, 38). Coexpression of GPC3 and β-catenin suggests a more direct role of these proteins in hepatoblastoma development, regardless, this interaction results in increased cell proliferation (34).
In contrast to the oncogenic role of GPC3 overexpression in liver cancer, loss of GPC3 function drives oncogenesis of breast cancer (39). In this tumor, GPC3’s interaction with canonical Wnt regulates cell–cell adhesion and cytoskeletal changes (40). Loss of GPC3 results in increased migration in breast cancer cell lines. There are several differences in GPC3 expression in breast tissue compared with other adult tissues, including evidence of expression at low levels in healthy adults (41, 42). Using the MCF-7 cell line, investigators demonstrated that interaction of GPC3 with the Wnt signaling pathway occurs through Wnt5a binding to GPC3’s HS chains, which has not been demonstrated in liver cancer (43). Therefore, a thorough understanding of unique GPC3 protein structures and modifications should be considered in individual cancer types that may provide insight into cancer-specific mechanisms and ultimately how GPC3 may or may not be therapeutically targeted.
HH interaction with GPC3 has also been implicated in oncogenesis (44). In normal development, the HH signaling pathway has a well-established role in embryogenesis (2). The Gli family of transcription factors are downstream of HH and regulate expression of cell-type–specific genes that control proliferation, migration, and differentiation (45). Wang and colleagues proposed that GPC3 promotes liver cancer via the HH pathway after in vitro analysis of HepG2 cells (46). GPC3 also acts as a competitive inhibitor of HH and by reducing Patched (Ptc) binding, regulates embryonic growth (2, 13, 17, 47). GPC3 also binds to sonic hedgehog (Shh) with high affinity (48). In addition, several groups demonstrated that the GPC3-HH complex undergoes endocytosis either by clathrin or LRP1-mediated endocytosis, resulting in reduced receptor and ligand for Ptc signaling (49). Taken together, it is possible that GPC3 plays a role in HH inhibition allowing cellular proliferation. Studies suggest this inhibitory action occurs via HS side chains as well as convertase cleavage, but more mechanistic study is required to dissect the precise pathways and mechanisms involved in carcinogenesis (50).
Recent advances in the understanding of HCC pathogenesis indicate that the Hippo-YAP pathway protects the liver from overgrowth and HCC development (51). This pathway has been implicated in the regulation of contact inhibition, organ size, and tumorigenesis, mediated through control of expression or localization of YAP (52). The oncogene YAP, a downstream effector, can be inactivated by phosphorylation; elevated YAP protein levels are strongly associated with HCC as well as deregulation of the Hippo pathway. In addition, single-cell RNA sequencing (RNA-seq) data for patients with hepatoblastoma samples shows upregulation of YAP in a tumor cluster with features resembling a potential cancer stem cell (53). Knockdown of GPC3 in Huh7 cells, an HCC cell line, resulted in reduced cell proliferation with YAP downregulation (54, 55). The exact mechanism by which GPC3 interacts with the Hippo/YAP pathway requires further investigation; however, there is support for cross-talk regulating cell proliferation in HCC. In addition, an antibody-targeting core GPC3 caused cell-cycle arrest and resulted in elevated levels of phosphorylated YAP. However, this antibody-mediated cell-cycle arrest was reversed by YAP overexpression (34). There is an overall lack of knowledge as to how YAP is regulated by GPC3 interactions, but the literature suggests it may occur through downstream activation from Wnt/GPC3 interactions or cross-talk with other pathways (3).
The insulin-like growth factor (IGF) signaling pathway is an additional growth-regulatory pathway that may interact with GPC3 in development, disease, and cancer (40). The IGF signaling pathway regulates cell proliferation, cell-cycle progression, prevention of apoptosis, and has been implicated in the transformation of cells to malignancy (56). In embryonal tumors, differential gene expression and correlation of GPC3 and IGF2 were evaluated. SGBS and Beckwith–Wiedemann syndrome (BWS) are similar overgrowth syndromes, with BWS resulting from overexpressed IGF2 via direct or indirect interaction with GPC3 (57). In HCC, studies performed in NIH3T3 cells overexpressing GPC3 or GPC3 knockdown demonstrated that GPC3 confers oncogenicity through interaction with IGF2/IGF2R, resulting in activation of the IGF-signaling pathway (56). Convertase-resistant RR-AA GPC3 was evaluated and it was found that convertase is required for optimal interaction with IGF2 (56). In ovarian clear cell carcinoma (CCC), GPC3 was found to interact with IGF2 and IGF-IR. Using a knockdown GPC3 cell line, investigators observed increased proliferation and concluded that GPC3 suppresses cell growth in ovarian CCC (58).
Other growth factors involved in normal development and activated by GPC3 include FGFs and BMPs (59). In HCC, GPC3 modulates proliferation through FGF2 binding in vivo and enhanced expression of GPC3 can inhibit BMP7 signaling (60). Given that many FGFs are reportedly upregulated in liver cancer it is crucial to understand how these growth factors interact to drive cell growth and carcinogenesis (61). Like other signaling pathways discussed above, specific structural features of GPC3 may mediate interaction with FGF and BMP and subsequent tumor proliferation. Overall, while evidence exists that GPC3 PTM can drive oncogenesis via misregulation of the above pathways, these PTMs are likely malignancy- and pathway-specific, and subsequent pathway cross-talk may also occur.
Complexity of GPC3
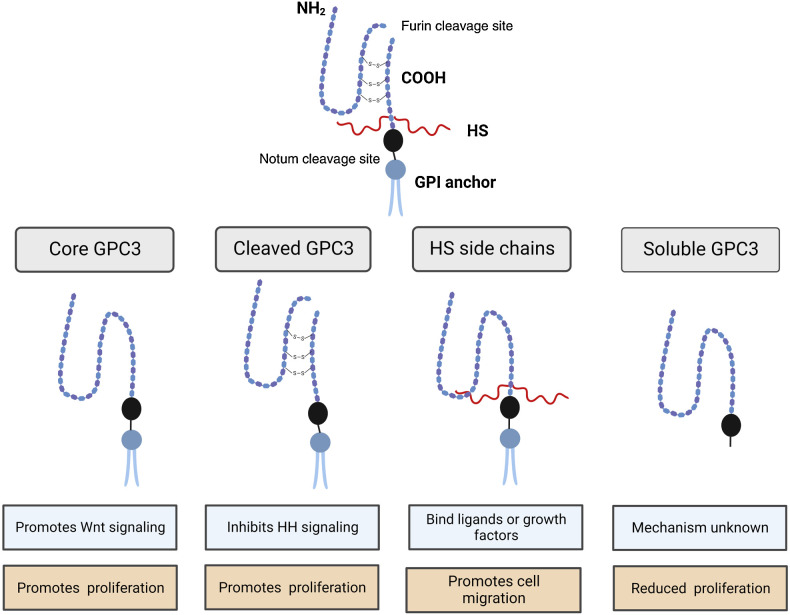
Glypicans have highly conserved core structures but undergo significant PTM including cleavage, addition of HS side chains, and linkage to the cell membrane by a GPI anchor (Fig. 2). Understanding the importance and impact of each of these processes can lead to further understanding of GPC3’s role in oncogenesis, epithelial–mesenchymal transition (EMT), and therapeutic targeting.
Figure 2.
Functional roles of PTM of GPC3. (Created with BioRender.com.)
Furin cleavage
In 2003, De Cat and colleagues demonstrated endoproteolytic cleavage of GPC3 in Madin-Darby canine kidney (MDCK) cells, a mammalian cell line and observed a 40 kDa, N-terminal fragment in addition to the known 70 kDa band corresponding to full-length GPC3 (5, 44). Processing of proteins from a larger precursor to an active form is well-established in molecular biology. Furin, a proprotein convertase, was the first discovered in its family and is found almost universally in mammalian cells, with numerous cleavage substrates in addition to GPC3, including matrix metalloproteinases (MMP), growth factors, receptors, and adhesion molecules (62, 63). Confirmation of cleavage by furin resulted in further understanding of the structure of GPC3. The C terminus contains a GPI anchor, HS attachment sites and a possible N-glycosylation site while the N terminus contains a cell surface signaling peptide and two additional N-glycosylation sites (5).
To answer whether furin processing was required for normal development, De Cat and colleagues evaluated GPC3 in zebrafish embryogenesis and generated a mutant, cleavage-resistant GPC3 (5). Inhibiting GPC3 cleavage blocked normal function including cell movement during gastrulation. They concluded that the aberrant development was related to altered interaction of GPC3 with canonical and noncanonical Wnt signaling (5). In breast and ovarian cell lines, processing by convertases was required for interaction with canonical Wnt1 ligand (5). Importantly, GPC3 transport to the cell-surface and subsequent glycosylation is independent of cleavage by furin (5, 64, 65). In contrast to De Cat and colleagues, Capurro and colleagues utilizing liver cancer cell lines (PLC/PRF/5), human lung fibroblasts (HLF), and human embryonic kidney (HEK) cells (293T), demonstrated that convertase processing is not required for interaction of GPC3 with Wnt7b and Wnt3a or potentiation of canonical Wnt-signaling (48). This group demonstrated that processing by convertase is required for inhibition of HH signaling by GPC3 through interaction with HS side chains and not the core GPC3 protein (66). In our lab, we investigated the effect of furin inhibition on cell proliferation and migration in liver cancer cell lines. Using a competitive furin inhibitor, we were able to reduce the cleaved GPC3 products and show reduced proliferation with increased cell migration (67). Ultimately, studies have concluded that cleavage of GPC3 is partial and dispensable to GPC3 interaction with Wnt and HH signaling pathways, promoting cell proliferation, and inhibiting apoptosis. We speculate that conformational changes of cleavage result in reduced binding of HS chains thereby disrupting interaction of GPC3 with cell signaling pathways that depend on the presence of these additional PTM. In terms of additional signaling pathways, cleavage does not appear necessary for BMP signaling, with further studies needed to evaluate cleavage in FGF and YAP signaling pathways (68).
HS side chains
As a HSPG, GPC3 contains a core protein with two HS insertion sites (S495 and S509) located on the C terminus. HS side chains mediate the majority of known biological functions of proteoglycans and facilitate interactions with the ECM, growth factors, and chemokines that influence cell growth, differentiation, and ultimately tumorigenicity (69, 70). The tools to evaluate the functional impact of modified HS side chains are still developing; however, the degree and pattern of sulfation impacts interaction and function of HS side chains (71).
To address this issue, Liu and colleagues developed a unique approach to interrogate the functional impact of modifying GAG in GPC3 (72). This group generated a soluble GPC3 model with modifiable HS side chains to explore effects of soluble and membrane-bound GPC3 with and without HS side chains on the HH signaling pathway (72). They found that GAG modification was required for membrane bound GPC3 to inhibit HH signaling while GPC3 without GAG had no impact. Furthermore, it is known that GPC3 core protein can interact with Wnt independent of HS chains (73, 74). Thus, another group evaluated the impact of modifying HS side chains on Wnt and HH signaling. They found that stimulation of Wnt signaling was proportional to the length of the attached HS side chain, and similar results were seen with the inhibitory effect of GPC3 on HH signaling (75).
In HCC cell lines, a mAb, HS20 that targets HS side chains of GPC3 blocked HCC tumor growth through Wnt3a-dependent mechanisms. Gao and colleagues determined that HS20 interaction with HS side chains is influenced by the pattern of sulfation. For instance, HS20 binding is dependent on 2-O and 6-O sulfation, with enhanced binding of Wnt and HS20 observed with 3-O sulfation (76). Further studies demonstrated that HS20 reduced cell migration and motility that reinforced data from GPC3 knockdown cells, suggesting that HS chains of GPC3 could mediate HCC cell migration and motility (77). Wang and colleagues explored the role of side chains on GPC3 to better define the role including interactions with frizzled, Wnt, and HH, but still more exploration is needed to define precise mechanisms. These studies highlight the functional importance of HS side chains on GPC3 and how modification of GAG can affect ligand affinity and downstream protein function.
GPI anchor and soluble GPC3
GPC3 localizes to the cell surface and is attached by a GPI anchor. GPC3 can be cleaved or removed from the cell surface either by endocytosis similar to other GPI-anchored proteins or cleavage by notum, a phospholipase (78, 79). Capurro and colleagues generated a mutant GPC3 that lacked a GPI anchor to further evaluate the interaction between GPC3 and Wnt. This soluble GPC3 (sGPC3) did not stimulate canonical Wnt activity or result in increased cell proliferation (44). In addition, sGPC3 protein is reported to inhibit HCC cell growth (55).
To further understand the mechanism of reduced proliferation with sGPC3, Capurro and colleagues treated several different HCC cell lines with sGPC3 using lentiviral transduction (73). They observed reduced proliferation of all cell types in both treatment arms, with cell-specific mechanisms including apoptosis, cell-cycle arrest and reduced proliferation in individual cell lines. They found increased apoptosis in Hep3B and HLE. sGPC3 treatment in Huh6, Huh7, and Li7 caused cell-cycle arrest. HepG2 cells underwent apoptosis and reduced proliferation of viable cells. HLF cells underwent apoptosis and cell-cycle arrest. Further studies are needed to evaluate the impact of sGPC3 on specific signaling pathways in other cancers, but taken together, sGPC3 appears to have an inhibitory function in contrast to the membrane-bound form.
GPC3 in Liver Cancer
We have discussed the role of GPC3 on cell signaling pathways demonstrating the structural, functional, and molecular implications. Hsu. and colleagues first reported GPC3 overexpression in liver cancers in 1997, focusing on overexpression of mRNA in HCC. These authors also found overexpression in one of two human specimens of hepatoblastoma (80). Studies have confirmed overexpression of GPC3 in HCC at the transcriptomic and protein level when compared with normal liver and nonmalignant liver lesions and clinically, pathologists currently utilize GPC3 IHC expression as a histologic marker (81, 82). In addition, soluble GPC3 is used as a serum marker for HCC with elevated levels in patients with HCC but undetectable in healthy patients and those with hepatitis (82–84). GPC3 overexpression is associated with undifferentiated, proliferating malignant hepatic cells, more aggressive HCC, poor prognosis, and overall survival (OS; refs. 55, 85). Shirakawa and colleagues found significantly lower 5-year survival in GPC3-positive patients with HCC compared with GPC3-negative patients (54.5% vs. 87.7%) with multivariate analysis as an independent prognostic factor for OS (86).
GPC3 acts as an oncofetal protein in HCC and hepatoblastoma. There is also evidence of GPC3 overexpression promoting cell migration and metastasis (87). In HCC, Wu and colleagues demonstrated a positive correlation between GPC3 expression in HCC samples with presence of metastatic disease in support of a role for GPC3 in migration and metastasis. In addition, these samples demonstrated an EMT phenotype with increased expression of mesenchymal markers and downregulation of epithelial cell markers. In vitro studies have shown that GPC3 contributes to EMT through Wnt or extracellular signal-regulated kinase (ERK) pathways, although the specific mechanism is likely organ and cancer specific (88). Another group evaluated motility in similar cell lines, including HepG2, a hepatoblastoma-derived cell line, and found reduced cell migration in GPC3 knockdown cells compared with normal controls (77). Although the precise mechanism by which GPC3 promotes EMT and subsequent migration and metastasis in cancer is unknown, there is sufficient evidence for further investigation, but given the complexity of the structure, function, and signaling, definition of cancer-specific mechanisms is necessary.
GPC3-Targeted Therapy
Interest in GPC3-based therapeutics began in the early 2000s and capitalized on the overexpression of GPC3 in cancer by creating targeted therapies (89). Since that time, numerous treatment approaches utilizing GPC3 have been developed including vaccines, mAbs, antibody–drug conjugates (ADC), bispecific antibodies, CTL, and chimeric antigen receptors (90–92). Of note, most current agents have an immunotherapeutic mechanism mediated through targeting of overexpressed GPC3, rather than targeting GPC3 function alone.
Current preclinical studies have utilized the protein structural components or modifications of GPC3 to design targeted antibodies. The C terminus and HS chains are commonly used to generate antibodies, but whole protein and bispecific approaches have also been reported (93). Gao and colleagues developed three different immunotoxins with alternate GPC3-binding sites to evaluate efficacy and toxicity of each potential site. YP7 recognizes the C terminus, similar to the binding site for the hGC33 mAb. Human mAb VH domain (HN3) targets a conformational epitope in GPC3 that inhibits YAP signaling and the human antibody HS20 recognizes HS chains on GPC3 blocking downstream Wnt signaling. These antibodies were conjugated with Pseudomonas exotoxin (PE38), a toxin that kills chemoresistant cancer cells. They found that both YP7-PE38 and NH3-PE38 were efficiently delivered to cells and capable of inducing cell death, concluding immunotoxin therapy can improve treatment of GPC3-positive tumors where antibody-targeted therapies have not shown significant tumor reduction (55, 94).
Codrituzumab (GC33) is a mAb targeting the C terminus of GPC3 that causes cell death by antibody-dependent cell cytotoxicity (95). It has undergone phase I and II clinical trials in HCC, and although well tolerated, failed to show therapeutic benefits (96). This drug is currently in phase I trials for management of pediatric solid tumors, including hepatoblastoma and is well tolerated with minimal toxicity (97). There is also a currently open phase I study of codrituzumab in children and young adults with recurrent or persistent solid tumors that has been open since June 2021 (NCT 04928677).
Targeting GPC3 alone will likely not be sufficient to induce tumor death (98). This issue can be overcome by utilizing GPC3 in combination with other chemotherapies or alternative therapies. With any immune-modulating drug, it is critical to design a drug that mitigates a robust, inappropriate immune response to minimize side effects. Finally, optimization of the drug-delivery construct to ensure optimal absorbance by the tumor is essential. Although GPC3-targeted treatments have shown modest survival benefits there are still numerous therapeutic approaches that take advantage of the overexpression of GPC3 in tumors.
Conclusions
GPC3 is a complex protein with a multifaceted role in liver cancer as well as many other cancer types with dysregulation contributing to aberrant proliferation, differentiation, adhesion, and migration. The PTM of GPC3 differentially regulates interactions with several cell signaling pathways. This review defines a knowledge gap in the evaluation of oncogenic pathways affected by PTM of GPC3, with significant literature on the impact of Wnt and HH signaling pathways but limited understanding in alternative pathways.
The field of GPC3 in cancer requires more extensive investigation of structural biology as much of the current work focuses on molecular approaches that have not yet effectively refined the functional mechanisms of this complex protein. GPC3 has profound cancer specificity and is a focus of targeted and immunotherapeutic treatments, but to date these therapies do not wholly kill cancer cells leaving the potential for adaptation and drug resistance. Disparate results are seen in a cancer- and tissue-specific manner further defining the complexity of GPC3. Future exploration of tissue-specific GPC3 function is needed to define precise GPC3 cancer pathogenesis.
An expanded knowledge of the PTMs of GPC3 as well as the functional interactions and downstream effectors are essential to understanding the role of GPC3 in organ specific carcinogenesis. The utilization of GPC3 as a therapeutic target requires insight into the role each protein structure plays as well as the spatial arrangement and interaction with other proteins in each cancer environment. Overall, GPC3 has been identified as a key player in cancer development, but to further utilize this knowledge to improve patient care, extensive research is still needed to understand structural complexity, modification, interaction, and pathway regulation. GPC3 has a diverse role in development and cancer with regulation through many cell signaling pathways. Rigorous exploration of the mechanism and functional role are essential to future work.
Authors' Disclosures
A. Bondoc reports grants from Stryker Corporation outside the submitted work. No disclosures were reported by the other authors.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
References
- 1. Chen C, Huang X, Ying Z, Wu D, Yu Y, Wang X, et al. Can glypican-3 be a disease-specific biomarker? Clin Transl Med 2017;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Filmus J, Capurro M. The role of glypicans in hedgehog signaling. Matrix Biol 2014;35:248–52. [DOI] [PubMed] [Google Scholar]
- 3. Kolluri A, Ho M. The role of glypican-3 in regulating Wnt, YAP and hedgehog in liver cancer. Front Oncol 2019;9:708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fico A, Maina F, Dono R. Fine-tuning of cell signaling by glypicans. Cell Mol Life Sci 2011;68:923–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. De Cat B, Muyldermans S-Y, Coomans C, Degeest G, Vanderschueren B, Creemers J, et al. Processing by proprotein convertases is required for glypican-3 modulation of cell survival, Wnt signaling, and gastrulation movements. J Cell Biol 2003;163:625–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Song HH, Filmus J. The role of glypicans in mammalian development. Biochim Biophys Acta 2002;1573:241–6. [DOI] [PubMed] [Google Scholar]
- 7. Watanabe K, Yamada H, Yamaguchi Y. K-glypican: a novel GPI-anchored heparan sulfate proteoglycan that is highly expressed in developing brain and kidney. J Cell Biol 1995;130:1207–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saunders S, Paine-Saunders S, Lander AD. Expression of the cell surface proteoglycan glypican-5 is developmentally regulated in kidney, limb, and brain. Dev Biol 1997;190:78–93. [DOI] [PubMed] [Google Scholar]
- 9. Song HH, Shi W, Xiang YY, Filmus J. The loss of glypican-3 induces alterations in Wnt signaling. J Biol Chem 2005;280:2116–25. [DOI] [PubMed] [Google Scholar]
- 10. Filmus J, Song HH. Glypicans. Proteoglycans: CRC Press; 2000. p.170–85. [Google Scholar]
- 11. Baumhoer D, Tornillo L, Stadlmann S, Roncalli M, Diamantis EK, Terracciano LM. Glypican 3 expression in human nonneoplastic, preneoplastic, and neoplastic tissues: a tissue microarray analysis of 4,387 tissue samples. Am J Clin Pathol 2008;129:899–906. [DOI] [PubMed] [Google Scholar]
- 12. Baeg G-H, Perrimon N. Functional binding of secreted molecules to heparan sulfate proteoglycans in Drosophila. Curr Opin Cell Biol 2000;12:575–80. [DOI] [PubMed] [Google Scholar]
- 13. Capurro MI, Xu P, Shi W, Li F, Jia A, Filmus J. Glypican-3 inhibits hedgehog signaling during development by competing with patched for hedgehog binding. Dev Cell 2008;14:700–11. [DOI] [PubMed] [Google Scholar]
- 14. Perrimon N, Bernfield M. Specificities of heparan sulphate proteoglycans in developmental processes. Nature 2000;404:725–8. [DOI] [PubMed] [Google Scholar]
- 15. Topczewski J, Sepich DS, Myers DC, Walker C, Amores A, Lele Z, et al. The zebrafish glypican knypek controls cell polarity during gastrulation movements of convergent extension. Dev Cell 2001;1:251–64. [DOI] [PubMed] [Google Scholar]
- 16. Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, et al. Identification of hedgehog pathway components by RNAi in drosophila cultured cells. Science 2003;299:2039–45. [DOI] [PubMed] [Google Scholar]
- 17. Bhave VS, Mars W, Donthamsetty S, Zhang X, Tan L, Luo J, et al. Regulation of liver growth by glypican 3, CD81, hedgehog, and Hhex. Am J Pathol 2013;183:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ochoa B, Syn WK, Delgado I, Karaca GF, Jung Y, Wang J, et al. Hedgehog signaling is critical for normal liver regeneration after partial hepatectomy in mice. Hepatology 2010;51:1712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pilia G, Hughes-Benzie RM, MacKenzie A, Baybayan P, Chen EY, Huber R, et al. Mutations in GPC3, a glypican gene, cause the Simpson-Golabi-Behmel overgrowth syndrome. Nat Genet 1996;12:241–7. [DOI] [PubMed] [Google Scholar]
- 20. Neri G, Gurrieri F, Zanni G, Lin A. Clinical and molecular aspects of the simpson-golabi-behmel syndrome. Am J Med Genet 1998;79:279–83. [DOI] [PubMed] [Google Scholar]
- 21. Tenorio J, Arias P, Martínez-Glez V, Santos F, García-Miñaur S, Nevado J, et al. Simpson-golabi-behmel syndrome Types I and II. Orphanet J Rare Dis 2014;9:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vuillaume ML, Moizard MP, Rossignol S, Cottereau E, Vonwill S, Alessandri JL, et al. Mutation update for the GPC3 gene involved in simpson-golabi-behmel syndrome and review of the literature. Hum Mutat 2018;39:790–805. [DOI] [PubMed] [Google Scholar]
- 23. Li M, Shuman C, Fei YL, Cutiongco E, Bender H, Stevens C, et al. GPC3 mutation analysis in a spectrum of patients with overgrowth expands the phenotype of simpson-golabi-behmel syndrome. Am J Med Genet 2001;102:161–8. [DOI] [PubMed] [Google Scholar]
- 24. Cano-Gauci DF, Song HH, Yang H, McKerlie C, Choo B, Shi W, et al. Glypican-3–deficient mice exhibit developmental overgrowth and some of the abnormalities typical of simpson-golabi-behmel syndrome. J Cell Biol 1999;146:255–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Capurro MI, Li F, Filmus J. Overgrowth of a mouse model of simpson–golabi–behmel syndrome is partly mediated by indian hedgehog. EMBO Rep 2009;10:901–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Filmus J. Glypicans in growth control and cancer. Glycobiology 2001;11:19R–23R. [DOI] [PubMed] [Google Scholar]
- 27. Hartwig S, Hu M-C, Cella C, Piscione T, Filmus J, Rosenblum ND. Glypican-3 modulates inhibitory Bmp2-Smad signaling to control renal development in vivo. Mech Dev 2005;122:928–38. [DOI] [PubMed] [Google Scholar]
- 28. Azizpour S, Ezati R, Saidijam M, Razavi AE, Jalilian FA, Mahdavinezhad A, et al. The expression of Glypican-3 in colorectal cancer. Cytology and Genetics 2019;53:430–40. [Google Scholar]
- 29. Moek KL, Fehrmann RS, van der Vegt B, de Vries EG, de Groot DJ. Glypican 3 overexpression across a broad spectrum of tumor types discovered with functional genomic mRNA profiling of a large cancer database. Am J Pathol 2018;188:1973–81. [DOI] [PubMed] [Google Scholar]
- 30. Toretsky JA, Zitomersky NL, Eskenazi AE, Voigt RW, Strauch ED, Sun CC, et al. Glypican-3 expression in Wilms tumor and hepatoblastoma. J Pediatr Hematol Oncol 2001;23:496–9. [DOI] [PubMed] [Google Scholar]
- 31. Richie JP. Gene expression profiling of early-and late-relapse nonseminomatous germ cell tumor and primitive neuroectodermal tumor of the testis. J Urol 2005;174:1826–7. [DOI] [PubMed] [Google Scholar]
- 32. Gilbert SF. Developmental biology. Sunderland, MA: Sinauer Associates; 2003. [Google Scholar]
- 33. Pez F, Lopez A, Kim M, Wands JR, de Fromentel CC, Merle P. Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol 2013;59:1107–17. [DOI] [PubMed] [Google Scholar]
- 34. Sha Y-L, Liu S, Yan W-W, Dong B. Wnt/β-catenin signaling as a useful therapeutic target in hepatoblastoma. Biosci Rep 2019;39:BSR20192466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li N, Wei L, Liu X, Bai H, Ye Y, Li D, et al. A frizzled-like cysteine-rich domain in Glypican-3 mediates wnt binding and regulates hepatocellular carcinoma tumor growth in mice. Hepatology 2019;70:1231–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Theocharis AD, Karamanos NK. Proteoglycans remodeling in cancer: underlying molecular mechanisms. Matrix Biol 2019;75:220–59. [DOI] [PubMed] [Google Scholar]
- 37. Satoh S, Daigo Y, Furukawa Y, Kato T, Miwa N, Nishiwaki T, et al. AXIN1 mutations in hepatocellular carcinomas, and growth suppression in cancer cells by virus-mediated transfer of AXIN1. Nat Genet 2000;24:245–50. [DOI] [PubMed] [Google Scholar]
- 38. Thompson MD, Monga SP. WNT/β-catenin signaling in liver health and disease. Hepatology 2007;45:1298–305. [DOI] [PubMed] [Google Scholar]
- 39. Peters M, Farias E, Colombo L, Filmus J, Puricelli L, Bal de Kier Joffe E. Inhibition of invasion and metastasis by glypican-3 in a syngeneic breast cancer model. Breast Cancer Res Treat 2003;80:221–32. [DOI] [PubMed] [Google Scholar]
- 40. Stigliano I, Puricelli L, Filmus J, Sogayar MC, de Kier Joffé EB, Peters MG. Glypican-3 regulates migration, adhesion and actin cytoskeleton organization in mammary tumor cells through Wnt signaling modulation. Breast Cancer Res Treat 2009;114:251–62. [DOI] [PubMed] [Google Scholar]
- 41. Fernández D, Guereño M, Huvelle MAL, Cercato M, Peters MG. Signaling network involved in the GPC3-induced inhibition of breast cancer progression: role of canonical Wnt pathway. J Cancer Res Clin Oncol 2018;144:2399–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Buchanan C, Stigliano I, Garay-Malpartida H, Rodrigues Gomes L, Puricelli L, Sogayar M, et al. Glypican-3 reexpression regulates apoptosis in murine adenocarcinoma mammary cells modulating PI3K/Akt and p38MAPK signaling pathways. Breast Cancer Res Treat 2010;119:559–74. [DOI] [PubMed] [Google Scholar]
- 43. Gao W, Ho M. The role of glypican-3 in regulating Wnt in hepatocellular carcinomas. Cancer Rep 2011;1:14. [PMC free article] [PubMed] [Google Scholar]
- 44. Capurro MI, Xiang Y-Y, Lobe C, Filmus J. Glypican-3 promotes the growth of hepatocellular carcinoma by stimulating canonical Wnt signaling. Cancer Res 2005;65:6245–54. [DOI] [PubMed] [Google Scholar]
- 45. Briscoe J, Thérond PP. The mechanisms of hedgehog signalling and its roles in development and disease. Nat Rev Mol Cell Biol 2013;14:416–29. [DOI] [PubMed] [Google Scholar]
- 46. Wang S, Chen N, Chen Y, Sun L, Li L, Liu H. Elevated GPC3 level promotes cell proliferation in liver cancer. Oncol Lett 2018;16:970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ding M, Wang X. Antagonism between hedgehog and Wnt signaling pathways regulates tumorigenicity. Oncol Lett 2017;14:6327–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Capurro MI, Shi W, Sandal S, Filmus J. Processing by convertases is not required for glypican-3-induced stimulation of hepatocellular carcinoma growth. J Biol Chem 2005;280:41201–6. [DOI] [PubMed] [Google Scholar]
- 49. Capurro MI, Shi W, Filmus J. LRP1 mediates Hedgehog-induced endocytosis of the GPC3–Hedgehog complex. J Cell Sci 2012;125:3380–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kaur SP, Cummings BS. Role of glypicans in regulation of the tumor microenvironment and cancer progression. Biochem Pharmacol 2019;168:108–18. [DOI] [PubMed] [Google Scholar]
- 51. Miao HL, Pan ZJ, Lei CJ, Wen JY, Li MY, Liu ZK, et al. Knockdown of GPC3 inhibits the proliferation of Huh7 hepatocellular carcinoma cells through down-regulation of YAP. J Cell Biochem 2013;114:625–31. [DOI] [PubMed] [Google Scholar]
- 52. Lee K-P, Lee J-H, Kim T-S, Kim T-H, Park H-D, Byun J-S, et al. The hippo–salvador pathway restrains hepatic oval cell proliferation, liver size, and liver tumorigenesis. Proc Natl Acad Sci U S A 2010;107:8248–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bondoc A, Glaser K, Jin K, Lake C, Cairo S, Geller J, et al. Identification of distinct tumor cell populations and key genetic mechanisms through single cell sequencing in hepatoblastoma. Commun Biol 2021;4:1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Feng M, Gao W, Wang R, Chen W, Man Y-G, Figg WD, et al. Therapeutically targeting glypican-3 via a conformation-specific single-domain antibody in hepatocellular carcinoma. Proc Natl Acad Sci U S A 2013;110:E1083–E91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gao W, Tang Z, Zhang Y-F, Feng M, Qian M, Dimitrov DS, et al. Immunotoxin targeting glypican-3 regresses liver cancer via dual inhibition of Wnt signalling and protein synthesis. Nat Commun 2015;6:6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheng W, Tseng C-J, Lin TT, Cheng I, Pan H-W, Hsu H-C, et al. Glypican-3-mediated oncogenesis involves the Insulin-like growth factor-signaling pathway. Carcinogenesis 2008;29:1319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Saikali Z, Sinnett D. Expression of glypican 3 (GPC3) in embryonal tumors. Int J Cancer 2000;89:418–22. [PubMed] [Google Scholar]
- 58. Sakurai M, Shibata K, Umezu T, Kajiyama H, Yamamoto E, Ino K, et al. Growth-suppressing function of glypican-3 (GPC3) via insulin like growth factor II (IGF-II) signaling pathway in ovarian clear cell carcinoma cells. Gynecol Oncol 2010;119:332–6. [DOI] [PubMed] [Google Scholar]
- 59. Grisaru S, Cano-Gauci D, Tee J, Filmus J, Rosenblum ND. Glypican-3 modulates BMP-and FGF-mediated effects during renal branching morphogenesis. Dev Biol 2001;231:31–46. [DOI] [PubMed] [Google Scholar]
- 60. Midorikawa Y, Ishikawa S, Iwanari H, Imamura T, Sakamoto H, Miyazono K, et al. Glypican-3, overexpressed in hepatocellular carcinoma, modulates FGF2 and BMP-7 signaling. Int J Cancer 2003;103:455–65. [DOI] [PubMed] [Google Scholar]
- 61. Wu Y, Liu H, Ding H. GPC-3 in hepatocellular carcinoma: current perspectives. J Hepatocell Carcinoma 2016;3:63–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Taylor NA, Van De Ven WJ, Creemers JW. Curbing activation: proprotein convertases in homeostasis and pathology. FASEB J 2003;17:1215–27. [DOI] [PubMed] [Google Scholar]
- 63. Bassi DE, Fu J, Lopez de Cicco R, Klein-Szanto AJ. Proprotein convertases:“master switches” in the regulation of tumor growth and progression. Mol Carcinog 2005;44:151–61. [DOI] [PubMed] [Google Scholar]
- 64. Gonzalez AD, Kaya M, Shi W, Song H, Testa JR, Penn LZ, et al. OCI-5/GPC3, a glypican encoded by a gene that is mutated in the Simpson-Golabi-Behmel overgrowth syndrome, induces apoptosis in a cell line–specific manner. J Cell Biol 1998;141:1407–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sun CK, Chua M-S, He J, Samuel KS. Suppression of glypican 3 inhibits growth of hepatocellular carcinoma cells through up-regulation of TGF-β2. Neoplasia 2011;13:735–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Capurro M, Shi W, Izumikawa T, Kitagawa H, Filmus J. Processing by convertases is required for glypican-3-induced inhibition of Hedgehog signaling. J Biol Chem 2015;290:7576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schepers EJ, Lake C, Glaser K, Bondoc AJ. Inhibition of Glypican-3 cleavage results in reduced cell proliferation in a liver cancer cell line. J Surg Res 2023;282:118–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yan D, Lin X. Opposing roles for glypicans in Hedgehog signalling. Nat Cell Biol 2008;10:761–3. [DOI] [PubMed] [Google Scholar]
- 69. Bernfield M, Götte M, Park PW, Reizes O, Fitzgerald ML, Lincecum J, et al. Functions of cell surface heparan sulfate proteoglycans. Annu Rev Biochem 1999;68:729–77. [DOI] [PubMed] [Google Scholar]
- 70. Sanderson RD, Yang Y, Kelly T, MacLeod V, Dai Y, Theus A. Enzymatic remodeling of heparan sulfate proteoglycans within the tumor microenvironment: growth regulation and the prospect of new cancer therapies. J Cell Biochem 2005;96:897–905. [DOI] [PubMed] [Google Scholar]
- 71. Lanzi C, Yates EA, Cassinelli G. Heparan sulfate proteoglycans and their endogenous modifying enzymes: cancer players, biomarkers and therapeutic targets. Front Oncol 2020;10:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Liu YC, Wierbowski BM, Salic A. Hedgehog pathway modulation by glypican 3–conjugated heparan sulfate. J Cell Sci 2022;135:jcs259297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zittermann SI, Capurro MI, Shi W, Filmus J. Soluble glypican 3 inhibits the growth of hepatocellular carcinoma in vitro and in vivo. Int J Cancer 2010;126:1291–301. [DOI] [PubMed] [Google Scholar]
- 74. Ho M, Kim H. Glypican-3: a new target for cancer immunotherapy. Eur J Cancer 2011;47:333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang W, Han N, Xu Y, Zhao Y, Shi L, Filmus J, et al. Assembling custom side chains on proteoglycans to interrogate their function in living cells. Nat Commun 2020;11:5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Gao W, Xu Y, Liu J, Ho M. Epitope mapping by a Wnt-blocking antibody: evidence of the Wnt binding domain in heparan sulfate. Sci Rep 2016;6:26245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Gao W, Kim H, Ho M. Human monoclonal antibody targeting the heparan sulfate chains of glypican-3 inhibits HGF-mediated migration and motility of hepatocellular carcinoma cells. PLoS One 2015;10:e0137664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kreuger J, Perez L, Giraldez AJ, Cohen SM. Opposing activities of dally-like glypican at high and low levels of Wingless morphogen activity. Dev Cell 2004;7:503–12. [DOI] [PubMed] [Google Scholar]
- 79. Shih T-C, Wang L, Wang H-C, Wan Y-JY. Glypican-3: a molecular marker for the detection and treatment of hepatocellular carcinoma. Liver Res 2020;4:168–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Hsu H-C, Cheng W, Lai P-L. Cloning and expression of a developmentally regulated transcript MXR7 in hepatocellular carcinoma: biological significance and temporospatial distribution. Cancer Res 1997;57:5179–84. [PubMed] [Google Scholar]
- 81. Zhu Z, Friess H, Wang L, Abou-Shady M, Zimmermann A, Lander A, et al. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut 2001;48:558–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Nakatsura T, Yoshitake Y, Senju S, Monji M, Komori H, Motomura Y, et al. Glypican-3, overexpressed specifically in human hepatocellular carcinoma, is a novel tumor marker. Biochem Biophys Res Commun 2003;306:16–25. [DOI] [PubMed] [Google Scholar]
- 83. Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, et al. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology 2003;125:89–97. [DOI] [PubMed] [Google Scholar]
- 84. Hippo Y, Watanabe K, Watanabe A, Midorikawa Y, Yamamoto S, Ihara S, et al. Identification of soluble NH2-terminal fragment of glypican-3 as a serological marker for early-stage hepatocellular carcinoma. Cancer Res 2004;64:2418–23. [DOI] [PubMed] [Google Scholar]
- 85. Cartier F, Indersie E, Lesjean S, Charpentier J, Hooks KB, Ghousein A, et al. New tumor suppressor microRNAs target glypican-3 in human liver cancer. Oncotarget 2017;8:41211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shirakawa H, Suzuki H, Shimomura M, Kojima M, Gotohda N, Takahashi S, et al. Glypican-3 expression is correlated with poor prognosis in hepatocellular carcinoma. Cancer Sci 2009;100:1403–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wu Y, Liu H, Weng H, Zhang X, Li P, Fan C-L, et al. Glypican-3 promotes epithelial-mesenchymal transition of hepatocellular carcinoma cells through ERK signaling pathway. Int J Oncol 2015;46:1275–85. [DOI] [PubMed] [Google Scholar]
- 88. Castillo LF, Tascón R, Huvelle MAL, Novack G, Llorens MC, Dos Santos AF, et al. Glypican-3 induces a mesenchymal to epithelial transition in human breast cancer cells. Oncotarget 2016;7:60133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ishiguro T, Sugimoto M, Kinoshita Y, Miyazaki Y, Nakano K, Tsunoda H, et al. Anti–glypican 3 antibody as a potential antitumor agent for human liver cancer. Cancer Res 2008;68:9832–8. [DOI] [PubMed] [Google Scholar]
- 90. Chen Z, Dong R. Advances in the conventional clinical treatment for hepatoblastoma and therapeutic innovation. World J Pediatr Surg 2021;4:e000220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zheng X, Liu X, Lei Y, Wang G, Liu M. Glypican-3: a novel and promising target for the treatment of hepatocellular carcinoma. Front Oncol 2022;12:824208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kim M, Wands JR. Points of therapeutic intervention along the Wnt signaling pathway in hepatocellular carcinoma. Advances in Cancer Drug Targets 2016;3:78. [DOI] [PubMed] [Google Scholar]
- 93. Guo M, Zhang H, Zheng J, Liu Y. Glypican-3: a new target for diagnosis and treatment of hepatocellular carcinoma. J Cancer 2020;11:2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Fleming BD, Ho M. Glypican-3 targeting immunotoxins for the treatment of liver cancer. Toxins 2016;8:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Feng M, Ho M. Glypican-3 antibodies: a new therapeutic target for liver cancer. FEBS Lett 2014;588:377–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Abou-Alfa GK, Puig O, Daniele B, Kudo M, Merle P, Park J-W, et al. Randomized phase II placebo controlled study of codrituzumab in previously treated patients with advanced hepatocellular carcinoma. J Hepatol 2016;65:289–95. [DOI] [PubMed] [Google Scholar]
- 97. Tsuchiya N, Hosono A, Yoshikawa T, Shoda K, Nosaka K, Shimomura M, et al. Phase I study of glypican-3-derived peptide vaccine therapy for patients with refractory pediatric solid tumors. Oncoimmunology 2018;7:e1377872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fu Y, Urban DJ, Nani RR, Zhang YF, Li N, Fu H, et al. Glypican-3-specific antibody drug conjugates targeting hepatocellular carcinoma. Hepatology 2019;70:563–76. [DOI] [PMC free article] [PubMed] [Google Scholar]