Abstract
Background
Changes in ABO blood type caused by a gradual decrease in antigen expression have been found in patients with acute myeloid leukemia (AML). Studies have indicated that alteration of ABO gene methylation accounts for 50% of acquired weak ABO antigen expression in patients with leukemia. However, the molecular mechanisms contributing to the remaining 50% of cases are unknown. We hypothesize that deregulation of miRNA is correlated with weak ABO antigen expression in patients with AML.
Methods
Blood samples of 19 patients with AML and 12 healthy controls were collected, in which the blood type was not changed in these AML patients. Flow cytometric analysis was applied to measure the ABO antigen expression titer among AML patients and controls. A total of 18 leukemia-related miRNAs were analyzed via quantitative real-time polymerase chain reactions.
Results
We found that miRNA profiles were correlated with the AML patients, especially in those who had constant or weakened ABO antigen expressions. Compared with healthy controls, the miR-16 and miR-451 expression were significantly lower in either AML cases with weak ABO antigen expressions (p = 0.003, p = 0.028, respectively) or AML cases with constant ABO antigen expressions (p = 0.043, p = 0.040, respectively). Although not statistically significant, decreasing trends in the miR-451 and miR-16 expressions in the AML patients with weakened ABO were observed compared to those with constant ABO antigens. The weak ABO antigen expression might correlate with miRNAs, especially miR-16 and miR-451.
Conclusion
This study indicated that decreasing in miR-16 and miR-451 was associated with AML and AML with weakened ABO expression. In the future, we will continue to include more cases and exclude the others factor influencing ABO antigen expression, promoter methylation and oxidative stress, to replicate the results of this study and investigate the underlying mechanism of decreasing miR-16 and miR-451 in AML patients with varied ABO antigen expression levels.
Keywords: ABO blood Type, microRNAs, Leukemia, Hematopoiesis
At a glance commentary
Scientific background on the subject
microRNAs (miRNAs) play a pivotal role in gene expression. Moreover, they participate in embryonic development. Thus, we hypothesized that the 30 % of patients with acute leukemia had very weak ABO antigen expression that were caused by miRNAs.
What this study adds to the field
We speculated that the weak ABO antigen expression might correlate with miRNAs, especially miR-16 and miR-451. This study provided a direction for investigating the underlying mechanism of the weakened ABO in acute leukemia patients.
It has been shown that A or B antigen expression had a 55% decrease in patients with acute myeloid leukemia (AML), which resulted in the alternation in ABO blood type [1]. Previous studies indicated that alteration of ABO promoter methylation accounts for 50% of leukemia patients who had acquired ABO antigen weak expression [2]. However, the molecular mechanisms contributing to the remaining 50% of the cases are unknown. The aim of this study was to find out the reason that influencing the weakened blood type in those 50% unknown casas.
miRNAs are a family of small non-coding RNAs (18–24 nucleotides), and recent studies demonstrated that microRNAs (miRNAs) play a pivotal role in gene expression by either degradation of the target mRNAs or blocking translation [3,4]. Moreover, miRNAs participate in regulation of various biological processes such as cell proliferation, differentiation, and embryonic development [5]. However, only a few miRNAs have been characterized at the functional level, and even less is known about the regulation of miRNA expression.
miRNAs also participate in hematopoiesis [6], and several miRNAs had been identified that control the differentiation of specific blood cell lineage, e.g. miR-130 in megakaryopoiesis [7], miR-223 in granulopoiesis [8], and miR-181a in T lymphopoiesis [9]. miR-221 and miR-222 were found to block kit receptor at mRNA level, and the finding indicated that miRNAs participate in early erythropoiesis [10]. The expression profiles of miRNAs at specific stages of erythropoiesis were identified by microarray studies [[11], [12], [13]]. miR-24 was proved to induce red cell maturation and restrict the activin-mediated accumulation of hemoglobin by targeting the activin type I receptor [14]. The abnormal regulation of miRNAs may also be a key point in abnormal erythropoiesis in polycythemia vera [13,15].
Because about 30% of patients with acute leukemia had very weak ABO antigen expression analyzed by flow cytometric analysis in our preliminary study, we hypothesize that deregulation of miRNA caused by AML attenuates the expression levels of ABO transferases and A/B antigen expression in the patients with acquired weak expression of ABO antigens.
Materials and methods
Subjects. This study was approved by the Institutional Review Board of Chang Gung Memorial Hospital (CGMH) with the approval ID of 103-6479B and 103-2419B. All the participants provided written informed consents to participate in this study, and all methods were performed in accordance with the relevant guidelines and regulations. Originally, a total of 53 AML patients with blood group A were collected and detected by flow cytometry. Among them, the A antigen of 7 patients were less than 90% compared to positive control. Additionally, a total of 32 AML patients with blood group B were collected and detected by flow cytometry, of which 15 patients had less than 90% of B antigen compared to positive control. After excluding subjects without complete data and unable to continue tracking, there were only 7 patients with blood type A and 12 patients with blood type B (19 in total). No blood type in these patients was found to be changed. These samples were collected between 2014 and 2017 at the Blood Bank of CGMH. The detailed information of final 19 AML patients was shown in Supplementary Table 1. Six negative control samples were included from healthy volunteers.
Flow cytometric analysis of A and B antigens. Flow cytometry was applied in A and B antigen detection, the healthy volunteers and patient's red blood cells were washed twice with 1X phosphate-buffered saline and 50 μl of a 1.5% red blood cell PBS suspension were incubated for 60 min at room temperature with 50 μl 1 : 700 dilutions of FITC-conjugated lectin from Helix pomatia for blood type A (L1034, Sigma) or 50 μl1:5 dilutions of FITC conjugated lectin from Bandeiraea simplicifolia (L2895, Sigma) for blood type B. The A cell (AC-0001B-01, Formasa Biomedical, Taiwan) and B cell (AC-0001B-02, Formasa Biomedical, Taiwan) were used as a reference control RBCs (positive control). As a negative control, 50 μl of a 1.5% reference RBCs was incubated with 50 μl of 1X PBS as a negative agglutination control. These samples were washed two times (3400 rpm/1 min) and suspend in 500 μl PBS, and immune phenotypes were analyzed on a flow cytometry Cytomics FC500 (Beckman Coulter) and CPX software (Beckman Coulter). Two variables, the relative percentage of antigen-expressing cells and the mean fluorescence index (MFI), were used for data analysis. MFI is a relative measurement for the amount of antigen expression on the cell surface and is defined as a statistical indicator, comparing changes of fluorescence intensity. In this study, the weakened ABO antigen expression was defined as those with the MFI lower than 90% of the standard blood cells based on the findings in our previous study [16] and subsequent work [[17], [18], [19]]. Defining weak ABO types with MFI was to avoid errors or inconsistencies of human judgment. In Chen et al. (2011), we found that the MFI of the A antigen expression for weak A subtype (A1v) were 65.71 ± 13.06% of the wild type A (A1); and the MFI of the B antigen expression for weak B (Bel) was 90.71 ± 1.72% of the wild type B (B1). Therefore, to include more subtle blood type changes, we used the MFI <90% (the upper limit) of the standard blood cells as the cutoff level for the weakened ABO antigen expression. And the histogram of flow cytometry showing the weakened antigen expression was shown in Supplementary Figure 1. The fluorescein isothiocyanate (FITC)-conjugated lectin from H. pomatia, and FITC-conjugated lectin from B. simplicifolia were purchased from Sigma (Saint Louis, MO).
miRNA quantitative real-time polymerase chain reaction (qRT-PCR)
miRNAs expression levels were assessed using the qRT-PCR method. 5 ng of total RNA was used per sample to synthesize cDNA. The expression of U6 was used as the internal control and for RNA template normalization. Total RNA was extracted from the peripheral blood RBC (3 × 106 cells) using the miRNeasy mini kit (cat no.217004, Qiagen, CA), cDNAs were synthesized using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen, #18080-051, CA USA) with a universal stem-loop primer method [20]. The 20 μl of reverse transcription reaction mixture contained 50 ng total RNA, 1 μl universal stem-loop primer USLP (5′-GAAAGAAGGCGAGGAGCAGATCGAGGAAGAAGACGGAAGAATGTGCGTCTCGCCTTCTTTCNNNNNNNN-3′, 10 μM), 1 μl U6RT primer (5′-CGCTTCACGAATTTGCGTGT CAB-3′, 10 μM), as per the manufacturer's instructions. The expression of miR-1, miR-9-5p, miR-10a, miR-16, miR-24, miR-125b, miR-145, miR-150, miR-155, miR-203-3p, miR-221, miR-222, miR-223, miR-331-3p, miR-451, miR-574-5p, miR-848-5p, and miR-1908-5p were measured by qRT-PCR. The primer sequences as described in Table 1, and the reasons for choosing the 18 miRNAs in study were based on the below literature. In a review paper from S Yendamuri (2009), he sorted out the role of miRNA in hematopoiesis from precursor cells to mature blood cells. Wherein miR-150 and miR-223 promoted cell differentiation; miR-150, miR-155, miR-221, and miR-222 inhibited cell differentiation; and miR-150, miR-155, miR-221, miR-222, miR-16, and miR-145 were related to red blood cell differentiation [21]. As for miR-9-5p, miR-155, and miR-203-3p, we referred to the research of Chuang MK et al. (2015) that predicted the prognosis of AML patients with 3-microrna scoring system [22]. miR-574-5p, miR-451, and miR-848-5p were selected based on the ABO gene 3′UTR sequence. Additionally, Kronstein Wiedemann R et al. reported that miR-331-3p and miR-1908-5p were related to the performance of tranferase A in 2015 ISBT annual meeting [23]. Furthermore, miR-1, miR-10a, miR-16, miR-24, miR-125b, miR-145 were often discussed as being associated with leukemia [[24], [25], [26]]. These miRNAs were included in the analysis and the clinical relevance of leukemia was summarized in Supplementary Table 2. But mir-848-5p has not been found to be related to leukemia yet.
Table 1.
miRNA qRT-PCR primer.
| Primer | Sequence |
|---|---|
| U6 qPCR Forward primer | 5′-GCTTCGGCAGCACATATACTAAAAT-3′ |
| U6 qPCR Reverse Primer | 5′-CGCTTCACGAATTTGCGTGTCAT.-3′ |
| miR-1 | 5′- GCGGGTGGAATGTAAAGAAGTATGTAT-3′ |
| miR-9-5p | 5′-GCGTCTTTGGTTATCTAGCTGT-3′ |
| miR-10a | 5′-CGTACCCTGTAGATCCGAATTTGTG |
| miR-16 | 5′-AGCAGCACGTAAATATTGGCG-3′ |
| miR-24 | 5′-GCTGCCTACTGAGCTGATATCAGT -3′ |
| miR-125b | 5′- GTCCCTGAGACCCTAACTTGTGA -3′ |
| miR-145 | 5′- GTCCAGTTTTCCCAGGAATCCCT -3′ |
| miR-150 | 5′-GC-TCTCCCAACCCTTGTACC-3′ |
| miR-155 | 5′-GCGG TTAATGCTAATCGTGATA-3′ |
| miR-203-3p | 5′-GTGAAATGTTTAGGACCACTAG-3′ |
| miR-221 | 5′- GCACCTGGCATACAATGTAGAT-3′ |
| miR-222 | 5′- CTCAGTAGCCAGTGTAGATC -3′ |
| miR-223 | 5′-CGTGTATTTGACAAGCTGAGTT -3′ |
| miR-331-3p | 5′-CCCTGGGCCTATCCTAGAA-3′ |
| miR-451 | 5′-CAAACCGTTACCATTACTGAGTT-3′ |
| miR-574-5p | 5′-TGAGTGTGTGTGTGTGAGTGT -3′ |
| miR-848-5p | 5′-CACACACACACACACACGTAT-3′ |
| miR-1908-5p | 5′-TATAGGGACGGCGATTGGTC-3′ |
| Reverse qPCR Primer | 5′-CGAGGAAGAAGACGGAAGAAT-3′ |
The level of miRNAs expression was measured using the quantification cycle (Cq) value. miRNAs expression assay was used and quantified by the comparative 2−ΔΔCq method [27] and normalized to U6 expression. We chose the small nuclear RNA (snRNA) U6 as a housekeeping gene. For a quantitative analysis of 18 miRNAs expression, the amplification of cDNAs by the qRT-PCR method was done using LightCycler FastStart DNA Master SYBR Green I on a LightCycler® 480 System (Roche, Sydney, Australia), according to the manufacturer's instructions, and each sample was analyzed in triplicate. The 20 μl real-time PCR volume included 0.5 μl of RT product, 1 μl of miRNA specific forward qPCR primer (4uM), 1 μl of universal reverse qPCR primer (4uM) which is derived from sequences within universal stem-loop primer, and 17.5 μl of SYBR Green real-time PCR Master Mix. The reactions were incubated at 95 °C for 10 min, followed by 45 cycles of 95 °C for 10s, 60 °C for 5s, 72 °C for 10s.
Statistical analysis
The relative amount of the miRNA expression level was calculated using the Cq value and presented as mean ± sd. The miRNA expression levels were first compared among the three groups of subjects: the healthy controls, the weak ABO antigen expression group, and the constant ABO antigen expression group using the Kruskal–Wallis test. Following three-group tests, pair-wise comparisons of the miRNA expression levels were then carried out using the Mann–Whitney U test between any two of the three groups. Linear regression was then performed to examine the linear correlation between any significantly identified miRNA and the level of antigen, measured by the multiparametric flow cytometry (FCM) %. All statistical analyses were performed using the SPSS 18.0 program for Windows. A test result was considered statistically significant if p<0.05.
Results
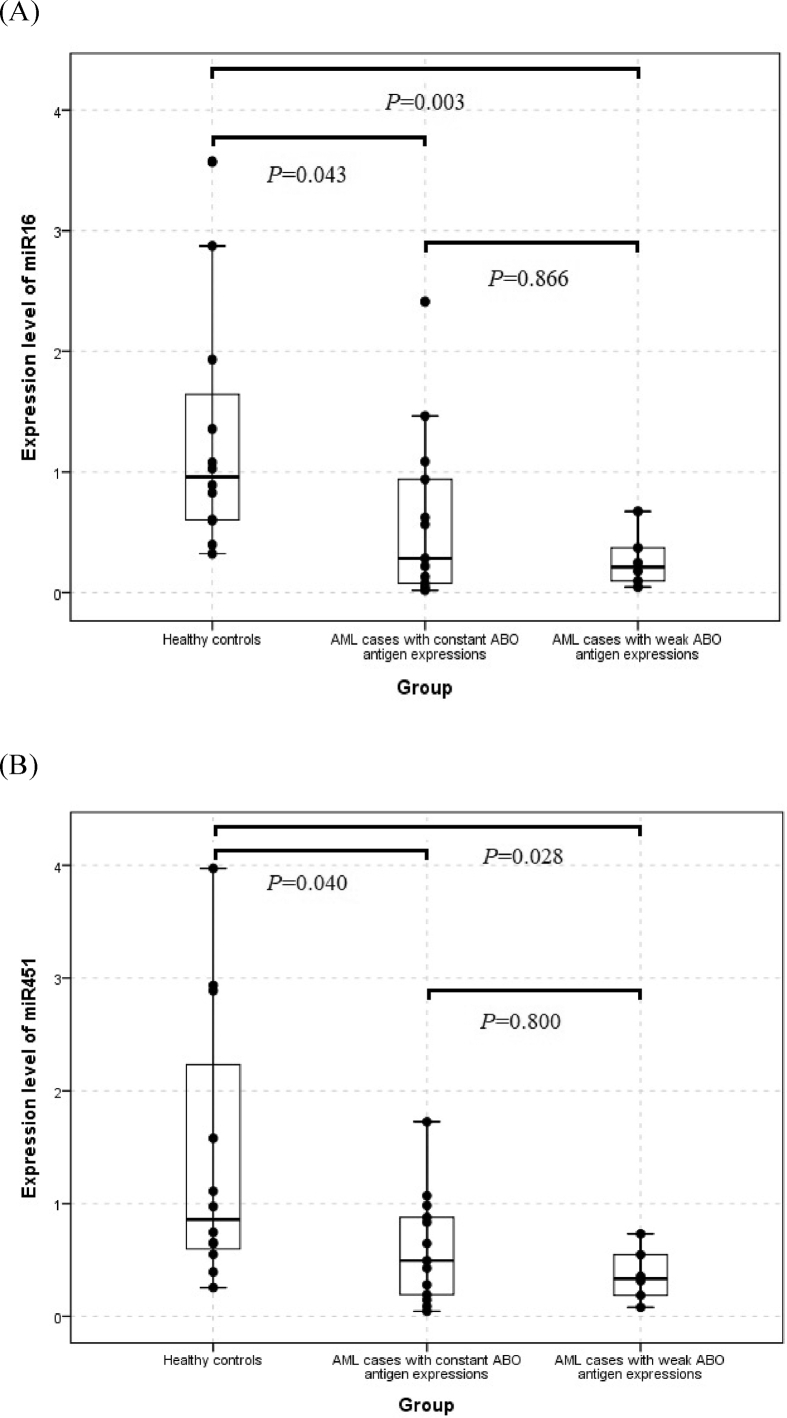
In the comparisons of the expression levels for 18 miRNAs among the control group, the AML group with weak ABO antigen expression and the AML group with constant ABO antigen expression, the results of the Kruskal–Wallis test revealed that the expression of miR-16 (p = 0.015) and miR-451 (p = 0.042) were significant among the three groups. In addition, both the quantity of miR-16 and miR-451 were the lowest in the AML cases with weak ABO antigen expression and the highest in the healthy controls (Table 2). Furthermore, the results of pair-wise comparisons using the Mann–Whitney U test suggested that, the miR-16 expression was significantly lower in either AML cases with weak ABO antigen expressions (p = 0.003) or AML cases with constant ABO antigen expression (p = 0.043), compared with the normal controls. Similarly, the expression level of miR-451 was significantly lower in both AML cases with weak ABO antigen expression (p = 0.028) and AML cases with constant ABO antigen expression (p = 0.040) than the healthy controls. However, the statistical differences of both miR-16 and miR-451 expression were not significant between these two AML sub-groups (Fig. 1). The target genes of miR-16 and miR-451 were summarized in Supplementary Table 3.
Table 2.
The miRNA expression levels (mean ± SD) in the AML sub-groups with weak/constant ABO antigen expression and in the control group.
| AML |
Control |
|||
|---|---|---|---|---|
| Weak (N = 7) | Constant (N = 12) | All (N = 19) | (N = 12) | |
| miR155 | 1.39 ± 1.17 | 1.98 ± 1.28 | 1.76 ± 1.24 | 1.46 ± 1.33 |
| miR150 | 0.65 ± 0.45 | 0.77 ± 0.58 | 0.73 ± 0.52 | 1.07 ± 0.40 |
| miR221 | 1.60 ± 1.41 | 1.58 ± 0.97 | 1.59 ± 1.11 | 1.31 ± 0.94 |
| miR222 | 1.66 ± 1.57 | 1.49 ± 1.09 | 1.55 ± 1.24 | 1.26 ± 0.92 |
| miR223 | 1.49 ± 1.68 | 1.44 ± 1.03 | 1.46 ± 1.26 | 1.39 ± 1.19 |
| miR95p | 2.59 ± 3.63 | 2.66 ± 5.17 | 2.64 ± 4.55 | 1.28 ± 1.01 |
| miR2033P | 3.93 ± 6.58 | 3.06 ± 4.58 | 3.38 ± 5.24 | 1.48 ± 1.37 |
| miR5745P | 3.83 ± 6.95 | 2.23 ± 2.76 | 2.82 ± 4.63 | 1.36 ± 1.14 |
| miR8485P | 4.25 ± 7.37 | 3.71 ± 7.21 | 3.91 ± 7.07 | 1.32 ± 0.98 |
| miR3313P | 3.78 ± 5.99 | 3.22 ± 4.99 | 3.43 ± 5.22 | 1.27 ± 0.97 |
| miR19085P | 1.26 ± 0.71 | 1.88 ± 3.17 | 1.65 ± 2.53 | 1.09 ± 0.48 |
| miR1 | 2.12 ± 1.44 | 3.15 ± 6.17 | 2.77 ± 4.92 | 1.55 ± 1.45 |
| miR10a | 1.39 ± 0.97 | 1.97 ± 3.12 | 1.76 ± 2.52 | 1.14 ± 0.58 |
| miR24 | 0.66 ± 0.39 | 1.45 ± 3.38 | 1.16 ± 2.68 | 2.67 ± 3.88 |
| miR125b | 1.48 ± 1.75 | 2.20 ± 1.55 | 1.94 ± 1.62 | 1.58 ± 1.71 |
| miR145 | 1.11 ± 0.74 | 0.82 ± 0.56 | 0.92 ± 0.63 | 1.17 ± 0.63 |
| miR16 | 0.31 ± 0.24a | 0.61 ± 0.74a | 0.50 ± 0.62a | 1.29 ± 1.01b |
| miR451 | 0.46 ± 0.32a | 0.57 ± 0.49a | 0.53 ± 0.43a | 1.39 ± 1.21b |
The Mann–Whitney U test compared with the control group had p< 0.05.
The Kruskal–Wallis test among the control group, the AML group with weak ABO antigen expressions and the AML group with constant ABO antigen expressions had p< 0.05.
Fig. 1.
Boxplots comparing the expression level of (A) miR-16 and (B) miR-451 between any two of the three groups. The Mann–Whitney U test p-values were indicated.
Discussion
The mechanism of weak ABO expression in patients with leukemia has not been comprehensively studied yet. This was the first report to demonstrate that miRNA expression was significantly different in AML patients with weakened ABO and healthy control. Studies have indicated that miRNA expression was regulated during hematopoietic differentiation, suggesting miRNA involvement in the hematopoietic differentiation process [8]. However, the relationship between the weakening of the ABO antigen and the regulation of miRNA has not been determined. Our results indicated that erythroid-specific miR-451 and miR-16 were downregulated in AML patients and especially in those with weak ABO expression.
In 2007, Zhan et al. examined the expression profiles of 295 miRNAs in an erythroid culture system using array hybridization and real-time PCR, and determined that miR-451 increased more than the other miRNAs in cellular content [28]. Rathjen et al. demonstrated miR-451 was abundant in human RBCs [29]. Moreover, reticulocytes are young RBCs and contain many types of messenger RNAs (mRNAs), which were still being translated for RBC function during the last stage of erythropoiesis [30]. In addition, miR-16 had a positive correlation to the expression of erythroid surface antigens and hemoglobin synthesis [31]. Consequently, some of the mRNAs in reticulocytes might be the target gene of miR-451 and miR-16 and then resulted in weakened ABO.
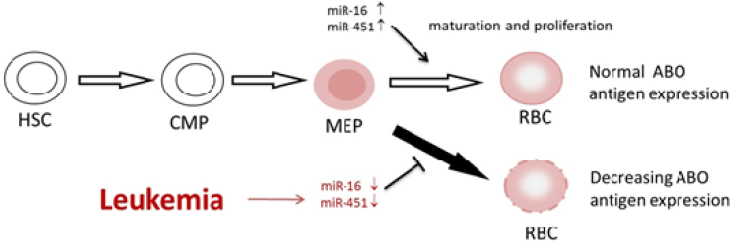
The reason why lower miR-16 and miR-451 expression levels were observed in the AML patients in contrast to the controls (Table 2 and Fig. 1) might be due to that the transcription level of ABO gene would be changed in AML patients [32], and that gene transcription level would be affected by miRNA regulation [33]. According to Havelange V and Garzon R's research [34], it indicated that miR-16 and miR-451 were increased during hematopoiesis from megakaryocyte–erythroid progenitor cell (MEP) to erythrocyte, and a study of Bruchova H et al. [35] showed that there was a significantly overexpression of miR-16 and miR-451 in reticulocytes, which is one of MEP. Consequently, we supposed that miR-16 and miR-451 were involved in the phase of MEP transform into RBC and then influenced the expression level of ABO antigen by transforming the sugars or proteins on RBCs membrane [36], and we created a concept map to illustrate that in Fig. 2.
Fig. 2.
The concept map that there might be a down-regulation of miR-16 and miR-451 during human erythroid differentiation. HSC: Hematopoietic stem cell; CMP: common myeloid progenitor; MEP: megakaryocyte–erythroid progenitor cell; RBC: red blood cell.
These findings were consistent with the finding regarding Plasmodium vivax infection. Goldberg DE et al. proposed that Plasmodium parasites eat, digest, and thereby receive nutrients from hemoglobin in RBCs [37]. Additionally, Rathjen et al. (2006) revealed that the level of miR-451 in Plasmodium-infected RBCs was lower than that in healthy RBCs [25]. Moreover, hypersplenism in malaria infection increases clearance of plasma miRNAs. In the pathophysiology of malaria infection, the spleen increases destruction of infected and noninfected RBCs [38]. This hypersplenism induces increased trapping of blood cells as well as the circulation of microparticles containing miRNAs. Therefore, malaria-induced hypersplenism might cause a decrease in plasma miR-451 and miR-16 levels [39]. The function of miRNA-16 is to regulate cell differentiation and apoptosis, and which had a positive association with the expression level of erythroid surface antigens [31]. Therefore, the reason of the weakened ABO antigen expression for the decrease of miRNA-16 expression might be like the mechanism of above-mentioned antigen.
The erythroid maturation was regulated by miR-451 according to repressing Cox10 and inhibiting mitochondrial activity [40]. Oxidative stress regulation plays an important role in erythropoiesis, which involved in the maturation of erythrocytes, differentiation of erythroid cell progenitors, and so on [41]. Consequently, the erythroid cell progenitors would be destroyed by the oxidative stress when miR-451 was decreased [42] his oxidative stress might be one of the reasons for the weakened expression of ABO antigen via modifying the lipids and proteins on the membrane of RBCs [43].
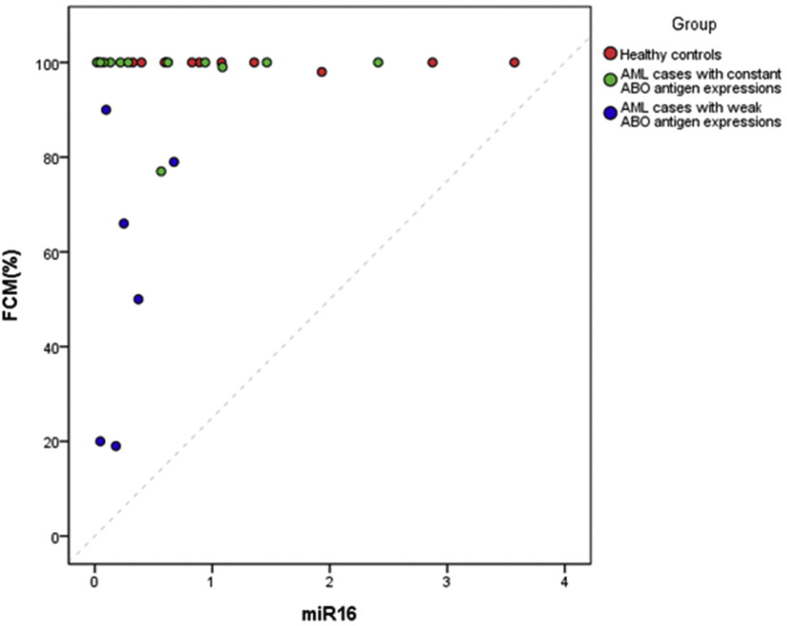
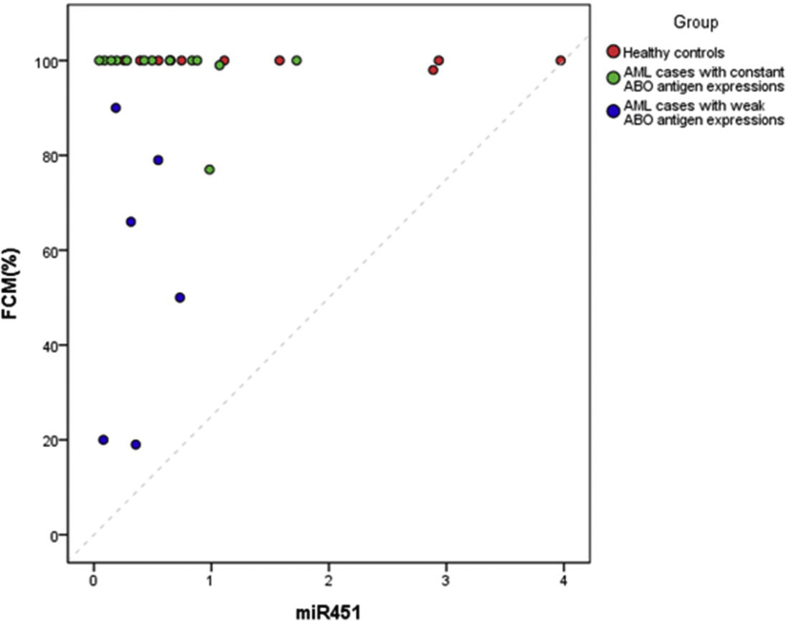
We have performed linear regression analyses to explore the relationship between the miR-16/miR-451 and the level of antigen, measured by the multiparametric flow cytometry (FCM) % in our case. We found that the linear correlation seemed not to be able to ideally delineate the relationship between miRNA and the level of antigen due to the frequency distribution of the level of antigen in our subjects (Supplementary Fig. 2(A) and 2(B)). For all the subjects (N = 31), the coefficient estimates of both miR-16 and miR-451 on the level of antigen were not significant with p = 0.1299 and 0.2391 and adjusted R-Square = 0.0471 and 0.0152, respectively. For the AML cases (N = 19), the condition was similar, and the coefficient estimates of both miR-16 and miR-451 on the level of antigen were not significant with p = 0.2924 and 0.4458 and adjusted R-Square = 0.0099 and −0.022, respectively.
The miR-16 and miR-451 expressions were not statistically different between the AML patients with and without decreased ABO antigens, possibly due to our limited sample size. The miR-16 and miR-451 were elevated in the final mature stage of red blood cells in healthy people, but in contrast, they were reduced in leukemia patients [30]. Therefore, it would be easier to identify statistically significant differences between leukemia patients and healthy people. According to our experimental data, the level of miR-16 and miR-451 were much lower in AML patients with weakened ABO than those with normal ABO expression (Table 2). However, a larger number of samples would be needed to identify statistical differences between leukemia patients with and without weakened ABO, even though we have seen an obvious trend in their proportions. Another possible reason was that other factors weakening the ABO antigen were not excluded, such as promoter methylation loss the ABO allelic expression [2,44], and oxidative stress modify lipid and proteins on RBCs [43]. However, there were several studies reporting that miR-16 and miR-451 were associated with late stage of erythroid differentiation [45], and Choong et al. showed a correlation of miR-16 expression with increase of erythroid surface antigens (Glycophorin A, MNS blood group) [31], which echoed our hypothesis. Nevertheless, there was large variations in the miR-16 and -451 expressions, which might have influenced the final statistical result with our limited sample size. We speculated that the possible reason of the outliers was that the microRNA or U6 might be degraded during the extraction of total RNA, which might have affected the results of RT-PCR. Although U6 was usually used as the internal control and normalization in the early quantitative analysis of microRNA, the stability of U6 was not satisfactory [46,47]. It was also reported that the expression of U6 might differ in different organizations [48], so the internal control should be carefully selected and employed in the future.
Conclusions
In conclusion, in addition to the observation that miR-16 and miR-451 decreased in AML patients, the miR-16 and miR-451 expression level had significance between healthy controls and AML with weakened ABO antigen expression. We speculated that the weak ABO antigen expression might correlate with miRNAs, especially miR-16 and miR-451, and the decreased miR-16 and miR-451 were related to the maturation and differentiation of erythrocytes and ABO antigen synthesis. Moreover, miRNA might be a factor in the expression of A or B antigen by transforming the sugars or proteins on RBCs membrane. In the future, we will continue to include more study samples and exclude the others factor influencing ABO weakening, promoter methylation and oxidative stress, to replicate the results of this study and investigate the underlying mechanism of decreasing miR-16 and miR-451 in the weakened ABO.
Funding
This work was supported by the Chang Gung Memorial Hospital at Linkou, Taoyuan, Taiwan. The grants CMRPG3D1731, CMRPG3D1732, and CMRPG3D1733 to D.P. Chen.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
The excellent consulting assistance and sample resources from molecular diagnosis laboratory of Chang-Gung Memorial Hospital are gratefully acknowledged.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2022.03.003.
Appendix A. Supplementary data
The following are the supplementary data to this article:
figs1.
figs2.
figs3.
References
- 1.Salmon C., Cartron J.P., Lopez M., Rahuel C., Badet J., Janot C. Level of the A, B and H blood group glycosyltransferases in red cell membranes from patients with malignant hemopathies. Rev Fr Transfus Immunohematol. 1984;27(5):625–637. doi: 10.1016/s0338-4535(84)80084-7. [DOI] [PubMed] [Google Scholar]
- 2.Bianco-Miotto T., Hussey D.J., Day T.K., O'Keefe D.S., Dobrovic A. DNA methylation of the ABO promoter underlies loss of ABO allelic expression in a significant proportion of leukemic patients. PLoS One. 2009;4(3):e4788. doi: 10.1371/journal.pone.0004788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moreno-Moya J.M., Vilella F., Simón C. MicroRNA: key gene expression regulators. Fertil Steril. 2014;l10(6):1516–1523. doi: 10.1016/j.fertnstert.2013.10.042. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 5.Gregory R.I., Chendrimada T.P., Cooch N., Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–640. doi: 10.1016/j.cell.2005.10.022. [DOI] [PubMed] [Google Scholar]
- 6.Lu J., Guo S., Ebert B.L., Zhang H., Peng X., Bosco J., et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14(6):843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelstein L.C., Bray P.F. MicroRNAs in platelet production and activation. Blood. 2011;117(20):5289–5296. doi: 10.1182/blood-2011-01-292011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vasilatou D., Papageorgiou S., Pappa V., Papageorgiou E., Dervenoulas J. The role of microRNAs in normal and malignant hematopoiesis. Eur J Hematol. 2010;84(1):1–16. doi: 10.1111/j.1600-0609.2009.01348.x. [DOI] [PubMed] [Google Scholar]
- 9.Chen C.Z., Li L., Lodish H.F., Bartel D.P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 10.Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Facchiano F., et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down- modulation. Proc Natl Acad Sci U S A. 2005;102(50):18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mraz M., Chen L., Rassenti L.Z., Ghia E.M., Li H., Jepsen K., et al. miR-150 influences B-cell receptor signaling in chronic lymphocytic leukemia by regulating expression of GAB1 and FOXP1. Blood. 2014;124(1):84–95. doi: 10.1182/blood-2013-09-527234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vasilescu C., Rossi S., Shimizu M., Tudor S., Veronese A., Ferracin M., et al. MicroRNA fingerprints identify miR-150 as a plasma prognostic marker in patients with sepsis. PLoS One. 2009;4(10) doi: 10.1371/journal.pone.0007405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Magrelli A., Azzalin G., Salvatore M., Viganotti M., Tosto F., Colombo T., et al. Altered microRNA expression patterns in hepatoblastoma patients. Transl Oncol. 2009;2(3):157–163. doi: 10.1593/tlo.09124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nilsen T.W. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 2007;23(5):243–249. doi: 10.1016/j.tig.2007.02.011. [DOI] [PubMed] [Google Scholar]
- 15.Mattes J., Collison A., Foster P.S. Emerging role of microRNAs in disease pathogenesis and strategies for therapeutic modulation. Curr Opin Mol Therapeut. 2008;10(2):150–157. [PubMed] [Google Scholar]
- 16.Chen D.P., Tseng C.P., Wang W.T., Sun C.F. Use of cell study models to confirm the weak ABO phenotypes caused by point mutations among Taiwanese. Ann Clin Lab Sci Autumn. 2011;41(4):346–352. [PubMed] [Google Scholar]
- 17.Chen D.P., Tseng C.P., Wang W.T., Sun C.F. Genetic and mechanistic evaluation for the mixed-field agglutination in B3 blood type with IVS3+5G>A ABO gene mutation. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0037272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D.P., Sun C.F., Ning H.C., Peng C.T., Wang W.T., Tseng C.P. Genetic and mechanistic evaluation for the weak A phenotype in Ael blood type with IVS6 + 5G>A ABO gene mutation. Vox Sang. 2015;108(1):64–71. doi: 10.1111/vox.12196. [DOI] [PubMed] [Google Scholar]
- 19.Chen D.P., Tseng C.P., Lin C.J., Wang W.T., Sun C.F. Mechanistic evaluation for mixed-field agglutination in the K562 cell study model with exon 3 deletion of A1 gene. Ann Clin Lab Sci Fall. 2015;45(6):674–679. [PubMed] [Google Scholar]
- 20.Yang L.H., Wang S.L., Tang L.L., Liu B., Ye W.L., Wang L.L., et al. Universal stem-loop primer method for screening and quantification of MicroRNA. PLoS One. 2014;9(12) doi: 10.1371/journal.pone.0115293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yendamuri S., Calin G.A. The role of microRNA in human leukemia: a review. Leukemia. 2009;23(7):1257–1263. doi: 10.1038/leu.2008.382. [DOI] [PubMed] [Google Scholar]
- 22.Chuang M.K., Chiu Y.C., Chou W.C., Hou H.A., Chuang E.Y., Tien H.F. A 3-microRNA scoring system for prognostication in de novo acute myeloid leukemia patients. Leukemia. 2015;29(5):1051–1059. doi: 10.1038/leu.2014.333. [DOI] [PubMed] [Google Scholar]
- 23.Kronstein-Wiedemann R., Milanov P., Gubbe K., SeifriedE Tonn T. Mirna regulation of blood group ABO genes. Blood. 2015:126–158. [Google Scholar]
- 24.Trino S., Lamorte D., Caivano A., Laurenzana I., Tagliaferri D., Falco G., et al. MicroRNAs as new biomarkers for diagnosis and prognosis, and as potential therapeutic targets in acute myeloid leukemia. Int J Mol Sci. 2018;19(2):E460. doi: 10.3390/ijms19020460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schotte D., Pieters R., Den Boer M.L. MicroRNAs in acute leukemia: from biological players to clinical contributors. Leukemia. 2012;26(1):1–12. doi: 10.1038/leu.2011.151. [DOI] [PubMed] [Google Scholar]
- 26.Dell'aversana C., Altucci L. miRNA-mediated deregulation in leukemia. Front Genet. 2012;3:252. doi: 10.3389/fgene.2012.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Zhan M., Miller C.P., Papayannopoulou T., Stamatoyannopoulos G., Song C.Z. MicroRNA expression dynamics during murine and human erythroid differentiation. Exp Hematol. 2007;35(7):1015–1025. doi: 10.1016/j.exphem.2007.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rathjen T., Nicol C., McConkey G., Dalmay T. Analysis of short RNAs in the malaria parasite and its red blood cell host. FEBS Lett. 2006;580(22):5185–5188. doi: 10.1016/j.febslet.2006.08.063. [DOI] [PubMed] [Google Scholar]
- 30.Lee E., Choi H.S., Hwang J.H., Hoh J.K., Cho Y.H., Baek E.J. The RNA in reticulocytes is not just debris: it is necessary for the final stages of erythrocyte formation. Blood Cells Mol Dis. 2014;53(1-2):1–10. doi: 10.1016/j.bcmd.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 31.Choong M.L., Yang H.H., McNiece I. MicroRNA expression profiling during human cord blood-derived CD34 cell erythropoiesis. Exp Hematol. 2007;35(4):551–564. doi: 10.1016/j.exphem.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Zhang W.L., Liu J.L., Zhang W., Zhuang Y.L. The potential association of the transcription levels of the ABO gene with the disease phases in AML patients. Transfus Apher Sci. 2017;56(5):719–722. doi: 10.1016/j.transci.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 33.Catalanotto C., Cogoni C., Zardo G. MicroRNA in control of gene expression: an overview of nuclear functions. Int J Mol Sci. 2016;17(10):E1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Havelange V., Garzon R. MicroRNAs: emerging key regulators of hematopoiesis. Am J Hematol. 2010;85(12):935–942. doi: 10.1002/ajh.21863. [DOI] [PubMed] [Google Scholar]
- 35.Bruchova H., Yoon D., Agarwal A.M., Mendell J., Prchal J.T. Regulated expression of miRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol. 2007;35(11):1657–1667. doi: 10.1016/j.exphem.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nambiar R.K., Narayanan G., Prakash N.P., Vijayalakshmi K. Blood group change in acute myeloid leukemia. Proc (Bayl Univ Med Cent) 2017;30(1):74–75. doi: 10.1080/08998280.2017.11929536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goldberg D.E., Slater A.F., Cerami A., Henderson G.B. Hemoglobin degradation in the malaria parasite Plasmodium falciparum: an ordered process in a unique organelle. Proc Natl Acad Sci U S A. 1990;87(8):2931–2935. doi: 10.1073/pnas.87.8.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buffet P.A., Safeukui I., Deplaine G., Brousse V., Prendki V., Thellier M., et al. The pathogenesis of Plasmodium falciparum malaria in humans: insights from splenic physiology. Blood. 2011;117(2):381–392. doi: 10.1182/blood-2010-04-202911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chamnanchanunt S., Fucharoen S., Umemura T. Circulating microRNAs in malaria infection: bench to bedside. Malar J. 2017;16(1):334. doi: 10.1186/s12936-017-1990-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu P., Palmer L.E., Lechauve C., Zhao G., Yao Y., Luan J., et al. Regulation of gene expression by miR-144/451 during mouse erythropoiesis. Blood. 2019;133(23):2518–2528. doi: 10.1182/blood.2018854604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Socolovsky M. Molecular insights into stress erythropoiesis. Curr Opin Hematol. 2007;14(3):215–224. doi: 10.1097/MOH.0b013e3280de2bf1. [DOI] [PubMed] [Google Scholar]
- 42.Yu D., dos Santos C.O., Zhao G., Jiang J., Amigo J.D., Khandros E., et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010;24(15):1620–1633. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carl H., Soumya R., Srinivas P., Vani R. Oxidative stress in erythrocytes of banked ABO blood. Hematology. 2016;21(10):630–634. doi: 10.1080/10245332.2016.1187824. [DOI] [PubMed] [Google Scholar]
- 44.Shao M., Lyu X.P., Tang P., Yang Q.K., Zhu W.T., Song J., et al. [Comparison of weak ABO antigen and normal ABO antigen in patients with acute leukemia] Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2017;25(5):1307–1313. doi: 10.7534/j.issn.1009-2137.2017.05.006. Chinese. [DOI] [PubMed] [Google Scholar]
- 45.Bruchova H., Yoon D., Agarwal A.M., Mendell J., Prchal J.T. Regulated expression of microRNAs in normal and polycythemia vera erythropoiesis. Exp Hematol. 2007;35(11):1657–1667. doi: 10.1016/j.exphem.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang M., Zeng Y., Yang R., Xu H., Chen Z., Zhong J., et al. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun. 2014;454(1):210–214. doi: 10.1016/j.bbrc.2014.10.064. [DOI] [PubMed] [Google Scholar]
- 47.Hirschberger S., Hübner M., Strauß G., Effinger D., Bauer M., Weis S., et al. Identification of suitable controls for miRNA quantification in T-cells and whole blood cells in sepsis. Sci Rep. 2019;9(1):15735. doi: 10.1038/s41598-019-51782-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lou G., Ma N., Xu Y., Jiang L., Yang J., Wang C., et al. Differential distribution of U6 (RNU6-1) expression in human carcinoma tissues demonstrates the requirement for caution in the internal control gene selection for microRNA quantification. Int J Mol Med. 2015;36(5):1400–1408. doi: 10.3892/ijmm.2015.2338. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.