Abstract
Background
During the early postnatal life, gut microbiota development experiences dynamic changes in their structural and functional composition. The postnatal period is the critical window to develop a host defense mechanism. The maturation of intestinal mucosal barrier integrity is one of the essential defense mechanisms to prevent the entry of pathogens. However, the co-development of intestinal microbial colonization, formation of barrier integrity, and intestinal epithelial cell layer is not entirely understood.
Methods
We studied the gut microbial composition and diversity using 16S rRNA marker gene-based sequencing in mice to understand postnatal age-dependent association kinetics between gut microbial and intestinal development. Next, we assessed the intestinal development by in vivo gut permeability assay, mRNA gene expression of different tight junction proteins and intestinal epithelial cell markers, goblet cells population, villus length, and cecal IgA quantification.
Results
Our results showed a significant shift in gut microbial structural and functional composition from postnatal day 14 onwards with early life Proteobacteria abundance. Relative abundance of Verrucomicrobia was maximum at postnatal day 14 and showed a gradual decrease over time. We also observed an age-dependent biphasic pattern in barrier integrity improvement and differentiation of intestinal epithelial cells (IECs). A significant improvement in barrier integrity between days 1 and 7 showed the host factor contribution, while that beyond day 14 revealed an association with changes in microbiota composition. Our temporal correlation analysis associated Bacteroidetes phylum with the mucosal barrier formation during postnatal development.
Conclusions
The present study revealed the importance and interplay of host factors and the microbiome in gut development and intestinal mucosal homeostasis.
Keywords: Gut barrier integrity, Intestinal epithelial cells (IECs), Gut microbiota, Postnatal period, Akkermansia muciniphila (A. muciniphila), Goblet cells
Graphical abstract
At a glance of commentary
Scientific background on this subject
Gut plays a vital role in defending the host from pathogenic infection. It is believed that gut maturity and resident microbes contribute to the defense mechanism. However, the relationship between gut development and gut microbial dynamics is still unknown during neonatal development.
What this study adds to the field?
The current work elucidated the cooperation between gut development and the microbial ecosystem. We reported the age-dependent pattern of the intestinal epithelium and barrier integrity development from the first day of postnatal birth. Specific intestinal microbial signatures associated with gut maturation will further strengthen the role of developing microbes.
Introduction
Following birth, the postnatal period is one of the most crucial windows to influence health [1]. During the postnatal period, the developing immune system is perhaps the primary reason to make newborns susceptible to pathogenic infections and other diseases. While most infections occur via impaired mucosal layer, gastrointestinal tract, or gut -the largest mucosal surface acts as an immuno-physiochemical barrier against luminal pathogens and antigens [2]. Intestinal epithelium prevents systemic translocation of pathogenic microorganisms and helps in selective absorption of nutrients and mucosal immunity development [3,4]. Additionally, the intestinal epithelium is a crucial hub of numerous immunological mediators, including chemokines and cytokines [5,] and is involved in antigen processing and recognizing microbial signals [6,7]. Therefore, proper structural and functional maturation of intestinal epithelium and associated immunity during the postnatal period shape individuals' health.
Along with other constituent members, which reinforce postnatal intestinal development, colonization of the microbial population in the mammalian intestine plays a vital role in modulating overall host immunity, physiology, and metabolism [8]. Microbial colonization starts early and undergoes dynamic changes until a stable gut microbial ecology is established [9]. Cross-talk between gut microbial ecosystem and host intestinal epithelium contributes to priming gut mucosal immunity and homeostasis.
Early life gut microbiota is majorly composed of Lactobacillus, Bifidobacterium, Streptococcus genus and experiences lower stability and bacterial richness [[10], [11], [12]]. Also, various factors, namely diet, delivery mode, antibiotic usage, and host genetics, shape the orchestrated succession of microbiota in the developing gut [[13], [14], [15], [16]]. Recent studies suggested that gut microbiota and their derived metabolites can regulate intestinal epithelial cells (IECs) transcriptomic profile [17], proliferation, and differentiation of different IECs [18,19] and also maintain gut barrier integrity [20]. Gut microbial dysbiosis could lead to various neonatal diseases such as necrotizing enterocolitis (NEC) [21] and late-onset sepsis (LOS) [22]. Previous reports indicated dynamic changes in different tight junction (TJ) proteins contributing to mature barrier formation [23] and global intestinal gene expressions [24] during the postnatal period in mice. Intestinal epithelium constantly experiences structural and functional changes during the postnatal period populated with i) absorptive cell-enterocytes, ii) secretory cells-paneth cells, goblet cells, and enteroendocrine cells [25].
Although gut microbiota's role in maintaining epithelial barrier integrity is known, how the neonatal shifts in the gut microbial composition and function influence gut development and mucosal immunity remain elusive. The current knowledge does not provide clear evidence of gut microbial association with temporal changes in barrier integrity formation during the neonatal to adulthood. Beaumont et al., in their recent study, showed the contribution of microbiota-derived metabolites in intestinal barrier maturation using the suckling rabbit model. But this study did not address the early postnatal days right after the birth [26]. We aimed to study the natural co-development of intestinal microbiota and gut mucosal barrier maturation at different postnatal age groups (1D, 7D, 14D, 21D & 28D) from right after the day of birth. We also tried to find specific microbial signatures at the particular postnatal days and correlate them with a significant structural and functional shift in intestinal development. We took advantage of a short gestation period of the mice model that allows the study of the pronounced change of the intestinal mucosa after birth, contrary to humans exhibiting already differentiated crypt-villus architecture at birth [27]. We used 16S rRNA-based characterization of postnatal gut microbiota composition and function in the current study. We employed various assays and evaluations, such as in vivo barrier function assay, gene and protein level expression of different markers from the host gut, goblet cell population, villi length, and cecal IgA production, to characterize the intestinal development and microbial interactions. The current results revealed an age-dependent pattern in intestinal microbiota's structural and functional development. Results also iterated specific microbial signatures that strongly coincide with postnatal mice's intestinal mucosal layer maturation.
Materials and methods
Animals, study design and sample collection
Institutional Animal Ethics Committee (IEAC), NISER, approved all laboratory animal experiments according to the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA), Govt. of India guidelines. We mated female 6-week-old C57BL/6 mice by housing two females and one adult male (8 weeks old) mice per cage. We housed breeding colonies in specific-pathogen-free (SPF), temperature and humidity-controlled (22 ± 3 °C and 55 ± 5% humidity) Individually ventilated cages (IVC) with a 12 h light and 12 h dark cycle. We provided a standard rodent chow diet (cat# AF6000B, Krishna Valley Agrotech LLP, India) (61.02% carbohydrate, 32.46% protein, 6.52% fat w/w) and water ad libitum. After birth, we pooled neonatal mice from multiple litters and redistributed them to standardize the litter size and minimize the in-cage variations [22]. We did not consider biological sex for practical reasons in selecting pups. We collected neonatal mice from litters of different dams to avoid dam-related bias while sacrificing at the experimental age groups. We weaned the neonatal mice 21 days after birth and housed them in separate cages till the 28th day. Maternal milk was the sole food and water source before weaning the pups, and we provided standard rodent chow after weaning of mice. Each week we recorded the body weight of individual neonatal mice. We harvested the entire cecum, colon, ileum, and blood during the sacrifice, as detailed in the following sections.
Cecal DNA extraction (n = 3 litters/age group)
We sacrificed mice by an overdose of CO2 asphyxiation with additional decapitation for 1D and 7D old mice. Due to the technical challenge of collecting adequate biosamples from individual neonatal mice, we pooled the entire cecum content with tissue from 3 to 6 neonatal mice from the same littermates per age group (1D-28D) and collected them in sterile micro-centrifuge tubes. Immediately we snap-frozen the samples and stored them at −80 °C freezer until further analysis. We extracted DNA from the cecal homogenate using QIAamp DNA Stool Mini Kit (Qiagen, Germany) as per the manufacturer's instructions. After that, we measured the concentration and purity of the extracted DNA using a NanoDrop (Thermo Fisher Scientific, USA) and Qubit4 fluorometer (Invitrogen, USA). We checked the DNA quality by running agarose gel electrophoresis.
FITC-dextran gut permeability assay (n = 3 litters/age group)
We determined the in vivo intestinal barrier integrity by measuring the translocation of FITC-dextran to serum described previously [23]. Briefly, neonatal mice were fasted by separating from their dams for 4 h. To minimize the maternal separation stress during the fasting, we placed home bedding and nesting material from the dam's cage. Subsequently, we orally administered mice with 22 μl g−1 (bodyweight) of 22 μg μl−1 3–5 kDa fluorescein isothiocyanate (FITC) conjugated dextran (Sigma–Aldrich, St. Louis, MO, USA) with a 24 G feeding needle. After 5 h of administration, we sacrificed mice and collected whole blood by decapitation (1D&7D) or cardiac puncture (14D-28D) and allowed it to clot at room temperature in the dark. We then prepared serum by centrifugation (1700 g, 4 °C) for 15 min and diluted in PBS. We loaded 100 μl serum samples in each well of a black 96-well microtiter plate (Corning, USA). We measured FITC fluorescence in serum using Varioskan Flash multimode reader (Thermo Fisher Scientific, USA) with excitation at 485 nm and emission at 528 nm. We calculated the serum FITC concentration from a calibration curve of known FITC-dextran dilutions. Serum FITC-dextran concentration is inversely proportional to the gut barrier integrity. Thus, we converted the serum FITC-dextran concentration to percentage gut barrier integrity.
RNA isolation from the neonatal mice tissue (n = 3 litters/age group)
After sacrifice, we dissected mice colon and ileum from the small intestine, washed with ice-cold PBS, cut into small pieces, and stored in RNAlater (Sigma–Aldrich, St. Louis, MO, USA) at −20 °C. We pooled tissue samples from 3 to 6 neonatal mice from the same littermates due to technical challenges. Then we extracted total RNA from tissue using the RNeasy Mini kit (Qiagen, Germany), as per the manufacturer's instructions. We homogenized the snap-frozen tissue samples in RLT buffer, then processed the tissue lysate in RNeasy mini-spin columns (Qiagen, Germany). We eluted the total RNA in nuclease-free water. We measured RNA concentration and absorbances at 230, 260, and 280 nm using a NanoDrop (Thermo Fisher Scientific, USA) to get the ratios at 260/280 and 260/230 to estimate the RNA quality.
Reverse Transcription-qPCR (RT-qPCR)
According to the manufacturer's protocol, we reverse-transcribed total RNA to cDNA using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, USA). We synthesized 2 μg of cDNA per 80 μl of reactions and stored it in −20 °C freezer till further use. Next, we performed a quantitative real-time PCR reaction with a total reaction volume of 10 μl containing 5 μl of GoTaq qPCR Master Mix (Promega, USA), 1 μl of cDNA template (25 ng per reactions), 1 μl of each 10 μM primers (IDT), and rest of nuclease-free water. We carried out qRT-PCR amplification in triplicate using fast 96-well optical plate format on QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems, USA) with the following conditions: 95 °C for 2 min, followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s and melt curve stage consisted of 95 °C for 15 s, 60 °C for 1 min, 95 °C for 15 s. As mentioned elsewhere, we designed and selected all the PCR primers using NCBI Primer-BLAST and OligoAnalyzer Tool (IDT) with exon-exon junction specificity [28]. We validated all the newly designed primer specificity (electrophoresis gel, melt curve profile, and non-template control). We have mentioned the primer sequences for the gene expression study in [Table 1]. Finally, we normalized the target gene expression to the housekeeping gene GAPDH as a reference gene and calculated the ΔCt value. We presented mRNA expression as % gene expression using the following formula.
| % gene expression = (ΔCt max - ΔCti)/(ΔCt max - ΔCt min) |
where, ΔCt max = maximum ΔCt value of a gene among the postnatal days; ΔCt min = minimum ΔCt value of a gene among the postnatal days and ΔCti = ΔCt value of a specific postnatal day.
Table 1.
List of gene names and respective PCR primers used in the gene expression study.
| Gene name | Primer sequences | Amplicon size (bp) | Reference | |
|---|---|---|---|---|
| Cldn3 | Forward | 5′-CCTCATCGTGGTGTCCATCC-3′ | 139 | This study |
| Reverse | 5′-CGCCAACAGGAAAAGCACTC-3′ | |||
| Ocln | Forward | 5′-TAGGCGACAGCGGTGGAGTT-3′ | 78 | This study |
| Reverse | 5′-ATAAGCGAACCTGCCGAGCC-3′ | |||
| ZO1 | Forward | 5′-TGTTTATGCGGACGGTGGCG-3′ | 106 | This study |
| Reverse | 5′-GGCCCTGGCGGACATCTTGT-3′ | |||
| Muc2 | Forward | 5′-GTGATTGTGTTTCAGGCTCC-3′ | 185 | Naik et al. (2019)[28] |
| Reverse | 5′-GTACATGGCAAAAGTCCCAC-3′ | |||
| Ki67 | Forward | 5′-CGACTGCGAGCTTCACCGAG-3′ | 164 | This study |
| Reverse | 5′-CAATACTCCTTCCAAACAGGCAGG-3′ | |||
| Lgr5 | Forward | 5′- GGCAACAGTGTGGACGACCT-3′ | 152 | This study |
| Reverse | 5′- GGCTTGAAGGGACCTGGGGA-3′ | |||
| ChgA | Forward | 5′- CAACCGCAGAGCAGAGGACC-3′ | 198 | This study |
| Reverse | 5′- AGGGGCAAAGGGGGTAACCT-3′ | |||
| Lyz1 | Forward | 5′- GGTCTACAATCGTTGTGAGTTGG-3′ | 92 | This study |
| Reverse | 5′- CTAAACACACCCAGTCAGCCA-3′ | |||
| GAPDH | Forward | 5′- TTCCAGGAGCGAGACCCCACT-3′ | 151 | This study |
| Reverse | 5′- CGGCAGAAGGGGCGGAGATG-3′ | |||
16S rRNA gene sequencing and microbiome analysis
We processed the genomic DNA samples with 16S rRNA library preparation and next-generation sequencing at the Eurofins Genomics Laboratory facility (Bangalore, India). We amplified V3–V4 region of the 16S rRNA gene using the following primer sequences: 16S_F = 5′-GCCTACGGGNGGCWGCAG-3′ and 16S_R = 5′-ACTACHVGGGTATCTAATCC-3′. Briefly, we prepared the (2 300 bp) amplicon libraries using Nextera XT Index kit (Illumina Inc., USA) as per the 16S Metagenomic Sequencing Library preparation protocol. We then purified the amplicon libraries by AMPure XP beads (Beckman Coulter, USA) and quantified them using a Qubit4 fluorometer (Invitrogen, USA). After obtaining a mean peak size from the 4200 Tape Station system (Agilent Technologies, USA), we pooled the libraries and sequenced them using the MiSeq Illumina platform (Illumina Inc., USA).
We processed the raw reads generated after sequencing through a bioinformatics pipeline. First, we obtained the high-quality (HQ) reads using Trimmomatic (v0.38) [29] to trim adapter sequences, ambiguous reads, and low-quality sequences (reads with more than 10% quality threshold <20 Phred score). Then we merged an average of 151,605 ± 61,312 (mean ± SD) HQ read using the FLASH (v1.2.11) program [30] and analyzed the pre-processed FASTQ files using Quantitative Insights into Microbial Ecology (QIIME) software pipeline [31]. We performed a closed-reference Operational Taxonomic Unit (OTU) picking for the SILVA 16S rRNA database [32] as reference using the UCLUST algorithm [33] with 97% similarity and assigned taxonomies. We rarefied all the sample reads to minimum sequencing depth and performed α-diversity, β-diversity, and OTU abundance analyses using a web-based platform- MicrobiomeAnalyst [34]. We calculated the relative abundance (%) of Rodentibacter and Akkermansia from the genus level sequencing reads. We used the Tax4Fun package [35] available in MicrobiomeAnalyst. First, we generated a KO counts table from the Tax4Fun function and incorporated the gene abundance table in the Shotgun Data Profiling (SDP) module. We selected KEGG Orthology (KO) as gene ID. After initial data integrity check and filtering, we used functional diversity profiling. We presented abundance data (total hits) by binning related genes to the KEGG metabolism functional category.
16S qPCR quantification of total bacterial DNA and Akkermansia muciniphila
To determine the total cecal bacterial load, we quantified the 16S rRNA gene from extracted DNA (described previously) by quantitative real-time PCR (qPCR) using universal bacterial primers [36] -Forward: 5′ TCCTACGGGAGGCAGCAGT-3′ and Reverse 5′-GGACTACCAGGGTA TCTAATCCTGTT-3′. We normalized the samples to the amount of input total DNA concentration. We performed qPCR reaction using PowerUp™ SYBR™ Green Master Mix (Applied Biosystems, USA) on QuantStudio™ 7 Flex Real-Time PCR System (Applied Biosystems, USA) with the following conditions: 50 °C for 2 min, 95 °C for 2 min followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Next, we constructed the standard curve of Ct value from serial dilutions of known concentration E.coli DH5α genomic DNA and calculated 16S rRNA gene concentration of cecal DNA [37]. Further, we calculated the copy number from DNA concentrations. We quantified A. muciniphila in the cecal sample with 16S rRNA qPCR using species-specific primer pairs (AM1: 5′-CAGCACGTGAAGGTGGGGAC-3′, AM2: 5′-CCTTGCGGTTGGCTTCAGAT-3′) as described previously [38] and calculated the proportion over total bacteria using the following formula [39].
| X = [(Eff. Univ.) Ct univ. / (Eff. Spec.) Ct spec.] x 100 |
where, Eff. Univ is the calculated efficiency of the universal primers and Eff. Spec refers to the efficiency of the Akkermansia specific primers. Primer efficiencies were obtained by the linear regression curve of the Ct value data points. “X” represents the percentage of 16S species-specific copy numbers in each cecal sample.
Histology and Alcian Blue/PAS staining (n = 3 litters/age group)
After sacrifice, we fixed PBS flushed mice ileum tissues in 4% paraformaldehyde for 48 h at room temperature, dehydrated, and embedded in paraffin following standard procedures. We made 4 μm sections from paraffin blocks using HistoCore MULTICUT rotary microtome (Leica Biosystems, Germany). We deparaffinized the slides in xylene and rehydrated them by serially decreasing alcohol concentrations. For morphometric analysis (villus height), we stained sections using Haematoxylin-eosin (H&E). We stained mucus-filled goblet cells with Alcian blue and periodic acid-Schiff (PAS) stain as mentioned elsewhere with some modifications [18]. Briefly, we stained slides with Alcian blue (pH 2.5), then inserted them into periodic acid (1%) solution for 5 min. After washing, we immersed slides in Schiff reagent for 15 min and washed them in warm water before counterstaining with hematoxylin. Next, we mounted the sections with DPX mounting media (Himedia, India) and obtained microscopic images using a CKX53 light microscope (Olympus Corporation, Japan) under 20× and 40× magnification. We quantified the total number of goblet cells per villus-crypt unit (10 villus-crypt units section/per section/3 sections per animal/3 animals per age group) according to Rodríguez-Daza et al. [39] with some modifications. Using ImageJ software, we measured the villus height (in μm) from approximately 20 well-oriented villus-crypt units (NIH, USA). We blinded the identity of the samples during the analysis of the images to the investigator.
Immunohistochemistry (n = 3 litters/age group)
We fixed the dissected tissues in 4% paraformaldehyde for 24 h at room temperature, then transferred to 30% sucrose in PBS overnight at 4 °C, frozen in liquid nitrogen, and stored at −80 °C until used. Next, we embedded the tissues in tissue freezing medium (Leica Biosystems, Germany) and cut frozen transverse sections of a 4.5 μm thickness using a cryostat (CM1860 UV, Leica Biosystems, Germany) and mounted them on positively charged glass slides. We then washed the sections three times with PBS to remove OCT, permeabilized with PBS containing 0.3% Triton X-100 and blocked with 5% BSA in PBS containing 0.1% Tween 20 for 1 h at room temperature. We stained the sections with the following primary antibodies: rabbit anti-mouse ChgA antibody (1:500 dilution, cat# NB120-15160; Novus biologicals), rabbit anti-mouse Lyz1 antibody (1:500 dilution, cat# PA5-16668; ThermoFisher Scientific), and rabbit anti-mouse Ki67 antibody (1:500 dilution, cat# PA5-19462; ThermoFisher Scientific) overnight at 4 °C. After washing, we used a goat anti-rabbit IgG secondary antibody conjugated with Alexa Fluor-555 (1:750 dilution, cat# A21428; ThermoFisher Scientific) for detection, followed by counterstaining the nuclei with DAPI. We then mounted the sections using Fluoromount-G (Invitrogen, USA) and acquired images on an inverted fluorescence microscope (Olympus CKX53, Japan) using 20× objective, fitted with a CCD camera (MicroPublisher 5 RTV, QImaging, Canada). We analyzed the images to count Ki67+, Lyz1+, and ChgA+ cells in the intestinal crypt/villus architecture using Cellsens (Olympus, Japan) and ImageJ software (NIH, USA).
IgA ELISA from the cecal homogenate (n = 3 litters/age group)
According to the manufacturer's instructions, we detected the cecal IgA concentration using RayBio® Mouse IgA ELISA Kit (RayBiotech, GA, USA). Briefly, we prepared the cecal homogenate (50 mg pooled cecal sample per 100 μl) in extraction buffer (1X PBS, 0.5% Tween-20, 0.05% sodium azide) by vigorous vortexing at 4 °C. We then centrifuged the samples (1500 g, 20 min) at 4 °C and added a 1X protease inhibitor cocktail (Sigma–Aldrich, St. Louis, MO, USA) to the supernatant. We then normalized the total protein concentration of the extracted supernatant using the Bradford microplate assay. We performed IgA ELISA with (1:50) sample dilution on the pre-coated 96-well plate and measured the OD at 450 nm. We calculated the IgA concentration of the cecal samples from the known IgA standard curve.
Lysozyme activity assay from the small intestinal tissue (n = 3 litters/age group)
We determined the lysozyme activity using a fluorescence-based lysozyme activity assay kit (BioVision, CA, USA) as per the manufacturer's protocol. We homogenized pooled (as mentioned earlier) snap-frozen small intestinal tissue (15 mg) rapidly in ice-cold assay buffer containing protease inhibitor cocktail (Sigma–Aldrich, St. Louis, MO, USA). We measured the hydrolyzed product's fluorescence (Ex/Em = 360/445 nm) by a fluorescence plate reader following 60 min of incubation with the homogenate. Next, we calculated the enzyme activity (mU/ml) from the 4-Methylumbelliferone standard curve.
Statistical analyses
We performed statistical analyses using Prism 7 software (GraphPad, USA). All data met the assumptions required for the statistical test used. We used one-way or two-way analysis of variance (ANOVA) followed by Tukey's or Dunnett's multiple comparisons tests as appropriate and represented data as mean ± SD for unpaired datapoints. We determined Pearson correlation coefficients (R) to find the association between the relative abundance of bacterial phyla and other parameters considering all the experimental age groups. The values ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001 were considered as statistically significant.
Results
Temporal changes in the gut microbial community during the postnatal development in mice
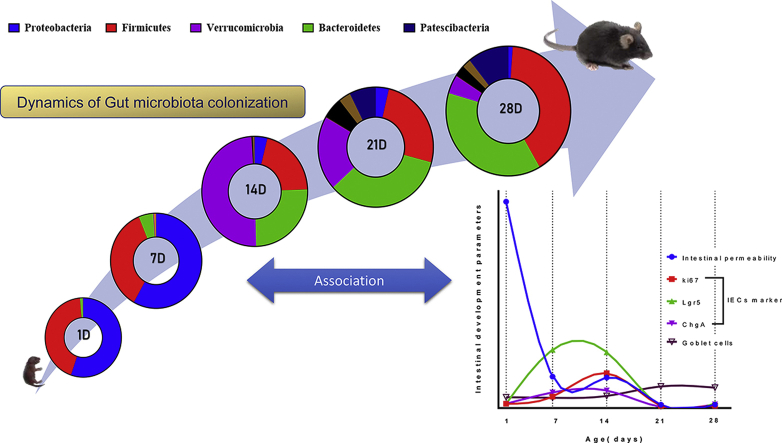
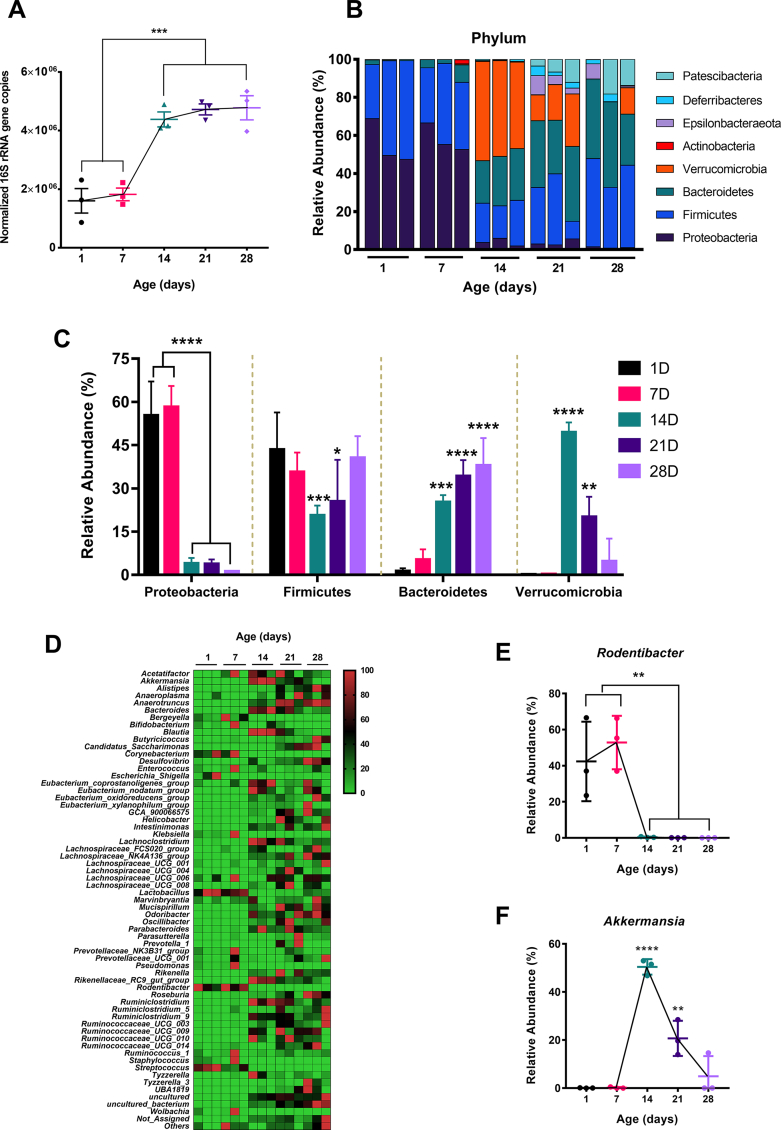
Previous reports showed an age-dependent change in the host gut microbial community following birth [22,26,40]. We performed 16S rRNA-based sequencing from the postnatal mice cecal content to explore the gut microbiota profile dynamics. Bodyweight data showed a gradual increase in postnatal mice [Fig. S1A]. We quantified the total bacterial population in the different postnatal age groups (1D, 7D, 14D, 21D & 28D) by 16S rRNA-based qPCR. We have observed a significant increase (more than 2-fold compared to postnatal days 1 & 7, p ≤ 0.001) in the copy number postnatal day 14 and onwards [Fig. 1A]. Although, we did not observe any significant changes in total bacterial load after postnatal day 14. We detected a prevalence of the Proteobacteria (relative abundance >50%), followed by Firmicutes (relative abundance >35%) phylum during the early postnatal life (1D & 7D) [Fig. 1B]. However, we observed the relative abundance of Proteobacteria is significantly reduced (relative abundance <6%, p ≤ 0.0001) and a transient increase in the Verrucomicrobia level (relative abundance >50%, p ≤ 0.0001) in the cecal microbiota population at postnatal day 14. Also, postnatal day 14 showed the lowest relative abundance of Firmicutes phylum (p ≤ 0.001) after birth.
Fig. 1.
Gut microbiota composition from postnatal day 1 till day 28. (A) Temporal abundance (presented as copy numbers of 16S rRNA gene by qPCR) of gut microbiota in neonatal mice, normalized to total DNA. (B) Stacked bar plots to show the relative taxonomic abundance of bacterial phyla. Each bar represents pooled cecal content from 3 to 6 neonatal mice of the same age. (C) Changes in the relative abundance of major bacterial phyla (Proteobacteria, Firmicutes, Bacteroidetes, and Verrucomicrobia) throughout experimental time points. (D) Heatmap of genus-level bacterial abundance from neonatal cecal microbiota as a function of age. (E, F) Changes in the relative abundance of Rodentibacter (E) and Akkermansia (F) genus (n = 3 biological replicates/group; each biological replicate consists of pooled cecal content from 3 to 6 neonatal mice from littermates of the same dam). (We represented data as mean ± SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001; one-way ANOVA with Dunnett's multiple comparisons test and compared with 1 day).
In contrast, the late postnatal days (21D & 28D) showed a gradual decrease (relative abundance ∼ 20% & 4.6% respectively) of Verrucomicrobia level, and bacterial phyla were majorly composed of Firmicutes and Bacteroidetes (combined relative abundance ∼ 59.5% & 78.3% respectively). Bacteroidetes phylum showed a gradual increase from postnatal day 1 with the highest relative abundance at postnatal day 28 (p ≤ 0.0001) [Fig. 1B, C]. Following, we performed a linear discriminant analysis (LDA) effect size (LEfSe) with LDA score >2 and FDR adjusted p-value > 0.05 to identify the most significant microbial features that are different among the postnatal age groups. Our analysis revealed that the Pasteurellaceae family from the Proteobacteria and Akkermansiaceae family from the Verrucomicrobia phylum (LDA score >6) are the most differentially abundant features [Fig. S1C]. We then characterized the temporal changes at the genus-level bacterial composition [Fig. 1D]. At the genus level also, we found Rodentibacter, a member of the Proteobacteria phylum, highly abundant (relative abundance >40%) at early postnatal days (1D & 7D) but significantly reduced (p ≤ 0.01) at the later development period [Fig. 1D, E]. Similarly, Akkermansia genera also showed a transient increase (relative abundance ∼ 50%) at postnatal day 14, followed by a gradual decrease (relative abundance ∼ 20% [21D] & 4% [28D]) [Fig. 1D, F].
We further validated the relative abundance of Akkermansia from the sequencing reads by a species-specific 16S based qPCR experiment [Fig. S1B]. The result showed a similar trend with the elevated A. muciniphila fold change (more than 2-fold increase than day 1, p ≤ 0.0001) at postnatal day 14. Notably, we have also observed enrichment of Lactobacillus, Streptococcus, Corynebacterium genus in the early days compared to late postnatal days [Fig. 1D]. We next intended to see the possible interactions between the gut microbial communities as a function of postnatal age. Thus, we did a correlation network analysis (Pearson correlation with a p-value threshold of 0.05) at the genus level taxon considering age as variables and focused on Akkermansia and Rodentibacter interaction [Fig. S1D]. Interestingly, we found a negative correlation (r = −0.5732). In summary, the postnatal shift in the cecal microbial community and total bacterial load is age-dependent with early life Proteobacterial relative abundance.
Gut microbial diversity and functional analysis during the postnatal development in mice
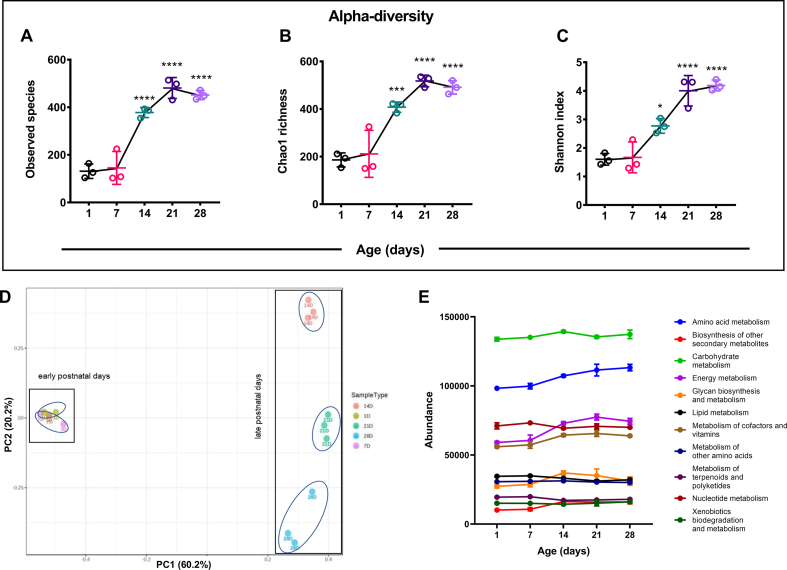
Significant shifts in the gut microbial abundance and community profile prompted us to investigate the diversity pattern of gut microbiota in neonatal mice. The microbial alpha diversity showed a significant shift after birth in postnatal mice. The species richness, calculated by observed species (p ≤ 0.0001) and Chao1 (p ≤ 0.001) index, significantly increased (∼2-fold increase compared to days 1 & 7) from postnatal day 14 onwards [Fig. 2A, B]. We noticed a similar trend for evenness metrics by Shannon index, with reduced microbial diversity at the early postnatal life [Fig. 2C]. Additionally, we performed beta diversity to understand the gut microbial community structure [Fig. 2D]. Principal coordinates analysis (PCoA) plot using Bray–Curtis dissimilarity index clearly separated microbial communities into two populations: early postnatal days (1D & 7D) and late postnatal days (14D, 21D and 28D) across the PC1 axis (60.2% variance) (PERMANOVA, R-squared: 0.84056; p-value < 0.001). Our analysis revealed that gut microbial communities from the 1-day and 7-day old postnatal mice are more similar (clustered together). In contrast, late postnatal days showed more inter-aged group divergent microbiota across the PC2 axis (20.2% variance) [Fig. 2D].
Fig. 2.
Diversity and functional analysis of temporal changes in gut microbiota during the neonatal mice development. (A, B, C) Alpha-diversity index of pooled cecal content from neonate mice; species richness represented as observed species (A), Chao1 index (B), and species evenness calculated using Shannon metrics (C). (D) Gut microbial beta-diversity presented as PCoA plot using the Bray–Curtis dissimilarity index (PERMANOVA, R-squared: 0.84056; p-value < 0.001). (E) Analysis of functional prediction of cecal microbiota using Tax4Fun package from SILVA annotated data. Data represented as an abundance (total hits) of KEGG metabolism (KEGG Orthology groups [KOs] as a functional category). (n = 3 biological replicates/group; each biological replicate consists of pooled cecal content from 3 to 6 neonatal mice from littermates of the same dam). (We represented data as mean ± SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001; one-way ANOVA with Dunnett's multiple comparisons test and compared with 1 day).
Beyond taxonomic composition, the microbial population present in the gut has enormous essential metabolic functions encoded in their genes that contribute to host physiology [41]. Thus, we did a prediction-based functional analysis in the Tax4Fun package from the SILVA annotated data. KEGG metabolism data were extracted from the KEGG Orthology groups [KOs] as a functional category, and their abundance was plotted [Fig. 2E]. We have also found a significant shift in the abundance of different metabolisms at postnatal day 14. We observed an upregulation in amino acid metabolism, energy metabolism, carbohydrate metabolism, and other secondary metabolites biosynthesis and downregulation in nucleotide metabolism at postnatal day 14. Collectively, these data suggested that the development of gut microbial diversity, structural composition, and function are influenced by the postnatal age of mice, showing notable changes at postnatal day 14.
Maturation of gut mucosal barrier integrity in neonatal mice development period
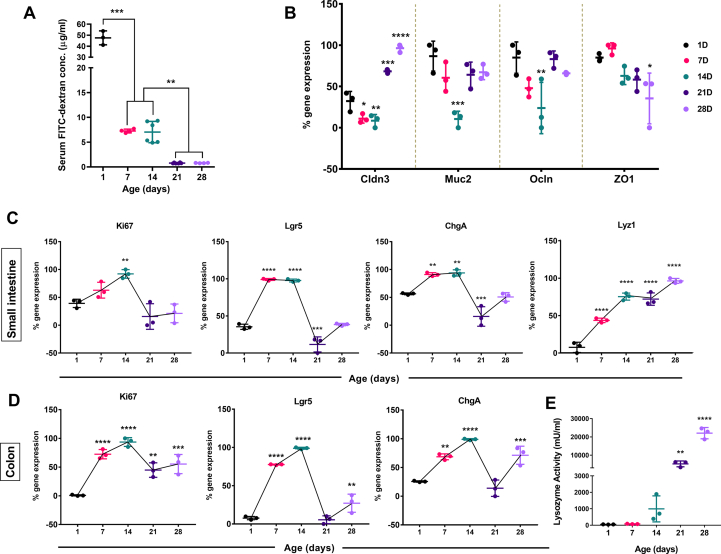
To monitor the maturation of gut barrier integrity postnatally, we performed the FITC-dextran based gut permeability assay. Serum FITC-dextran concentration provides an in vivo picture of functional gut barrier integrity [42]. Serum FITC-dextran concentration 5 h post-administration showed the highest level at postnatal day 1 [Fig. 3A]. We found a significant decline (∼85% reduction in intestinal permeability, p ≤ 0.001) at postnatal day 7, although postnatal days 7 & 14 showed comparable permeability. We observed another decrease in permeability at postnatal day 21 (∼90% reduction in intestinal permeability, p ≤ 0.01) with no change at postnatal 28 days [Fig. 3A]. These data indicate a stepwise gut barrier maturation after birth in neonatal mice. We focused on the small intestinal ileum since ileum was densely colonized with microbiota that interacts with Peyer's patches to induce antigen-specific responses [43,44]. We analyzed the mRNA level gene expression in neonatal mice's small intestinal tissue to understand the role of different tight junction protein markers- Claudin 3 (Cldn3), Occludin (Ocln) & Zonula occludens-1(ZO1) that contribute to barrier integrity [45]. Cldn3 showed a significant upregulation in gene expression at postnatal days 21(p ≤ 0.001) & 28 (p ≤ 0.0001) compared to postnatal day 1. Other markers (Ocln & ZO1) showed no significant upregulation in gene expression over the postnatal age groups [Fig. 3B]. Surprisingly, we found a significant decrease (p ≤ 0.05) in the relative mRNA expression of Mucin 2 (Muc2), an essential component of the intestinal mucus layer [46], only at the postnatal day 14 [Fig. 3B]. Altogether, these results indicate improvement of gut barrier integrity is an age-dependent phenomenon.
Fig. 3.
Development of gut barrier integrity and different intestinal cells marker in neonate mice. (A) In vivo gut integrity development was assessed using serum FITC-dextran concentration (μg/ml) in neonate mice (n = 3–6 mice/experimental age groups) following 5 h of oral gavage. (We represented data as mean ± SD. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001; one-way ANOVA with Tukey's multiple comparisons test). (B) % mRNA gene expression values of different tight junction proteins (Cldn3, Ocln, ZO1) and Muc2 from neonatal mice small intestines using quantitative real-time PCR. (C & D) % mRNA gene expression values of different intestinal cells marker; proliferation marker (Ki67), intestinal stem cell marker (Lgr5), enteroendocrine cell marker (ChgA), and paneth cell marker (Lyz1) in neonatal mice small intestine (C) and colon (D) over experimental age groups. (E) Measurement of lysozyme enzyme activity (mU/ml) from the neonatal mice small intestinal tissue. (n = 3 biological replicates/group; We pooled tissue samples from 3 to 6 neonatal mice from littermates of the same dam for each biological replicate). (We represented data as mean ± SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001; two-way ANOVA [B] and one-way ANOVA [C, D & E] with Dunnett's multiple comparisons test and compared with 1 day).
Differentiation pattern of intestinal epithelium cells in postnatal mice
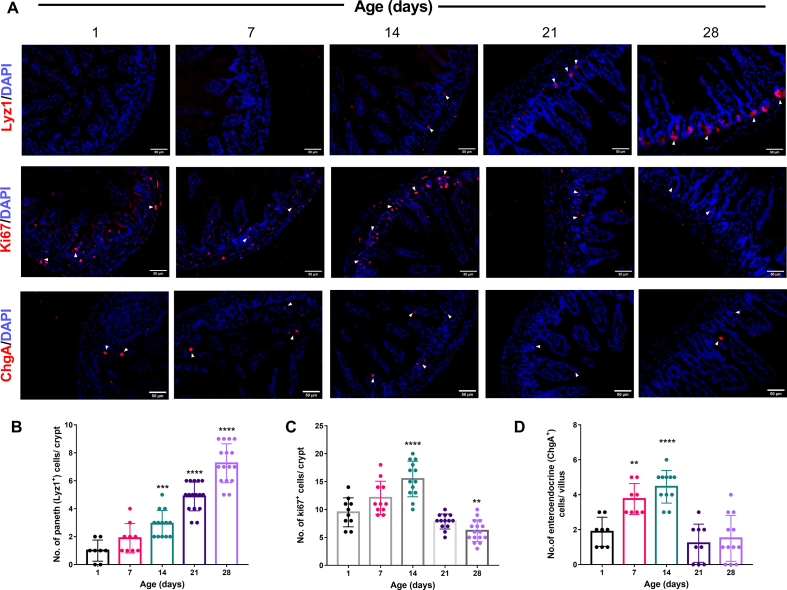
Intestinal epithelial cells (IECs) communicate between commensal microbiota and different immune cells to improve barrier integrity and promote host defense [47]. To understand how the intestinal epithelium layer matures postnatally, we looked into various cell types such as i) enteroendocrine cells, ii) paneth cells, and iii) intestinal stem cells. We checked mRNA level gene expression of markers for each cell type: proliferation marker (Ki67), intestinal stem cell marker (Lgr5), enteroendocrine cell marker (ChgA), and paneth cell marker (Lyz1) [18,25,43] in neonatal mice small intestine and colon over experimental age groups. We observed a continuous increase in Ki67, Lgr5, and ChgA expression from postnatal day 1. These markers reached the maximum expression on postnatal day 14 in small intestinal (Ki67: p ≤ 0.01, Lgr5: p ≤ 0.0001, ChgA: p ≤ 0.01) and colonic (Ki67: p ≤ 0.0001, Lgr5: p ≤ 0.0001, ChgA: p ≤ 0.0001) tissues. At later postnatal days (21D & 28D), we observed a decline in the mentioned markers gene expression [Fig. 3C, D]. In contrast, Lyz1 (paneth cell marker) showed a different pattern with a sustained increase of expression till postnatal day 28 in the small intestine isolated from neonatal mice [Fig. 3C]. We observed a similar trend in the total lysozyme enzymatic activity secreted by the paneth cells from the small intestine as in RT-qPCR data [Fig. 3E]. Further to support the gene expression data, we quantified the Ki67+ proliferative cells, Lyz1+ paneth cells in the small intestinal crypts, and ChgA+ enteroendocrine cells in the villus. Our protein level IHC data showed a similar trend with a significantly increased number of Ki67+ and ChgA+ cells on postnatal day 14 and the highest number of Lyz1+ cells on day 28 [Fig. 4A–D]. Overall, we noticed an age-dependent differentiation pattern of intestinal epithelial cells from the above results.
Fig. 4.
Postnatal age-dependent changes in the number of different types of small intestinal epithelial cells and proliferation markers in mice. (A) Representative fluorescent images for immunostained frozen sections of Lyz1-positive paneth cells, Ki67-positive proliferative cells, ChgA-positive enteroendocrine cells (red), and DAPI stained nuclei (blue), obtained from the postnatal mice small intestine (ileum). (20× magnification, scale bar = 50 μm). Quantification of paneth cells (B), proliferative cells (C) in the intestinal crypts, and enteroendocrine cells (D) in the villus of neonate mice. (Data are represented as means ± SD from ≥8 crypts or villus from 3 animals per age group. ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001; one-way ANOVA with Dunnett's multiple comparisons test and compared with 1 day).
Goblet cell dynamics, villus length, and cecal IgA production during the postnatal period in mice
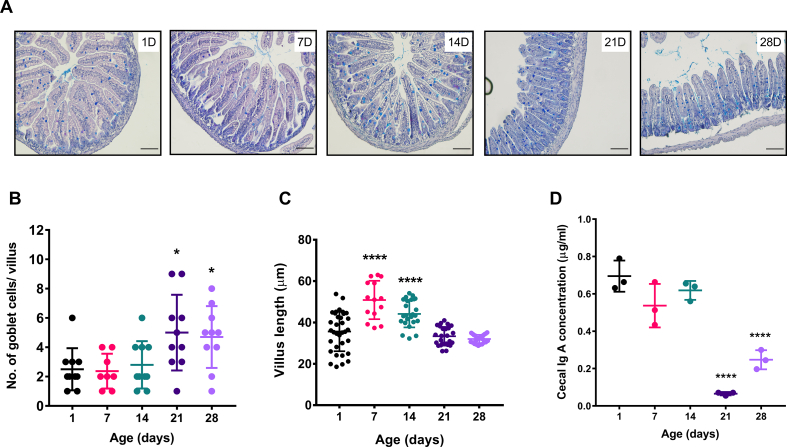
Goblet cells are the key producer of gel-forming mucins [48]. Recent reports indicated that gut commensals (e.g., A. muciniphila) utilize mucin as a significant carbon source and are also involved in host mucous turnovers [49]. We specifically stained the mucin-enriched goblet cells to see whether the increased abundance of A. muciniphila at postnatal days 14 & 21 is associated with goblet cells number. We performed Alcian blue/PAS staining to the neonatal mice's small intestine histological sections. Blinded analysis of the microscopic images revealed a significant increase (p ≤ 0.05) in mucin-producing goblet cell number per villus (blue) at postnatal days 21 and 28 [Fig. 5A, B]. However, we did not notice any significant difference in goblet cell number at postnatal day 14, where Akkermansia abundance was highest. Next, we have also determined the villus length at each experimental age group of neonatal development. We found the most extended villi structure at postnatal days 7 and 14 (average villus length = 50.8 μm) (p ≤ 0.0001), with a gradual decrease in the following days [Fig. 5C].
Fig. 5.
Mucus producing goblet cells, villus length, and cecal IgA production during neonatal mice developmental period. (A) Microscopic images of histological sections (small intestine) stained with Alcian blue/PAS (goblet cells in blue color) of neonate mice. Images are representatives of at least three independent experiments. (20× magnification, scale bar = 50 μm). Scatter plots indicate the (B) number of goblet cells per villus and (C) average villus length (10 villus-crypt units per section/3 sections per animal/3 animals per age group) from 1 day to 28-day old neonate mice. (D) Cecal IgA (μg/ml) level from pooled cecal samples in the experimental age groups. (We represented data as mean ± SD. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001; one-way ANOVA with Dunnett's multiple comparisons test and compared with 1 day).
Secretory IgA (sIgA) is a crucial component of the neonatal gut that regulates epithelial barrier integrity and shapes intestinal microbial homeostasis [50,51]. Thus, to examine how cecal IgA production dynamics are associated with postnatal gut microbiota development, we quantified the level of secreted IgA from the neonatal cecal homogenate by ELISA. Early postnatal 1,7 & 14 days showed a significantly high abundance of cecal IgA (p ≤ 0.0001 and p ≤ 0.0001) compared to late postnatal days (21D & 28D) [Fig. 5D]. Together, these results suggest a gut microbiota-associated pattern in the goblet cell dynamics, villus length, and cecal IgA production as a function of postnatal age.
Association between gut microbiota and intestinal development during the postnatal period
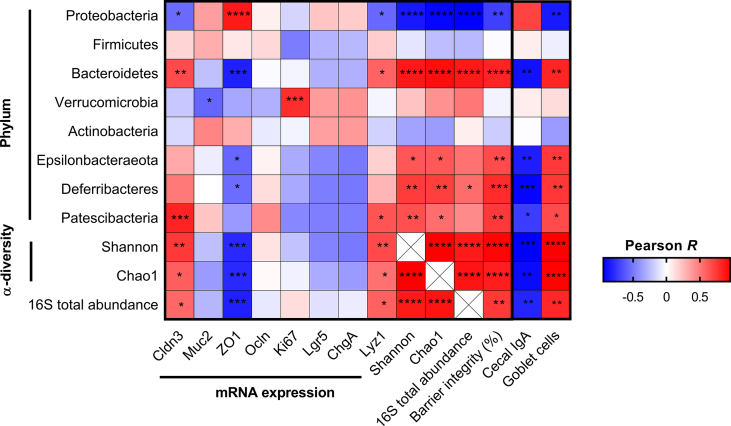
Finally, we intended to evaluate the associations between gut microbiota development and intestinal gene expression of different tight junction proteins (Cldn3, ZO1, Ocln), IECs markers (Ki67, Lgr5, ChgA, Lyz1), barrier integrity (%), cecal IgA, and goblet cells. Thus, we performed a Pearson correlation analysis at the phylum level of gut microbial composition in postnatal mice [Fig. 6] & [Table 2]. Proteobacteria level correlated negatively with Cldn3 (Pearson R = −0.555, p = 0.032), Lyz1 (Pearson R = −0.564, p = 0.028), barrier integrity (%) (Pearson R = −0.715, p = 0.003) and goblet cells (Pearson R = −0.727, p = 0.002). Whereas, Bacteroidetes level strongly correlated with Cldn3 (Pearson R = 0.669, p = 0.006), Lyz1 (Pearson R = 0.581, p = 0.023), barrier integrity (%) (Pearson R = 0.817, p = 0.0001), goblet cells (Pearson R = 0.734, p = 0.002) and negatively correlated with cecal IgA abundance (Pearson R = 0.754, p = 0.001). Interestingly, Verrucomicrobia abundance was positively associated with proliferation marker Ki67 (Pearson R = 0.783, p = 0.001). We did not find any significant correlations for ChgA and Lgr5 with any bacterial phyla. Gut microbiota alpha-diversity (Shannon and Chao1 richness) also significantly negatively correlated with Proteobacteria level and positively correlated with Bacteroidetes level. Additionally, we have found positive correlations between alpha-diversity (Shannon and Chao1 richness), 16S total abundance, and Cldn3, barrier integrity (%). Overall, the correlation analysis revealed specific gut microbial phylum is associated with specific features of neonatal intestinal development.
Fig. 6.
Correlation analysis between gut microbial phyla, alpha-diversity, total bacterial abundance, barrier integrity, and gene expression. Heatmap (correlation matrix) of Pearson correlation coefficients (R) between the relative abundance (%) of bacterial phyla, small intestinal gene expression of tight junction proteins and intestinal epithelial cells (IECs) marker, alpha-diversity metrics, gut barrier integrity (%), 16S rRNA total abundance, cecal IgA and goblet cells during the postnatal development period in mice. (∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001 and ∗∗∗∗p ≤ 0.0001).
Table 2.
Significant correlations between gut microbial phyla, alpha-diversity, total bacterial abundance, barrier integrity, and gene expression.
| Cldn3 |
Muc2 |
ZO1 |
Ki67 |
Lyz1 |
Shannon |
Chao1 |
16S total abundance |
Barrier integrity (%) |
|
|---|---|---|---|---|---|---|---|---|---|
| Pearson R (p-value) | |||||||||
| Proteobacteria | −0.555 (p = 0.032) | 0.360 (p = 0.188) | 0.853 (p = 0.00005) | −0.162 (p = 0.565) | −0.564 (p = 0.028) | −0.864 (p = 0.00003) | −0.928 (p = 5.92e-7) | −0.947 (p = 2.858e-7) | −0.715 (p = 0.003) |
| Bacteroidetes | 0.669 (p = 0.006) | −0.234 (p = 0.401) | −0.836 (p = 0.0001) | −0.042 (p = 0.881) | 0.581 (p = 0.023) | 0.859 (p = 0.00004) | 0.898 (p = 0.00005) | 0.857 (p = 0.00008) | 0.817 (p = 0.0001) |
| Verrucomicrobia | −0.205 (p = 0.465) | −0.572 (p = 0.026) | −0.328 (p = 0.233) | 0.783 (p = 0.001) | −0.037 (p = 0.895) | 0.222 (p = 0.425) | 0.409 (p = 0.129) | 0.504 (p = 0.066) | −0.046 (p = 0.870) |
| Epsilonbacteraeota | 0.314 (p = 0.254) | −0.070 (p = 0.805) | −0.569 (p = 0.026) | −0.319 (p = 0.247) | 0.173 (p = 0.537) | 0.626 (p = 0.012) | 0.621 (p = 0.013) | 0.527 (p = 0.052) | 0.671 (p = 0.006) |
| Deferribacteres | 0.497 (p = 0.059) | −0.001 (p = 0.997) | −0.549 (p = 0.033) | −0.322 (p = 0242) | 0.273 (p = 0.324) | 0.726 (p = 0.002) | 0.716 (p = 0.002) | 0.547 (p = 0.042) | 0.783 (p = 0.001) |
| Patescibacteria | 0.817 (p = 0.0002) | 0.214 (p = 0.444) | −0.381 (p = 0.161) | −0.447 (p = 0.095) | 0.634 (p = 0.011) | 0.653 (p = 0.008) | 0.542 (p = 0.036) | 0.434 (p = 0.120) | 0.734 (p = 0.002) |
| Shannon | 0.741 (p = 0.002) | −0.241 (p = 0387) | −0.795 (p = 0.0003) | −0.228 (p = 0.414) | 0.672 (p = 0.006) | 1.000 | 0.951 (p = 5.701e-8) | 0.852 (p = 0.0001) | 0.929 (p = 5.506e-7) |
| Chao1 | 0.598 (p = 0.019) | −0.375 (p = 0.168) | −0.799 (p = 0.0003) | −0.056 (p = 0.844) | 0.532 (p = 0.041) | 0.951 (p = 5.701e-8) | 1.000 | 0.878 (p = 0.00003) | 0.836 (p = 0.0001) |
| 16S total abundance | 0.563 (p = 0.036) | −0.272 (p = 0.347) | −0.834 (p = 0.0002) | 0.126 (p = 0.667) | 0.586 (p = 0.028) | 0.852 (p = 0.0001) | 0.878 (p = 0.00003) | 1.000 | 0.739 (p = 0.003) |
Discussions
The postnatal period is one of the critical windows for gastrointestinal immunity's structural and functional maturation, which protects against pathogenic infections and shapes up an individual's health [52]. Along with various intrinsic and extrinsic factors, microbial populations in the gut act as major contributors to priming host mucosal barrier immunity [53]. This study characterized the development of gut microbial colonization at different ages and the co-development of the host mucosal barrier integrity in postnatal C57BL/6 mice. We considered suckling neonates right after the day of birth (1D) and continued to wean young adults (28D) to monitor the gut microbiota profile transition. We found that Proteobacteria phylum is the primary colonizer of early neonatal mice gut (1D & 7D), corroborating the previous report [22]. Our genus-level analysis also showed a high relative abundance of Rodentibacter sp. of the Proteobacteria phylum and facultative anaerobes Lactobacillus and Streptococcus of the Firmicutes phylum, which is among well-known early colonizers of mammalian neonatal gut [Fig. 1B, D] [26,[54], [55], [56]]. Consistent with gut microbiota structure, we also observed a reduction in total abundance and intra-species alpha diversity (based on Chao1, Shannon, and observed species), indicating immature gut microbiota at early postnatal days [Figs. 1A, 2A-C]. These findings align with our correlation analysis [Fig. 6], where we found a negative association between Proteobacteria abundance and alpha-diversity metrics. Our results showed a substantial shift in the total bacterial abundance and intestinal microbial alpha diversity at postnatal day 14, suggesting postnatal day 14 as a crucial window in neonatal gut microbiota development [Figs. 1A, 2A-C]. On postnatal day 14, Phylum level analysis revealed Verrucomicrobia as the most abundant phylum [Fig. 1B, C]. Genus level microbiota profile, species-specific 16S qPCR data, and LDA effect size analysis at the family level strongly support the transient increase in Akkermansia of Verrucomicrobia phylum postnatal day 14 [Fig. 1D, S1B & C]. A. muciniphila, a strict anaerobic gut bacterium, recently gained attention because of its several beneficial effects, e.g., glucose homeostasis and gut barrier integrity and mucous turnover [49,57,58]. Late postnatal days (21D & 28D) showed a gradual reduction in Verrucomicrobia relative abundance and comprised majorly of Firmicutes and Bacteroidetes. Other minor bacterial phyla- Actinobacteria, Epsilonbacteraeota, Deferribacteres, and Patescibacteria, were also present at the late postnatal days, which indicate increased diversity and complex microbial structure [Fig. 1B, C]. We observed a significant difference in gut microbial community structure from the Beta-diversity analysis, where again divergence was visible from postnatal day 14, suggesting a dynamic nature of gut microbiota [Fig. 2D]. Longitudinal studies in non-human primate and calf gut microbiota also indicate a shift in bacterial composition such as Bifidobacterium, Lactobacillus, Streptococcus, Bacteroides, Clostridium, and Prevotella associated with social environment and immune system development [[59], [60], [61]]. Consistent with the microbiota composition, predictive functional analysis using the Tax4Fun package strongly supports our observation of a significant shift in neonatal gut microbiota development at postnatal day 14 [Fig. 2E]. Network analysis at the Genus level showed a negative correlation between Akkermansia and Rodentibacter sp. during the neonatal development period, which could explain a competitive microbial interaction in the mammalian gut [Fig. S1D] [62,63]. We observed the drastic change in gut microbiota profile before the diet switched to solid food or weaning. At the same time, other recent temporal studies showed a suckling-weaning gut microbiota transition, where experimental sampling was done from or after postnatal day 14 [26,40]. So we can speculate that host-associated factors such as anti-microbial peptides (AMPs) production, a gradual decrease in luminal oxygen availability in the neonatal gut may be responsible for the shift, rather than exogenous factors [48,64].
Notably, the dynamic shift in the intestinal microbial ecology is associated with altered mucosal barrier integrity. The Gut microbial population overtly shapes the appropriate structure and function of host epithelial barrier integrity, which is a crucial step in neonatal mucosal immunity development [23,43,[65], [66], [67]]. We observed a marked biphasic decrease in serum FITC-dextran concentration on postnatal days 7, 14, and again on postnatal days 21, 28 suggesting maturation of postnatal in vivo barrier integrity [Fig. 3A]. We observed a significant shift in the barrier permeability following postnatal 14 days (∼90% reduction), indicating a subsequent improvement in the barrier function following gut microbial dynamics. However, we did not notice any alteration in the microbiota profile associated with the evident decrease in permeability from postnatal day 1–7. Thus, the biphasic nature of FITC-dextran flux in serum may indicate initial gut barrier formation is associated with the host factors, independent of microbiota composition. Current findings suggest host epithelia-associated factors, particularly anti-microbial peptides (AMPs), mucus barrier, secretory IgAs (sIgAs), tight epithelial junctions, also could modulate gut microbiota shaping [48,68,69]. So, it may be possible that early mucus barrier formation regulates microbiota development, which helps in further barrier improvement after postnatal day 14, suggesting host and microbiota regulate each other's development. An earlier report showed that among a panel of tight junction (TJ) proteins, Cldn3, which acts as a sealing component in TJs, is significantly upregulated during the postnatal maturation period [23,70]. We also found a transcriptional upregulation of Cldn3 from postnatal day 21 in small intestinal tissue, whereas other TJ proteins Ocln and ZO1 showed no significant upregulation [Fig. 3B]. Although, intestinal permeability did not correlate well with the timing of Cldn3 upregulation at early postnatal days. This may be due to confounding factors such as high blood microcirculation [71] and apical canalicular system (ACS) that contribute to increased intestinal permeability at the neonatal period. The renewal of ACSs cells with adult enterocytes as a function of age also contributes in gradual improvement in barrier integrity [72]. Several other studies support that neonatal intestinal maturation needs an appropriate gut ecosystem. Any dysbiosis leads to inflammatory diseases, such as inflammatory bowel diseases, necrotizing enterocolitis, etc. [21,73]. Our study also suggests a possible microbial link where early life abundance of Proteobacteria phylum is associated with an underdeveloped barrier function [Fig. 6] & [Table 2]. Previous reports from our lab and others showed the high quantity of Proteobacteria phylum associated with compromised barrier integrity, intestinal inflammation, and endotoxin in the blood [67,74,75]. Recent studies elucidate the role of A. muciniphila of Verrucomicrobia phylum in barrier integrity improvement [57]. Although the Verrucomicrobia level was notably high at postnatal 14 days, it did not show any significant association with barrier integrity [Fig. 6] & [Table 2]. These findings suggest the overabundance of A. muciniphila may not help improve barrier function in neonates. Our correlation analysis revealed Bacteroidetes as the most strongly correlated phyla with barrier integrity and Cldn3 expression. This observation is in line with previous reports that demonstrate communities of Bacteroidetes phylum improve barrier protection [76] and prevent pathogenic colonization in the intestine [77]. Recent studies have reported that intestinal epithelium encounters significant structural and functional developmental changes during the neonatal to adulthood transition [24,25]. Here we aimed to establish an association between gut microbiota development and kinetics of different intestinal epithelial cells (IECs) at the postnatal period in mice. Strikingly, the transcriptional profiling of different intestinal cells type markers (Ki67, Lgr5 & ChgA) showed upregulation until postnatal day 14, followed by a gradual decrease in the small intestine and colon [Fig. 3C, D]. Intestinal stem cells (ISCs) are responsible for the renewal, repair, and differentiation of intestinal epithelial cells [78,79] and enteroendocrine cells-an intestinal secretory cell plays a vital role in the secretion of hormones such as ghrelin, PYY, serotonin, etc. [80]. To integrate with our microbiome data, we found a positive correlation of Verrucomicrobia with Ki67, Lgr5, and ChgA. However, the association with proliferation marker Ki67 was only statistically significant [Fig. 6] & [Table 2]. These may indicate that the high abundance of Verrucomicrobia is related to intestinal epithelial proliferation and differentiation [81]. We observed a gradual increase in Lyz1-paneth cell marker till postnatal day 28 [Fig. 3C, E], the primary producer of different antimicrobial peptides [25,82]. Gut microbiota or derived metabolites regulate intestinal cell proliferation and functions [18,83]. Also, experiments with germ-free animals showed delayed epithelial cells transition [84], reduced the number of paneth cells and associated proteins [43,85], impaired development of villus capillary network during weaning [86]. Previous reports showed that treatment with A. muciniphila increased mucin-producing goblet cell density [57,58]. In this regard, we have also observed a rise in Alcian blue-PAS positive goblet cell number at postnatal day 21 in the small intestinal sections [Fig. 5A, B]. Furthermore, goblet cells can increase the gut barrier integrity by producing different molecules such as intestinal trefoil factor and resistin-like molecule β [58]. Reports showed that the -high Proteobacteria abundance in different intestinal diseases is negatively associated with host antimicrobial responses [[87], [88], [89]]. Present findings also observed a high Proteobacteria abundance linked with low density of antimicrobial peptide producing cells and goblet cells during postnatal leaky gut before day 7. Even though we have not profiled the mucosal microbiota during neonatal development, which is distinct from stool microbiota, we cannot overlook their importance in establishing intestinal development. Finally, we observed an abundance of cecal IgA concentration in early postnatal life consistent with the previous report [Fig. 5D] [26]. The increased amount of sIgA before the weaning may also be a possible alternative cause of the initial decrease in gut permeability from postnatal day 1–7, in our study [50].
Although our study did not deal with the genetic or disease model to understand the causal effect of microbiota, the identified microbial taxa from our findings are closely associated with the host physiology and metabolism. Identified bacteria, A. muciniphila, could be a promising probiotic to improve neonatal health. Other reports have shown the beneficial role of A. muciniphila in improving gut barrier integrity and glucose metabolism [49,58]. The Bacteroides sp. from Bacteroidetes phylum, associated with neonatal development, helps metabolize carbohydrates and produce short-chain fatty acids [90].
However, the present study has its limitations. Firstly, while pooling mice from multiple litters, we did not consider the biological sex of neonate mice. Thus, our study could not reveal whether the relationship between gut microbiota and intestinal development was sex-dependent. Secondly, we pooled samples from multiple neonates due to technical limitations in sample collection and considered them an experimental unit. So, our data did not resolve the microbial composition changes in individual neonate mice. Finally, the gut microbiome composition depended on vivarium, specific mouse lines, and suppliers [91]. Furthermore, the potential maternal stress induced due to the FITC-dextran experiment in neonatal mice should not be overlooked.
Conclusions
In conclusion, the present study elucidated postnatal gut microbiota and gut barrier integrity co-development in C57BL/6 mice. The results revealed that neonatal gut development was associated with age-dependent differential abundance, composition, and gut microbial diversity. Further studies by perturbing the postnatal gut microbiota will help understand the causal role of the age-dependent microbial profile in regulating intestinal mucosal homeostasis. In summary, the present study provides newer insights into neonate to adulthood gut microbial ecology and evolution. This knowledge could help improve health.
Data availability
All 16S rRNA microbiome sequence reads generated from this study are available in the NCBI SRA database under the Bioproject ID PRJNA647429.
Funding
The intra-mural core funding from DAE, GoI supported this work through parent institute National Institute of Science Education and Research (NISER) to PA. DAE provides Ph.D. fellowship to UP. The funding agency had no role in experimental design, data acquisition, analysis, manuscript preparation, or publication.
Conflicts of interest
The authors declare no conflicts of interest.
Acknowledgment
We want to acknowledge the Animal Research and Experimentation Facility, NISER and Central Instrumentation Facility (CIF), School of Biological Sciences, NISER. We thank lab member Swati.S.Panda for helping in the blind analysis of histological sections.
Footnotes
Peer review under responsibility of Chang Gung University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bj.2022.03.004.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Robertson R.C., Manges A.R., Finlay B.B., Prendergast A.J. The human microbiome and child growth – first 1000 Days and beyond. Trends Microbiol. 2019;27(2):131–147. doi: 10.1016/j.tim.2018.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Takiishi T., Fenero C.I.M., Câmara N.O.S. Intestinal barrier and gut microbiota: shaping our immune responses throughout life. Tissue Barriers. 2017;5(4) doi: 10.1080/21688370.2017.1373208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischoff S.C., Barbara G., Buurman W., Ockhuizen T., Schulzke J.-D., Serino M., et al. Intestinal permeability – a new target for disease prevention and therapy. BMC Gastroenterol. 2014;14:189. doi: 10.1186/s12876-014-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turner J.R. Intestinal mucosal barrier function in health and disease. Nat Rev Immunol. 2009;9(11):799–809. doi: 10.1038/nri2653. [DOI] [PubMed] [Google Scholar]
- 5.Okumura R., Takeda K. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med. 2017;49(5):e338. doi: 10.1038/emm.2017.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henderson P., van Limbergen J.E., Schwarze J., Wilson D.C. Function of the intestinal epithelium and its dysregulation in inflammatory bowel disease. Inflamm Bowel Dis. 2011;17(1):382–395. doi: 10.1002/ibd.21379. [DOI] [PubMed] [Google Scholar]
- 7.Peterson L.W., Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014;14(3):141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 8.Cho I., Blaser M.J. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13(4):260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moore R.E., Townsend S.D. Temporal development of the infant gut microbiome. Open Biol. 2019;9(9):190128. doi: 10.1098/rsob.190128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka M., Nakayama J. Development of the gut microbiota in infancy and its impact on health in later life. Allergol Int. 2017;66(4):515–522. doi: 10.1016/j.alit.2017.07.010. [DOI] [PubMed] [Google Scholar]
- 11.Arrieta M.C., Stiemsma L.T., Amenyogbe N., Brown E.M., Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014;5:427. doi: 10.3389/fimmu.2014.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spor A., Koren O., Ley R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. 2011;9(4):279–290. doi: 10.1038/nrmicro2540. [DOI] [PubMed] [Google Scholar]
- 13.Fallani M., Amarri S., Uusijarvi A., Adam R., Khanna S., Aguilera M., et al. Determinants of the human infant intestinal microbiota after the introduction of first complementary foods in infant samples from five European centres. Microbiology. 2011;157(Pt 5):1385–1392. doi: 10.1099/mic.0.042143-0. [DOI] [PubMed] [Google Scholar]
- 14.Tanaka S., Kobayashi T., Songjinda P., Tateyama A., Tsubouchi M., Kiyohara C., et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol. 2009;56(1):80–87. doi: 10.1111/j.1574-695X.2009.00553.x. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Bello M.G., Costello E.K., Contreras M., Magris M., Hidalgo G., Fierer N., et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fulde M., Sommer F., Chassaing B., van Vorst K., Dupont A., Hensel M., et al. Neonatal selection by Toll-like receptor 5 influences long-term gut microbiota composition. Nature. 2018;560(7791):489–493. doi: 10.1038/s41586-018-0395-5. [DOI] [PubMed] [Google Scholar]
- 17.Schumann A., Nutten S., Donnicola D., Comelli E.M., Mansourian R., Cherbut C., et al. Neonatal antibiotic treatment alters gastrointestinal tract developmental gene expression and intestinal barrier transcriptome. Physiol Genom. 2005;23(2):235–245. doi: 10.1152/physiolgenomics.00057.2005. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y.S., Kim T.Y., Kim Y., Lee S.H., Kim S., Kang S.W., et al. Microbiota-derived lactate accelerates intestinal stem-cell-mediated epithelial development. Cell Host Microbe. 2018;24(6) doi: 10.1016/j.chom.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 19.Park J., Kotani T., Konno T., Setiawan J., Kitamura Y., Imada S., et al. Promotion of intestinal epithelial cell turnover by commensal bacteria: role of short-chain fatty acids. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0156334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly C.J., Zheng L., Campbell E.L., Saeedi B., Scholz C.C., Bayless A.J., et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe. 2015;17(5):662–671. doi: 10.1016/j.chom.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Patel R.M., Denning P.W. Intestinal microbiota and its relationship with necrotizing enterocolitis. Pediatr Res. 2015;78(3):232–238. doi: 10.1038/pr.2015.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deshmukh H.S., Liu Y., Menkiti O.R., Mei J., Dai N., O'Leary C.E., et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat Med. 2014;20(5):524–530. doi: 10.1038/nm.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel R.M., Myers L.S., Kurundkar A.R., Maheshwari A., Nusrat A., Lin P.W. Probiotic bacteria induce maturation of intestinal claudin 3 expression and barrier function. Am J Pathol. 2012;180(2):626–635. doi: 10.1016/j.ajpath.2011.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rakoff-Nahoum S., Kong Y., Kleinstein S.H., Subramanian S., Ahern P.P., Gordon J.I., et al. Analysis of gene–environment interactions in postnatal development of the mammalian intestine. Proc Natl Acad Sci U S A. 2015;112(7):1929–1936. doi: 10.1073/pnas.1424886112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Navis M., Martins Garcia T., Renes I.B., Vermeulen J.L., Meisner S., Wildenberg M.E., et al. Mouse fetal intestinal organoids: new model to study epithelial maturation from suckling to weaning. EMBO Rep. 2019;20(2) doi: 10.15252/embr.201846221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beaumont M., Paes C., Mussard E., Knudsen C., Cauquil L., Aymard P., et al. Gut microbiota derived metabolites contribute to intestinal barrier maturation at the suckling-to-weaning transition. Gut Microb. 2020;11(5):1268–1286. doi: 10.1080/19490976.2020.1747335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tourneur E., Chassin C. Neonatal immune adaptation of the gut and its role during infections. Clin Dev Immunol. 2013;2013 doi: 10.1155/2013/270301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Naik A.K., Pandey U., Mukherjee R., Mukhopadhyay S., Chakraborty S., Ghosh A., et al. Lactobacillus rhamnosus GG reverses mortality of neonatal mice against Salmonella challenge. Toxicol Res. 2019;8(3):361–372. doi: 10.1039/c9tx00006b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Magoc T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K., et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pruesse E., Quast C., Knittel K., Fuchs B.M., Ludwig W., Peplies J., et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35(21):7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 34.Chong J., Liu P., Zhou G., Xia J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat Protoc. 2020;15(3):799–821. doi: 10.1038/s41596-019-0264-1. [DOI] [PubMed] [Google Scholar]
- 35.Asshauer K.P., Wemheuer B., Daniel R., Meinicke P. Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics. 2015;31(17):2882–2884. doi: 10.1093/bioinformatics/btv287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadkarni M.A., Martin F.E., Jacques N.A., Hunter N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology. 2002;148(Pt 1):257–266. doi: 10.1099/00221287-148-1-257. [DOI] [PubMed] [Google Scholar]
- 37.Fachi J.L., Felipe J. de S., Pral L.P., da Silva B.K., Corrêa R.O., de Andrade M.C.P., et al. Butyrate protects mice from Clostridium difficile-induced colitis through an HIF-1-Dependent mechanism. Cell Rep. 2019;27(3) doi: 10.1016/j.celrep.2019.03.054. :750–61.e7. [DOI] [PubMed] [Google Scholar]
- 38.Collado M.C., Derrien M., Isolauri E., de Vos W.M., Salminen S. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol. 2007;73(23):7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodríguez-Daza M.-C., Daoust L., Boutkrabt L., Pilon G., Varin T., Dudonné S., et al. Wild blueberry proanthocyanidins shape distinct gut microbiota profile and influence glucose homeostasis and intestinal phenotypes in high-fat high-sucrose fed mice. Sci Rep. 2020;10(1):2217. doi: 10.1038/s41598-020-58863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hughes K.R., Schofield Z., Dalby M.J., Caim S., Chalklen L., Bernuzzi F., et al. The early life microbiota protects neonatal mice from pathological small intestinal epithelial cell shedding. FASEB J. 2020;34(5):7075–7088. doi: 10.1096/fj.202000042R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heintz-Buschart A., Wilmes P. Human gut microbiome: function matters. Trends Microbiol. 2018;26(7):563–574. doi: 10.1016/j.tim.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Woting A., Blaut M. Small intestinal permeability and gut-transit time determined with low and high molecular weight fluorescein isothiocyanate-dextrans in C3H mice. Nutrients. 2018;10(6):685. doi: 10.3390/nu10060685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu Y., Lu L., Sun J., Petrof E.O., Claud E.C. Preterm infant gut microbiota affects intestinal epithelial development in a humanized microbiome gnotobiotic mouse model. Am J Physiol Gastrointest Liver Physiol. 2016;311(3):G521–G532. doi: 10.1152/ajpgi.00022.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mowat A.M., Agace W.W. Regional specialization within the intestinal immune system. Nat Rev Immunol. 2014;14(10):667–685. doi: 10.1038/nri3738. [DOI] [PubMed] [Google Scholar]
- 45.Ulluwishewa D., Anderson R.C., McNabb W.C., Moughan P.J., Wells J.M., Roy N.C. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011;141(5):769–776. doi: 10.3945/jn.110.135657. [DOI] [PubMed] [Google Scholar]
- 46.Johansson M.E., Larsson J.M., Hansson G.C. The two mucus layers of colon are organized by the MUC2 mucin, whereas the outer layer is a legislator of host-microbial interactions. Proc Natl Acad Sci U S A. 2011;108(Suppl 1):4659–4665. doi: 10.1073/pnas.1006451107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allaire J.M., Crowley S.M., Law H.T., Chang S.Y., Ko H.J., Vallance B.A. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 2018;39(9):677–696. doi: 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 48.Chang C.S., Kao C.Y. Current understanding of the gut microbiota shaping mechanisms. J Biomed Sci. 2019;26(1):59. doi: 10.1186/s12929-019-0554-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Everard A., Belzer C., Geurts L., Ouwerkerk J.P., Druart C., Bindels L.B., et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013;110(22):9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogier E.W., Frantz A.L., Bruno M.E., Wedlund L., Cohen D.A., Stromberg A.J., et al. Secretory antibodies in breast milk promote long-term intestinal homeostasis by regulating the gut microbiota and host gene expression. Proc Natl Acad Sci U S A. 2014;111(8):3074–3079. doi: 10.1073/pnas.1315792111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fransen F., Zagato E., Mazzini E., Fosso B., Manzari C., El Aidy S., et al. BALB/c and C57bl/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity. 2015;43(3):527–540. doi: 10.1016/j.immuni.2015.08.011. [DOI] [PubMed] [Google Scholar]
- 52.Renz H., Brandtzaeg P., Hornef M. The impact of perinatal immune development on mucosal homeostasis and chronic inflammation. Nat Rev Immunol. 2012;12(1):9–23. doi: 10.1038/nri3112. [DOI] [PubMed] [Google Scholar]
- 53.Sommer F., Bäckhed F. The gut microbiota — masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 54.Arboleya Silvia, Binetti Ana, Salazar Nuria, Fernández Nuria, Gonzalo Solís Ana Hernández-Barranco, Abelardo Margolles, Clara G. de los Reyes-Gavilán MG. Establishment and development of intestinal microbiota in preterm neonates. FEMS Microbiol Ecol. 2012;79(3):763–772. doi: 10.1111/j.1574-6941.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 55.Gritz E.C., Bhandari V. The human neonatal gut microbiome: a brief review. Front Pediatr. 2015;3:17. doi: 10.3389/fped.2015.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bittinger K., Zhao C., Li Y., Ford E., Friedman E.S., Ni J., et al. Bacterial colonization reprograms the neonatal gut metabolome. Nat Microbiol. 2020;5(6):838–847. doi: 10.1038/s41564-020-0694-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plovier H., Everard A., Druart C., Depommier C., Van Hul M., Geurts L., et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med. 2017;23(1):107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 58.Shin N.R., Lee J.C., Lee H.Y., Kim M.S., Whon T.W., Lee M.S., et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014;63(5):727–735. doi: 10.1136/gutjnl-2012-303839. [DOI] [PubMed] [Google Scholar]
- 59.Dettmer A.M., Allen J.M., Jaggers R.M., Bailey M.T. A descriptive analysis of gut microbiota composition in differentially reared infant rhesus monkeys ( Macaca mulatta ) across the first 6 months of life. Am J Primatol. 2019;81(10-11) doi: 10.1002/ajp.22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nunez N., Réot L., Menu E. Neonatal immune system ontogeny: the role of maternal microbiota and associated factors. How might the non-human primate model enlighten the path? Vaccines. 2021;9(6):584. doi: 10.3390/vaccines9060584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lyons T., Jahns H., Brady J., O'Hara E., Waters S.M., Kenny D., et al. Integrated analyses of the microbiological, immunological and ontological transitions in the calf ileum during early life. Sci Rep. 2020;10(1):21264. doi: 10.1038/s41598-020-77907-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Coyte K.Z., Rakoff-Nahoum S. Understanding competition and cooperation within the mammalian gut microbiome. Curr Biol. 2019;29(11):R538–R544. doi: 10.1016/j.cub.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Figueiredo A.R.T., Kramer J. Cooperation and conflict within the microbiota and their effects on animal hosts. Front Ecol Evol. 2020;8 [Google Scholar]
- 64.Chong C., Bloomfield F., O'Sullivan J. Factors affecting gastrointestinal microbiome development in neonates. Nutrients. 2018;10(3):274. doi: 10.3390/nu10030274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Halpern M.D., Denning P.W. The role of intestinal epithelial barrier function in the development of NEC. Tissue Barriers. 2015;3(1-2) doi: 10.1080/21688370.2014.1000707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chelakkot C., Ghim J., Ryu S.H. Mechanisms regulating intestinal barrier integrity and its pathological implications. Exp Mol Med. 2018;50(8):103. doi: 10.1038/s12276-018-0126-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ray P., Pandey U., Aich P. Comparative analysis of beneficial effects of vancomycin treatment on Th1- and Th2-biased mice and the role of gut microbiota. J Appl Microbiol. 2021;130(4):1337–1356. doi: 10.1111/jam.14853. [DOI] [PubMed] [Google Scholar]
- 68.Yu L.C.H. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J Biomed Sci. 2018;25(1):79. doi: 10.1186/s12929-018-0483-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Darnaud M., Dos Santos A., Gonzalez P., Augui S., Lacoste C., Desterke C., et al. Enteric delivery of regenerating family member 3 alpha alters the intestinal microbiota and controls inflammation in mice with colitis. Gastroenterology. 2018;154(4) doi: 10.1053/j.gastro.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 70.Milatz S., Krug S.M., Rosenthal R., Günzel D., Müller D., Schulzke J.-D., et al. Claudin-3 acts as a sealing component of the tight junction for ions of either charge and uncharged solutes. Biochim Biophys Acta. 2010;1798(11):2048–2057. doi: 10.1016/j.bbamem.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 71.Watkins D.J., Besner G.E. The role of the intestinal microcirculation in necrotizing enterocolitis. Semin Pediatr Surg. 2013;22(2):83–87. doi: 10.1053/j.sempedsurg.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Garcia T.M., van Roest M., Vermeulen J.L.M., Meisner S., Smit W.L., Silva J., et al. Early life antibiotics influence in vivo and in vitro mouse intestinal epithelium maturation and functioning. Cell Mol Gastroenterol Hepatol. 2021;12(3):943–981. doi: 10.1016/j.jcmgh.2021.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huang X.Z. Bacterial colonization and intestinal mucosal barrier development. World J Clin Pediatr. 2013;2(4):46. doi: 10.5409/wjcp.v2.i4.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mirpuri J., Raetz M., Sturge C.R., Wilhelm C.L., Benson A., Savani R.C., et al. Proteobacteria-specific IgA regulates maturation of the intestinal microbiota. Gut Microb. 2014;5(1):28–39. doi: 10.4161/gmic.26489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hiippala K., Jouhten H., Ronkainen A., Hartikainen A., Kainulainen V., Jalanka J., et al. The potential of gut commensals in reinforcing intestinal barrier function and alleviating inflammation. Nutrients. 2018;10(8):988. doi: 10.3390/nu10080988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang W., Zhu B., Xu J., Liu Y., Qiu E., Li Z., et al. Bacteroides fragilis protects against antibiotic-associated diarrhea in rats by modulating intestinal defenses. Front Immunol. 2018;9:1040. doi: 10.3389/fimmu.2018.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sequeira R.P., McDonald J.A.K., Marchesi J.R., Clarke T.B. Commensal Bacteroidetes protect against Klebsiella pneumoniae colonization and transmission through IL-36 signalling. Nat Microbiol. 2020;5(2):304–313. doi: 10.1038/s41564-019-0640-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15(1):19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 79.Baulies A., Angelis N., Li V.S.W. Hallmarks of intestinal stem cells. Development. 2020;147 doi: 10.1242/dev.182675. [DOI] [PubMed] [Google Scholar]
- 80.Peck B.C.E., Shanahan M.T., Singh A.P., Sethupathy P. Gut microbial influences on the mammalian intestinal stem cell niche. Stem Cell Int. 2017;2017 doi: 10.1155/2017/5604727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim S., Shin Y.C., Kim T.Y., Kim Y., Lee Y.S., Lee S.H., et al. Mucin degrader Akkermansia muciniphila accelerates intestinal stem cell-mediated epithelial development. Gut Microbs. 2021;13(1):1892441. doi: 10.1080/19490976.2021.1892441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Clevers H.C., Bevins C.L. Paneth cells: maestros of the small intestinal crypts. Annu Rev Physiol. 2013;75:289–311. doi: 10.1146/annurev-physiol-030212-183744. [DOI] [PubMed] [Google Scholar]
- 83.Ye L., Mueller O., Bagwell J., Bagnat M., Liddle R.A., Rawls J.F. High fat diet induces microbiota-dependent silencing of enteroendocrine cells. Elife. 2019;8 doi: 10.7554/eLife.48479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Savage D.C., Siegel J.E., Snellen J.E., Whitt D.D. Transit time of epithelial cells in the small intestines of germfree mice and ex-germfree mice associated with indigenous microorganisms. Appl Environ Microbiol. 1981;42(6):996–1001. doi: 10.1128/aem.42.6.996-1001.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hooper L.V., Stappenbeck T.S., Hong C.V., Gordon J.I. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003;4(3):269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 86.Stappenbeck T.S., Hooper L.V., Gordon J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc Natl Acad Sci U S A. 2002;99(24):15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lo Presti A., Zorzi F., Del Chierico F., Altomare A., Cocca S., Avola A., et al. Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease. Front Microbiol. 2019;10:1655. doi: 10.3389/fmicb.2019.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jalanka J., Cheng J., Hiippala K., Ritari J., Salojärvi J., Ruuska T., et al. Colonic mucosal microbiota and association of bacterial taxa with the expression of host antimicrobial peptides in pediatric ulcerative colitis. Int J Mol Sci. 2020;21(17):6044. doi: 10.3390/ijms21176044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yu L.C., Wei S.C., Li Y.H., Lin P.Y., Chang X.Y., Weng J.P., et al. Invasive pathobionts contribute to colon cancer initiation by counterbalancing epithelial antimicrobial responses. Cell Mol Gastroenterol Hepatol. 2022;13(1):57–79. doi: 10.1016/j.jcmgh.2021.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zafar H., Saier M.H., Jr Gut Bacteroides species in health and disease. Gut Microb. 2021;13(1):1–20. doi: 10.1080/19490976.2020.1848158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Long L.L., Svenson K.L., Mourino A.J., Michaud M., Fahey J.R., Waterman L., et al. Shared and distinctive features of the gut microbiome of C57BL/6 mice from different vendors and production sites, and in response to a new vivarium. Lab Anim (NY) 2021;50(7):185–195. doi: 10.1038/s41684-021-00777-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All 16S rRNA microbiome sequence reads generated from this study are available in the NCBI SRA database under the Bioproject ID PRJNA647429.