Abstract
Because the organization of snoRNA genes in vertebrates, plants and yeast is diverse, we investigated the organization of snoRNA genes in a distantly related organism, Trypanosoma brucei. We have characterized the second example of a snoRNA gene cluster that is tandemly repeated in the T.brucei genome. The genes encoding the box C/D snoRNAs TBR12, TBR6, TBR4 and TBR2 make up the cluster. In a genomic organization unique to trypanosomes, there are at least four clusters of these four snoRNA genes tandemly repeated in the T.brucei genome. We show for the first time that the genes encoding snoRNAs in both this cluster and the SLA cluster are transcribed in an unusual way as a polycistronic RNA.
INTRODUCTION
Eukaryotic pre-ribosomal RNA (rRNA) processing and ribosome assembly occurs primarily in the cell nucleolus. Here, rDNA is transcribed by RNA polymerase I into a long 35–40S precursor rRNA which undergoes several processing steps and nucleotide modifications to form the mature rRNAs of the ribosome (1–3). The 18S rRNA comprises the small ribosomal subunit; the 5.8S and 28S rRNAs comprise the large ribosomal subunit. The 5S rRNA is transcribed separately but is also part of the large ribosomal subunit. Unlike higher eukaryotes, the trypanosome 28S rRNA is further processed into the 28Sα, 28Sβ, sr1, sr2, sr4 and sr6 RNAs (4–6). Factors that may be involved in this unusual form of 28S rRNA processing are currently unknown.
Small nucleolar RNAs play a role in both pre-rRNA processing and rRNA nucleotide modification (reviewed in 7–11). In yeast and vertebrates, there are two major classes of snoRNAs, the box C/D and box H/ACA snoRNAs (12). The snoRNAs in both families are defined by conserved sequence elements and by associated nucleolar proteins (reviewed in 8,11,13). A few snoRNAs in each class are involved in pre-rRNA processing but the majority of them are involved in nucleotide modification. Most box C/D snoRNAs are involved in 2′-O-ribose methylation of specific rRNA nucleotides and most box H/ACA snoRNAs are involved in the conversion of specific uridine nucleotides to pseudouridine (14–21). The box C/D snoRNAs involved in methylation, known as methylation guide RNAs, contain complementary guide sequences to rRNA, and the rRNA nucleotide to be modified is always opposite the fifth nucleotide upstream to a box D or D′ sequence. This is referred to as the box D-plus-five rule. Trypanosomes have the U3 box C/D snoRNA homolog (22,23) which in other studied eukaryotes has been shown to be involved in pre-rRNA processing (24–27). Recently, many other trypanosome box C/D snoRNAs have been identified (28–30). Most of the newly identified trypanosome snoRNAs appear to be methylation guide RNAs to rRNA and obey the box D-plus-five rule of methylation (30). Based on the conservation of the guide sequence, several of the snoRNAs appear to be homologs to methylation guide snoRNAs found in yeast and vertebrates (30). Interestingly, no box H/ACA snoRNAs have yet been identified in trypanosomes.
Eukaryotic genomes house snoRNA genes several different ways. The majority of vertebrate snoRNAs are encoded within introns of a host gene (7). Several vertebrate pre-mRNA transcripts bear multiple snoRNAs that are processed from introns, with each intron containing a single snoRNA (31,32). In yeast, the majority of snoRNAs are transcribed from independent promoters although a few are processed from introns of pre-mRNA transcripts (19). Yeast cells also contain three clusters of snoRNA genes that are transcribed by upstream promoters as polycistronic RNAs which are processed by Rnt1p and Rat1p (33–36). Plants also contain clusters of snoRNA genes that contain box C/D snoRNA genes as well as a box H/ACA snoRNA gene (37,38). The plant snoRNA gene clusters are transcribed as polycistronic RNAs by an upstream promoter (38). Trypanosomes contain a cluster of three snoRNA genes located within the spliced leader RNA (SLA) locus (29). The SLA locus is itself tandemly repeated about 11 times in the trypanosome genome (39).
We have characterized the second example of a snoRNA gene cluster that is tandemly repeated in the T.brucei genome. The genes that make up the cluster are the box C/D snoRNA genes TBR12, TBR6, TBR4 and TBR2. So far, clusters of snoRNA genes that are tandemly repeated are unique to trypanosomes. We also show for the first time that both the SLA cluster containing box C/D snoRNA genes TBR17, TBR7 and TBR5 and the box C/D snoRNA gene cluster identified in this report are produced as polycistronic RNAs.
MATERIALS AND METHODS
Growth of T.brucei
The procyclic form of T.brucei rhodesience (YTaT1.1) strain was used in this study (C. Tschudi and E. Ullu, Yale School of Medicine). Cells were grown at 28°C in SM medium supplemented with 20% fetal calf serum (40).
Production of probes to T.brucei snoRNA genes and Southern hybridizations
Plasmids containing snoRNA clones were used in PCR reactions with clone-specific 5′ and 3′ oligonucleotide primers and were α-32P-labeled (3000 Ci/mmol), as described by Dunbar et al. (30). Blots were hybridized to 1 × 107 c.p.m. of labeled probes in 5× SSPE/10× Denhardt’s/7% SDS at 65°C. Blots were washed twice in 3× SSC–0.1% SDS at room temperature for 15 min per wash and once in 3× SSC–0.1% SDS at 65°C for 10 min and exposed to X-ray film.
Identification of a T.brucei P1 phage clone containing the TBR6 snoRNA gene
A library SM10#47 consisting of 1900 single P1 colonies robotically arrayed onto a high density filter was obtained from the Molteno Institute for Parasitology at the University of Cambridge, Cambridge, UK. Each P1 colony contained phage DNA with inserts of T.brucei genomic DNA ranging in size from 20 to 100 kb. The phage DNA also conferred kanamycin resistance. The average insert size in this library is 65 kb and covers ∼3–3.5 times the haploid genome. The filter was hybridized to a PCR-generated probe to the full-length TBR6 snoRNA gene. After hybridization to the P1 phage genomic library, several P1 clones were positive for the TBR6 gene. The positive clones were obtained from the Molteno Institute for Parasitology. The clones were rescreened by gridding each onto an LB agar plate containing 25 µg/ml kanamycin, transferring the colonies to a nitrocellulose membrane and hybridizing the membrane to the labeled TBR6 probe. One P1 phage clone positive for the TBR6 gene was further characterized.
Subcloning and sequencing of P1 phage fragments containing snoRNA genes
P1 phage DNA that was positive for the TBR6 gene was obtained using the following method. The P1 phage clone was grown overnight at 37°C in 100 ml of LB medium containing 25 µg/ml kanamycin. Cells were pelleted and resuspended in 10 ml of gET buffer (50 mM glucose, 10 mM EDTA, 25 mM Tris–HCl pH 8.0). A 2.0 ml solution of 0.2 M NaOH, 1% SDS were added and mixed followed by the addition of 1.5 ml of 3 M potassium acetate. The sample was extracted with phenol/chloroform and precipitated with ethanol. The isolated P1 phage DNA was digested with various restriction enzymes and electrophoresed on a 0.8% agarose gel. The DNA fragments were then transferred onto a Zeta-probe membrane by Southern blotting. The membrane was hybridized with the TBR6 probe and a 0.9 kb RsaI fragment positive for the TBR6 gene was selected and subcloned into the SmaI site of the pGEM-3Z vector.
The 0.9 kb RsaI cloned fragment was completely sequenced on an Applied Biosystems 373 Stretch Sequencer with vector SP6 and T7 primers. A larger BamHI fragment positive for the TBR6 gene was also isolated from the P1 clone and subcloned into the pGEM-3Z vector. Partial sequencing of the 8 kb insert was obtained by primer walking using oligonucleotides SP6, T7, a, b, DD7SP1, DD7SP2 and DD7SP3 (Table 1). Seqman (DNA STAR) was used to consolidate the internal sequences obtained using oligonucleotides a and b with the complete sequence of the 0.9 kb RsaI fragment and the sequence from the ends of the 8 kb insert obtained by sequencing with primers T7, SP6 and DD7SP1–3. Contiguous sequences were generated and a full restriction map was obtained using DNA Strider.
Table 1. Oligonucleotides used in this study.
| a | 5′-CGCGTGATGAGGTGCAGAAGG-3′ |
| b | 5′-CTGCATGATGTGCTCAACTGGAATTAC-3′ |
| c | 5′-AGATGGTAATTCCAGTTGAGC-3′ |
| d | 5′-CGCGTGATGACATACAAAGTT-3′ |
| e | 5′-CGGTTGATTAGCAGTGCGTCTTCCACCTAA-3′ |
| f | 5′-CATCAGAGATTGTTCACCCAT-3′ |
| g | 5′-CCGCGACAAGGTCAGCCTGAGGGCACACCT-3′ |
| h | 5′-AAAGCTCTTTTATGTAGTGTGCGT-3′ |
| i | 5′-TGATGACTGACAAAACATCAC-3′ |
| j | 5′-GAAGTGATTGACACCTAGGCC-3′ |
| k | 5′-GATCAGTCAGGGCATAAAATA-3′ |
| DD7SP1 | 5′-CGTTACTTCGCGTACTATCTCTGTGA-3′ |
| DD7SP2 | 5′-TCGGTACTACAAGAATACGGATGTGT-3′ |
| DD7SP3 | 5′-CGGCGGCGCAGCAGAAACT-3′ |
| TBR2.2 | 5′-ACGCGCACAGCCAGACGGCTA-3′ |
| SP6 | 5′-GATTTAGGTGACACTATAG-3′ |
| T7 | 5′-TAATACGACTCACTATAGGG-3′ |
Determination of the number of clusters within the TBR12, TBR6, TBR4 and TBR2 locus
To determine the approximate number of gene clusters contained within the BamHI 8 kb fragment, single copy reconstruction experiments were performed. In order to generate data for comparison purposes, 1.2 pmol of the 8 kb BamHI fragment and 1.2 pmol of the 0.9 kb RsaI fragment were generated using the appropriate plasmids and restriction enzymes. The fragments were electrophoresed on a 0.8% agarose gel, transferred onto a Zeta-probe membrane, and hybridized with a probe to the TBR6 gene. The experiment was then repeated for PCR-generated probes to the TBR12, TBR4 and TBR2 snoRNA genes. The TBR2 probe is a partial probe to the TBR2 gene made with the a and TBR2.2 oligonucleotides (Table 1) because the plasmid containing the 0.9 kb RsaI fragment contains only a partial TBR2 gene. The blots were analyzed by autoradiography and the number of gene clusters in the 8 kb fragment compared to the 0.9 kb fragment containing single copies of each gene was determined by Molecular Dynamics PhosphorImager analyses.
Southern blot hybridization of genomic and P1 phage DNA
To isolate T.brucei genomic DNA the following strategy was used. Cells (200 ml) were grown to a concentration of 2–3 × 107 cells/ml. The cells were pelleted and washed in trypanosome washing buffer (20 mM HCl pH 7.5, 100 mM NaCl, 3 mM MgCl2) and resuspended in 400 µl of breaking buffer (2% v/v Triton X-100, 2% v/v SDS, 100 mM NaCl, 10 mM Tris–HCl pH 8.0 and 1 mM EDTA). A half volume of glass beads (0.45–0.5 mm diameter) and 400 µl of phenol/chloroform/isoamyl alcohol (PCA) were added and the contents were vortexed for 3 min after which 200 µl of TE were added and vortexed briefly. The contents were then PCA extracted three times, ethanol precipitated and resuspended in 400 µl of TE. The sample was treated with 30 µl of 1 mg/ml RNase A for 5 min at 37°C. The DNA was subsequently ethanol precipitated and resuspended in 100 µl of dH2O. Samples of 10 µg of genomic and 10 µg of P1 phage DNA were digested with EcoRI, BamHI and BstXI restriction endonucleases and electrophoresed on a 0.8% agarose gel. The DNA fragments were transferred to a Zeta-probe membrane and hybridized with PCR-generated probes to TBR12, TBR6, TBR4 and TBR2. The results were analyzed by autoradiography and PhosphorImager quantitation.
RNA isolation and RT–PCR mapping
Trypanosome total RNA was isolated using a guanadinium thiocyanate/acid phenol extraction technique outlined by Dunbar et al. (30). About 30 µg of total RNA in 100 µl of DNase I buffer was incubated with 50 U RNase-free DNase I (Gene Trapper) for 1 h at 37°C and subsequently extracted with PCA and ethanol precipitated. RT was performed using Superscript II reverse transcriptase on 15 µg of DNase treated total RNA using either an oligonucleotide complementary to the 3′ end of TBR2 (oligonucleotide f, Table 1) or an oligonucleotide complementary to the 3′ end of TBR5 (oligonucleotide k, Table 1). PCR was then performed on an aliquot of the reactions with one of the oligonucleotides used in RT and corresponding forward oligonucleotides with these cycling steps: 94, 55 and 72°C (30 s each cycle for 20 cycles). The PCR products were analyzed on 1.2% agarose gels.
RESULTS
Identification of a gene cluster containing four snoRNA genes in the T.brucei genome
Because the organization of snoRNA genes in vertebrates, plants and yeast is diverse, we investigated the organization of snoRNA genes in a distantly related organism, T.brucei. Previously we identified 17 T.brucei box C/D snoRNAs (30). We have analyzed the genomic organization of the genes encoding four of them in the following manner. A genomic library containing 3–3.5 times the haploid T.brucei genome was screened with a PCR-generated probe complementary to the TBR6 snoRNA gene. One positive P1 phage clone was chosen for further characterization. A Southern blot of the P1 phage DNA digested with the RsaI restriction endonuclease produced a band of 0.9 kb that hybridized to the TBR6 probe. The 0.9 kb fragment was cloned into the PGEM-3Z vector and sequenced in both directions using vector primers. Sequencing indicated that the 0.9 kb fragment contained, from 5′ to 3′, the TBR12, TBR6, TBR4 snoRNA genes and a partial sequence of TBR2 interrupted by the RsaI cloning site (Fig. 1).
Figure 1.
The TBR12, TBR6, TBR4 and TBR2 gene cluster. Sequence of the 1.2 kb BstXI cluster unit containing snoRNA genes TBR12 (blue), TBR6 (green), TBR4 (red) and TBR2 (yellow) in order from 5′ to 3′. This sequence was obtained by first sequencing the 0.9 kb RsaI fragment (containing a partial TBR2 gene) and extending that sequence with data from the sequence of an 8 kb BamHI fragment. Underlined sequences represent conserved boxes C (consensus TGATGA) and D (consensus CTGA). The DDBJ/EMBL/GenBank accession number for the complete sequence is AF232064.
To determine sequences upstream and downstream of the 0.9 kb fragment a Southern blot was performed on digests of the P1 phage DNA to identify a larger TBR6 hybridizing fragment. An 8 kb fragment was generated by digestion with the restriction endonuclease BamHI and subcloned into the PGEM-3Z vector. To sequence the 8 kb BamHI fragment, primer walking was performed, beginning with a primer to the vector SP6 promoter sequence and proceeding further downstream until the first snoRNA gene was reached. Sequencing revealed ∼2 kb of genomic sequence before the first snoRNA gene (TBR12; Fig. 2). Sequencing was also performed starting with a primer to the vector T7 promoter sequence until the last recognizable gene was reached. This latter sequence demonstrated the presence of a partial TBR12 gene sequence at the 3′ end of the 8 kb fragment (Fig. 2). Sequencing was continued from both the vector SP6 and T7 promoter regions until an identical repeated cluster that contained the TBR12, TBR6, TBR4 and TBR2 snoRNA genes was reached. We conclude from these results that the cluster of four snoRNA genes identified by sequencing the 0.9 kb RsaI fragment is tandemly repeated within the larger 8 kb fragment.
Figure 2.
The genes for TBR12, TBR6, TBR4 and TBR2 are clustered and tandemly repeated. Schematic diagram of the 8 kb BamHI subclone illustrating multiple snoRNA gene clusters. The 0.9 kb RsaI fragment was cloned and sequenced completely. A larger 8 kb BamHI fragment was then cloned and sequenced. Also shown are the 1.5 and 1.2 kb BstXI fragments that each contain one cluster of snoRNA genes.
To determine the number of repeated snoRNA genes in our subcloned genomic DNA fragment, single copy reconstruction was performed. The 0.9 kb RsaI fragment contains a single copy of the TBR12, TBR6 and TBR4 genes as well as the partial sequence of the TBR2 gene. Equal number of moles of the 8 kb BamHI and 0.9 kb RsaI fragments were resolved on an agarose gel, transferred onto a Zeta-probe membrane and hybridized individually with PCR-generated probes to TBR12, TBR6, TBR4 and TBR2 (data not shown). Direct comparisons of quantitation data obtained via PhosphorImager analyses indicate that there are approximately four snoRNA gene clusters within the 8 kb fragment (Fig. 2).
A calculation confirms the number of clusters contained within the 8 kb BamHI fragment. The DNA length that is comprised of snoRNA gene clusters in the 8 kb BamHI fragment is ∼5 kb. Since the average snoRNA gene cluster including extragenic regions is ∼1.2 kb, it follows that there are approximately four clusters of the four snoRNA genes each within the 8 kb BamHI subclone.
Experiments were designed to search for novel box C/D snoRNAs in this cluster; we have not detected any other box C/D snoRNA genes within clusters and between clusters using labeled PCR-generated probes hybridized to RNA blots containing T.brucei total RNA and fibrillarin immunoprecipitable RNA. Using the same strategy, we have also not detected any box C/D snoRNA genes upstream of the first snoRNA gene cluster in the repeat (data not shown).
Genomic organization of the TBR12, TBR6, TBR4 and TBR2 snoRNA genes
In order to investigate whether the gene organization shown in Figure 2 is representative of that in the trypanosome genome, digests of T.brucei genomic and the P1 phage clone DNA were compared by Southern blotting with probes complementary to the four snoRNA genes in the cluster. Autoradiography showed identical hybridization patterns for each of the snoRNA gene probes for both the T.brucei genomic and P1 phage DNAs (Fig. 3), indicating that these four genes are located in close proximity to each other along the ~65 kb insert sequence of the P1 phage clone and within the T.brucei genome. Figure 3 thus indicates that the organization of these snoRNA genes in a cluster is not only true of the P1 phage clone, but also of the entire T.brucei genome.
Figure 3.
The genomic organization of the genes for TBR12, TBR6, TBR4 and TBR2. Genomic and P1 phage DNA were digested with the indicated enzymes, resolved on a 0.8% agarose gel and transferred to a Zeta-probe membrane. Quantitation of the 1.2 and 1.5 kb BstXI fragments for each of the blots indicates that the ratio of 1.5 to 1.2 kb fragments in the genome is 1:5.
Analysis of the Southern blot reveals that both the genomic and P1 phage DNA digested with BstXI yield fragments of 1.2 and 1.5 kb in length that each contain all four snoRNA genes (Fig. 3, lanes 3 and 6 of each panel). Digestion of the 8 kb BamHI subclone with the restriction endonuclease BstXI also generates fragments of 1.2 and 1.5 kb that contain all four snoRNA genes (Fig. 2 and data not shown). These results suggest that the cluster organization found within the 8 kb BamHI fragment, i.e. snoRNA gene clusters contained within the 1.2 and 1.5 kb BstXI fragments, is that of the entire genome. We have therefore determined the genomic organization of TBR12, TBR6, TBR4 and TBR2 genes in the T.brucei genome.
PhosphorImager quantitation data from Figure 3 (lanes 3 and 6 of each panel) indicates that the ratio of 1.5 to 1.2 kb fragments is ∼1:5 for both the P1 phage clone and the genome. This result indicates that the 1.5 kb piece containing a single cluster may constitute the first cluster unit of a series of tandemly repeated 1.2 kb cluster units, all of which are generated by digestion with BstXI (Fig. 2). Moreover, given the fact that the genomic DNA yields two positive bands when digested with BamHI (Fig. 3, lane 2 of each panel) whereas the PI phage DNA yields only one band (the 8 kb band that was cloned) when digested with BamHI (Fig. 3, lane 5 of each panel) and the end of the 8 kb subclone contains the presence of a partial TBR12 gene, it is likely that the 1.2 kb cluster units continue to be tandemly repeated beyond what has been determined here. This suggests that we have located the beginning of a series of tandemly repeated snoRNA gene clusters that each contain four snoRNAs.
Trypanosoma brucei snoRNA genes are transcribed as polycistronic RNAs
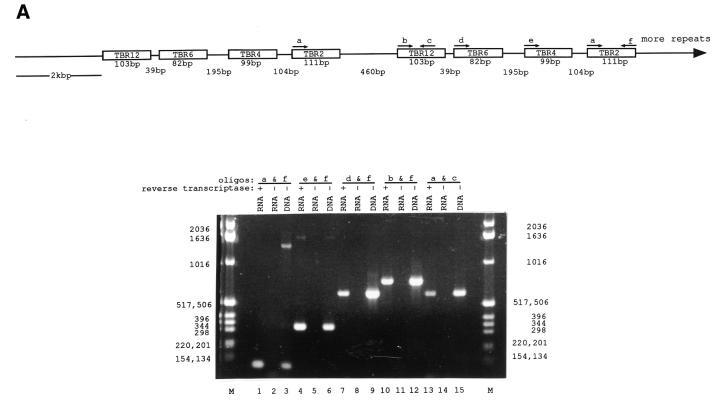
Using RT–PCR, we investigated whether transcription of the snoRNA gene clusters is polycistronic. That is, do the snoRNAs within a cluster and between clusters form a contiguous RNA transcript? By RT–PCR and different oligonucleotide combinations, we could amplify RNA transcripts containing TBR2 alone (Fig. 4A, lane 1), TBR2 and TBR4 (Fig. 4A, lane 4), TBR2 and TBR6 (Fig. 4A, lane 7), and between TBR2 and TBR12 (Fig. 4A, lane 10). By RT–PCR we could also amplify an RNA containing the spacer region that separates snoRNA gene clusters (Fig. 4A, lane 13). The sizes of the RT–PCR products were identical to those produced using the same oligonucleotides on T.brucei genomic DNA shown in lanes 3, 6, 9, 12 and 15 of Figure 4A but were not present if reverse transcriptase was omitted from the RNA samples (Fig. 4A, lanes 2, 5, 8, 11 and 14). Using oligonucleotide primers e and f, a PCR product was obtained of ∼1.5 kb that was also present when oligonucleotides e and f were used on genomic DNA (Fig. 4A, lanes 4 and 6). This amplified fragment is the correct size if the RNA transcript spanned the TBR4 sequence from an upstream cluster to the TBR2 sequence in a downstream cluster. These results indicate that these snoRNAs within a cluster and between clusters form a contiguous RNA transcript.
Figure 4.
Clustered, tandemly repeated snoRNA genes are transcribed as polycistronic RNAs. (A) Detection of polycistronic snoRNA transcripts containing TBR12, TBR6, TBR4 and TBR2 snoRNAs. (B) Detection of a polycistronic snoRNA transcript containing TBR17, TBR7 and TBR5 snoRNAs and the SLA RNA. Both (A) and (B) show control RT–PCR reactions carried out without reverse transcriptase, and PCR reactions carried out on T.brucei genomic DNA. MspI-digested PBR322 DNA markers are also shown in (A) and (B).
To see if the polycistronic nature of these snoRNA genes was common to other snoRNA clusters, we investigated whether the spliced leader associated RNA (SLA) gene locus is transcribed as a polycistronic transcript. The SLA locus contains the SLA gene as well as the TBR17, TBR7 and TBR5 snoRNA genes (29,30,39). By RT–PCR and different oligonucleotide combinations, we could amplify RNA transcripts containing TBR5 alone (Fig. 4B, lane 1), TBR5 and TBR7 (Fig. 4B, lane 4), TBR5 and the SLA RNA (Fig. 4B, lane 7) and TBR5 and TBR17 (Fig. 4B, lane 10). The sizes of the RT–PCR products were identical to those produced using the same oligonucleotides on T.brucei genomic DNA, shown in Figure 4B (lanes 3, 6, 9 and 12), but were not present if reverse transcriptase was omitted from the RNA samples (Fig. 4B, lanes 2, 5, 8 and 11). By RT–PCR and oligonucleotides i and k a fragment with a higher molecular weight than expected was also produced (Fig. 4B, lane 4). This fragment was also present when oligonucleotides i and k were used on genomic DNA (Fig. 4B, lane 6). This suggests that the SLA gene clusters which are tandemly repeated in the T.brucei genome (39) are transcribed as a contiguous transcript. Taken together, these results indicate that transcription of at least two clusters of snoRNAs in T.brucei proceeds in a polycistronic manner.
DISCUSSION
We have characterized the genomic organization of the genes encoding four snoRNAs in T.brucei. The TBR12, TBR6, TBR4 and TBR2 genes are found in multiple, tandemly arrayed gene clusters. It is estimated that there are at least four clusters of these four genes in the T.brucei genome. Using RT–PCR we have also demonstrated, for the first time, that these four snoRNA genes are transcribed as a polycistronic RNA transcript. This is not unique to this particular cluster as we have also shown that the genes in the SLA cluster containing SLA, TBR17, TBR7 and TBR5 are produced as a polycistronic RNA.
The cluster bearing TBR12, TBR6, TBR4 and TBR2 represents the second example of a snoRNA cluster that is tandemly repeated in the T.brucei genome. The first example is the SLA locus which contains the TBR17, TBR7 and TBR5 snoRNA genes along with the SLA gene (29,30,39) which was estimated to be tandemly repeated about 11 times in the genome (39). Levitan et al. (28) have recently reported that the snoRNA2 snoRNA gene from the related trypanosomatid Leptomonas collosoma is tandemly repeated at least five times in the genome based on Southern blot analysis. It appears that the genomic organization of these snoRNAs is similar to that of the trypanosome spliced leader RNA gene which is encoded by approximately 200 copies of tandemly arrayed genes (41) in that they are all repeated multiple times.
The genomic organization of these clusters of snoRNAs is different from that of the genes encoding other trypanosome small RNAs. The U2, U4, U6 and 7SL small RNAs and U3 snoRNA are all single copy genes (22,42). The U6 and 7SL small RNAs and the U3 snoRNA are transcribed by RNA polymerase III, and are flanked by tRNA genes which contain conserved box elements required for their expression (43,44). We could not detect any tRNA genes located within the DNA that we have sequenced, which includes 2 kb upstream of the first gene in the cluster suggesting that different sequence elements are important for expression of these box C/D snoRNAs.
Clusters of snoRNA genes have also been identified in both yeast and plant cells. In yeast, there are five small clusters of box C/D snoRNA genes that contain two to seven snoRNA genes per cluster (19,34,36,45). Plants possess snoRNA gene clusters that contain several tandemly arrayed U14 box C/D snoRNA genes as well as other box C/D snoRNA genes and a box H/ACA snoRNA gene that are all tightly linked to the U14 genes (37,38). Neither the yeast nor the plant snoRNA gene clusters are tandemly repeated. Thus, the organization of snoRNA gene clusters in tandem repeats is so far unique to trypanosomes.
We have shown, for the first time, that two different clusters of snoRNA genes (the TBR12, TBR6, TBR4 and TBR2 cluster and the SLA cluster) are produced as polycistronic RNAs in T.brucei. Polycistronic transcription of snoRNAs is also found in both yeast and plants. In yeast, the snR190 and U14 box C/D snoRNAs are produced as a dicistronic RNA which is processed by the Rnt1p endonuclease and by the Rat1p exonuclease to produce mature snR190 and U14 box C/D snoRNAs (33,35). Chanfreau et al. (34) also showed that the Z2–Z8 box C/D snoRNAs and the snR41, snR70 and snR51 box C/D snoRNAs are produced as polycistronic RNAs which are processed by Rnt1p. Qu et al. (36) has shown that the Z2–Z8 polycistronic RNA is processed by both Rnt1p and Rat1p to produce mature snoRNAs. The Z2–Z8 polycistronic RNA is synthesized by an independent promoter, probably by RNA polymerase II, since the promoter region contains polymerase II sequence elements (36). The plant snoRNA gene clusters are also transcribed as a polycistronic RNA from an upstream promoter (38) but the RNA polymerase responsible for polycistronic transcription has not been identified. It will be interesting to determine whether the trypanosome polycistronic snoRNAs are processed by trypanosome homologs of Rnt1p and Rat1p or if trypanosomes have evolved different strategies to process their polycistronic snoRNAs.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Elisabetta Ullu and Chris Tschudi for suggestions on the single copy reconstruction experiments used in this study. We thank Vanessa Leech and Sara Melville at the Molteno Insitute for Parasitology, University of Cambridge, for the T.brucei genome robotic arrays and the corresponding positive clones. This work was supported by the Council for Tobacco Research.
DDBJ/EMBL/GenBank accession no. AF232064
REFERENCES
- 1.Eichler D.C. and Craig,N. (1995) Prog. Nucleic Acid Res. Mol. Biol., 49, 179–239. [DOI] [PubMed] [Google Scholar]
- 2.Venema J. and Tollervey,D. (1999) Annu. Rev. Genet., 33, 261–311. [DOI] [PubMed] [Google Scholar]
- 3.Maden B.E.H. (1990) Prog. Nucleic Acid Res. Mol. Biol., 39, 241–303. [DOI] [PubMed] [Google Scholar]
- 4.White T.C., Rudenko,G. and Borst,P. (1986) Nucleic Acids Res., 14, 9471–9489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spencer D.F., Collings,J.C., Schnare,M.N. and Gray,M.W. (1987) EMBO J., 6, 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell D.A., Kubo,K., Clark,C.G. and Boothroyd,J.C. (1987) J. Mol. Biol., 196, 113–124. [DOI] [PubMed] [Google Scholar]
- 7.Maxwell E.S. and Fournier,M.J. (1995) Annu. Rev. Biochem., 64, 897–934. [DOI] [PubMed] [Google Scholar]
- 8.Smith C.M. and Steitz,J.A. (1997) Cell, 89, 669–672. [DOI] [PubMed] [Google Scholar]
- 9.Tollervey D. and Kiss,T. (1997) Curr. Opin. Cell Biol., 9, 337–342. [DOI] [PubMed] [Google Scholar]
- 10.Bachellerie J. and Cavaille,J. (1998) In Grosjean,H. and Benne,R. (eds), Modification and Editing of RNA. ASM Press, Washington, DC, pp. 255–272.
- 11.Lafontaine D. and Tollervey,D. (1998) Trends Biol. Sci., 23, 383–388. [DOI] [PubMed] [Google Scholar]
- 12.Balakin A., Smith,L. and Fournier,M. (1996) Cell, 86, 823–834. [DOI] [PubMed] [Google Scholar]
- 13.Weinstein L.B. and Steitz,J.A. (1999) Curr. Opin. Cell Biol., 11, 378–384. [DOI] [PubMed] [Google Scholar]
- 14.Kiss-Laszlo Z., Henry,Y., Bachellerie,J.P., Caizergues-Ferrer,M. and Kiss,T. (1996) Cell, 85, 1077–1088. [DOI] [PubMed] [Google Scholar]
- 15.Cavaille J., Nicoloso,M. and Bachellerie,J.P. (1996) Nature, 383, 732–735. [DOI] [PubMed] [Google Scholar]
- 16.Tycowski K.T., Smith,C.M., Shu,M.D. and Steitz,J.A. (1996) Proc. Natl Acad. Sci. USA, 93, 14480–14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu Y.-T., Shu,M.-D. and Steitz,J.A. (1997) RNA, 3, 324–331. [PMC free article] [PubMed] [Google Scholar]
- 18.Dunbar D.A. and Baserga,S.J. (1998) RNA, 4, 195–204. [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe T.M. and Eddy,S.R. (1999) Science, 283, 1168–1171. [DOI] [PubMed] [Google Scholar]
- 20.Ni J., Tien,A. and Fournier,M. (1997) Cell, 89, 565–573. [DOI] [PubMed] [Google Scholar]
- 21.Ganot P., Bortolin,M.-L. and Kiss,T. (1997) Cell, 89, 799–809. [DOI] [PubMed] [Google Scholar]
- 22.Mottram J., Perry,K.L., Lizardi,P.M., Luhrmann,R., Agabian,N. and Nelson,R. (1989) Mol. Cell. Biol., 9, 1212–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hartshorne T. and Agabian,N. (1993) Mol. Cell. Biol., 13, 144–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes J.M.X. and Ares,M.,Jr (1991) EMBO J., 10, 4231–4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kass S., Tyc,K., Steitz,J.A. and Sollner-Webb,B. (1990) Cell, 60, 897–908. [DOI] [PubMed] [Google Scholar]
- 26.Mougey E.B., Pape,L.K. and Sollner-Webb,B. (1993) Mol. Cell. Biol., 13, 5990–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Savino R. and Gerbi,S.A. (1990) EMBO J., 9, 2299–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levitan A., Xu,Y., Ben-Dov,C., Ben-Shlomo,H., Zhang,Y. and Michaeli,S. (1998) Nucleic Acids Res., 26, 1775–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Roberts T.G., Sturm,N.R., Yee,B.K., Michael,C.Y., Hartshorne,T., Agabian,N. and Campbell,D.A. (1998) Mol. Cell. Biol., 18, 4409–4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dunbar D.A., Wormsley,S., Lowe,T.M. and Baserga,S.J. (2000) J. Biol. Chem., 275, 14767–14776. [DOI] [PubMed] [Google Scholar]
- 31.Tycowski K., Shu,M.D. and Steitz,J.A. (1996) Nature, 379, 464–466. [DOI] [PubMed] [Google Scholar]
- 32.Smith C.M. and Steitz,J.A. (1998) Mol. Cell. Biol., 18, 6897–6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chanfreau G., Rotondo,G., Legrain,P. and Jacquier,A. (1998) EMBO J., 17, 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chanfreau G., Legrain,P. and Jacquier,A. (1998) J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]
- 35.Petfalski E., Dandekar,T., Henry,Y. and Tollervey,D. (1998) Mol. Cell. Biol., 18, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qu L.H., Henras,A., Lu,Y.J., Zhou,H., Zhou,W.X., Zhu,Y.Q., Zhao,J., Henry,Y., Caizergues-Ferrer,M. and Bachellerie,J.P. (1999) Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leader D., Sanders,J.F., Waugh,R., Shaw,P. and Brown,J.W. (1994) Nucleic Acids Res., 22, 5196–5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leader D.J., Clark,G.P., Watters,J., Beven,A.F., Shaw,P.J. and Brown,J.W.S. (1997) EMBO J., 16, 5742–5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roberts T.G., Dungan,J.M., Watkins,K.P. and Agabian,N. (1996) Mol. Biochem. Parisitol., 83, 163–174. [DOI] [PubMed] [Google Scholar]
- 40.Cunningham I. (1977) J. Protozool., 24, 325–329. [DOI] [PubMed] [Google Scholar]
- 41.Nelson R., Parsons,M., Selkirk,M., Newport,G., Barr,P.J. and Agabian,N. (1984) Nature, 308, 665–667. [DOI] [PubMed] [Google Scholar]
- 42.Ben-Shlomo H., Levitan,A., Beja,O. and Michaeli,S. (1997) Nucleic Acids Res., 25, 4977–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fantoni A., Dare,O.A. and Tschudi,C. (1994) Mol. Cell. Biol., 14, 2021–2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nakaar V., Dare,O.A., Hong,D., Ullu,E. and Tschudi,C. (1994) Mol. Cell. Biol., 14, 6736–6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zagorski J., Tollervey,D. and Fournier,M.J. (1988) Mol. Cell. Biol., 8, 3282–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]