Abstract
The 5′-cap structure and poly(A) tail of eukaryotic mRNAs function synergistically to promote translation initiation through a physical interaction between the proteins that bind to these regulatory elements. In this study, we have examined the effect of leader length and the presence of secondary structure on the translational competence and the function of the cap and poly(A) tail for mRNAs microinjected into Xenopus oocytes. Increasing the length of the 5′-leader from 17 to 144 nt resulted in a 2- to 4-fold increase in expression from an mRNA containing an unstructured leader but increased expression up to 20-fold for an mRNA containing 5′-proximal structure. Consequently, the presence of secondary structure was less inhibitory for those mRNAs with a longer 5′-leader. Co-injection of poly(A)-binding protein (PABP) mRNA increased the function of the cap and poly(A) tail in promoting translation from poly(A)+ but not poly(A)– mRNAs, particularly for mRNAs containing secondary structure. In the absence of an internal ribosome entry site, expression from the distal cistron of a dicistronic mRNA increased as a function of the length of the intercistronic region and the concentration of PABP. The inhibitory effect of intercistronic located secondary structure on translation was position-dependent. Indeed, the effect of secondary structure was abolished if positioned 134 nt upstream of the distal cistron. These data suggest that the length of a leader, the presence of secondary structure and the concentration of PABP determine the extent to which the cap and poly(A) tail regulate translation.
INTRODUCTION
The translation of most eukaryotic mRNAs begins with binding of the 40S ribosomal subunit close to the 5′-cap structure of an mRNA followed by its scanning until the initiation codon is located. Numerous initiation factors (eIFs) assist in each step during the initiation process. The 5′-cap serves as the binding site for eIF4E, the small subunit of eIF4F. eIF4G, the large subunit of eIF4F, interacts with eIF3, which in turn promotes 40S ribosomal subunit binding to an mRNA (reviewed in 1–3). Consequently, the 5′-cap plays a critical role in initiating the assembly of those factors required to assist in the earliest steps of translation initiation. The poly(A) tail serves as the binding site for poly(A)-binding protein (PABP), which assists in assembly of the initiation complex through a physical interaction with eIF4G, an interaction that is conserved in animals, plants and yeast (4–7). The interaction between PABP and eIF4G increases the poly(A)-binding activity of PABP by over an order of magnitude by reducing its rate of disassociation (4) and increasing the affinity of eIF4F for the 5′-cap structure by 40-fold (8). As the rate limiting step, the initiation of translation is the step most often targeted for control. The regulation of one or a few mRNAs is often achieved through a specific trans-acting regulatory factor that binds to a site within the leader, e.g. regulation of ferritin mRNA by the iron-response element binding protein (reviewed in 9). The regulated phosphorylation of some initiation factors, e.g. eIF4E, eIF4G and eIF2, is thought to play a role in the global control of cellular protein synthesis.
In addition to these examples, the structural characteristics of individual mRNAs can influence the initiation process. The length of the leader, the presence of secondary structure either upstream or downstream of the initiation codon and the length of the poly(A) tail have been shown to affect the translatability of an mRNA in higher eukaryotes (10–15). Stable secondary structure within a leader can impede scanning of the 40S ribosomal subunit in its search for the initiation codon and thereby inhibit translation (10,11,16,17).
Because the 5′-cap directs 40S ribosomal subunit binding close to the 5′-terminus of an mRNA and most ribosomes are not competent to reinitiate translation following translational termination, most eukaryotic cellular mRNAs are monocistronic. However, several viral and some cellular mRNAs have evolved regulatory elements [i.e. internal ribosome entry site (IRES) elements] within the leader that function through cap-independent mechanisms to promote 40S ribosomal subunit binding internally to an mRNA (18,19). Dicistronic mRNA constructs, in which a sequence is introduced intercistronically to determine whether it can promote expression from the 5′-distal cistron, have been used extensively to establish whether a sequence contains an IRES (reviewed in 20).
In this study, we have examined how the structural characteristics of an mRNA influence the translatability of monocistronic or dicistronic mRNAs in Xenopus oocytes. For monocistronic constructs, the presence of even moderately stable secondary structure was highly inhibitory when present near the 5′-terminus of an mRNA and was more inhibitory for capped mRNAs than for uncapped mRNAs. The inhibitory effect of secondary structure was substantially relieved when it was moved 134 nt upstream from the initiation codon. A similar effect was observed for translation from the 5′-distal cistron in a dicistronic mRNA: inhibition imposed by secondary structure was abolished when it was moved 134 nt upstream from the distal cistron. The addition of a cap promoted translation from an mRNA with an unstructured leader to a greater extent than it did from an mRNA with a structured leader regardless of the length of the leader. In oocytes overexpressing PABP, an increase in the function of the cap and poly(A) tail and an increase in absolute expression from capped and polyadenylated mRNAs was observed, particularly when the mRNA contained a structured leader. These data indicate that the presence of secondary structure and its distance from the initiation codon influence the translatability of an mRNA and alter the function of the cap and poly(A) tail. These results also indicate that the concentration of PABP influences the function of the cap and poly(A) tail in overcoming the inhibitory effects of a structured 5′-leader.
MATERIALS AND METHODS
mRNA constructs
Leader constructs were designed to contain one or two copies (both in a forward orientation) of a 60% AT-rich, 72 nt sequence (AATATCTTATTGCCGGGAAAAGTGTACGTATCACCGTTTGTGTGAACAACGAACTGAACTGGCAGACTATAA) introduced into the HindIII and SalI sites of pT7-luc-A50 (21), resulting in Con72-luc-A50 or Con144-luc-A50 mRNAs containing a 72 or 144 nt 5′-leader, respectively. The free energy calculated by the fold algorithm for these mRNA leaders is ΔG = –11.5 kcal/mol. A third control mRNA, Con17-luc-A50, was constructed with the 17 nt 5′-leader sequence GCCTAAGCTTGTCGACC, representing a free energy of –0.9 kcal/mol.
pT7-SL24-luc-A50 [containing a 24 bp stem–loop (SL) structure of ΔG = –42.9 kcal/mol] was produced by inserting the following 52 bp palindromic oligonucleotide into the HindIII site, allowing the formation of a stem–loop positioned 4 nt downstream of the cap of the mRNA:
HindIII ApaI BglII MluI SnaBI MluI BglII ApaI HindIII
AAGCTTGGGCCCAGATCTACGCGTACGTACGCGTAGATCTGGGCCCAAGCTT
Introduction of the 24 bp stem–loop structure into the 17, 72 or 144 nt leader constructs described above resulted in the mRNA constructs SL-Con7-luc-A50, SL-Con62-luc-A50 and SL-Con134-luc-A50, in which the structure was positioned 7, 62 and 134 nt upstream of the initiation codon, respectively. To produce pT7-SL19-luc-A50, pT7-SL13-luc-A50 and pT7-SL7-luc-A50 [containing a 19, 13 or 7 bp SL structure of ΔG = –31.8, –21.3 and –4.5 kcal/mol, respectively], pT7-SL24-luc-A50 was digested with MluI, BglII and ApaI, respectively.
Dicistronic constructs were generated by inserting the uidA gene [composed of the coding region for β-glucuronidase (GUS) and 73 nt of sequence 3′ to the uidA termination codon] upstream of the luc constructs described above, resulting in the mRNA constructs GUS-Con17-luc-A50, GUS-SL-Con7-luc-A50, GUS-Con72-luc-A50, GUS-SL-Con62-luc-A50, GUS-Con144-luc-A50 and GUS-SL-Con134-luc-A50.
The Xenopus PABP construct, pSP64T-ABP, has been described (22) and was kindly provided by Dr Michael Wormington.
In vitro transcription and translation
RNAs were synthesized using template plasmids linearized immediately upstream or downstream of the poly(A)50 sequence to produce poly(A)– and polyadenylated mRNAs, respectively. Uncapped mRNAs were synthesized in vitro as described previously (23) using 3 µg template DNA in 40 mM Tris–HCl, pH 7.9, 6 mM MgCl2, 2 mM spermidine, 100 µg/ml bovine serum albumin, 500 µM each ATP, CTP, UTP and GTP, 10 mM dithiothreitol (DTT), 0.5 U RNase inhibitor RNasin (Promega, Madison, WI) and 0.5 U/ml T7 RNA polymerase (New England Biolabs, Beverly, MA). Capped RNAs were synthesized using 3 µg template in the same reaction mix as described above except that GTP was used at 160 µM and 1 mM m7GpppG was included. Under these conditions >95% of the mRNA was capped. The yield of each mRNA was determined by northern analysis.
Equal amounts of mRNA (150 ng/25 µl reaction) were translated using rabbit reticulocyte lysate as described by the manufacturer except that all amino acids were unlabeled. The reactions were incubated for 2 h and aliquots assayed for luciferase activity. Each mRNA construct was translated in triplicate and each in vitro translation was assayed in duplicate for luciferase activity. The average value and standard deviation for each construct is reported.
Microinjection of RNA into Xenopus oocytes
For monocistronic mRNAs, 2 ng GUS mRNA and 5 ng each luc mRNA construct were co-injected into the cytoplasm of stage VI oocytes in batches of 30 oocytes as described (24). Five nanograms of Xenopus PABP mRNA was co-injected into oocytes for those experiments in which elevated PABP was used. For the dicistronic experiments, 10 ng of each GUS–luc dicistronic mRNA construct was injected into stage VI oocytes. Oocytes were incubated for 18 h in modified Barth’s saline solution prior to harvesting. Oocyte extracts were broken in 25 mM Tricine, pH 8, 5 mM MgCl2, 0.1 mM EDTA buffer and the insoluble material removed by centrifugation for 10 min. The supernatant was used for the GUS and luciferase assays. Each sample was assayed in triplicate and each experiment was repeated a minimum of three times.
Western analysis
Buffer or capped mRNA encoding Xenopus PABP (20 ng/oocyte) was microinjected into Xenopus stage VI oocytes as indicated. After 3 h, oocytes were lysed in buffer A (20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 µM microcystin, 40 mM β-glycerophosphate, 50 mM NaF, 2 mM EGTA, 2 mM EDTA, 2 mM benzamidine, 7 mM 2-mercaptoethanol) and clarified in a cooled microfuge. Total soluble protein or protein purified using m7GTP–Sepharose chromatography was resolved using standard SDS–PAGE and the protein transferred to PVDF membrane by electroblotting. Following transfer, the membranes were blocked in 5% milk in TBS–Tween and decorated with antiserum specific for eIF4E, eIF4G or PABP as described (25). The proteins were visualized using alkaline phosphatase-conjugated antibodies (Sigma, St Louis, MO) as described (25).
Luciferase and GUS assays
Oocyte extract or rabbit reticulocyte lysate was diluted in luciferase assay buffer (25 mM Tricine, pH 8, 5 mM MgCl2, 0.1 mM EDTA, supplemented with 33.3 mM DTT, 270 µM coenzyme A and 500 µM ATP) and assayed for luciferase activity following injection of 0.5 mM luciferin using a Monolight 2010 Luminometer (Analytical Luminescence Laboratory, San Diego, CA).
GUS activity was assayed in a 100 µl reaction as described (26) using 1 mM 4-methylumbelliferyl-β-d-glucuronide as the substrate. The assay was performed for 30 min at 37°C whereupon the reaction was terminated by addition of 900 µl of 0.2 M NaCO2. The amount of the fluorescent product produced in each assay was measured in a TKO 100 Fluorometer (Hoefer Scientific, San Francisco, CA) using excitation at 365 nm and emission at 455 nm.
RESULTS
The inhibitory effect of secondary structure in the 5′-leader is relieved by increasing its distance from the initiation codon
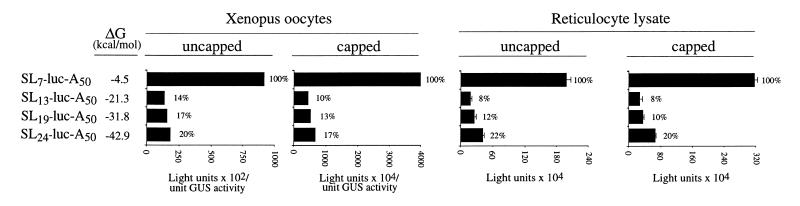
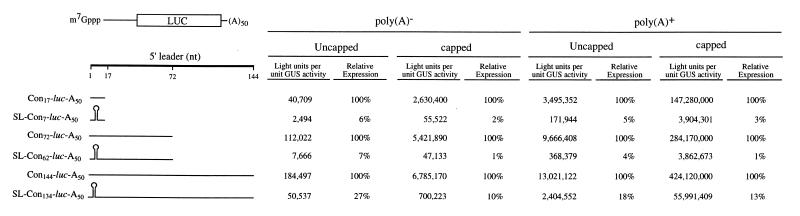
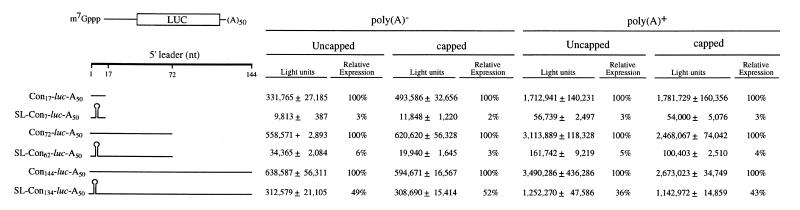
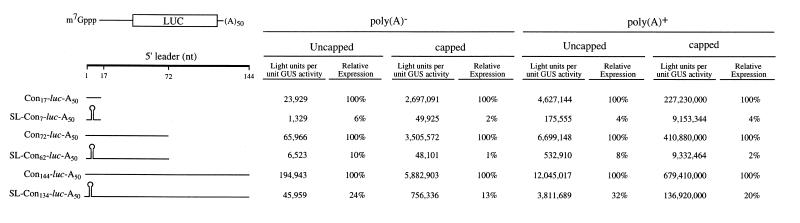
In order to investigate how the presence of secondary structure affects the translatability of a mRNA as a function of its distance from the initiation codon, a stable stem–loop structure containing a 24 bp stem was introduced 4 nt downstream of the 5′-terminus of the luciferase (luc) reporter mRNA (referred to as SL24-luc-A50). In addition, deletions were made within the 24 bp stem–loop to generate less stable secondary structures with a 19, 13 or 7 bp stem (referred to as SL19-luc-A50, SL13-luc-A50 and SL7-luc-A50, respectively). Each construct was synthesized in vitro as a capped or uncapped mRNA [each terminating in a poly(A50) tail] and injected into stage VI Xenopus oocytes. Capped GUS-A25 mRNA was co-injected with each luciferase mRNA construct to serve as an internal control. The oocytes were allowed to translate the mRNAs for 18 h and the degree of translation from each luc mRNA construct was measured and normalized to expression from the GUS internal control. Introduction of a 24 bp stem–loop structure (ΔG = –42.9 kcal/mol) reduced expression ∼5- to 6-fold relative to SL7-luc-A50 mRNA (ΔG = –4.5 kcal/mol) regardless of whether the mRNA was capped or uncapped (Fig. 1). Similar levels of inhibition were observed when either the 19 or 13 bp stem–loop structure (ΔG = –31.8 and –21.3 kcal/mol, respectively) was introduced. A similar degree of inhibition was observed when the same mRNA constructs were translated in vitro in rabbit reticulocyte lysate (Fig. 1). In these constructs, only seven bases separated the stem–loop structure from the initiation codon of the luc coding region. In order to examine the effect of secondary structure on translation when positioned further upstream of the start site, constructs in which the 24 bp stem–loop was positioned 7, 62 or 134 nt upstream of the start site (referred to as SL-Con7-luc-A50, SL-Con62-luc-A50 and SL-Con134-luc-A50, respectively) and corresponding control constructs with a 17, 72 or 144 nt leader (referred to as Con17-luc-A50, Con72-luc-A50 and Con144-luc-A50, respectively) were synthesized as uncapped poly(A)–, capped poly(A)–, uncapped poly(A)+ and capped poly(A)+ mRNAs and each injected into oocytes. For all mRNAs, whether or not possessing a cap and/or a poly(A) tail, introduction of the secondary structure 7 nt upstream from the initiation codon reduced expression to ∼2–6% of that observed for the Con17-luc-A50 mRNA control construct (Fig. 2), a greater degree of inhibition than the 5- to 6-fold reduction observed relative to the SL7-luc-A50 mRNA in which a 7 bp stem–loop was present 4 nt from the 5′-terminus (Fig. 1). The same structure positioned 62 nt upstream of the initiation codon exhibited a similar degree of inhibition (1–7% of the Con72-luc-A50 control mRNA) as that observed when the structure was present just 7 nt upstream from the start codon. However, when the structure was 134 nt upstream from the initiation codon, expression was inhibited to 10–27% of the Con144-luc-A50 control mRNA, a level of inhibition that was considerably less than that observed when the stem–loop was positioned closer to the initiation codon. These data indicate that secondary structure in a 5′-leader is less inhibitory when present sufficiently upstream from an initiation codon. A similar but even more pronounced trend was observed during translation of the same mRNAs in rabbit reticulocyte lysate. Introduction of the 24 bp stem–loop reduced expression to 2–6% of control levels when positioned 7 or 62 nt upstream of the initiation codon but reduced expression to 36–52% of control levels when 134 nt upstream from the start codon (Fig. 3), suggesting that, similar to translation in vivo in oocytes, secondary structure is less inhibitory when positioned at least 134 nt upstream from an initiation codon.
Figure 1.
5′-Proximal secondary structure is inhibitory to translation in Xenopus oocytes. A stem–loop (SL) with a 7, 13, 19 or 24 bp stem was introduced 4 nt downstream of the 5′-terminus of luc-(A)50 mRNA. The free energy (ΔG) of the control leader and each stem–loop is indicated. The luc mRNA constructs were synthesized in vitro as uncapped or capped polyadenylated mRNAs and co-injected with capped and polyadenylated uidA mRNA (encoding GUS, which served as an internal control) into stage VI Xenopus oocytes. Each mRNA was injected into 25 oocytes and each luciferase assay was performed in triplicate. Luciferase expression (normalized to the internal control) is reported and is also indicated as a percentage (indicated to the right of each histogram) of the SL-luc-(A)50 mRNA. The same luc mRNAs were translated in triplicate in rabbit reticulocyte lysate.
Figure 2.
The inhibition imposed by 5′-proximal secondary structure on expression in oocytes can be partially relieved by increasing its distance from the initiation codon. The 24 bp stem–loop (SL) used in Figure 1 was introduced 7, 62 or 134 nt upstream of the luc initiation codon. The luc mRNA constructs were synthesized in vitro as uncapped poly(A)–, uncapped poly(A)+, capped poly(A)– or capped poly(A)+ mRNAs and injected into stage VI Xenopus oocytes. Capped poly(A)+ uidA mRNA was co-injected to serve as an internal control. Each mRNA was injected into 25 oocytes and each luciferase assay was performed in triplicate. Luciferase expression (normalized to the internal control) is reported. The level of expression of each mRNA containing 5′-proximal secondary structure is also reported as a percentage of the corresponding control (i.e. the same construct without 5′-proximal secondary structure) which is set at 100%. The length of the 5′-leader and presence of the stem–loop are shown schematically to the left of the expression data.
Figure 3.
The inhibition imposed by 5′-proximal secondary structure on expression in vitro can be partially relieved by increasing its distance from the initiation codon. The same mRNAs tested in Figure 2 were translated in triplicate in rabbit reticulocyte lysate and each luciferase assay performed in triplicate. The average and standard deviation of expression from each mRNA is reported. The level of expression of each mRNA containing 5′-proximal secondary structure is also reported as a percentage of the corresponding control (i.e. the same construct without 5′-proximal secondary structure) which is set at 100%. The length of the 5′-leader and presence of the stem–loop are shown schematically to the left of the expression data.
The introduction of secondary structure was consistently more inhibitory for capped mRNAs than for uncapped mRNAs in oocytes, regardless of whether the mRNA was polyadenylated or not (Fig. 2). The greater degree of inhibition observed for capped mRNAs is consistent with the observation that translation is strongly cap-dependent for non-polyadenylated as well as polyadenylated mRNAs in Xenopus oocytes (compare expression from capped mRNAs to uncapped mRNAs, Fig. 2). When the same mRNAs were translated in vitro, the preferential inhibition of expression from capped mRNAs by the introduction of secondary structure was not observed (Fig. 3).
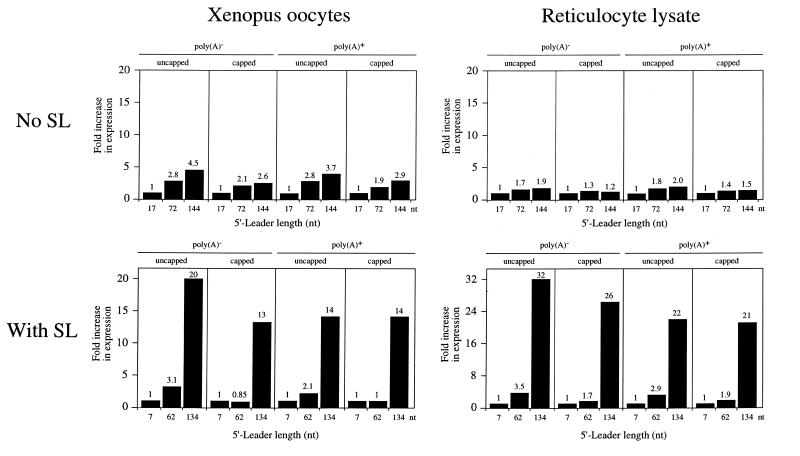
Expression from mRNAs with an unstructured 5′-leader of 17, 72 or 144 nt increased as a function of the length of the 5′-leader in oocytes, particularly for uncapped mRNAs, an increase which was smaller when the mRNAs were translated in vitro (Fig. 4). Expression from uncapped mRNAs with a structured leader increased moderately as the length was increased from 7 to 62 nt, whereas expression from capped mRNAs did not (Fig. 4), suggesting that 40S subunit binding was directed upstream of the secondary structure in a capped mRNA. Moreover, the data suggest that increasing the distance between the stem–loop and the initiation codon of an uncapped mRNA permits a moderate increase in 40S subunit binding downstream of the secondary structure. Expression from mRNAs with a structured leader increased substantially as the length was increased from 62 to 134 nt, an effect observed in both oocytes and in vitro translation lysate (Fig. 4), suggesting that this additional distance between the stem–loop and the initiation codon is required to overcome the inhibitory effect of secondary structure when present in capped or uncapped mRNAs.
Figure 4.
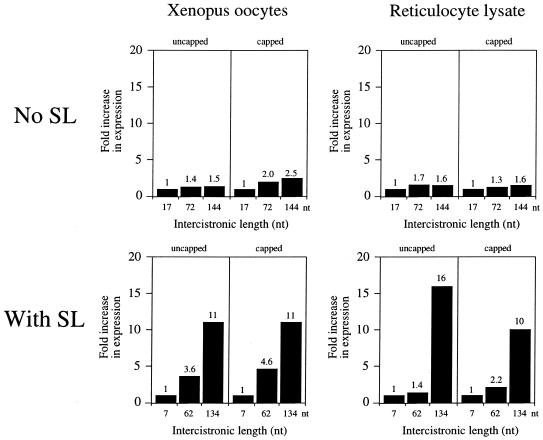
Expression increases as a function of the length of the 5′-leader or distance between secondary structure and the initiation codon for monocistronic mRNAs in Xenopus oocytes and in vitro. Expression from the constructs in Figures 2 and 3 is reported relative to the constructs with the shortest 5′-leader for mRNA without a structured leader (i.e. no SL) or with secondary structure present (i.e. with SL). Expression from the constructs with a 17 nt 5′-leader (i.e. no SL) or 7 nt 5′-leader (i.e. with SL) is set at a value of 1 and the fold increase in expression from the constructs with longer 5′-leaders is reported. The fold increase in expression is shown above each histogram. Each construct is designated by the length of its 5′-leader (i.e. no SL) or distance between the secondary structure and the initiation codon (i.e. with SL) below each histogram.
The function of the cap and the poly(A) tail increases in oocytes expressing an elevated level of PABP
Xenopus oocytes synthesize a large quantity of polyadenylated mRNA but the level of PABP at stage VI is less than one molecule of PABP for each poly(A)-binding site (27). PABP expression increases substantially during embryo development (22,27). Overexpression of PABP prevents maturation-specific poly(A) shortening and maintains the translational activity of maternal mRNAs that would otherwise be translationally repressed following maturation (28,29), observations suggesting that increasing PABP expression increases the function of the poly(A) tail. This was supported by the observation that polyadenylated luciferase mRNA was translated to a greater degree in progesterone-matured oocytes expressing an elevated level of PABP (29). To examine the effect of increasing the expression of PABP on the translatability of mRNAs with a structured or unstructured leader, the mRNAs used in the experiment described in Figures 2 and 3 were co-injected into stage VI oocytes with Xenopus PABP mRNA and incubated for 18 h prior to assaying for luciferase expression. The same batch of oocytes that were used for Figure 2 were injected on the same day to allow direct comparison between the two data sets. Overexpression of PABP was confirmed following western analysis of injected and control oocytes (Fig. 5, left). Interestingly, the increase in PABP expression resulted in a greater incorporation of PABP into the eIF4F complex, as demonstrated by probing for co-purification of PABP with eIF4F using m7GTP–Sepharose chromatography (Fig. 5, right). No change in the level of expression of eIF4E or eIF4G was observed between control oocytes and oocytes injected with PABP mRNA (Fig. 5, right), suggesting that the low concentration of PABP in uninjected stage VI oocytes is a limiting factor in determining its association with eIF4F.
Figure 5.
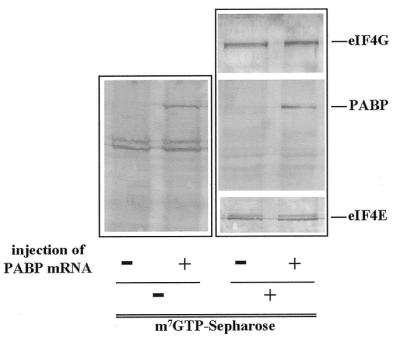
Overexpression of PABP and its association with eIF4F in Xenopus oocytes. Buffer or capped mRNA encoding Xenopus PABP (20 ng/oocyte) was microinjected into Xenopus stage VI oocytes as indicated. After 3 h, oocytes were lysed in buffer A (20 mM Tris–HCl, pH 7.5, 100 mM NaCl, 1 µM microcystin, 40 mM β-glycerophosphate, 50 mM NaF, 2 mM EGTA, 2 mM EDTA, 2 mM benzamidine, 7 mM 2-mercaptoethanol) and clarified in a cooled microfuge. (Left) Total soluble protein was resolved by SDS–PAGE and expression of PABP was monitored directly by immunoblotting using anti-PABP antiserum (a gift from Dan Schoenberg). Each lane was loaded with protein equivalent to 0.5 oocytes. (Right) Oocyte extracts prepared in the left panel were subjected to m7GTP–Sepharose chromatography. eIF4E and associated proteins were recovered, resolved by SDS–PAGE and eIF4E, eIF4G and PABP visualized by immunoblotting with the antisera indicated.
The introduction of secondary structure exerted a similar trend of inhibition to that observed for mRNAs translated in the absence of co-injected PABP mRNA (Fig. 6), i.e. secondary structure was most inhibitory when positioned from 7 to 62 nt upstream from the initiation codon and its inhibitory effect was significantly relieved when the stem–loop was positioned 134 nt upstream of the initiation codon. Overexpression of PABP resulted in an increase in absolute expression from mRNAs that were both capped and polyadenylated: expression from those with an unstructured 5′-leader increased by ∼50%, whereas expression from those containing 5′-proximal secondary structure increased by ∼2.4-fold (compare expression level from luc mRNAs that are both capped and polyadenylated in Fig. 6 with Fig. 2). Much smaller or no increases were observed for those uncapped polyadenylated mRNAs with 5′-proximal secondary structure, whereas expression from those mRNAs lacking a poly(A) tail, whether capped or not, was little changed or reduced in oocytes overexpressing PABP relative to control oocytes. These data suggest that those mRNAs that are both capped and polyadenylated preferentially benefit from an increase in the level of PABP and the increased availability of PABP helps to reduce the inhibitory effect of secondary structure within a leader.
Figure 6.
Inhibition imposed by 5′-proximal secondary structure in oocytes overexpressing PABP. The same mRNAs were injected into the same batch of stage VI Xenopus oocytes used in Figure 2. The oocytes were incubated, harvested and assayed for reporter gene expression at the same time as those in Figure 2. Capped poly(A)+ uidA mRNA was co-injected to serve as an internal control. Each mRNA was injected into 25 oocytes and each luciferase assay was performed in triplicate. Luciferase expression (normalized to the internal control) is reported. The level of expression of each mRNA containing 5′-proximal secondary structure is also reported as a percentage of the corresponding control (i.e. the same construct without 5′-proximal secondary structure) which is set at 100%. The length of the 5′-leader and presence of the stem–loop are shown schematically to the left of the expression data.
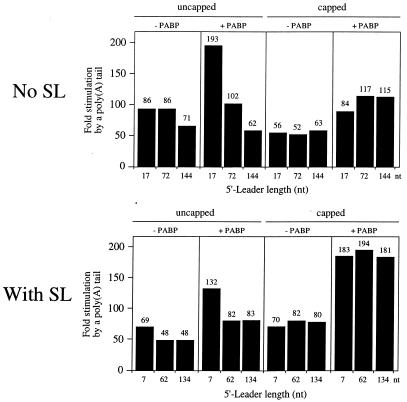
The effect of overexpressing PABP on the function of the poly(A) tail could be determined by calculating the extent to which addition of the poly(A)50 tail increased expression. Addition of a poly(A)50 tail to a capped mRNA with an unstructured 5′-leader increased expression 52- to 63-fold in control oocytes but 84- to 117-fold in oocytes overexpressing PABP (Fig. 7). The increase in poly(A) tail function was the result of an increase in absolute expression from the capped and polyadenylated mRNAs in oocytes overexpressing PABP (compare the expression levels from capped and polyadenylated luc mRNAs in Fig. 6 with Fig. 2). An even greater increase in the function of the poly(A) tail was observed for a capped mRNA with a 5′-proximal secondary structure in oocytes overexpressing PABP (Fig. 7). The addition of a poly(A)50 tail to a capped mRNA containing 5′-proximal secondary structure increased expression 70- to 82-fold in control oocytes but 181- to 194-fold in oocytes overexpressing PABP, which, like the observation with the corresponding unstructured mRNAs, was the result of an increase in absolute expression from the capped and polyadenylated form of the mRNAs. An increase in poly(A) tail function was also observed for an uncapped mRNA with a short leader, regardless of whether it was structured or not (Fig. 7), however, this was the result, in part, of a decrease in expression from the uncapped poly(A)– mRNA as well as an increase in expression from the uncapped poly(A)+ mRNA in oocytes overexpressing PABP (compare expression levels in Fig. 6 with Fig. 2). These data indicate that overexpressing PABP increases the function of the poly(A) tail for polyadenylated mRNAs, especially for those that are also capped. The data also indicate that the translation of mRNAs containing 5′-proximal secondary structure benefit preferentially from an increase in the concentration of PABP.
Figure 7.
Increasing the concentration of PABP increases the function of the poly(A) tail for capped monocistronic mRNAs in Xenopus oocytes. The degree to which the addition of a poly(A)50 tail increases expression from the constructs in Figures 2 and 6 is reported as the fold increase over the corresponding poly(A)– mRNA for constructs without a structured leader (i.e. no SL) or with secondary structure present (i.e. with SL). The fold increase in expression is shown above each histogram. Each construct is designated by the length of its 5′-leader (i.e. no SL) or distance between the secondary structure and the initiation codon (i.e. with SL) below each histogram.
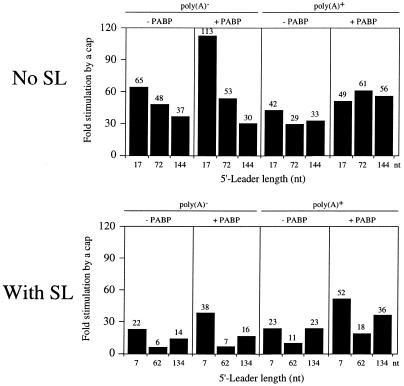
The function of the cap for mRNAs with a structured or unstructured leader could also be determined in control oocytes or in oocytes overexpressing PABP. The cap consistently stimulated translation of mRNAs with an unstructured leader to a greater extent than it did of mRNAs with a structured leader, regardless of whether the mRNA was polyadenylated or not (Fig. 8). Overexpression of PABP resulted in a moderate increase in the function of the cap for polyadenylated mRNAs whether or not the leader was structured (Fig. 8). The increase in cap function was the result of an increase in absolute expression from the capped and polyadenylated form of the mRNAs (compare expression levels in Fig. 6 with Fig. 2). An increase in cap function was also observed for a poly(A)– mRNA with a short leader, regardless of whether it was structured or not (Fig. 8). However, this resulted more from a decrease in absolute expression from the uncapped poly(A)– mRNA (presumably due to the greater competition exerted by endogenous polyadenylated mRNAs when PABP was overexpressed) rather than an increase in expression from the capped poly(A)– mRNA in oocytes overexpressing PABP [compare expression levels from uncapped and poly(A)– luc mRNAs in Fig. 6 with Fig. 2]. These data indicate that overexpressing PABP increases the function of the cap for polyadenylated mRNAs, especially for those that contain 5′-proximal secondary structure.
Figure 8.
Increasing the concentration of PABP increases the function of the cap for polyadenylated monocistronic mRNAs in Xenopus oocytes. The degree to which the addition of a cap increases expression from the constructs in Figures 2 and 6 is reported as the fold increase over the corresponding uncapped mRNA for constructs without a structured leader (i.e. no SL) or with secondary structure present (i.e. with SL). The fold increase in expression is shown above each histogram. Each construct is designated by the length of its 5′-leader (i.e. no SL) or distance between the secondary structure and the initiation codon (i.e. with SL) below each histogram.
Intercistronic length and structure affect translation from the distal cistron of a dicistronic mRNA
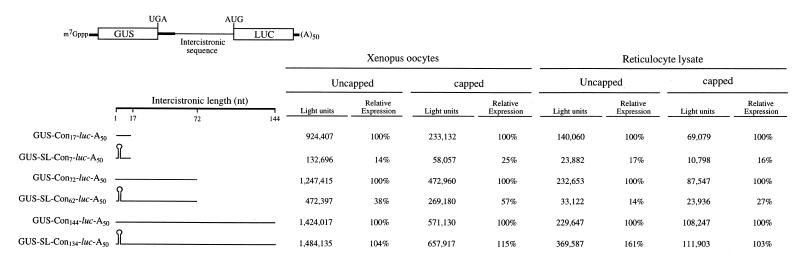
The previous experiments suggest that translation increases with length of the 5′-leader, particularly for mRNAs containing 5′-proximal secondary structure. To examine whether the length of the intercistronic region of a dicistronic mRNA might have a similar effect on translation from the distal cistron, the uidA coding region, which encodes the GUS reporter enzyme (and 73 bp of sequence downstream from the uidA stop codon), was introduced upstream of the luc constructs described in Figure 2. This resulted in a series of dicistronic GUS–luc mRNA constructs in which the intercistronic region (in addition to the 73 bp from the uidA fragment) was 17, 72 or 144 nt in length. In addition, dicistronic GUS–luc mRNA constructs were made in which the 24 bp stem–loop structure used in Figure 2 was present 7, 62 or 134 nt upstream from the distal cistron. The stem–loop was positioned 73 bp downstream of the uidA stop codon in order to prevent it from repressing translational termination from the 5′-proximal cistron (30). Capped and uncapped dicistronic mRNAs were synthesized to contain a poly(A)50 tail and injected into oocytes. Expression from the 5′-distal cistron was reduced 2- to 4-fold when the mRNA was capped (Fig. 9), suggesting that presence of the cap, which increased translation from the proximal cistron by ∼60-fold (data not shown), diverted the translational machinery to the 5′-terminus of the dicistronic mRNA. When the mRNA was uncapped, expression from the constructs in which secondary structure was introduced 7, 62 or 134 nt upstream of the distal cistron was 14, 38 or 104%, respectively, of that from the corresponding control dicistronic constructs in which no stem–loop was present (Fig. 9). The presence of secondary structure in the intercistronic region was also inhibitory in vitro and could be relieved by increasing the distance between it and the initiation codon to at least 134 nt (Fig. 9). Increasing the intercistronic region from 17 to 144 nt resulted in a moderate increase in expression from the distal cistron in both oocytes and in vitro translation lysate (Fig. 10) similar to that observed for monocistronic mRNA constructs (see Fig. 5). Increasing the length between the secondary structure and the distal cistron increased expression substantially and to a similar extent regardless of whether the mRNA was capped or not (Fig. 10).
Figure 9.
The inhibition imposed by secondary structure on expression from the distal cistron of a dicistronic mRNA is abolished by increasing its distance from the initiation codon. Dicistronic constructs, in which uidA (encoding GUS) served as the 5′-proximal cistron and luc served as the 5′-distal cistron, were synthesized in vitro as capped or uncapped mRNAs that terminated in a poly(A)50 tail and injected into stage VI Xenopus oocytes. The 24 bp stem–loop (SL) used in Figure 1 was introduced into the intercistronic region 7, 62 or 134 nt upstream of the luc initiation codon. Seventy-six nucleotides from the uidA gene were present in all constructs upstream of the site into which the secondary structure was introduced. Each mRNA was injected into 25 oocytes and each luciferase assay using an equal amount of oocyte extract was performed in triplicate. The level of expression of each mRNA containing secondary structure is also reported as a percentage of the corresponding control (i.e. the same construct without the intercistronic secondary structure) which is set at 100%. The length of the intercistronic region and presence of the stem–loop are shown schematically to the left of the expression data.
Figure 10.
Expression increases as a function of the length of the intercistronic region or distance between secondary structure and the distal cistron of a dicistronic mRNA. Expression from the constructs in Figure 9 is reported relative to the constructs with the shortest intercistronic region in the absence (i.e. no SL) or presence of secondary structure (i.e. with SL). Expression from the constructs with a 17 nt intercistronic region (i.e. no SL) or 7 nt intercistronic region (i.e. with SL) is set at a value of 1 and the fold increase in expression from the constructs with longer intercistronic regions is reported. The fold increase in expression is shown above each histogram. Each construct is designated by the length of its intercistronic region (i.e. no SL) or distance between the secondary structure and the initiation codon (i.e. with SL) below each histogram.
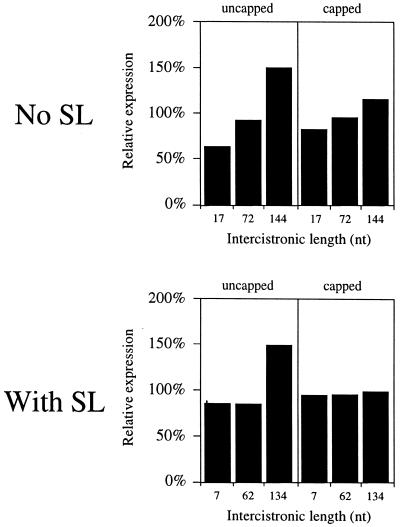
To examine whether increasing the concentration of PABP affected expression from the distal cistron of a dicistronic mRNA, the same dicistronic mRNAs were co-injected with PABP mRNA into the same batch of oocytes used for the data in Figure 9 and expression from the distal cistron measured at the same time as the control oocytes. For uncapped mRNAs, an increase in expression from the distal cistron in oocytes overexpressing PABP was observed only for the construct with the longest intercistronic region, regardless of whether secondary structure was present in the intercistronic region or not (Fig. 11). For dicistronic mRNAs with shorter intercistronic regions, expression from the distal cistron did not benefit or was reduced in oocytes with an elevated level of PABP (Fig. 11). Increasing the concentration of PABP did not result in a significant increase in expression from the 5′-distal cistron of any of the dicistronic constructs when the mRNAs were capped (Fig. 11). These data indicate that expression from a distal cistron increases with the intercistronic distance and that the inhibition imposed by secondary structure in the intercistronic region can be completely reversed if positioned 134 nt upstream from the distal cistron. The data also indicate that an increase in the level of PABP benefits expression from a distal cistron only when the mRNA does not contain a cap at the 5′-terminus.
Figure 11.
Expression from the distal cistron of a dicistronic mRNA increases in oocytes overexpressing PABP. Expression from the 5′-distal luc cistron of the dicistronic constructs used in Figure 9 in oocytes overexpressing PABP is reported as the percent change relative to expression from the same constructs in control oocytes. A value <100% represents a decrease in expression from the distal luc cistron in oocytes overexpressing PABP relative to expression in control oocytes, whereas a value >100% represents an increase in expression from the distal luc cistron in oocytes overexpressing PABP relative to expression in control oocytes. Each construct is designated by the length of its intercistronic region (i.e. no SL) or distance between the secondary structure and the initiation codon (i.e. with SL) below each histogram.
DISCUSSION
In this study, we have demonstrated that the structural features of an mRNA affect its expression in oocytes, in part through affecting the function of the cap and poly(A) tail. The cap and poly(A) tail are bifunctional in that they can affect the stability as well as the translatability of a message. Although transcript stability in stage VI oocytes is not substantially altered by changes in the length of the poly(A) tail (31), addition of a cap can increase transcript stability by 3- to 4-fold (31,32). Northern analysis of the constructs used in this study confirmed these observations in that no significant difference in the level of poly(A)+ or poly(A)– mRNAs was detected 18 h following their delivery to oocytes, whereas capped mRNAs were several-fold more stable than uncapped mRNAs (data not shown). Consequently, the higher expression levels observed from capped monocistronic mRNAs would be expected to result from increases in translatability as well as mRNA stability.
Expression from a monocistronic mRNA increased with the length of the 5′-leader up to at least 144 nt. The increase was greater for uncapped mRNAs than for capped mRNAs. Expression was substantially inhibited by the presence of 5′-proximal secondary structure, which could be partially relieved by increasing the distance (by at least 134 nt in length) between the secondary structure and the initiation codon. Secondary structure that was positioned 62 nt from the initiation codon inhibited expression to an extent virtually identical to that imposed by the structure when just 7 nt from the start site. As the 40S ribosomal subunit covers ∼25–30 nt when bound to an mRNA (33), the inhibitory effect of secondary structure 62 nt from the initiation codon cannot be explained solely by steric hindrance of 40S ribosomal subunit binding to the initiation codon. 5′-Proximal secondary structure was somewhat less inhibitory for uncapped mRNAs than for capped mRNAs (see Fig. 2), as might be expected for an mRNA whose translation must occur independently of a cap. Nevertheless, the presence of secondary structure substantially inhibited translation from uncapped mRNAs, even when positioned 134 nt upstream from the initiation codon. In contrast, positioning the secondary structure 134 nt upstream from the initiation codon of the distal cistron of a dicistronic mRNA had no effect on expression from the distal cistron (see Fig. 9), although it inhibited translation from the distal cistron if positioned more closely to the initiation codon. These data suggest that 134 nt is sufficient to permit internal initiation to the extent that it can occur and, therefore, the inhibition observed by secondary structure 134 nt upstream from the initiation codon of an uncapped, monocistronic mRNA indicates the degree to which 40S subunit binding remains 5′-end-dependent. These data indicate that translation in stage VI oocytes is highly cap-dependent and, even in the absence of a cap, remains largely 5′-end-dependent. 5′-End-independent translation initiation is observed to some extent, but only when sufficient distance separates the secondary structure from the initiation codon. 5′-Proximal secondary structure was also inhibitory to translation in vitro, but its effect could be substantially relieved by positioning the structure 134 nt upstream from the initiation codon. Although the in vitro translations were performed in reticulocyte lysate that contained a full complement of endogenous mRNAs, these data suggest that the lysate contains an amount of translational machinery sufficient to promote a greater degree of internal initiation when secondary structure is positioned sufficiently upstream of an initiation codon to allow unhindered ribosome binding than that observed in oocytes. Increasing the concentration of PABP in oocytes did not increase expression from monocistronic mRNAs that lacked a cap or a poly(A) tail but did increase expression from monocistronic mRNAs that possessed both a cap and a poly(A) tail by 2- to 3-fold regardless of the length of the 5′-leader (compare the absolute levels of expression in Fig. 6 with those in Fig. 2). This effect was seen with both structured and unstructured mRNAs, but was more pronounced for the former. These data are in good agreement with previous observations (29) that expression from mRNAs in Xenopus oocytes is limited by the amount of available PABP and, furthermore, they indicate that only those mRNAs that can bind PABP, i.e. polyadenylated mRNAs, can benefit from an increase in PABP concentration. They also suggest that an increase in PABP concentration preferentially benefits expression from mRNAs containing a structured leader. Because the concentration of PABP increases substantially by the neurula stage of embryo development, the preferential increase in expression from structured mRNAs following an elevation in the level of PABP suggests a possible developmental role for PABP that may result in a greater level of translation from structured mRNAs during embryogenesis.
The function of the cap was substantially diminished by the presence of 5′-proximal secondary structure and a decrease in cap function was observed in control oocytes regardless of the distance between the structure and the initiation codon (Fig. 8). Because the close proximity of secondary structure to the cap may have inhibited binding of eIF4F or binding or scanning by the 40S ribosomal subunit, increasing the distance between the secondary structure and the initiation codon would not be expected to relieve the inhibition of cap function. The preferential increase in expression from structured mRNAs in oocytes expressing an elevated level of PABP likely results from the increased availability of PABP to bind the poly(A) tail as well as its increased association with eIF4G as observed in Figure 5. It is possible that this increased association between PABP and eIF4G promotes internal initiation from structured mRNAs downstream of the 5′-proximal structure. However, as increased association between PABP and eIF4G is observed when the latter is present as part of eIF4F (see Fig. 5) and the eIF4E subunit would be expected to direct the eIF4F complex to the 5′-cap, it is more likely that the increased association of PABP with eIF4G would stabilize eIF4F binding to the cap, which, in turn, would assist in unwinding the secondary structure. That PABP can perform this function, resulting in an increase in RNA helicase activity by the cap-associated initiation factors, has been demonstrated in wheat (34).
The function of the cap was lowest when the secondary structure was positioned 62 nt upstream from the initiation codon (Fig. 8). This was a consequence of the fact that increasing the distance between the secondary structure and the coding region from 7 to 62 nt increased expression from the uncapped form of the mRNA by several-fold without significantly increasing expression from the capped form of the mRNA (see Fig. 2). As a result, the degree to which the cap stimulated expression from SL-Con62-luc-A50 mRNA, i.e. the ratio of expression from the capped mRNA to that from the uncapped mRNA, was lower than that observed for SL-Con7-luc-A50.
Increasing the concentration of PABP increased the function of the cap for polyadenylated monocistronic mRNAs as well as for the non-polyadenylated form of Con7-luc mRNA (see Fig. 8). The increase in cap function for capped, polyadenylated monocistronic mRNAs was a consequence of an increase in absolute expression from the capped form of each mRNA. In contrast, the increase in cap function for the non-polyadenylated monocistronic mRNA was the result of a decrease in expression from the uncapped form of Con7-luc mRNA (compare expression levels in Fig. 6 to Fig. 2). This decreased expression was disproportionately large for mRNAs lacking both a cap and poly(A) tail, which may result from the increased competition provided by endogenous mRNAs in oocytes in which the concentration of PABP has been elevated. These results indicate that the increase in PABP concentration may benefit those messages that are capped and polyadenylated at the expense of poorly competitive mRNAs, e.g. those that lack a cap and poly(A) tail, and suggest that translation from competitive mRNAs may be increasingly favored with the increase in PABP expression that follows fertilization.
The function of the poly(A) tail was not significantly altered by changes in the length of the 5′-leader of capped or uncapped monocistronic mRNAs (see Fig. 7). However, increasing the concentration of PABP increased the function of the poly(A) tail for capped polyadenylated monocistronic mRNAs which, similar to the effect on the function of the cap, was the result of an increase in absolute expression from the polyadenylated form of each mRNA (compare expression levels in Fig. 6 with Fig. 2). This effect was particularly evident for mRNAs containing a structured leader. An increase in poly(A) tail function was also observed for the uncapped form of Con7-luc mRNA (see Fig. 7). Although this reflects a small stimulation by PABP of translation of the polyadenylated uncapped transcript, it is mainly due to a decrease in expression of the mRNA lacking both a cap and a poly(A) tail in oocytes overexpressing PABP (compare expression levels in Fig. 6 with Fig. 2). These results indicate that the increase in PABP concentration benefits expression specifically from polyadenylated mRNAs, and particularly from those that are also capped.
Dicistronic mRNAs are frequently used to diagnose the presence of specific IRES in a 5′-untranslated region. Putative IRES elements are introduced intercistronically to test their ability to promote translation of the downstream cistron (reviewed in 20). While archetypal IRES elements, such as those from encephalomyocarditis viral RNAs, promote high rates of translation in dicistronic RNA assays, the efficiency of translation driven by potential IRES elements derived from cellular mRNAs is often lower (35,36). The data presented here (see Figs 9 and 10) indicate that, even in the absence of a true IRES element, in vivo as well as in vitro translation systems can support limited levels of internal initiation and that this initiation can be enhanced by increasing the length of unstructured sequence upstream of the 5′-distal cistron. Moreover, this occurred for both uncapped and capped mRNAs, suggesting that translation of the second cistron of a dicistronic mRNA is not directly dependent on the rate of translation of the first cistron. Similar observations were made when the intercistronic region contained secondary structure. However, the absolute level of expression from the distal cistron decreased 2- to 4-fold when the mRNA was capped relative to when the mRNA was uncapped (see Fig. 9), suggesting that the cap diverted the translational machinery to the 5′-terminus of the dicistronic mRNA. The reduction in translation of the distal cistron under circumstances in which translation of the 5′-proximal cistron is increased suggests that translation of the second cistron occurs in a cap-independent and 5′-end-independent manner. The observation that the introduction of secondary structure 134 nt upstream from an initiation codon was inhibitory to expression of a monocistronic mRNA but not to the distal cistron of a dicistronic mRNA supports the conclusion that translation of the second cistron occurs through internal initiation. An elevation in the concentration of PABP resulted in an increase in translation of the distal cistron, but only when the dicistronic mRNA was uncapped and polyadenylated. This observation indicates that the increase in PABP expression promoted internal initiation when no cap was present that would otherwise divert the translational machinery to the 5′-proximal cistron. It is possible that the increased association of PABP with eIF4G may be responsible for this increase in internal initiation when a mRNA lacks a cap. The lack of a similar increase when the discistronic mRNA was capped supports the conclusion that the increase in expression from a capped monocistronic mRNA with a 5′-proximal structure in oocytes overexpressing PABP was more likely a result of stabilization of eIF4F to the cap than a result of PABP promoting internal initiation within a capped mRNA.
In conclusion, these data suggest that translation from an mRNA in stage VI Xenopus oocytes is highly dependent on the presence of a cap and a poly(A) tail and that expression becomes more cap- and poly(A) tail-dependent when the concentration of PABP is increased. They also suggest that expression from poorly competitive mRNAs, i.e. uncapped and non-polyadenylated, decreases following an increase in the concentration of PABP, suggesting that the translational machinery is diverted to translationally competitive mRNAs. Finally, these studies support the conclusion that the level of PABP in oocytes, which is subject to developmental regulation (22), controls expression directly from translationally competitive mRNAs (i.e. those mRNAs that are both capped and polyadenylated) whereas it affects expression from less competitive mRNAs indirectly by increasing the competition provided by competitive mRNAs.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Dr Michael Wormington for the Xenopus PABP construct pSP64T-ABP and Dr Daniel Schoenberg for the anti-Xenopus PABP antiserum. This work was supported by a grant from the US Department of Agriculture (NRICGP 99-35301-7866) to D.R.G. and grants (040800, 045619 and 056778) to V.M.P and S.M. from the Wellcome Trust. S.J.M. is a Senior Research Fellow of the Wellcome Trust.
REFERENCES
- 1.Jackson R.J. and Wickens,M. (1997) Curr. Opin. Genet. Dev., 7, 233–241. [DOI] [PubMed] [Google Scholar]
- 2.Morley S.J., Curtis,P.S. and Pain,V.M. (1997) RNA, 3, 1085–1104. [PMC free article] [PubMed] [Google Scholar]
- 3.Gallie D.R. (1998) Gene, 216, 1–11. [DOI] [PubMed] [Google Scholar]
- 4.Le H., Tanguay,R.L., Balasta,M.L., Wei,C.-C., Browning,K.S., Metz,A.M., Goss,D.J. and Gallie,D.R. (1997) J. Biol. Chem., 272, 16247–16255. [DOI] [PubMed] [Google Scholar]
- 5.Piron M., Vende,P., Cohen,J. and Poncet,D. (1998) EMBO J., 17, 5811–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imataka H., Gradi,A. and Sonenberg,N. (1998) EMBO J., 17, 7480–7489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tarun S.Z. and Sachs,A.B. (1996) EMBO J., 15, 7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 8.Wei C.-C., Balasta,M.L., Ren,J. and Goss,D.J. (1998) Biochemistry, 37, 1910–1916. [DOI] [PubMed] [Google Scholar]
- 9.Klausner R.D., Rouault,T.A. and Harford,J.B. (1993) Cell, 72, 19–28. [DOI] [PubMed] [Google Scholar]
- 10.Pelletier J. and Sonenberg,N. (1985) Cell, 40, 515–526. [DOI] [PubMed] [Google Scholar]
- 11.Kozak M. (1986) Proc. Natl Acad. Sci. USA, 83, 2850–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kozak M. (1988) Mol. Cell. Biol., 8, 2737–2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kozak M. (1990) Proc. Natl Acad. Sci. USA, 87, 8301–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gallie D.R., Lucas,W.J. and Walbot,V. (1989) Plant Cell, 1, 301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallie D.R., Feder,J.N., Schimke,R.T. and Walbot,V. (1991) Mol. Gen. Genet., 228, 258–264. [DOI] [PubMed] [Google Scholar]
- 16.Kozak M. (1991) J. Biol. Chem., 266, 19867–19870. [PubMed] [Google Scholar]
- 17.Kozak M. (1991) J. Cell Biol., 115, 887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pelletier J. and Sonenberg,N. (1988) Nature, 334, 320–325. [DOI] [PubMed] [Google Scholar]
- 19.Jang S.K., Krausslich,H.-G.,Nicklin,M.J.H., Duke,G.M., Palmenberg,A.C. and Wimmer,E. (1988) J. Virol., 62, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson R.J. and Kaminski,A. (1995) RNA, 1, 985–1000. [PMC free article] [PubMed] [Google Scholar]
- 21.Gallie D.R. (1991) Genes Dev., 5, 2108–2116. [DOI] [PubMed] [Google Scholar]
- 22.Zelus B.D., Giebelhaus,D.H., Eib,D.W., Kenner,K.A. and Moon,R.T. (1989) Mol. Cell. Biol., 9, 2756–2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yisraeli J.K. and Melton,D.A. (1989) Methods Enzymol., 180, 42–50. [DOI] [PubMed] [Google Scholar]
- 24.Colman A. (1984) In Hames,B.D. and Higgins,S.J. (eds), Transcription and Translation: A Practical Approach, IRL Press, Oxford, UK, pp. 271–302.
- 25.Fraser C.S., Pain,V.M. and Morley,S.J. (1999) J. Biol. Chem., 274, 196–204. [DOI] [PubMed] [Google Scholar]
- 26.Everett J.G. and Gallie,D.R., (1992) Yeast, 8, 1007–1014. [DOI] [PubMed] [Google Scholar]
- 27.Stambuk R.A. and Moon,R.T. (1992) Biochem. J., 287, 761–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheets M.D., Fox,C.A., Hunt,T., Vande Woude,G. and Wickens,M. (1994) Genes Dev., 8, 926–938. [DOI] [PubMed] [Google Scholar]
- 29.Wormington M., Searfoss,A.M. and Hurney,C.A. (1996) EMBO J., 15, 900–909. [PMC free article] [PubMed] [Google Scholar]
- 30.Niepel M., Ling,J. and Gallie,D.R. (1999) FEBS Lett., 462, 79–84. [DOI] [PubMed] [Google Scholar]
- 31.Drummond D.R., Armstrong,J. and Colman,A. (1985) Nucleic Acids Res., 13, 7375–7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Furuichi Y., LaFiandra,A. and Shatkin,A.J. (1977) Nature, 266, 235–239. [DOI] [PubMed] [Google Scholar]
- 33.Kozak M. (1977) Nucleic Acids Res., 269, 390–394. [Google Scholar]
- 34.Bi X.P., Ren,J.H. and Goss,D.J. (2000) J. Biol. Chem., 275, 17740–17746. [DOI] [PubMed] [Google Scholar]
- 35.Kim J.G., Armstrong,R.C., Berndt,J.A., Kim,N.W. and Hudson,L.D. (1998) Mol. Cell. Neurosci., 12, 119–140. [DOI] [PubMed] [Google Scholar]
- 36.Sella O., Gerlitz,G., Le,S.-Y. and Elroy-Stein,O. (1999) Mol. Cell. Biol., 19, 5429–5440. [DOI] [PMC free article] [PubMed] [Google Scholar]