Significance
Phagocytes efficiently clear sick or damaged neuronal branches by engulfing them, while leaving healthy branches untouched. How phagocytes recognize degenerating neurites remains poorly understood. Here, we identified a key role for the secreted protein Orion in the detection and engulfment of degenerating neurites in Drosophila. Using multiple models of dendrite degeneration, we found that Orion acts as a bridging molecule between the neuronal “eat-me” signal phosphatidylserine (PS) and the engulfment receptor Draper on phagocytes, enabling phagocytosis. Our study reveals a missing link in PS-mediated phagocytosis in vivo, sheds light on factors determining the sensitivity of phagocytes, and implies the potential for manipulating the detection of neuronal “eat-me” signals in neurodegenerative diseases.
Keywords: phosphatidylserine, phagocytosis, da neurons, epidermal cells, Orion
Abstract
Phagocytic clearance of degenerating neurons is triggered by “eat-me” signals exposed on the neuronal surface. The conserved neuronal eat-me signal phosphatidylserine (PS) and the engulfment receptor Draper (Drpr) mediate phagocytosis of degenerating neurons in Drosophila. However, how PS is recognized by Drpr-expressing phagocytes in vivo remains poorly understood. Using multiple models of dendrite degeneration, we show that the Drosophila chemokine–like protein Orion can bind to PS and is responsible for detecting PS exposure on neurons; it is supplied cell-non-autonomously to coat PS-exposing dendrites and to mediate interactions between PS and Drpr, thus enabling phagocytosis. As a result, the accumulation of Orion on neurons and on phagocytes produces opposite outcomes by potentiating and suppressing phagocytosis, respectively. Moreover, the Orion dosage is a key determinant of the sensitivity of phagocytes to PS exposed on neurons. Lastly, mutagenesis analyses show that the sequence motifs shared between Orion and human immunomodulatory proteins are important for Orion function. Thus, our results uncover a missing link in PS-mediated phagocytosis in Drosophila and imply conserved mechanisms of phagocytosis of neurons.
Phagocytosis of apoptotic and degenerative neurons is essential for the development and homeostasis of the nervous system (1, 2). Abnormal phagocytosis is also associated with neuroinflammation and neurodegenerative diseases (3). Neuronal debris is recognized and cleared by resident phagocytes of the nervous system through “eat-me” signals exposed on the neuronal surface. A conserved “eat-me” signal is phosphatidylserine (PS), a negatively charged phospholipid normally kept in the inner leaflet of the plasma membrane by P4-ATPase flippases (4). During neurite degeneration and apoptosis, PS is externalized to the outer surface of neuronal membranes (5–8). The exposed PS dominantly triggers phagocytosis of neurons (6, 9, 10). Besides clearing neuronal debris, PS-mediated phagocytosis also drives the degeneration of injured neurites and neurons with certain genetic perturbations (6, 11). In the central nervous system (CNS), local PS exposure enables microglia-mediated synaptic elimination (12–14). Thus, the regulation and recognition of neuronal PS exposure are critical for the development and homeostasis of the nervous system.
Drosophila has been an important model organism for studying neuronal phagocytosis. In Drosophila, Draper (Drpr) is the best-known receptor responsible for phagocytosis of neurons (15). As a homolog of the Caenorhabditis elegans engulfment receptor CED-1 (16) and the mammalian engulfment receptors Jedi-1 and MEGF10 (17), Drpr is involved in many contexts of neuronal phagocytosis, including the clearance of apoptotic neurons during embryonic development (15, 18), axon and dendrite pruning during neuronal remodeling (19, 20), injury-induced neurite degeneration (21, 22), and removal of destabilized boutons at neuromuscular junctions (23). Despite the well-known importance of Drpr in sculpting the nervous system, how Drpr recognizes degenerating neurons in vivo is still unclear.
Recently, the secreted protein Orion was discovered as being required for the developmental pruning and clearance of Drosophila mushroom body (MB) axons (24). Orion shares a CX3C motif with mammalian CX3CL1 (also known as fractalkine), which is required for the elimination of synapses in the mouse barrel cortex (25). CX3CL1 is known as a chemokine because of its ability to direct migration of leukocytes and microglia (26, 27). Thus, Orion represents the first known chemokine-like molecule in Drosophila and shares conserved functions with CX3CL1 in the remodeling of the nervous system. However, how Orion and CX3CL1 function in the phagocytosis of neurons is still unknown.
In this study, we examined Orion’s function in the phagocytosis of Drosophila class IV dendritic arborization (C4da) neurons, a well-established in vivo model for studying PS-mediated phagocytosis (6, 11, 28). Here, we present in vivo evidence that the transmembrane engulfment receptor Drpr relies on Orion to sense PS on neurons. Orion is secreted by peripheral nonneural tissues and serves as a nonautonomous permissive factor for phagocytosis of neurons. Orion binds to PS-exposing neurons and is also enriched on phagocytes overexpressing Drpr. Strikingly, a membrane-tethered Orion expressed by neurons dominantly induces phagocytosis even in the absence of PS exposure, while Orion accumulation on the surface of phagocytes makes phagocytes blind to PS-exposing neurons. Importantly, the dosage of Orion determines the sensitivity of phagocytes to neuronal PS exposure. Lastly, we establish that the motifs Orion shares with human chemokines and neutrophil peptides are critical for engulfment of PS-exposing neurons. These findings reveal key mechanisms of PS recognition in Drosophila and imply potentially conserved roles of chemokines in PS-mediated phagocytosis of neurons.
Results
orion Is Required for Phagocytosis of Dendrites.
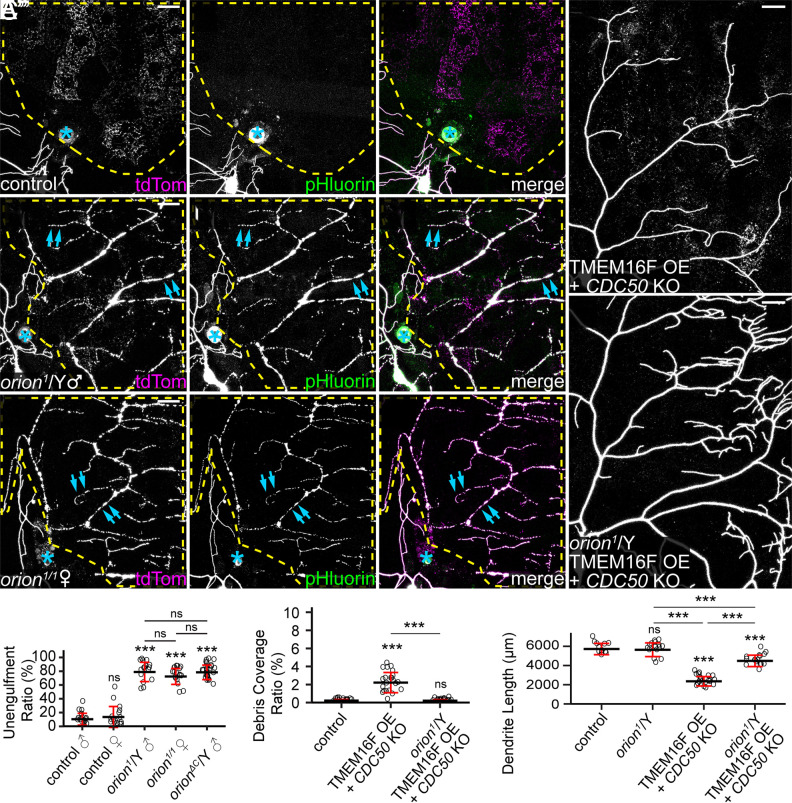
To determine whether orion is involved in the phagocytosis of degenerating dendrites of da neurons, we first examined phagocytosis of injured C4da dendrites in the orion1 mutant, which contains a G to D mutation in the C termini of both Orion protein isoforms (SI Appendix, Fig. S1A) and abolishes clearance of pruned axons during MB remodeling (24). C4da neurons grow elaborate sensory dendrites on larval epidermal cells, which act as the primary phagocytes during dendrite degeneration and remodeling (28). Dendrites were severed from the cell body using laser, and engulfment of the injured dendrites was visualized using MApHS, a pH-sensitive dendritic marker consisting of extracellular pHluorin and intracellular tdTom (28). Engulfment was signified by the loss of pHluorin signal due to the drop of pH in early phagosomes (28, 29). Injured dendrites of C4da neurons in the control larvae were completely engulfed by epidermal cells 24 h after injury (AI), as indicated by the loss of pHluorin signals on tdTom-positive dendritic debris dispersed in epidermal cells (Fig. 1 A–A” and D). Because orion is located on the X chromosome, we examined both hemizygous male larvae and homozygous female larvae of orion1. In contrast to the wild type (WT), both groups of orion1 mutants showed little engulfment of injured dendrites by epidermal cells (Fig. 1 B, C”, and D), as indicated by the presence of pHluorin signals on tdTom-labeled debris of injured dendrites. The debris remained in the original dendritic patterns, which is another sign of impaired engulfment by epidermal cells. These results suggest that orion is required for the engulfment of injured dendrites. Since male and female orion mutants exhibited similar degrees of engulfment defects (Fig. 1D), we used hemizygous male mutants (orion1/Y) only in subsequent assays.
Fig. 1.
orion is required for phagocytosis of dendrites in multiple dendrite degeneration paradigms. (A–C”) Partial dendritic fields of ddaC neurons in control (A–A”), orion1 hemizygous (B–B”), and orion1 homozygous (C–C”) larvae at 22 to 24 h after injury (AI). Yellow dash outlines: territories originally covered by injured dendrites; blue asterisks: injury sites; blue arrows: injured but unengulfed dendrite fragments. (D) Quantification of unengulfment ratio of injured dendrites (pHluorin-positive debris area/tdTom-positive debris area). n = number of neurons and N = number of animals: control males (n = 18, N = 10); control females (n = 16, N = 9); orion1/Y (n = 16, N = 8); orion1/1 (n = 18, N = 9); orionΔC/Y (n = 25, N = 12). One-way ANOVA and Tukey’s test. (E and F) Partial dendritic fields of TMEM16F OE + CDC50 KO neurons in the wild-type background (E) and orion1 hemizygous background (F). (G and H) Quantification of debris coverage ratio (percentage of debris area normalized by dendrite area) (G) and dendrite length (H) at 140 h after egg laying (AEL). n = number of neurons and N = number of animals: control (n = 14, N = 7); orion1/Y (n = 15, N = 8); TMEM16F OE + CDC50 KO (n = 22, N = 11); TMEM16F OE + CDC50 KO in orion1/Y (n = 16, N = 8). For (G), Kruskal–Wallis (one-way ANOVA on ranks) and Dunn’s test, P-values adjusted with the Benjamini–Hochberg method; for (H), one-way ANOVA and Tukey’s test. In all image panels, neurons were labeled by ppk-MApHS. (Scale bars, 25 μm.) For all quantifications, ***P ≤ 0.001; n.s., not significant. The significance level above each genotype is for comparison with the control. Black bar, mean; red bar, SD. See also SI Appendix, Fig. S1.
In addition to phagocytosis after injury, C4da dendrites are all pruned and engulfed by epidermal cells during metamorphosis (28). Consistent with the role of orion in axonal pruning of MB neurons (24), we also confirmed that orion1 exhibits strong defects in the clearance of pruned dendrites of C4da neurons during metamorphosis (SI Appendix, Fig. S1 B–D).
The engulfment of injured dendrites by epidermal cells is mediated by PS exposure on dendrites (6, 11). To test directly whether orion is required for PS exposure–induced phagocytosis, we examined a paradigm in which PS exposure was ectopically induced in otherwise healthy neurons using a combination of CDC50 knockout (KO) and TMEM16F overexpression (OE). CDC50 encodes a chaperone protein required for the activity of P4-ATPases that keep PS in the inner leaflet of the plasma membrane (30). TMEM16F is a mammalian scramblase that mixes PS between the two leaflets of the plasma membrane (31, 32). Dendrites of CDC50 KO + TMEM16F OE neurons expose PS and shed membrane in a phagocytosis-dependent manner (6). Whereas these neurons showed reduced dendrite length and elevated debris levels in wandering 3rd instar larvae with wild-type orion (Fig. 1 E, G, and H), orion1/Y exhibited almost normal dendritic length and no dendritic debris in epidermal cells (Fig. 1 F–H), indicating a lack of engulfment by phagocytes. The slight dendrite reduction of CDC50 KO + TMEM16F OE neurons in orion1/Y is likely caused by TMEM16F OE (6), instead of the loss of orion, since dendrite length in orion1/Y was comparable to that of the wild-type control (Fig. 1H).
Because orion1 carries a missense mutation that may not completely abolish Orion function, we compared its properties to those of orionΔC, a predicted null mutation that lacks all three common C-terminal exons of both orion isoforms (SI Appendix, Fig. S1A) (24). orionΔC showed similar levels of phagocytosis defects as orion1 in injury-induced degeneration (SI Appendix, Fig. S1 E–E” and D), suggesting that orion1 has lost most, if not all, of its function. We next asked whether one or both isoforms contribute to phagocytosis. Mutations targeting each of the two orion isoforms separately (SI Appendix, Fig. S1A) showed no phagocytosis defects in injury-induced degeneration (SI Appendix, Fig. S1 F–G”), suggesting that OrionA and OrionB isoforms are redundant in the phagocytosis of injured dendrites. These results are consistent with the redundant effects of the same mutations in MB neuron axon pruning (24).
Orion Functions Cell-Non-Autonomously.
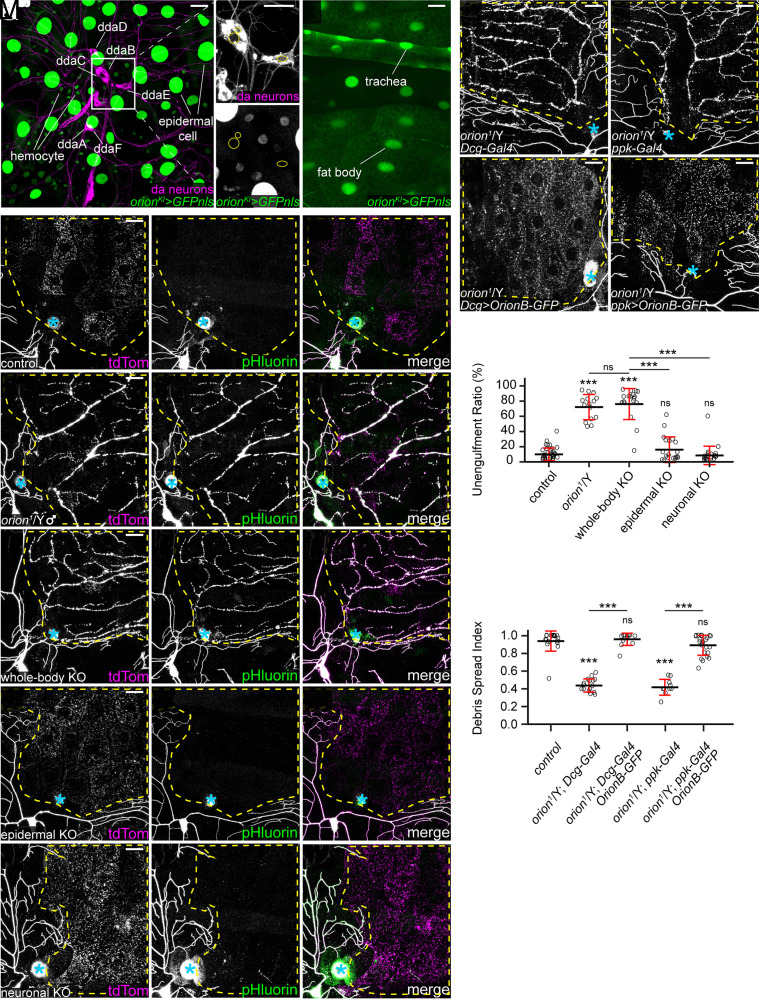
To determine which tissues express Orion, we generated an orion knock-in (KI) allele in which the C terminus of orion is fused in-frame with 4 copies of mNeonGreen211 (mNG211) (33), a V5 tag, a self-cleaving F2A sequence (34), and LexA::VP16 (35) (SI Appendix, Fig. S2A). This allele, abbreviated as orionKI, has several purposes: detection of orion-expressing cells by LexA::VP16 transcription activity, detection of endogenous Orion proteins by V5 staining, and visualization of endogenous Orion proteins in a tissue-specific manner by reconstitution of mNG2 fluorescence with tissue-specific expression of mNG21-10 (33).
By using orionKI with a LexAop-GFPnls or a LexAop-GFP reporter, we found that orion transcripts are expressed in many peripheral tissues in larvae, including epidermal cells, hemocytes, trachea, and the fat body (Fig. 2 A and B). However, orion transcription is missing in larval da neurons (Fig. 2 A, Inset), even though a weak orion transcription activity was later detected in a subset of da neurons at 5 to 12 h after puparium formation (APF) (SI Appendix, Fig. S2 B–B”) when some da neurons die and others undergo dendritic pruning (36). In the CNS, we found that orion was transcribed in most glial cells and in a small subset of neurons at the wandering 3rd instar larval stage (SI Appendix, Fig. S2 C–D”). Consistent with a role of orion in axonal pruning of MB neurons, orion is expressed in Kenyon cells at 8 h APF (SI Appendix, Fig. S2E). Despite the robust LexA::VP16 activity in orionKI, Orion proteins appear to be expressed at a low level. Using V5 staining, we could observe only weak Orion signals on the larval epidermis (SI Appendix, Fig. S2 F and G). By expressing secreted mNG21-10 in a patch of epidermal cells, we also detected weak Orion-mNG2 signals in epidermal cells (SI Appendix, Fig. S2 H and I).
Fig. 2.
Orion functions cell-non-autonomously. (A and B) GFPnls expression driven by orion-LexA (orionKI) in epidermal cells, hemocytes (A), trachea, and fat body (B), but not in da neurons (A, Insets). Yellow circles indicate nuclei of ddaB, ddaC, and ddaE neurons (A). [Scale bars, 25 μm (A), 10 μm (A Insets), and 50 μm (B).] (C–G”) Partial dendritic fields of neurons in control (C–C”, same images as Fig. 1 A–A”), orion1 hemizygotes (D–D”, same images as Fig. 1 B–B” for clarity), whole-body orion KO (E–E”), epidermal orion KO (F–F”), and neuronal orion KO (G–G”) at 22 to 24 h AI. (H–K) Partial dendritic fields of ddaC neurons in orion1 hemizygotes with fat body Gal4 (H), with OrionB-GFP OE in the fat body (I), with neuron Gal4 (J), and with OrionB-GFP OE in neurons (K) at 22 to 24 h AI. (L) Quantification of unengulfment ratio of injured dendrites. n = number of neurons and N = number of animals: control and orion1/Y (same dataset as in Fig. 1D); whole-body orion KO (n = 18, N = 10); epidermal orion KO (n = 19, N = 11); neuronal orion KO (n = 21, N = 11). One-way ANOVA and Tukey’s test. (M) Quantification of debris spread index of injured dendrites (area ratio of all 15 pixel × 15 pixel squares that contained dendrite debris in the region of interest). n = number of neurons and N = number of animals: orion1/Y (n = 18, N = 10); Dcg-Gal4 in orion1/Y (n = 16, N = 9); Dcg>OrionB-GFP in orion1/Y (n = 13, N = 7); ppk-Gal4 in orion1/Y (n = 10, N = 6); ppk>OrionB-GFP in orion1/Y (n = 24, N = 12). One-way ANOVA and Tukey’s test. Neurons were labeled by 21-7>UAS-CD4-tdTom (A and B), ppk-MApHS (C–G”), ppk-CD4-tdTom (H and I), and ppk>CD4-tdTom (J and K). In (C–K), yellow dash outlines: territories originally covered by injured dendrites; blue asterisks: injury sites; (scale bars, 25 μm.) For all quantifications, ***P ≤ 0.001; n.s., not significant. The significance level above each genotype is for comparison with the control. Black bar, mean; red bar, SD. See also SI Appendix, Fig. S2.
To investigate whether orion is required in specific tissues for phagocytosis of injured dendrites, we knocked out orion in either neurons or epidermal cells using tissue-specific Cas9s and ubiquitously expressed guide RNAs (gRNAs) (37). As a control, knocking out orion with a ubiquitous Cas9 (Act5C-Cas9) (38) faithfully replicated the phagocytosis defects of orion1 (Fig. 2 C–E” and L), demonstrating the effectiveness of orion KO. However, orion KO in either da neurons alone [with SOP-Cas9 (37)] or in epidermal cells alone [with shot-Cas9 (39)] did not interfere with the engulfment of injured dendrites (Fig. 2 F–G” and L).
The lack of effect of orion KO in da neurons or in epidermal cells suggests that Orion functions cell-non-autonomously. We further tested this idea by asking whether supplying Orion in the extracellular space is sufficient to rescue the impaired engulfment of injured dendrites in orion mutants. Extracellular Orion supply was achieved by overexpressing Orion in the fat body, which can efficiently secrete proteins into the hemolymph (6). Because OE of OrionA in the fat body caused early larval lethality, we chose to overexpress a GFP-tagged OrionB (OrionB-GFP) in these rescue experiments. While the debris of injured dendrites remained in the original dendrite patterns in orion1/Y (Fig. 2H), which indicates a lack of engulfment by epidermal cells, OrionB OE in the fat body of orion1/Y resulted in even spreading of the debris in the underlying epidermal cells (Fig. 2I), suggesting successful rescue of engulfment. Similarly, neuronal expression of OrionB-GFP also fully rescued the engulfment defects as measured by the spread of debris (Fig. 2 J, K, and M).
Together, these results indicate that Orion is primarily expressed in nonneural tissues in the periphery and functions cell-non-autonomously for the engulfment of dendrites.
PS Exposure Induces Orion Binding to the Cell Surface.
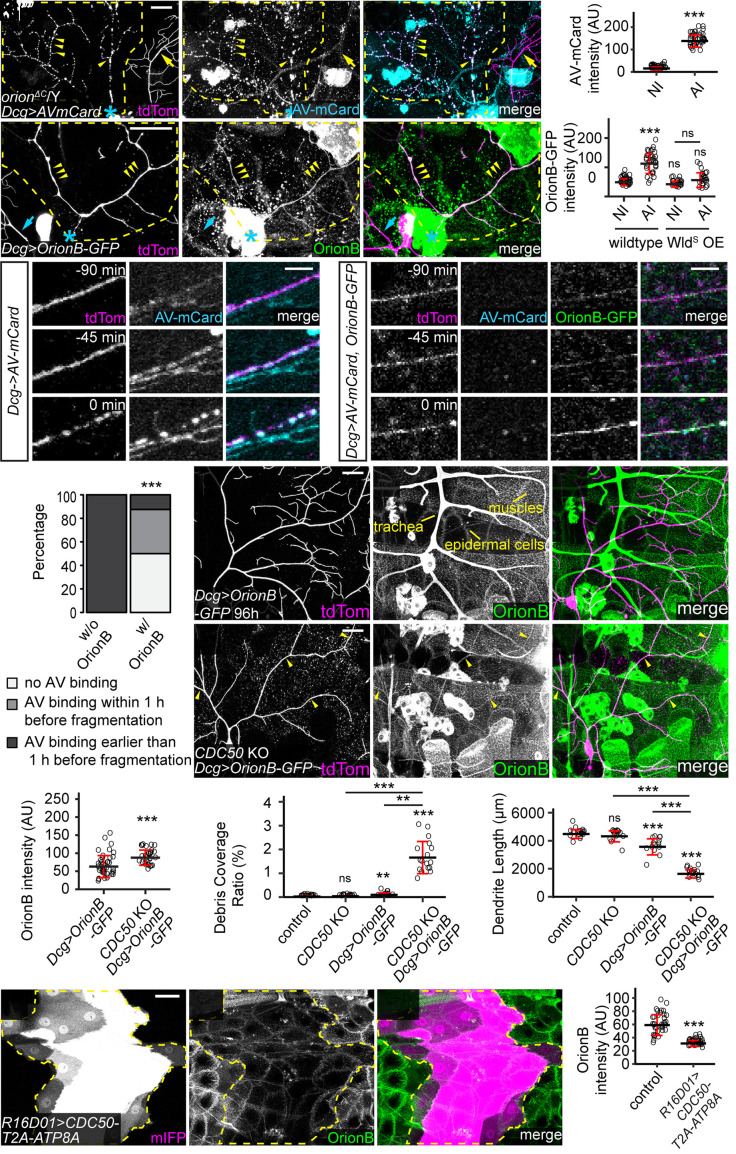
To further understand the engulfment defects in orion mutants, we examined whether PS exposure on injured dendrites is affected in orion mutants. We used fat body–derived Annexin V-mCardinal (AV-mCard) as a PS sensor; it labels injured but not healthy dendrites (6). AV-mCard robustly labeled dendrite fragments in orionΔC mutant larvae at 24 h AI (Fig. 3 A and B), suggesting that orion LOF does not interfere with PS exposure on injured dendrites.
Fig. 3.
PS exposure induces Orion binding to the cell surface. (A–A”) Labeling of injured dendrites of a ddaC neuron by AV-mCard at 23 h AI in orionΔC hemizygous larvae. Yellow arrowheads: injured dendrites with AV-mCard labeling; yellow arrows: uninjured dendrites lacking AV-mCard labeling. (B) Quantification of AV-mCard binding on dendrites. AV-mCard intensities were measured on both uninjured (NI) and injured (AI) dendrites. n = number of measurements and N = number of animals: NI (n = 39, N = 8); AI (n = 39, N = 8). Welch’s t test. (C–C”) Labeling of injured dendrites of a ddaC neuron by Orion-GFP at 6 h AI in wild-type larvae. Yellow arrowheads: injured dendrites with OrionB-GFP labeling. (D) Quantification of Orion-GFP binding on dendrites of wild-type and WldS OE neurons. n = number of measurements and N = number of animals: wild-type NI (n = 39, N = 10); wild-type AI (n = 36, N = 10); WldS OE NI (n = 22, N = 6); WldS OE AI (n = 23, N = 6). Kruskal–Wallis (one-way ANOVA on ranks) and Dunn’s test, P-values adjusted with the Benjamini–Hochberg method. (E and F) Time series of injured dendrites of ddaC neurons from 90 min before fragmentation to the moment of fragmentation, with only AV-mCard expressed (E) or both OrionB-GFP and AV-mCard coexpressed (F) by the fat body. Time stamps are relative to the frame of dendrite fragmentation. (G) Percentages of injured dendrites showing no AV binding, AV binding within 1 h before fragmentation, AV binding earlier than 1 h before fragmentation. n = number of measurements and N = number of animals: w/o OrionB OE (n = 9, N = 5); w/ OrionB OE (n = 8, N = 3). Fisher’s exact test. (H–I”) Distribution of fat body–derived OrionB-GFP with wild-type (H–H”) and CDC50 KO (I–I”) dendrites at 96 h AEL. Peripheral tissues showing OrionB binding are labeled in (H’). Yellow arrowheads indicate OrionB binding on CDC50 KO dendrites (I–I”). (J) Quantification of OrionB-GFP binding on wild-type and CDC50 KO dendrites. n = number of measurements: Dcg>OrionB-GFP (n = 41, N = 7); CDC50 KO + Dcg>OrionB-GFP (n = 29, N = 6). Welch’s t-test. (K and L) Quantification of debris coverage ratio (K) and dendrite length (L) at 96 h AEL. n = number of neurons and N = number of animals: control (n = 17, N = 9); CDC50 KO (n = 15, N = 8); Dcg>OrionB-GFP (n = 13, N = 8); CDC50 KO + Dcg>OrionB-GFP (n = 17, N = 9). For (K), Kruskal–Wallis (one-way ANOVA on ranks) and Dunn’s test, P-values adjusted with the Benjamini–Hochberg method; for (L), one-way ANOVA and Tukey’s test. (M–M”) OrionB-GFP binding on epidermal cells that expressed CDC50-T2A-ATP8A. Yellow dash outlines: CDC50-T2A-ATP8A overexpressing region. (N) Quantification of OrionB-GFP binding on wild-type epidermal cells and CDC50-T2A-ATP8A OE epidermal cells. n = number of measurements and N = number of animals: control (n = 36, N = 9); R16D01>CDC50-T2A-ATP8A (n = 36, N = 9). Welch’s t test. In (A–A”) and (C–C”), yellow dash outlines: territories originally covered by injured dendrites; blue asterisks: injury sites. Neurons were labeled by ppk-MApHS (A–A” and E–E”) and ppk-CD4-tdTom (C–C”, F–F”, and H–I”). [Scale bars, 25 μm (A–A”, C–C”, and H–I”), 5 μm (E and F), and 50 μm (M–M”).] For all quantifications, **P ≤ 0.01, ***P ≤ 0.001; n.s., not significant. The significance level above each genotype is for comparison with the control. Black bar, mean; red bar, SD. See also Movies S1–S3 and SI Appendix, Fig. S3.
We next asked where Orion is located during engulfment. We observed strong enrichment of fat body–derived OrionB-GFP on injured dendrites at 4 to 6 h AI (Fig. 3 C and D and Movie S1), a timepoint when injured dendrites expose high levels of PS (6). OE of WldS, a transgene that suppresses fragmentation and PS exposure of injured dendrites when overexpressed in neurons (6, 11), efficiently suppressed OrionB-GFP enrichment on injured dendrites SI Appendix, Fig. S3 A–A” and Fig. 3 D, indicating that PS exposure may be required for Orion binding to injured dendrites.
To test the affinity of Orion for PS-exposing injured dendrites in vivo, we examined whether OrionB can compete with AV-mCard for binding to injured dendrites, given that AV binds to PS directly (4). Using long-term time-lapse imaging (40), we found that AV-mCard accumulated on injured dendrites at least 60 min before dendrite fragmentation (9/9 movies) (Fig. 3 E and G and Movie S2). However, when both OrionB-GFP and AV-mCard were coexpressed by the fat body, OrionB-GFP was detected on injured dendrites long before fragmentation (8/8 movies), while AV-mCard did not bind the same injured dendrite in half of the cases (4/8 movies) (Fig. 3 F and G and Movie S3). In most cases where AV-mCard was observed on injured dendrites (3/4 movies), the labeling only appeared right at the time of dendrite fragmentation (Fig. 3G and SI Appendix, Fig. S3E) when PS exposure is at its peak (6). These results suggest that Orion has a higher affinity to injured dendrites compared with AV.
To test whether neuronal PS exposure is sufficient to induce Orion binding, we ectopically induced dendritic PS exposure by knocking out CDC50 in neurons (6). At 96 h AEL, CDC50 KO alone was not yet sufficient to cause membrane loss of dendrites (Fig. 3 K and L). While OrionB-GFP showed little binding to wild-type dendrites at this stage (Fig. 3 H–H” and J), it bound robustly to CDC50 KO dendrites (Fig. 3 I and J). Interestingly, the presence of OrionB-GFP caused appreciable degeneration of CDC50 KO dendrites, as indicated by the drastically increased dendrite debris and reduced dendrite length (Fig. 3 K and L). These results suggest that Orion is recruited to PS-exposing dendrites and OrionB binding on dendrites potentiates epidermal engulfment.
The PS-binding C1C2 domain of mouse lactadherin (LactC1C2) has been used as a PS sensor in multiple systems (5, 6, 41, 42). We previously reported that fat body–derived LactC1C2 not only labels degenerating dendrites but also promotes degeneration of CDC50 KO dendrites (6) (SI Appendix, Fig. S3 B–B”). Surprisingly, this degeneration was completely suppressed in orion1/Y (SI Appendix, Fig. S3 C–C”), suggesting that the effects of LactC1C2 are mediated by Orion.
To further ask whether Orion directly interacts with PS in vitro, we purified fat body–derived OrionB-GFP protein from larvae and conducted liposome sedimentation assays (43). As positive and negative controls, we also tested AV-GFP and AV(mut)-GFP (6) which carries mutations that abolish AV/PS interactions (41, 44), respectively. As expected, AV-GFP coprecipitated with PS-containing liposomes but not PC-only liposomes (SI Appendix, Fig. S3 F and G). In contrast, AV(mut)-GFP did not coprecipitate with either liposomes (SI Appendix, Fig. S3 F and H). Surprisingly, about half OrionB-GFP protein in our assay coprecipitated with PC-only liposomes, and liposome-bound OrionB-GFP increased to 62% when PS was added to the liposomes (SI Appendix, Fig. S3 F and I). These data suggest that OrionB may have an intrinsic affinity to phospholipid bilayer and that this interaction is enhanced by the presence of PS.
In vivo, we noticed that OrionB-GFP also binds to the surface of several other peripheral tissues, including epidermal cells, muscles, and trachea (Fig. 3 H–H”). The OrionB-GFP binding on epidermal cells was enhanced after we gently pinched the larval body wall (SI Appendix, Fig. S3 D and D’), which is expected to mildly disrupt the epidermal cell membrane. To understand how much the binding of OrionB-GFP on epidermal cells depends on PS exposure, we overexpressed ATP8A, an ortholog of the PS-specific flippase TAT-1 (9, 45). OE of ATP8A in da neurons is sufficient to suppress PS exposure and the associated dendrite degeneration caused by genetic perturbations of the NAD+ pathway (11). The P4-ATPase chaperone CDC50 was coexpressed with ATP8A in a patch of epidermal cells to facilitate ATP8A trafficking. This manipulation drastically suppressed OrionB-GFP binding on the surface of epidermal cells (Fig. 3 M and N), suggesting that Orion binding on epidermal cells is largely mediated by PS exposure.
Together, our results suggest that PS exposure induces Orion binding to the cell surface of both neuronal and nonneural cells.
Orion Recruits Epidermal Drpr to Injured Dendrites.
Because drpr is the only other gene known to be required for epidermal engulfment of PS-exposing dendrites (6, 11), we wondered whether Orion and Drpr act in the same pathway. We found that orion1 hemizygous and drpr mutant larvae exhibited similar degrees of near-complete phagocytosis deficiency in injury-induced dendrite degeneration, as indicated by the portion of unengulfed MApHS-labeled debris (SI Appendix, Fig. S4 A and B). We further asked whether removing both drpr and orion produces stronger phagocytosis defects than the loss of either. We induced whole-body KO of orion and drpr individually and together. Injured dendrites showed indistinguishable levels of near-complete blockage of engulfment in all the three genotypes at 24 h AI (SI Appendix, Fig. S4 C–E), suggesting that orion and drpr function in the same genetic pathway.
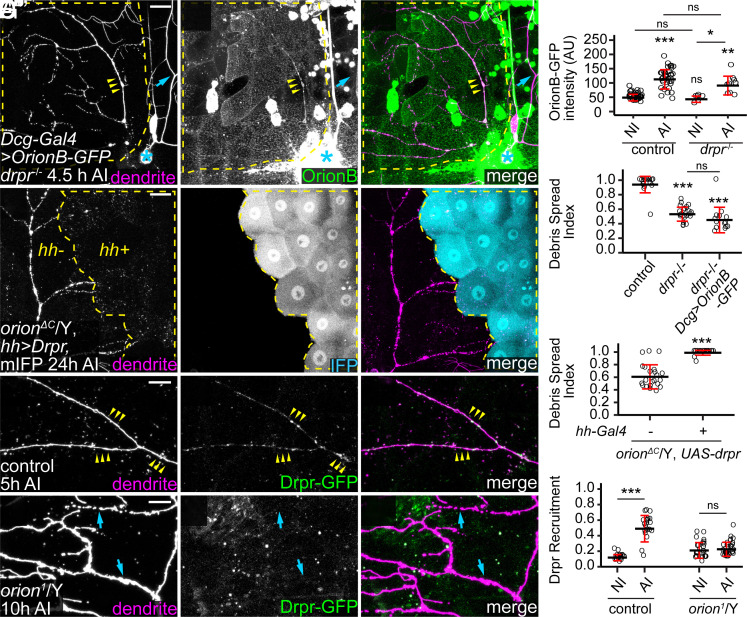
To elucidate the epistatic relationship between orion and drpr, we tested whether the Orion–PS interaction depends on Drpr and whether gain of function (GOF) of one gene can compensate for the loss of the other. At 4 to 5 h AI, OrionB-GFP bound to injured dendrites in wild-type and drpr null larvae at similar levels (Fig. 4 A and B), demonstrating that Orion does not need Drpr for binding to PS. Two observations further suggest that Orion GOF cannot compensate for the loss of drpr in engulfment: First, OE of Orion in the fat body did not rescue the engulfment of injured dendrites in drpr mutants at 24 h AI (SI Appendix, Fig. S4F and Fig. 4C); second, the degeneration of CDC50 KO dendrites induced by OrionB-GFP (Fig. 3 I–L) was absent in drpr mutants (SI Appendix, Fig. S4 G–I). To test the effects of drpr GOF in orion LOF, we overexpressed Drpr in a patch of epidermal cells in the posterior hemisegment (driven by hh-Gal4) of orionΔC hemizygotes. Interestingly, Drpr OE restored engulfment of injured dendrites, as indicated by the dispersion of dendrite debris specifically in the hh domain (Fig. 4 D and E). These data suggest that phagocytes with overexpressed Drpr does not absolutely need Orion to detect injured dendrites, which could be due to the ability of Drpr extracellular domain to directly interact with PS (18). However, our data also show that, at physiological levels of Drpr, Orion is indispensable for engulfment.
Fig. 4.
Orion recruits epidermal Drpr to injured dendrites. (A–A”) Labeling of injured dendrites of a ddaC neuron by OrionB-GFP in drpr−/− at 4.5 h AI. Yellow arrowheads: injured dendrites with OrionB-GFP labeling; blue arrows: uninjured dendrites lacking OrionB-GFP binding. (B) Quantification of Orion-GFP binding on dendrites in the wild-type and the drpr−/− larvae. n = number of measurements and N = number of animals: wild-type NI and wild-type AI (same dataset as in Fig. 3D); drpr−/− NI (n = 6, N = 2); drpr−/− AI (n = 9, N = 3). Kruskal–Wallis (one-way ANOVA on ranks) and Dunn’s test, P-values adjusted with the Benjamini–Hochberg method. (C) Quantification of debris spread index of injured dendrites at 22 to 24 h AI. n = number of neurons and N = number of animals: control (same dataset as in Fig. 2M); drpr−/− (n = 23, N = 10); drpr−/− + Dcg>OrionB-GFP (n = 14, N = 7). One-way ANOVA and Tukey’s test. (D–D”) Engulfment of injured dendrites in an orionΔC hemizygous larva with Drpr overexpressed in the hh domain. Yellow dash outlines: hh>Drpr, mIFP region. (E) Quantification of debris spread index of injured dendrites at 22 to 24 h AI in region without Drpr OE and region with Drpr OE in orionΔC/Y. n = number of neurons and N = number of animals: region without Drpr OE (n = 22, N = 12) and region with Drpr OE (n = 24, N = 12). Welch’s t test. (F–F”) Distribution of Drpr-GFP in the presence of injured dendrites in control at 5 h AI (F–F”) and in orion1/Y at 10 h AI (G–G”). Yellow arrowheads (F–F”): injured dendrites with Drpr-GFP recruitment; blue arrowheads (G–G”): injured dendrites lacking Drpr-GFP recruitment. (H) Quantification of Drpr-GFP recruitment (Drpr-GFP-positive area on dendrites/total dendrite area). n = measurements: control NI (n = 20, N = 12); control AI (n = 21, N = 12); orion1/Y NI (n = 27, N = 8); orion1/Y AI (n = 31, N = 8). Welch’s t test. For all image panels, neurons were labeled by ppk-CD4-tdTom. Yellow dash outlines: territories originally covered by injured dendrites; blue asterisks: injury sites. [Scale bars, 25 μm (A–A”, D–D”) and 10 μm (F–G”).] For all quantifications, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; n.s., not significant. The significance level above each genotype is for comparison with the control. Black bar, mean; red bar, SD. See also SI Appendix, Fig. S4.
To further understand how orion LOF affects Drpr, we examined the distribution of Drpr protein by live imaging. For this purpose, we generated a KI allele of drpr so that the C terminus of endogenous Drpr is tagged by mNeonGreen (mNG) (46) (SI Appendix, Fig. S4J). To further boost the Drpr signal, we also made a Drpr-GFP transgene that contains a genomic fragment of the drpr locus tagged with GFP at the Drpr C terminus (SI Appendix, Fig. S4J). Combining a copy of each of drpr-mNG and drpr-GFP (referred to as simply drpr-GFP because both fluorescent proteins are green), we were able to see robust Drpr recruitment to injured dendrites prior to dendrite fragmentation (Fig. 4 F–F” and H). However, this recruitment was abolished in the orion1/Y (Fig. 4 G and H). These results suggest that Orion regulates Drpr’s response to degenerating dendrites after injury.
Orion Mediates the Interaction between PS and Drpr.
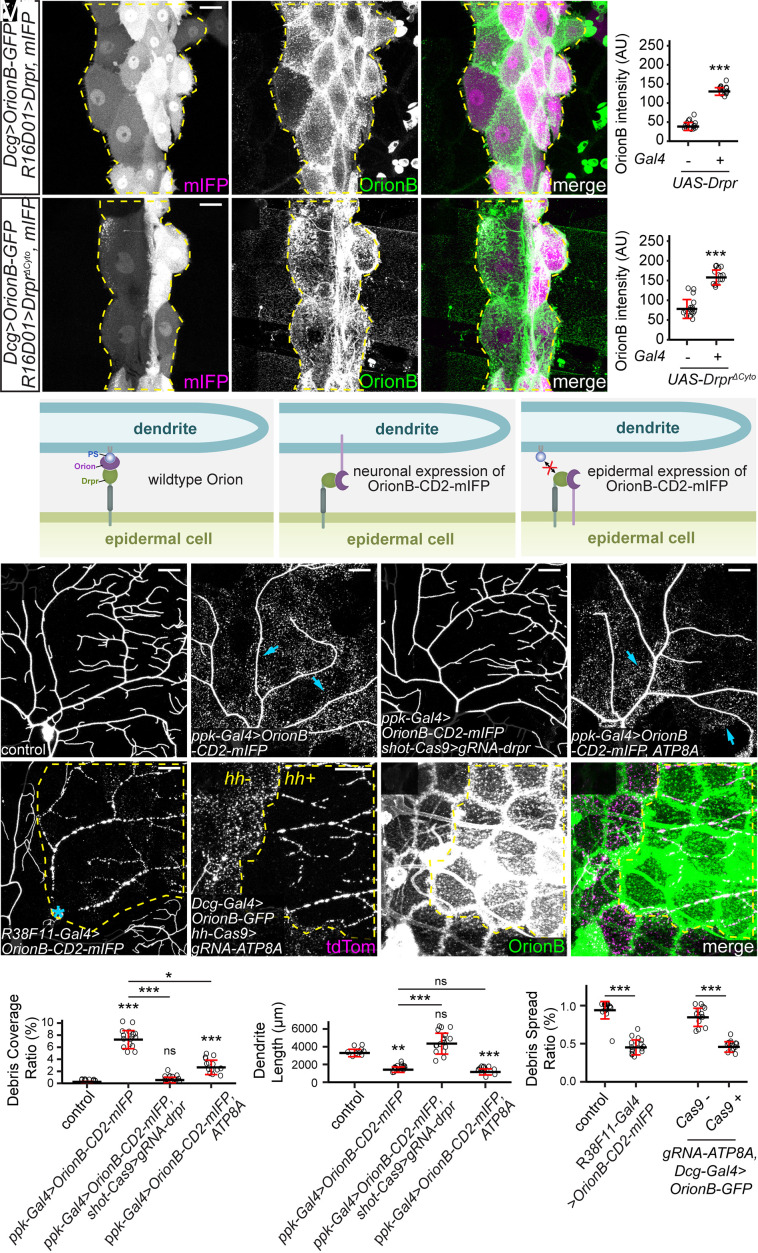
Because Orion interacts with PS and functions genetically upstream of Drpr, we wondered whether Orion mediates PS recognition by interacting with Drpr. We first tested whether Orion could bind to Drpr in vivo by expressing Drpr in a patch of epidermal cells (driven by R16D01-Gal4). Fat body–derived OrionB-GFP was found to accumulate specifically on the surface of these Drpr OE cells in wandering 3rd instar larvae (Fig. 5 A and B). Interestingly, some epidermal cells with higher OrionB-GFP enrichment (due to higher Gal4 activities) extruded into neighboring epidermal cells that had low or no OrionB-GFP enrichment (SI Appendix, Fig. S5 A–A”). We interpret these extrusions as engulfment of high-OrionB cells by low-OrionB cells. To exclude the possibility that OrionB-GFP was recruited by other cell-surface molecules on epidermal cells as a result of Drpr OE, we also tested a truncated Drpr without the intracellular domain (DrprΔCyto), which should be defective in intracellular signaling. OE of DrprΔCyto also caused drastic OrionB-GFP accumulation on epidermal cells (Fig. 5 C and D). These results suggest that OrionB may directly interact with the extracellular domain of Drpr.
Fig. 5.
Orion mediates the interaction between PS and Drpr. (A–A”) Distribution of fat body–derived OrionB-GFP with Drpr overexpressed in a patch of epidermal cells driven by R16D01-Gal4. mIFP was coexpressed with Drpr and thus the mIFP intensity is correlated with the Gal4 activity. Yellow dash outlines: Drpr OE region. (B) Quantification of Orion-GFP binding on epidermal cells with or without Drpr OE. n = number of measurements and N = number of animals: without Drpr OE (n = 16, N = 8); with Drpr OE (n = 16, N = 8). Welch’s t test. (C-C") Distribution of fat body–derived OrionB-GFP with DrprΔCyto overexpressed in a patch of epidermal cells (labeled by mIFP). Yellow dash outlines: DrprΔCyto OE region. (D) Quantification of Orion-GFP binding on epidermal cells with or without DrprΔCyto OE. n = number of measurements and N = number of animals: without DrprΔCyto OE (n = 16, N = 8); with DrprΔCyto OE (n = 14, N = 7). Welch’s t test. (E) Models of Orion function in mediating PS–Drpr interaction (Left), interactions between Drpr and Orion-CD2-mIFP overexpressed by neurons (Middle) or epidermal cells (Right). (F–I) Partial dendritic fields of a control ddaC neuron (F), an OroinB-CD2-mIFP OE neuron (G), an OroinB-CD2-mIFP OE neuron with drpr KO in epidermal cells (H), and an OroinB-CD2-mIFP + ATP8A OE neuron (I). (J–K) Quantification of debris coverage ratio (J) and dendrite length (K) at 96 h AEL. n = number of neurons and N = number of animals: control (n = 10, N = 6); ppk-Gal4>OroinB-CD2-mIFP (n = 17, N = 9); ppk-Gal4>OroinB-CD2-mIFP + shot-Cas9>gRNA-drpr (n = 17, N = 9); ppk-Gal4>OroinB-CD2-mIFP, ATP8A (n = 14, N = 7). Kruskal–Wallis (one-way ANOVA on ranks) and Dunn’s test, P-values adjusted with the Benjamini–Hochberg method. (L) Partial dendritic field of an injured ddaC neuron at 25 h AI with OroinB-CD2-mIFP expressed in all epidermal cells. (M–M”) Partial dendritic field of an injured ddaC neuron at 9 h AI when OroinB-GFP was expressed in fat body and ATP8A was knocked out in hh-epidermal cells. (N) Quantification of debris spread index of injured dendrites. n = number of neurons and N = number of animals: control (n = 18, N = 10); R38F11>OroinB-CD2-mIFP (n = 16, N = 8); Dcg-Gal4>OrionB (n = 14, N = 8); Dcg-Gal4>OrionB + hh-Cas9>ATP8A (n = 13, N = 8). Welch’s t test. Neurons were labeled by ppk-MApHS (F–I) and ppk-CD4-tdTom (L and M–M”). For all image panels, (scale bars, 25 μm.) For all quantifications, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; n.s., not significant. The significance level above each genotype is for comparison with the control. Black bar, mean; red bar, SD. See also SI Appendix, Fig. S5.
To further test the hypothesis that Orion mediates the interaction between Drpr and PS, we asked whether expression of an Orion that is permanently tethered to the surface of otherwise wild-type dendrites could bypass the requirement of PS exposure and induce Drpr-dependent phagocytosis. For this purpose, we made an OrionB-CD2-mIFP [mIFP: monomeric infrared fluorescent protein (47)] transgene in which Orion is located on the extracellular side of the CD2 transmembrane domain (48). As expected, OE of OrionB-CD2-mIFP in neurons caused robust dendrite degeneration (Fig. 5 E–G, J, and K). This degeneration was completely suppressed by epidermis-specific KO of drpr (Fig. 5 H, J, and K) but was unaffected by suppressing PS exposure in neurons via ATP8A OE (Fig. 5 I–K). These data suggest that membrane-tethered Orion is sufficient to induce PS-independent and Drpr-dependent phagocytosis.
Meanwhile, if Orion mediates the recognition of PS by Drpr, we predict that excessive Orion on the surface of epidermal cells would interact with Drpr on the same membrane and interfere with the sensing of PS on dendrites. We first tested this idea by overexpressing OrionB-CD2-mIFP in all epidermal cells (Fig. 5E). Indeed, this manipulation fully blocked the engulfment of injured dendrites at 25 h AI (Fig. 5 L and N). Drpr was robustly detected on the cell membranes of OrionB-CD2-mIFP-expressing cells (SI Appendix, Fig. S5 C and D), suggesting that the impaired engulfment was not due to defects in Drpr subcellular localization. We then tested whether accumulation of secreted Orion on the surface of epidermal cells has a similar effect in blocking engulfment. Consistent with the idea that Orion binds PS, ATP8A KO in epidermal cells resulted in a drastically increased surface level of OrionB-GFP (Fig. 5M'). This OrionB-GFP accumulation was associated with phagocytosis deficiency, as indicated by the lack of spread of dendritic debris at 9 h AI specifically in the ATP8A KO cells (Fig. 5 M and N).
Together, our results show that interactions between Orion and Drpr from the same versus apposing membranes produce opposite phenotypes (defective versus dominant dendrite engulfment, respectively) and support the idea that Orion functionally bridges PS and Drpr.
The Orion Dosage Determines the Sensitivity of Epidermal Cells to PS-Exposing Dendrites.
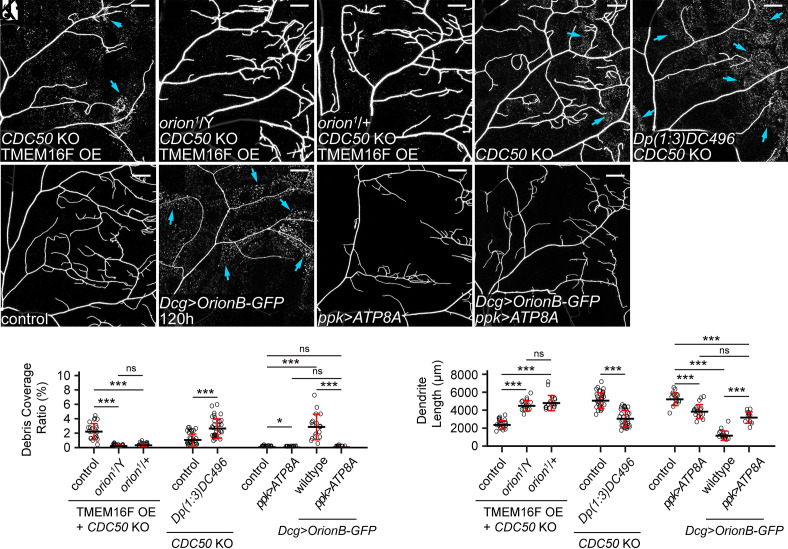
While examining the phenotypes of the orion1 mutant, we noticed impaired phagocytosis in orion1/+ heterozygous larvae: CDC50 KO + TMEM16F OE neurons showed no signs of membrane loss or degeneration in these animals (Fig. 6 A–C, J, and K), comparable to those in orion1 hemizygotes. However, the engulfment of injured dendrites was normal in orion1/+ (SI Appendix, Fig. S6 A and B). Considering that CDC50 KO and TMEM16F OE cause milder PS exposure than does injury (6), these data suggest that removing half the dosage of Orion reduces the sensitivity of epidermal cells to PS exposure on dendrites but does not block phagocytosis when PS exposure is high. To further test whether the Orion dosage determines the ability of epidermal cells to sense PS-exposing dendrites, we increased the Orion dosage by adding an orion duplication (Dp(1;3)DC496) to the WT. The orion duplication strongly enhanced the debris level and dendrite loss in a CDC50 KO background (Fig. 6 E, J, and K), which by itself only causes weak PS exposure (6) and low levels of debris (Fig. 6 D, J, and K). Thus, extra Orion can indeed increase the sensitivity of epidermal cells to neuronal PS.
Fig. 6.
The Orion dosage determines the sensitivity of epidermal cells to PS-exposing dendrites. (A–C) Partial dendritic fields of a TMEM16F OE + CDC50 KO ddaC neuron in the WT background (A, same image as Fig. 1E), in the orion1 hemizygous background (B, same image as Fig. 1F), and in the orion1 heterozygous background (C). (D and E) Partial dendritic fields of CDC50 KO neurons in the control (D) and Dp(1;3)DC496 (E) at 120 h AEL. Blue arrows: debris shed from dendrites. (F–I) Partial dendritic fields of ddaC neurons in the control (F), with fat body–derived OrionB-GFP (G), with ATP8A OE in the neuron (H), and with fat body–derived OrionB-GFP and ATP8A OE in the neuron (I) at 120 h AEL. (J and K) Quantification of debris coverage ratio (J) and dendrite length (K). n = number of neurons and N = number of animals: for TMEM16F OE + CDC50 KO, control and orion1/Y (same dataset as in Fig. 1G), orion1/+ (n = 18, N = 9); for (J), Kruskal–Wallis (one-way ANOVA on ranks) and Dunn’s test, P-values adjusted with the Benjamini–Hochberg method; for (K), one-way ANOVA and Tukey’s test. For CDC50 KO, control (n = 33, N = 17), Dp(1;3)DC496 (n = 33, N = 17), Welch’s t test. For effects of Dcg>OrionB-GFP and ppk>ATP8A at 120 h AEL, control (n = 17, N = 9), ppk>ATP8A (n = 17, N = 9), Dcg>OrionB-GFP (n = 17, N = 9), ppk>ATP8A + Dcg>OrionB-GFP (n = 12, N = 7); for (J), Kruskal–Wallis (one-way ANOVA on ranks) and Dunn’s test, P-values adjusted with the Benjamini–Hochberg method; for (K), one-way ANOVA and Tukey’s test. Neurons were labeled by ppk-MApHS (A–C and H), ppk-CD4-tdTom (D–G), and ppk-Gal4>CD4-tdTom (I). For all image panels, (scale bars, 25 μm.) For all quantifications, *P ≤ 0.05, ***P ≤ 0.001; n.s., not significant. The significance level above each genotype is for comparison with the control. Black bar, mean; red bars, SD. See also SI Appendix, Fig. S6.
Further supporting this notion, we found that fat body–derived OrionB bound to wild-type dendrites at 120 h AEL (SI Appendix, Fig. S6C) and induced pronounced degeneration (Fig. 6 F, G, J, and K). This degeneration is PS dependent because it was completely suppressed by ATP8A OE in neurons (Fig. 6 H–K). This surprising result demonstrates that, in late larval stages, wild-type dendrites expose low levels of PS that cannot be detected by common PS sensors such as AV and LactC1C2, but can be bound by overexpressed Orion.
CX3C and RRY Motifs Are Important for Orion Function.
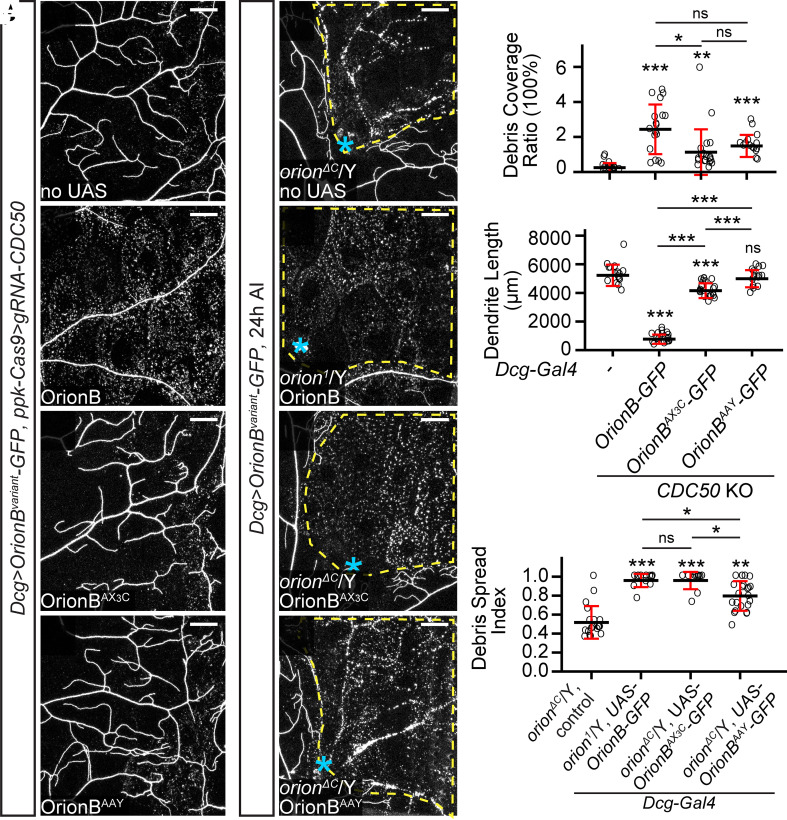
Orion shares a CX3C motif with the human chemokine CX3CL1 and an RRY motif with several human neutrophil peptides. The CX3C motif is required for Orion’s function in MB remodeling (24), but the importance of the RRY motif in Orion has not been investigated. To test whether these motifs play any role in PS-mediated dendrite engulfment, we generated UAS-OrionB-GFP variants carrying mutations in them (OrionBAX3C and OrionBAAY). We first compared these OrionB-GFP variants in their abilities to potentiate dendrite degeneration of CDC50 KO neurons, which would be reflected by increased debris level and reduction of dendrite length. Although fat body–derived OrionBAX3C and OrionBAAY both potentiated the debris level of CDC50 KO neurons (Fig. 7 A–D and I), only OrionBAX3C, but not OrionBAAY, caused a weak (21%) dendrite reduction of CDC50 KO neurons (Fig. 7J). In comparison, WT OrionB caused 85% reduction of CDC50 KO dendrites (Fig. 7J). We next tested OrionB-GFP variants in rescuing orionΔC hemizygotes with the dendrite injury assay. OrionAX3C restored the engulfment of injured dendrites to the same level as WT OrionB-GFP, while OrionBAAY only partially restored the engulfment (Fig. 7 E–I). These results suggest that these mutations impair, but do not abolish, Orion activity.
Fig. 7.
CX3C and RRY motifs are important for Orion function. (A–D) Partial dendritic fields of CDC5O KO ddaC neurons without OrionB variants (A), with WT OrionB-GFP (B), OrionBAX3C-GFP (C), and OrionBAAY-GFP (D) expressed in the fat body at 120 h AEL. (E–H) Partial dendritic fields of ddaC neurons in orionΔC or orion1 hemizygous larvae without OrionB variants (E), with WT OrionB-GFP (F), OrionBAX3C-GFP (G), and OrionBAAY-GFP (H) expressed in the fat body at 22 to 24 h AI. Yellow dash outlines: territories originally covered by injured dendrites; blue asterisks: injury sites. (I and J) Quantification of debris coverage ratio (I) and dendrite length (J) at 120 h AEL. n = number of neurons and N = number of animals: CDC50 KO control (n = 16, N = 8); CDC50 KO + Dcg>OrionB-GFP (n = 18, N = 9); CDC50 KO + Dcg>OrionBAX3C-GFP (n = 20, N = 10); CDC50 KO + Dcg>OrionBAAY-GFP (n = 16, N = 8). For (I), Kruskal–Wallis (one-way ANOVA on ranks) and Dunn’s test, P-values adjusted with the Benjamini–Hochberg method; for (J), one-way ANOVA and Tukey’s test. (K) Quantification of debris spread index of injured dendrites. n = number of neurons and N = number of animals: orionΔC/Y control (n = 19, N = 9); orion1/Y + Dcg>OrionB-GFP (same dataset as in Fig. 2M); orionΔC/Y + Dcg>OrionBAX3C-GFP (n = 12, N = 7); orionΔC/Y + Dcg>OrionBAAY-GFP (n = 23, N = 12). Kruskal–Wallis (one-way ANOVA on ranks) and Dunn’s test, P-values adjusted with the Benjamini–Hochberg method. For all image panels, neurons were labeled by ppk-CD4-tdTom. (Scale bars, 25 μm.) For all quantifications, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; n.s., not significant. The significance level above each genotype is for comparison with the control. Black bar, mean; red bars, SD.
Discussion
Orion Mediates Interactions between PS and Drpr.
By triggering phagocytosis, the recognition of PS exposed on neurons is a critical event during neurodegeneration and clearance. Although several studies have implied the involvement of Drpr in this process in Drosophila (6, 11, 15, 19–21, 28, 39), how Drpr mediates PS recognition in vivo is unclear. In this study, we present several lines of in vivo evidence that strongly indicate that the Drosophila chemokine–like Orion is a PS-binding bridging molecule that enables Drpr to respond to neuronal PS exposure. First, Orion is required for multiple scenarios of Drpr-dependent phagocytosis of sensory dendrites and functions upstream of Drpr. Second, Orion binds to PS-exposing cell surfaces. We show that Orion binds to neurons and epidermal cells that expose PS as a result of tissue-specific KO of the PS flippase ATP8A. In addition, ATP8A OE, which retains PS in the inner membrane leaflet, eliminates Orion binding on epidermal cells and late-larval dendrites, suggesting that this binding is PS-dependent. Importantly, Orion outcompetes Annexin V for binding to injured dendrites, suggesting that Orion may directly interact with PS in vivo. Third, overexpressed Drpr proteins can trap Orion on the cell surface, suggesting that Drpr interacts with Orion in vivo. Lastly, when expressed in neurons, membrane-tethered Orion bypasses the requirement for PS in inducing Drpr-dependent engulfment, but when expressed in phagocytes, membrane-tethered Orion blocks PS-induced engulfment. Based on these observations, we propose that Orion functions as a bridging molecule between PS and Drpr.
Previously, SIMU, a PS-binding transmembrane protein expressed by Drosophila embryonic phagocytes, was proposed to be a bridging molecule (49, 50). Orion and SIMU contribute to phagocytosis through distinct mechanisms. First, SIMU is expressed by phagocytes to allow them to tether apoptotic cells (49), while Orion is secreted from many peripheral tissues and functions as an opsonin to enable phagocytosis. Second, SIMU is a membrane protein that shares homology with Drpr but functions at a different step in apoptotic neuron clearance compared to Drpr (49). In contrast, as a secreted protein, Orion functions at the same step of phagocytosis as Drpr. Therefore, SIMU behaves more like a tethering receptor (50), while Orion is functionally analogous to PS-bridging molecules in other species (42, 51). Although we focus our analyses on the engulfment of somatosensory dendrites, the ubiquitous roles of PS and Drpr in phagocytosis and the broad expression patterns of Orion suggest that Orion may be widely involved in PS-mediated phagocytosis in Drosophila. This view is supported by our findings that Orion deposited in the hemolymph can mediate phagocytosis in distant tissues and that the accumulation of Orion on epidermal cells turns these cells into targets of phagocytosis.
The Level of Orion Modulates Phagocyte Sensitivity to PS.
Although the role of PS exposure in inducing phagocytosis has been well documented (7, 52), what determines the sensitivity of phagocytes to PS is much less understood. In this study, we discovered that the available level of Orion is a determinant of phagocyte sensitivity to PS in Drosophila. We show that reducing the dosage of functional Orion by one half makes phagocytes blind to dendrites with moderate levels of PS exposure (i.e., CDC50 KO + TMEME16F OE neurons), but the reduced Orion does not affect the ability of phagocytes to engulf dendrites that display high levels of PS exposure (i.e., injury). Conversely, an extra copy of the orion locus enhances the ability of phagocytes to engulf dendrites that have mild PS exposure (i.e., CDC50 KO). These results suggest that endogenous Orion is likely expressed at a finely balanced level to enable the right amount of phagocytosis: Too much Orion may cause unintended phagocytosis of stressed cells that display mild PS exposure, while too little Orion may interfere with efficient clearance of sick cells or structures that are beyond rescue. Consistent with this idea, endogenous Orion is expressed at a low level during larval development (SI Appendix, Fig. S2 G and I), but is dramatically up-regulated during metamorphosis (24), a time when large-scale tissue remodeling and clearance take place (53).
Orion Has Distinct Roles in Neurite Maintenance and Remodeling.
Orion is required for axonal pruning and clearance of MB γ Kenyon neurons during metamorphosis (24). In that context, Orion is expressed in the remodeling MB neurons and functions as a “find-me” signal for glia to penetrate the axon bundles and engulf axonal debris. In contrast, in the larval peripheral nervous system (PNS), Orion is supplied by many nonneural tissues and functions as a permissive signal for phagocytosis of sick or broken dendrites. This distinction is likely due to two differences between the larval PNS and the remodeling CNS. First, in the PNS, the dendrites of da neurons are exposed to the hemolymph and are readily accessible to Orion secreted from other tissues, whereas in the CNS, the axons are more tightly packed and may be harder for extrinsic Orion to access. Neuron-derived Orion would thus be more effective than extrinsic Orion for promoting phagocytosis in the CNS. Intriguingly, we detected Orion expression in a small number of neurons in the larval ventral nerve cord. It will be interesting to find out whether these neurons are particularly subject to degeneration. Second, compared to the larval PNS where degenerative events are rare, the nervous system undergoing metamorphosis has a much greater demand for phagocytosis. Turning on Orion expression in neurons may thus be required for efficient clearance of all pruned neurites. Consistent with this idea, we detected Orion expression also in a subset of da neurons during metamorphosis.
Orion Possesses Unique Properties Compared to Other PS-Binding Molecules.
Fluorescent PS probes based on AV and Lact are widely used to visualize PS exposure in cell culture and live animals and have been crucial for many discoveries in PS biology (6, 41, 42, 54, 55). LactC1C2 is known to have a higher affinity to PS than AV (56–58). We previously observed additional differences between these two proteins. Compared to LactC1C2, which coats the dendrite surface well, AV tends to traffic to endocytic vesicles inside PS-exposing dendrites, consistent with the ability of the AV complex to induce endocytosis upon PS binding (59, 60). Importantly, unlike AV, which does not alter the kinetics of neurite degeneration, LactC1C2 binding to PS-exposing dendrites potentiates Drpr-dependent degeneration (6). This latter observation previously led us to hypothesize that LactC1C2 may contain unknown sequences that interact with Drpr (6). In this study, we show that the effect of LactC1C2 in exacerbating dendrite degeneration depends on Orion, suggesting instead that LactC1C2 may indirectly promote phagocyte/dendrite interactions by enhancing Orion function. One possible mechanism is that LactC1C2 binding on the plasma membrane further disrupts the membrane and causes more PS exposure that can subsequently be detected by endogenous Orion.
Compared to AV and LactC1C2, Orion displays unique lipid-binding properties. Our in vivo evidence suggests that Orion may have a higher affinity for PS than AV, as Orion efficiently outcompetes AV in binding to injured dendrites. In addition, overexpressed Orion binds to healthy epidermal cells and dendrites (albeit the latter only in wandering 3rd instar larvae), while AV and LactC1C2 do not (6). Although our in vitro evidence suggests that Orion may have an intrinsic affinity for phospholipid bilayers, in vivo Orion binding to epidermal cells and WT dendrites largely depends on PS exposure. One possibility is that other endogenous factors modify Orion–lipid interactions and make Orion more specific to PS in vivo.
The surprising finding that overexpressed Orion binds to peripheral tissues and dendrites suggests that these cells may expose PS under physiological conditions, perhaps at a level too low to detect by AV and LactC1C2. Thus, low levels of PS exposure may be much more prevalent in vivo than previously thought. Intriguingly, binding of overexpressed Orion induces degeneration of wild-type dendrites but does not cause obvious phagocytosis of other nonneural tissues, suggesting that neurons may be more vulnerable to PS-induced phagocytosis than other cell types.
Functional Conservation between Orion and Human Immunomodulatory Proteins.
Recently, many human chemokines were found to bind to PS exposed on apoptotic vesicles and serve as “find-me” signals to attract phagocytes (61). Orion shares the CX3C motif with the mammalian chemokine CXC3L1 and has also three glycosaminoglycan putative binding sequences, a hallmark of chemokine activity (62). Even though direct binding to PS has not been demonstrated for CXC3L1, this chemokine is required for microglia-mediated synapse elimination after whisker lesioning (25), a process likely involving PS exposure (63). In addition, Orion contains a RRY motif commonly found in human neutrophil peptides, small antimicrobial peptides important for innate immunity (64). We found that both CX3C and RRY motifs are important for Orion function in mediating phagocytosis of neurons. Although Orion does not show global sequence homologies to mammalian immunomodulatory proteins, its interaction partner Drpr has mammalian homologs that are involved in the phagocytosis of neurons through unknown mechanisms (17). Thus, the common features between Orion and human proteins indicate that a functional conservation may exist between PS-sensing mechanisms in insects and humans.
Methods
Fly Strains.
The details of fly strains used in this study are listed in SI Appendix, Table S1 (Key Resource Table). For labeling of C4da neurons, we used ppk-MApHS, ppk-CD4-tdTom, and ppk-Gal4 UAS-CD4-tdTom. For labeling of all da neurons, we used 21-7-Gal4 UAS-CD4-tdTom. For labeling PS exposure on dendrites, we used dcg-Gal4 UAS-AnnexinV-mCard and dcg-Gal4 UAS-GFP-LactC1C2. For visualizing OrionB labeling on cell surface, we used dcg-Gal4 UAS-OrionB-GFP, Dcg-LexA LexAop-OrionB-GFP, and R16A03-LexA LexAop-OrionB-GFP. dcg-LexA was converted from dcg-Gal4 (65).
See SI Appendix, Methods for details of molecular cloning and transgenic flies, generation of KI flies, CRISPR-TRiM, live imaging, immunohistochemistry, protein purification, liposome sedimentation assay, image analysis and quantification, and statistical analysis. See SI Appendix, Table S2 for gRNA target sequences.
Supplementary Material
Appendix 01 (PDF)
OrionB-GFP labels degenerating dendrites after injury, related to Figure 3. Time-lapse movie of laser-injured ddaC dendrites from 2.5 to 11 hrs AI, showing OrionB-GFP labeling on injured dendrites. The dendrite arbor on the bottom right was injured while the ones at the top were not. The mobile cells in the OrionB-GFP channel are hemocytes. Time stamp is relative to the first frame.
AV-mCard labels injured dendrites before dendrite fragmentation, related to Figure 3. Time-lapse movie of laser-injured ddaC dendrites from 2.5 to 5 hrs AI, showing AV-mCard labeling on injured dendrites as early as 2 hrs before fragmentation. The AV-mCard signals that do not colocalize with tdTom signals indicate AV-mCard labeling on injured dendrites of other types of neurons. Time stamp is relative to the frame of dendrite fragmentation.
AV-mCard fails to label injured dendrites when OrionB-GFP is coexpressed, related to Figure 3. Time-lapse movie of laser-injured ddaC dendrites from 1 to 3 hrs AI showing OrionB-GFP labeling but not AV-mCard labeling on injured dendrites before fragmentation. Time stamp is relative to the frame of dendrite fragmentation.
Acknowledgments
We thank Larry Zipursky, Loren Looger, Xinhua Lin, Bacterial Artificial Chromosome and P1-derived Artificial Chromosome (BACPAC) Resources Center, Drosophila Genomics Resource Center, the National Cancer Institute, and Addgene for plasmids; Marc Freeman and Bloomington Stock Center for fly stocks; Cornell Biotechnology Resource Center (BRC) Imaging facility for access to microscopes (funded by NIH grant S10OD018516); Cornell Statistical Consulting Unit (CSCU) for advice on statistics; lab members of Chris Fromme, Jeremy Baskin, and Yuxin Mao for technical assistance and advice on biochemistry; Shaogeng Tang for experimental tests; and Mike Goldberg for critical reading and suggestions on the manuscript. This work was supported by NIH grants (R01NS099125 and R21OD023824) awarded to C.H. and by Appalachian Regional Commission (ARC) grant (PJA 20151203422) and Fondation pour la Recherche Médicale (FRM) grant (DEQ20160334870) awarded to J.-M.D.
Author contributions
H.J., B.W., and C.H. designed research; H.J., B.W., Y.S., D.L., J.L., X.C., and A.B. performed research; H.J., B.W., M.S., A.B., J.-M.D., and C.H. contributed new reagents/analytic tools; H.J., Y.S., D.L., J.L., and X.C. analyzed data; and H.J. and C.H. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information. Constructs and fly strains will be available from Addgene and Bloomington Drosophila Stock Center, respectively, or upon request.
Supporting Information
References
- 1.Galloway D. A., Phillips A. E. M., Owen D. R. J., Moore C. S., Phagocytosis in the brain: Homeostasis and disease. Front. Immunol. 10, 790 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frost J. L., Schafer D. P., Microglia: Architects of the developing nervous system. Trends Cell Biol. 26, 587–597 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salter M. W., Stevens B., Microglia emerge as central players in brain disease. Nat. Med. 23, 1018–1027 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Leventis P. A., Grinstein S., The distribution and function of phosphatidylserine in cellular membranes. Annu. Rev. Biophys. 39, 407–427 (2010). [DOI] [PubMed] [Google Scholar]
- 5.Shacham-Silverberg V., et al. , Phosphatidylserine is a marker for axonal debris engulfment but its exposure can be decoupled from degeneration. Cell Death Dis. 9, 1116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sapar M. L., et al. , Phosphatidylserine externalization results from and causes neurite degeneration in Drosophila. Cell Rep. 24, 2273–2286 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagata S., Apoptosis and clearance of apoptotic cells. Annu. Rev. Immunol. 36, 489–517 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Sievers C., Platt N., Perry V. H., Coleman M. P., Conforti L., Neurites undergoing Wallerian degeneration show an apoptotic-like process with Annexin V positive staining and loss of mitochondrial membrane potential. Neurosci. Res. 46, 161–169 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Darland-Ransom M., et al. , Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science 320, 528–531 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Zhu X., et al. , Mutations in a P-type ATPase gene cause axonal degeneration. PLoS Genet. 8, e1002853 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji H., Sapar M. L., Sarkar A., Wang B., Han C., Phagocytosis and self-destruction break down dendrites of Drosophila sensory neurons at distinct steps of Wallerian degeneration. Proc. Natl. Acad. Sci. U.S.A. 119, e2111818119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li T., et al. , A splicing isoform of GPR56 mediates microglial synaptic refinement via phosphatidylserine binding. EMBO J. 39, e104136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scott-Hewitt N., et al. , Local externalization of phosphatidylserine mediates developmental synaptic pruning by microglia. EMBO J. 39, e105380 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J., et al. , Microglial MERTK eliminates phosphatidylserine-displaying inhibitory post-synapses. EMBO J. 40, e107121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freeman M. R., Delrow J., Kim J., Johnson E., Doe C. Q., Unwrapping glial biology: Gcm target genes regulating glial development, diversification, and function. Neuron 38, 567–580 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Zhou Z., Hartwieg E., Horvitz H. R., CED-1 is a transmembrane receptor that mediates cell corpse engulfment in C. elegans. Cell 104, 43–56 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Wu H. H., et al. , Glial precursors clear sensory neuron corpses during development via Jedi-1, an engulfment receptor. Nat. Neurosci. 12, 1534–1541 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tung T. T., et al. , Phosphatidylserine recognition and induction of apoptotic cell clearance by Drosophila engulfment receptor Draper. J. Biochem. 153, 483–491 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Awasaki T., et al. , Essential role of the apoptotic cell engulfment genes draper and ced-6 in programmed axon pruning during Drosophila metamorphosis. Neuron 50, 855–867 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Williams D. W., Kondo S., Krzyzanowska A., Hiromi Y., Truman J. W., Local caspase activity directs engulfment of dendrites during pruning. Nat. Neurosci. 9, 1234–1236 (2006). [DOI] [PubMed] [Google Scholar]
- 21.MacDonald J. M., et al. , The Drosophila cell corpse engulfment receptor draper mediates glial clearance of severed axons. Neuron 50, 869–881 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Tao J., Rolls M. M., Dendrites have a rapid program of injury-induced degeneration that is molecularly distinct from developmental pruning. J. Neurosci. 31, 5398–5405 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuentes-Medel Y., et al. , Glia and muscle sculpt neuromuscular arbors by engulfing destabilized synaptic boutons and shed presynaptic debris. PLoS Biol. 7, e1000184 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boulanger A., et al. , Axonal chemokine-like Orion induces astrocyte infiltration and engulfment during mushroom body neuronal remodeling. Nat. Commun. 12, 1849 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunner G., et al. , Sensory lesioning induces microglial synapse elimination via ADAM10 and fractalkine signaling. Nat. Neurosci. 22, 1075–1088 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bazan J. F., et al. , A new class of membrane-bound chemokine with a CX3C motif. Nature 385, 640–644 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Maciejewski-Lenoir D., Chen S., Feng L., Maki R., Bacon K. B., Characterization of fractalkine in rat brain cells: Migratory and activation signals for CX3CR-1-expressing microglia. J. Immunol. 163, 1628–1635 (1999). [PubMed] [Google Scholar]
- 28.Han C., et al. , Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila. Neuron 81, 544–560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Botelho R. J., Grinstein S., Phagocytosis. Curr. Biol. 21, R533–538 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Segawa K., Nagata S., An apoptotic “Eat Me” signal: Phosphatidylserine exposure. Trends Cell Biol. 25, 639–650 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Nagata S., Suzuki J., Segawa K., Fujii T., Exposure of phosphatidylserine on the cell surface. Cell Death Differ. 23, 952–961 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suzuki J., Umeda M., Sims P. J., Nagata S., Calcium-dependent phospholipid scrambling by TMEM16F. Nature 468, 834–838 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Feng S., et al. , Improved split fluorescent proteins for endogenous protein labeling. Nat. Commun. 8, 370 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szymczak A. L., et al. , Correction of multi-gene deficiency in vivo using a single “self-cleaving” 2A peptide-based retroviral vector. Nat. Biotechnol. 22, 589–594 (2004). [DOI] [PubMed] [Google Scholar]
- 35.Chen Y., et al. , Cell-type-specific labeling of synapses in vivo through synaptic tagging with recombination. Neuron 81, 280–293 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams D. W., Truman J. W., Cellular mechanisms of dendrite pruning in Drosophila: Insights from in vivo time-lapse of remodeling dendritic arborizing sensory neurons. Development 132, 3631–3642 (2005). [DOI] [PubMed] [Google Scholar]
- 37.Poe A. R., et al. , Robust CRISPR/Cas9-mediated tissue-specific mutagenesis reveals gene redundancy and perdurance in Drosophila. Genetics 211, 459–472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Port F., Chen H. M., Lee T., Bullock S. L., Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 111, E2967–E2976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ji H., Sapar M. L., Sarkar A., Wang B., Han C., Phagocytosis and self-destruction break down dendrites of Drosophila sensory neurons at distinct steps of Wallerian degeneration. Proc. Natl. Acad. Sci. U.S.A. 119, e2111818119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ji H., Han C., LarvaSPA, A method for mounting Drosophila larva for long-term time-lapse imaging. J. Vis. Exp. 156, (2020), 10.3791/60792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mapes J., et al. , CED-1, CED-7, and TTR-52 regulate surface phosphatidylserine expression on apoptotic and phagocytic cells. Curr. Biol. 22, 1267–1275 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanayama R., et al. , Identification of a factor that links apoptotic cells to phagocytes. Nature 417, 182–187 (2002). [DOI] [PubMed] [Google Scholar]
- 43.Shami Shah A., et al. , PLEKHA4/kramer attenuates dishevelled ubiquitination to modulate Wnt and planar cell polarity signaling. Cell Rep. 27, 2157–2170.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dubois T., et al. , Annexin V inhibits protein kinase C activity via a mechanism of phospholipid sequestration. Biochem. J. 330, 1277–1282 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wehman A. M., Poggioli C., Schweinsberg P., Grant B. D., Nance J., The P4-ATPase TAT-5 inhibits the budding of extracellular vesicles in C. elegans embryos. Curr. Biol. 21, 1951–1959 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shaner N. C., et al. , A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10, 407–409 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu D., et al. , A naturally monomeric infrared fluorescent protein for protein labeling in vivo. Nat. Methods 12, 763–765 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan D., Wu Y., Feng Y., Lin S. C., Lin X., The core protein of glypican Dally-like determines its biphasic activity in wingless morphogen signaling. Dev. Cell 17, 470–481 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kurant E., Axelrod S., Leaman D., Gaul U., Six-microns-under acts upstream of Draper in the glial phagocytosis of apoptotic neurons. Cell 133, 498–509 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shklyar B., Levy-Adam F., Mishnaevski K., Kurant E., Caspase activity is required for engulfment of apoptotic cells. Mol. Cell Biol. 33, 3191–3201 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang X., et al. , Caenorhabditis elegans transthyretin-like protein TTR-52 mediates recognition of apoptotic cells by the CED-1 phagocyte receptor. Nat. Cell Biol. 12, 655–664 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lemke G., How macrophages deal with death. Nat. Rev. Immunol. 19, 539–549 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Williams D. W., Truman J. W., Remodeling dendrites during insect metamorphosis. J. Neurobiol. 64, 24–33 (2005). [DOI] [PubMed] [Google Scholar]
- 54.Andersen M. H., Graversen H., Fedosov S. N., Petersen T. E., Rasmussen J. T., Functional analyses of two cellular binding domains of bovine lactadherin. Biochemistry 39, 6200–6206 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Koopman G., et al. , Annexin V for flow cytometric detection of phosphatidylserine expression on B cells undergoing apoptosis. Blood 84, 1415–1420 (1994). [PubMed] [Google Scholar]
- 56.Shi J., Heegaard C. W., Rasmussen J. T., Gilbert G. E., Lactadherin binds selectively to membranes containing phosphatidyl-L-serine and increased curvature. Biochim. Biophys. Acta 1667, 82–90 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Tait J. F., Gibson D., Phospholipid binding of annexin V: Effects of calcium and membrane phosphatidylserine content. Arch. Biochem. Biophy.s 298, 187–191 (1992). [DOI] [PubMed] [Google Scholar]
- 58.Andree H. A., et al. , Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J. Biol. Chem. 265, 4923–4928 (1990). [PubMed] [Google Scholar]
- 59.Oling F., et al. , Structure of membrane-bound annexin A5 trimers: A hybrid cryo-EM - X-ray crystallography study. J. Mol. Biol. 304, 561–573 (2000). [DOI] [PubMed] [Google Scholar]
- 60.Kenis H., et al. , Cell surface-expressed phosphatidylserine and annexin A5 open a novel portal of cell entry. J. Biol. Chem. 279, 52623–52629 (2004). [DOI] [PubMed] [Google Scholar]
- 61.Pontejo S. M., Murphy P. M., Chemokines act as phosphatidylserine-bound “find-me” signals in apoptotic cell clearance. PLoS Biol. 19, e3001259 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Monneau Y., Arenzana-Seisdedos F., Lortat-Jacob H., The sweet spot: How GAGs help chemokines guide migrating cells. J. Leukoc. Biol. 99, 935–953 (2016). [DOI] [PubMed] [Google Scholar]
- 63.Faust T. E., Gunner G., Schafer D. P., Mechanisms governing activity-dependent synaptic pruning in the developing mammalian CNS. Nat. Rev. Neurosci. 22, 657–673 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klotman M. E., Chang T. L., Defensins in innate antiviral immunity. Nat. Rev. Immunol. 6, 447–456 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Karuparti S., Yeung A. T., Wang B., Guicardi P. F., Han C., A toolkit for converting Gal4 into LexA and Flippase transgenes in Drosophila. G3 (Bethesda). 13, jkad003 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
OrionB-GFP labels degenerating dendrites after injury, related to Figure 3. Time-lapse movie of laser-injured ddaC dendrites from 2.5 to 11 hrs AI, showing OrionB-GFP labeling on injured dendrites. The dendrite arbor on the bottom right was injured while the ones at the top were not. The mobile cells in the OrionB-GFP channel are hemocytes. Time stamp is relative to the first frame.
AV-mCard labels injured dendrites before dendrite fragmentation, related to Figure 3. Time-lapse movie of laser-injured ddaC dendrites from 2.5 to 5 hrs AI, showing AV-mCard labeling on injured dendrites as early as 2 hrs before fragmentation. The AV-mCard signals that do not colocalize with tdTom signals indicate AV-mCard labeling on injured dendrites of other types of neurons. Time stamp is relative to the frame of dendrite fragmentation.
AV-mCard fails to label injured dendrites when OrionB-GFP is coexpressed, related to Figure 3. Time-lapse movie of laser-injured ddaC dendrites from 1 to 3 hrs AI showing OrionB-GFP labeling but not AV-mCard labeling on injured dendrites before fragmentation. Time stamp is relative to the frame of dendrite fragmentation.
Data Availability Statement
All study data are included in the article and/or supporting information. Constructs and fly strains will be available from Addgene and Bloomington Drosophila Stock Center, respectively, or upon request.