Significance
The evolutionary history of the organisms that make up an ecosystem profoundly influences vegetation distributions and the trajectory of ecosystem function and biodiversity under climate change. Current methods to monitor and predict biodiversity can be improved by leveraging this critical factor. We formulated a unique analysis approach to remotely sense dominant phylogenetic lineages. Our approach allowed us to use imaging spectroscopy to accurately represent phylogenetic biogeographic patterns in a biodiversity hot spot. This indicates promise for incorporating similar biogeographic patterns into ecological forecasting with emerging hyperspectral data sources.
Keywords: evolutionary biology, biogeography, hyperspectral, plant ecology, lineage functional types
Abstract
Biogeographic history can set initial conditions for vegetation community assemblages that determine their climate responses at broad extents that land surface models attempt to forecast. Numerous studies have indicated that evolutionarily conserved biochemical, structural, and other functional attributes of plant species are captured in visible-to-short wavelength infrared, 400 to 2,500 nm, reflectance properties of vegetation. Here, we present a remotely sensed phylogenetic clustering and an evolutionary framework to accommodate spectra, distributions, and traits. Spectral properties evolutionarily conserved in plants provide the opportunity to spatially aggregate species into lineages (interpreted as “lineage functional types” or LFT) with improved classification accuracy. In this study, we use Airborne Visible/Infrared Imaging Spectrometer data from the 2013 Hyperspectral Infrared Imager campaign over the southern Sierra Nevada, California flight box, to investigate the potential for incorporating evolutionary thinking into landcover classification. We link the airborne hyperspectral data with vegetation plot data from 1372 surveys and a phylogeny representing 1,572 species. Despite temporal and spatial differences in our training data, we classified plant lineages with moderate reliability (Kappa = 0.76) and overall classification accuracy of 80.9%. We present an assessment of classification error and detail study limitations to facilitate future LFT development. This work demonstrates that lineage-based methods may be a promising way to leverage the new-generation high-resolution and high return-interval hyperspectral data planned for the forthcoming satellite missions with sparsely sampled existing ground-based ecological data.
The evolutionary history of the organisms that make up an ecosystem profoundly constrains the attributes and responses of that ecosystem. The trait patterns that result from evolutionary relatedness have a tangible impact on vegetation climate responses at global scales (e.g., ref. 1). Patterns of plant distributions that stem from evolutionary and biogeographic history influence disturbance regimes (e.g., ref. 2), determine critical biodiversity features (3, 4), and impact the trajectory of ecosystem responses to environmental change (5). Yet ecological forecasting at broad extents (e.g., land surface models (LSMs); ref. 6) is often coarse and disconnected from the evolutionary history and biogeographic contingencies that shape ecological communities (7–9).
Remote sensing has the potential to identify a vast array of vegetation properties (e.g., from biodiversity to biomass) from plots to landscapes to global extents (e.g., ref. 10). Numerous studies have indicated that VSWIR (visible-to-short wavelength infrared; 400-2,500 nm) reflectance properties of vegetation capture evolutionarily conserved biochemical, structural, and other functional attributes of plant species (e.g., refs. (11–14)). Yet, phylogenetic turnover and diversity have not been fully explored as alternative methods for assessing biodiversity from space. Lineage functional types (LFTs), or vegetation types informed by evolutionary information, have been proposed as an alternate paradigm to the use of plant functional types (PFT) in LSMs and thereby incorporate critical biogeographic history into ecological forecasts (15, 16). In this study, we explore the potential to map this aspect of plant functional diversity remotely.
The LFT concept is a natural extension of the increased inclusion of “tree-thinking” in biology that has produced significant advancements in community ecology, biogeography, and trait ecology over the last several decades (e.g., refs. 17–20). We envision LFTs as a balance between broadly defined PFTs and local ecology. On the one end, LFTs would be explicitly linked through phylogenetic relatedness (e.g., ref. 21), which provides implicit inclusion of evolutionary patterns that result from history and a better representation of ecological and biogeographic patterns than physiognomic classification. On the other end, leveraging phylogeny in creating vegetation types prevents models from being overly specified for large-scale prediction. The goal of including evolution in the creation/development of vegetation types is not to simply provide more cover types (and to some degree, PFTs already include LFTs, e.g., gymnosperms); instead, the aim is to provide more ecologically realistic and flexible groupings with the potential to link model parameters directly to remote sensing and/or field-observed traits at different phylogenetic scales (15, 16). LFTs might capture large-scale groupings that work for modeling, improve the ability to classify them with remotely sensed data, and reveal interesting biogeographic patterns that could otherwise be missed. In this sense, we define LFTs as related species that, with a given scale of analysis, can be grouped for the purpose of improved detection or modeling. Distantly related species or groups of species often co-occur locally, and the manner in which these assemblages can be represented by phylogenetic clusters will likely vary by application. For instance, a process model might allow for fractional representations, or remote sensing approaches might only detect one or more dominant groups. It remains an open question how generally applicable LFT-based approaches can be, especially in hyperdiverse tropical ecosystems where aggregating by lineage might either be powerful or impossible. Overall, evolutionary relatedness provides a potentially fruitful framework for integrating modeling efforts with the expanding availability of phylogenies, species distributions, traits, and remotely sensed data.
Evolutionary lineage–based functional types present a means to assimilate data from a wide range of datasets (e.g., traits, remote sensing data) and ask questions incorporating, for example, the evolutionary drivers that lead to dominance or high endemism (22). There is also a critical need to better understand and predict the distribution of functional types and their functional attributes and to better represent the physiological dynamics of vegetation. These are critical questions that limit our ability to use LSMs for ecological forecasts. The 2018 Decadal Survey outlines a set of important scientific questions related to the “structure, function, and biodiversity of Earth’s ecosystems,” many of which can be addressed by the development of LFTs (23). The Decadal Survey also calls for the development of a wide range of remote sensing systems, together now called the Earth System Observatory and including the Surface Biology and Geology (SBG)–designated observable comprising planned satellites with full VSWIR coverage with 16-d return intervals at 30-meter resolution and a thermal sensor with 60-meter resolution. Furthermore, numerous hyperspectral imagers are planned or already deployed. As such, LFTs mapped via VSWIR remote sensing will have the potential to leverage temporal dynamics in the function, distribution, and diversity of plant lineages. These data would form the basis for spatially explicit and high-resolution ecological forecasting in the context of the whole Earth System (10).
Biodiversity hot spots cover a small proportion of the land surface, but they contain irreplaceable biodiversity and represent a global priority for conservation efforts (24, 25). The California Floristic Province (CFP) is one of the world’s biodiversity hot spots, characterized by threats to a large number of endemic species (24, 25), and is an ideal model system for these questions because of its high diversity of ecosystem types and rich data availability. The CFP is a focal point for many influential lines of inquiry in biodiversity research (e.g., refs. 7, 9, and 26) and the evolutionary origins of CFP biodiversity (8, 22, 27) and community assembly dynamics (7, 9). The CFP hot spot is characterized largely by a Mediterranean climate and includes a range of biotic subregions that together support nearly 7,000 native species or subspecies of plants (28). Vegetation types within the CFP are a mosaic of ecosystems ranging from serpentine chaparral to coniferous forests and exhibit high spatial variation distributed across a topographically diverse landscape (26). This highlights the importance of Mediterranean ecosystems as islands of diversity and the importance of evolutionary age of lineages in understanding the biogeographic origins of diversity patterns (8). Here, we present an approach to creating LFTs for the southern Sierra Nevada mountains in the CFP to explore high-resolution patterns of biodiversity with the goal of mapping phylogenetic clusters.
Methods & Results
Rationale of the Approach.
We adopted an approach that uses existing ground reference data (e.g., vegetation surveys) to link with remote sensed data, which we consider potentially informative for future regional or global-scale remote sensing hyperspectral products. As such, we began with an exploratory analysis of remote sensing data, used data reduction approaches to annotate the spectral variation (e.g., ref. 4), and then moved toward a supervised classification approach informed by structure and phylogeny assessed from field data. We started the analysis with the examination of spectral variation in the imagery, focusing on what can feasibly be retrieved from space. Then, we linked this variation to abundance and evolutionary relatedness data. We selected lineages to represent functional types that can be mapped with remote sensing and that balance evolutionary distinctiveness with abundance.
Study Area.
We focused our study in the southern Sierra Nevada flight box flown in 2013 by Airborne Visible/Infrared Imaging Spectrometer (AVIRIS) as part of the hyperspectral preparatory data campaign from the Hyperspectral Infrared Imager program (HyspIRI; ref. 29). The flight lines and vegetation surveys (described below) are shown in SI Appendix, Fig. S1. These data have an 18-m resolution and span 360 to 2,500 nm wavelengths with 224 spectral bands.
Image Processing.
All 11 flight lines from June 12, 2013, were georeferenced in ref. 29 to a coregistration accuracy within a half-pixel. Ref. 29 compensated for brightness differences from sun-angle geometry and bidirectional reflectance distribution function (BRDF) across flight lines by applying a continuum removal, a process which normalizes spectra to a convex hull (30). Other approaches exist ranging from simple spectral normalization to more complex flight-specific approaches (e.g., ref. 31). However, we adopted an approach similar to that described in ref. 32 where albedo information is maintained (i.e., not normalized) and where transferability to future studies would be greater. The impact of these effects, which lead to the banding in SI Appendix, Figs. S1 and S2, is considered further in the Discussion. Water vapor absorption features from 1,810 to 1,950 nm and from 1,350 to 1,450 nm were removed. We used the early-June 2013 flights because mid-year images would have less snow and because other AVIRIS collections are more consistently collected in June. June 2013 represents a time period of relatively low disturbance to the study area compared with later AVIRIS flights (i.e., prior to the August 2013 Rim Fire) and avoids larger fires and the bulk of tree mortality that occurred later in the decade (33, 34). Furthermore, although observations from the vegetation surveys used in this study exist from 2,000 to present, over 98% of plots have observations prior to June 2013.
Plot Data.
We used existing vegetation survey data from three distinct sources that overlapped with the flight box: the Forest Inventory and Analysis (FIA) Program of the U.S. Forest Service (https://apps.fs.usda.gov/fia/datamart/datamart.html) (36) (n = 544), the Vegetation Classification and Mapping Program (VegCAMP) of the California Department of Fish and Wildlife (https://wildlife.ca.gov/Data/VegCAMP) (37) (n = 180), and VegBank (38) (n = 542). FIA data provide exhaustive enumeration of trees with diameter at breast height larger than 12.7 cm within a cluster plot comprising four fix-radius (7.32 m) subplots (https://www.fia.fs.usda.gov/program-features/basic-forest-inventory/). FIA data also include detailed descriptors of near-ground vegetation and coarse woody material and a plethora of other parameters. We obtained true coordinate information for FIA plot location (instead of fuzzed and swapped data). Surveys from VegCAMP represent manually delineated homogenous polygon areas of 100 m2 to 1,000 m2 depending on the vegetation type (from herbaceous to wooded) taken in representative stands including visual estimates of species cover. The VegBank data come from a range of independent sources and also ranged from 100 to 1,000 m2 in size, and we used surveys reporting cover abundances (i.e., the VegBank “cover” field). Plots were observed from 2000 to 2019, but where data exist before and after 2013, only data prior to 2013 were used to avoid impacts from disturbance. VegCAMP canopy cover and VegBank variable cover metrics were then compiled for the study area. The distribution of species-relative abundances followed expected patterns of dominance (39, 40), with a few species dominating 95% of the relative abundance estimates (SI Appendix, Fig. S3).
Plots were not collected at the same time as the hyperspectral flyovers and so we used the National Land Cover Database (NLCD) (41) to filter out all plots that had any history of vegetation cover change noted from 2000 to 2019 (SI Appendix, Fig. S4). This would include fires that led to a vegetation change (e.g., a shift from forested to grass) but may not detect mortality that shifts species abundances within an NLCD cover class.
Phylogeny.
We used the dated phylogeny developed and tested by (22). We chose to use this phylogenetic tree because it was developed specifically for this type of biogeographic study and the original authors found that their biogeographic analyses of CFP flora were robust given this phylogeny. In this phylogeny, the backbone relationships among plant lineages are well supported, dated, and provide excellent coverage of our study species. The phylogeny represents species and groups of species as operational taxonomic units (OTUs) conducive to generating a robust tree. In essence, OTUs are mathematical definitions of taxonomic units defined by the similarity of molecular sequences. We scrubbed (cleaned and matched to accepted binomials) the species names in the vegetation survey data using http://www.theplantlist.org as implemented in the Taxonstand R package (42). This process aided the connection of the scrubbed species table to OTUs in ref. 22, and we found that 71% of species were linked to the phylogeny automatically. We manually classified the remaining species into the OTUs, and only 0.13% of surveyed species could not be confidently assigned to an OTU (Dataset S1). The OTU tree from ref. 22 is fully bifurcated, and because manual OTU assignment was only conducted when inclusion was unambiguous (e.g., red versus white Oak clades), this process did not create polytomies.
Initial Classification of Remote Sensing Data.
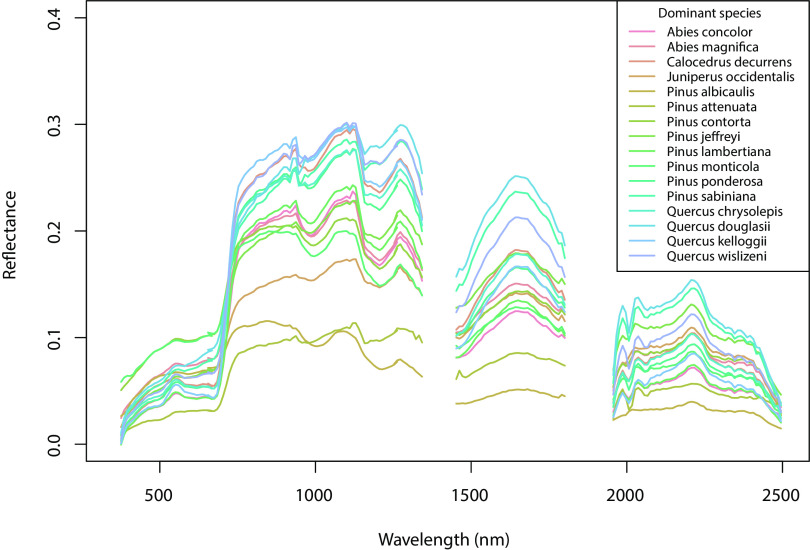
First, we identified the dominant spectral classes in the study area using unsupervised classification. We extracted the reflectance spectra from the AVIRIS data (examples for dominant woody species in FIA data shown in Fig. 1) for each vegetation survey location based on plot coordinate data; location data had a mean error of 5.4 m for VegCamp, our VegBank subset did not report location accuracy, and for the FIA data, we used the actual plot coordinates (i.e., not fuzzed or swapped). The majority of FIA plots in the study area were georeferenced using HighPrecision Global Navigation Satellite System devices and postprocessing with resulting precision between 1 and 2 m (43). We performed a feature selection by canonical discriminant analysis (CDA) in the R package “candisc” (44); CDA reduces the dimensions of the spectral data, producing orthogonal axes that most distinguish groups (e.g., dominant species in each plot). We kept the CDA axes that collectively accounted for two-thirds of the spectral variation, a total of 35 (SI Appendix, Fig. S5). This was similar to the approach successfully taken in ref. 33 in a study of vegetation classification of AVIRIS data for Santa Barbara, California area.
Fig. 1.
Mean spectral traces for each of the woody species that are dominant in the FIA plot data in the southern Sierra Nevada mountains, California, USA. Species show a wide range of variation across the spectra and in overall albedo.
We applied the K-means algorithm to cluster the CDA-transformed spectra (SI Appendix, Fig. S2) into groups that minimize spectral variation within each of k groups. The elbow method (a common approach to selecting the number of clusters based on diminishing return in variance explanation when adding clusters) was used to select 15 spectral clusters, nine of which were logical clusters of vegetation and the remaining were rare groupings or urban areas. This analysis was performed using the RcppArmadillo package with KMeans_rcpp() function (45).
To annotate the clusters, we identified the most dominant OTUs in each cluster (Fig. 2) that collectively accounted for 95% of the cover in that cluster and performed an indicator species analysis (ISA) in the R package indicspecies (46, 47). ISA is a community ecology analysis that finds species that are associated with communities and our purpose in using it was to create a list of species that represented each spectral cluster. Statistics for balancing abundance, species features, phylogeny, and spectral distinctiveness do not exist. ISA based on (47) worked better than other preexisting methods we tried as it allowed indicator taxa to exist across spectral clusters to better represent their distributions. As such, this approach is appropriate for phylogenetic analysis because in this study, we focus on dominant LFTs that emerge at the plot scale. The ISA also allowed us to visualize the turnover in important species across the gradient from West to East across the Sierra Nevada flight box (Fig. 2) and across the phylogeny.
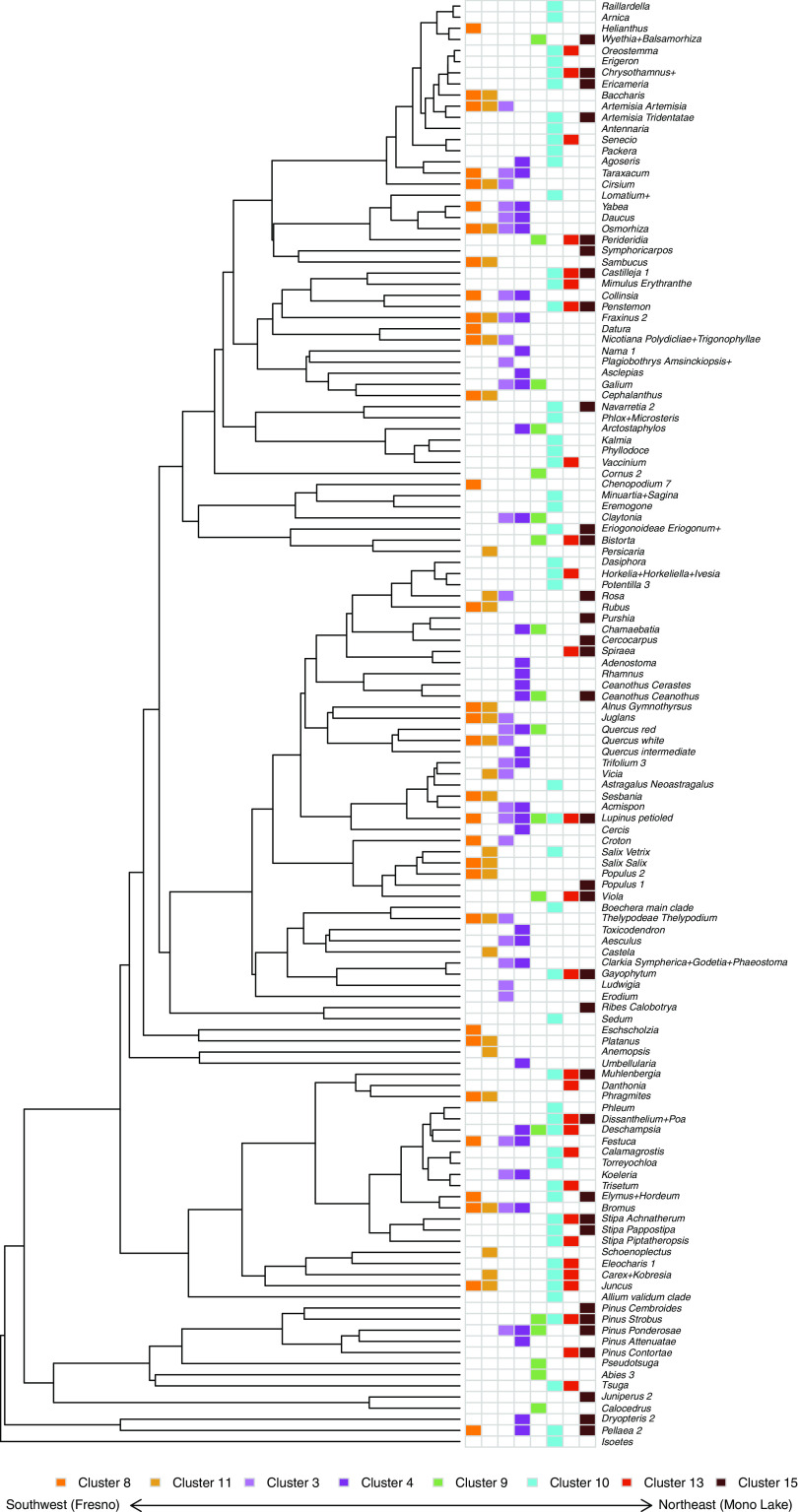
Fig. 2.
Indicator species analysis for the spectral clusters in SI Appendix, Fig. S2, sorted from West to East and by phylogenetic relatedness. This phylogeny represents the 129 OTUs that comprise over 95% of the relative abundance in the community survey data. Community similarity between the neighboring clusters is apparent, especially in Spectral Cluster 8 and 11, 3, and 4 as well as similarity in 13 and 15. From West to East, expected patterns of turnover are apparent (especially for canopy species) as the occurrence of broadleaf species peaks in Spectral Cluster 4 and then shifts toward Gymnosperms in 10, 13, and 15.
LFT Generation.
Next, our goal was to produce remotely detectable vegetation types. We developed a simple method that balanced the relevance of each species (i.e., their abundance and association with spectral clusters) with the tree topology. We compared this method to several other approaches described in SI Appendix, Supplementary Methods S1. In short, clustering the phylogeny based on evolutionary distinctiveness (lineage age alone) or functional distinctiveness does not appropriately prioritize the need for functional types to capture the diversity among the most dominant species on the landscape and resulted in suboptimal LFT classifications. Our LFT generation method is outlined as follows:
Step 1 – Ordinate community data.
We started with the presence–absence matrix from the ISA, meaning that the analysis focused on 129 indicator OTUs (across eight spectral clusters) that accounted for over 95% of relative abundance. We chose ISA as our approach because we wanted to organize our analysis around suites of species that would be associated with diagnostic spectral features but also allow these species to exist across spectral classes. We used nonmetric multidimensional scaling (SI Appendix, Fig. S6) to summarize the community variation into two axes. The ordination algorithm found a stable solution with a very low stress value (0.05), which indicates that species were well sorted along the reduced axes.
Step 2 – Identify communities.
To identify community clusters, we conducted a second K-means clustering to group the community variation into five communities (again, using the elbow method). The purpose of this step is to discretize the species into communities that could map to the phylogenetic tree.
Step 3 – Intersect communities and phylogeny.
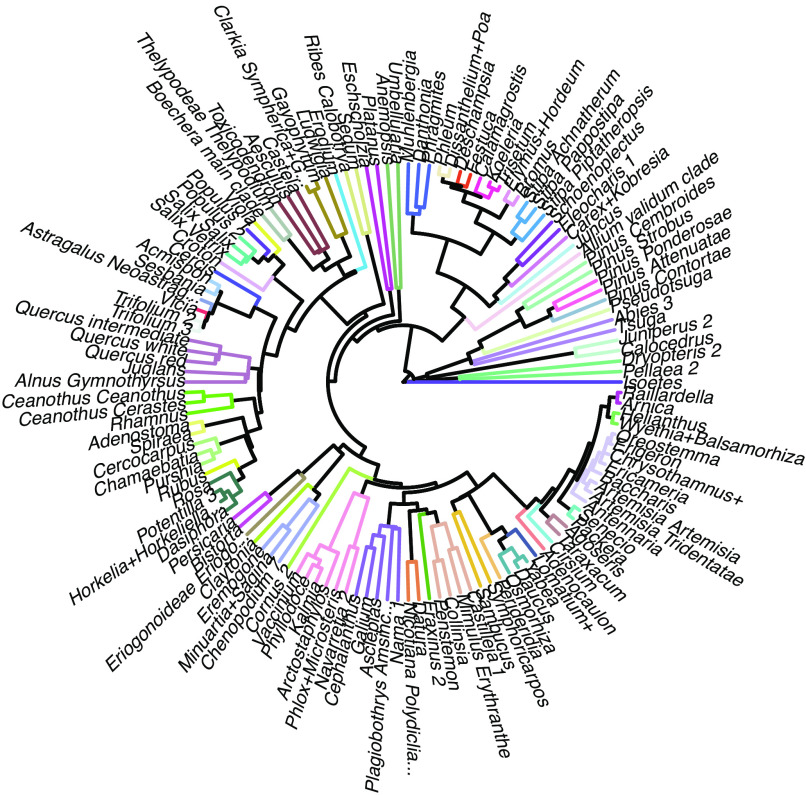
With each OTU across the phylogeny mapped to 1 of 5 community clusters, we grouped lineages where the majority of the descendants of a common ancestor shared the same community cluster association. We labeled these groups of related species from the same ecological associations as LFTs (e.g., LFT #1, #2, and #3). Some lineages included an internal branch of one OTU associated with a different community cluster, and we assigned these instead to the ancestral LFT to avoid the generation of three groups where one was more parsimonious. This process allowed the LFTs to emerge from the fusion of the phylogeny with the combinations of species that were observed to associate with specific communities (Fig. 3). This intersection resulted in 60 LFTs from across the full phylogeny.
Fig. 3.
Lineage functional types (LFTs) classified as the intersection of community indicator species, spectral clustering, and phylogeny (Methods). Different colors indicate 60 different LFTs that were produced by our method, creating fewer, spectrally similar clusters from the original 1572 species. In Fig. 4, we classify the six dominant woody canopy LFTs that most influence the spectral signals for the FIA survey data.
LFT Classification.
Finally, we created a supervised classification model for LFTs, trained on CDA-transformed reflectance data at vegetation plots using a support vector machine (SVM). Because of the added complication of creating a supervised classifier from multistrata community data, we focused this part of our work on the woody species based on FIA basal area data, which should dominate spectral signatures of airborne imagery and which were collected in a consistent way. The woody species of the FIA data comprises just six LFTs from the 60 that were produced in the LFT generation steps. Models were developed using a training subset and validated on a stratified random 10% subset of the data.
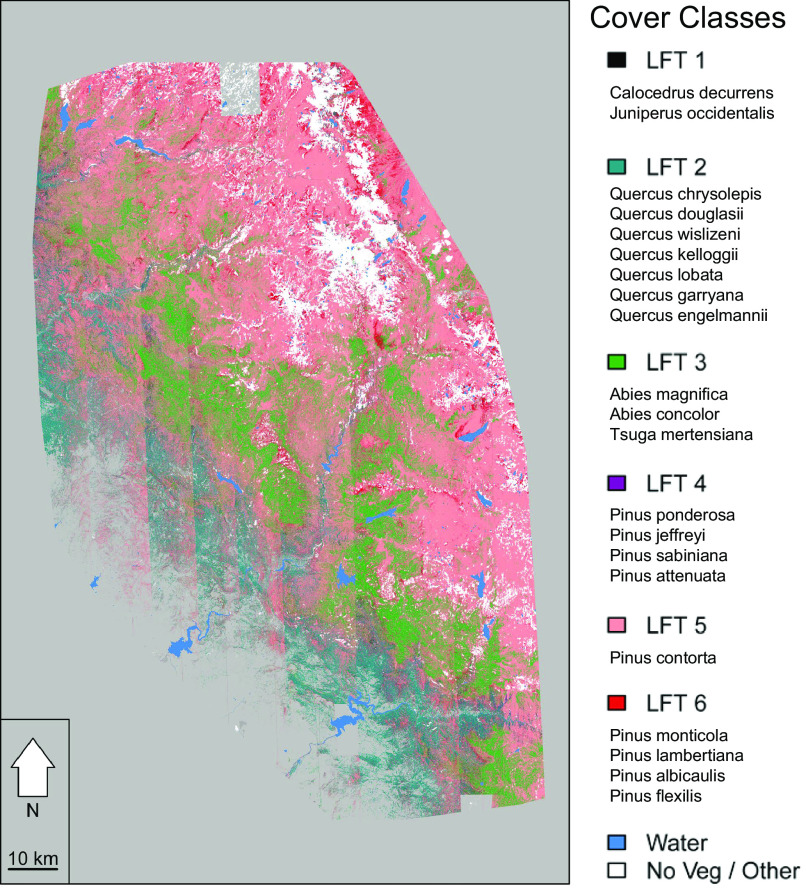
We felt most confident in the sample size per LFT (n > 25) and comparability of the FIA data for woody vegetation, and so when projecting the model outputs across the landscape, we restricted these classifications to the spatial boundary of the FIA data (specifically, the convex hull of the coordinates) (Fig. 4). Our simple method identified six LFTs to represent the woody canopy cover (from the 60 LFTs that constitute the entire species pool including herbaceous species) in the southern Sierra Nevada flight box. These LFTs were derived from the original 1,572 species in the dataset. We found that our SVM provided moderate calibration (0.76) and validation (0.72) Kappa values for these LFTs. Mathews correlation coefficient was also 0.76, suggesting that Kappa was a reliable metric to assess our classifications in this instance (48). The overall accuracy was 80.9%. Accuracy statistics and a confusion matrix can be found in Table 1. We performed a spatial classification error assessment that showed that commission errors were not clustered based on a join-count test (P = 0.43) and do not visually associate with BRDF banding (SI Appendix, Fig. S9). We also calculated phylogenetic dispersion statistics for spectral clusters (SI Appendix, Fig. S10).
Fig. 4.
Supervised classification of six woody LFTs built from FIA data using a support vector machine classifier. Classification statistics are available in Table 1. Species associated with each LFT are listed in the legend under each cover class, and they are ordered by abundance highest to lowest from top to bottom. While the classifications performed well, flight line artifacts related to solar illumination and hyperspectral sensor geometry are apparent and discussed in text.
Table 1.
Classification statistics for the final SVM model
| LFT 1 | LFT 2 | LFT 3 | LFT 4 | LFT 5 | LFT 6 | ||
|---|---|---|---|---|---|---|---|
| User’s accuracy | 43.5 | 97.9 | 88.6 | 59.2 | 96.8 | 44.4 | |
| Producer’s accuracy | 90.9 | 87.9 | 77.8 | 76.3 | 73.2 | 94.1 | |
| Confusion matrix | LFT 1 | LFT 2 | LFT 3 | LFT 4 | LFT 5 | LFT 6 | Total |
| LFT 1 | 10 | 1 | 0 | 0 | 0 | 0 | 11 |
| LFT 2 | 1 | 94 | 0 | 12 | 0 | 0 | 107 |
| LFT 3 | 5 | 0 | 70 | 7 | 2 | 6 | 90 |
| LFT 4 | 5 | 1 | 2 | 29 | 0 | 1 | 38 |
| LFT 5 | 2 | 0 | 6 | 1 | 60 | 13 | 82 |
| LFT 6 | 0 | 0 | 1 | 0 | 0 | 16 | 17 |
| Total | 23 | 96 | 79 | 49 | 62 | 36 | 345 |
Discussion
We found that evolutionary relatedness increased the ability to classify a hyperspectral image with diverse training data, resulting in a logical number of vegetation types that could be used in ecological modeling, and that the classification scheme was rooted in a framework that captured clusters of related species that have resulted from biogeographic, evolutionary, and ecological processes (Fig. 2). Vegetation plots with related species had similar spectral signatures which enabled enhanced classification of the land surface. This finding aligns with research indicating broad-scale biome conservatism (49–51), patterns of phylogenetic trait and habit conservatism (52, 53), and spectral similarity of related species (11, 13). A major next step for LFTs is to explicitly include trait data and further ecological context. As such, LFTs may increase the likelihood that ecological forecasts and landscape classifications capture, for example, conserved attributes of trees that determine their drought responses (54), although there are limitations to a completely automated approach and the inclusion of expert knowledge or improved phylofunctional clustering methods may be desired.
We identified patterns of phylogenetic clustering within unsupervised classifications of the spectral data. We mapped the locations associated with these lineages and processes at a high spatial resolution using hyperspectral data. Our LFT generation process produced vegetation clusters that qualitatively agree with visual expectations for the ecological distributions of plants in the Sierra Nevada (Figs. 2 and 3). In general, broadleaf vegetation types in the Southwest transition, with elevation, toward the Northeast into areas dominated by needleleaf lineages. The oaks were a particularly interesting LFT, as the major evolutionary oak groups become one LFT. This makes sense in the context of the community analysis, but potentially misses key attributes within the oaks that might not be captured (54, 55) or discussed in ref. 22. This possible simplification highlights the need to include remote sensing of traits directly (e.g., leaf mass per area and nitrogen) (56) as well as traits from online vegetation databases to generate parameter values to potentially also pull out those unique branches that might be important (57). The impact of analysis extent (spatial or phylogenetic), as a more focused study (or one allowing for more community clusters), might divide lineages more finely. Similarly, within the grasses included in this analysis, the primarily C4 lineage comes out as a separate LFT from the solely C3 grass clade which is more common in the region (58). Then, process-based models could be run in a spatially explicit way to model ecosystem function and distributional change (59). Inclusion of trait data would also enable testing of hypotheses about why the woody vegetation LFTs mapped in Fig. 4 are organized the way they are. Species groups may have similar leaf types, canopy structure, and albedo, that results in similar spectra.
Past research on remote sensing of evolutionary history with spectral information relied primarily on field or leaf spectroscopy (11–14, 60). Field-collected spectra represent nearly optimal data, are collected to represent pure spectral signatures of a target, and do not have positional error, or atmospheric interference. Depending on how they are collected, they can minimize canopy effects such as shading, architecture, and multiple scattering that might make this work difficult in many regions (61). This study represents real-world application of airborne remote sensing data combined with diverse vegetation plot data collected at different dates, scales, with different methods and locational accuracy to map LFTs across a region. In addition to these nonuniformities in the plot data, the 18-m imagery contains perennial remote sensing challenges such as mixed pixels and BRDF correction issues. Despite these challenges, our approach showed that a majority of spectral clusters from moderate-resolution airborne imagery represented species groups that were more phylogenetically related than expected, and we succeeded in mapping LFTs with an overall accuracy of 80.9%. We note that these classifications represent dominant LFTs and that it is likely that distantly related species or secondary LFTs co-occur within mixed pixels. Roth et al. (62), who looked at spectral classification of plant species across a range of ecosystems in North America, found that in the Sierra Nevada, it was particularly difficult to classify conifer species because they are so heavily mixed at relatively fine scales (for example, Pinus lambertiana exists typically as individual crowns). Aggregating at a higher taxonomic level that is linked by relatedness was a more effective classification strategy in this complex mixture because clusters of LFTs are probably more likely to occur than clusters of individual species. These results indicate promise for scaling these analyses to larger areas with emerging hyperspectral satellite imagery.
These results also point to the future possibility of scalability and creating integrated datasets that allow the generation of LFTs at different spatial scales. Datasets exist to generate phylogenies (e.g., Open Tree of Life: https://opentreeoflife.github.io/), request functional traits (e.g., TRY: www.try-db.org), and acquire distribution and abundance data (e.g., https://mol.org/ and BIEN which also includes a draft phylogeny and traits). Integration of these data linked through LFTs to remotely sensed hyperspectral data (such as SBG: https://sbg.jpl.nasa.gov/, EMIT—Earth Surface Mineral Dust Source Investigation on ISS: https://earth.jpl.nasa.gov/emit/, HISUI—Hyperspectral Imager Suite on ISS: www.meti.go.jp, DESIS—DLR Earth Sensing Imaging Spectrometer on ISS: www.dlr.de, or CHIME and EnMAP—Environmental Mapping and Analysis Program: https://www.esa.int/ and www.enmap.org) could generate model parameters and biogeographic knowledge dynamically across a range of scales (spatial and phylogenetic). We suggest that more advanced statistics could be employed to cluster phylogenies in conjunction with functional distinctiveness, spectral distinctiveness, and abundances to produce better or tunable groupings.
This work has several important limitations and only represents a starting point based on best-available data. As discussed, we decided not to normalize reflectance data and instead look toward methodological improvements such as planned BRDF corrections that will produce uniform spectra for these flight boxes. These BRDF corrections are an integral part of the data-processing pipeline being prepared for future satellites which will have inherently reduced BRDF sun-angle effects compared to airborne data due to altitude and collection speed. BRDF correction would provide moderate improvements to calibration accuracy. In support of this, our spatial error assessment did not suggest that errors were clustered or associated with BRDF striping (SI Appendix, Fig. S9), and more likely were associated with issues of scale disparity between plots and pixels, variation in stand structure, or species richness and phylogenetic overdispersion (SI Appendix, Fig. S10).
While the LFT approach did help somewhat with harmonizing the disparate vegetation datasets, our work highlights the common classification problem that remotely sensed data and ground-based training data often differ considerably in spatial extent, temporal coverage, and information content. For example, classifying the diversity of the herbaceous layer in mixed pixels with airborne data remains a major challenge. Some pixels are represented by both herbaceous understory and woody vegetation or have extremely high richness, and it is possible that using an approach such as the Multiple-Endmember Spectral Mixture Analysis or convolutional autoencoder for subpixel classification or hierarchical random forests (e.g., HieRanFor) where cover classes could be nested to reflect evolutionary relatedness might produce improved results. Subpixel unmixing technique might be increasingly important for lineage-based approaches if applied to forthcoming satellite missions that will have lower resolution than AVIRIS data. Our study also benefited from the use of a fully bifurcated phylogeny based on groups of species (22) that was able to accommodate the full breadth of our vegetation plots at regional scales. However, for future studies requiring higher taxonomic resolution, it is unclear exactly how sensitive LFT generation will be to phylogenies with polytomies. Studies could also explore the use of a wider range of approaches for arriving at the optimal number of clusters to use for analysis (e.g., NbClust R package; ref. 63). Similarly, we were not able to quantitatively assess the impact that species richness and pixel size played in how LFTs become discretized. This is an opportunity for improvement, especially if approaches can be developed that better included pixels and plots with mixed LFT composition (and the vegetation data do not necessarily represent true absences in all cases). Future hyperspectral imagery with improved revisit times will also allow for improved multitemporal assessment of LFTs and the inclusion of LFT-specific phenology (33, 53). Furthermore, this highlights a potential opportunity for data fusion approaches where other instruments like LiDAR could be used to first estimate the woody canopy cover and partition woody LFTs to the canopy accordingly (although stature may not always be a conserved attribute). For example, the Global Ecosystem Dynamics Investigation aboard the International Space Station or the NEON AOP (National Ecological Observatory Network, Airborne Observing Platform) could be tested for this purpose. As a combined consequence of these limitations, we were ultimately only able to map six total LFTs whereas we might hope to distinguish more. This constraint primarily stems from constricting the analysis to woody vegetation for classification because our approach and data cannot capture understory vegetation or mixed vegetation very well. The approach also does not provide a means to easily test the sensitivity of the classification to changes at each step of the LFT generation process, or how these errors compound. Another obvious limitation is that this study does not explicitly bring in trait data, as the current focus was the development of the classification approach based on spectral distinctiveness, phylogeny, and abundance. Finally, another alternative might be to scale the classification accuracy assessment by the evolutionary distance to the actual cover class. Ultimately though, our results suggest that increased availability of hyperspectral remote sensing data might enable monitoring of short- and long-term LFT changes induced by disturbance or the changing climate.
In conclusion, we present remotely sensed phylogenetic clustering and an evolutionary framework to accommodate spectra, distributions, and traits of plants. Future iterations of this approach hold promise for elucidating unique biodiversity patterns [e.g., rapidly identifying at risk endemism (22), monitoring lineage turnover as a dimension of biodiversity (4, 16), or generating parameters for vegetation models used in climate modeling, thus incorporating patterns produced by the trajectory of biogeographic history into ecological forecasting.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (CSV)
Acknowledgments
D.M.G. acknowledges support from NASA under the auspices of the Surface Biology and Geology Study and from United States Geological Survey through the National Innovation Center. K.B.B. was funded by the United States Geological Survey Land Change Science Program and United States Geological Survey National Land Imaging Program. We appreciate the California Department of Fish and Wildlife’s vegetation program’s (https://wildlife.ca.gov/Data/VegCAMP) collection, aggregation, and publication of vegetation data. We also acknowledge the Forest Inventory and Analysis Program of the United States Department of Agriculture Forest Service for providing data and thank them for their collaboration and support. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. government.
Author contributions
D.M.G., K.B.B., L.D.L.A., and R.R.N. designed research; D.M.G. and E.A. performed research; D.M.G., K.B.B., D.G., D.R., and R.Y. contributed new reagents/analytic tools; D.M.G., K.B.B., and L.D.L.A. analyzed data; K.B.B. and L.D.L.A. editing; and D.M.G., D.G., D.R., and R.R.N. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
The FIA and VegCAMP vegetation data used in this study were obtained through interagency data-sharing agreements. FIA data can be obtained online (https://apps.fs.usda.gov/fia/datamart/datamart.html) (36), with exact coordinates fuzzed and swapped to protect landowner privacy per the Food Security Act. Details for VegCAMP are online (https://wildlife.ca.gov/Data/VegCAMP). VegBank data are openly available online (38). Code for completing these analyses is available in Zenodo repository (64) and has been stripped of components that would reveal protected information.
Supporting Information
References
- 1.Lehmann C. E. R., et al. , Savanna vegetation-fire-climate relationships differ among continents. Science 343, 548–552 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Archibald S., Lehmann C. E. R., Gomez-Dans J. L., Bradstock R. A., Defining pyromes and global syndromes of fire regimes. Proc. Natl. Acad. Sci. U.S.A. 110, 6442–6447 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jetz W., et al. , Monitoring plant functional diversity from space. Nat. Plants 2, 16024 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Jetz W., et al. , Essential biodiversity variables for mapping and monitoring species populations. Nat. Ecol. Evol. 3, 539–551 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Moncrieff G. R., Bond W. J., Higgins S. I., Revising the biome concept for understanding and predicting global change impacts. J. Biogeogr. 43, 863–873 (2016). [Google Scholar]
- 6.Still C. J., Cotton J. M., Griffith D. M., Assessing earth system model predictions of C4 grass cover in North America: From the glacial era to the end of this century. Global Ecol. Biogeogra. 28, 145–157 (2019). [Google Scholar]
- 7.Harrison S., Cornell H., Toward a better understanding of the regional causes of local community richness. Ecol. Lett. 11, 969–979 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Ackerly D. D., Evolution, origin and age of lineages in the californian and mediterranean floras. J. Biogeography 36, 1221–1233 (2009). [Google Scholar]
- 9.Anacker B. L., Harrison S. P., Historical and ecological controls on phylogenetic diversity in californian plant communities. Am. Naturalist 180, 257–269 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Cawse-Nicholson K., et al. , NASA’s surface biology and geology designated observable: A perspective on surface imaging algorithms. Remote Sensing Environ. 257, 112349 (2021). [Google Scholar]
- 11.Cavender-Bares J., et al. , Associations of leaf spectra with genetic and phylogenetic variation in oaks: prospects for remote detection of biodiversity. Remote Sensing 8, 221 (2016). [Google Scholar]
- 12.Schweiger A. K., et al. , Plant spectral diversity integrates functional and phylogenetic components of biodiversity and predicts ecosystem function. Nat. Ecol. Evol. 2, 976–982 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Meireles J. E., et al. , Leaf reflectance spectra capture the evolutionary history of seed plants. New Phytol. 228, 485–493 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffith D. M., et al. , Variation in leaf reflectance spectra across the california flora partitioned by evolutionary history, geographic origin, and deep time. JGR Biogeosci. 128, e2022JG007160 (2023). [Google Scholar]
- 15.Griffith D. M., et al. , Lineage-based functional types: Characterising functional diversity to enhance the representation of ecological behaviour in land surface models. New Phytol. 228, 15–23 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Anderegg L. D. L., et al. , Representing plant diversity in land models: An evolutionary approach to make “Functional Types” more functional. Global Change Biol. 28, 2541–2554 (2022). [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J., Phylogenies and the comparative method. Am. Naturalist 125, 1–15 (1985). [Google Scholar]
- 18.Webb C. O., Ackerly D. D., McPeek M. A., Donoghue M. J., Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505 (2002). [Google Scholar]
- 19.Diniz-Filho J. A. F., et al. , Mapping the evolutionary twilight zone: Molecular markers, populations and geography. J. Biogeogr. 35, 753–763 (2008). [Google Scholar]
- 20.Cavender-Bares J., Kozak K. H., Fine P. V. A., Kembel S. W., The merging of community ecology and phylogenetic biology. Ecol. Lett. 12, 693–715 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Slik J. W. F., et al. , Phylogenetic classification of the world’s tropical forests. Proc. Natl. Acad. Sci. U.S.A. 115, 1837–1842 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thornhill A. H., et al. , Spatial phylogenetics of the native california flora. BMC Biol. 15, 96 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.National Academies of Sciences, Engineering, and Medicine, Thriving on our changing planet: A decadal strategy for earth observation from space. 10.17226/24938. Accessed 13 April 2019. [DOI]
- 24.Myers N., Mittermeier R. A., Mittermeier C. G., da Fonseca G. A. B., Kent J., Biodiversity hotspots for conservation priorities. Nature 403, 853–858 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Mittermeier R., “Biodiversity hotspots” in Reference Module in Earth Systems and Environmental Sciences (Elsevier, 2018), 10.1016/B978-0-12-409548-9.09962-0. April 15, 2019. [DOI] [Google Scholar]
- 26.Keeley J. E., VTM plots as evidence of historical change: Goldmine or landMINE? Madroño 51, 8 (2004). [Google Scholar]
- 27.Baldwin B. G., Origins of plant diversity in the california floristic province. Annu. Rev. Ecol. Evol. Syst. 45, 347–369 (2014). [Google Scholar]
- 28.Burge D. O., et al. , Plant diversity and endemism in the california floristic province. Madroño 63, 3–206 (2016). [Google Scholar]
- 29.Lee C. M., et al. , An introduction to the NASA hyperspectral infrared imager (HyspIRI) mission and preparatory activities. Remote Sens. Environ. 167, 6–19 (2015). [Google Scholar]
- 30.Tane Z., Roberts D., Koltunov A., Sweeney S., Ramirez C., A framework for detecting conifer mortality across an ecoregion using high spatial resolution spaceborne imaging spectroscopy. Remote Sens. Environ. 209, 195–210 (2018). [Google Scholar]
- 31.Clark R. N., Roush T. L., Reflectance spectroscopy: Quantitative analysis techniques for remote sensing applications. J. Geophys. Res. 89, 6329–6340 (1984). [Google Scholar]
- 32.Wetherley E. B., McFadden J. P., Roberts D. A., Megacity-scale analysis of urban vegetation temperatures. Remote Sens. Environ. 213, 18–33 (2018). [Google Scholar]
- 33.Meerdink S. K., et al. , Classifying california plant species temporally using airborne hyperspectral imagery. Remote Sens. Environ. 232, 111308 (2019). [Google Scholar]
- 34.Asner G. P., et al. , Progressive forest canopy water loss during the 2012–2015 California drought. Proc. Natl. Acad. Sci. U.S.A. 113, E249–E255 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huesca M., Ustin S. L., Shapiro K. D., Boynton R., Thorne J. H., Detection of drought-induced blue oak mortality in the sierra nevada mountains, California. Ecosphere 12, e03558 (2021). [Google Scholar]
- 36.Forest Inventory and Analysis Database, U.S. Department of Agriculture, Forest Service, Northern Research Station. St. Paul, MN. https://apps.fs.usda.gov/fia/datamart/datamart.html. Accessed 8 February 2023.
- 37.Vegetation Classification and Mapping Program (VegCAMP), Fine-scale Vegetation Sampling, Classification, and Mapping of the Southern Sierra Nevada Foothills (Vegetation Classification and Mapping Program, California Department of Fish and Game, Sacramento, CA, 2023), https://wildlife.ca.gov/Data/VegCAMP. [Google Scholar]
- 38.Peet R., Lee M., Jennings M., Faber-Langendoen D., VegBank – a permanent, open-access archive for vegetation-plot data. Biodiversity Ecol. 4, 233–241 (2012). [Google Scholar]
- 39.Smith M. D., Knapp A. K., Dominant species maintain ecosystem function with non-random species loss. Ecol. Lett. 6, 509–517 (2003). [Google Scholar]
- 40.ter Steege H., et al. , Hyperdominance in the amazonian tree flora. Science 342, 1243092–1243092 (2013). [DOI] [PubMed] [Google Scholar]
- 41.Wickham J., Stehman S. V., Sorenson D. G., Gass L., Dewitz J. A., Thematic accuracy assessment of the NLCD 2016 land cover for the conterminous United States. Remote Sens. Environ. 257, 112357 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cayuela L., Granzow-de la Cerda Í., Albuquerque F. S., Golicher D. J., Taxonstand: An r package for species names standardisation in vegetation databases: Taxonstand. Methods Ecol. Evol. 3, 1078–1083 (2012). [Google Scholar]
- 43.Andersen H.-E., Strunk J., McGaughey R. J., Using high-performance global navigation satellite system technology to improve Forest Inventory and Analysis plot coordinates in the Pacific region (Gen. Tech. Rep. PNW-GTR-1000, Portland, OR, U.S. Department of Agriculture, Forest Service, Pacific Northwest Research Station, 2022), p. 38. [Google Scholar]
- 44.Friendly M., Fox J., Friendly M. M., Package ‘candisc’ (2021).
- 45.Eddelbuettel D., Sanderson C., RcppArmadillo: Accelerating R with high-performance C++ linear algebra. Comput. Statist. Data Analysis 71, 1054–1063 (2014). [Google Scholar]
- 46.Dufrêne M., Legendre P., Species assemblages and indicator species:the need for a flexible asymmetrical approach. Ecol. Monographs 67, 345–366 (1997). [Google Scholar]
- 47.De Caceres M., Jansen F., De Caceres M. M., Package ‘indicspecies’. Indicators 8, 1 (2016). [Google Scholar]
- 48.Delgado R., Tibau X.-A., Why Cohen’s Kappa should be avoided as performance measure in classification. PLoS One 14, e0222916 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crisp M. D., et al. , Phylogenetic biome conservatism on a global scale. Nature 458, 754–756 (2009). [DOI] [PubMed] [Google Scholar]
- 50.Gagnon E., Ringelberg J. J., Bruneau A., Lewis G. P., Hughes C. E., Global succulent biome phylogenetic conservatism across the pantropical caesalpinia group (Leguminosae). New Phytol. 222, 1667–1669 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Donoghue M. J., Edwards E. J., Biome shifts and niche evolution in plants. Annu. Rev. Ecol. Evol. Systemat. 45, 547–572 (2014). [Google Scholar]
- 52.Ackerly D., Conservatism and diversification of plant functional traits: Evolutionary rates versus phylogenetic signal. Proc. Natl. Acad. Sci. U.S.A. 106, 19699–19706 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Davies T. J., et al. , Phylogenetic conservatism in plant phenology. J. Ecol. 101, 1520–1530 (2013). [Google Scholar]
- 54.Skelton R. P., et al. , Evolutionary relationships between drought-related traits and climate shape large hydraulic safety margins in western North American oaks. Proc. Natl. Acad. Sci. U.S.A. 118, e2008987118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sapes G., et al. , Canopy spectral reflectance detects oak wilt at the landscape scale using phylogenetic discrimination. Remote Sensing Environ. 273, 112961 (2022). [Google Scholar]
- 56.Wang Z., et al. , Foliar functional traits from imaging spectroscopy across biomes in eastern North America. New Phytol. 228, 494–511 (2020). [DOI] [PubMed] [Google Scholar]
- 57.Cornwell W. K., et al. , Functional distinctiveness of major plant lineages. J. Ecol. 102, 345–356 (2014). [Google Scholar]
- 58.Edwards E. J., Still C. J., Climate, phylogeny and the ecological distribution of C4 grasses. Ecol. Lett. 11, 266–276 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Coffield S. R., Hemes K. S., Koven C. D., Goulden M. L., Randerson J. T., Climate-driven limits to future carbon storage in california’s wildland ecosystems. AGU Adv. 2, e2021AV000384 (2021). [Google Scholar]
- 60.Griffith D. M., Anderson T. M., The ‘plantspec’ r package: A tool for spectral analysis of plant stoichiometry. Methods Ecol. Evol. 10, 673–679 (2019). [Google Scholar]
- 61.Roberts D. A., et al. , Spectral and structural measures of northwest forest vegetation at leaf to landscape scales. Ecosystems 7, 545–562 (2004). [Google Scholar]
- 62.Roth K. L., et al. , Differentiating plant species within and across diverse ecosystems with imaging spectroscopy. Remote Sens. Environ. 167, 135–151 (2015). [Google Scholar]
- 63.Charrad M., Ghazzali N., Boiteau V., Niknafs A., NbClust: An R package for determining the relevant number of clusters in a data set. J. Stat. Soft. 61, 1–36 (2014). [Google Scholar]
- 64.Griffith D. M., griffithdan/Griffith-et-al-SNLFTs: Code for Griffith et al PNAS (LFTs). Zenodo. 10.5281/zenodo.7938124. Deposited 15 May 2023. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (CSV)
Data Availability Statement
The FIA and VegCAMP vegetation data used in this study were obtained through interagency data-sharing agreements. FIA data can be obtained online (https://apps.fs.usda.gov/fia/datamart/datamart.html) (36), with exact coordinates fuzzed and swapped to protect landowner privacy per the Food Security Act. Details for VegCAMP are online (https://wildlife.ca.gov/Data/VegCAMP). VegBank data are openly available online (38). Code for completing these analyses is available in Zenodo repository (64) and has been stripped of components that would reveal protected information.