Abstract
Objectives
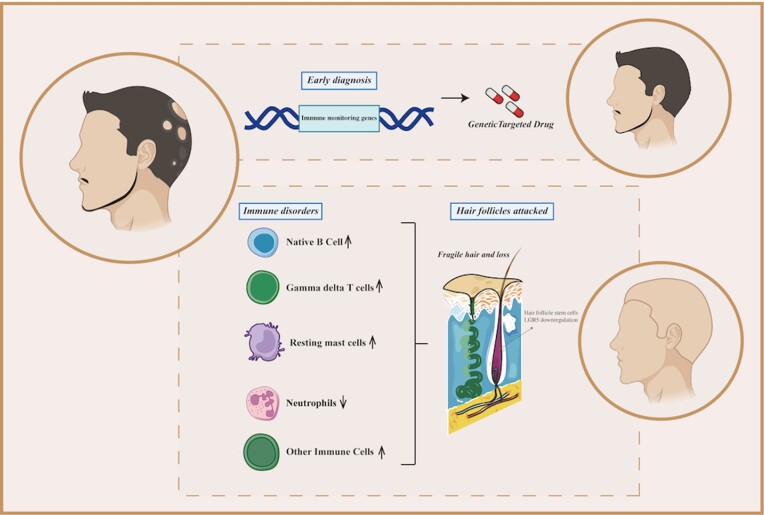
Alopecia areata (AA) is an autoimmune-related non-cicatricial alopecia, with complete alopecia (AT) or generalized alopecia (AU) as severe forms of AA. However, there are limitations in early identification of AA, and intervention of AA patients who may progress to severe AA will help to improve the incidence rate and prognosis of severe AA.
Methods
We obtained two AA-related datasets from the gene expression omnibus database, identified the differentially expressed genes (DEGs), and identified the module genes most related to severe AA through weighted gene co-expression network analysis. Functional enrichment analysis, construction of a protein–protein interaction network and competing endogenous RNA network, and immune cell infiltration analysis were performed to clarify the underlying biological mechanisms of severe AA. Subsequently, pivotal immune monitoring genes (IMGs) were screened through multiple machine-learning algorithms, and the diagnostic effectiveness of the pivotal IMGs was validated by receiver operating characteristic.
Results
A total of 150 severe AA-related DEGs were identified; the upregulated DEGs were mainly enriched in immune response, while the downregulated DEGs were mainly enriched in pathways related to hair cycle and skin development. Four IMGs (LGR5, SHISA2, HOXC13, and S100A3) with good diagnostic efficiency were obtained. As an important gene of hair follicle stem cells stemness, we verified in vivo that LGR5 downregulation may be an important link leading to severe AA.
Conclusion
Our findings provide a comprehensive understanding of the pathogenesis and underlying biological processes in patients with AA, and identification of four potential IMGs, which is helpful for the early diagnosis of severe AA.
Keywords: alopecia areata, immune response, machine learning, immune monitoring genes, diagnosis
Introduction
Alopecia areata (AA) is an autoimmune-related non-cicatricial alopecia that often manifests as rounded patches of alopecia (AAP), and will gradually develop into total alopecia (AT) that involves hair loss on the entire head, or universal alopecia (AU) that involves hair loss all over the body in severe cases.1 AT and AU belong to the progressive type of AA (severe AA), which are refractory hair diseases and seriously damage the morphology of patients. Although AA is not life-threatening, it has a huge impact on the patient's image and can be a significant influence on mental illnesses such as anxiety and depression.2 The etiology of AA is still unclear, but scholars believe that it is caused by genetics, pathogen infection, and autoimmunity.3–5 Previous studies have reported that immune cells infiltrating around the hair follicles of AA patients and secreting many cytokines such as interferon-γ, interleukin 17 (IL-17), and IL-2 leads to the collapse of the immune privilege, miniaturization of the hair follicle, and hair loss.6,7 Thus, immune disorders play an important role in the onset of AA.
For mild AAP, some patients can recover without any treatment, or can be cured by topical application of glucocorticoid. However, the current clinical treatment methods for severe AA are limited, and combined immunosuppressive therapy is often required, but its adverse reactions are relatively large, and the recurrence rate after drug withdrawal is high, so it is not the first choice for AA treatment.8 Therefore, it is particularly important to identify and intervene early in AA patients who may progress to severe AA, which will help improve the incidence of severe AA and its prognosis.
Machine learning is gradually becoming more widely used in bioinformatics and can be used to identify potential mechanisms and diagnostic biomarkers of various diseases.9 At present, there is limited research on the use of immune monitoring genes (IMGs) for early severe AA identification and the application of machine learning to screen severe AA-related immune genes (IGs) to obtain IMGs for severe AA diagnosis. In this study, two AA-related datasets from the gene expression omnibus (GEO) database, identified the differentially expressed genes (DEGs) through the classical Bayesian method in limma, and identified the module genes most related to severe AA through weighted gene co-expression network analysis (WGCNA). Functional enrichment analysis, construction of a protein–protein interaction (PPI) network and competing endogenous RNA (ceRNA) network, and immune cell infiltration analysis were performed to clarify the underlying biological mechanisms of severe AA. Subsequently, pivotal IMGs were screened through multiple machine-learning algorithms, including least absolute shrinkage and selection operator (LASSO), random forest (RF), and short time-series expression miner (STEM) algorithms, and the diagnostic effectiveness of the pivotal IMGs was validated by receiver operating characteristic (ROC). This study will help identify potential diagnostic markers related to immunity in severe AA patients.
Materials and methods
Microarray data collection
The research flowchart of this study is shown in supplementary Fig. S1, see online supplementary material. The gene expression datasets were collected from the GEO public database (https://www.ncbi.nlm.nih.gov/geo).10 The AA-related datasets (GSE68801, GSE45512, and GSE80342) were download from GEO for further analysis. The AA-related datasets were based on the GPL570 platform (Affymetrix Human Genome U133 Plus 2.0 Array), in which GSE68801 includes samples from 36 normal controls, 54 AAP patients, 9 AT patients, and 23 AU patients. The immune-related genes were obtained from the ImmPort and MSigDB databases. The details of datasets are shown in supplementary Table S1, see online supplementary material.
Probe re-annotation and identification of DEGs and DE-lncRNAs
The affymetrix was used to obtain all chip probe sequences, the human reference genome (GRCh38) was downloaded from the GENCODE database, and seqmap software was used to compare all probe sequence ratios with the reference genome. First, the unique map probe was retained, and then its position on the chromosome and positive and negative chain information was used to obtain the corresponding gene of each probe according to the human gene annotation file (release 25) provided by GENCODE.11 The probe with the annotation information of “protein_coding” as the corresponding probe of mRNA, and the probes with the annotation information of “antisense”, “sense_intronic”, “lincRNA”, “sense_overlapping”, or “processed_transcript” were reserved as the corresponding probe of long non-coding RNA (lncRNA). Finally, by mutual matching between the probe number and gene symbol, the probes that do not match to the gene symbol were removed, and for different probes mapping to the same gene, the average value of different probes was taken as the final expression value of this mRNA/lncRNA.
The classical Bayesian method in “limma” R package was performed for the differential analysis of datasets. For GSE68801, AAP, AT, and AU were uniformly regarded as disease groups. Gene expression profiles of the disease and control groups in the datasets were compared to identify DEGs and DE-lncRNAs. A gene with a P-value < 0.05 and a fold-change value > ± 0.585 (1.5-fold change) was defined as a DEG and DE-lncRNA. In this study, the intersection of DEG and DE-lncRNA of GSE45512 and GSE68801 was performed to obtain the common upregulated and downregulated DEGs and DE-lncRNA for follow-up analysis.
WGCNA
WGCNA is a systems biology method to characterize patterns of gene association between different samples and can be used to identify highly synergistic sets of genes and the most relevant modular genes based on the endogeneity of the gene set and the association between gene sets and phenotypes.12 The median absolute deviation (MAD) of each gene in the dataset was first determined, and the top 50% of genes with the smallest MAD were excluded. The goodSamplesGenes function of the “WGCNA” R package was used to remove unqualified genes and samples, and a further scale-free co-expression network was constructed. The “soft” threshold power (β) calculates the adjacency between genes and transforms the adjacency into a topological overlap matrix (TOM), which is used to measure network connectivity and similarity. Based on the TOM dissimilarity and the minimum genome size of the gene number map (n = 30), genes with similar expression profiles are classified as gene modules using the average linkage hierarchical clustering and dynamic tree-cutting function detection module. To further analyze the module, the dissimilarity of module eigen genes was calculated, and a cut line for module dendrogram was chosen, and some modules were combined. In this study, important modules related to AT and AU were identified for follow-up investigation.
Enrichment analysis of DEGs
Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed by the DAVID database (https://david.ncifcrf.gov/). GO annotation enrichment analysis involves simple gene annotations of molecular function (MF), biological process (BP), and cell component (CC). A P-value < 0.05 was considered significant.
ceRNA interaction network construction
The matched sample mRNA and lncRNA data were used to calculate the Pearson correlation coefficient of each of the DEGs and DE-lncRNAs, respectively. Then, the correlation test was carried out to screen DEG and DE-lncRNA relationship pairs that may have a synergistic effect. The correlation coefficient value (r) > 0.05 and a P-value < 0.05 were considered to have a synergistic effect. The construction of lncRNA–mRNA co-expression network was visualized by Cytoscape (version 3.7.2). Subsequently, miRWalk2.0 was used to synthesize the results of the four databases (miRWalk, miRanda, RNA22, and Targetscan) to predict the miRNA–mRNA relationship.13 If the predicted results appear in the above databases, it is considered that the corresponding miRNA regulates the corresponding mRNA. The miRanda (v3.3a) software was used to predict miRNAs targeted by lncRNA and filter out miRNA–lncRNA relationship pairs with score ≥ 140, energy ≤ −20.14 The lncRNA–mRNA and miRNA–lncRNA relationship pairs were combined and further screened to construct the ceRNA network and visualized with Cytoscape.
PPI network construction and integration analysis
The complex regulatory network between proteins constructed by STRING version 11.5 database (https://cn.string-db.org/) was used to expand the PPI network. The PPI networks were visualized by Cytoscape, and the hub genes were calculated by using the cytoHubba plug-in. In order to reduce the bias caused by a single algorithm, five common algorithms, including MCC, Degree, Closeness, Radiality, and EPC in the cytoHubba plug-in were used to calculate and identify the hub genes.
Evaluation of immune cell infiltration
CIBERSORTx analytical tool was used to evaluate the input gene expression profiles and 22 immune cell enrichment abundances, and LM22 (22 immune cell types) and 1000 permutations were selected as the signature matrix file for significance analysis. Pearson correlation analysis was used for immune cell infiltration correlation and a P-value < 0.05 was filtered. In addition, the single sample Gene Set Enrichment Analysis (ssGSEA) algorithm was used to detect the immune infiltration status of 28 kinds of immune cells enrichment abundances in AA patients. The relationship between hub genes and infiltrating immune cells was performed by Pearson correlation analysis, and the results were visualized by “ggplot2” R package.
Machine learning and STEM algorithm
The Lasso regression method, RF method and STEM algorithm were performed to filter monitoring genes for severe AA diagnosis. Based on the IMGs related to AA screened from the ImmPort database, we divided the GSE68801 dataset into two features: AAP and severe AA. Subsequently, we used the “glmnet” R package to integrate the gene expression data and performed regression analysis using the Lasso–Cox method. Additionally, we also set up 5-fold cross-validation to obtain the optimal model.15 RF has better sensitivity and specificity and can be used to predict selection continuous variables.16 Based on the above data grouping, the IMGs can be classified by using the “randomForest” R packages and all IMGs are used as a training set. After comparison, it is determined that the gene belongs to AAP or severe AA. The STEM algorithm arranges the number of changes in genes in the process of disease trends, randomly disrupts the time point, re-analyzes the trend, counts the number of genes in each trend, and performs a large number of random rearrangements.17 After a large number of random rearrangements, a desired number of genes can be obtained in each trend, and finally the hypergeometric distribution algorithm is used to calculate the P-value of the trend; a P-value < 0.05 was considered significant. STEM software performed trend analysis of the IMGs. Notably, the intersection genes of LASSO, RF, and STEM were considered as monitoring genes in severe AA diagnosis.
Analyses of diagnostic value and genetic-targeted drug
The diagnostic effectiveness of biomarkers was performed by ROC analysis, and the value of area under curve (AUC) was used to evaluate the diagnostic effectiveness in severe AA. The Drug Gene Interaction Database (DGIdb) (https://dgidb.org/) was used to explore the drug–gene interaction or potentially druggable category. The query score and the interaction score were utilized to assess the confidence of gene and drug interactions; higher values of both scores indicate higher correlation between genes and drugs.
Construction of AA model and immunohistochemistry
Imiquimod cream (Mingxin Pharmaceutical Co., Ltd, Sichuan, China) was used in C3H/HeJ mice to generate AA as our previous study described.18 In brief, a total of four 6-week-old male C3H/HeJ mice were used to establish the AA model, and a cleaning cotton swab was used to dip about 0.05 g of imiquimod cream and evenly smear it on the mouse neck skin (an area ≈1.5 cm × 1.5 cm, three times a week). After 3 weeks of administration, the mouse neck skin showed obvious depilation, indicating that the model was successful. The control group (n = 4) was treated with a mixture of vaseline and lanolin in equal proportion, and the treatment method was the same as that of the AA group. All animal experiments have been approved by the ethics committee of the Naval Medical University.
One week after the formation of obvious depilation in the AA group, the full-thickness skin of the depilated area was taken, and the full-thickness skin of the same area was taken in the control group. The skin samples were fixed with 4% paraformaldehyde and embedded in paraffin. Then, paraffin embedded with the skin sample was cut into 10 micron tissue slices and stained with hematoxylin-eosin (HE). Immunohistochemistry was performed as previously reported.19 The primary antibodies used in immunohistochemistry were LGR5 (1:800, Abcam, USA) and CK19 (1:800, Abcam, UnSA). Photomicrographs were collected under an optical microscope (Leica, Germany). Four areas in each section were randomly selected and analyzed by Image-Pro Plus 6.
Statistical analysis
Comparison between the two group was performed by using Student's sample t-test, and a P-value < 0.05 was considered significant.
Results
Identification of DEGs and DE-lncRNAs
A total of 644 upregulated (308 in GSE68801 and 336 in GSE45512) and 633 downregulated (231 in GSE68801 and 372 in GSE45512) DEGs were identified (supplementary Fig. S2A, C, E and G). A total of 61 upregulated (41 in GSE68801 and 20 in GSE45512) and 68 downregulated (18 in GSE68801 and 50 in GSE45512) DE-lncRNAs were identified (supplementary Fig. S2B, D, F and H, see online supplemenatary material). After the intersection of the Venn diagram, 88 common upregulated DEGs and 172 common downregulated DEGs were obtained (supplementary Fig. S3A, see online supplemehntary material), and 6 common upregulated DE-lncRNAs and 10 common downregulated DE-lncRNAs were obtained (supplementary Fig. S3B).
WGCNA and key module identification
WGCNA was used to identify the modules in AA that were most relevant to severe AA. Based on scale independence and average connectivity, β = 20 (scale-free R2 = 1.24) was selected as the best “soft” threshold (supplementary Fig. S4A, B, see online supplementary material), and supplementary Fig. S4C shows the clustering dendrogram of normal, AAP, AT, and AU. Based on this power, 12 gene co-expression modules (GCMs) of different colors were produced (supplementary Fig. S4D, E). The correlation between different clinical phenotypes of AA and GCMs is shown in supplementary Fig. S4F, and the purple (AT, correlation coefficient = 0.19, P = 0.04; AU, correlation coefficient = 0.49, P = 1.1×10−8), dark green (AT, correlation coefficient = 0.22, P = 0.01; AU, correlation coefficient = 0.2, P = 0.03), dark grey (AT, correlation coefficient = 0.26, P = 3.6×10−3; AU, correlation coefficient = 0.33, P = 2.0×10−4) and light cyan (AT, correlation coefficient = 0.21, P = 0.02; AU, correlation coefficient = 0.31, P = 4.4×10−4) modules were significantly positively correlated with severe AA, and the brown module (AT, correlation coefficient = −0.32, P = 4.0×10−4; AU, correlation coefficient = −0.57, P = 9.3×10−12) was significant negatively correlated with severe AA, and the genes in these modules were regarded as pivotal genes for subsequent analysis.
Functional correlation analysis of severe AA-related DEGs
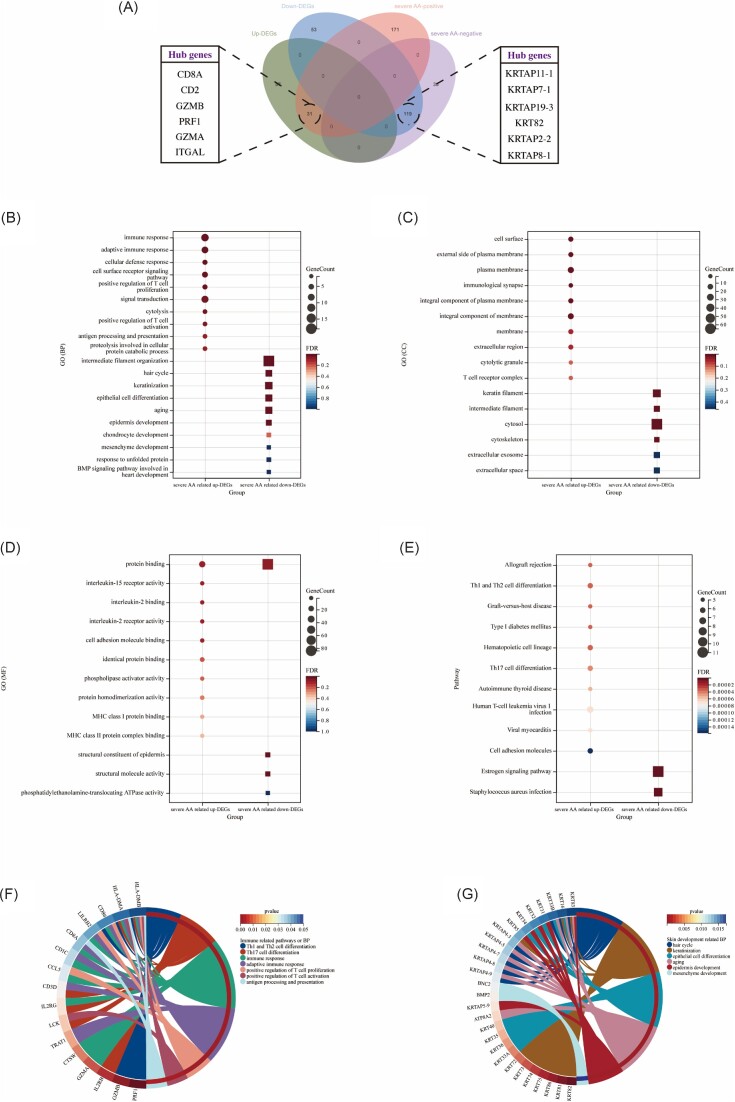
To understand the biological functions and pathways involved in severe AA-related DEGs, a total of 31 common upregulated severe AA-related DEGs and 119 common downregulated severe AA-related DEGs were obtained through a Venn diagram (Fig. 1A). For biological process (BP) (Fig. 1B), the upregulated severe AA-related DEGs were mainly enriched in immune response, adaptive immune response, and cellular defense response, while the downregulated severe AA-related DEGs were mainly involved in intermediate filament organization, keratinization, and epithelial cell differentiation. For cell component (CC) (Fig. 1C), the majority of upregulated severe AA-related DEGs were components of cell surface, external side of plasma membrane, and plasma membrane, whereas the downregulated severe AA-related DEGs were mainly components of keratin filament, intermediate filament, and cytosol. For molecular function (MF) (Fig. 1D), the upregulated severe AA-related DEGs were mainly involved in protein binding, IL-15 receptor activity, and IL-2 binding, whereas the downregulated severe AA-related DEGs were mainly involved in protein binding, structural constituent of epidermis, and structural molecule activity. In terms of KEGG pathway (Fig. 1E), the upregulated severe AA-related DEGs were significantly enriched in allograft rejection, Th1 and Th2 cell differentiation, and graft-versus-host disease, while the downregulated severe AA-related DEGs were mainly involved in the estrogen signaling pathway and Staphylococcus aureus infection. At the same time, the enrichment analysis results of severe AA-related DEGs and common DEGs were basically similar, mainly showing that the upregulated genes participate in a variety of immune responses, while the downregulated genes were enriched in the process of skin development and hair development (supplementary Fig. S3C–F). Figure 1F and G respectively show the immune-related pathways and BPs involving specific upregulated severe AA-related DEGs and the BPs of skin development involving downregulated severe AA-related DEGs.
Figure 1.
Functional correlation analysis of severe AA-related DEGs. (A) Venn diagram displays 31 upregulated and 119 downregulated severe AA-related DEGs. (B–D) GO analysis of the DEGs, including BP, CC, and MF, respectively. (E) KEGG pathway analysis of the DEGs. (F) Circle diagram shows the immune-related pathways, BPs, and related enriched genes. (F) Circle diagram shows the skin development related BPs and related enriched genes.
PPI network construction and hub genes identification of severe AA-related DEGs
PPI network is an important means of understanding functional links between proteins. The PPI networks of severe AA-related DEGs was constructed by Cytoscape (supplementary Fig. S5A, B, see online supplementary material). Hub genes are regulatory genes with high node degree in the PPI network. In this study, the top 10 hub genes were calculated using five algorithms in the cytoHubba plug-in and intersected by Venn diagram (supplementary Fig. S5C, D), and the expression levels were verified in the GSE80342 dataset (supplementary Fig. S5E, F). A total of 6 upregulated hub genes (CD8A, CD2, GZMB, PRF1, GZMA, and ITGAL) and 6 downregulated genes (KRTAP11-1, KRTAP7-1, KRTAP19-3, KRT82, KRTAP2-2, and KRTAP8-1) were identified (Fig. 1A).
Construction of the ceRNA regulatory network
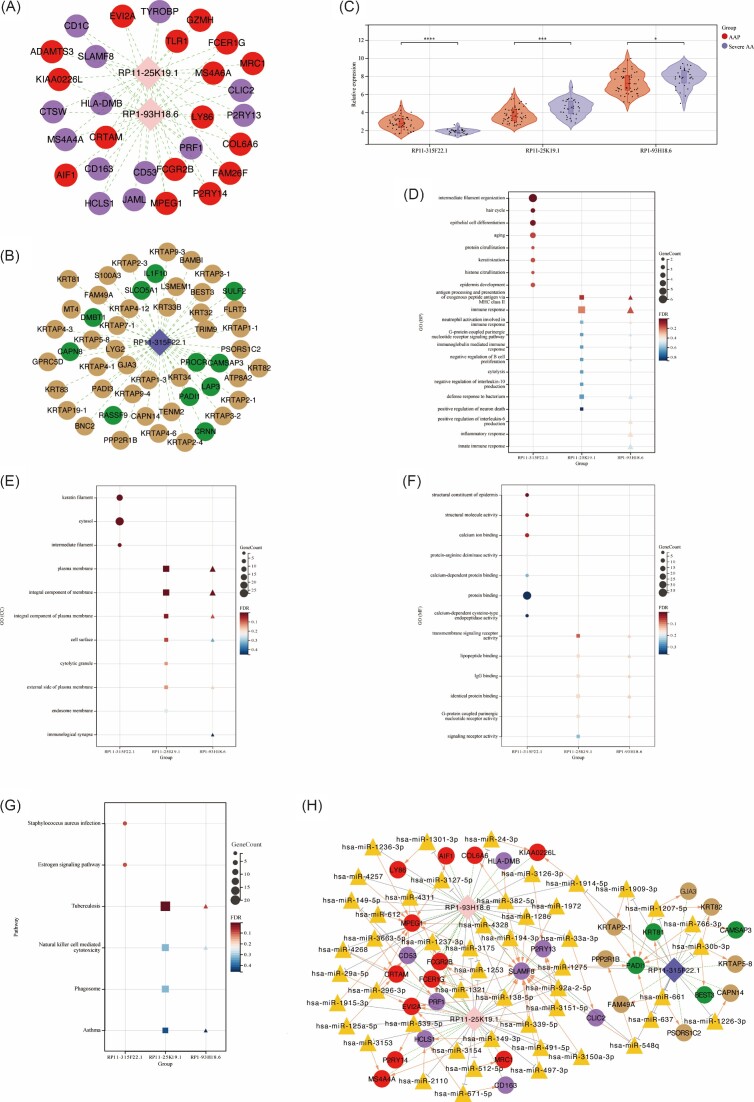
According to the set threshold value, a total of 103 synergistically expressed mRNA-LncRNA relationships were screened, including 80 mRNAs and 3 lncRNAs (Fig. 2A, B). Enrichment analysis was performed to each mRNA with synergistic effects of lncRNA. Compared with AAP group, RP11-315F22.1 was significantly lower expressed in severe AA group, with its synergistic mRNAs mainly enriched in hair cycle, keratin filament and structural constituent of epidermis. RP11-25K19.1 and RP1-93H18.6 were significantly higher expressed in severe AA group (Fig. 2C). Their synergistic mRNAs were mainly enriched in antigen processing and presentation of exogenous peptide antigen via major histocompatibility complex (MHC) class II, plasma membrane, transmembrane signaling receptor activity and tuberculosis (Fig. 2D-G). Subsequently, 91 LncRNA-miRNA-mRNA regulatory relationships were screened in the miRWalk2.0 database, including 57 LncRNA-miRNA relationship pairs, 79 miRNA-mRNA relationship pairs, and 41 LncRNA-mRNA co-expression relationship pairs. The ceRNA regulatory network was constructed using Cytoscape (Fig. 2H).
Figure 2.
Construction of the ceRNA regulatory network and enrichment analysis. (A, B) Regulatory network of synergistically expressed mRNA–lncRNA. (C) The expression trend of DE-lncRNAs (RP11-315F22.1, RP11-25K19.1, and RP1-93H18.6) in the AAP group and severe AA group. (D–F) GO analysis of synergistic mRNAs, including BP, CC, and MF, respectively. (G) KEGG pathway analysis of the synergistic mRNAs. (H) The ceRNA network was constructed through Cytoscape. Pink and dark blue rhombuses represent upregulated and downregulated DE-lncRNAs, respectively. Red and green dots represent upregulated and downregulated AA-related DEGs, respectively. Purple and brown dots represent upregulated and downregulated severe AA-related DEGs, respectively. Orange triangle represents potentially regulated miRNAs. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Immune cell infiltration analysis
The enrichment analysis results strongly suggest a crucial role of immune response in the pathogenesis of AA, especially severe AA. Therefore, this study explored the immune cell infiltration status of patients with AA of different disease courses. Based on the CIBERSORT algorithm, we simultaneously analyzed the infiltration of 22 immune cell types between the four groups (Fig. 3A), and the results showed that native B cells, CD8 T cells, native CD4 T cells, resting CD4 memory T cells, gamma delta T cells, M1 macrophages, M2 macrophages, resting mast cells, and neutrophils have different enrichment levels in different disease courses of AA (Fig. 3B). Notably, resting CD4 memory T cells, gamma delta T cells, resting mast cells, and neutrophils are immune cells with high relative expression level and significant enrichment difference in severe AA patients. Compared with the AAP group, resting CD4 memory T cells and neutrophils were significantly downregulated in severe AA patients (Fig. 3C, F), while gamma delta T cells (Fig. 3D) and resting mast cells (Fig. 3E) were upregulated in AU and AT patients, respectively. The correlation analysis of immune cells revealed that native CD4 T cells were positively associated with M1 macrophages (r = 0.50), whereas activated dendritic cells were negatively related to M1 macrophages (r = −0.56), and that resting mast cells were negatively related to resting dendritic cells (Fig. 3G). In addition, the correlation analysis showed that six upregulated hub genes were significant positively correlated with gamma delta T cells and M1 macrophages, and significant negatively correlated with resting CD4 memory T cells and resting mast cells, whereas six downregulated hub genes were significant positively correlated with neutrophils, and significant negatively correlated with gamma delta T cells and M2 macrophages (Fig. 3H). Moreover, the ssGSEA algorithm was used to detect the immune infiltration status of 28 kinds of immune cells in AA patients (supplementary Fig. S6, see online supplementary material), and the results showed that many types of T cells and B cells were involved in immune infiltration in severe AA patients, which is consistent with the function enrichment results.
Figure 3.
Immune cell infiltration status in AA patients. (A) Histogram showed the composition of 22 kinds of immune cells in normal, AAP, AT, and AU samples. (B) Box diagram of the infiltration of immune cells between the four groups of samples. (C–F) Violin diagram shows the specific enrichment status of resting CD4 memory T cells, gamma delta T cells, resting mast cells, and neutrophils in the four groups. (G) Correlation heat map of 22 types of immune cells. (H) Correlation heat map between severe AA-related hub genes and immune cells in GSE68801; the top and bottom numbers represent correlation coefficients and P-values, respectively. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Screening and comprehensive analysis of severe AA-related IGs
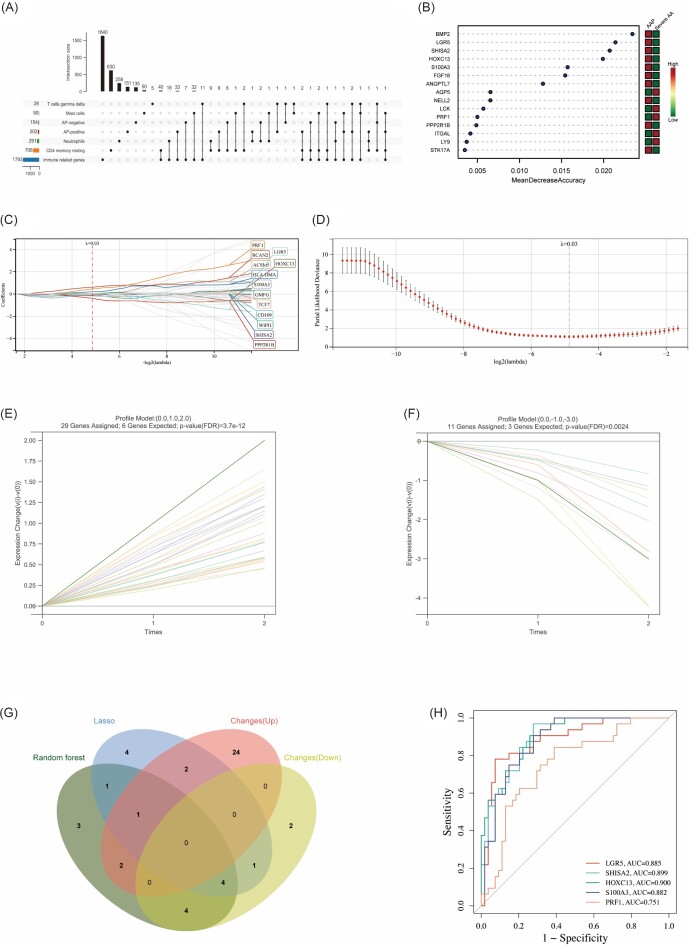
Numerous studies have shown that severe AA is due to hair loss caused by the attack of immune cells on the hair follicles.8,20 Therefore, early identification and intervention may be able to effectively prevent the incidence of severe AA. We obtained 2483 IGs from ImmPort database, and 706 resting CD4 memory T cells-related genes, 291 neutrophils-related genes, 26 gamma delta T cells-related genes and 90 mast cells-related genes from the MSigDB database. A total of 70 severe AA-related IGs were obtained by intersection of these IGs and severe AA-related genes (Fig. 4A), and were verified in GSE68801 and it was found that ESRG and CD8B did not show significant differences in the process of AAP progressing to severe AA (supplementary Fig. S7A, B, see online supplementary material), so 68 severe AA-related immune genes were finally identified. The enrichment analysis of these severe AA-related IGs showed that they were significantly enriched in the immune response, external side of plasma membrane, cytokine receptor activity, and Th17 cell differentiation pathway (supplementary Fig. S8A, B, see online supplementary material). The PPI networks of severe AA-related IGs was constructed by Cytoscape (supplementary Fig. S8C), and hub genes were also obtained by five algorithms; a total of seven hub severe AA-related IGs (CD4, CD8A, PRF1, GZMB, IL2RG, CD86, ITGAX) were identified (supplementary Fig. S8D). Correlation analysis showed that seven hub severe AA-related IGs were significantly positively correlated with gamma delta T cells and M1 macrophages, and significantly negatively correlated with resting CD4 memory T cells and resting mast cells (supplementary Fig. S8E).
Figure 4.
Machine learning in screening immune diagnostic biomarkers for severe AA and the diagnostic value evaluation. (A) Upset diagram shows the intersection of immune genes and severe AA-related genes. (B) Random forest algorithm shows the top 15 important genes, which are ranked based on the mean decrease accuracy. (C, D) Biomarker's screening in the LASSO regression model. The number of genes (n = 13) corresponding to the lowest point of the curve is the most suitable for severe AA diagnosis. (E, F) Trend line graph shows that STEM algorithm screens out the most upregulated and downregulated change genes, respectively. (G) Venn diagram shows that five potential immune monitoring genes are identified via the above three algorithms including random forest algorithm, LASSO regression algorithm, and STEM algorithm. (H) ROC curves of each immune monitoring gene (PRF1, LGR5, SHISA2, HOXC13 and S100A3) show the significant severe AA diagnostic value. AUC, area under the curve.
Identification and diagnostic effectiveness assessment of IMGs
LASSO regression, RF, and STEM algorithms were performed to filter IMGs for severe AA diagnosis. The RF algorithm calculates the importance of each gene and ranks them, and selects the top 15 genes as potential IMGs (Fig. 4B and supplementary Table S2, see online supplementary material). The Lambda value is set to 0.0342824422648852 in the LASSO regression algorithm, and 13 potential IMGs were finally identified (Fig. 4C, D). The STEM algorithm conducted trend analysis on severe AA-related IGs. All data were filtered by mathematical model to remove data with insignificant time-gradient expression difference. Finally, four significant modules were screened, and we selected 40 genes in the most significant changes modules as potential IMGs (Fig. 4E, F). The intersection of the potential IMGs from the RF, LASSO regression, and STEM algorithms were visualized via the Venn diagram (Fig. 4G), and five genes (LGR5, SHISA2, HOXC13, S100A3, and PRF1) were identified for the final validation. Based on the dataset where the IMGs are located and used to validate the diagnostic effectiveness for severe AA by ROC analysis, AUC > 0.8 was used as a threshold for diagnostic capability with excellent specificity and sensitivity. Except for PRF1 (AUC = 0.751), LGR5 (AUC = 0.885), SHISA2 (AUC = 0.899), HOXC13 (AUC = 0.900), and S100A3 (AUC = 0.882) had the capability to diagnose severe AA with excellent specificity and sensitivity (Fig. 4H).
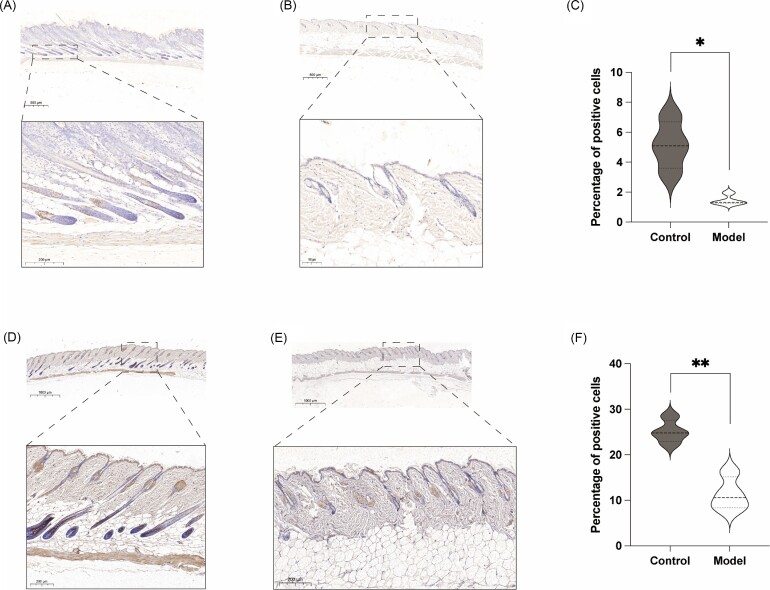
Previous studies have reported that LGR5 is an important marker of hair follicle stem cells (HFSCs).21,22 LGR5+ HFSCs can produce new hair follicles and maintain all cell lines of hair follicles for a long time.23 Therefore, the significantly low expression of LGR5 in severe AA patients has aroused our strong interest. As previously described, the model of AA at week 3 was successfully established, and the area of hair removal in the mice was consistent with the area of drug intervention (Fig. 5A, B). The HE section staining of skin tissue showed that the number of hair follicles in the model group was significantly less than that in the control group (Fig. 5C-E). Notably, a large number of LGR5-positive cells can be found in the outer root sheath of hair follicle in the control group, but it is quite difficult to find positive cells in the model group (Fig. 6A–C). CK19 was expressed in stem cells and plays an important role in the differentiation of epidermal cells.24 Similarly, the number of CK19-positive cells in the control group was much higher than that in the model group (Fig. 6D–F).
Figure 5.
Construction of the AA model and section HE staining. (A) Brief schematic diagram shows the process of model establishment. (B) A model of AA at week 3 was successfully established and the area of hair removal in the mice was consistent with the area of drug intervention (n = 4). (C) HE staining of skin tissue sections in control group. (D) HE staining of skin tissue sections in model group. (E) HE staining revealed that the number of hair follicles in the model group was significantly less than that in the control group. *P < 0.05; **P < 0.01; ***P < 0.001.
Figure 6.
The expression of LGR5 and CK19 were verified in vivo. (A) LGR5 immunohistochemistry staining of skin tissue sections in control group. (B) LGR5 immunohistochemistry staining of skin tissue sections in model group. (C) LGR5 immunohistochemistry staining revealed that LGR5-positive cells in the control group was significantly more than that in the model group. (D) CK19 immunohistochemistry staining of skin tissue sections in control group. (E) CK19 immunohistochemistry staining of skin tissue sections in model group. (F) CK19 immunohistochemistry staining revealed that CK19-positive cells in the control group was significantly more than that in the model group. *P < 0.05; **P < 0.01.
Identification of potential genetic-targeted drugs
The DGIdb database was applied to identify the potential gene-targeted therapy drugs that modulate the immune response, which may become an effective strategy for the treatment of severe AA. All hub genes were used for drug screening. As shown in supplementary Fig. S9 (see online supplementary material), various drugs such as ALEFACEPT, SIPLIZUMAB, TREGALIZUMAB, and ABATACEPT can target and regulate IGs to become candidate gene-targeted drugs for severe AA treatment.
Discussion
Hair loss, one of the most common disease complications, severely damages the patient's appearance and psychological state,25 and AA is the second most common non-scarring alopecia disease, with a prevalence rate of 0.1%–0.2% worldwide.26 If AAP progresses to AT/AU, the difficulty and cost of treatment increases significantly. Therefore, early identification and diagnosis of severe AA is particularly important.
Previously, Zhang and Nie27 explored potential biomarkers for AA, including significantly upregulated CD28 and significantly downregulated HOXC13, KRTAP1-3, and GPRC5D, and suggested early identification and intervention for high-risk patients through these potential biomarkers. However, there is currently little research on constructing the internal regulatory network of AA patients. Therefore, this study first constructed a ceRNA regulatory network for AA patients, where two lncRNAs (RP11-25K19.1 and RP1-93H18.6) were significantly overexpressed in severe AA, which may be important regulatory nodes leading to the production of severe AA. In addition, a study on the immune enrichment status of AA patients was conducted and it was found that there is a significant enrichment of immune cells in AA patients (especially severe AA). If key IMGs can be screened and altered, then this is expected to provide early recognition and intervention for severe AA. By identifying IMGs in high-risk patients through various machine-learning algorithms and conducting partial validation, it was found that LGR5 is a good biomarker for predicting severe AA. Overall, we explored the transcriptional features of AA to reveal the intrinsic mechanisms, and identified the most relevant module genes of severe AA via WGCNA. We first performed a comprehensive analysis of the potential mechanisms of severe AA, including functional enrichment analysis, PPI network construction, and ceRNA network construction. Interestingly, the upregulated DEGs of AA were mainly enriched in immune response, adaptive immune response, and positive regulation of T cell proliferation, while the downregulated DEGs were mainly enriched in pathways related to hair cycle, keratinization, and epithelial cell differentiation (Fig. 1). Under normal physiological conditions, hair follicle tissue cells produce cytokines that inhibit the expression of major histocompatibility complex (MHC) to enjoy immune privilege.28 We found that severe AA-related upregulated DEGs were significantly enriched in the MF of MHC class I protein binding and MHC class II protein complex binding, which is one of the important reasons for the loss of hair follicle immune privilege.
Hub genes are regulatory genes with high node degree in the PPI network, which is of great significance in the progress of the disease. Severe AA-related upregulated hub genes were closely related to immune response. Numerous studies have shown that inflammatory tissue will chemotactically recruit T lymphocytes, leading to the formation of spontaneous AA.29–31 CD8A is the surface glycoprotein of most cytotoxic T lymphocytes and mediates mutual recognition between cells in the immune system.32 CD2 is the surface antigen of all peripheral blood T cells and has the effect of immune recognition.33 GZMA and PRF1 are common components necessary for cytotoxic T lymphocytes and natural killer cells to lysis target cells.34,35 ITGAL is involved in inter-leukocyte adhesion and lymphocyte co-stimulation signals.36 The expression of GZMB increases in a variety of human autoimmune skin diseases, and many studies have found that it will lead to the destruction of the extracellular matrix, with numerous immune cells entering the hair follicles and leading to the collapse of hair follicle immune privilege.37 Severe AA-related downregulated hub genes mainly belong to a member of the keratin gene family or keratin-associated protein family, and are closely related to hair follicle regeneration and skin development, and their significant downregulation may be the core link leading to hair loss in severe AA patients.38
The enrichment analysis results strongly suggest a crucial role of immune response in the pathogenesis of AA, especially severe AA. Therefore, we explored the immune cells enrichment status of AA patients. Compared with the AAP group, resting CD4 memory T cells and neutrophils were significantly downregulated in severe AA patients, while gamma delta T cells and resting mast cells were upregulated in AU and AT patients, respectively (Fig. 3B). McElwee et al.39 found that non-cultured CD4+ T cells alone were sufficient to induce AA. Hashimoto et al.40 further showed that effector memory CD4+ T cells were able to stimulate native and CD8+cells through antigen-presenting cells in axillary lymph nodes and direct T cells to attack hair follicles, leading to AA. In addition, although a significant decrease in neutrophils was observed in the severe AA group in this study, previous studies have shown that the ratio of neutrophils to lymphocytes was not a good indicator for evaluating the severity of AA.41,42 Gamma delta T cells in human skin play the function of outposts of physiological stress, and their excessive activity will promote the overexpression of various cytokines such as CD1d, CXCL12, and MICA, inducing the collapse of the immune privilege of hair follicles.43 Yuan et al.44 found an infiltration of gamma delta T cells and M1 macrophages in the AA tissues, which is consistent with the analysis in this study. Mast cells are important immunoregulatory cells involved in T cell-dependent immunity, immune privilege, and hair growth. Bertolini et al.45 found that hair follicles of AA patients showed more infiltration of mast cells and physical MC/CD8+ T cells than controls, which may be an important mechanism of severe AA.
Attack of immune response of hair follicles is the core factor of AA onset and progression. Therefore, if these immune genes can be monitored early, it may be possible to identify high-risk patients who may progress to severe AA in time. In this study, we screened the IMGs through LASSO regression, RF, and STEM machine-learning algorithms, and the diagnostic effectiveness of the pivotal IMGs was validated by ROC. Finally, four IMGs, including LGR5, SHISA2, HOXC13, and S100A3, with good diagnostic efficiency were obtained. As shown in supplementary Fig. S7A, the expression of LGR5, SHISA2, HOXC13, and S100A3 in the severe AA group was significantly lower than that in the AAP group (P < 0.0001), which also confirmed the effectiveness of HOXC13 as one of the IMGs. Previous studies have found that HOXC13 is particularly important for the differentiation of hair follicles, and HOXC13-targeted mice grow completely hairless with severe nail dystrophy.46,47 Guan et al.48 observed the delay of hair follicles entering the growth period, the reduction of hair elongation, and the reduction of the number of hair follicles in subcutaneous tissue by blocking the expression of S100A3, and the downregulation of hair growth induction-related genes in vivo. LGR5 is a stem marker of HFSCs, and HFSCs maintain the growth, shedding, and replacement of hair.49,50 Therefore, the low expression of LGR5 in severe AA patients aroused our strong interest. We first established an animal model of AA, and found that the number of hair follicles in the animal model was significantly reduced and hair follicles were atrophied. At the same time, the number of LGR5- and CK19-positive stem cells also decreased significantly, and it was quite difficult to observe positive cells. The above results indicate that LGR5 is a good biomarker for predicting severe AA.
Subsequently, we used the DGIdb database to find that ALEFACEPT, SIPLIZUMAB, TREGALIZUMAB, and ABATACEPT are good gene-targeted drugs to modulate the hub genes in severe AA-related PPI network, including ITGAL, GZMA, GZMAB, CD2, PRF1, IL2RG, CD4, and CD86, which may become effective target drugs for severe AA. However, further clinical studies are needed to verify the effectiveness and safety of these drugs for severe AA.
Conclusions
In sum, our findings not only help to improve our understanding of the pathogenesis and underlying biological processes in patients with severe AA, but also use various machine-learning algorithms to identify potential IMGs, which is helpful for the early diagnosis of severe AA (Fig. 7).
Figure 7.
Schematic diagram showing the potential biological processes and early diagnosis of severe AA.
Supplementary Material
Acknowledgement
This work was supported by the National Natural Science Foundation of China (Grant No. 32071186).
Contributor Information
Jiachao Xiong, Department of Histology and Embryology, Naval Military Medical University, Shanghai 200433, China; Department of Plastic Surgery, Shanghai East Hospital, Tongji University School of Medicine, Shanghai 200120, China.
Guodong Chen, Institute of Translational Medicine, Naval Military Medical University, Shanghai 200433, China.
Zhixiao Liu, Department of Histology and Embryology, Naval Military Medical University, Shanghai 200433, China.
Xuemei Wu, Department of Plastic Surgery, Shanghai East Hospital, Tongji University School of Medicine, Shanghai 200120, China.
Sha Xu, Institute of Translational Medicine, Naval Military Medical University, Shanghai 200433, China.
Jun Xiong, Department of Histology and Embryology, Naval Military Medical University, Shanghai 200433, China.
Shizhao Ji, Department of Burn Surgery, The First Affiliated Hospital of Naval Medical University, Shanghai 200433, China.
Minjuan Wu, Department of Histology and Embryology, Naval Military Medical University, Shanghai 200433, China.
Ethics approval and consent to participate
The authors declare human ethics approval was not needed for this study, and animal experiments have been approved by the Ethics Committee of the Naval Medical University.
Conflict of interest
None declared.
Author contributions
J.X., G.C., and Z.L. contributed to project conception and design, helped in the collection of data, analyzed and interpreted data, and wrote the manuscript. S.X. and J.X. provided study material and helped in the collection of data. M.W. and S.J. contributed to project conception and design, analyzed and interpreted data, provided administrative support, and approved the final manuscript. X.W. revised the manuscript. All authors have read and approved the final manuscript.
Availability of data and material
Publicly available datasets were analyzed in this study. These data can be obtained from the GEO database.
References
- 1. Pratt CH, King LE Jr., Messenger AGet al. Alopecia areata. Nat Rev Dis Primers. 2017;3:17011. doi: 10.1038/nrdp.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kim JC, Lee ES, Choi JW.. Impact of alopecia areata on psychiatric disorders: A retrospective cohort study. J Am Acad Dermatol. 2020;82:484–6.. doi: 10.1016/j.jaad.2019.06.1304. [DOI] [PubMed] [Google Scholar]
- 3. Simakou T, Butcher JP, Reid Set al. Alopecia areata: A multifactorial autoimmune condition. J Autoimmun. 2019;98:74–85.. doi: 10.1016/j.jaut.2018.12.001. [DOI] [PubMed] [Google Scholar]
- 4. Rudnicka L, Lukomska M.. Alternaria scalp infection in a patient with alopecia areata. Coexistence or causative relationship?. J Dermatol Case Rep. 2012;6:120–4.. doi: 10.3315/jdcr.2012.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Betz RC, Petukhova L, Ripke Set al. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat Commun. 2015;6:5966. doi: 10.1038/ncomms6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Whiting DA. Histopathologic features of alopecia areata: A new look. Arch Dermatol. 2003;139:1555–9.. doi: 10.1001/archderm.139.12.1555. [DOI] [PubMed] [Google Scholar]
- 7. Gilhar A, Laufer-Britva R, Keren Aet al. Frontiers in alopecia areata pathobiology research. J Allergy Clin Immunol. 2019;144:1478–89.. doi: 10.1016/j.jaci.2019.08.035. [DOI] [PubMed] [Google Scholar]
- 8. Kim JE, Lee YJ, Lee KJet al. Ex vivo treatment with allogenic mesenchymal stem cells of a healthy donor on peripheral blood mononuclear cells of patients with severe alopecia areata: Targeting dysregulated T cells and the acquisition of immunotolerance. Int J Mol Sci. 2022;23:13228. doi: 10.3390/ijms232113228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Y, Shi W, Zhao Det al. Identification of immune-associated genes in diagnosing aortic valve calcification with metabolic syndrome by Integrated bioinformatics analysis and machine learning. Front Immunol. 2022;13:937886. doi: 10.3389/fimmu.2022.937886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiong J, Wu B, Hou Qet al. Comprehensive analysis of LncRNA AC010789.1 delays androgenic alopecia progression by targeting MicroRNA-21 and the wnt/β-catenin signaling pathway in hair follicle stem cells. Front Genet. 2022;13:782750. doi: 10.3389/fgene.2022.782750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Harrow J, Frankish A, Gonzalez JMet al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74.. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Langfelder P, Horvath S.. WGCNA: An R package for weighted correlation network analysis. BMC Bioinf. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12:697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 14. Enright AJ, John B, Gaul Uet al. MicroRNA targets in drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang C, Delcher C, Shenkman Eet al. Machine learning approaches for predicting high cost high need patient expenditures in health care. Biomed Eng Online. 2018;17:131. doi: 10.1186/s12938-018-0568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alderden J, Pepper GA, Wilson Aet al. Predicting pressure injury in critical care patients: A machine-learning model. Am J Crit Care. 2018;27:461–8.. doi: 10.4037/ajcc2018525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ernst J, Bar-Joseph Z.. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinf. 2006;7:191. doi: 10.1186/1471-2105-7-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu M, Xu C, Jiang Jet al. JAM-A facilitates hair follicle regeneration in alopecia areata through functioning as ceRNA to protect VCAN expression in dermal papilla cells. Precis Clin Med. 2022;5:pbac020. doi: 10.1093/pcmedi/pbac020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiong J, Ji B, Wang Let al. Human adipose-derived stem cells promote seawater-immersed wound healing by activating skin stem cells via the EGFR/MEK/ERK pathway. Stem Cells Int. 2019;2019:7135974. doi: 10.1155/2019/7135974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Żeberkiewicz M, Rudnicka L, Malejczyk J.. Immunology of alopecia areata. Cent Eur J Immunol. 2020;45:325–33.. doi: 10.5114/ceji.2020.101264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Polkoff KM, Gupta NK, Green AJet al. LGR5 is a conserved marker of hair follicle stem cells in multiple species and is present early and throughout follicle morphogenesis. Sci Rep. 2022;12:9104. doi: 10.1038/s41598-022-13056-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joost S, Jacob T, Sun Xet al. Single-cell transcriptomics of traced epidermal and hair follicle stem cells reveals rapid adaptations during wound healing. Cell Rep. 2018;25:585–97.e7.. doi: 10.1016/j.celrep.2018.09.059. [DOI] [PubMed] [Google Scholar]
- 23. Jaks V, Barker N, Kasper Met al. Lgr5 marks cycling, yet long-lived, hair follicle stem cells. Nat Genet. 2008;40:1291–9.. doi: 10.1038/ng.239. [DOI] [PubMed] [Google Scholar]
- 24. El Sakka D, Gaber MA, Abdou AGet al. Stem cell markers (Cytokeratin 17 and Cytokeratin 19) in scarring and nonscarring alopecia. J Cutan Aesthet Surg. 2016;9:165–71.. doi: 10.4103/0974-2077.191650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu Q, Wang C, Jing Qet al. Follow-up of patients with COVID-19 by the Delta variant after hospital discharge in Guangzhou, Guandong, China. Rev Inst Med Trop Sao Paulo. 2022;64:e31. doi: 10.1590/s1678-9946202264031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ramírez-Marín HA, Tosti A.. Emerging drugs for the treatment of alopecia areata. Expert Opin Emerg Drugs. 2022;27:379–87.. doi: 10.1080/14728214.2022.2149735. [DOI] [PubMed] [Google Scholar]
- 27. Zhang T, Nie Y.. Prediction of the risk of Alopecia Areata progressing to Alopecia Totalis and Alopecia Universalis: Biomarker development with bioinformatics analysis and machine learning. Dermatology. 2022;238:386–96.. doi: 10.1159/000515764. [DOI] [PubMed] [Google Scholar]
- 28. Rajabi F, Drake LA, Senna MMet al. Alopecia areata: A review of disease pathogenesis. Br J Dermatol. 2018;179:1033–48.. doi: 10.1111/bjd.16808. [DOI] [PubMed] [Google Scholar]
- 29. Zainodini N, Hassanshahi G, Arababadi MKet al. Differential expression of CXCL1, CXCL9, CXCL10 and CXCL12 chemokines in alopecia areata. Iran J Immunol. 2013;10:40–6. [PubMed] [Google Scholar]
- 30. Maouia A, Sormani L, Youssef Met al. Differential expression of CXCL9, CXCL10, and IFN-γ in vitiligo and alopecia areata patients. Pigment Cell Melanoma Res. 2017;30:259–61.. doi: 10.1111/pcmr.12559. [DOI] [PubMed] [Google Scholar]
- 31. Bilgic O, Sivrikaya A, Unlu A, et al. Serum cytokine and chemokine profiles in patients with alopecia areata. J Dermatolog Treat. 2016;27:260–3.. doi: 10.3109/09546634.2015.1093591. [DOI] [PubMed] [Google Scholar]
- 32. Zheng L, Han X, Yao Set al. The CD8α-pilrα interaction maintains CD8(+) T cell quiescence. Science. 2022;376:996–1001.. doi: 10.1126/science.aaz8658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Romain G, Strati P, Rezvan Aet al. Multidimensional single-cell analysis identifies a role for CD2-CD58 interactions in clinical antitumor T cell responses. J Clin Invest. 2022;132(17):e159402. doi: 10.1172/JCI159402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Díaz-Basabe A, Burrello C, Lattanzi Get al. Human intestinal and circulating invariant natural killer T cells are cytotoxic against colorectal cancer cells via the perforin-granzyme pathway. Mol Oncol. 2021;15:3385–403.. doi: 10.1002/1878-0261.13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cheuk S, Martini E, Bergh Ket al. Granzyme A potentiates chemokine production in IL-17-stimulated keratinocytes. Exp Dermatol. 2017;26:824–7.. doi: 10.1111/exd.13284. [DOI] [PubMed] [Google Scholar]
- 36. Shulman Z, Shinder V, Klein Eet al. Lymphocyte crawling and transendothelial migration require chemokine triggering of high-affinity LFA-1 integrin. Immunity. 2009;30:384–96.. doi: 10.1016/j.immuni.2008.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Duncan FJ, Silva KA, Johnson CJet al. Endogenous retinoids in the pathogenesis of alopecia areata. J Invest Dermatol. 2013;133:334–43.. doi: 10.1038/jid.2012.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heid HW, Werner E, Franke WW.. The complement of native alpha-keratin polypeptides of hair-forming cells: A subset of eight polypeptides that differ from epithelial cytokeratins. Differentiation. 1986;32:101–19.. doi: 10.1111/j.1432-0436.1986.tb00562.x. [DOI] [PubMed] [Google Scholar]
- 39. McElwee KJ, Freyschmidt-Paul P, Hoffmann Ret al. Transfer of CD8(+) cells induces localized hair loss whereas CD4(+)/CD25(-) cells promote systemic alopecia areata and CD4(+)/CD25(+) cells blockade disease onset in the C3H/HeJ mouse model. J Invest Dermatol. 2005;124:947–57.. doi: 10.1111/j.0022-202X.2005.23692.x. [DOI] [PubMed] [Google Scholar]
- 40. Hashimoto K, Yamada Y, Sekiguchi Ket al. Induction of alopecia areata in C3H/HeJ mice using cryopreserved lymphocytes. J Dermatol Sci. 2021;102:177–83.. doi: 10.1016/j.jdermsci.2021.04.009. [DOI] [PubMed] [Google Scholar]
- 41. İslamoğlu ZGK, Demirbaş A.. Evaluation of complete blood cell and inflammatory parameters in patients with alopecia areata: Their association with disease severity. J Cosmet Dermatol. 2020;19:1239–45.. doi: 10.1111/jocd.13131. [DOI] [PubMed] [Google Scholar]
- 42. Dere G, Gündoğdu M.. Investigation of the relationship between alopecia areata and inflammatory blood parameters. J Cosmet Dermatol. 2021;20:4048–51.. doi: 10.1111/jocd.14048. [DOI] [PubMed] [Google Scholar]
- 43. Uchida Y, Gherardini J, Pappelbaum Ket al. Resident human dermal γδt-cells operate as stress-sentinels: Lessons from the hair follicle. J Autoimmun. 2021;124:102711. doi: 10.1016/j.jaut.2021.102711. [DOI] [PubMed] [Google Scholar]
- 44. Yuan X, Tang Y, Zhao Zet al. Identification and verification of EOMEs regulated network in Alopecia areata. Int Immunopharmacol. 2020;84:106544. doi: 10.1016/j.intimp.2020.106544. [DOI] [PubMed] [Google Scholar]
- 45. Bertolini M, Zilio F, Rossi Aet al. Abnormal interactions between perifollicular mast cells and CD8+ T-cells may contribute to the pathogenesis of alopecia areata. PLoS One. 2014;9:e94260. doi: 10.1371/journal.pone.0094260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Potter CS, Pruett ND, Kern MJet al. The nude mutant gene Foxn1 is a HOXC13 regulatory target during hair follicle and nail differentiation. J Invest Dermatol. 2011;131:828–37.. doi: 10.1038/jid.2010.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Qiu W, Lei M, Tang Het al. Hoxc13 is a crucial regulator of murine hair cycle. Cell Tissue Res. 2016;364:149–58.. doi: 10.1007/s00441-015-2312-7. [DOI] [PubMed] [Google Scholar]
- 48. Guan W, Deng Q, Yu XLet al. Blockade of S100A3 activity inhibits murine hair growth. Genet Mol Res. 2015;14:13532–44.. doi: 10.4238/2015.October.28.14. [DOI] [PubMed] [Google Scholar]
- 49. Yang H, Adam RC, Ge Yet al. Epithelial-mesenchymal micro-niches govern stem cell lineage choices. Cell. 2017;169:483–496.e13.. doi: 10.1016/j.cell.2017.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang X, Chen H, Tian Ret al. Macrophages induce AKT/β-catenin-dependent Lgr5(+) stem cell activation and hair follicle regeneration through TNF. Nat Commun. 2017;8:14091. doi: 10.1038/ncomms14091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. These data can be obtained from the GEO database.