Abstract
Objectives:
The aim was to determine the accuracy of cell-free DNA testing (cfDNA) for detecting sex chromosome aneuploidies (SCA) in singleton pregnancies.
Methods:
A systematic review and meta-analysis was performed to assess cfDNA accuracy for prenatal detection of 45,X, 47,XXY, 47,XXX and 47,XYY. Inclusion was restricted to studies published between January 2010 and December 2021 reporting both cfDNA and confirmatory diagnostic test results.
Results:
For 45,X, the sensitivity was 98.8% (95%CI 94.6%–100%), specificity 99.4% (95%CI 98.7%–99.9%) and positive predictive value (PPV) 14.5% (95%CI 7.0%–43.8%). For 47,XXY, the sensitivity was 100% (95%CI 99.6%–100%), specificity 100% (95%CI 99.9%–100%) and PPV 97.7% (95%CI 78.6%–100%). For 47,XXX, the sensitivity was 100% (95%CI 96.9%–100%), specificity 99.9% (95%CI 99.7%–100%) and PPV 61.6% (95%CI 37.6%–95.4%). For 47,XYY, the sensitivity was 100% (95%CI 91.3%–100%), specificity 100% (95% CI 100%–100%) and PPV 100% (95%CI 76.5%–100%). All four SCAs had estimated negative predictive values (NPV) exceeding 99.99%, though false negatives were reported.
Conclusions:
This analysis suggests that cfDNA is a reliable screening test for SCA, though both false negatives and false positives were reported. These estimates of test performance are derived from pregnancies at high pretest risk for aneuploidy, limiting the generalisability to average risk pregnancies.
1 |. INTRODUCTION
Cell-free DNA (cfDNA) analysis enables non-invasive prenatal screening for sex chromosome aneuploidies (SCA), common trisomies (trisomy 13, 18 and 21), and certain monogenic disorders, copy number variants and rare autosomal trisomies.1–4 Since the discovery of circulating cell-free fetal DNA in 1997, its use has rapidly expanded and cfDNA is now recommended to be offered to all pregnant individuals in the United States.5,6 However, cfDNA remains a screening test, and diagnostic testing with chorionic villus sampling (CVS) or amniocentesis is recommended after a high-risk screening result. The test performance of cfDNA varies by the genetic disorder tested, with higher positive predictive values for trisomy 21 with increasing ovum age, in contrast to lower positive predictive values for more rare conditions such as 22q11.2 deletion syndrome.1,7–9
SCAs are the most frequent aneuploidies in humans, with an estimated incidence of 1/440 live births. SCAs detectable with cfDNA include Klinefelter syndrome (47,XXY), Turner syndrome (45,X), Jacobs syndrome (47,XYY) and triple X syndrome (47,XXX). Klinefelter syndrome (47,XXY) is the most common of the SCAs, with a frequency of 0.16%–0.24% among pregnancies in those 35 years and older.6,10,11
Prior studies have assessed the overall test performance of cfDNA, but few publications have focused on the detection of SCA specifically.1,12–19 Sensitivities of cfDNA for the common trisomies have been reported as 95.8%–99.7%, with specificities above 99%1. In contrast, sensitivities of cfDNA for SCA have been more variable, reported as 83.4%–93.9% for monosomy X and 76.3%–93.0% for XXY, XYY and XXX (1,12,14). Prior analyses suggest that test performance may vary based on the underlying assay with, for example, higher sensitivity for detecting monosomy X by using massively parallel shotgun sequencing (MPSS) compared to targeted massively parallel sequencing (TMPS).14 Further, until 2020, the American College of Obstetricians and Gynecologists (ACOG) recommended cfDNA only for high-risk pregnancies, so most existing studies of cfDNA performance include populations at higher risk for aneuploidy. Additionally, among larger published cohorts of unselected pregnancies at low or average risk undergoing cfDNA screening, detection of common trisomies has usually been the primary focus.
It is important to acknowledge though, that interpretation of cfDNA results for SCA can pose challenges for patients and providers. Both placental and maternal mosaicism are known contributors of false negative and false positive results.20,21 SCA phenotypes vary significantly between affected individuals, and many with SCA may be mildly affected. It is estimated, for example, that only 25% of the patients with 47,XXY receive a clinical diagnosis during their lifetime.22 Given this variable expressivity and the potential for mild ultimate outcomes, the utility of cfDNA testing for SCA has been questioned.23 However, prenatal screening for SCA is available through most laboratories and detection of SCA in utero can be important for pregnancy decision-making for many individuals, for preparation for the needs of the neonate and child, and for early interventions to improve outcomes.24,25
Through this systematic review and meta-analysis, we used the currently available evidence to determine the accuracy of cfDNA for detecting SCA (45,X; 47,XXY; 47,XYY; and 47,XXX) in singleton pregnancies. We hypothesised that cfDNA would perform well for the detection of SCA and that variations in performance would be observed by specific SCA disorders.
2 |. METHODS
This systematic review was performed according to PRISMA guidelines. The search protocol and aims were registered a priori on PROSPERO (CRD42022292376). As this study included only de-identified data from previously published manuscripts, this study was exempt from IRB approval.
2.1 |. Study identification
PubMed, MEDLINE, EMBASE, Google Scholar, CINAHL, LILACS and ClinicalTrials.gov were queried for eligible studies using the following search terms: “cfDNA,” “cffDNA,” “ffDNA,” “Free fetal DNA,” “Free fetal DNA,” “Cell free DNA,” “Cell-free DNA,” “Cell-free fetal DNA,” “Cell-free fetal DNA,” “Sex chromosome aneuploidy,” “Sex chromosome aneuploid,” “Non-invasive prenatal testing,” “Non-invasive prenatal screening,” “Noninvasive prenatal testing,” “Noninvasive prenatal screening,” “NIPT,” “NIPS,” “Monosomy X,” “XO,” “Turner syndrome,” “Turners syndrome,” “Klinefelter Syndrome,” “XXY,” “Jacobs Syndrome,” “XYY,” “Triple X Syndrome,” and “XXX”. Due to technical limitations with study identification by using the above search terms, the results from Google Scholar were excluded.
2.2 |. Study selection
The exclusion of duplicate studies was performed electronically by assessing PubMed ID, digital object identifier (DOI) or exact title matches using Microsoft excel. The initial cohort of titles was cross-referenced against previously published meta-analyses evaluating cfDNA for SCA to ensure inclusion of potentially relevant studies. MAS performed this title screen independently. A random selection of 10% of candidate abstracts were then screened independently by a combination of two researchers (MAS and either KS or RG), with >95% agreement in terms of inclusion demonstrated between researchers, so the remainder of abstracts were screened independently. The full manuscripts for these potentially relevant abstracts were then independently reviewed, and a random selection of 10% was screened by a combination of two researchers (MAS and either KS or RG). Greater than 95% agreement was demonstrated and any disagreements regarding inclusion or exclusion were adjudicated by a third researcher (TS).
2.3 |. Eligibility criteria
Inclusion was restricted to studies reporting singleton pregnancies that underwent cfDNA testing and were published online or in print in English from January 1, 2010 to December 1, 2021. Studies were eligible only if they utilised diagnostic genetic testing during pregnancy or after delivery to confirm cfDNA results for SCA. Acceptable diagnostic test results included karyotype, fluorescence in situ hybridisation (FISH), microarray or polymerase chain reaction (PCR) performed prenatally using samples from CVS, amniotic fluid, cord blood, products of conception (POC) or postnatally on any neonatal tissue source. Studies were excluded if the results of diagnostic testing were known to researchers prior to undergoing cfDNA, or if the results of diagnostic testing were either unknown or not available for >20% of the cohort to avoid the risk of reporting bias.17 Preimplantation genetic testing, prenatal ultrasound and/or postnatal clinical exam alone were not considered an acceptable diagnostic confirmation of fetal sex chromosome complement. Studies of experimental methods of cfDNA, proof-of-concept studies of novel laboratory methods of cfDNA analysis, case reports and conference abstracts were excluded.
2.4 |. Data extraction
The cfDNA results for SCA and corresponding confirmatory diagnostic reference test results were extracted and recorded in Excel. These were converted into 2 × 2 table format representing true positive, true negative, false positive or false negative results. Additional data were extracted where available including platform used (Single Nucleotide Polymorphism (SNP), MPSS, other), study design, pretest risk for aneuploidy (high/low), percent of the cohort with reportable diagnostic test results, diagnostic test (karyotype, FISH microarray, or PCR), tissue source (CVS, amniocentesis, cord blood and POC). High pretest risk of aneuploidy was defined as ≥50% of the reported cohort with one or more of the following characteristics: abnormal serum analyte screening, maternal age ≥35 years, or fetal abnormality detected by ultrasound including, but not limited to: absent or hypoplastic nasal bone, renal pyelectasis, echogenic bowel, ventriculomegaly, short femur length, echogenic intracardiac foci or choroid plexus cyst, enlarged nuchal translucency, cystic hygroma or thickened nuchal fold.6 Clarification regarding missing or incomplete data was requested from the corresponding authors of included studies as appropriate.
2.5 |. Bias assessment
All studies included in the final analysis were assessed for publication bias by two independent reviewers by using the QUADAS-2.26 Bias assessments were compared between reviewers and any discordance between these assessments was resolved by a third reviewer (TS).
2.6 |. Data analysis
Meta-analysis of the 2 × 2 table summary statistics for cfDNA detection of SCA was performed using exact binomial methods (Clopper-Pearson) via the metaprop package in Stata version 17.0 (StataCorp; College Station, TX).27 Forest plots with pooled estimated sensitivity and specificity were calculated in aggregate with 95% confidence intervals (CI). Separate analyses were carried out for individual SCA, including 45,X, 47,XXY, 47,XYY and 47,XXX. Joint meta-analysis of sensitivity and specificity using MIDAS and metandi Stata packages was not possible due to high sensitivity and specificity levels.
Positive predictive values (PPV) and negative predictive values (NPV) were calculated using standard formulas (supplementary appendix). The estimated population prevalence of each individual SCA was derived from previously published cohorts.10,11,28,29 The 95% confidence interval upper and lower boundary estimates for PPV and NPV were derived by using varying estimates of the population prevalence of SCA along with the upper and lower boundaries of the estimated sensitivity and specificity.
3 |. RESULTS
A total of 31,718 studies resulted from the database search, and 10,877 of these were duplicates. 20,831 titles were assessed for relevance, of which 19,563 were excluded and 1268 underwent abstract or full text screening. Ultimately, after applying the exclusion criteria as outlined in the methods, 21 articles were included in the final meta-analysis30–50 (Figure 1). Characteristics of included studies are shown in Table 1.
FIGURE 1.
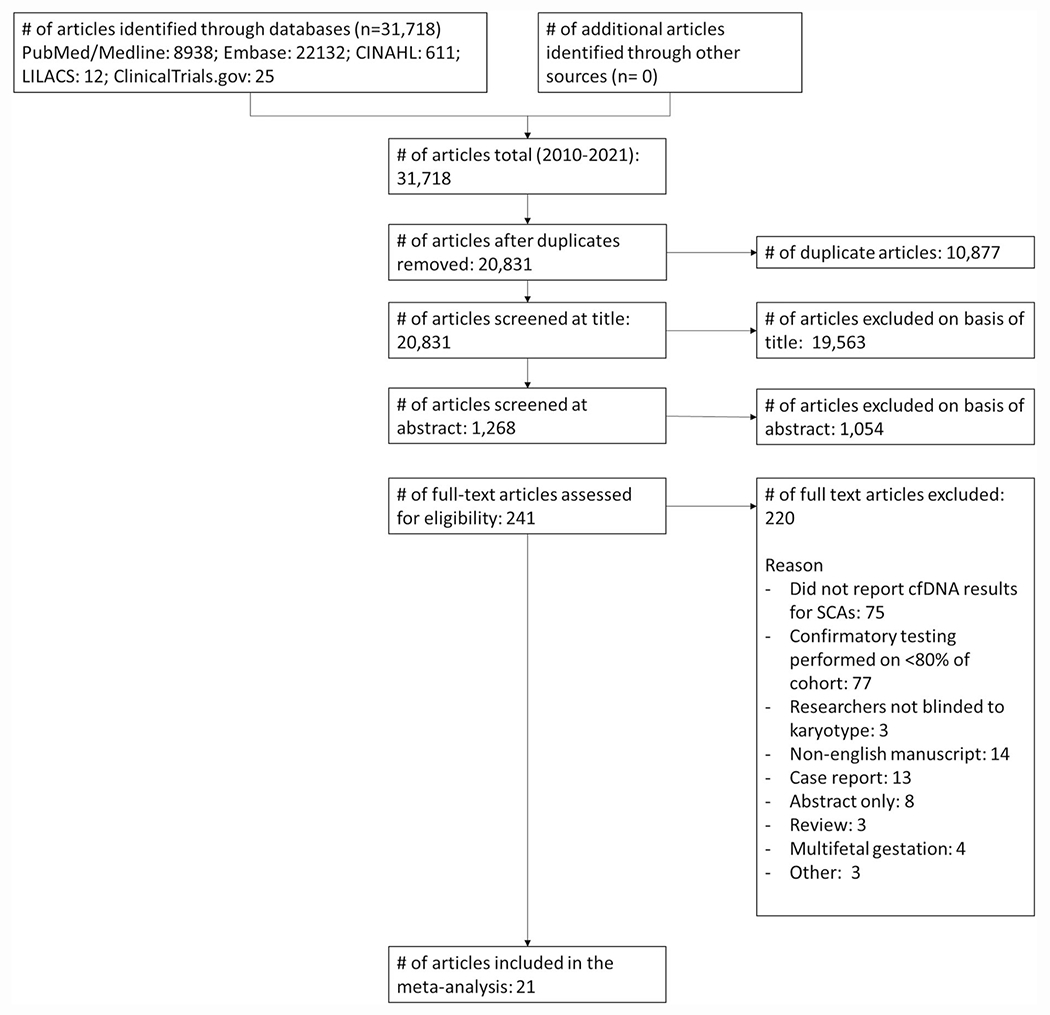
PRISMA diagram of study selection.
TABLE 1.
Characteristics of studies included for meta-analysis of cfDNA testing for fetal SCA
| Study author | Year | Pretest risk | Test description | Diagnostic reference | Study design |
|---|---|---|---|---|---|
| Bianchi | 2012 | High | MPSS | Karyotype, FISH | Prospective, multicenter, observational |
| Bianchi | 2013 | High | MPSS | Karyotype | Prospective, multicenter, observational |
| Guseh | 2021 | High | MPSS | Karyotype, microarray | Prospective, single center, observational |
| Hooks | 2014 | High | MPSS | Karyotype | Prospective, single center, observational |
| Lau | 2012 | High | MPSS | Karyotype | Prospective, single center, observational |
| Lefkowitz | 2016 | High | MPSS | Karyotype, microarray, sequencing | Retrospective, multicenter, observational |
| Li | 2018 | High | MPSS | Karyotype | Retrospective, single center, observational |
| Liang | 2013 | High | MPSS | Karyotype | Prospective, multicenter, observational |
| Mazloom | 2013 | High | MPSS | Karyotype | Retrospective, single center, observational |
| Meck | 2015 | High | Multiple | Karyotype, microarray | Retrospective, multi center, observational |
| Nicolaides | 2013 | High | TMPS | Karyotype | Prospective, single center, observational |
| Nicolaides | 2014 | High | SNP | Karyotype | Retrospective, single center, case-control |
| Pergament | 2014 | High | SNP | Karyotype | Prospective, multicenter, observational |
| Persico | 2016 | High | SNP | Karyotype, microarray | Prospective, multicenter, observational |
| Petersen | 2017 | High | Multiple | Karyotype, FISH, microarray | Retrospective, single center/multisite, observational |
| Porreco | 2014 | High | MPSS | Karyotype | Prospective, multicenter, observational |
| Porreco | 2020 | High | SNP | Karyotype | Prospective and retrospective, single center, observational |
| Sehnert | 2011 | High | MPSS | Karyotype | Retrospective, multicenter, observational |
| Shubina | 2017 | High | MPSS | Karyotype, FISH | Retrospective and prospective, single center, observational |
| Song | 2015 | High | MPSS | Karyotype | Prospective, single center, observational |
| Zhu | 2019 | High | MPSS | Karyotype | Prospective, single center, observational |
Data are presented as first author’s last name, year of publication, pretest risk (high/low), test description, diagnostic reference standard.
Abbreviations: FISH, fluorescence in situ hybridization; MPSS, massively parallel shotgun sequencing; SNP, single nucleotide polymorphism; TMPS, targeted massively parallel sequencing.
3.1 |. 45,X
All 21 included studies reported cfDNA results for monosomy X(30–50). This represented 12,926 cell-free DNA tests with 254 confirmed affected pregnancies, 133 false positive tests and 18 false negative tests (Supplemental Table 1). The pooled estimated sensitivity was 98.8% (95%CI 94.6%–100%), and specificity was 99.4% (95%CI 98.7%–99.9%) (Table 2). The forest plots of included studies are reported in Figure 2. The estimated PPV was 14.5% (95%CI 7.0%–43.8%), and the negative predictive value (NPV) was 100% (95% CI 99.995%–100%). The PPV and NPV for the detection of 45,X based on estimates of SCA prevalence are included in Table 3. Two studies reported results for 45,X from 74 duplicate cases.30,31 A sensitivity analysis was performed excluding one of these studies,31 yielding a pooled estimated sensitivity of 98.8% (95%CI 94.3%–100%), specificity of 99.4% (95%CI 98.7%–99.9%), PPV 13.7% (95%CI 7.0%–43.8%) and NPV 100% (95%CI 99.994%–100%). The results of the sensitivity analysis are included in Supplemental Table 5 and Supplemental Table 6.
TABLE 2.
cfDNA accuracy for prenatal detection of sex chromosome aneuploidy
| SCA | Pooled estimated sensitivity | 95% CI lower | 95% CI upper | Pooled estimated specificity | 95% CI lower | 95% CI upper |
|---|---|---|---|---|---|---|
| 45,X | 98.754% | 94.640% | 100% | 99.413% | 98.723% | 99.870% |
| 47,XXY | 100% | 99.559% | 100% | 99.994% | 99.935% | 100% |
| 47,XXX | 100% | 96.901% | 100% | 99.887% | 99.709% | 99.991% |
| 47,XYY | 100% | 91.284% | 100% | 100% | 99.979% | 100% |
Abbreviations: SCA, sex chromosome aneuploidy, 95% CI, 95% confidence interval; 100%, 100.000%.
FIGURE 2.
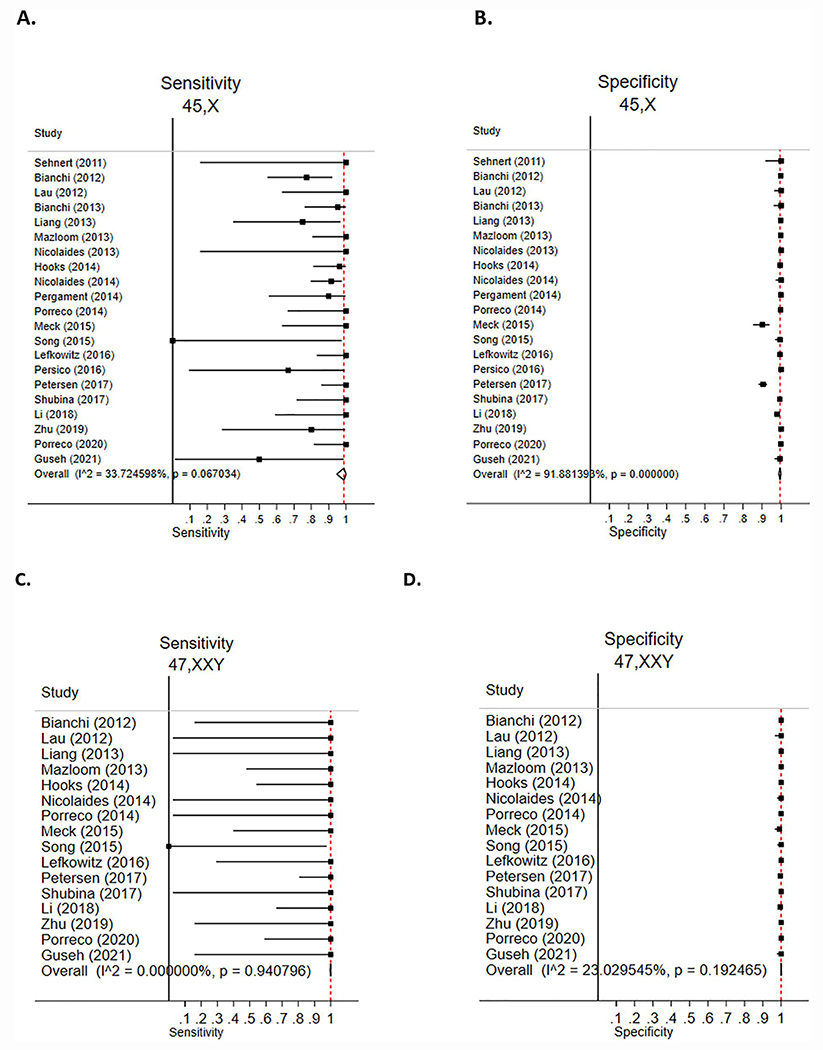

cfDNA accuracy for prenatal detection of sex chromosome aneuploidy. Data are presented as author (year) with calculated sensitivity and specificity displayed as point estimates with 95% confidence intervals. Forest plots for sensitivity and specificity are included for 45,X (A,B), 47,XXY (C,D), 47,XXX (E,F) and 47,XYY(G,H). The overall estimated pooled summary statistic. I^2 = variation in the estimated pooled sensitivity or specificity attributable to heterogeneity.
TABLE 3.
The predictive value of cfDNA for the prenatal detection of sex chromosome aneuploidy
| SCA | Estimated Prevalencea | PPV | 95% CI lower | 95% CI upper | NPV | 95% CI lower | 95% CI upper |
|---|---|---|---|---|---|---|---|
| 45,X | 176/100,000 | 22.9% | 11.6% | 57.6% | 99.998% | 99.990% | 100% |
| 101/100,000 | 14.5% | 7.0% | 43.8% | 99.999% | 99.995% | 100% | |
| 30/100,000 | 4.8% | 2.2% | 18.8% | 100% | 99.998% | 100% | |
|
| |||||||
| 47,XXY | 240/100,000 | 97.7% | 78.6% | 100% | 100% | 99.999% | 100% |
| 91/100,000 | 94.1% | 58.2% | 100% | 100% | 99.9996% | 100% | |
|
| |||||||
| 47,XXX | 180/100,000 | 61.6% | 37.6% | 95.4% | 100% | 99.994% | 100% |
| 54/100,000 | 32.4% | 15.3% | 86.3% | 100% | 99.998% | 100% | |
|
| |||||||
| 47,XYY | 75/100,000 | 100% | 76.5% | 100% | 100% | 99.993% | 100% |
| 32/100,000 | 100% | 58.2% | 100% | 100% | 99.997% | 100% | |
3.2 |. 47,XXY
A total of 16 included studies reported cfDNA results for Klinefelter Syndrome (47,XXY).30,32–39,41,44,46,48–50 This represented 11,248 cell-free DNA tests with 62 confirmed affected pregnancies, 10 false positive tests and 10 false negative tests (Supplemental Table 2). The pooled estimated sensitivity was 100% (95%CI 99.6%–100%) and specificity was 100% (95%CI 99.9%–100%) (Table 2). The forest plots of included studies are reported in Figure 2. The estimated PPV was 97.7% (95%CI 78.6%–100%), and NPV was 100% (95%CI 99.999%–100%). The PPV and NPV for detection of 47,XXY based on estimates of SCA prevalence are included in Table 3.
3.3 |. 47,XXX
A total of 13 included studies reported cfDNA results for triple X syndrome (47,XXX).30,33,35–39,41,44,46,48,49 This represented 10,255 cell-free DNA tests with 38 confirmed affected pregnancies, 18 false positive tests and one false negative test (Supplemental Table 3). The pooled estimated sensitivity was 100% (95%CI 96.9%–100%) and specificity was 99.9% (95%CI 99.7%–99.99%) (Table 2). The forest plots of included studies are reported in Figure 2. The estimated PPV was 61.6% (95%CI 37.6%–95.4%), and the NPV was 100% (95% CI 99.994%–100%). The PPV and NPV for the detection of 47,XXX based on estimates of SCA prevalence are included in Table 3.
3.4 |. 47,XYY
A total of nine included studies reported cfDNA results for Jacobs syndrome (47,XYY).30,35–38,41,44–46 This represented 8473 cell-free DNA tests and with 22 confirmed affected pregnancies, two false positive tests and no false negative tests (Supplemental Table 4). The pooled sensitivity was 100% (95% CI 91.3%–100%). The pooled specificity was 100% (95% CI 100%–100%) (Table 2). The forest plots of included studies are reported in Figure 2. The estimated PPV was 100% (95%CI 76.5%–100%), and the NPV is 100% (95% CI 99.993%–100%). The PPV and NPV for detection of 47,XYY based on the estimated prevalence of SCA by age are included in Table 3.
3.5 |. Bias assessment
The results of the QUADAS-2 assessment for bias are summarised in Supplemental Figure 1. The majority of studies (81%, 17/21) had concerns for introduction of bias based on the included patient populations, as the majority of published studies on cfDNA to date have included high-risk populations for aneuploidy. Two studies (10%, 2/21) had a high concern for bias based on the nature of the index test used, as one or more authors stated a potential conflict of interest in an association with a company or commercial laboratory performing prenatal genetic testing.
4 |. DISCUSSION
Cell-free DNA screening is increasingly utilised for prenatal risk assessment of aneuploidies in the United States and globally.6 Overall, we found high sensitivity, specificity and high NPV for cfDNA in the detection of SCA. PPVs were more variable across SCAs due to differing underlying disease prevalence, with often wide confidence intervals. This meta-analysis adds to the literature in this area as few prior studies have focused specifically on cfDNA performance for the detection of SCAs. However, it is important to emphasise that as studies meeting inclusion criteria were based primarily on the application of cfDNA to populations with greater a priori risk for aneuploidy, this limits generalisability to average risk pregnancies.
Estimates of PPVs varied from 14.5% (95%CI 7.0%–43.8%) for 45,X to 100% (95%CI 76.5%–100%) for 47,XYY. All four SCAs investigated had NPVs exceeding 99.99% regardless of the estimated population prevalence and reproductive age. We acknowledge that variations in risk of SCA may exist with increasing ovum age6,10,11,28,29 and that such changes can affect estimates of PPV as reflected in Table 3, although when present these are generally attenuated relative to the common trisomies. Our findings support that cfDNA is a reliable test for ruling out SCA, though false negatives have been reported. These findings are overall consistent with previously published studies.1,14,17 However, it is important to highlight that positive screening results should be confirmed with diagnostic testing considering the population prevalence of SCA and variable PPV, and that false positives have been reported.
Many published analyses of the accuracy of cfDNA screening for the common trisomies have relied on clinical exams or ultrasound for confirmation of screening results, as the common trisomies have strong patterns of phenotypic differences at birth and are usually readily diagnosed at delivery.51 Reliance on neonatal exam or ultrasound has limitations for SCA though, as these may manifest with milder or less characteristic features in the newborn relative to the common trisomies.52 While 45,X may present with a prenatal phenotype such as cystic hygroma or bicuspid aortic valve, the prenatal phenotypes for 47,XXY, 47,XYY or 47,XXX are more likely to be normal.53
Prenatal detection of SCAs is important, as it allows for genetic counseling and preparation for an infant that may have physical differences and unique health needs. For patients with 47,XXX, this may include assessment for hypotonia, monitoring of developmental milestones or evaluation for seizure activity.54 For pregnancies affected by 47,XXY, prenatal diagnosis offers the opportunity for genetic counseling and education on associated health outcomes such as androgen deficiency and neurocognitive differences. Implications of prenatal detection of SCAs and other genetic disorders extend beyond discussions of termination of pregnancy to detailed conversations about expected prognosis for those who choose to continue, as well as recurrence risk. Prenatal diagnosis also allows early referral to appropriate subspecialists at birth. In cases of monosomy X, for example, there is a life-long schedule of screening and treatment including early echocardiography.55 A 2019 survey of parents who underwent cfDNA with a confirmed SCA found that 88% of respondents felt the early referral and interventions were made possible by prenatal diagnosis, which had a beneficial effect on their child’s health.25
4.1 |. Strengths
There are several strengths of our approach to note. Data extraction, bias assessment and abstract and manuscript review were confirmed by two independent reviewers and adjudicated by a third reviewer. Unlike some previously published studies, we included only studies with confirmatory diagnostic testing. To further minimise reporting bias, we included only studies in which diagnostic testing was performed on >80% of study participants.17 Despite these restrictive selection criteria, the final analysis included over 12,000 cfDNA test results, significantly adding to the literature beyond recent published analysis.14
4.2 |. Limitations
There are also important limitations of our work to highlight. Because the existing literature on cfDNA has primarily included pregnancies at high pretest risk of aneuploidy, our results are less generalisable to pregnancies with low pretest risk of aneuploidy. The PPV for each SCA resulted in wide 95% confidence intervals, and PPV varied significantly based on estimated prevalence. Application of these results to post-test counseling may therefore pose challenges.23 We did not assess accuracy by sequencing the platform, although some evidence does suggest this may impact accuracy of the test.14 Finally, we did not include studies of multifetal gestations, although reporting of SCA for multifetal gestations was generally not available in the commercial setting during the time period defined in the study selection criteria of this analysis.
5 |. CONCLUSION
This systematic review and meta-analysis demonstrate that cfDNA testing for SCA has high sensitivity and specificity in the prenatal detection of 45,X, 47,XXY, 47,XYY, and 47,XXX, with NPV exceeding 99.99% for all SCA included. Further confirmatory studies in average risk pregnancies are needed, as these estimates of test performance are derived from pregnancies at high pretest risk for aneuploidy.
Supplementary Material
Key points.
What’s already known about this topic?
The test performance of cfDNA for SCAs are incompletely understood, with prior studies often based upon small cohorts and varying technologies.
What does this study add?
cfDNA for prenatal detection of sex chromosome aneuploidies is both highly sensitive and specific, but with differences in positive predictive value due to varying disease prevalence.
Further confirmatory studies in average risk pregnancies are needed, as these estimates of test performance are derived from the existing literature which largely reflects pregnancies at high pretest risk for aneuploidy.
ACKNOWLEDGMENTS
Dr. Sparks is supported by grant R01HD107190 from the NIH/NICHD, as well as by grants from the Doris Duke Charitable Foundation, the Chan Zuckerberg Biohub, and the Brianna Marie Foundation in collaboration with the Fetal Health Foundation. Dr. Norton is supported by grant U01HG009599 from the NIH/NHGRI. Dr. Jelin is supported by K23DK119949 from the NIH/NIDDK. The contents of the publication are solely the responsibility of the authors and do not necessarily represent the official views of the NIH.
Footnotes
CONFLICT OF INTEREST
The following authors report no conflict of interest: MAS, TS, ACJ, RG, KS, JB. MN has received prior research funding from Natera and is a clinical advisor to Luna Genetics.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
DATA AVAILABILITY STATEMENT
Data referenced in this article is freely available upon email request to the corresponding author matthew.shear@ucsf.edu.
REFERENCES
- 1.Badeau M, Lindsay C, Blais J, et al. Genomics-based Non-invasive Prenatal Testing for Detection of Fetal Chromosomal Aneuploidy in Pregnant Women. Cochrane Pregnancy and Childbirth Group; 2015. Cochrane Database Syst Rev [Internet] [cited 2022 Feb 2]; Available from:. 10.1002/14651858.CD011767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ge Y, Li J, Zhuang J, et al. Expanded noninvasive prenatal testing for fetal aneuploidy and copy number variations and parental willingness for invasive diagnosis in a cohort of 18, 516 cases. BMC Med Genom. 2021;14(1):106. 10.1186/s12920-021-00955-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu EKL, Hui WWI, Chiu RWK. cfDNA screening and diagnosis of monogenic disorders - where are we heading?: cfDNA screening and diagnosis of monogenic disorders. Prenat Diagn. 2018;38(1):52–58. 10.1002/pd.5207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu YJ, Zheng YR, Li L, et al. Diagnostic accuracy of non-invasive fetal RhD genotyping using cell-free fetal DNA: a meta analysis. J Matern Fetal Neonatal Med. 2014;27(18):1839–1844. 10.3109/14767058.2014.882306 [DOI] [PubMed] [Google Scholar]
- 5.Lo YMD, Corbetta N, Chamberlain PF, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350(9076):485–487. 10.1016/s0140-6736(97)02174-0 [DOI] [PubMed] [Google Scholar]
- 6.American College of Obstetricians and Gynecologists’ Committee on Practice Bulletins–Obstetrics. Committee on genetics, society for maternal-fetal medicine. Screening for fetal chromosomal abnormalities: ACOG practice bulletin, number 226. Obstet Gynecol. 2020;136(4):e48–e69. [DOI] [PubMed] [Google Scholar]
- 7.Familiari A, Boito S, Rembouskos G, et al. Cell-free DNA analysis of maternal blood in prenatal screening for chromosomal microdeletions and microduplications: a systematic review. Prenat Diagn. 2021;41(10):1324–1331. 10.1002/pd.5928 [DOI] [PubMed] [Google Scholar]
- 8.Dar P, Jacobsson B, Clifton R, et al. Cell-free DNA screening for prenatal detection of 22q11.2 deletion syndrome. Am J Obstet Gynecol. 2022. S0002937822000060 [DOI] [PubMed] [Google Scholar]
- 9.Dar P, Jacobsson B, MacPherson C, et al. Cell-free DNA screening for trisomies 21, 18, and 13 in pregnancies at low and high risk for aneuploidy with genetic confirmation. Am J Obstet Gynecol. 2022. S0002937822000412. [DOI] [PubMed] [Google Scholar]
- 10.Forabosco A, Percesepe A, Santucci S. Incidence of non-age-dependent chromosomal abnormalities: a population-based study on 88965 amniocenteses. Eur J Hum Genet. 2009;17(7):897–903. 10.1038/ejhg.2008.265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravholt CH, Juul S, Naeraa RW, Hansen J. Prenatal and postnatal prevalence of Turner’s syndrome: a registry study. BMJ. 1996;312(7022):16–21. 10.1136/bmj.312.7022.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mackie F, Hemming K, Allen S, Morris R, Kilby M. The accuracy of cell-free fetal DNA-based non-invasive prenatal testing in singleton pregnancies: a systematic review and bivariate meta-analysis. BJOG Int J Obstet Gynaecol. 2017;124(1):32–46. 10.1111/1471-0528.14050 [DOI] [PubMed] [Google Scholar]
- 13.Gil MM, Akolekar R, Quezada MS, Bregant B, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: meta-analysis. Fetal Diagn Ther. 2014;35(3):156–173. 10.1159/000358326 [DOI] [PubMed] [Google Scholar]
- 14.Soukkhaphone B, Lindsay C, Langlois S, Little J, Rousseau F, Reinharz D. Non-invasive prenatal testing for the prenatal screening of sex chromosome aneuploidies: a systematic review and meta-analysis of diagnostic test accuracy studies. Mol Genet Genomic Med. 2021;9(5). [Internet] [cited 2022 Feb 2];9(5). 10.1002/mgg3.1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liao H, Liu S, Wang H. Performance of non-invasive prenatal screening for fetal aneuploidy in twin pregnancies: a meta-analysis: NIPS for fetal trisomy 21, 18, and 13 screening for twins. Prenat Diagn. 2017;37(9):874–882. 10.1002/pd.5118 [DOI] [PubMed] [Google Scholar]
- 16.Geppert J, Stinton C, Johnson S, Clarke A, Grammatopoulos D, Taylor-Phillips S. Antenatal screening for fetal trisomies using microarray-based cell-free DNA testing: a systematic review and meta-analysis. Prenat Diagn. 2020;40(4):454–462. 10.1002/pd.5621 [DOI] [PubMed] [Google Scholar]
- 17.Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis: cell-free DNA in screening for aneuploidies. Ultrasound Obstet Gynecol. 2017;50(3):302–314. 10.1002/uog.17484 [DOI] [PubMed] [Google Scholar]
- 18.Gil MM, Galeva S, Jani J, et al. Screening for trisomies by cfDNA testing of maternal blood in twin pregnancy: update of the Fetal Medicine Foundation results and meta-analysis. Ultrasound Obstet Gynecol. 2019:uog.20284. 10.1002/uog.20284 [DOI] [PubMed] [Google Scholar]
- 19.Taylor-Phillips S, Freeman K, Geppert J, et al. Accuracy of non-invasive prenatal testing using cell-free DNA for detection of Down, Edwards and Patau syndromes: a systematic review and meta-analysis. BMJ Open. 2016;6(1):e010002. 10.1136/bmjopen-2015-010002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang B, Zhou Q, Chen Y, et al. High false-positive non-invasive prenatal screening results for sex chromosome abnormalities: are maternal factors the culprit? Prenat Diagn. 2020;40(4):463–469. 10.1002/pd.5529 [DOI] [PubMed] [Google Scholar]
- 21.Grati FR. Implications of fetoplacental mosaicism on cell-free DNA testing: a review of a common biological phenomenon. Ultrasound Obstet Gynecol Off J Int Soc Ultrasound Obstet Gynecol. 2016;48(4):415–423. 10.1002/uog.15975 [DOI] [PubMed] [Google Scholar]
- 22.Nieschlag E. Klinefelter syndrome: the commonest form of hypogonadism, but often overlooked or untreated. Dtsch Arzteblatt Int. 2013;110(20):347–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston M, Warton C, Pertile MD, et al. Ethical issues associated with prenatal screening using non-invasive prenatal testing for sex chromosome aneuploidy. Prenat Diagn. 2022. [DOI] [PubMed] [Google Scholar]
- 24.Samango-Sprouse C, Keen C, Sadeghin T, Gropman A. The benefits and limitations of cell-free DNA screening for 47, XXY (Klinefelter syndrome): Benefits and limitations of cell-free DNA screening for XXY. Prenat Diagn. 2017;37(5):497–501. 10.1002/pd.5044 [DOI] [PubMed] [Google Scholar]
- 25.Samango-Sprouse CA, Porter GF, Lasutschinkow PC, Tran SL, Sadeghin T, Gropman AL. Impact of early diagnosis and noninvasive prenatal testing (NIPT): Knowledge, attitudes, and experiences of parents of children with sex chromosome aneuploidies (SCAs). Prenat Diagn. 2020;40(4):470–480. 10.1002/pd.5580 [DOI] [PubMed] [Google Scholar]
- 26.Whiting PF. QUADAS-2: a Revised Tool for the Quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155(8):529. 10.7326/0003-4819-155-8-201110180-00009 [DOI] [PubMed] [Google Scholar]
- 27.Nyaga VN, Arbyn M, Aerts M. Metaprop: a Stata command to perform meta-analysis of binomial data. Arch Publ Health. 2014;72(1):39. 10.1186/2049-3258-72-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira JCP, Grati FR, Bajaj K, et al. Frequency of fetal karyotype abnormalities in women undergoing invasive testing in the absence of ultrasound and other high-risk indications. Prenat Diagn. 2016;36(12):1146–1155. 10.1002/pd.4951 [DOI] [PubMed] [Google Scholar]
- 29.Ferguson-Smith MA, Yates JR. Maternal age specific rates for chromosome aberrations and factors influencing them: report of a collaborative european study on 52 965 amniocenteses. Prenat Diagn. 1984;4(7):5–44. Spec No:5–44. 10.1002/pd.1970040704 [DOI] [PubMed] [Google Scholar]
- 30.Bianchi DW, Platt LD, Goldberg JD, Abuhamad AZ, Sehnert AJ, Rava RP. Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol. 2012;119(5):890–901. 10.1097/aog.0b013e318258c419 [DOI] [PubMed] [Google Scholar]
- 31.Bianchi DW, Prosen T, Platt LD, et al. Massively parallel sequencing of maternal plasma DNA in 113 cases of fetal nuchal cystic hygroma. Obstet Gynecol. 2013;121(5):1057–1062. 10.1097/aog.0b013e31828ba3d8 [DOI] [PubMed] [Google Scholar]
- 32.Guseh S, Wilkins-Haug L, Kaimal A, et al. Utility of noninvasive genome-wide screening: a prospective cohort of obstetric patients undergoing diagnostic testing. Genet Med. 2021;23(7):1341–1348. 10.1038/s41436-021-01147-4 [DOI] [PubMed] [Google Scholar]
- 33.Hooks J, Wolfberg AJ, Wang ET, et al. Non-invasive risk assessment of fetal sex chromosome aneuploidy through directed analysis and incorporation of fetal fraction. Prenat Diagn. 2014;34(5):496–499. 10.1002/pd.4338 [DOI] [PubMed] [Google Scholar]
- 34.Lau TK, Chen F, Pan X, et al. Noninvasive prenatal diagnosis of common fetal chromosomal aneuploidies by maternal plasma DNA sequencing. J Matern Fetal Neonatal Med. 2012;25(8):1370–1374. 10.3109/14767058.2011.635730 [DOI] [PubMed] [Google Scholar]
- 35.Lefkowitz RB, Tynan JA, Liu T, et al. Clinical validation of a noninvasive prenatal test for genomewide detection of fetal copy number variants. Am J Obstet Gynecol. 2016;215(2):227.e1–227.e16. 10.1016/j.ajog.2016.02.030 [DOI] [PubMed] [Google Scholar]
- 36.Li H, Lei Y, Zhu H, et al. The application of NIPT using combinatorial probe-anchor synthesis to identify sex chromosomal aneuploidies (SCAs) in a cohort of 570 pregnancies. Mol Cytogenet. 2018;11(1):59. 10.1186/sl3039-018-0407-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liang D, Lv W, Wang H, et al. Non-invasive prenatal testing of fetal whole chromosome aneuploidy by massively parallel sequencing: whole chromosome NIPT by MPS. Prenat Diagn. 2013;33(5): 409–415. 10.1002/pd.4033 [DOI] [PubMed] [Google Scholar]
- 38.Mazloom AR, Džakula Ž, Oeth P, et al. Noninvasive prenatal detection of sex chromosomal aneuploidies by sequencing circulating cell-free DNA from maternal plasma: noninvasive prenatal detection of sex chromosome aneuploidy by MPS. Prenat Diagn. 2013;33(6):591–597. 10.1002/pd.4127 [DOI] [PubMed] [Google Scholar]
- 39.Meck JM, Kramer Dugan E, Matyakhina L, et al. Noninvasive prenatal screening for aneuploidy: positive predictive values based on cytogenetic findings. Am J Obstet Gynecol. 2015;213(2):214.e1–214.e5. 10.1016/j.ajog.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 40.Nicolaides KH, Syngelaki A, Gil M, Atanasova V, Markova D. Validation of targeted sequencing of single-nucleotide polymorphisms for non-invasive prenatal detection of aneuploidy of chromosomes 13, 18, 21, X, and Y: validation study of cfDNA testing using SNPs. Prenat Diagn. 2013;33(6):575–579. 10.1002/pd.4103 [DOI] [PubMed] [Google Scholar]
- 41.Nicolaides KH, Musci TJ, Struble CA, Syngelaki A, del Mar Gil M. Assessment of fetal sex chromosome aneuploidy using directed cell-free DNA analysis. Obstet Gynecol Surv. 2014;69(5):249–250. 10.1097/01.ogx.0000450115.09778.f5 [DOI] [PubMed] [Google Scholar]
- 42.Pergament E, Cuckle H, Zimmermann B, et al. Single-nucleotide polymorphism–based noninvasive prenatal screening in a high-risk and low-risk cohort. Obstet Gynecol. 2014;124(2):210–218. 10.1097/aog.0000000000000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Persico N, Boito S, Ischia B, et al. Cell-free DNA testing in the maternal blood in high-risk pregnancies after first-trimester combined screening: cell-free DNA testing and combined screening. Prenat Diagn. 2016;36(3):232–236. 10.1002/pd.4773 [DOI] [PubMed] [Google Scholar]
- 44.Petersen AK, Cheung SW, Smith JL, et al. Positive predictive value estimates for cell-free noninvasive prenatal screening from data of a large referral genetic diagnostic laboratory. Am J Obstet Gynecol. 2017;217(6):691.e1–691.e6. 10.1016/j.ajog.2017.10.005 [DOI] [PubMed] [Google Scholar]
- 45.Porreco RP, Garite TJ, Maurel K, et al. Noninvasive prenatal screening for fetal trisomies 21, 18, 13 and the common sex chromosome aneuploidies from maternal blood using massively parallel genomic sequencing of DNA. Am J Obstet Gynecol. 2014;211(4):365.e1–365.e12. 10.1016/j.ajog.2014.03.042 [DOI] [PubMed] [Google Scholar]
- 46.Porreco RP, Sekedat M, Bombard A, et al. Evaluation of a novel screening method for fetal aneuploidy using cell-free DNA in maternal plasma. J Med Screen. 2020;27(1):1–8. 10.1177/0969141319873682 [DOI] [PubMed] [Google Scholar]
- 47.Sehnert AJ, Rhees B, Comstock D, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin Chem. 2011;57(7):1042–1049. 10.1373/clinchem.2011.165910 [DOI] [PubMed] [Google Scholar]
- 48.Shubina J, Trofimov DY, Barkov IY, et al. In silico size selection is effective in reducing false positive NIPS cases of monosomy X that are due to maternal mosaic monosomy X. Prenat Diagn. 2017;37(13):1305–1310. 10.1002/pd.5178 [DOI] [PubMed] [Google Scholar]
- 49.Song Y, Huang S, Zhou X, et al. Non-invasive prenatal testing for fetal aneuploidies in the first trimester of pregnancy. Ultrasound Obstet Gynecol. 2015;45(1):55–60. 10.1002/uog.13460 [DOI] [PubMed] [Google Scholar]
- 50.Zhu Y, Shan Q, Zheng J, et al. Comparison of Efficiencies of non-invasive prenatal testing, karyotyping, and chromosomal Microarray for diagnosing fetal chromosomal anomalies in the Second and third Trimesters. Front Genet. 2019;10:69. 10.3389/fgene.2019.00069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones K, Jones M, del Campo M. Smith’s Recognizable Patterns of Human Malformation. 8th ed. Elsevier, Inc; 2021. [Google Scholar]
- 52.Linden MG, Bender BG, Robinson A. Genetic counseling for sex chromosome abnormalities. Am J Med Genet. 2002;110(1):3–10. 10.1002/ajmg.10391 [DOI] [PubMed] [Google Scholar]
- 53.Swanson K, Bishop JC, Al-Kouatly HB, et al. Prenatal phenotype of 47, XXY (Klinefelter syndrome). Prenat Diagn. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Otter M, Schrander-Stumpel CT, Curfs LM. Triple X syndrome: a review of the literature. Eur J Hum Genet. 2010;18(3):265–271. 10.1038/ejhg.2009.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gravholt CH, Andersen NH, Conway GS, et al. Clinical practice guidelines for the care of girls and women with Turner syndrome: proceedings from the 2016 Cincinnati International Turner Syndrome Meeting. Eur J Endocrinol. 2017;177(3):G1–G70. 10.1530/eje-17-0430 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data referenced in this article is freely available upon email request to the corresponding author matthew.shear@ucsf.edu.


