ABSTRACT
Clostridioides difficile infection (CDI) is associated with high recurrence rates that have substantial effects on patients’ quality of life. To investigate the risk factors and potential mechanisms contributing to recurrent CDI (rCDI), a total of 243 cases were enrolled in this study. The history of omeprazole (OME) medication and ST81 strain infection were considered the two independent risks with the highest odds ratios in rCDI. In the presence of OME, we detected concentration-dependent increases in the MIC values of fluoroquinolone antibiotics against ST81 strains. Mechanically, OME facilitated ST81 strain sporulation and spore germination by blocking the pathway of purine metabolism and also promoted an increase in cell motility and toxin production by turning the flagellar switch to the ON state. In conclusion, OME affects several biological processes during C difficile growth, which have fundamental impacts on the development of rCDI caused by ST81 strains. Programmed OME administration and stringent surveillance of the emerging ST81 genotype are matters of considerable urgency and significance in rCDI prevention.
KEYWORDS: omeprazole, Clostridioides difficile infection, recurrence, ST81, regulatory mechanism
INTRODUCTION
Clostridioides difficile, a Gram-positive spore-forming anaerobic bacillus, is among the most common pathogens responsible for antibiotic-associated diarrhea and pseudomembranous colitis. During the initial episode, between 15% and 30% of infected patients experience recurrent C. difficile infection (rCDI) within 8 weeks of successful antimicrobial therapy, and this proportion increases significantly with an increase in the number of recurrences (1). Consequently, identifying risk factors for rCDI is essential for early detection, treatment, and prevention.
In the past few decades, the incidence and mortality associated with CDI have increased markedly worldwide, which can be attributed to multiple contributory factors, including the aging population, prolonged hospital stays, and the abuse of antibiotics and immunosuppressive drugs; the latter have contributed, at least in part, to the massive outbreak of hypervirulent BI/NAP1/027 strains in Europe and North America (2). In China, however, CDI has been characterized by somewhat different epidemiological trends. According to the statistical data from multilocus sequence typing (MLST) in a retrospective study, substantial changes in the proportions of the sequence types (STs) of CDI-prevalent strains have occurred during the past 10 years, with ST81 gradually replacing ST35, ST3, and ST37 and becoming established as the predominant ST in the majority of cities throughout mainland China (3, 4). In addition to efficient colonization, survival, sporulation, and motility (5), ST81 strains have also been demonstrated to exhibit higher resistance to several antibiotics than other strains (6), which has presumably contributed to the current prevalence of ST81 in most rCDI cases.
Proton pump inhibitors (PPIs) are a class of drugs that are commonly used for the treatment of peptic ulcers, bleeding, and gastroesophageal reflux diseases, based on their irreversible inhibition of the activities of H+/K+-ATPase in gastric parietal cells. Compared with other PPIs, omeprazole (OME) is universally acknowledged to be among the most frequently administered PPIs and is currently recommended by most relevant guidelines (7). However, although PPIs have long been used in clinical practice, the findings of a growing number of studies have indicated a close association between certain adverse effects, including infections, fractures, dementia, and renal impairment, and the prolonged use of these agents (8). In recent years, there has been a growth in interest regarding the effects of PPIs on the bactericidal potency of antibiotics. For example, it has been reported that the long-term administration of OME can increase the MIC values of tigecycline against Enterococcus spp. by 4- to 128-fold (9). Contrastingly, however, the findings of another study have indicated that OME and rabeprazole interact synergistically with fluconazole to promote the eradication of Candida albicans by inhibiting biofilm formation, drug efflux pumps, and phospholipase activity (10).
Given the current increase in the rates of CDI and its recurrence caused by ST81 strains nationwide, as well as the diverse effects of PPIs on the bactericidal efficacy of antimicrobial drugs, in this study, we assessed changes in the MIC values of common antibiotics against ST81 strains with or without OME treatment. In addition, we aimed to elucidate the underlying mechanisms with respect to the effects of OME on the sporulation, spore germination, motility, and toxin production of C. difficile, which we anticipate will provide an experimental foundation for further in-depth analysis of the recurrence of CDI in the clinical setting.
RESULTS
Recurrence and survival rates of patients are related to ST81 strain infection and OME administration.
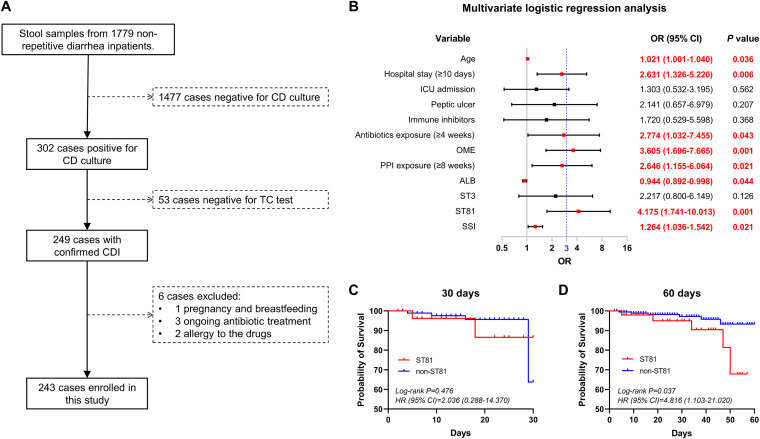
A total of 249 patients with confirmed CDI were included from among 1,779 consecutive patients with diarrhea, excluding those negative by C. difficile culture and toxigenic culture (TC) tests. After excluding 6 patients with pregnancy, breastfeeding, ongoing antibiotic treatment, and drug allergy, 243 patients were eventually enrolled in this study (Fig. 1A). These patients were subsequently divided into rCDI and non-rCDI groups according to the outcomes within 8 weeks after discontinuing anti-CDI treatment. On the basis of the crude odds ratio (OR) and P values obtained in univariate analysis, 12 of 51 variables were incorporated into a multivariate logistics regression model (see Table S1 in the supplemental material). As shown in the forest plot, patients with rCDI were more likely to be characterized by an advanced age, long hospital stay (≥10 days), low concentration of serum albumin, high severity score index, and ST81 strain infection. In addition, the histories of antibiotic exposure (≥4 weeks), OME treatment, and PPI exposure (≥8 weeks) were also regarded as independent risk factors associated with CDI recurrence. Of these, a history of OME treatment (OR = 3.605, P = 0.001) and ST81 strain infection (OR = 4.175, P = 0.001) were the only two variables with OR values greater than 3 (Fig. 1B). Constructed Kaplan-Meier curves further confirmed that patients infected with an ST81 strain were more likely to have lower survival rates than those infected with non-ST81 strains over a 60-day observation period, whereas we detected no statistically significant difference between patients from the two groups with respect to 30-day survival (Fig. 1C and D).
FIG 1.
Analysis of the medical records of 243 patients with Clostridioides difficile infection. (A) Flow diagram of the procedures for patient inclusion. (B) The odds ratio (OR) and 95% confidence interval (CI) were calculated based on binary logistics regression analysis and then used to screen the independent risk factors underlying the recurrence of Clostridioides difficile infection (CDI). Variables differing significantly (P < 0.05) are highlighted in bold red font. OME, omeprazole; ALB, serum albumin; SSI, severity score index. (C and D) Kaplan-Meier survival curves of patients with or without ST81 strain infection up to 30 and 60 days after the initial CDI diagnosis. The hazard ratio (HR), 95% CI, and P value were calculated based on a log rank test and then used to determine the statistical difference between two populations.
OME attenuates the bactericidal effects of fluoroquinolone antibiotics on ST81 strains.
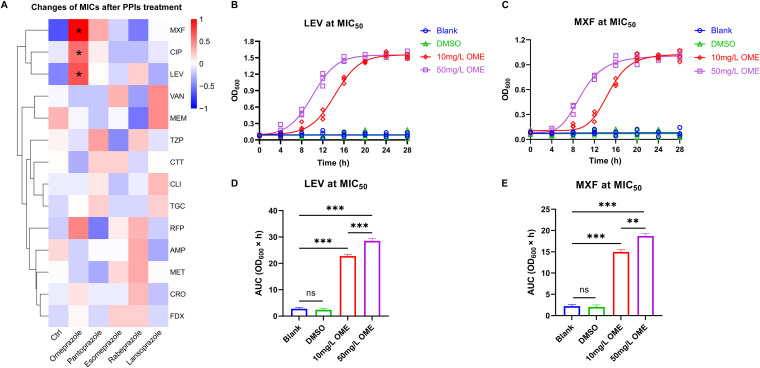
To investigate the bactericidal effects of antibacterial drugs on ST81 strains subsequent to PPI treatment, we initially performed an in vitro combined antimicrobial susceptibility test. As shown in Fig. 2A, among the 14 assessed antimicrobial agents, only the fluoroquinolones (moxifloxacin [MXF; t = 2.626, P = 0.047], ciprofloxacin [CIP; t = 3.162, P = 0.025], and levofloxacin [LEV; t = 2.926, P = 0.033]) promoted a marked increase in the MICs against ST81 strains after OME treatment at a concentration of 50 mg/L. Furthermore, in terms of the results of growth kinetics assays, we detected significant increases in the area under the curve (AUC) of LEV-sensitive (Fig. 2B and D) and MXF-sensitive (Fig. 2C and E) ST81 strains in response to OME treatment, thereby implying that OME also endows ST81 strains with an enhanced resistance to fluoroquinolone antibiotics in a dose-dependent manner. Nevertheless, apart from the triple-site missense mutation of GyrA (T82I) plus GyrB (S366A, D426V) inherent in all ST81 strains, we identified no other new mutation site in response to OME treatment.
FIG 2.
Omeprazole induces ST81 strain resistance to fluoroquinolone antibiotics. (A) Changes in the MICs of antimicrobial agents against ST81 strains following PPI treatment are displayed in a heatmap. Data shown in the heatmap were converted to the log2 geometric mean values of MICs and then clustered by the average-linkage hierarchical algorithm based on Euclidean distance measurements. (B and C) The growth curves of ST81 strains were plotted by recording the OD600 values at fixed time points under LEV (B) and MXF (C) treatment at MIC50. (D and E) The quantitative results in the bar graphs (D and E) show the area under the curve (AUC) of each group in panels B and C, respectively. Statistical analysis was performed using Student’s t test. All data are presented as the mean ± SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
OME stimulates the sporulation and spore germination of ST81 strains.
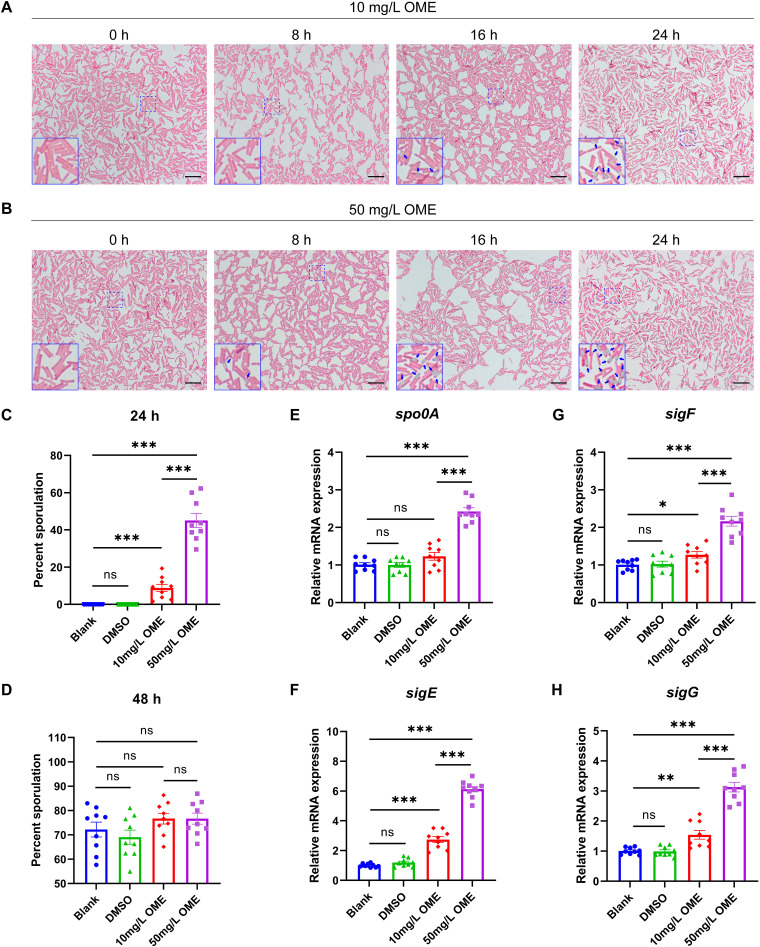
To examine the influence of OME on the spore homeostasis of ST81 strains, we initially compared the spore-forming capacities of ST81 strains from different groups based on Schaeffer-Fulton staining. In line with expectations, OME was found to accelerate spore maturation, although it had no appreciable influence on the gross morphology of bacterial cells. Mature spores were observed at 16 and 8 h after the administration of OME at 10 and 50 mg/L, respectively (Fig. 3A and B). Given that some of the visual bodies with strong light refractivity were not viable spores, we subsequently assessed the effects of OME treatment on the sporulation efficiency of ST81 strains for a more precise calculation. Results obtained from the heat resistance assay revealed a clear increase in the percentage of sporulation among bacterial cells that had been exposed to OME treatment for 24 h in a concentration-dependent manner (Fig. 3C; Fig. S1A). However, given that the culture time was extended to 48 h, we detected no significant differences between the groups in this regard (Fig. 3D; Fig. S1B). Thus, these findings indicate that in ST81 strains, although OME shortens the period of spore maturation, it does not have any pronounced effects on overall sporulation capacity. In terms of transcriptional regulation, we established that OME has no significant effect on the expression of spo0A, a master regulator of sporulation, at 10 mg/L, although it did promote a marked upregulation in mRNA levels at 50 mg/L (Fig. 3E). Similarly, we detected significant increases in the mRNA expression of sigE, sigF, and sigG, the target genes of phosphorylated Spo0A, in response to OME intervention at both 10 and 50 mg/L (Fig. 3F to H).
FIG 3.
Omeprazole promotes spore formation in ST81 strains. (A and B) Spore formation was examined using Schaeffer-Fulton staining following treatment with 10 and 50 mg/L omeprazole (OME) for the indicated times. The spores are stained light green, whereas the remainder of the bacterial cells and the vegetative cells are stained pink. The blue arrows in the magnified image of the bottom left corner indicate mature spores. Bar, 10 μm. (C and D) Percent sporulation of the ST81 strains with or without OME treatment was calculated based on a heat resistance assay after incubation for 24 and 48 h. (E to H) Relative mRNA levels of spo0A, sigE, sigF, and sigG were analyzed by qRT-PCR, with quantitative analysis being performed following normalization using the rrn gene. Nine ST81 strains were selected as biological replicates. Statistical analysis was performed using Student’s t test. All data are presented as the mean ± SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
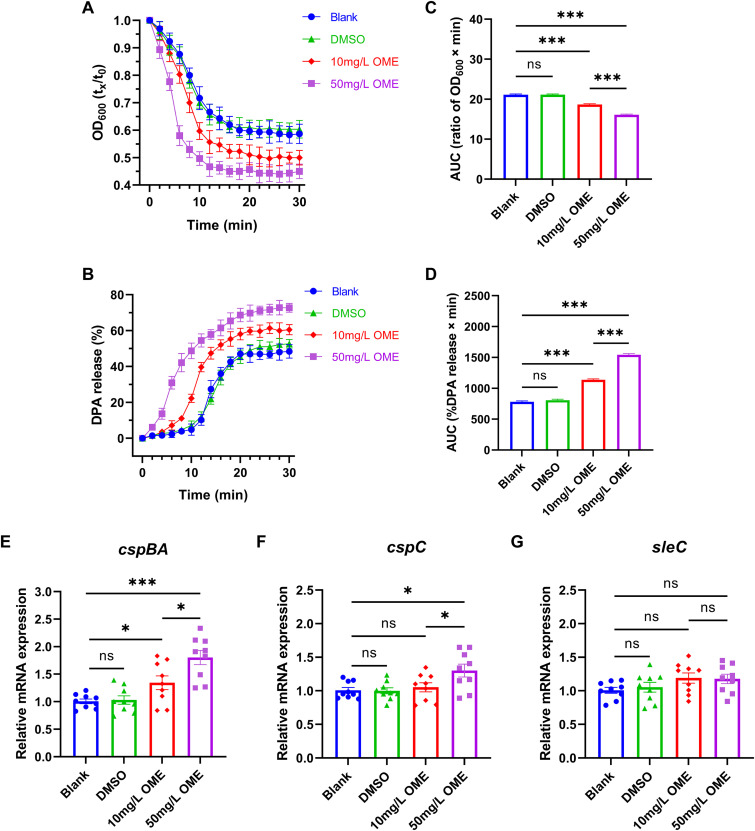
We also monitored the overall spore germination process by measuring the values of optical density at 600 nm (OD600) of pure spore suspensions incubated under different conditions. Not surprisingly, in response to OME treatment, we detected a rapid reduction in optical density (OD) within the first 10 min, which is indicative of spore revival. This reduction was most evident in the group treated with 50 mg/L OME (F = 631.7, P < 0.001) (Fig. 4A and C). Meanwhile, we established that OME promoted a more complete release of dipicolinic acid (DPA) during spore germination, which indicates that OME-treated spores germinated more readily than those subjected to the blank and solvent (dimethyl sulfoxide [DMSO]) control treatments (F = 1332.0, P < 0.001) (Fig. 4B and D). Furthermore, we found that 50 mg/L OME administration promoted a significant upregulation of the gene expression of cspBA and cspC, two members of the cspBAC locus that are primarily involved in signal transduction for spore germination, whereas it had no demonstrable effects on the mRNA levels of the cortex lytic enzyme-encoding gene sleC. Comparatively, we observed that only the mRNA level of cspBA was elevated in response to OME administered at 10 mg/L, which tends to provide evidence indicating that the effects of OME in activating spore germination of ST81 strains are mainly mediated by the amino acid cogerminant receptor CspBA (Fig. 4E to G).
FIG 4.
Facilitation of spore germination in ST81 strains by omeprazole. (A) Spore germination was monitored by plotting the ratio of the OD600 at a given time to the OD600 at time zero. (B) The dipicolinic acid (DPA) release rate of germinating spores was determined using Tb3+ fluorescence in the indicated time frame and then calculated according to the given formula. (C and D) The quantitative results presented in the bar graphs (C and D) show the area under the curve (AUC) of each group in panels A and B, respectively. (E to G) Relative mRNA levels of cspBA, cspC, and sleC were analyzed by qRT-PCR, with quantitative analysis performed following normalization using the rrn gene. Nine ST81 strains were selected as biological replicates. Statistical analysis was performed using Student’s t test. All data are presented as the mean ± SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
OME influences the purine metabolism of ST81 strains.
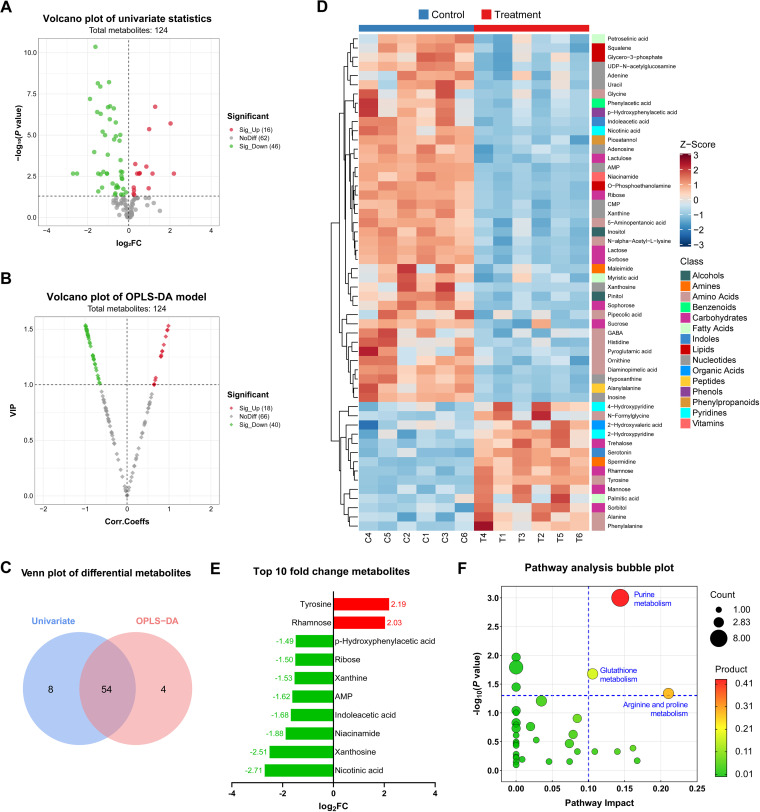
We sought to determine the potential mechanisms of how OME regulates the sporulation and spore germination of ST81 strains by performing untargeted metabolomics profiling. The quality control (QC) results revealed that the fluctuating dots in a multivariate control chart (MCC) were restricted to within the control limits (Fig. S2A) and were distinctly clustered into groups corresponding to the different treatments (Fig. S2B and C). The results of a permutation test further confirmed that no overfitting was detected in this model (Fig. S2D). In this study, we identified a total of 124 metabolites, which were distributed among 10 general classes, of which amino acids (n = 28, 22.58%), carbohydrates (n = 18, 14.52%), and fatty acids (n = 16, 12.90%) were the most abundant (Fig. S2E). For differential metabolite selection, we selected 62 and 58 candidate metabolites with threshold P values of less than 0.05 based on univariate analysis (Fig. 5A) and variable importance in projection (VIP) scores greater than 0 based on an orthogonal partial least-square discriminant analysis (OPLS-DA) model (Fig. 5B), respectively. Following pairwise comparisons based on Venn diagram analysis, 54 overlapping differential metabolites were filtered (Fig. 5C), which were considered potential biomarkers for the response to OME treatment, the expression and classification of which are presented in Fig. 5D. As shown, the majority of these metabolites (n = 40, 74.07%) were downregulated in the OME treatment group, most of which were nucleotides (n = 10, 25.00%), amino acids (n = 9, 22.50%), and carbohydrates (n = 6, 15.00%). Furthermore, among the top 10 metabolites, we detected significant reductions in four-fifths, with the largest absolute values of log2 fold change. Among these, eight components with reduced expression, namely, AMP, adenosine, adenine, inosine, hypoxanthine, xanthine, xanthosine, and glycine, were detected in the purine metabolic pathway (Fig. 5E). According to our phylogenetic analysis based on 16S rRNA sequences, the ST81 strain of C. difficile and IAM 12118 strain of Bacillus subtilis have a relatively close evolutionary relationship. Thus, given the absence of C. difficile accessions in the KEGG database species library, we used Bacillus subtilis as a substitute in subsequent metabolic pathway analyses (Fig. S3). The results shown in Fig. 5F indicate that three pathways were enriched based on the cutoff values (pathway impact > 0.10 and P < 0.05) that are widely applied in multiple comparisons to exclude confounders (11, 12). Compared with the other two pathways, we identified purine metabolism as the pathway most susceptible to OME treatment, based on the highest product of values corresponding to the horizontal and vertical coordinates.
FIG 5.
Mechanistic studies of omeprazole in the regulation of sporulation and spore germination based on metabonomic analysis. (A and B) Differential metabolites were filtered by a volcano plot of univariate statistics and the orthogonal partial least-square discriminant analysis (OPLS-DA) model. Red, green, and gray dots denote metabolites with significant upregulation, significant downregulation, and nonsignificant change, respectively, after omeprazole (OME) treatment. log2FC, log2 fold change. (C) The Venn plot illustrates the numbers of differential metabolites identified using a single model and their cooperation. (D) The 54 potential biomarkers are displayed in a Z-score heatmap combined with hierarchical cluster analysis. The annotated blocks in columns and rows indicate the groups (control and OME treatment) and the classes of metabolites, respectively. GABA, gamma-aminobutyric acid. (E) Histogram of the top 10 metabolites with the largest absolute fold change values among the 54 potential biomarkers after the binary logarithm conversion. Red and green represent up- and downregulation, respectively. (F) Pathway analysis bubble plot of the Bacillus subtilis (KEGG) set. The size of the bubbles is proportional to the number of enriched metabolites in the corresponding pathway. The color represents the products of the values in the abscissa and ordinate. Only those bubbles in the upper-right quadrant (pathway impact > 0.1 and P < 0.05) are labeled with pathway names.
A bidirectional flagellar switch was identified in ST81 strains.
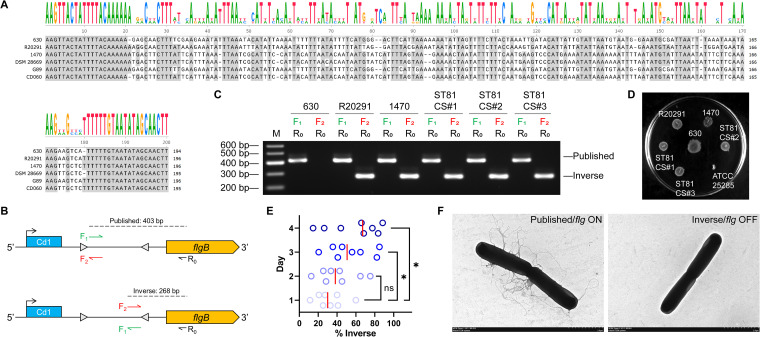
It has been reported that the flagellum and toxin biosynthesis are phase variables mediated via site-specific recombination that inverts a DNA element termed the flagellar switch, which is involved in the regulation of the flgB operon (the early-stage flagellar genes) and downstream toxin gene expression via complex mechanisms involving posttranscriptional initiation (13). Multiple sequence alignments (shown in Fig. 6A) indicate that the flagellar switch consists of 20-bp inverted repeats (IRs) on each flank and a central sequence of ~150 bp. Whereas the IRs are evolutionarily highly conserved, the intervening sequences are characterized by strain-dependent variation. Interestingly, even though strains G89 and CD060 are classified as ST81, they have different intervening sequences. On the basis of these in silico findings, we defined sequences in the published genome of the given strains as the “published orientation” and the sequences with an inversion between the IRs as the “inverse orientation.” In orientation-specific PCR analyses, we used a common reverse primer, R0, complementary to the coding sequence of the flgB gene, in conjunction with the forward primer F1 (annealing immediately 3′ of the left-hand IRs) or F2 (annealing immediately 5′ of the right-hand IRs) to distinguish the flagellar switch in the published and inverse orientations, respectively (Fig. 6B). In line with expectations, all strains, with the exception of 630, yielded products with both orientations when grown in brain heart infusion-supplemented (BHIS) broth (Fig. 6C) and exhibited rough colonies with filamentous edges when grown on motility medium for 48 h (Fig. 6D). On the basis of these observations, we speculated that the switch orientation might influence the formation of flagella. To confirm this assumption, we next quantified the frequencies of switch orientation in the enriched flagellar phase-variant populations. Compared with day 1 (median = 30.04%), the proportion of colonies with the flagellar switch in the inverse orientation rose steadily after 2 (median = 38.12%, t = 0.960, P = 0.354), 3 (median = 50.60%, t = 2.242, P = 0.042), and 4 (median = 67.07%, t = 2.392, P = 0.031) days of cultivation, which indicates that growth on a solid agar surface favors the proliferation of C. difficile with the flagellar switch in the inverse orientation (Fig. 6E). The results of transmission electron microscopy (TEM) further confirmed that most bacteria from colonies with the flagellar switch in the published orientation developed flagella (termed “flg ON” here), whereas bacteria from colonies with the switch in the inverse orientation were largely aflagellate (termed “flg OFF” here) (Fig. 6F). Collectively, these findings indicate that both the flg ON and OFF states are detected in ST81 strains irrespective of the growth medium.
FIG 6.
ST81 strains have the flagellar switch in both ON and OFF phases. (A) Nucleotide sequences corresponding to the 5′ untranscribed region (UTR) of the flgB operon from genome sequences available for the indicated strains were aligned using the T-Coffee server. Shading indicates identical nucleotides. The height of the letters is positively associated with the conservation of the residues at the respective positions. (B) Diagram of the PCR strategy applied to determine the putative flagellar switch orientation. Cd1 indicates the c-di-GMP riboswitch. The reverse primer R0 was used with either the forward primer F1 or the forward primer F2 to characterize the flagellar switch in published or inverse orientation, respectively. The predicted product sizes are based on published sequences of ST81 strain G89. The diagram is not drawn to scale. (C) Orientation-specific PCR products using the genomic DNA of three standard strains (630, R20291, and 1470) from different STs and three clinical strains from ST81 (CS#1 to CS#3) as the templates. M represents DNA ladders. (D) Discrepancy in swimming motilities among the indicated strains after incubation for 48 h. ATCC 25285 served as a negative control. The image is representative of three technical replicates. (E) The percentage of bacteria in each colony with the flagellar switch in the inverse orientation was calculated based on quantitative orientation-specific PCR during 4 days of cultivation. The red lines indicate medians. Statistical analysis was performed using Student’s t test. *, P < 0.05; ns, not significant. (F) Representative transmission electron micrographs show the presence and absence of flagella with the switch in the ON and OFF orientation, respectively. Bar, 2 μm.
The OME-mediated switch to flagellar phase ON increases the motility and toxin production of ST81 strains.
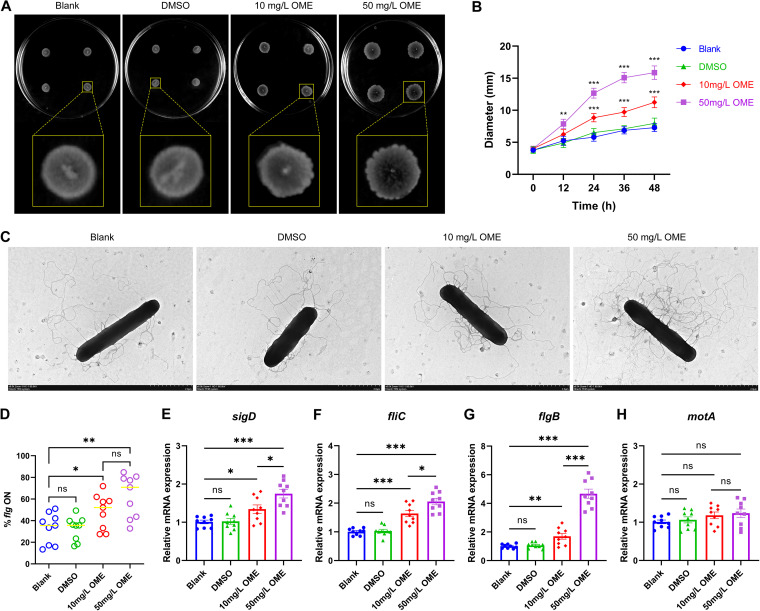
To elucidate the association, if any, between the administration of OME and the flagellar phase switch in ST81 strains, we initially compared the swimming motilities of ST81 strains in the presence and absence of OME. We found that bacteria exposed to OME had a significant increase in the diameter of expanding colonies, with a corresponding change in colony morphology from smooth and circular to spreading and filamentous during the 48-h time frame of the assay (Fig. 7A and B). Additionally, with an increase in the concentration of OME, flagellar growth tended to be more prolific, and perhaps more corkscrew-like, as revealed in transmission electron micrographs (Fig. 7C). Given that bacteria in the flg ON state were sufficient with respect to flagellar biosynthesis (Fig. 6F), we speculated that the vigorous motility of bacteria treated with OME may have been mediated via a switch to flagellar phase ON. Indeed, we found that OME treatment coincided with higher frequencies of the bacterial population in the flg ON state (Fig. 7D). In a subsequent quantitative real-time PCR (qRT-PCR) assay, we found that OME promoted a significant upregulation in the relative transcript abundances of sigD (flagellar alternative sigma factor), fliC (flagellin filament), and flgB (rod of flagellar basal body), particularly when administered at a concentration of 50 mg/L. Conversely, we detected no comparable effects on the mRNA levels of motA (stator of flagellar motor) (Fig. 7E to H).
FIG 7.
The motility of ST81 strains is enhanced in response to omeprazole treatment. (A) Motility of the ST81 strains in swimming agar following incubation for 48 h. Four technical replicates were stab inoculated into each plate. Representative images of the partial magnification below show the bacterial spot in detail. (B) The swimming diameters on the plate were measured at 12-h intervals over a 2-day period. (C) The flagella of ST81 strains were observed using transmission electron microscopy after negative staining. Bar, 2 μm. (D) Frequencies of bacterial population with flg ON were measured based on quantitative orientation-specific PCR. The yellow lines indicate medians. (E to G) Relative mRNA levels of sigD, fliC, flgB, and motA were analyzed by qRT-PCR, with quantitative analysis being performed following normalization using the rrn gene. Nine ST81 strains were selected as biological replicates. Statistical analysis was performed using Student’s t test. All data are presented as the mean ± SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
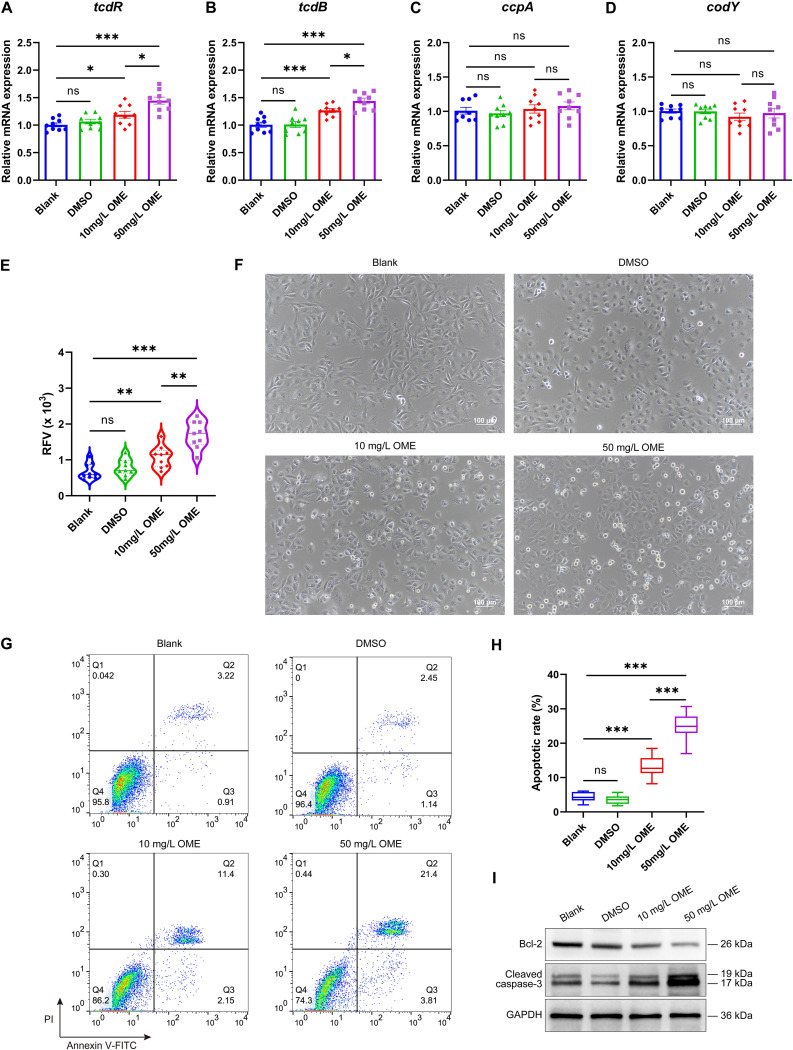
To further establish whether the upregulation of sigD mRNA mediated by OME also influences the toxin production of ST81 strains, we subsequently compared the gene expression levels of tcdR (toxin-specific sigma factor), tcdB (encoding cytotoxin of C. difficile), ccpA (global regulator of carbon catabolite), and codY (global regulator of virulence and nutrition) before and after OME treatment using qRT-PCR. As anticipated, we detected significant increases in the abundances of tcdR and tcdB transcripts in response to treatment with either 10 or 50 mg/L OME; in contrast, the abundances of ccpA and codY transcripts remained unchanged (Fig. 8A to D). As in the case of tcdR and tcdB transcripts, we detected significant increases in the protein levels of toxins A and B following OME treatment (Fig. 8E). An apparent cytopathic effect (CPE) on Vero cells was observed following the exposure of these cells to diluted medium supernatants of bacteria treated with OME (Fig. 8F). Specifically, we detected apoptotic rates of 13.19% ± 3.21% and 24.92% ± 3.93% among cells exposed to 10 and 50 mg/L OME, respectively, which were considerably higher than those detected in blank (4.33% ± 1.39%) and solvent (DMSO) (3.58% ± 1.29%) control groups (both P < 0.01) (Fig. 8G and H). Consistently, Western blot analysis revealed that the toxins secreted by OME-treated bacteria downregulated expression of the antiapoptotic protein Bcl-2 and upregulated expression of the proapoptotic protein cleaved caspase-3 (Fig. 8I). Collectively, these findings provide evidence to indicate that OME is a positive driver of the biosynthesis of flagella and toxins, which is mediated by the turning on of the flagellar switch.
FIG 8.
Omeprazole promotes the toxin production of ST81 strains. (A to D) Relative mRNA levels of tcdR, tcdB, ccpA, and codY were analyzed by qRT-PCR, with quantitative analysis being performed following normalization using the rrn gene. (E) Protein levels of toxins A and B were determined based on relative fluorescence values (RFVs) using an enzyme-linked fluorescent assay. Quantitative analysis was performed after normalization with the standard RFVs obtained from calibration. (F) The cellular morphology of Vero cells was observed using phase-contrast microscopy. Bar, 100 μm. (G and H) The apoptotic rates were examined using flow cytometry, and the statistical results are presented in the box plot. PI, propidium iodide; FITC, fluorescein isothiocyanate. (I) The expression of apoptosis-related proteins was determined by Western blotting. Nine ST81 strains were selected as biological replicates. Shown are representative images of three independent experiments. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. Statistical analysis was performed using Student’s t test. All data are presented as the mean ± SD from three independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not significant.
DISCUSSION
C. difficile infection is a condition associated with high morbidity and mortality, as well as representing a considerable financial burden to both health care systems and societies in general, which can mainly be attributed to the high rate of recurrence. With recent rapid advances in medical technology, a progressive reduction in the mortality of those with CDI has been achieved over the past decade, although the rates of CDI recurrence have remained relatively stable (14). In the past, considerable effort was expended in attempts to establish the factors contributing to the persistent recurrence of this disease. A breakthrough, however, came in 2010, with the findings of a study conducted by Linsky et al., who were the first to consider the possibility that PPI therapy might represent a potential risk factor for rCDI (15). Since then, numerous studies focusing on this issue have been published (16–18). According to the medical records of the 243 CDI patients enrolled in this study, elderly patients with prolonged periods of hospitalization were at a particularly high risk of rCDI, largely as a consequence of the continuous use of antibiotics and PPIs (particularly OME). In addition to being the variable with the highest OR among the independent risk factors for rCDI, infection with ST81 strains was also associated with a lower probability of survival during long-term (60 days) follow-up (Fig. 1), which is consistent with the conclusions drawn in a previous study conducted by Yang et al. (3).
In recent years, ST81 has emerged as the predominant C. difficile clone and is characterized by a high level of fluoroquinolone resistance in northern and eastern China (3, 6, 19). In addition to the amino acid mutation of GyrA and GyrB, it has been reported that a sodium-dependent efflux pump of the multidrug and toxic compound extrusion (MATE) subfamily, encoded by the cdeA gene, also confers resistance to fluoroquinolones in C. difficile (20). In the present study, we found that the administration of OME contributed to suppressing the bactericidal effects of fluoroquinolone antibiotics against ST81 strains, in the absence of any novel mutation sites other than the intrinsic GyrA (T82I) and GyrB (S366A and D426V) mutations (Fig. 2). Based on these findings, we speculate that as an antiacid targeting human parietal cells, OME may also function as an agent that effectively disables proton pumps in the cell walls of ST81 strains, thereby promoting the elimination of K+ together with the uptake of Na+ via the Na+/K+-ATPase and eventually activating MATE to expel antibiotics. This specific inhibitory effect exclusive to OME can plausibly be attributed to its particular chemical structure that distinguishes it from other PPIs; however, further studies will be necessary to substantiate this conjecture.
Although the precise mechanisms whereby PPIs contribute to an increased prevalence of rCDI have yet to be sufficiently determined, they appear to be multifactorial. It is well established that prolonged PPI usage induces an increment in gastric pH, thereby promoting a proliferation of bacteria and enhancing spore survival, which results in the deterioration, and even death, of patients with chronic disease due to intestinal dysbacteriosis (21–23). Thus, to gain further insights in this regard, we proceeded to examine in vitro whether OME influences ST81 strain spore formation and germination in a non-pH-related manner.
We established that OME promoted an increase in the transcriptional levels of spo0A, a key sporulation regulator, as well as its downstream targets (Fig. 3) and interfered with purine metabolism (Fig. 5), which enhances the sporulation of ST81 strains, in the absence of any significant effects on the gross morphology of bacterial cells or overall sporulation capacity. These observations are consistent with those reported by Fawcett et al., who verified in Bacillus subtilis that Spo0A contributes to establishing a self-reinforcing loop for entry into sporulation by repressing purine biosynthetic genes (24). In this regard, in addition to being an end product of purine catabolism, glycine is also assumed to be the most effective amino acid cogerminant with respect to the initiation of spore germination in C. difficile (25, 26). In the present study, we showed that the expression of glycine was downregulated in response to the inhibitory effect of OME on purine metabolism (Fig. 5). To compensate for this loss, biosynthesis of cogerminant receptors was stimulated by upregulating the mRNA levels of the coding gene cspBA, which eventually promoted spore germination in ST81 strains (Fig. 4).
Flagellar and toxin phase variation in C. difficile can contribute to the generation of population heterogeneity that can potentially influence disease diagnosis and treatment in clinical practice. Specifically, during gastrointestinal infection, such variation enables C. difficile to adopt a hedging strategy that either promotes colonization and inflammation in certain tissue sites or facilitates evasion from attack by the host immune system (27). In the present study, we demonstrated for the first time that the flagellar switch of ST81 strains was characterized by two interconvertible states that influence flagellar development in a very different way, irrespective of the type of medium (Fig. 6), which has also been validated in other mucosal pathogens, including Salmonella enterica (28) and Campylobacter jejuni (29).
On the basis of the previously observed inhibitory effects of OME on the in vitro motility of Helicobacter pylori (30), we next investigated whether this activity is also evident in C. difficile. Surprisingly, we found that in contrast, OME promoted the motility of ST81 strains by increasing the frequencies of bacterial populations characterized by the flg ON state (Fig. 7), indicating that these discrepant findings could be ascribed to the species-specific binding of OME to bacterial surface receptors and modification of downstream signaling. In addition to altering the abundance and morphology of flagella, OME also promoted the upregulated mRNA expression of sigD, as well as its target genes fliC and flgB, although not motA, even at a high dose (Fig. 7). Therefore, this implies that the OME-mediated promotion of motility in ST81 strains may influence the biosynthesis of the filament and rod rather than the stator of flagellar elements.
As an alternative sigma factor encoded within the flgB operon, SigD is essential not only with respect to the transcription of late-stage flagellar genes but also for virulence by influencing toxin gene expression (31). Consistent with evidence presented in previously published studies (32, 33), we found that OME promoted an increase in the toxin production of ST81 strains in vitro without influencing the expression of ccpA and codY mRNA (Fig. 8). As a pleiotropic regulator of carbohydrate metabolism, CcpA not only directly represses key factors during the early stages of sporulation (spo0A and sigF) and the expression of toxin genes (tcdA and tcdB) in C. difficile but also inhibits those enzymes that regulate the degradation of glycine to acetate in the Stickland reaction (34). Given that we detected no significant changes in ccpA mRNA expression in response to OME treatment (Fig. 8), we believe the conclusion that OME promotes spore formation (Fig. 3) and toxin production (Fig. 8) in ST81 strains is reasonable, although no appreciable effects on glycine catabolism have been observed. It is plausible that the observed reduction in glycine is attributable to a reduction in its synthesis following the inhibition of purine metabolism (Fig. 5).
Despite these important findings, the study has some limitations. First, the patients and strains examined in this study were all derived from a single hospital, which may to a certain extent limit our ability to make a generalized assessment of the epidemiology of CDI. Second, the phenotypes described were not substantiated by in vivo experimental data. By taking into consideration these limitations, in further research in progress, we are seeking to establish a cooperative venture involving multiple centers and are undertaking in vivo studies designed to validate the effects of OME on rCDI in a mouse model.
To summarize, in this study, we initially established that a history of OME medication and ST81 strain infection were the two independent risk factors for rCDI with the highest ORs. Exposure to OME was found to endow ST81 strains with an enhanced resistance to fluoroquinolone antibiotics in a concentration-dependent manner. In terms of the biological processes of ST81 strains, OME not only accelerated spore formation and germination by disrupting purine metabolism but also promoted bacterial motility and toxin production by enhancing the proliferation of bacterial populations in the flg ON state. Collectively, these findings provide persuasive evidence that OME promotes recurrence of CDI induced by ST81 strains, which, if confirmed, will broaden our perception of the common side effects of this drug observed in clinical practice. Accordingly, we predict that in the near future, standardized use of OME and epidemiological monitoring of ST81 strains will become the new focus of research on the prevention and treatment of rCDI.
MATERIALS AND METHODS
Ethics.
This study was approved by the Ethics Committee of Beijing Friendship Hospital, Capital Medical University (no. 2019-P2-273-01), in accordance with the Declaration of Helsinki. Written informed consent was obtained from patients prior to the use of samples in this research.
Bacterial isolates and patients.
This retrospective study was carried out at Beijing Friendship Hospital (Xicheng and Tongzhou campus) during the period from September 2020 to June 2022. The enrolled cases were required to simultaneously meet the following criteria: adults, inpatients, initial CDI, and the occurrence of diarrhea more than three times within 24 h. Exclusion criteria were pregnancy, breastfeeding, any ongoing antibiotic treatment, and any allergy to the drugs assessed in this study. The methods for screening, identification, MLST, examination of the mutation sites in fluoroquinolone resistance genes (gyrA and gyrB), and preservation of C. difficile isolated from clinical specimens have been described in detail in our previous study (6). Unless otherwise specified, all strains examined in this study were cultured in BHIS broth (BHI broth [Oxoid, Basingstoke, UK] supplemented with 5 g/L of yeast extract [Oxoid]). The results of TC (a reference method of toxin detection [35]) served as a basis for determining colonization or infection when patients were indicated to be positive for C. difficile culture. The C. difficile standard strains 630 (DSM 27543), R20291 (DSM 27147), and 1470 (DSM 27544) were purchased from the Leibniz Institute DSMZ (Brunswick, Germany) and maintained under the same conditions as the clinical strains. Clinical characteristics of the ST81 strains and their usage in each experiment are summarized in Data Set S1 in the supplemental material.
PPI combined antimicrobial susceptibility test.
In vitro susceptibilities to antimicrobial agents alone and in combination with PPIs were determined using the agar dilution method. The operation details and breakpoint interpretation were in line with the criteria established by the Clinical and Laboratory Standards Institute and previous studies (6, 36, 37). Briefly, six ST81 strains were initially selected as biological replicates. Thereafter, approximately 104 CFU of each strain was inoculated onto reduced Brucella blood agar (Solarbio, Beijing, China) containing a series of 2-fold increments in the concentrations of antibiotics applied alone and in combination with PPIs at a concentration of 50 mg/L. MICs were determined following a 48-h anaerobic incubation at 37°C. The 14 assessed antimicrobial agents (metronidazole [MET], vancomycin [VAN], ciprofloxacin [CIP], levofloxacin [LEV], moxifloxacin [MXF], ampicillin [AMP], piperacillin-tazobactam [TZP], clindamycin [CLI], meropenem [MEM], ceftriaxone [CRO], cefotetan [CTT], fidaxomicin [FDX], rifaximin [RFP], and tigecycline [TGC]) and 5 PPIs (omeprazole, pantoprazole, esomeprazole, rabeprazole, and lansoprazole) were purchased from Solarbio, SelleckChem (Houston, TX, USA), and the National Institutes for Food and Drug Control of China.
Growth kinetics assay.
Prior to experimentation, modified medium consisting of BHIS broth containing LEV or MXF at a concentration of one-half the MIC (BHIS-FQs), according to the results of antimicrobial susceptibility testing (Table S2), was prepared and reduced in an anaerobic environment. Subsequently, three ST81 strains (all susceptible to LEV and MXF) were harvested during the respective logarithmic phases and then inoculated into the fresh BHIS-FQs medium at a concentration of 1.5 × 108 CFU/mL with or without OME treatment. Thereafter, OD values were monitored at 4-h intervals for 28 h by shaking the plates for 10 s prior to each reading at an absorption wavelength of 600 nm. The corresponding AUCs were calculated and served as a statistical tool for comparisons of the differences between groups.
Spore preparation and staining (Schaeffer-Fulton method).
BHIS agar plates used for spore preparation and staining were prepared using BHIS broth supplemented with 10 g/L agar. All assessed C. difficile strains were streaked on the reduced medium with or without OME treatment under anaerobic conditions. For spore preparation, following incubation for 72 h, all surface growth on plates was harvested and placed in microcentrifuge tubes containing 1 mL of sterile ice-cold water and incubated at 4°C overnight. Following a brief centrifugation, the resulting pellets were washed at least five times and resuspended in 3 mL of ice-cold sterile water. A 1.5-mL aliquot of each suspension was then carefully layered on top of a 10-mL bed volume of 50% (wt/vol) sucrose aqueous solution, and the spores were separated from cell debris by centrifugation at 3,200 × g for 20 min. The pellets thus obtained were rinsed five times with water to remove the residual sucrose, and the purity of spore preparations was examined by phase-contrast microscopy (Olympus, Tokyo, Japan), prior to storage at −20°C until required for subsequent use.
For spore staining, nine ST81 strains were chosen as biological replicates and then treated with OME (at 10 and 50 mg/L) for 0 (blank control), 8, 16, and 24 h. Then, colonies were collected to prepare smears. Staining was performed using a Schaeffer and Fulton spore stain kit (Solarbio) according to the manufacturer’s instructions. Following microscopic examination, images were obtained using a Spot Diagnostic charge-coupled device camera (Sterling Heights, MI, USA).
Heat resistance assay.
Cells were harvested from BHIS agar plates (with or without OME supplementation) after incubation for 24 or 48 h and then resuspended in 1 mL of phosphate-buffered saline (PBS). For total CFU counts, serial dilutions were prepared by diluting an aliquot of resuspended culture in PBS followed by culturing on BHIS agar plates supplemented with 0.1% (wt/vol) sodium taurocholate (Solarbio). For spore counts, an aliquot of resuspended culture was heated at 65°C for 30 min, serially diluted, and cultured on BHIS agar plates supplemented with 0.1% (wt/vol) sodium taurocholate. The numbers of heat-resistant spores (N1) and total cells (N2) were determined from the CFU on plates after incubation for 24 h in an anaerobic environment. To reduce statistical error, only a number between 30 and 300 on each plate was considered valid (38). Percent sporulation was calculated as the ratio of N1 to N2 (39). Images were obtained using a ChemiDoc XRS+ system (Bio-Rad, Hercules, CA, USA) following incubation for the indicated times. Nine ST81 strains were selected as biological replicates.
Germination assay for loss of OD.
Germination assays were performed by initially suspending purified spores in a germination buffer containing 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 100 mM glycine, and 10 mM sodium taurocholate. The spores were then heat shocked at 65°C for 30 min followed by cooling on ice. Thereafter, 5 μL of spore suspension was diluted with 995 μL of germination buffer and mixed thoroughly. The OD at 600 nm was recorded spectrophotometrically at 37°C over a 30-min period, with readings at 2-min intervals. The values thus obtained were plotted as a ratio relative to the initial OD600 (tx/t0). Nine ST81 strains were selected as biological replicates. None of the operations performed in this assay were influenced by the presence of oxygen (40).
DPA release assay.
Heat-shocked spores (described above) were pelleted at 14,000 × g for 1 min and then resuspended in an equal volume of water. Thereafter, 5 μL of spore suspension and 125 μL of the abovementioned germination buffer supplemented with 800 μM terbium chloride (TbCl3; Sigma-Aldrich, Saint Louis, MO, USA) were mixed and added to the wells of an opaque 96-well plate. The release of DPA was detected using a SpectraMax M3 fluorescence plate reader (Molecular Devices, Sunnyvale, CA, USA). The excitation, emission, and cutoff wavelengths were set as 270, 545, and 420 nm, respectively. Spores without germination buffer treatment and spores autoclaved at 121°C for 20 min served as negative and positive controls, respectively. The DPA release ratio was calculated using the formula DPA % = [(F1 − F0)/(F2 − F0)] × 100%, where F0, F1, and F2 represent the fluorescence values of the negative-control, experimental sample, and positive-control groups, respectively (41). Nine ST81 strains were selected as biological replicates. None of the operations performed in this assay were influenced by the presence of oxygen (40).
RNA extraction, cDNA synthesis, and qRT-PCR.
Nine ST81 strains were selected as biological replicates and then cultured in BHIS broth supplemented with or without OME for 24 h, following which total RNA was extracted using TRIzol reagent (TaKaRa Bio Inc., Kusatsu, Japan) and reverse transcribed to cDNA using a PrimeScript RT reagent kit (TaKaRa) according to the manufacturer’s instructions. The expression of spo0A, sigE, sigF, sigG, cspBA, cspC, sleC, sigD, fliC, flgB, motA, tcdR, tcdB, codY, ccpA, and rrn was quantified based on qRT-PCR analyses. All reactions (20-μL reaction mixtures) were performed in triplicate using a SLAN-96P real-time PCR system (Hongshi, Shanghai, China) and Hieff qPCR SYBR green master mix (No Rox; Yeasen, Shanghai, China). The primers used to amplify the abovementioned genes were synthesized by Sangon Biotech (Shanghai, China), and their specific sequences (QP01F to QP16R) are listed in Table S3. The thermal profile used was 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. Data were analyzed using the comparative cycle threshold method (ΔΔCT, where CT is the threshold cycle), with the constitutively expressed rrn gene (an operon encoding 16S rRNA) used to normalize the amounts of target gene transcripts. Samples in the blank group were used as reference samples.
16S rRNA gene sequencing and phylogenetic analysis.
Genomic DNA extraction and gene amplification were performed as previously described (42). 16S rRNA sequences of C. difficile ST81 strains were obtained using Sanger sequencing (Sangon Biotech) and subsequently aligned with the NCBI reference sequences of other strains. Evolutionary history was inferred using the neighbor-joining method in MEGA 7 software. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Kimura 2-parameter method and expressed as the number of base substitutions per site.
Metabolomics.
Untargeted metabolomics profiling was performed using the XploreMET platform (Metabo-Profile, Shanghai, China). The procedures used for sample preparation were based on previously published methods with minor modifications (43). Briefly, ST81 strains were cultured in BHIS broth with (treatment group) or without (control group) OME supplementation at a concentration of 50 mg/L. Following anaerobic incubation, approximately 1 × 108 bacterial cells in the logarithmic phase of growth were harvested by centrifugation in Safe-Lock Eppendorf tubes (Sigma-Aldrich) at 1,000 × g for 10 min. The resulting pellets were subsequently washed twice with precooled PBS, after which metabolism was suppressed by immediately immersing the bottom of the tubes in liquid nitrogen. Six biological replicates were prepared for each group. Samples used for QC were prepared by mixing aliquots of the test samples for each group and injecting these at the beginning and end of each analytical run. QC analyses (MCC and principal-component analysis [PCA]), multivariate statistical analyses (OPLS-DA and permutation test), univariate statistical analyses (Student’s t test, Mann-Whitney U test, and Z-score), biomarker screening, and pathway analysis were all performed using an in-house iMAP system (v1.0, Metabo-Profile). Statistical algorithms were adapted from the statistical analysis software packages in R Studio (http://cran.r-project.org/).
Orientation-specific PCR and quantitative analysis.
Flagellar switch orientation was determined based on PCR analysis using two primer pairs that distinguish the orientation of the switch. The primer sequences targeting the genomic templates of standard strains (630 [GenBank accession no. AM180355.1], R20291 [GenBank accession no. FN545816.1], and 1470 [missing from GenBank data, so the assembly from the American Type Culture Collection {ATCC, Manassas, VA, USA} {43598} was used as the reference]) and ST81 clinical strains (G89 [GenBank accession no. CP081021.1] and CD060 [GenBank accession no. CP081023.1]) (OS01F to OS00R) are listed in Table S3 (OS01F to OS00R). PCR products were separated on 2% agarose gels and stained with GelRed (Mei5 Biotech, Beijing, China) for imaging using the ChemiDoc XRS+ system. For quantitative analysis, we used Hieff qPCR SYBR green master mix (Yeasen), with primers at a final concentration of 200 nM and an annealing temperature of 60°C. The specific sequences of each primer (QOS01F to QOS00R) are listed in Table S3. DNA copy number was calculated using the ΔΔCT method with the rrn gene (QP16F and QP16R) serving as an internal control (44).
Enrichment of flagellar phase-variant populations.
The spores of ST81 strains were plated on BHIS agar supplemented with 0.1% (wt/vol) sodium taurocholate. Following a 24-h incubation, six to 10 individual colonies from each plate were suspended in BHIS broth, spotted onto individual BHIS agar plates, and grown for 1 to 4 days as indicated. From the resulting colonies, at least eight single colonies were selected (days 1 to 4) for subsequent passaging on fresh BHIS agar plates and used to calculate the frequency of flagellar switching using a quantitative orientation-specific PCR assay. It has been established that the orientation of the flagellar switch has little influence on growth and remains stable for at least 24 h during growth on an agar surface (13). These results were finally verified by examining the presence of flagella under the TEM.
Motility assay.
The swimming agar used for motility assays was prepared using BHIS broth and 0.3% (wt/vol) Difco Bacto agar (BD Biosciences, San Diego, CA, USA). Prior to inoculation, the medium in plates was orientated vertically at room temperature overnight, after which the shaped soft agar was air dried for 15 min in a hood and then transferred to a GENbag anaerobic gas generator (bioMérieux, Marcy l’Etoile, France) for prereduction at least 4 h prior to use. In this assay, bacteria in the exponential growth phase were suspended in BHIS broth containing a 0.5 McFarland turbidity standard and then stab inoculated (3 μL) into the reduced swimming agar. Subsequent motility was quantitatively determined by measuring the diameters of colonies at 12-h intervals over a span of 2 days under oxygen-deprived conditions. Images were obtained after incubation for 48 h using the ChemiDoc XRS+ system.
Flagellar negative staining.
Cells from indicated groups were fixed in a buffer containing 2.5% glutaraldehyde, 2% paraformaldehyde, and 0.1 M sodium cacodylate (Solarbio) at 4°C for 24 h. Thereafter, the suspension was adsorbed onto 150-mesh copper grids coated with carbon film, rinsed with water, and stained with 2% uranyl acetate for 1 min. Finally, samples were observed using a transmission electron microscope (HT7800; Hitachi, Japan) operating at 80 kV.
Determination of toxin production.
The Vero cell line, originally derived from the kidney of a normal adult African green monkey and widely used in the detection of CPE, was obtained from the ATCC. Cells were cultivated in Eagle’s minimum essential medium (ATCC) supplemented with 10% fetal bovine serum (Gibco, Rockville, MD, USA). For CPE assays, cells were initially seeded in the wells of 96-well plates at a density of 1 × 104/well and left to stand at 37°C in a 5% CO2 atmosphere for 24 h. Subsequently, following a 48-h incubation in BHIS broth, the supernatants of C. difficile cultures were harvested and then filtered through a sterile membrane. To maintain control group cells in a viable condition (apoptosis rate below 5%), serial dilutions of the supernatant were also required prior to treatment. Finally, Vero cell monolayers were treated with the diluted supernatant for 24 h. CPE was determined by visualizing cell shrinkage and rounding under a bright-field microscope (Olympus), with images of at least three fields of vision being obtained for each biological replicate per group. Quantitative analyses of toxin production, apoptosis, and expression of apoptosis-related protein were performed using a C. difficile toxin A and B (CDAB) assay, flow cytometry, and Western blotting, respectively. The abovementioned experiments were performed according to previously published protocols (45, 46).
Statistical analysis.
Values shown graphically are presented as the mean ± standard deviation (SD). Student’s t test was used to compare the data obtained for two independent groups. One-way analysis of variance (ANOVA) was applied to determine the statistically significant differences between the means of three or more unrelated groups. Univariate analysis was performed in the preliminary screening of risk factors for each variable. Variables showing a statistically significant difference at a P value of <0.05 were retained in the multivariate logistic regression model to determine the potential independent risk factors for rCDI. The entry and removal probability for the stepwise-backward method of multivariate logistic regression were set as 0.05 and 0.10, respectively. ORs and 95% confidence intervals (CIs) were calculated to evaluate the association between the factors and outcomes. Kaplan-Meier analysis with log rank test and hazard ratio (HR) calculation was used to determine the survival rates of patients. All statistical analyses were performed using SPSS 25.0 (IBM Corp., Armonk, NY, USA) and GraphPad Prism 9 (GraphPad Software, Inc., La Jolla, CA, USA) software. Heatmaps and volcano plots were drawn using online tools at https://www.omicstudio.cn. For all tests, a P value of <0.05 was considered statistically significant. All experiments were performed independently three times.
ACKNOWLEDGMENTS
This work was supported by grants from Capital Health Research and Development of Special Fund (2018-2-1114) and National Key Research and Development Program of China (2021YFC2301000).
We thank Zhi Xu from the First Affiliated Hospital of Zhejiang University School of Medicine for his insightful suggestions on data analysis and Editage (www.editage.cn) for English language editing.
Footnotes
Supplemental material is available online only.
[This article was published in 24 May 2023 with an error in the affiliation line and with content missing from reference 6. The affiliation line and reference were updated in the current version, posted on 26 May 2023.]
REFERENCES
- 1.Singh T, Bedi P, Bumrah K, Singh J, Rai M, Seelam S. 2019. Updates in treatment of recurrent Clostridium difficile infection. J Clin Med Res 11:465–471. doi: 10.14740/jocmr3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hong S, Putsathit P, George N, Hemphill C, Huntington PG, Korman TM, Kotsanas D, Lahra M, McDougall R, Moore CV, Nimmo GR, Prendergast L, Robson J, Waring L, Wehrhahn MC, Weldhagen GF, Wilson RM, Riley TV, Knight DR. 2020. Laboratory-based surveillance of Clostridium difficile infection in Australian health care and community settings, 2013 to 2018. J Clin Microbiol 58:e01552-20. doi: 10.1128/JCM.01552-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang Z, Huang Q, Qin J, Zhang X, Jian Y, Lv H, Liu Q, Li M. 2020. Molecular epidemiology and risk factors of Clostridium difficile ST81 infection in a teaching hospital in eastern China. Front Cell Infect Microbiol 10:578098. doi: 10.3389/fcimb.2020.578098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu XS, Li WG, Zhang WZ, Wu Y, Lu JX. 2018. Molecular characterization of Clostridium difficile isolates in China from 2010 to 2015. Front Microbiol 9:845. doi: 10.3389/fmicb.2018.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su T, Chen W, Wang D, Cui Y, Ni Q, Jiang C, Dong D, Peng Y. 2021. Complete genome sequencing and comparative phenotypic analysis reveal the discrepancy between Clostridioides difficile ST81 and ST37 isolates. Front Microbiol 12:776892. doi: 10.3389/fmicb.2021.776892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Ma L, Sun W, Cheng J, Wang Y, Su J. 2023. Molecular epidemiology, antimicrobial susceptibility, and toxin production of clinical Clostridioides difficile isolates from a teaching hospital in Northern China. Diagnostic Microbiology and Infectious Disease 106:115972. doi: 10.1016/j.diagmicrobio.2023.115972. [DOI] [PubMed] [Google Scholar]
- 7.Koyyada A. 2021. Long-term use of proton pump inhibitors as a risk factor for various adverse manifestations. Therapie 76:13–21. doi: 10.1016/j.therap.2020.06.019. [DOI] [PubMed] [Google Scholar]
- 8.Chinzon D, Domingues G, Tosetto N, Perrotti M. 2022. Safety of long-term proton pump inhibitors: facts and myths. Arq Gastroenterol 59:219–225. doi: 10.1590/S0004-2803.202202000-40. [DOI] [PubMed] [Google Scholar]
- 9.Hassan RM, Ghaith DM, Ismail DK, Zafer MM. 2018. Reduced susceptibility of Enterococcus spp. isolates from Cairo University Hospital to tigecycline: highlight on the influence of proton pump inhibitors. J Glob Antimicrob Resist 12:68–72. doi: 10.1016/j.jgar.2017.12.005. [DOI] [PubMed] [Google Scholar]
- 10.Lu M, Yan H, Yu C, Yuan L, Sun S. 2020. Proton pump inhibitors act synergistically with fluconazole against resistant Candida albicans. Sci Rep 10:498. doi: 10.1038/s41598-019-57174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu G, Lee DP, Schmidt E, Prasad GL. 2019. Pathway analysis of global metabolomic profiles identified enrichment of caffeine, energy, and arginine metabolism in smokers but not moist snuff consumers. Bioinform Biol Insights 13:1177932219882961. doi: 10.1177/1177932219882961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee J, Kim S, Kim YH, Park U, Lee J, McKee AC, Kim KH, Ryu H, Lee J. 2022. Non-targeted metabolomics approach revealed significant changes in metabolic pathways in patients with chronic traumatic encephalopathy. Biomedicines 10:1718. doi: 10.3390/biomedicines10071718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anjuwon-Foster BR, Tamayo R. 2017. A genetic switch controls the production of flagella and toxins in Clostridium difficile. PLoS Genet 13:e1006701. doi: 10.1371/journal.pgen.1006701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, Luo Y, Grinspan AM. 2021. Epidemiology of community-acquired and recurrent Clostridioides difficile infection. Therap Adv Gastroenterol 14:17562848211016248. doi: 10.1177/17562848211016248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linsky A, Gupta K, Lawler EV, Fonda JR, Hermos JA. 2010. Proton pump inhibitors and risk for recurrent Clostridium difficile infection. Arch Intern Med 170:772–778. doi: 10.1001/archinternmed.2010.73. [DOI] [PubMed] [Google Scholar]
- 16.McDonald EG, Milligan J, Frenette C, Lee TC. 2015. Continuous proton pump inhibitor therapy and the associated risk of recurrent Clostridium difficile infection. JAMA Intern Med 175:784–791. doi: 10.1001/jamainternmed.2015.42. [DOI] [PubMed] [Google Scholar]
- 17.Villafuerte-Gálvez JA, Kelly CP. 2018. Proton pump inhibitors and risk of Clostridium difficile infection: association or causation? Curr Opin Gastroenterol 34:11–18. doi: 10.1097/MOG.0000000000000414. [DOI] [PubMed] [Google Scholar]
- 18.D’Silva KM, Mehta R, Mitchell M, Lee TC, Singhal V, Wilson MG, McDonald EG. 2021. Proton pump inhibitor use and risk for recurrent Clostridioides difficile infection: a systematic review and meta-analysis. Clin Microbiol Infect 27:697–703. doi: 10.1016/j.cmi.2021.01.008. [DOI] [PubMed] [Google Scholar]
- 19.Wang B, Peng W, Zhang P, Su J. 2018. The characteristics of Clostridium difficile ST81, a new PCR ribotype of toxin A− B+ strain with high-level fluoroquinolones resistance and higher sporulation ability than ST37/PCR ribotype 017. FEMS Microbiol Lett 365:fny168. doi: 10.1093/femsle/fny168. [DOI] [PubMed] [Google Scholar]
- 20.Marcos P, Whyte P, Burgess C, Ekhlas D, Bolton D. 2022. Detection and genomic characterisation of Clostridioides difficile from spinach fields. Pathogens 11:1310. doi: 10.3390/pathogens11111310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freedberg DE, Toussaint NC, Chen SP, Ratner AJ, Whittier S, Wang TC, Wang HH, Abrams JA. 2015. Proton pump inhibitors alter specific taxa in the human gastrointestinal microbiome: a crossover trial. Gastroenterology 149:883–885.e9. doi: 10.1053/j.gastro.2015.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clooney AG, Bernstein CN, Leslie WD, Vagianos K, Sargent M, Laserna-Mendieta EJ, Claesson MJ, Targownik LE. 2016. A comparison of the gut microbiome between long-term users and non-users of proton pump inhibitors. Aliment Pharmacol Ther 43:974–984. doi: 10.1111/apt.13568. [DOI] [PubMed] [Google Scholar]
- 23.Lin CY, Cheng HT, Kuo CJ, Lee YS, Sung CM, Keidan M, Rao K, Kao JY, Hsieh SY. 2022. Proton pump inhibitor-induced gut dysbiosis increases mortality rates for patients with Clostridioides difficile infection. Microbiol Spectr 10:e0048622. doi: 10.1128/spectrum.00486-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fawcett P, Eichenberger P, Losick R, Youngman P. 2000. The transcriptional profile of early to middle sporulation in Bacillus subtilis. Proc Natl Acad Sci USA 97:8063–8068. doi: 10.1073/pnas.140209597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Méndez-Salazar EO, Martínez-Nava GA. 2022. Uric acid extrarenal excretion: the gut microbiome as an evident yet understated factor in gout development. Rheumatol Int 42:403–412. doi: 10.1007/s00296-021-05007-x. [DOI] [PubMed] [Google Scholar]
- 26.Zhu D, Sorg JA, Sun X. 2018. Clostridioides difficile biology: sporulation, germination, and corresponding therapies for C. difficile infection. Front Cell Infect Microbiol 8:29. doi: 10.3389/fcimb.2018.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anjuwon-Foster BR, Maldonado-Vazquez N, Tamayo R. 2018. Characterization of flagellum and toxin phase variation in Clostridioides difficile ribotype 012 isolates. J Bacteriol 200:e00056-18. doi: 10.1128/JB.00056-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang H, Tang Z, Xue B, Lu Q, Liu X, Zou Q. 2022. Salmonella regulator STM0347 mediates flagellar phase variation via Hin invertase. Int J Mol Sci 23:8481. doi: 10.3390/ijms23158481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cayrou C, Barratt NA, Ketley JM, Bayliss CD. 2021. Phase variation during host colonization and invasion by Campylobacter jejuni and other Campylobacter species. Front Microbiol 12:705139. doi: 10.3389/fmicb.2021.705139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsutsui N, Taneike I, Ohara T, Goshi S, Kojio S, Iwakura N, Matsumaru H, Wakisaka-Saito N, Zhang HM, Yamamoto T. 2000. A novel action of the proton pump inhibitor rabeprazole and its thioether derivative against the motility of Helicobacter pylori. Antimicrob Agents Chemother 44:3069–3073. doi: 10.1128/AAC.44.11.3069-3073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anjuwon-Foster BR, Tamayo R. 2018. Phase variation of Clostridium difficile virulence factors. Gut Microbes 9:76–83. doi: 10.1080/19490976.2017.1362526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trzilova D, Warren MAH, Gadda NC, Williams CL, Tamayo R. 2022. Flagellum and toxin phase variation impacts intestinal colonization and disease development in a mouse model of Clostridioides difficile infection. Gut Microbes 14:2038854. doi: 10.1080/19490976.2022.2038854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart DB, Hegarty JP. 2013. Correlation between virulence gene expression and proton pump inhibitors and ambient pH in Clostridium difficile: results of an in vitro study. J Med Microbiol 62:1517–1523. doi: 10.1099/jmm.0.059709-0. [DOI] [PubMed] [Google Scholar]
- 34.Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, Rodionov DA, Martin-Verstraete I, Dupuy B. 2012. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res 40:10701–10718. doi: 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirzaei EZ, Rajabnia M, Sadeghi F, Ferdosi-Shahandashti E, Sadeghi-Haddad-Zavareh M, Khafri S, Davoodabadi A. 2018. Diagnosis of Clostridium difficile infection by toxigenic culture and PCR assay. Iran J Microbiol 10:287–293. [PMC free article] [PubMed] [Google Scholar]
- 36.Reigadas E, Alcalá L, Gómez J, Marín M, Martin A, Onori R, Muñoz P, Bouza E. 2018. Breakthrough Clostridium difficile infection in cirrhotic patients receiving rifaximin. Clin Infect Dis 66:1086–1091. doi: 10.1093/cid/cix918. [DOI] [PubMed] [Google Scholar]
- 37.Dai W, Yang T, Yan L, Niu S, Zhang C, Sun J, Wang Z, Xia Y. 2020. Characteristics of Clostridium difficile isolates and the burden of hospital-acquired Clostridium difficile infection in a tertiary teaching hospital in Chongqing, Southwest China. BMC Infect Dis 20:277. doi: 10.1186/s12879-020-05014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qin J, Dai Y, Ma X, Wang Y, Gao Q, Lu H, Li T, Meng H, Liu Q, Li M. 2017. Nosocomial transmission of Clostridium difficile genotype ST81 in a general teaching hospital in China traced by whole genome sequencing. Sci Rep 7:9627. doi: 10.1038/s41598-017-09878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahmed UKB, Shadid TM, Larabee JL, Ballard JD. 2020. Combined and distinct roles of Agr proteins in Clostridioides difficile 630 sporulation, motility, and toxin production. mBio 11:e03190-20. doi: 10.1128/mBio.03190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francis MB, Sorg JA. 2016. Dipicolinic acid release by germinating Clostridium difficile spores occurs through a mechanosensing mechanism. mSphere 1:e00306-16. doi: 10.1128/mSphere.00306-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li J, Sun Y, Chen F, Hu X, Dong L. 2021. Pressure and temperature combined with microbial supernatant effectively inactivate Bacillus subtilis spores. Front Microbiol 12:642501. doi: 10.3389/fmicb.2021.642501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dos Santos HRM, Argolo CS, Argôlo-Filho RC, Loguercio LL. 2019. A 16S rDNA PCR-based theoretical to actual delta approach on culturable mock communities revealed severe losses of diversity information. BMC Microbiol 19:74. doi: 10.1186/s12866-019-1446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohd Kamal K, Mahamad Maifiah MH, Abdul Rahim N, Hashim YZH, Abdullah Sani MS, Azizan KA. 2022. Bacterial metabolomics: sample preparation methods. Biochem Res Int 2022:9186536. doi: 10.1155/2022/9186536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sekulovic O, Mathias Garrett E, Bourgeois J, Tamayo R, Shen A, Camilli A. 2018. Genome-wide detection of conservative site-specific recombination in bacteria. PLoS Genet 14:e1007332. doi: 10.1371/journal.pgen.1007332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, Jiang Y, Wang N, Jin Q, Ji F, Zhong C, Zhang Z, Yang J, Ye X, Chen T. 2017. Invalidation of mitophagy by FBP1-mediated repression promotes apoptosis in breast cancer. Tumour Biol 39:1010428317708779. doi: 10.1177/1010428317708779. [DOI] [PubMed] [Google Scholar]
- 46.Kim N, Lee SY, Park J, Lee J. 2022. Comparative evaluation of three immunoassays for the simultaneous detection of Clostridioides difficile glutamate dehydrogenase and toxin A/B. Microorganisms 10:947. doi: 10.3390/microorganisms10050947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1. Download aac.00221-23-s0002.xls, XLS file, 2.8 MB (2.8MB, xls)
Tables S1 to S3 and Fig. S1 to S3. Download aac.00221-23-s0001.pdf, PDF file, 1.1 MB (1.1MB, pdf)