Abstract
A sensitive NMR spectroscopic method for detection of duplex forms of self-complementary nucleic acid sequences has been implemented. The G·U wobble base pair formed between a 15N-labeled strand and an unlabeled probe strand is used to identify the duplex. The guanine imino resonance, with its characteristic chemical shift, is detected using a 2D 15N–1H heteronuclear multiple quantum coherence (HMQC) spectrum and provides a sensitive and unambiguous route to hairpin–duplex discrimination. The method has been used to identify the duplex and hairpin forms of an RNA oligonucleotide at concentrations of ∼20 µM. This method has also been used to rule out possible duplex formation of an RNA oligonucleotide corresponding to the unmodified anticodon stem–loop of Escherichia coli tRNAPhe and suggests that this hairpin has a 3 nt loop.
INTRODUCTION
Self-complementarity is a property often built into RNA and DNA oligonucleotide sequences designed for solution-state biophysical studies. This property permits the generation of self-structured unimolecular species and simplifies the preparation of bimolecular species with one-to-one stoichiometric ratios of the individual strands. A drawback of this sequence design is that the molecules possess the inherent potential to form either a hairpin or a duplex conformation, sometimes making NMR structure studies problematic. The structural features that set these species apart from one another, the loop of the hairpin and the internal loop of the duplex, tend to have similar nuclear Overhauser enhancement (NOE) patterns and thus do not provide a reliable basis for structure discrimination. However, strategies employing a mixture of unlabeled and 15N-labeled oligonucleotide strands now exist than can be used to distinguish the hairpin and duplex conformations of oligonucleotides. These strategies are based on the ability to differentiate intra-molecular and inter-molecular NOEs using 15N filters or characteristic NOE cross-peak splitting patterns (1,2). However, the effectiveness of these NOE-based strategies depends upon adequate sample concentration and minimal spectral overlap.
The X-ray crystal structure of fully modified yeast tRNAPhe shows that the anticodon arm forms a 5 bp stem and a 7 nt loop (3–5). We recently began solution NMR studies of the unmodified anticodon stem–loop of Escherichia coli tRNAPhe and found that imino (NH) spectra of the oligonucleotide contained peaks not expected to be produced by the RNA hairpin. However, due to spectral overlap and low sensitivity, attempts to determine the monomer–dimer state of this molecule using the NOE-based methods yielded ambiguous results. To unambiguously determine the oligomeric state of the molecule, we have used a chemical shift-based method that relies on the characteristic chemical shift of the G NH proton resonance of a G·U wobble base pair to distinguish hairpin and duplex conformations. The experiment is simple to interpret: a new wobble cross peak in the NH region of the 15N–1H heteronuclear multiple quantum coherence (HMQC) spectrum indicates the presence of a duplex molecule. The method is highly sensitive and has been used to identify the RNA duplex at concentrations as low as 20 µM.
MATERIALS AND METHODS
All enzymes were purchased (Sigma) except for T7 RNA polymerase, which was prepared as described (6). Deoxyribonuclease I type II, pyruvate kinase, adenylate kinase and nucleotide monophosphate kinase were obtained as powders, dissolved in solutions of 15% glycerol, 1 mM dithiothreitol and 10 mM Tris–HCl, pH 7.4, and then stored at –20°C. Guanylate kinase and nuclease P1 were obtained as solutions and stored at –20°C. Unlabeled 5′ nucleoside triphosphates (5′-NTPs), phosphoenolpyruvate (potassium salt) (Bachem) and 99% [15N] ammonium sulfate (Cambridge Isotope Labs) were obtained as powders.
Preparation of RNA samples
RNA molecules (Fig. 1) were prepared by in vitro transcription using T7 RNA polymerase and synthetic DNA templates (7). Isotopically enriched 5′ nucleoside monophosphates (5′-NMPs) were prepared and converted to 5′-NTPs as described (8,9). Unlabeled oligonucleotides (RNA IU and RNA IIU) were prepared from 10 ml transcription reactions using 4 mM 5′-NTPs. 15N-labeled oligonucleotides (RNA I and RNA II) were prepared similarly except that the transcription volumes were 16 ml and the concentration of NTPs was 3 mM. RNA molecules were purified by passage through 20% (w/v) preparative polyacrylamide gels, electroeluted (Schleicher & Schuell) and precipitated with ethanol. The purified oligonucleotides were dissolved in 1.0 M NaCl, 20 mM sodium phosphate (pH 6.8) and 2.0 mM EDTA and dialyzed extensively against 2.5 mM sodium phosphate (pH 6.8) and 0.1 mM EDTA using a Centricon-3 concentrator (Amicon Inc.). The samples were lyophilized and suspended (90% H2O/10% D2O) under final buffer conditions that favor hairpin formation [2.5 mM sodium phosphate (pH 6.8) and 0.1 mM EDTA] or duplex formation [100 mM NaCl, 2.5 mM sodium phosphate (pH 6.8) and 0.1 mM EDTA]. The final sample concentrations of 15N-labeled RNA I oligonucleotide were 0.40 and 0.078 mM (20 and 4 A260 OD units, respectively, in 500 µl). An equal amount of RNA IU was added to prepare the RNA I + RNA IU mixed samples. For the RNA II sample, 30 A260 OD units each of RNA II and RNA IIU were mixed in 200 µl (≈1.0 mM 15N-labeled RNA II).
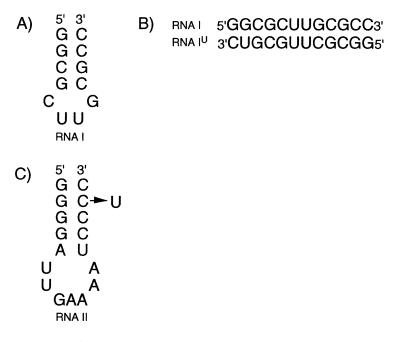
Figure 1.
Sequences and secondary structures of (A) RNA I hairpin, (B) RNA I–RNA IU heterodimer duplex and (C) RNA II hairpin. The RNA IIU molecule has the substitution C42U and is numbered according to full-length tRNAPhe.
NMR spectroscopy
Experiments were acquired at 500 MHz (Bruker, AMX) using a 1H-{X} inverse detection probe. Quadrature detection was achieved using the States-TPPI method and acquisition was delayed by a half-dwell in the indirectly detected dimension. Acquisition times for the 15N–1H HMQC (10) and NOESY (11) experiments were ω1 = 36–50 ms and ω2 = 168 ms and the 1H and 15N carriers were positioned at 4.85 and 150 p.p.m., respectively. The spectra were collected at 12°C and the solvent signal was suppressed using the binomial 11 method (12). Typically, the data points were extended by 33% using linear prediction for the indirectly detected dimensions and the data were apodized using 1 Hz line broadening and 65° shifted sinebell functions. The NH resonances of RNA I and RNA II were assigned by sequential correlation of the NH protons using NOESY spectra (τm = 360 ms) (5,13). All spectra were processed and analyzed with Felix 98.0 (Molecular Simulations, Inc.).
RESULTS AND DISCUSSION
The NH protons of G·U wobble base pairs resonate in a characteristic region of the NMR spectrum, shifted 2–3 p.p.m. upfield from the NH resonances of G·C and A·U base pairs, and are readily identified in the 15N–1H HMQC spectrum. The chemical shift method is based on the detection of the wobble base pair using the HMQC experiment. The principal advantages of the HMQC experiment to distinguish monomer and dimer forms of oligonucleotides are improved sensitivity relative to filtered NOESY methods and improved sensitivity and spectral dispersion relative to 15N non-decoupled NOESY methods (1,2). Also, since the cross peaks in the HMQC spectrum are generated from the one-bond 15N–1H scalar coupling, it is possible to accurately quantify the ratio of duplex-to-hairpin conformations in a sample.
The chemical shift-based method to distinguish hairpin and duplex forms of oligonucleotides employs a mixture of labeled and unlabeled strands, analogous to NOE-based methods. However, instead of using strands of identical sequence, a C→U base substitution is introduced into the unlabeled strand in a region predicted to form a stem in both the hairpin and duplex. Thus, if conditions favor hairpin formation, the 15N-labeled strand will give rise to an NH cross peak in the HMQC spectrum corresponding to the G·C base pair at the marker position in the stem. Since the G·U base pair forms in the stem of the unlabeled strand, it will be ‘silent’ and will not give rise to an NH cross peak. If conditions favor duplex formation, three different strand combinations can exist: 15N:15N and 14N:14N homodimers and an 15N:14N heterodimer in a ratio of 1:1:2. The 14N:14N homodimer contains two G·U base pairs, but will not contribute cross peaks to the HMQC spectrum. The 15N:15N homodimer contains two symmetrically equivalent G·C base pairs and so will contribute one NH cross peak to the HMQC spectrum. The two 15N:14N heterodimers have a marker G·U base pair that will give rise to one G NH cross peak in the HMQC spectrum and one G·C base pair that is silent and will not contribute to the spectrum. Thus, the presence of a G NH cross peak corresponding to the marker G·U base pair in the spectrum of the duplex and the absence of that cross peak from the spectrum of the hairpin distinguish the hairpin and duplex conformations. If the exchange rates of the NH protons with the solvent are equal at all base pairs in the duplex, the cross-peak intensities of the marker G·C (homodimer) and G·U (heterodimer) base pairs will be equal but each will be one-half the intensity of the other NH resonances in the spectrum. For the hairpin, the NH cross-peak intensity of the G·C base pair at the marker position will be equal to the intensity of the other NH resonances in the spectrum.
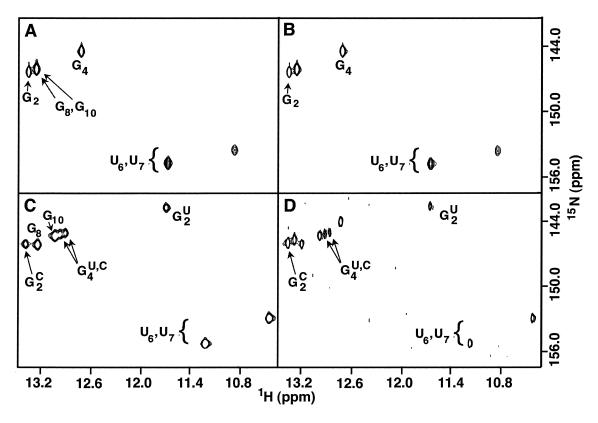
The strategy is demonstrated using 15N-labeled RNA I (Fig. 1) and unlabeled RNA IU, which contains a C11→U substitution. RNA I forms a hairpin in the absence of NaCl (14) and gives rise to three G NH cross peaks in the HMQC spectrum (Fig. 2A) (the G8 and G10 NH resonances of the hairpin are nearly degenerate and appear as a single cross peak in these spectra). The addition of an equimolar amount of unlabeled RNA IU does not lead to appearance of a G·U base pair G NH resonance, confirming the monomeric state of the RNA molecule (Fig. 2B). However, when NaCl is added to a concentration of 0.1 M, a G NH cross peak at 11.5 p.p.m. characteristic of a G·U base pair appears, clearly indicating that the RNA I·RNA IU duplex has formed (Fig. 2C). Of the six G NH resonances that characterize the spectrum of the duplex, two correspond to G2 and two correspond to G4. The doubling of the G4 NH resonance results from the proximity of G4 to the G2·C11 (homodimer) or G2·U11 (heterodimer) base pairs. The G4 NH cross-peak intensities indicate that the ratio of the homodimeric to heterodimeric duplex is ≈1.2:1. This difference presumably results from the thermodynamic penalty incurred by replacing the penultimate G·C base pair with a G·U base pair.
Figure 2.
Imino regions from 15N–1H HMQC spectra of (A) 15N–RNA I hairpin, (B) a mixture of 15N–RNA I and RNA IU harpins, (C) a mixture of duplexes formed by equimolar amounts of 15N–RNA I and RNA IU strands (0.40 mM each), and (D) a mixture of hairpins and duplexes formed by equimolar amounts (0.078 mM each) of 15N–RNA I and RNA IU strands. Resonance assignments for the hairpin and duplex conformations were determined using NOESY spectra. In (D), the low sample concentration and solvent exchange of the imino protons leads to very weak signals for the U6 and U7 NH resonances of the hairpin and so are not observed at this contour level. In (C) and (D), the imino resonances of G2 and G4 are labeled with C and U for the homodimer and heterodimer, respectively. The buffer conditions for (C) and (D) are identical but the 5-fold lower oligonucleotide concentration in (D) leads to partitioning between hairpin and duplex conformations.
The chemical shift-based method is very sensitive and can be used to quantify the relative populations of monomer and dimer molecules. The RNA I strands partition between hairpin and duplex forms at low concentration in the presence of 100 mM NaCl and lead to two sets of cross peaks in the NMR spectrum. Figure 2D shows the spectrum of a solution containing RNA I and RNA IU (78 µM each) under conditions that both hairpin and duplex conformations are present. The relative amounts of hairpin and duplex can be quantified by comparing the intensity of the G4 NH resonance of the hairpin with the sum of the intensities of the two G4 resonances of the duplex. For the case shown in Figure 2D, these intensities are approximately equal, indicating that 50% (or 39 µM) of RNA I is in duplex form. Since one-half of the RNA I duplex strands are the heteroduplex, the concentration of RNA I·RNA IU duplex, which gives rise to the G2·U11 NH marker cross peak, is ≈20 µM. This illustrates that very small quantities of oligonucleotide are sufficient to perform the experiment. Thus an additional benefit of this method over the NOE-based methods is that less labeled material is required. Loss of 1H signal due to 15N coupling during acquisition (2) or 1H transverse relaxation during 15N filter periods (1) requires that much more concentrated samples be used to perform NOE-based experiments that distinguish hairpin and duplex conformations. Further, since this method is based on the one-bond 15N–1H scalar coupling constant (1JHN ≈92 Hz), the population of hairpin and duplex species can be more accurately quantified.
In the experiments described above, a cytidine nucleotide in the original sequence was replaced with a uridine nucleotide and the G NH resonance of the newly formed G·U base pair serves as the marker to identify duplex molecules. If the original sequence had a G·U base pair appropriately located, then combining a 15N-labeled strand containing a U→C substitution with the unlabeled original strand would yield the same spectral results for duplex formation [the presence of a new HMQC-observable (G)·U NH resonance in a previously G·C rich spectrum]. The alternative strategy of combining the U-containing original strand that is 15N-labeled with a C-substituted strand that is unlabeled will yield a spectrum of the hairpin that will have equally intense G and U NH cross peaks corresponding to the G·U base pair. The spectrum of the duplex will have G and U NH cross peaks from the G·U base pair (homodimer) but also will contain a G NH cross peak from the marker G·C base pair (heterodimer). The intensities of each of the G NH cross peaks from the G·U and G·C base pairs in the dimer will be one-half the intensity of the U NH cross peak from the marker G·U base pair in the dimer.
During the course of our studies of a 17 nt RNA molecule containing the unmodified anti-codon stem–loop from E.coli tRNAPhe (Fig. 1C) (15), three uridine NH resonances corresponding to base pairs A31·U39, U32·A38 and U33·A37 were identified, although the latter is weak. The addition of Mg2+ leads to the disappearance of the U33 NH resonance and significant weakening of the U32 NH resonance. The presence of the U32 and U33 NH resonances suggests that the loop of the hairpin is either more structured than expected based on crystallographic studies of fully modified yeast tRNAPhe (3–5) or that the oligonucleotide forms a duplex in the absence of Mg2+. However, the poor sensitivity and spectral overlap of the NOE based methods for distinguishing hairpin and duplex conformations prevented determination of the monomer–dimer state of RNA II.
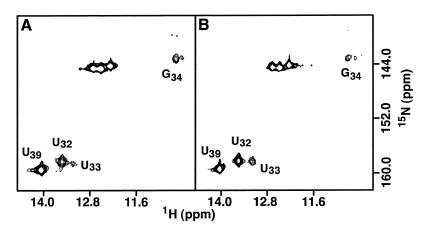
To determine the oligomeric state of the RNA molecule, 15N-labeled RNA II was mixed with unlabeled RNA IIU, which contains a C42→U substitution (Fig. 1C). In the absence of Mg2+, no G NH proton resonance diagnostic of a G·U pair (heteroduplex) could be observed in the HMQC spectrum (Fig. 3). The one-dimensional 1H spectrum of the mixed sample contains imino resonances characteristic of a G·U base pair and acquisition of this spectrum with and without 15N decoupling confirms that only RNA IIU contributes to the wobble base pair (spectrum not shown). These results demonstrate that RNA II forms a hairpin and that the stem contains two base pairs not found in the anticodon stem–loop of fully modified yeast tRNAPhe (3–5). The modified nucleotides in the anticodon stem and loop of E.coli tRNAPhe (15) may prevent formation of the additional secondary structure observed in the unmodified molecule. Experiments are now under way to determine the influence of the base modifications on the structure of this RNA hairpin.
Figure 3.
Imino regions from 15N–1H HMQC spectra of (A) a mixture of 15N–RNA II and RNA IIU and (B) 15N–RNA II. The absence of a wobble G resonance indicates that RNA II and RNA IIU adopt only the hairpin conformation and supports participation of U33 in an intra-molecular base pair.
The chemical shift method distinguishes unambiguously hairpin and duplex conformations of oligonucleotides. The advantages of this method over NOE-based approaches are higher sensitivity, shorter acquisition time, simple interpretation and substantially reduced spectral overlap. The high sensitivity of the method can also permit the discrimination of multiple conformations of a hairpin from duplex–hairpin mixtures, even when the duplex is very scarce (approaching the sub-micromolar range). Additionally, this method has potential as an accurate tool to quantify hairpin-to-duplex ratios since the cross-peak intensities primarily depend upon the NH proton–solvent proton exchange rate. The only additional step that is required (compared to other hairpin–duplex discrimination techniques) is the purification of the second DNA oligonucleotide template for in vitro transcription of the unlabeled U-substituted RNA. We believe that this is a minor inconvenience given the unambiguous results generated by the chemical shift method.
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. Michnicka for preparation of the T7 RNA polymerase and Dr S. Moran for critical reading of the manuscript. J.C.V. also wishes to thank Consejo Nacional de Ciencia y Tecnología for partial support. This work was supported by grant C-1277 from the Robert A. Welch Foundation to E.P.N.
REFERENCES
- 1.Aboul-ela F., Nikonowicz,E.P. and Pardi,A. (1994) FEBS Lett., 347, 261–264. [DOI] [PubMed] [Google Scholar]
- 2.Sich C., Flemming,J., Ramachandran,R. and Brown,L.R. (1996) J. Magn. Reson. Series B, 112, 275–281. [DOI] [PubMed] [Google Scholar]
- 3.Kim S.-H., Suddath,F.L., Quigley,G.J., McPherson,A., Sussman,J.L., Wang,A.H.J., Seeman,N.C. and Rich,A. (1974) Science, 185, 435–440. [DOI] [PubMed] [Google Scholar]
- 4.Robertus J.D., Ladner,J.E., Finch,J.R., Rhodes,D., Brown,R.S., Clark,B.F.C. and Klug,A. (1974) Nature, 250, 546–551. [DOI] [PubMed] [Google Scholar]
- 5.Hingerty B., Brown,R.S. and Jack,A. (1978) J. Mol. Biol., 124, 523–534. [DOI] [PubMed] [Google Scholar]
- 6.Davanloo P., Rosenburg,A.H., Dunn,J.J. and Studier,F.W. (1984) Proc. Natl Acad. Sci. USA, 81, 2035–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Nucleic Acids Res., 15, 8783–8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikonowicz E.P., Sirr,A., Legault,P., Jucker,F.M., Baer,L.M. and Pardi,A. (1992) Nucleic Acids Res., 20, 4507–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batey R.T., Inada,M., Kujawinski,E., Puglisi,J.D. and Williamson,J.R. (1992) Nucleic Acids Res., 20, 4515–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Griffey R.H., Poulter,D.C., Bax,A., Hawkins,B.L., Yamaizumi,Z. and Nishimura,S. (1983) Proc. Natl Acad. Sci. USA, 80, 5895–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.States D.J., Haberkorn,R.A. and Ruben,D.J. (1982) J. Magn. Reson., 48, 286–292. [Google Scholar]
- 12.Hore P.J. (1983) J. Magn. Reson., 55, 283–300. [Google Scholar]
- 13.Kime M.J. (1984) FEBS Lett., 175, 259–262. [DOI] [PubMed] [Google Scholar]
- 14.Jucker F.M. and Pardi,A. (1995) Biochemistry, 34, 14416–14427. [DOI] [PubMed] [Google Scholar]
- 15.Björk G.R. (1996) In Neidhardt,F.C., Ingraham,J.L., Resnikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella. Cellular and Molecular Biology, Vol. 1. ASM Press, Washington, DC, pp. 861–886.