Abstract
Highly active antiretroviral therapy (HAART) has revolutionized human immunodeficiency virus (HIV) healthcare, turning it from a terminal to a potentially chronic disease, although some patients can develop severe comorbidities. These include neurological complications, such as HIV associated neurocognitive disorders (HAND), which result in cognitive and or motor function symptoms. We now describe the discovery, synthesis, and evaluation of a new class of N-phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazol-3-amine HIV-1 non-nucleoside reverse transcriptase inhibitors (NNRTI) aimed at avoiding HAND. The most promising molecule, 12126065, exhibited antiviral activity against wild-type HIV-1 in TZM cells (EC50 = 0.24 nM) with low in vitro cytotoxicity (CC50 = 4.8 µM) as well as retained activity against clinically relevant HIV mutants. 12126065 also demonstrated no in vivo acute or subacute toxicity, good in vivo brain penetration, minimal neurotoxicity in mouse neurons up to 10 µM, with a 50% toxicity concentration (TC50) of >100 µM, well below its EC50.
Keywords: Human immunodeficiency virus, non-nucleoside reverse transcriptase inhibitors, NNTRI, HIV-associated neurocognitive disorder, HAND, Drug discovery
Graphical Abstract
Introduction
Human immunodeficiency virus (HIV) infection, the virus causing acquired immune deficiency syndrome (AIDS), is one of the most important pathogens affecting mankind. Finding new treatments to cure HIV is therefore one of the most pressing challenges in contemporary virology and medicinal chemistry. The WHO reports that HIV has claimed more than 40.1 million (33.6 – 48.6 million) lives globally since the start of the epidemic, with approximately 650,000 (510,000 – 860,000) people dying from HIV-related illnesses in 2021 alone. The numbers of new global HIV infections are still immoderate, with 1.5 million (1.1 – 2.0 million) new cases in 2021. As of 2021, 38.4 million are living with HIV and among them 2.73 million (2.06 – 3.47 million) were children between 0–19 years of age with 850 new daily infections in children 1–3. HIV is prevalent in the developing world, representing two thirds of current infections (25.6 million) in the WHO African Region, but it is also resurfacing in wealthy countries with the CDC reporting 30,635 newly diagnosed cases in the US in 2020 with a 2019 estimate of over 1.04 million living with HIV in the USA 2, 4. While these statistics are grim, there is reason for cautious optimism as HIV can be managed primarily with a cocktail of 3–4 antiretroviral drugs that need to be taken regularly.
The implementation of combination antiretroviral therapy (cART) or the synonymous highly active antiretroviral therapy (HAART) can delay the progression of the most severe symptoms of HIV for decades by restoring the immune system and controlling viral load 5. cART/HAART is a cocktail of multiple HIV-targeting drugs, with the most common regimens being comprised of nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTI), integrase stand transfer inhibitors (INSTI) and/or boosted protease inhibitors. Prior to the initiation of cART/HAART, genotypic drug-resistance testing is recommended based on the current rates of the transmission of drug-resistant HIV and focuses on finding mutations in HIV reverse transcriptase, protease and integrase that are known to imbue resistance. The initial combination drug regime includes two NRTI and a third compound from one of the previously described classes 6. When multiple drugs are available therapies are often tailored to the patient’s needs, based on virological efficacy, drug-drug interactions and treatment cost. cART/HAART is common intervention in HIV-infected children as well, with approximately 54% of children living with HIV receiving cART/HAART in 2018 globally 2. It has been well established that early cART/HAART in children improves their immune reconstitution drastically reducing AIDS-related mortality 7, 8. While cART/HAART has overall been effective, low-level viremia (LLV) are common even among those undergoing cART/HAART, though this prevalence is strongly variable between populations (0.4%-38.7%) 9–12 and with the added complexity of no standard definition of LLV. LLV has clinical relevance due to it leading to an increased risk of virological failure, transmission and antiviral resistance 13. This is exacerbated in individuals with persistent LLV, specifically regarding an increased risk of virological failure 14, 15. Some recent publications have further analyzed the relationship between LLV and or virological failure, with the appearance of drug-resistant mutants in 42.6% (Northern Taiwan), 70.2% (Cameroon) and 92% (Northern Tanzania) of patients with either LLV or virological failure. These clinical isolates were found to have at least one drug resistant mutation 16–18. Many of these mutations had resistance to NRTI(s) and or NNRTI(s). Mutations imbuing resistance against NNRTIs were the most common, with K103N occurring with the highest frequency, followed by Y181C. These are common clinical HIV I reverse transcriptase (RT) mutations that are resistant to established drugs, with K103N, Y181C, and G190A accounting for more than 90% of NNRTI resistance in the United States 19.
Side effects are also an important concern for drugs that need to be taken for decades 20. A large number of patients develop HIV associated neurocognitive disorders (HAND), which results in symptoms from minor problems with memory to severe dementia-like symptoms. Autopsy studies have been reported showing white matter changes and demyelination in these cases 21. Even though small molecule treatments are highly effective in the periphery, virus can remain in the CNS and replicate which then results in the neurological disorders. This may in some cases be due to the inability of HIV medications to cross the blood brain barrier (BBB) 22 and inhibit HIV in the brain. Several reasons may explain why cART is inefficient at preventing HAND, such as poor CNS penetration and incomplete inhibition of HIV replication in this anatomical compartment, drug resistance, cART neurotoxicity, or irreversible brain damage prior to initiation of cART. So far the data is mixed on CNS-targeted cART 23, 24 and there have been several studies assessing CSF concentrations of drugs and viral suppression in adults 25 and children 26 showing adequate viral suppression for some drugs and suboptimal CSF concentrations for others. High CNS penetration of cART is also important to limit tissue injury and recovery in those at risk of cerebrovascular disease 27. NNRTIs such as efavirenz (EFV) are well documented from the perspective of CNS effects, which may be due to multiple mechanisms 28. Macrophages, particularly in the CNS, are likely components of the persistent reservoir that resist HIV eradication. However, cART has limited effect in macrophages, due to their scarce phosphorylation activity, which limits the activity of nucleoside analogs, and the expression of P-gp transporters 29, which pump out protease inhibitors. There are also issues relating to drug-drug interactions between HIV treatments (due to P450’s 30) or other CNS side effects 28, 31–36. It is therefore our aim to optimize novel NNRTI that cross the BBB 20, are safe and effective at preventing viral replication in the brain and the periphery. New anti-HIV compounds, selected specifically for their ability to overcome the growing list of HIV RT strains (e.g. K103N, Y181C and G190A19) that are resistant to established drugs, are beginning to populate a small pipeline of potential future drugs 19, 37, 38. We therefore need to have more diversity in the types of chemical structures assessed in order to stand the best chance of addressing both drug resistance and HIV CNS dysfunction in future.
To date, six 1st and 2nd generation NNTRI have been approved by the FDA (nevirapine (NVP), efavirenz (EFV), delavirdine (DLV), etravirine (ETR), doravirine (DOR) and rilpivirine (RPV); Figure S1) 39. This class of drug binds in an allosteric pocket of HIV RT inhibiting the progression of viral DNA synthesis 40. Since these drugs target a protein not found in eukaryotes, off target interaction is likely reduced as compared to nucleoside analog class of inhibitors (NRTIs). Currently, the three NNRTIs DOR, EFV, and RPV are recommended by the Panel on Antiretroviral Guidelines for Adults and Adolescents for use in cART therapy 6. The selection of a drug regime is based on multiple factors, including but not limited to the presence of drug-resistant mutations. In 2021 the FDA approved the first extended-release, injectable drug Cabenuva (cabotegravir and RPV) which requires monthly injections in lieu of daily oral dosing, providing an alternative treatment option. While potentially a revolutionary treatment option, recent recommendations by the same panel have suggested that those taking Cabenuva or who may have been infected by an individual taking Cabenuva undergo genotypic resistance testing to probe for resistance against the integrase strand transfer inhibition class, suggesting that more treatment options are still needed 6.
First generation NNRTIs have a low genetic barrier to resistance and only require one mutation to confer resistance, while second generation NNRTIs have a higher genetic barrier 41. These compounds are highly potent with low toxicity yet are still hampered by rapid viral drug resistance, as HIV is highly prone to develop mutational-based drug resistance due to the lack of a proofreading activity of RT 42. Individual clinical isolates have been identified which have resistance to one or more of each of the FDA-approved NNTRIs 39, strongly supporting the need for future compounds which retain effectiveness against these drug-resistant mutants.
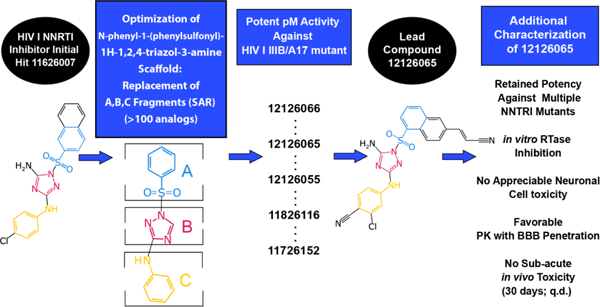
We have identified the N-phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazol-3-amine scaffold as possessing potent activity as an NNRTI. Based on an initial hit found through phenotypic screening, we have developed >100 analogs using a classical medicinal chemistry structure activity relationship (SAR) to optimize activity against wild-type, A17 mutant (K103N/Y181C) as well as other common mutant strains resistant to approved NNRTIs. This has led to the discovery of a series of optimized compounds with picomolar activity against wild-type HIV that retain activity against these clinically relevant mutants.
Results
Synthesis:
We explored the optimization of the N-phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazol-3-amine scaffold via a classical SAR, where we made a detailed study of the role and importance of atoms from fragments “A”, “B” and “C” (Figure 1). The target compounds were synthesized by the reaction of the aminotriazole with a corresponding aryl sulphonyl chloride to yield a mixture of sulfonamides. The major product resulted from reaction of the triazole NH with a minor product resulting from sulfonylation of the adjacent nitrogen. This mixture is a common result with 3-amino-1,2,4-triazoles, with selectivity resulting from the increased acidity of the N-H group due to inductive effects from the neighboring amino moiety 43–45. Essential atoms were identified as well as positions for future derivatization.
Figure 1.
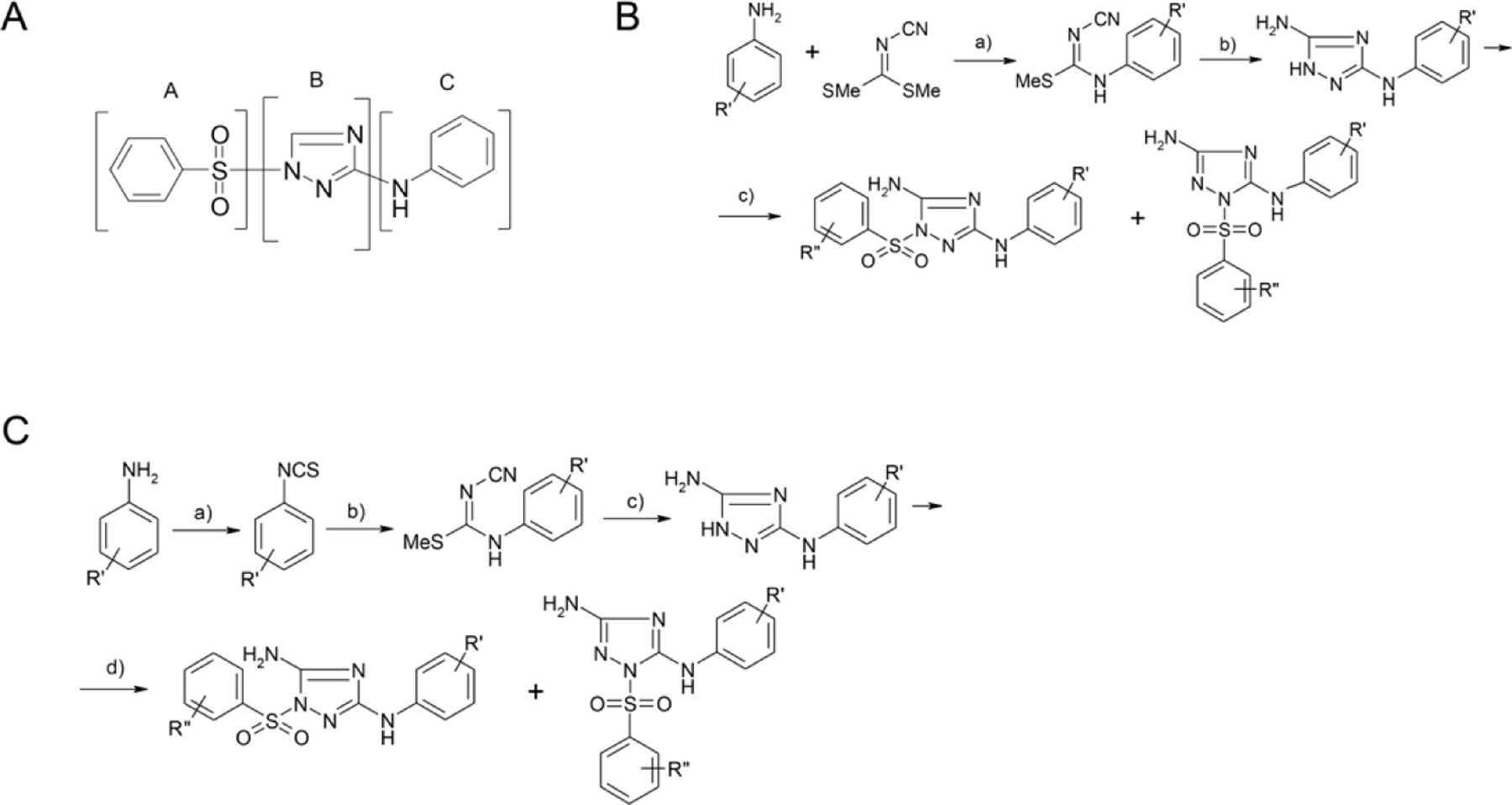
General synthetic scheme. (A) N-phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazol-3-amine scaffold used in this study. (B) a) EtOH, reflux; b) hydrazine hydrate, EtOH, reflux; c) PhSO2Cl, THF, Et3N. (C) a) toluene, dimethiocarbamoyl chloride; b) NaOEt, EtOH, NH2CN, MeI, boiling; c) hydrazine hydrate, EtOH, boiling; d) PhSO2Cl, THF, Et3N.
SAR:
The N-phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazol-3-amine scaffold was investigated by initially testing a panel of synthetic derivatives against a wild-type (IIIB) and K103N/Y181C (A17) mutant serially using TZM-bl cells. A representative group of compounds is displayed in Table 1. The N1-sulfonamide (fragment “A”) group was modified by introducing substituents of various size, polarity, and positions. A variety of structurally diverse heterocycles were well tolerated, including 6-chloro-imidazothiazole (IMTZ), 2,4-dimethylthiazole, and substituted naphthalenes (Nap), without significant loss in activity against wild-type, however, simple methylsulfonamide, larger aliphatic or cycloaliphatic sulfonamides were completely inactive. In general, the replacement of the phenyl fragment “A” by a naphthalene ring resulted in a sharp increase in activity. This led us to investigate various substituents on the naphthalene fragment. Compounds containing cyano, acetonitrile, or acrylonitrile groups in the 6-position had the highest activity against at least wild-type. Many such compounds, including 12126055, 12126065 and 12126066, exhibited improved potencies relative to 11626007. Sterically bulky, electron-deficient rings at this location also seemed to be required for retaining potent activity against the A17 mutant.
Table 1.
A representative sampling of the tested compounds exemplifying a substantial change in activity, either in wild-type (WT), A17 or both, with functional group substitutions at positions R1 and R2.

| ||||
|---|---|---|---|---|
|
| ||||
| Molecule | R1 | R2 | EC50 (TZM, WT)a | EC50 (TZM, WT)a |
| 11926331 | 4-CN | IMTZ | 1.77 | 705 |
| 12026113 | 4-CN, 2-CI | IMTZ | >1000 | >1000 |
| 12026123 | 4-CN, 3-CI | IMTZ | 0.980 | 175 |
| 12126060 | 4-CN, 3-CI | 6-CH2CN Nap | 0.598 | 153 |
| 11826116 | 4-CN, 3-CI | 6-CN Nap | 0.420 | 39.6 |
| 12026124 | 4-CN, 3-CI |
|
0.879 | 306 |
| 11826313 | 4-CN, 3-CI | 6-NMe2 Nap | 10.6 | >1000 |
| 12126065 | 4-CN, 3-CI | 6-CH=CHCN Nap | 0.263 | 0.689 |
| 11926326 | 4-CI | mesityl | 32.0 | >1000 |
All EC50 values displayed in nM.
Among the tested 1,2-substituted triazoles (modified fragment “B”), only 8 displayed activities against the wild-type, with three derivatives exhibiting an EC50 below 100 nM (11826370, 11926095, 11926104). Of these active compounds, there was no general trend for activity with regard to structure. Moreover, all of the active 1,2-substituted triazoles displayed no activity against the A17 mutant, leading us to focus our efforts on the 1,3-substituted triazoles.
In general, electron-deficient aromatics in the aniline fragment “C” were found to be more active against wild-type, with the exception of 2-chloro substituents which result in a complete loss of activity. The introduction of one or two additional methylene groups between the nitrogen atom and the aryl rings in fragment “C” resulted in compounds that were inactive or much less active. Removal of the 4-chloro substituent in 11626007 led to a substantial (>10–40 fold) reduction in potency, while 4-cyano or 4-acetonitrile groups increased activity significantly. Finally, the addition of a 3-chloro substituent resulted in a small increase in potency in the wild-type and a dramatic increase in potency against the A17 mutant when paired with a bulky, electron-deficient ring in domain “A”.
The compounds which retained activity against the A17 mutant (11726152, 12126055, and 12126065) were also tested against other clinically relevant NNRTI-resistant mutants (Figure 2). While all three tested compounds retained high potency in most of these mutants, the inhibition of only 12126065 was not drastically mitigated in the L100I, K103N mutant. We attribute this retained potency to the extra vinyl group between the naphthalene ring and the nitrile group.
Figure 2.
Structure and dose response versus WT, A17 (K103N/Y181C), Y181C, L100I/K103N, K103N HIV mutants and toxicity in TZM-bl cells for 11726152, 12126055 and 12126065. (D) Calculated IC50/CC50 (±SD) and (E) fold-difference versus wild-type using a 3-parameter fit in Graphpad Prism 9.2. Min/Max values were constrained to be shared for each mutant dataset as full inhibition was not reached for all compounds in some mutants. A minimum of 6 replicates were performed for each HIV mutant and for cytotoxicity. Error bars represent SEM.
Antiviral screening:
Preliminary screening was performed versus HIV I (IIIB) in TZM-bl cells and RT activity (details below) was confirmed for selected compounds. Qualitative cytotoxicity was also assessed for each compound in the ranges used to test inhibition using a MTT assay. Concentration ranges for TZM-bl were 0.05–500 nM. Dose response curves identified potent HIV inhibitory activity in TZM-bl cells as well as in RT for an initial hit, 11626007, with whole cell (EC50) and RT (IC50) inhibition of 9.3 ± 2.3 nM and 0.28 ± 0.1 µM, respectively (Table 2, Table S1–S2). No observable toxicity was indicated using an MTS assay in TZM-bl cells within the dose range tested (data not shown). Our early screening also assessed the inhibition of a NNRTI-resistant double mutant (K103N/Y181C; A17). No appreciable inhibition was seen for this compound in cells infected with the HIV-1 A17 mutant (Table S1), so we continued to evaluated analogs of this compound. Many of these have nearly equivalent or improved activity versus the EFV control with a comprehensive list of all of the IC50’s from these in vitro inhibition experiments against the wild-type (IIIB) and Y181C/K103N (A17) IIIB variant (Table S1). As the potency was high for many of our best compounds an “extended concentration range” was also performed against both wild-type and A17 in order to reach plateaus for each normalized inhibition extreme. These “extended concentration ranges” were performed with concentrations between 0.028 – 5000 nM (15 point). This confirmed each of these compounds as potent inhibitors of both wild-type and A17 HIV. Cytotoxicity was assessed more rigorously and CC50’s were determined when possible (Table 3 with individual curves shown in Figure S2). The compounds that retained potent A17 activity (11726152, 12126055 and 12126065) were also followed up with additional testing versus clinically relevant NNRTI-resistant mutants Y181C, L100I/K103N, K103N. All mutants, including A17, were (re)tested at a concentration range of 0.05–500 nM with a minimum of 6 replicates performed over multiple days (Figure 2).
Table 2.
in vitro inhibition of HIV reverse transcriptase for select compounds. EC50 was calculated using 10-fold dilutions in Graphpad Prism 9.2.1 and “±” represents SEM.
| Name | EC50 (µM) | |
|---|---|---|
| EFV | 0.03 ± 0.01 | |
| RPV | 0.16 ± 0.06 | |
| Nevirapine | 0.40 ± 0.14 | |
| 11626007 | 0.28 ± 0.1 | |
| 11726152 | 0.7 ± 0.22 | |
| 12126055 | 0.35 ± 0.13 | |
| 12126065 | 0.23 ± 0.07 |
Table 3.
Inhibition of HIV (IIIB) and HIV (A17) in TZM-bl cells using an extended dosing range (0.028–5000 nM; 15 points). Cytotoxicity dilution range exceeded the maximum inhibition range with concentration of 0.062–5.0 µM. Individual data points and curve fits are shown in Figure S1. (SI = selectivity Index (CC50/IC50))
| WT | A17 | Cytotoxicity | |||
|---|---|---|---|---|---|
| Compound | EC50 (nM) | SI | EC50 (nM) | SI | CC50 (µM) |
| 11726152 | 0.35 ± 0.05 | 2.2E+04 | 49 ± 7.8 | 1.6E+02 | 7.8 ± 5.2 |
| 11826116 | 0.34 ± 0.04 | 1.7E+04 | 25 ± 6.6 | 2.4E+02 | 6.0 ± 1.1 |
| 12126055 | 0.21 ± 0.01 | 1.3E+05 | 49 ± 9.5 | 5.7E+02 | 28 ± 66 |
| 12126065 | 0.47 ± 0.05 | 1.0E+04 | 3.4 ± 1.1 | 1.2E+03 | 4.8 ± 1.2 |
| 12126066 | 0.22 ± 0.03 | 4.1E+03 | 24.5 ± 5.1 | 3.7E+01 | 0.91 ± 0.08 |
| RPV | 0.35 ± 0.03 | >1.4E+04 | 2.5 ± 1.9 | >2.0E+03 | >5 |
| EFV | 0.68 ± 0.03 | >7.3E+03 | 18 ± 4.5 | >2.7E+02 | >5 |
NNRTI assay:
Our results confirm HIV reverse transcriptase as a target of this series of compounds (Table 2, Figures 3 and S3). We found nM inhibition, on par with RPV, for the compounds which retained potent A17 inhibition using a PicoGreen dsDNA quantitation reagent (Table 2). This confirmed that the target for these select compounds is HIV RT (Table 2). Interestingly, we had initially discovered a significant disparity in the EC50’s between cell (TZM-bl) and reverse transcriptase HIV I inhibition using a colormetric absorbance assay, so we expanded our testing using this assay to include several clinically used NNRTIs. This observation was also confirmed in nearly all NNRTI tested apart from nevirapine and doravirine (Figure S4). The fold difference varied from ~50–8500, with both 12126065 and RPV having over a 3000-fold difference. This suggests that this large difference is not unique for multiple bonafide NNRTIs, but it is assay dependent. This trend was identified previously by us during a large-scale comparison of HIV I whole cell and RT inhibition AC50’s using literature data 47. We also found that as compounds displayed increased potency, either in the RT or whole cell assay, the significance of this correlation was reduced, and compounds often showed orders of magnitude differences in potency. In addition, the physiochemical properties were also calculated for each compound using ChemAxon software, but no relationship was found that that would explain the difference in assay potency (Figure S4).
Figure 3.
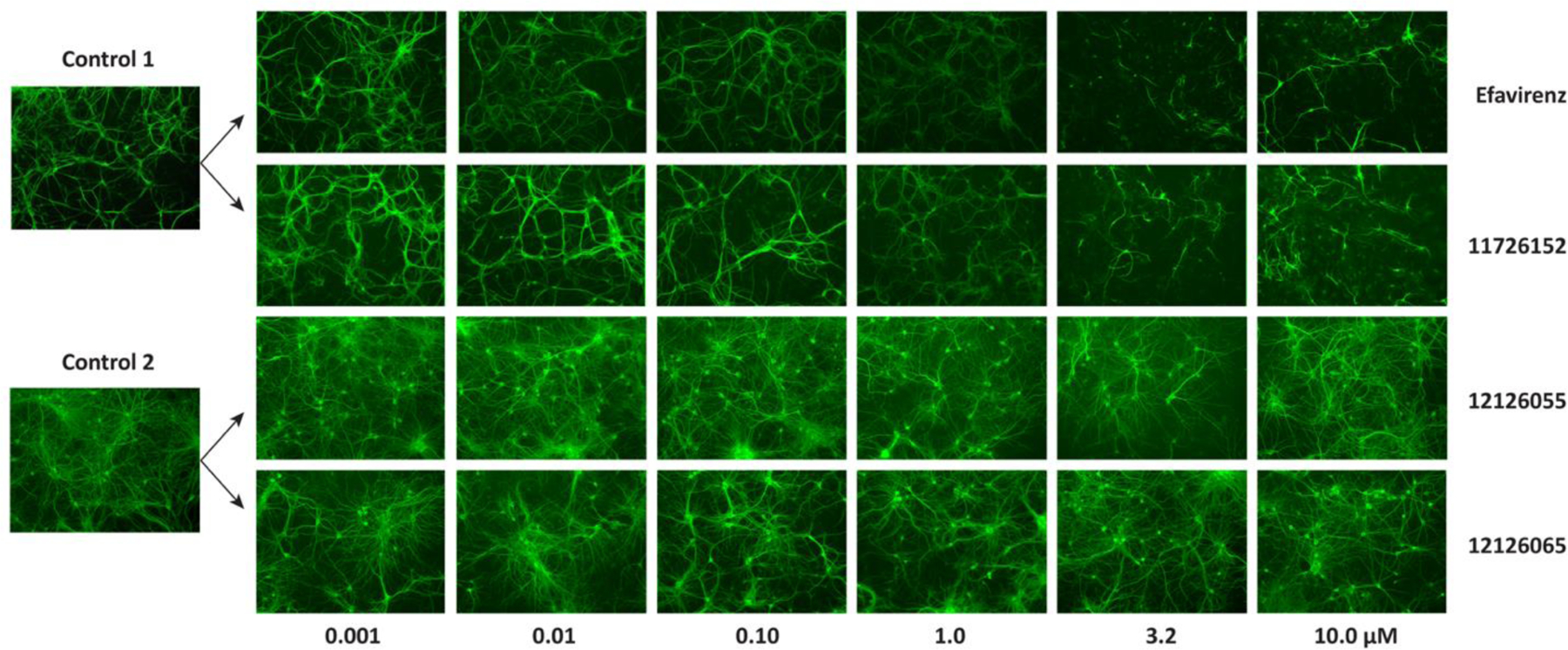
Representative images showing the dose-dependent changes in MAP-2 staining. Compounds were arbitrarily chosen as representative from each experiment as indicated by the controls.
In vitro ADME/Tox:
We assessed the in vitro ADME properties for our lead compound 12126065 (Table S3). Apart from the poor solubility, which is common for NNRTI’s and does not appear to impact the mouse pharmacokinetics (PK, described below), this compound had good metabolic stability in mouse and human liver microsomes, relatively low levels of CYP inhibition compared to expected required clinical levels (CYP2C9 IC50 1.42 µM), high protein binding and no indication of efflux in Caco-2 cells. While weak hERG inhibition was suggested for 12126065 (IC50 11.52 µM) based on a fluorescent polarization assay (Figure S5 and Table S2), the controls suggest that this may be due to non-specific, non-hERG binding. This inhibition was also less potent than the RPV control (IC50 3.31 µM).
In vitro Neurotoxicity Studies
MAP-2 Staining:
Neuronal toxicity was determined by the dose-dependent changes in MAP-2 staining in primary mouse neurons and was quantified using two methods: using relative MAP-2 intensity or area. Both approaches yielded very similar TC50’s, suggesting these are essentially equivalent. There was an exception with 12126065, where a TC50 was only able to be determined for relative area, but the deviation from the mean suggests there is very little confidence in this value regardless. Of the compounds tested, 3 showed a dose-dependent response: 11726152, RPV and EFV with TC50’s of 1.0 ± 0.5, 4.4 ± 1.5 and 1.5 ± 0.9 µM, respectively, as measured by MAP-2 intensity. Some toxicity was also shown in the other compounds tested, but these were significantly less than 11726152, RPV and EFV (Table 4, representative images shown in Figure 3 and curve fits shown in Figure S6). Using the methods previously described 48, no treatments resulted in complete loss of MAP-2 staining at the concentrations tested. As a complete loss of MAP-2 staining was not observed, an independent, semi-quantitative approach was also completed to assess neuronal damage (Figure S7). As neuronal density can vary significantly between experiments and within a well, samples chosen for imaging highlighting these differences are shown (Figure S8).
Table 4.
Summary of neurotoxicity of select molecules. TC50 = median toxic concentration (±SD), TC10 is the estimated concentration where we would see a 10% loss of MAP-2, and the “safety index” (s-index) attempts to quantify potential neurotoxicity. More specifically, the s-index looks at whether a drug at its maximum therapeutic concentration (assume 100xEC50) may begin to have some toxic effects as suggest by the TC10, which stringently shows the likelihood of toxicity. An s-index ≤1 indicates there is some risk of toxic effects, assuming full penetration of the blood brain barrier (i.e., plasma conc = brain conc). S-index = TC10/(100*EC50). An s-index was unable to be estimated for 12126065 as the TC10 could not be calculated due to low and no predicted toxicity for relative MAP-2 area and intensity, respectively. N.C.= Not able to be calculated due to low confidence
| Compound | TC50 (µM) Relative MAP-2 intensity | Estimated TC10 (µM) Relative MAP-2 Intensity | Safety Index | TC50 (µM) Relative MAP-2 Area | Estimated TC10 (µM) Relative MAP-2 Area | IC50 (nM) HIV IIIB | Safety Index | |
|---|---|---|---|---|---|---|---|---|
| 11626007 | 23 ± 18 | 6.1 | 6.5 | 34 ± 46 | N.C. | 9.3 | N.C. | |
| 11726152 | 1.0 ± 0.5 | 0.17 | 5.4 | 1.2 ± 0.6 | 0.050 | 0.32 | 1.6 | |
| 12126055 | 50 ± 140 | 4.5 | 140 | 48 ± 86 | N.C. | 0.32 | N.C. | |
| 12126065 | >10 | >10 | N/A | 100 ± 410 | N.C. | 0.31 | N.C. | |
| RPV | 4.4 ± 1.5 | 0.93 | 27 | 3.9 ± 2.3 | 0.60 | 0.35 | 17 | |
| EFV | 1.5 ± 0.9 | N.C. | N.C. | 0.9 ± 0.8 | 0.07 | 0.66 | 1.1 |
To rigorously quantify neurotoxicity potential, we introduce a “safely index” (s-index) metric (Table 4). Firstly, the TC10 is the estimated concentration where we would see an estimated 10% loss of MAP-2. The s-index looks at whether a drug at its maximum therapeutic concentration (assume 100xEC50) may begin to have some toxic effects as suggest by the TC10, which stringently shows the likelihood of toxicity. A s-index ≤1 indicate there some risk of toxic effects, assuming full penetration of the blood brain barrier (i.e., plasma conc = brain conc). By this measure both EFV and 11726152 suggest a neurotoxic risk potential, while the other tested compounds (>1) do not fall within the range. The TC10 could not be determined for 12126065 as there is little confidence to any neuronal toxicity within the concentrations tested.
Calcium Accumulation Assay:
The calcium accumulation shows the average for all neurons and indicates whether the compounds activate calcium signaling (acute stage) and or provoke a delayed rise. All compounds were tested at 1 µM. In the delayed phase, EFV showed a pronounced increase in calcium accumulation (Figure 4A). For reference, a toxic challenge that affects calcium regulation would provoke a delayed increase of about 800–1000 or more. Anything under about 200 would be considered in the normal range. Overall, only EFV seemed to have some toxic effects by this measure.
Figure 4.
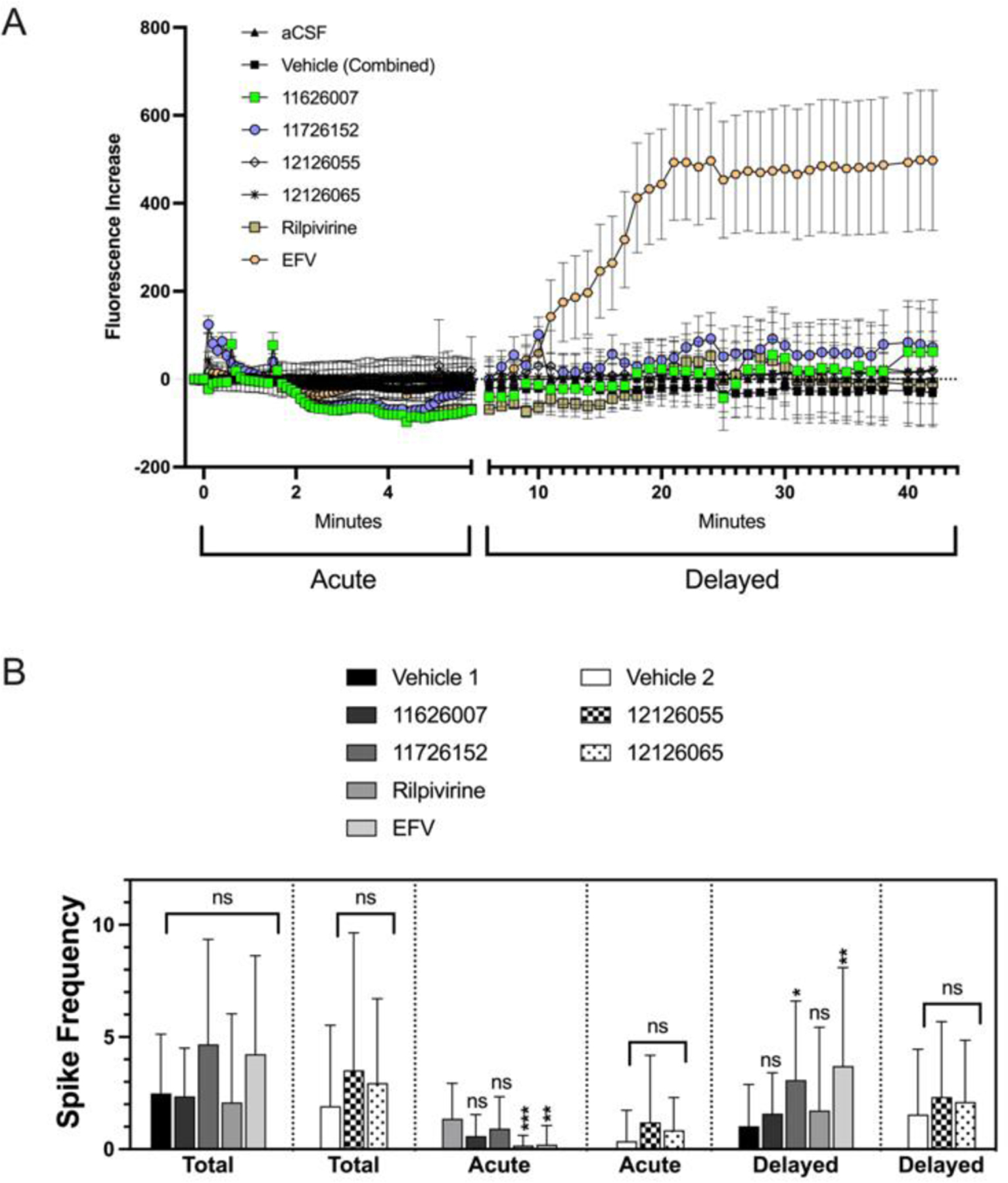
Calcium accumulation in primary mouse neuron cultures. (A) Shows the increase in the average calcium signaling for all neurons with a segmented x-axis indicating signaling stage (acute stage; depicted as axis with no additional ticks). (B) Calcium spiking shows the calcium transients for individual neurons and indicates whether the compounds activate calcium signaling. Average spikes are assessed in the acute or delayed phase as well as a summation average. All compounds were tested at 1 µM. Due to variability in control spiking, the tested compounds were compared to their corresponding control. Statistical significance was determined by a one-way ANOVA test (Brown-Forsythe and Welch) followed by Dunnett’s T3 multiple comparison tests as performed in Prism 9.2.0 for Mac OS (GraphPad; San Diego, CA). Error bars represent SEM (A) or SD (B) (n ≥ 23). SEM is shown to enhance visibility.
In addition to calcium accumulation for the entire population, a more sensitive approach was also taken. Calcium spiking shows the calcium transients for individual neurons, which gives an indication of signaling activity that may not necessarily translate to changes in the average calcium accumulation in the entire population, though this can help elucidate less pronounced effects. Due to the variability in the neuron activity by preparation, comparisons were made between their respective controls. The average number of spikes in total (acute + delayed phases) shows that there is no statistically significant overall elevated calcium signaling (Figure 4B). From the patterns shown in the raw spike summaries there is an acute suppression in RPV and EFV spiking frequency in the acute phase, which also led to increases in the delayed phase (Figure 4B). Based on previous data from our lab, the normal range is typically about 2–4 calcium spikes per neuron, so these increases are relatively minor though statistically significant. This suggests that the compounds are not totally benign, but the effects are small (Figure S9).
In vivo studies
Pharmacokinetics:
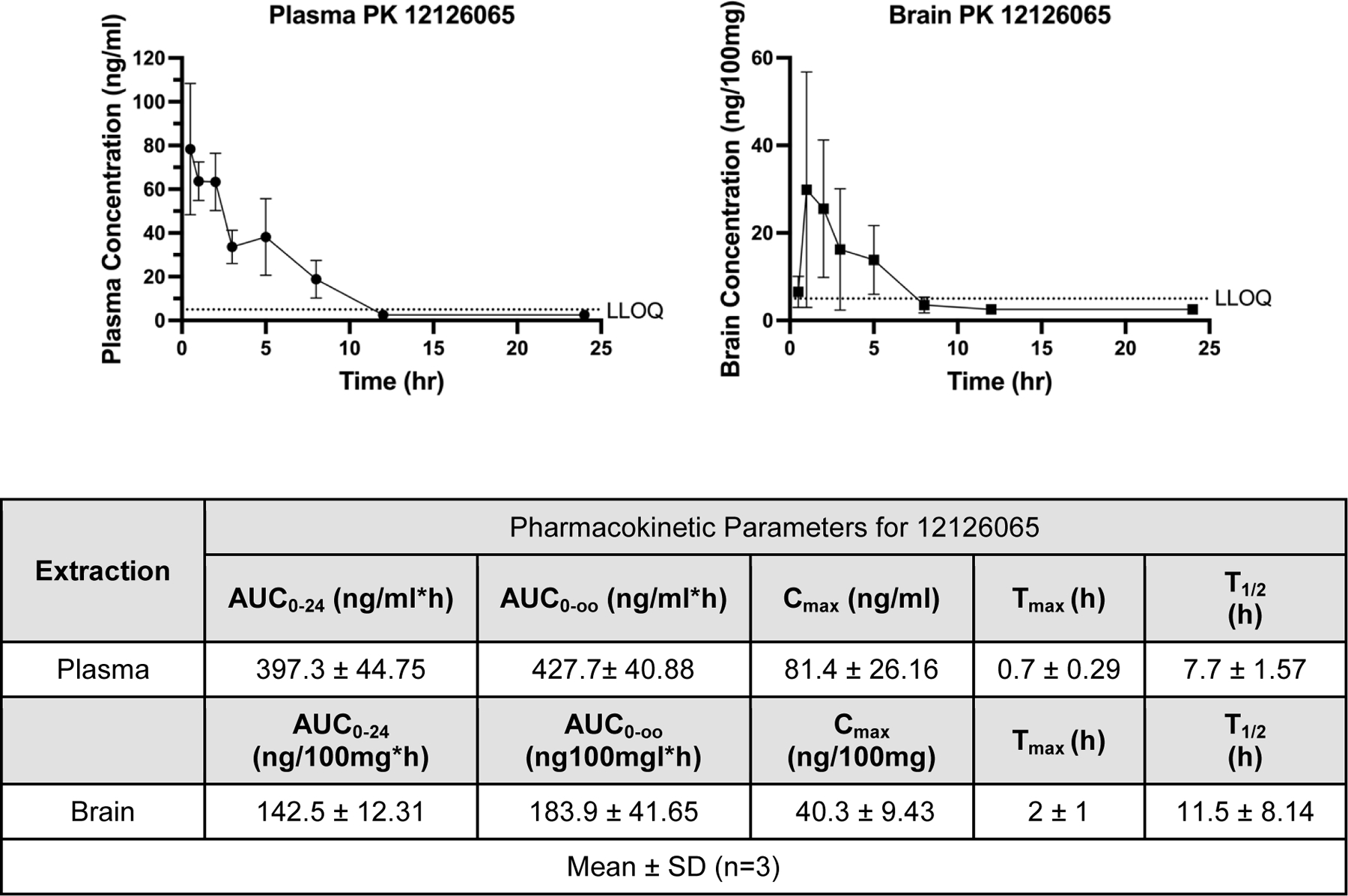
An intragastrically administered 250 mg/kg single-dose PK study in mice with 12126065 showed a T1/2 of 7.7 ± 1.57 hr and a Cmax 81.4 ± 26.16 ng/ml (Figure 5). Our data also suggests high concentrations of the compound in the brain (Cmax ~40.3 ± 9.43 ng/100 mg) and a similar half-life (11.5 ± 8.14 hr) suggesting it can readily penetrate the BBB.
Figure 5.
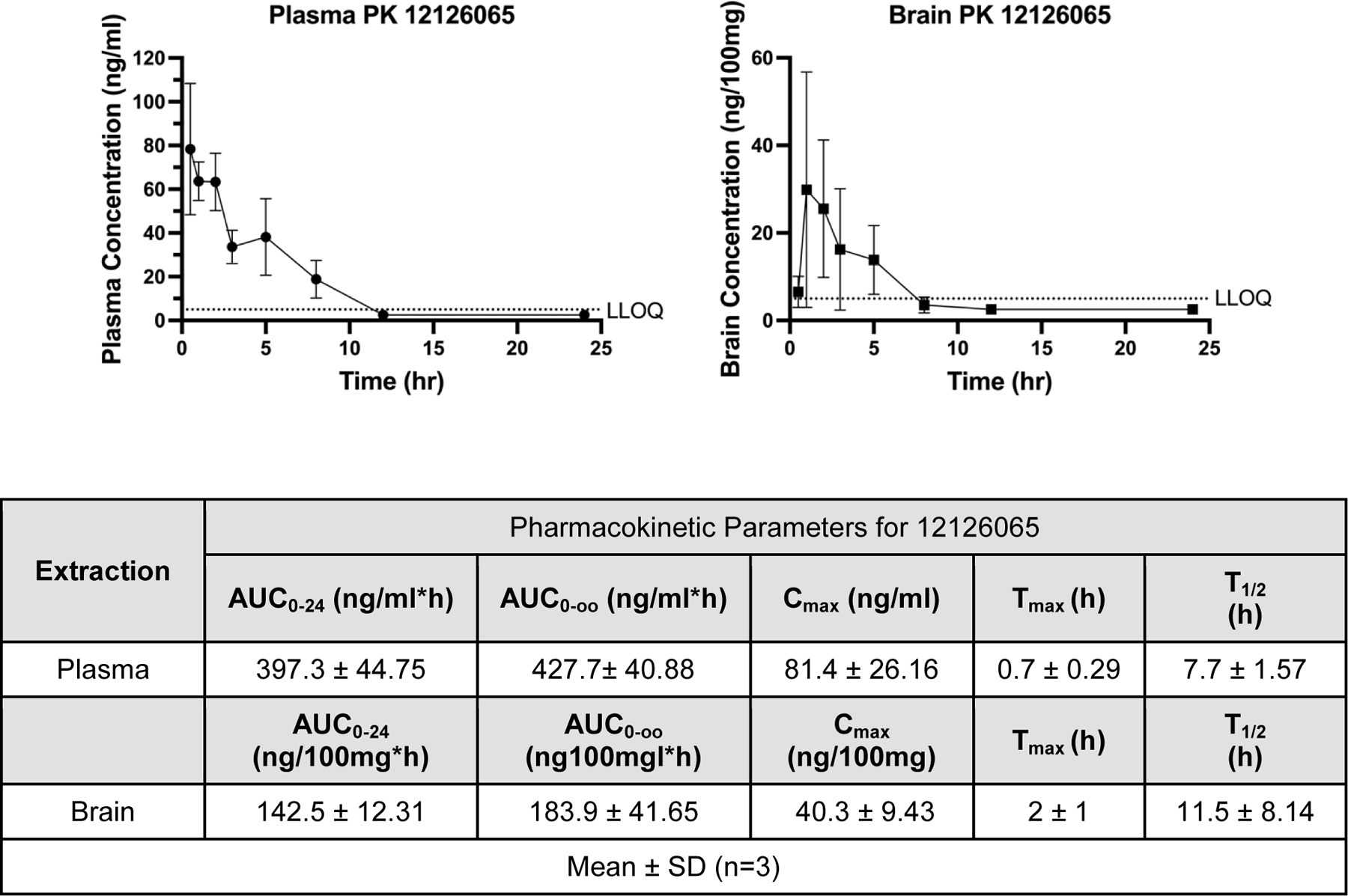
Pharmacokinetic data for mice dosed with 12126065 via intragastric intubation administration (250 mg/kg) in either plasma (left) or brain (right). Pharmacokinetic parameters were calculated with a Noncompartmental Pharmacokinetics Analysis method. LLOQ = 5 ng/ml; any calculated values below LLOQ were set to or . Error bars represent SD.
Acute Toxicity Study:
No visible signs of intoxication were observed with mice in the 2000 mg/kg oral dosing group over the course of the experiment. Over the 14-day timeline there were not any deviations in appearance, coat condition, excretion patterns or behavioural response of the experimental animals. The dynamics of weight gain of the experimental mice group also did not differ from the control animals (Figure S10). The statistical analysis of the measured results of weight dynamics of these animals was performed using the Šídák’s multiple comparisons test in Prism for Mac 9.3.1. The acute toxicity study showed no visible sign of toxicity with a 2000 mg/kg dose over a 14-day period.
Subacute Toxicity Study:
Neither experimental group (25 or 250 mg/kg) had any deviations in appearance, coat condition or excretion compared to the control group (each group n=10) following the 30-day study. The macroscopic examination of the internal organs (heart, lungs, liver, kidneys, spleen, stomach, intestinal canal) showed the experimental group had no visible pathological changes compared to the control group, apart from the liver. Heart, liver, kidneys, spleen had normal colour and were moderately full-blooded. Lungs were of pink colour at the section, adrenal glands were of normal size and moderately full-blooded. Stomach and intestinal canal were normal. The liver phenotype exception was found in the 250 mg/kg group only, with the majority of these animals having multiple microfocal and inflammatory infiltrates in their liver tissue. Expanded descriptions of each pathological analysis are shown in the Supplementary Text. Additional results presented in Figure S11 also show that this compound had no influence on the dynamics of weight gain or patterns of behavioural response in the experimental mice at either dosing regimen. The distribution of behavioural response did appear to show some variation between the groups, but this was not shown to be statistically significant (Dunnett’s multiple comparisons and Kolmogorov-Smirnov tests within Prism for Mac 9.3.11).
The analysis of the results comparing multiple biochemical and peripheral blood factors, differential blood count and relative organ weight found only one significant difference between the groups (n=7/group). These groups were compared statistically as a whole (i.e., biochemical factors, peripheral blood factors, etc. for 25 mg/kg or 250 mg/kg versus control) and no significant differences were found for any of categories apart from GOT (aminotransferase) differing in the 250 mg/kg dosing group only. This is based on a “Mann-Whitney rank comparisons test” using a Holm-Šídák method” with a statistically significant threshold of p < 0.05 done within Prism for Mac 9.3.11 (Figure S12). Total protein levels, alanine aminotransferase and alkaline phosphatase in the blood serum, which characterize the functional condition of liver in the mice, did not differ from the same factors in the control animals (Figure S12). No differences were found in urea or creatinine levels, suggesting normal kidney function. In addition, no significant differences were shown in either the peripheral blood or differential blood counts between these groups. The performed morphological analysis also did not reveal any differences in the relative weight of internal organs of animals.
Docking:
Docking was done in several of the numerous HIV RT protein structures deposited in the PDB to elucidate the improved activity of the most potent compound. Initially we docked 11626007 in a HIV RT (PDB 3MEC) crystal structure using the commercial software LibDock (Biovia, San Diego, CA). 11626007 was shown to fit very well (Figure S12C; Libdock score 140.36) and overlapped onto RPV with similar predicted interactions (Figure S13A). Docking of 12126065 also showed similar positioning as RPV in both wild-type (PDB 3MEC) and the K103N/Y181C mutant (PBD 4G1Q) with Libdock scores of 180.457 and 160.344, respectively, with the proposed interacting residues shown in Figures S13B,E and S14B. As a control, docking studies were also performed with RPV in the wild-type RT structure (PBD 4G1Q) with a Libdock score of 142.101 (Figure S12D) and with a similar pose to 12126065. For 12126065 it was found that several important residue interactions were conserved between the docking pose and those found in the crystal structure with RPV including: hydrophobic interactions between the acrylonitrile group and residues W229, Y188, F227 and L234, π-stacking with Y181/Y188, and a hydrogen bond with the main chain of K101 49. Our docking studies suggest these same types of interactions may also be formed by 12126065 and RT even though they are structurally distinct. Superimposition of RPV and 12126065 also show significant overlap of π-systems, hydrogen bond donor and acceptor, as well as hydrophobic functional groups (Figure S14C). In addition, the docking of 12126065 in the K103N/Y181C mutant shows that in the absence of Y181 a counterbalancing interaction is formed with Y183 in a similar manner to RPV in the same mutant 49.
t-SNE visualization:
A visualization technique, which compressed molecular descriptors (ECFP6) to a lower dimensional space, was used to assess the novelty of the chemical property space explored by the NNRTI molecules in this study. We have compared the chemical property space of two large libraries of compounds tested for HIV inhibition that had been previously assembled from multiple sources (primary literature, ChEMBL, NIAID dataset 47. These compounds assessed general HIV inhibition (Figure 6A) or HIV reverse transcriptase inhibition (Figure 6B). The t-SNE plots suggest our tested compounds occupy a unique property space as compared to the dataset compiled in 2019 from various sources. The whole cell and RT datasets have ~19,000 and ~5700 compounds, respectively. We have also highlighted all the currently FDA-approved NNRTIs in these plots, which do not overlap with our novel molecules.
Figure 6.
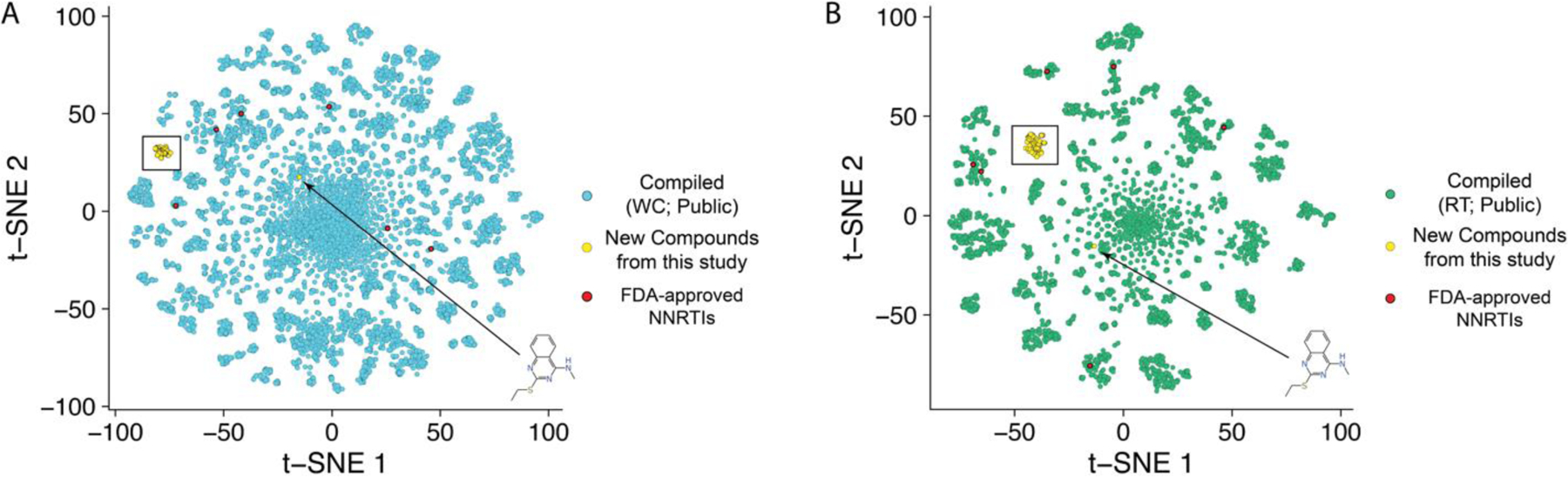
t-SNE plots of a previously compiled dataset of compounds tested against HIV in vitro versus the dataset of “new compounds from this study”. The “public” datasets are assembled from multiple sources (primary literature, ChEMBL, NIAID dataset (circa 2017)). These compounds were assessing (A; blue) general viral inhibition or (B, green) in assays specifically looking at HIV reverse transcriptase inhibition (various methods). As a reference, FDA-approved NNRTIs (red) are highlighted to show a completely distinct chemical space for the new compounds from this study. The new compounds are shown in yellow. A black box encompasses all of the new compounds from this study except for the structure indicated in the lower righthand side of each graph, which is a different structural class.
Discussion and Conclusions
HIV remains a pronounced global threat with the prevalence of drug resistant mutations representing a significant portion of infections. While current therapies are transformative in terms of life expectancy and quality of life, the need for additional treatments in preparation for drug resistant strains remains of utmost importance. Additionally, current therapies have yet to eradicate the prevalence of HAND; therefore, this still represents an important therapeutic goal.
One of the current therapies for HIV includes a cocktail of multiple HIV-targeting drugs with regimens including but not limited to a nucleoside RT inhibitor (NRTI), a non-nucleoside RT inhibitor (NNRTI), and/or protease inhibitors 50. While cART therapy has drastically changed the survivability and quality of life for those infected with HIV, a significant portion (10–30%) are still unable to control viral replication completely 51, 52. This has led to a myriad of drug-resistance mutants, with the most common being associated with approved NNRTIs. It is also common for cART naïve infected individuals to also have drug resistant mutations, suggesting that the virus is often drug-resistant prior to infection 17.
Several groups have focused on developing new NNRTI’s 53–72. Substituted imidazoles were developed by GSK (cpd 43, EC50 0.1nM) and progressed to the clinic 53. Substituted pyridinones (MK-1439 Doravirine, IC50 11 nM) were developed by Merck 54, demonstrated good activity in combination with other drugs 55 and showed activity against mutant forms of HIV-RT (FDA approved in 2018 56). Other classes of compounds include: 2,4,5-trisubstituted thiazole derivatives (Compound 24, IC50 0.046 μM) 57, diarylnicotinamide derivatives (compound 6b11, EC50 0.027μM) 58, diaryltriazine derivatives (compound 1, Ki 9 nM) 59, triazine derivatives (Compound DSC-a4, EC50 7.8 nM) 60, piperidin-4-yl-aminopyrimidine derivatives (EC50 single digit nM) 61, bicyclic arylaminoazines (e.g. Compound 8f, EC50 1.1 nM)62, aryl-phospho-indole (IDX899, EC50 1 nM)63, arylazolyl(azinyl)thioacetanilides derivatives (LAD-8, EC50 0.78 μM)64, diarylnicotinamide 1,4-disubstituted 1,2,3-triazoles (3b9, EC50 0.02 μM) 65 and diarylbenzopyrimidines (12z, EC50 3.4nM) 66. These are select examples of the earlier extensive efforts in NNRTI chemistry 67–72. In the last few years we have also identified several studies focused on NNRTI (including 2,4,5-trisubstituted pyrimidines 73, hydroxyl-substituted biphenyl-diarylpyrimidines 74, thiophene-biphenyl-DAPY 75, biphenyl-substituted diarylpyrimidines 76, hydrazone-substituted thiophene[3,2-d]pyrimidines 77, 78, diarylpyrimidines 79–83, indolylarylsulfones 84, 85, dihydrofuro[3,4-d]pyrimidines 86, triazoles 87, 3-(1,3,4-thiadiazol-2-yl)thiazolidin-4-one 88, rilpivirine analogs 89, 2-(((5-akly/aryl-1 H-pyrazol-3-yl)methyl)thio)-5-alkyl-6-(cyclohexylmethyl)-pyrimidin-4(3H)-ones 90, morpholine-substituted diarylpyrimidines 91, (2-hydroxyethoxy)methyl]-6-(phenylthio)thymine (HEPT) analogs 92, biphenyl -substituted pyridone analogs 93 and indolylarylsulfone (IAS) analogs 94).
In the course of this work, we focused on optimizing the antiviral activity of a N-phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazol-3-amine core with various substituents on fragments “A” and “C” (Figure 1). From these data we can now generalize the requirements of this scaffold to retain its activity: (1) the secondary amine in the linker to fragment “C” was required, (2) the fragment “C” phenyl group requires a halogen, cyano or acetonitrile substituent, (3) naphthalene in fragment “A” is responsible for higher antiviral activity and (4) the naphthalene containing a cyano, acetonitrile or acrylonitrile group in the 6-position substantially enhanced the potency and was the most effective substituent. Our data agrees with essentially all known RT inhibitors, which are comprised of a central azaheterocycle core between substituted arenes, with the vast majority of active compounds also having a cyano group as a substituent.
In the process of this work, we also identified other heterocycles which are well tolerated in the “A” fragment, including imidazothiophene (11926331), 1,3-thiazole (11926332) and thiophenes (11926333), etc., with these compounds showing both excellent RT and whole cell virus activity (Table S1). Further, the replacement of the phenyl ring with a heterocyclic moiety provides new possibilities for the future design of analogs with promising biological and physicochemical properties.
We also discovered the cyano-substituted naphthalene rings in the “A” position resulted in the most potent activity against the tested mutants, with the most potent compound having an extra vinyl group between the naphthalene ring and the nitrile group. We propose that the extra flexibility in the domain of the molecule likely enables a more favorable steric interaction within the binding site in these mutants than the aryl nitriles, which is a similar hypothesis for the retained potency of RPV and ETR 95, 96.
It has been well established that EFV, a clinically used NNRTI, has negative neurological side effects in patients 28, so one focus of this study was to assess neuronal toxicity of our new class of NNRTIs to avoid this clinical pitfall. Primary murine neuronal cell toxicity, assessed by both a reduction in MAP-2 staining and effects on calcium signaling, confirmed the toxicity of EFV (TC50 = 0.99 – 1.57 µM with significant reduction of acute signaling). Recommended intake for adults for EFV typically results in plasma levels of between 3.17 – 12.67 µM, though variability in quantified plasma levels vary substantially and concentrations of 30 – 50 µM are common and up to 80 µM has been recorded. EFV also readily passes the BBB with typical median EFV CSF levels of between 35 – 90 nM with recommended intake 28. Our in vitro data shows an estimated TC10 of 70 nM in primary neurons, which coincides with typical EFV CSF levels, suggesting that a TC10 may represent levels associated with neurotoxicity. Interestingly, RPV also showed in vitro neurotoxicity, with a TC10 of ~600–930 nM and statistically significant reduced acute signaling at 1 µM. Since there is an in vitro potency difference of RPV over EFV, this translates to a reduced in vivo effective concentration requirement for RPV, with a proposed plasma target concentration of 12.1 ng/ml or 33 nM (protein binding-adjusted EC90). There are recorded CSF concentrations of up to 2.9 ng/ml (7.9 nM) for RPV 97, well below the estimated TC10 found in our study. Nonetheless neurological adverse events are also common in RPV-treated patients 28, though this frequency is lower than with EFV. Based on this, it is ambiguous if the in vitro toxicity directly relates to the recorded neurological events or not, but the correlation is still there. Regardless, 12126065 showed much lower in vitro neurotoxicity as compared to EFV or RPV, suggesting that at therapeutic levels, based on an estimated EC90 of 19 nM, neurotoxicity is not expected.
Previously, RPV has been shown to inhibit hERG in vitro with a relatively low concentration (300 nM) and an IC50 3.1 µM in cardiac myocytes 98. In addition to this in vitro data, a prolonged QT interval was seen prior to Phase 3 clinical trials prompting a change in the proposed therapeutic dosing regimen from 75 mg to 25 mg (qd). A follow-up QT study suggested that 25 mg qd did not demonstrate a substantial effect on the QT interval and did not present a cardiac risk. Current RPV dosing is close to a threshold which may prevent increased doses required to treat future novel NNRTI drug-resistant mutants. We have independently assessed hERG binding for RPV and several of our compounds (Figure S5 and Table S2). RPV (IC50 3.31 ± 0.29µM) was similar to the value calculated from the patch clamp assay (IC50 3.1 µM). Our lead compounds show significantly reduced in vitro hERG inhibition compared to RPV (Figure S5 and Table S2) suggesting potential benefit to increase the dose without subsequent cardiac risks.
Like other NNRTIs, these molecules have low solubility, but pharmacokinetics results show this does not completely mitigate oral bioavailability as they have high stability in plasma and microsomes (Table S3) with an in vivo half-life of 7.7 ± 1.57 hr (Figure 6). 12126065 has also shown significant brain penetration following oral administration, suggesting this NNRTI readily passes the BBB and may be used to target HIV that has permeated this compartment. 12126065 also demonstrated a significantly lower in vitro neurotoxicity as compared to RPV and EFV and therefore may avoid the neurotoxic effects observed with these earlier NNRTI’s.
Our acute and subacute in vivo studies have shown no apparently toxicity at the concentrations tested. For the acute study, no deviations from the control group were shown after the single, oral administration of a 2000 mg/kg dose in a 14-day study. For our subacute studies, we evaluated two, daily dosing concentrations of 25 and 250 mg/kg. Over this 30-day study only one of the measured metrics showed a significant difference between the experimental groups as compared to the control group, which was a reduced aspartate aminotransferase activity in the 250 mg/kg group only. As far as we are aware, a reduced aspartate aminotransferase activity is not related to any type of pathogenesis. Overall, these studies suggest that toxicity is unlikely to be observed even with chronic treatment in humans as the therapeutic dose is likely to be significantly lower.
In summary, we have described the synthesis of a series of potent HIV-1 inhibitors with a N-phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazol-3-amine core, many with picomolar activity (EC50) against HIV I in TZM-bl cells as well as low cytotoxicity producing high SIs (typically exceeding 160). A t-SNE analysis showed these compounds are distinct from both the current FDA-approved NNRTIs and from other compounds assessed previously (Figure 6). Several of our compounds retained activity against a K103N/Y181C double mutant (Figure 2D) with the most potent of these, 12126065, having an EC50 1.33 ± 0.16 nM, which is comparable with RPV. 12126065 also retained activity against additional clinically relevant mutants (Y181C, L100I/K103N, K103N).
In conclusion, we have developed a class of NNRTIs with supporting data suggesting potency against drug resistant mutants, no apparent neurotoxicity, BBB-penetration, and no subacute toxicity, thus potentially representing an important addition to the catalog of compounds used in the treatment of HIV (Figure S15). Following future in vivo efficacy confirmation in mouse 99 this body of work shows that these N-phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazol-3-amines may provide an additional class of molecules to help combat drug resistance as well as comorbidities for this on-going global threat.
Experimental Section
Chemistry Synthesis
An effective method for synthesizing derivatives of N-phenyl-1-(phenylsulfonyl)-1H-1,2,4-triazoles was developed, enabling introduction of targeted replacements into the molecule structure. The key intermediate for the preparation of the final compound was substituted methyl N’-cyano-N-phenylimidothiocarbamate, which can be prepared by different methods. One method is based on the reaction of anilines with dimethyl cyanothioimidocarbonate with elimination of methylmercaptane. This method works for anilines having either electron donors or neutral substitutes.
All reagents and solvents were purchased from commercial suppliers and used without further purification. 1H and 13C Spectra were measured on Bruker AC-500 (500 MHz, 1H) or Bruker AC-200 (75 MHz, 13C). Chemical shifts were measured in DMSO-d6 or CDCl3, using tetramethylsilane as an internal standard, and reported as units (ppm) values. The following abbreviations are used to indicate the multiplicity: s, singlet; d, doublet; t, triplet; m, multiplet; dd, doublet of doublets; brs, broad singlet; brm, broad multiplet.
HRMS: spectra were recorded on an Agilent 1290 Infinity II HPLC system coupled to Agilent 6460 triple-quadrupole HRMS: spectrometer equipped with an electrospray ionization source. The chromatographic separation was carried out on Agilent Eclipse Plus C18 RRHD column (2.1 × 50 mm, 1.8 µm) at 40 ºC with sample injection volume of 0.2 µL. The mobile phase comprising 0.1 % formic acid / water (A), and 0.1 % formic acid and 85 % acetonitrile / water (B) was programmed to do a gradient elution (0.0–3.0 min, 60 % B; 3.0–4.0 min, 60 % to 97 % B; 4.0–6.0 min, 97 % B; 6.0–6.1 min, 97 % to 60 % B) at a flow rate of 0.4 mL/min. The HRMS: spectrometric detection was operated in a positive ion mode. Optimal parameters were capillary voltages of 3500 V, a nebulizer pressure of 35 psi, a gas temperature of 350 ºC, a gas flow rate of 12 L/min.
Purity was measured by analytical high-performance liquid chromatography (HPLC) on an Elute HPLC system (Bruker Daltonik) equipped with Azura UVD 2.1S UV detector (Knauer) using Acquity HSS T3 column (2.1 × 100 mm, 1.3 µm, 100 Å) at 30 °C, 2 µL injection, 250 µL/min gradient elution 30─95% B (A: 0.1% FA in H2O, B: 0.1% FA in MeCN) over 9 min with 1 min gradient delay, 1 Hz acquisition rate at 254 nm. Data were processed with Compass DataAnalysis 5.1 (Bruker Daltonik). Purity is > 95% of all final compounds.
Melting points were determined by Electrothermal 9001 (10 ºC per min) and are uncorrected. Merck KGaA silica gel 60 F254 plates were used for analytical thin-layer chromatography. Column chromatography was performed on Merck silica gel 60 (70–230 mesh). Yields refer to purified products and are not optimized. The starting materials and the intermediates of the synthetic reaction schemes were also isolated and purified as desired using conventional techniques, including but not limited to, filtration, distillation, crystallization, chromatography, etc. Such materials were characterized using conventional means, including physical constants and spectral data.
In order to synthesize the compounds having an electron-withdrawing group in the aniline fragment we successfully developed two alternative methods. Both these methods are based on the initial reaction of an aniline, which is independent of whether it contains either electron-donor or electron-acceptor substituents. In one case, we used dimethylaminothiocarbamoyl chloride in an anhydrous medium. The corresponding aryl isothiocyanate is obtained in quantitative yield, which is subsequently reacted with cyanamide in the presence of sodium ethoxide, followed by treatment with methyl iodide, forming the key intermediate N’-cyano-N-phenylimidothiocarbamate. The same intermediate can be synthesized by the reaction of different substituted anilines with thiophosgene under alkaline conditions. For example, in the presence of triethylamine, diisopropylethylamine, sodium methoxide or calcium carbonate etc. in an anhydrous solvent causes the formation of the corresponding isothiocyanate and its following conversion to N’-cyano-N-phenylimidothiocarbamate by the reaction with cyanamide. All 3 methods work well with high yield and purity of the target compounds.
The reaction of the resulting methyl N’-cyano-N-phenylimidothiocarbamate with hydrazine hydrate leads to the closure of the 1,2,4-triazole ring. The last step is the reaction with an arylsulfonyl chloride, which usually leads to a mixture of the two isomers. However, we were able to select the conditions for the reaction and crystallization of the resulting mixture at which the concentration of the desired isomer N3-phenyl-1- (phenylsulfonyl)-1H-1,2,4-triazole-3,5-diamine significantly prevails. Alternatively, we can separate the isomers by chromatography.
All compounds were synthesized according to Figure 1 with details described below.
Step a).
A mixture of the corresponding aniline (1.0–1.2 mmol) and dimethyl cyanodithioiminocarbonate (1.0 mmol) in a small volume of n-butanol was refluxed for 3–4 hours. The reaction mixture was cooled, the formed precipitate was filtered off and then washed by hexane. The desired methyl N’-cyano-N-R-phenylimidothiocarbamate was recrystallized from ethanol.
Step b).
A solution of the aniline derivative (1.0 mmol) in dry toluene or benzene was treated with solid dimethylthiocarbamoyl chloride (1.0–1.1 mmol) and was refluxed for 2–3 hours. The reaction mixture was cooled, dissolved in hexane and the resulting solid was filtered off. The mother solution was evaporated in a vacuum and the desired isothiocyanate derivative was used without additional purification.
Step c).
A mixture of substituted anilines (1.0 mmol) with the addition of a suitable alkali, such as triethylamine, diisopropylethylamine, calcium carbonate, potassium carbonate etc., (1.8–2.2 mmol) in either toluene, benzene or CH2Cl2, was treated with thiophosgene (1.0 mmol) and stirred for 2 hours at room temperature or refluxed for 4–6 hours. The reaction mixture was diluted by water and the organic phase was separated, concentrated in vacuum, and then chromatographed (preferably benzene) to give the desired isothiocyanate derivative as a light yellow or white solid.
Step d).
A mixture of sodium ethoxide (2.0–2.2 mmol) in 20 mL of ethanol and cyanamide (2.0 mmol) was stirred at room temperature for 30–40 minutes. The corresponding isothiocyanatobenzene (2.0–2.2 mmol) was added to the reaction mixture and stirred for 1.5 hr, followed by the addition of iodomethane (4.0–4.5 mmol), and finally refluxed for 1–2 hours and stored overnight at room temperature. The resulting residue was filtered off and dried to give the methyl N’-cyano-N-R-phenylimidothiocarbamate.
Step e).
A solution of hydrazine in water (3.0–5.0 mmol) was added to a solution of methyl N-4-bromo-3,5-dichlorophenyl-N’-cyanocarbamimidothioate (1.0–1.5 mmol) in ethanol and heated at 70 °C for 3–4 hours. The reaction mixture was cooled to room temperature and suspended in ice water. The precipitate was collected and recrystallized from ethanol to give the N5-phenyl-1H-1,2,4-triazole-3,5-diamine as an off-white solid.
Step f)
A high quality sulfonyl chloride derivative (1.0–1.2 mmol) was added to the suspension of the corresponding N5-phenyl-1H-1,2,4-triazole-3,5-diamine (1.0 mmol) in a small volume of pyridine. The reaction mixture was stored overnight at room temperature, then diluted with water, cooled to 4 °C for 6–24 hours and then the precipitate was filtered off. The desired N3-(4-phenyl)-1-(2-R6-sulfonyl)-1,2,4-triazole-3,5-diamine isomer was separated from the byproduct by recrystallization (EtOH or EtOH/DMF) or by chromatography (often chloroform:methanol 10/1).
Examples of synthesis and spectra are provided in the Supplemental. Analytical data is only presented for 53 of the most relevant compounds described herein. Additional synthetic details for the additional compounds are available upon request.
Analytical data of synthesized compounds
Figure S16 shows HPLC traces of 6 representative compounds and Figure S17 describes 1H and 13C NMR for all tested compounds.
N3-(4-chlorophenyl)-1-(2-naphthylsulfonyl)-1,2,4-triazole-3,5-diamine 11626007
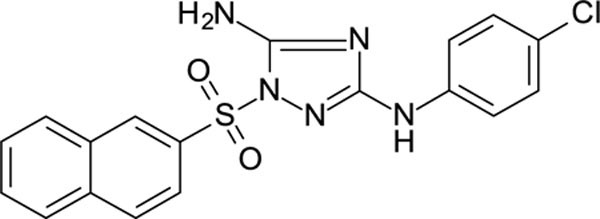
White solid in yield 55%. Mp 211–2 oC. HRMS: calc. for C18H14ClN5O2S [M]+ 399.8550, found 399.8552. Mp = 88.7 °C. 1H NMR (500 MHz, DMSO-d6) δ 9.28 (s, 1H, NH), 8.72 (s, 1H, HC(1`)), 8.23 (d, J = 7.8 Hz, 1H, HC(8`)), 8.18 (d, J = 8.8 Hz, 1H, C(4`)), 8.05 (d, J = 8.0 Hz, 1H, HC(5`)), 7.90 (dd, J = 8.7, 1.8 Hz, 1H, HC(3`)), 7.82 – 7.67 (m, 2H, HC(6`,7`)), 7.45 (d, J = 8.9 Hz, 2H, HC(3``,5``)), 7.40 (s, 2H, NH2), 7.26 (d, J = 8.9 Hz, 2H, HC(2``,6``)). 13C NMR (75 MHz, DMSO-d6) δ 159.12 (C-3), 156.89 (C-5), 139.64 (C-1``), 134.89 (C-4à), 133.10 (C-2`), 131.65 (C-8à), 129.76 (C-3`), 129.86 (C-4`), 128.97 (C-5`), 128.35 (C-3``, 5``), 128.25 (C-1`), 127.97 (C-6`), 123.69 (C-4``), 122.12 (C-7`), 121.65 (C-8`), 118.12 (C-2``, 6``).
5-[[5-amino-3-(4-cyanoanilino)-1,2,4-triazol-1-yl]sulfonyl]naphthalene-2-carbonitrile 11726152
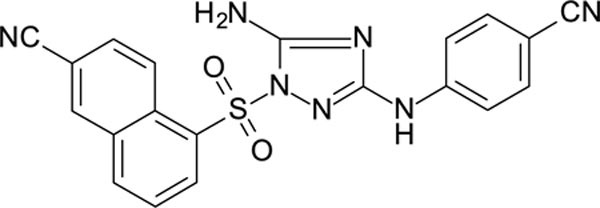
White solid in yield 47%. Mp 268–9 oC. HRMS: calc. for C20H13N7O2S [M]+ 415.4292, found 415.4291. Mp = 88.7 °C. 1H NMR (200 MHz, DMSO): δ 9.79 (s, 1H, NH), 9.12 (d, J = 9.1 Hz, 1H, HC(4`)), 8.76 (d, J = 1.7 Hz, 1H, HC(1`)), 8.64 (d, J = 7.2 Hz, 1H, HC(6`)), 8.50 (d, J = 8.0 Hz, 1H, HC(8`)), 8.15 (dd, J = 9.1, 1.9 Hz, 1H, HC(3`)), 7.92 (t, J = 7.9 Hz, 1H, HC(7`)), 7.67 (d, J = 8.8 Hz, 2H, HC(3``), HC(5``)), 7.56 (s, 2H, NH2), 7.51 (d, J = 8.9 Hz, 2H, HC(2``), HC(6``)). 13C NMR (50 MHz, DMSO): δ 158.51 (C-3), 156.33 (C-5), 144.53 (C-1``), 136.98 (C-5`), 135.32 (C-1`), 133.56 (C-4à), 133.01 (C-3``, C-5``), 132.86 (C-8à), 132.28 (C-3`), 128.92 (C-8`), 126.77 (C-6`), 126.55 (C-7`), 126.31 (C-4`), 119.49 (CN-C2`), 118.12 (CN-C4``), 116.61 (C-2``, C-6``), 110.18 (C-1`), 101.60 (C-4``).
4-[[5-amino-1-(2-naphthylsulfonyl)-1,2,4-triazol-3-yl]amino]-2-chloro-benzonitrile 11726158
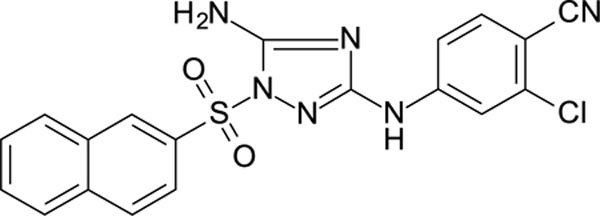
White solid in yield 45%. HRMS: calc. for C19H13ClN6O2S [M]+ 424.8645, found 424.8646. 1H NMR (500 MHz, DMSO): δ 10.09 (s, 1H, NH), 8.75 (s, 1H, HC(1`)), 8.35–7.32 (m, 11H). 13C NMR (126 MHz, DMSO): δ 158.58 (C-3), 157.26 (C-5), 145.82 (C-4``), 135.96 (C-2``), 135.10 (C-4à), 134.94 (C-6``), 132.88 (C-2`), 131.37 (C-8à), 130.03 (C-3`), 129.96 (C-4`), 129.57 (C-5`), 128.18 (C-1`), 127.97 (C-6`), 121.65 (C-8`), 116.64 (CN), 116.35 (C-3``), 115.37 (C-5``), 101.77 (C-1’’).
N3-(3,4-dichlorophenyl)-1-(1-naphthylsulfonyl)-1,2,4-triazole-3,5-diamine 11726159
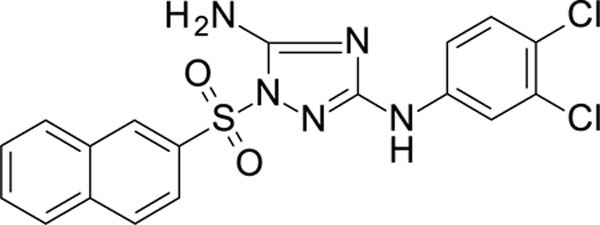
White solid in yield 43%. HRMS: calc. for C18H13Cl2N5O2S [M]+ 434.2997, found 434.2997. 1H NMR (200 MHz, DMSO): δ 9.54 (s, 1H, NH), 8.72 (d, J = 2.0 Hz, 1H, HC(2``)), 8.21 (d, J = 8.2 Hz, 1H, HC(2`)), 8.18 (d, J = 8.5 Hz, 1H, HC(4`)), 8.06 (d, J = 8.9 Hz, 1H, HC(8`), 7.90 (dd, J = 8.7, 1.9 Hz, 1H, HC(5`)), 7.82–7.65 (m, 3H, HC(3`,5`,6`)), 7.48 (s, 2H, NH2), 7.43 (d, J = 8.8 Hz, 1H, HC(5``)), 7.36 (dd, J = 8.8, 2.0 Hz, 1H, HC(6``)). 13C NMR (50 MHz, DMSO): δ 158.90 (C-3), 157.00 (C-5), 141.05 (C-1``), 135.36 (C-1`), 132.77 (C-3``), 131.16 (C-4à), 130.65 (C-8à), 130.04 (C-2`), 129.57 (C-5``), 129.27 (C-4`, C-5`), 127.79 (C-8`, C-6`), 127.70 (C-4``), 121.52 (C-3`), 121.12 (C-7`), 117.43 (C-6``), 116.54 (C-2``).
5-[[5-amino-3-(4-cyano-2-methyl-anilino)-1,2,4-triazol-1-yl]sulfonyl]naphthalene-2-carbonitrile 11826114
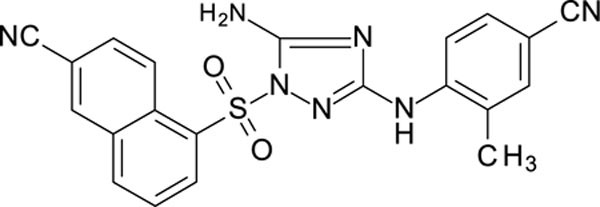
White solid in yield 61%. HRMS: calc for C21H15N7O2S [M]+ 429.4558, found 429.4558. 1H NMR (200 MHz, DMSO) δ 9.12 (s, 1H, NH), 9.08 (d, J = 9.1 Hz, 1H, HC(4`)), 8.74 (d, J = 1.7 Hz, 1H, HC(1`)), 8.62 (d, J = 7.2 Hz, 1H, HC(6`)), 8.48 (d, J = 8.0 Hz, 1H, HC(8`)), 8.10 (dd, J = 9.1, 1.9 Hz, 1H, HC(3`)), 7.93 (t, J = 7.9 Hz, 1H, HC(7`)), 7.60 (d, J = 8.2 Hz, 1H, HC(6``)), 7.52 (s, 2H, NH2), 7.43 (dd, J = 1.2, 8.2 Hz, 1H HC(5``), 7.40 (d, J = 1.2 Hz, 1H, HC(2``)), 2.30 (s, 3H, CH3). 13C NMR (50 MHz, DMSO) δ 158.71 (C-3), 156.43 (C-5), 141.43 (C-1``), 136.28 (C-5`), 135.62 (C-1`), 133.86 (C-4à), 134.71 (C-3``), 132.66 (C-8à), 132.48 (C-3`), 131.21 (C-5``), 128.91 (C-2``), 128.72 (C-8`), 126.87 (C-6`), 126.45 (C-7`), 126.51 (C-4`), 120.21 (C-6``), 119.49 (CN-C2`), 116.12 (CN-C4``), 110.18 (C-2`), 105.60 (C-4``), 17.81 (CH3).
5-[[5-amino-3-(3-chloro-4-cyano-anilino)-1,2,4-triazol-1-yl]sulfonyl]naphthalene-2-carbonitrile 11826116
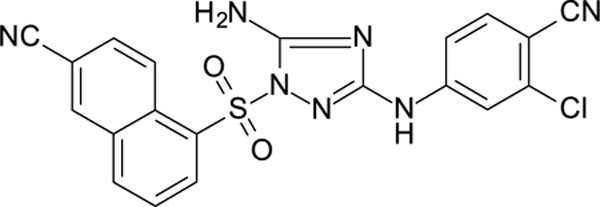
White solid in yield 74%. Mp 236–7 oC. HRMS: calc. for C20H12ClN7O2S [M]+ 449.8741, found 449.8742. Mp = 88.7 °C. 1H NMR (500 MHz, DMSO): δ 10.07 (s, 1H, NH), 9.04 (d, J = 9.0 Hz, 1H, HC(4`)), 8.79 (d, J = 1.8 Hz, 1H, HC(1`), 8.65 (d, J = 7.5 Hz, 1H, HC(6`)), 8.52 (d, J = 8.2 Hz, 1H, HC(8`)), 8.11 (dd, J = 9.0, 1.8 Hz, 1H, HC(3`)), 7.94 (t, J = 7.9 Hz, 1H, HC(7`)), 7.78 (d, J = 8.7 Hz, 1H, HC(5``)), 7.66 (s, 2H, NH2), 7.64 (d, J = 2.2 Hz, 1H, HC(2``)), 7.41 (dd, J = 8.7, 2.2 Hz, 1H, HC(5``)). 13C NMR (126 MHz, DMSO): δ 158.15 (C-3), 156.40 (C-5), 145.60 (C-1``), 137.12 (C-3``), 135.93 (C-5`), 135.47 (C-5``)), 134.91 (C-1`), 133.77 (C-6`), 132.79 (C-8à), 132.01 (C-4à), 129.17 (C-8`), 129.08 (C-3`), 126.64 (C-7`), 125.72 (C-4`), 118.11 (CN-C4``), 116.62 (CN-C2`), 116.26 (C-2``), 115.32 (C-6``), 110.16 (C-2`), 101.90 (C-4``).
N3-(4-chlorophenyl)-1-[[5-(dimethylamino)-1-naphthyl]sulfonyl]-1,2,4-triazole-3,5-diamine 11826313
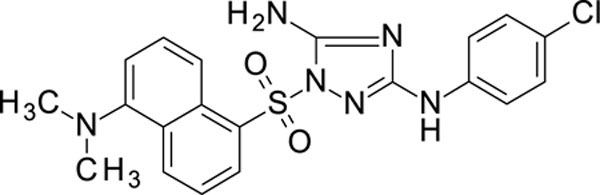
White solid in yield 52%. HRMS: calc. for C20H19ClN6O2S [M]+ 442.9228, found 442.9226. 1H NMR (200 MHz, DMSO): δ 9.28 (brs, 1H, NH), 8.59 (d, J = 8.2 Hz, 1H, HC(2`)), 8.55 (d, J = 8.3 Hz, 1H, HC(4`)), 8.43 (d, J = 7.3 Hz, 1H, HC(8`)), 7.72 (t, J = 8.2 Hz, 1H, HC(3`)), 7.66 (t, J = 8.1 Hz, 1H, HC(7`)), 7.41 (d, J = 8.8 Hz, 2H, HC(3``, 5``)), 7.40 (s, 2H, NH2), 7.23 (d, J = 8.8 Hz, 2H, HC(2``, 6``)), 2.80 (s, 6H, N(CH3)2). 13C NMR (50 MHz, DMSO): δ 158.47 (C-3), 156.07 (C-5), 151.31 (C-5`), 139.49 (C-1``), 132.39 (C-1`), 131.96 (C-8à), 130.57 (C-4`), 129.26 (C-2`), 128.99 (C-7`), 128.53 (C-4à), 128.14 (C-3``, 5``), 123.47 (C-3`, С-4``), 118.83 (C-8`), 117.86 (C-2``, 6``), 115.58 (C-6`), 44.91 (N(CH3)2).
N-[4-[[5-amino-3-(4-cyanoanilino)-1,2,4-triazol-1-yl]sulfonyl]phenyl]-N-methyl-prop-2-enamide 11826317
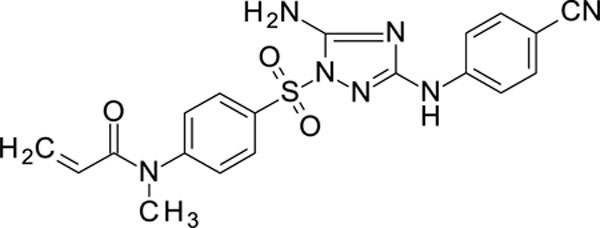
White solid in yield 47%. HRMS: calc. C19H17N7O3S [M]+ C19H17N7O3S 423.4497, found 423.4498. 1H NMR (200 MHz, DMSO): δ 9.86 (brs, 1H, NH), 8.03 (d, J = 8.6 Hz, 2H, HC(3`,5`)), 7.65 (d, J = 8.5 Hz, 2H, HC(3``, 5``)), 7.60 (d, J = 8.5 Hz, 2H, HC2`, 6`)), 7.59 (d, J = 8.6 Hz, 2H, HC(2``,6``)), 7.46 (brs, 2H, NH2), 6.18 (d, J = 4.5 Hz, 1H, (Z)-HC=), 6.17 (d, J = 7.8 Hz, 1H, (E)-HC=), 5.61 (dd, J = 7.7, 4.8 Hz, 1H, HC=CO), 3.30 (s, 3H, NCH3). 13C NMR (50 MHz, DMSO): δ 164.42 (C=O), 158.90 (C-3), 156.99 (C-5), 148.60 (C-1`), 144.69 (C-1``), 133.71 (C3``, 5``), 132.98 (C-4`), 128.67 (C-3`,5`), 128.55 (H2C=), 128.04 (CCO), 127.43 (C2`, 6`), 119.37 (CN), 116.64 (C-2``, 6``), 101.45 (C-4``), 36.50 (N(CH3)2).
1-[(6-chloro-2-naphthyl)sulfonyl]-N3-(4-chlorophenyl)-1,2,4-triazole-3,5-diamine 11826320
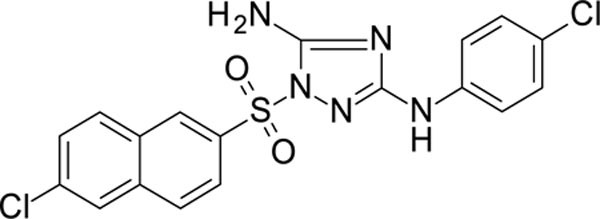
White solid in yield 62%. HRMS: calc. for C18H13Cl2N5O2S [M]+ 434.2997, found 434.2997. 1H NMR (200 MHz, DMSO): δ 9.32 (brs, 1H, NH), 8.77 (d, J = 1.9 Hz, 1H, HC(1`)), 8.29 (d, J = 8.7 Hz, 1H, HC(4`)), 8.19 (d, J = 1.9 Hz, 1H, HC(5`)), 8.16 (d, J = 9.2 Hz, 1H, HC(8`), 7.95 (dd, J = 8.7, 2.0 Hz, 1H, HC(3`)), 7.71 (dd, J = 8.8, 2.1 Hz, 1H, HC(7`)), 7.45 (d, J = 8.9 Hz, 2H, HC(3``, 5``)), 7.41 (s, 2H, NH2), 7.24 (d, J = 8.9 Hz, 2H, HC(2``,6``)). 13C NMR (50 MHz, DMSO): δ 159.57 (C-3), 157.33 (C-5), 139.56 (C-1``), 135.80 (C4à), 134.62 (C-2`), 133.47 (C-6`), 131.68 (C-8à), 129.92 (C-8`), 129.49 (C-7`), 129.07 (C-3`), 128.65 (C-5), 128.34 (C-3``, 5``), 126.74 (C-4`), 123.73 (C-4``, 123.10 (C-1`), 118.15 (C-2``, 6``).
N-[4-[[5-amino-3-(4-chloroanilino)-1,2,4-triazol-1-yl]sulfonyl]-3,5-dimethyl-phenyl]prop-2-enamide 11826322
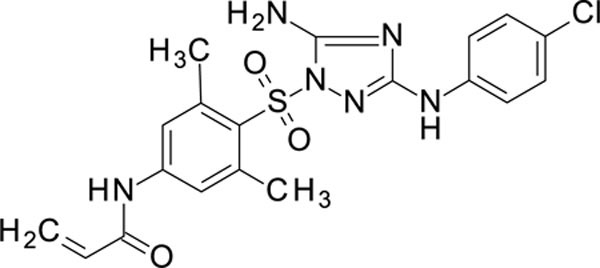
White solid in yield 57%. HRMS: calc. for C19H19ClN6O3S [M]+ 446.9115, found 446.9113. 1H NMR (500 MHz, DMSO): δ 10.42 (s, 1H, NHCO), 9.33 (s, 1H, NH), 7.60 (s, 2H, HC(2`,6`)), 7.39 (d, J = 8.9 Hz, 2H, HC(3``, 5``)), 7.25 (s, 2H, NH2), 7.21 (d, J = 8.9 Hz, 2H, HC(2``,6``)), 6.42 (dd, J = 17.0, 10.1 Hz, 1H, (E)-HC=), 6.30 (dd, J = 17.0, 2.0 Hz, 1H, (Z)-HC=), 5.81 (dd, J = 10.1, 2.0 Hz, 1H, HCCO), 2.65 (s, 6H, 2CH3). 13C NMR (126 MHz, DMSO): δ 163.74 (CO), 158.56 (C-3), 155.69 (C-5), 143.29 (C-1`), 141.82 (C-3`,5`), 139.78 (C-1``), 131.26 (=CCO), 128.25 (C-4`), 128.22 (C-3``, 5``), 128.08 (H2C=), 123.45 (C-4``), 120.82 (C-2`, 6`), 117.87 (C-2``, 6``), 22.80 (2CH3).
N3-(4-chlorophenyl)-1-(1-methylimidazol-4-yl)sulfonyl-1,2,4-triazole-3,5-diamine 11826369
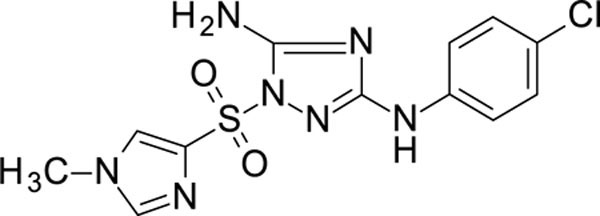
White solid in yield 32%. HRMS: calc. for C12H12ClN7O2S [M]+ 353.7884, found 353.7884. 1H NMR (200 MHz, DMSO): δ 9.28 (s, 1H, NH), 8.15 (s, 1H, HC(5`)), 7.81 (s, 1H, HC(2`)), 7.45 (d, J = 8.8 Hz, 2H, HC(3``,5``)), 7.23 (d, J = 8.6 Hz, 2H, HC(2``,6``)), 7.14 (s, 2H, NH2), 3.72 (s, 3H, NCH3). 13C NMR (50 MHz, DMSO): δ 158.87 (C-3), 157.17 (C-5), 140.45 (C-2`), 139.74 (C-1``), 135.13 (C-4`), 128.19 (C- 3``,5``), 127.46 (C-5`), 123.40 (C-4``), 117.95 (C-2``, 6``), 33.67 (NCH3).
1-[(5-bromo-6-chloro-3-pyridyl)sulfonyl]-N3-(4-chlorophenyl)-1,2,4-triazole-3,5-diamine 11826371
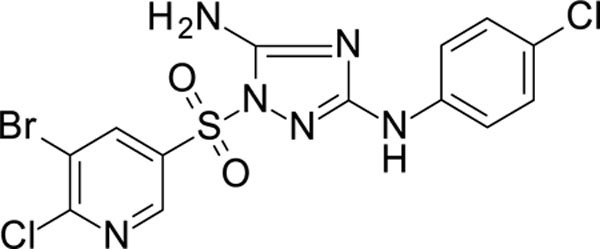
White solid in yield 45%. HRMS: calc. C13H9BrCl2N6O2S [M]+ 464.1252, found 464.1251. 1H NMR (300 MHz, DMSO): δ 9.50 (s, 1H, NH), 8.93 (d, J = 2.2 Hz, 1H, HC(2`)), 8.72 (d, J = 2.2 Hz, 1H, HC(4`)), 7.49 (d, J = 8.9 Hz, 2H, HC(3``,5``), 7.48 (brs, 2H, NH2), 7.30 (d, J = 8.9 Hz, 2H, HC(2``,6``). 13C NMR (50 MHz, DMSO): δ 160.09 (C-3), 157.36 (C-5), 155.41 (C-6`), 146.32 (C-2`), 141.05 (C-1``), 139.31 (C-4`), 132.19 (C-3`), 128.51 (C-3``,5``), 124.01 (C-4``), 120.67 (C-5`), 118.31 (C-2``, 6``).
1-(4-tert-butylphenyl)sulfonyl- N3-(4-chlorophenyl)-1,2,4-triazole-3,5-diamine 11826372
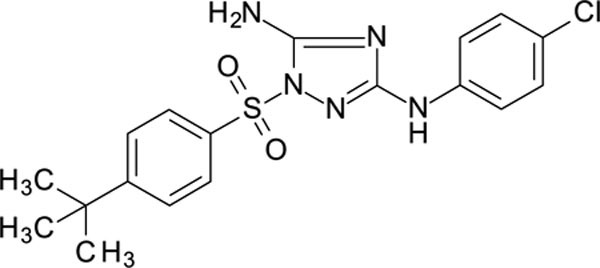
White solid in yield 56%. HRMS: calc. for C18H20ClN5O2S [M]+ 405.9026, found 405.9026. 1H NMR (200 MHz, DMSO): δ 9.36 (brs, 1H, NH), 7.89 (d, J = 8.2 Hz, 2H, HC(2`,6`)), 7.67 (d, J = 8.2 Hz, 2H, HC(3`, 5`)), 7.49 (d, J = 8.2, 2H, HC(2``, 6``), 7.35 (brs, 2H, NH2), 7.26 (d, J = 8.2 Hz, 2H, HC(3``, 5``)), 1.27 (s, 9H, (CH3)3). 13C NMR (50 MHz, DMSO): δ 159.31 (C-3), 158.03 (C-4`), 157.02 (C-5), 139.68 (C-1``), 133.46 (C-1`), 128.35 (C-3``, 5``), 127.31 (C-2`, 6`), 126.50 (C-3`, 5`), 123.69 (C-4``), 118.12 (C-2``, 6``), 35.08 (CMe3), 30.60 (CH3).
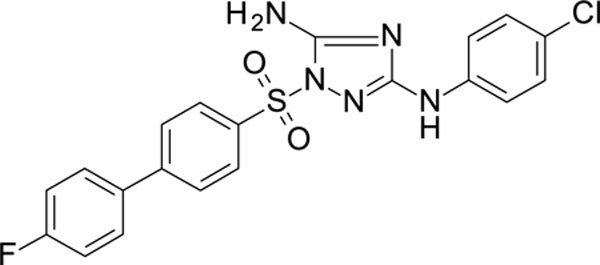
N3-(4-chlorophenyl)-1-[4-(4-fluorophenyl)phenyl]sulfonyl-1,2,4-triazole-3,5-diamine 11826378
White solid in yield 41%. HRMS: calc for C20H15ClFN5O2S [M]+ 443.8827, found 443.8828 1H NMR (200 MHz, DMSO): δ 9.35 (brs, 1H, NH), 8.02 (d, J = 8.5 Hz, 2H, HC(2`,6`)), 7.90 (d, J = 8.5 Hz, 2H, HC(3`,5`)), 7.76 (dd, J = 8.8, 5.5 Hz, 2H, HC(3```,5```)), 7.49 (d, J = 8.9 Hz, 2H, HC(3``,5``)), 7.38 (s, 2H, NH2), 7.28 (d, J = 8.8 Hz, 2H, HC(2```, 6```)),7.26 (d, J = 8.9 Hz, 2H, HC(2``, 6``). 13C NMR (50 MHz, DMSO): δ 165.9 (d, J = 98 Hz, (C-4```)), 159.45 (C-3), 157.17 (C-5), 145.05 (C-4`), 139.53 (C-1``), 134.61 (C-1`), 134.41 (d, J = 11 Hz, C-1```), 129.30 (d, J = 21.5 Hz, С-3``, 5``), 128.28 (d, J = 38 Hz, C-2```,6```), 127.97 (C-3`,5`), 127.55 (C-2`,6`), 123.67 (C-4``), 118.06 (C-2``,6``), 115.80 (d, J = 53 Hz, (C-3```,5```)).
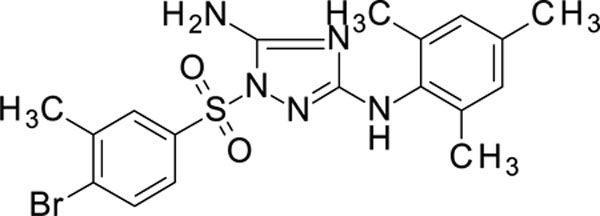
1-(4-bromo-3-methyl-phenyl)sulfonyl-N3-(2,4,6-trimethylphenyl)-1,2,4-triazole-3,5-diamine 11926099
White solid in yield 64%. HRMS: calc. for C18H20BrN5O2S [M]+ 450.3539, found 450.3539. 1H NMR (300 MHz, DMSO): δ 8.52 (s, 1H, HC(1`)), 8.18 (d, J = 8.6 Hz, 1H, HC(8`)), 8.18 (d, J = 8.5 Hz, 1H, HC(3`)), 8.08 (d, J = 7.8 Hz, 1H, HC(5`)), 7.83–7.68 (m, 3H, HC(6`), HC(7`), HC(4`)), 7.66 (brs, 1H, NH), 7.23 (brs, 2H, NH2), 6.75 (s, 2H, HC(3``,5``)), 2.18 (s, 3H, C(2``)-CH3, C(6``)-CH3), 1.79 (s, 6H, C(4``)-CH3). 13C NMR (50 MHz, DMSO): δ 162.51 (C-3), 158.82 (C-5), 135.26 (C-1``), 134.92 (C-2`), 134.62 (C-4à), 134.07 (C-8à), 132.91 (C-3`), 131.37 (C-4`), 129.70 (C-5`), 129.45 (C-2``, 6``), 129.32 (C-4``), 128.19 (C-3``, 5``), 127.95 (C-1`), 127.89 (C-6`), 121.70 (C-8`), 20.40 ((C-4``)-CH3), 17.58 (C(2``)-CH3, C(6``)-CH3).
N3-(4-chlorophenyl)-1-[1-[5-(trifluoromethyl)-2-pyridyl]imidazol-4-yl]sulfonyl-1,2,4-triazole-3,5-diamine 11926103
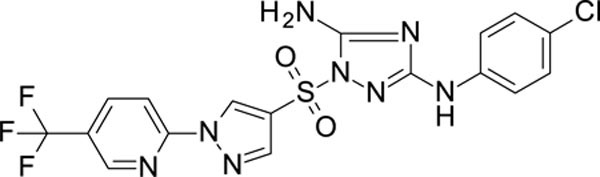
White solid in yield 37%. HRMS: calc. for C17H12ClF3N8O2S [M]+ 484.8438, found 484.8439. 1H NMR (200 MHz, DMSO): δ 9.39 (brs, 1H, NH), 9.28 (s, 1H, HC(5`)), 8.93 (d, J = 2.2 Hz, 1H, HC(6```)), 8.44 (dd, J = 8.7, 2.5 Hz, 1H, HC(4```), 8.39 (s, 1H HC(2`)), 8.14 (d, J = 8.7 Hz, 1H, HC(3```)), 7.52 (d, J = 8.8 Hz, 2H, HC(3``, 5``)), 7.40 (brs, 2H, NH2), 7.26 (d, J = 8.8 Hz, 2H, HC(2``, 6``)). 13C NMR (50 MHz, DMSO) δ 159.75 (C-3), 157.24 (C-5), 151.80 (C-2```), 145.95 (q, J = 5 Hz, C-6```), 141.29 (C-2`), 139.50 (C-1``), 137.69 (q, J = 6 Hz, C-4```), 130.25 (C-5`), 128.28 (C-3``, 5``), 125.10 (q, J = 265 Hz, CF3), 124.43 (q, J = 30 Hz, C-5```), 123.73 (C-4``), 121.01 (C-4`), 118.18 (C-2``, 6``), 113.22 (C-3`).
N3-(4-chlorophenyl)-1-[1-[4-(trifluoromethyl)pyrimidin-2-yl]imidazol-4-yl]sulfonyl-1,2,4-triazole-3,5-diamine 11926105
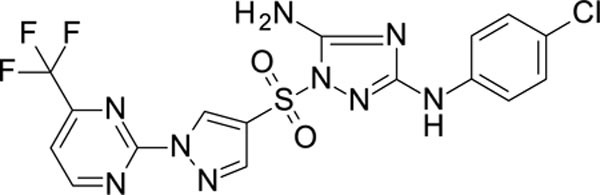
White solid in yield 55%. HRMS: calc. for C16H11ClF3N9O2S [M]+ 485.8319, found 485.8319. 1H NMR (500 MHz, DMSO): δ 9.42 (brs, 1H, NH), 9.29 (s, 1H, HC(5`)), 9.28 (d, J = 5.0 Hz, 1H, HC(6```)), 8.39 (s, 1H, HC(2`)), 8.09 (d, J = 5.0 Hz, 1H, HC(5````)), 7.52 (d, J = 8.9 Hz, 2H, HC(3``,5``)), 7.44 (brs, 2H, NH2), 7.28 (d, J = 8.9 Hz, 2H, HC(2``,6``)). 13C NMR (126 MHz, DMSO): δ 163.39 (C-2```), 159.89 (C-3), 157.41 (C-5), 155.40 (q, J = 48 Hz, C-4```), 154.50 (C-6```), 141.58 (C-2`), 139.52 (C-1``), 132.66 (C-4`), 128.39 (C-3``, 5``), 123.82 (C-4``), 121.19 (C-5`), 118.89 (q, J = 274 Hz, CF3), 118.24 (C-2``, 6``), 116.88 (C-5```).
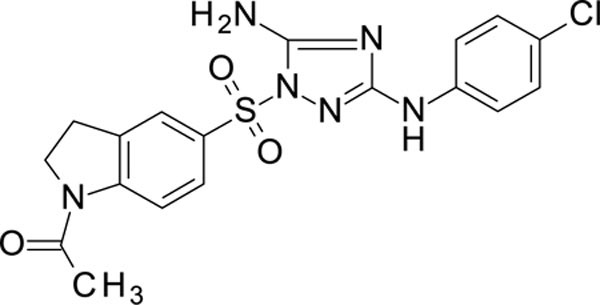
1-[5-[[5-amino-3-(4-chloroanilino)-1,2,4-triazol-1-yl]sulfonyl]indolin-1-yl]ethanone 11926106
White solid in yield 37%. HRMS: calc. for C18H17ClN6O3S [M]+ 432.8849, found 432.8849. 1H NMR (200 MHz, DMSO): δ 9.32 (brs, 1H, NH), 8.16 (brd, J = 8.6 Hz, 1H, HC(7`)), 7.79 (d, J = 8.6 Hz, 1H, HC(6`), 7.77 (brs, 1H, HC(4`)), 7.47 (d, J = 8.9 Hz, 2H, HC(3``, 5``)), 7.28 (brs, 2H, NH2), 7.26 (d, J = 8.9 Hz, 2H, HC(2``, 6``)), 4.13 (t, J = 8.67 Hz, 2H, H2C(2`)), 3.18 (t, J = 8.7 Hz, 2H, H2C(3`)), 2.17 (s, 3H, CH3CO). 13C NMR (50 MHz, DMSO): δ 169.51 (CO), 159.25 (C-3), 157.07 (C-5), 148.09 (C-7à), 139.62 (C-1``), 133.41 (C-3à), 129.25 (C-5`), 128.25 (C-3``, 5``), 128.11 (C-4`), 123.90 (C-6`), 123.55 (C-4``), 117.99 (C-2``, 6``), 115.13 (C-7`), 48.64 (C-2`), 26.79 (C-3`), 23.91 (CH3CO).
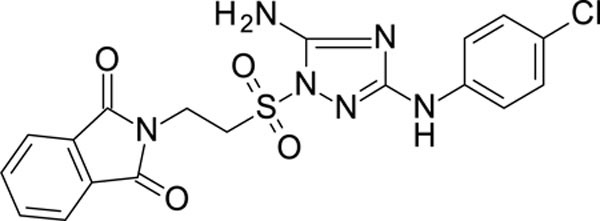
2-[2-[[5-amino-3-(4-chloroanilino)-1,2,4-triazol-1-yl]sulfonyl]ethyl]isoindoline-1,3-dione 11926108
White solid in yield 64%. HRMS: calc. for C18H15ClN6O4S [M]+ 446.8684, found 446.8685. 1H NMR (500 MHz, DMSO): δ 9.26 (s, 1H, NH), 7.77–7.72 (m, 4H, HC(4`, 5`, 6`, 7`), 7.43 (d, J = 8.9 Hz, 2H, HC(3``, 5``)), 7.25 (d, J = 8.9 Hz, 2H, HC(2``, 6``)), 7.11 (s, 2H, NH2), 4.07–3.99 (m, 4H, 2CH2). 13C NMR (126 MHz, DMSO) δ 167.03 (CO), 159.00 (C-3) 156.31 (C-5), 139.62 (C-1``), 134.23 (C-4`, 5`), 131.37 (C-3à, 7à), 128.23 (C-3``, 5``), 123.56 (C-4`, 7`), 122.93 (C-4``), 118.12 (C-2``, 6``), 49.30 (CS), 31.68 (CH2N).
N3-(4-chlorophenyl)-1-(2,4-dimethylthiazol-5-yl)sulfonyl-1,2,4-triazole-3,5-diamine 11926109
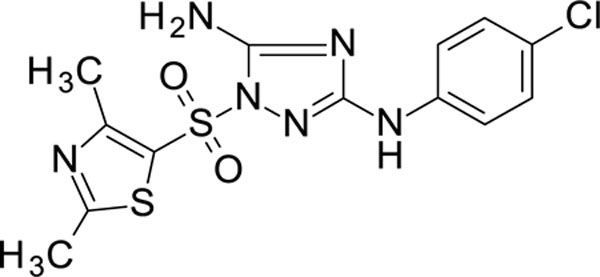
White solid in yield 57%. HRMS: calc. for C13H13ClN6O2S2 [M]+ 384.8663, found 384.8664. 1H NMR (200 MHz, DMSO): δ 9.42 (s, 1H, NH), 7.51 (d, J = 8.9 Hz, 2H, HC(3``, 5``)), 7.39 (brs, 2H, NH2), 7.26 (d, J = 8.9 Hz, 2H, HC(2``, 6``)), 2.65 (d, J = 1.8 Hz, 6H, 2CH3).
13C NMR (50 MHz, DMSO): δ 171.73 (C-2`), 159.79 (C-3), 158.88 (C-4`), 157.15 (C-5), 139.41 (C-1``), 128.25 (C-3``, 5``), 124.61 (C-5`), 123.85 (C-4``), 118.09 (C-2``, 6``), 19.09 (CH3-C(2`)), 16.54 (CH3-C(4`)).
1-(2,1,3-benzoxadiazol-4-ylsulfonyl)-N3-(4-chlorophenyl)-1,2,4-triazole-3,5-diamine 11926110
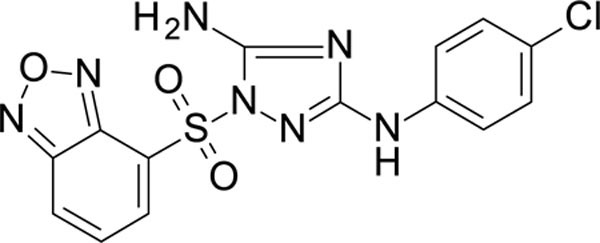
White solid in yield 24%. HRMS: calc. for C14H10ClN7O3S [M]+ 391.7933 found 391.7933. 1H NMR (500 MHz, DMSO): δ 9.36 (s, 1H, NH), 8.49 (d, J = 9.0 Hz, 1H, HC(7`)), 8.44 (d, J = 6.8 Hz, 1H, HC(5`)), 7.85 (dd, J = 9.1, 6.9 Hz, 1H, HC(6`)), 7.50 (brs, 2H, NH2), 7.35 (d, J = 8.9 Hz, 2H, HC(3``, 5``)), 7.20 (d, J = 8.9 Hz, 2H, HC(2``, 6``)). 13C NMR (126 MHz, DMSO): δ 159.51 (C-3), 157.14 (C-5), 149.22 (C-3à), 143.80 (C-7à), 139.38 (C-1``), 137.49 (C-6`), 131.52 (C-5`), 128.27 (C-3``, 5``), 124.58 (C-4`), 123.77 (C-4``), 123.69 (C-7`), 118.09 (C-2``, 6``).
1-(4-bromo-2-fluoro-phenyl)sulfonyl-N3-(4-chlorophenyl)-1,2,4-triazole-3,5-diamine 11926326
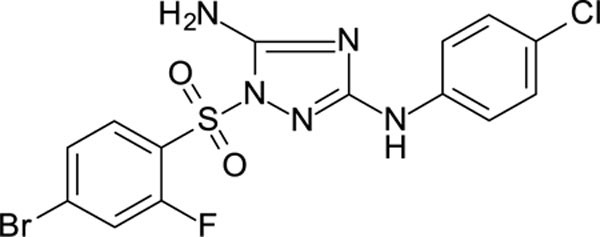
White solid in yield 26%. HRMS: calc. for C14H10BrClFN5O2S [M]+ 446.6828, found 446.6827.
1H NMR (200 MHz, DMSO): δ 9.35 (brs, 1H, NH), 7.96 (d, J = 8.4 Hz, 1H, HC(6`), 7.88 (d, J = 9.2 Hz, 1H, HC(3`)), 7.72 (d, J = 8.4 Hz, 1H, HC(5`)), 7.41 (d, J = 8.7 Hz, 2H, HC(3``, 5``)), 7.33 (brs, 2H, NH2), 7.21 (d, J = 8.6 Hz, 2H, HC(2``, 6``). 13C NMR (50 MHz, DMSO): δ 159.46 (C-3), 158.03 (d, J = 262.0 Hz, C-2`), 156.92 (C-5), 139.42 (C-1``), 131.96 (C-4`), 130.17 (d, J = 9.3 Hz, C-6`), 128.66 (d, J = 4.0 Hz, C-5`), 128.21 (C-3``, C-5``), 123.73 (C-4``), 123.53 (d, J = 13.8 Hz, C-1`), 121.17 (d, J = 24.3 Hz, C-3`), 118.05 (C-2``, C-6``).
1-(5-chloro-2-methoxy-phenyl)sulfonyl-N3-(4-chlorophenyl)-1,2,4-triazole-3,5-diamine 11926323
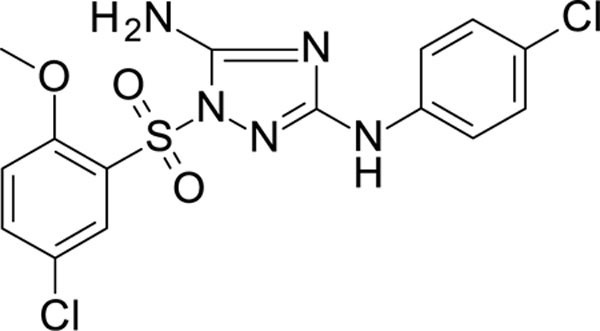
White solid in yield 40%. HRMS: calc. for C15H13Cl2N5O3S [M]+ 414.2670, found 414.2671. 1H NMR (200 MHz, DMSO): δ 9.33 (s, 1H, NH), 7.89 (d, J = 2.6 Hz, 1H, HC(6`), 7.76 (dd, J = 9.0, 2.6 Hz, 1H, HC(4`)), 7.36 (d, J = 9.1 Hz, 2H, HC(3``, 5``)), 7.28 (d, J = 9.0 Hz, 1H, HC(3`)), 7.22 (s, 2H, NH2), 7.20 (d, J = 9.1 Hz, 2H, HC(2``, 6``)), 3.82 (s, 3H, OCH3). 13C NMR (50 MHz, DMSO): δ 158.97 (C-3), 157.95 (C-2`), 155.79 (C-5), 139.66 (C-1``), 136.22 (C-4`), 129.56 (C-5`), 128.22 (C-3``, 5``), 125.73 (C-6`), 124.14 (C-1`), 123.54 (C-4``), 117.91 (C-2``, 6``), 115.33 (C-3`), 56.62 (OCH3).
N3-(4-chlorophenyl)-1-(2,3,4-trifluorophenyl)sulfonyl-1,2,4-triazole-3,5-diamine 11926328
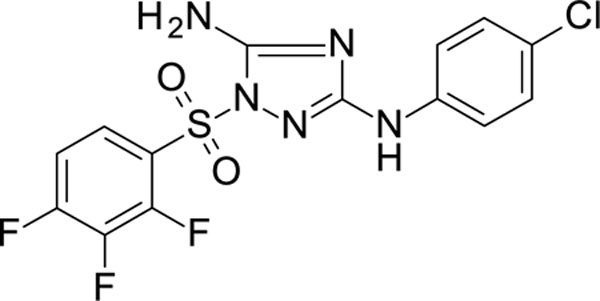
White solid in yield 38%. HRMS: calc. for C14H9ClF3N5O2S [M]+ 403.7677, found 403.7677. 1H NMR (200 MHz, DMSO): δ 9.44 (s, 1H, NH), 8.02–7.83 (m, J = 2.3, 5.6, 8.1, 9.3 Hz, 1H, HC(6`)), 7.62 (tdd, J = 9.3, 6.7, 2.1 Hz, 1H, HC(5`)), 7.45 (s, 2H, NH2), 7.43 (d, J = 8.7 Hz, 2H, HC(3``, 5``)), 7.23 (d, J = 8.8 Hz, 2H, HC(2``, 6``)). 13C NMR (50 MHz, DMSO): δ 159.72 (C-3), 156.94 (C-5), 151.00 (ddd, J = 152.0, 8.0, 3.0 Hz, (C-4`)), 145.57 (ddd, J = 4.6, 12.0, 146.0 Hz, (C-2`)), 142.26 (dt, J = 255.0, 15.0 Hz, (C-3`)), 139.47 (C-1``), 128.31 (C-3``, 5``), 125.94 (dd, J = 4.6, 9.0 Hz, C-6`), 123.92 (C-4``), 121.90 (dd, J = 3.0, 12.0 Hz, C-1`), 118.22 (C-2``, 6``), 113.90 (dd, J = 3.0, 19.0 Hz, C-5`).
1-[(5-bromo-2-thienyl)sulfonyl]-N3-(4-chlorophenyl)-1,2,4-triazole-3,5-diamine 11926329
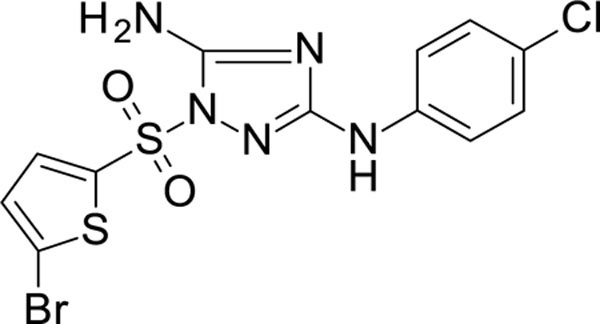
White solid in yield 47%. HRMS: calc. C12H9BrClN5O2S2 [M]+ 434.7211, found 434.7211. 1H NMR (200 MHz, DMSO): δ 9.47 (s, 1H, NH), 7.72 (d, J = 4.1 Hz, 1H, HC(3`)), 7.53 (d, J = 9.0 Hz, 2H, HC(3``, 5``)), 7.44 (brs, 2H, NH2), 7.42 (d, J = 4.1 Hz, 1H, HC(4`)), 7.29 (d, J = 9.0 Hz, 2H, HC(2``, 6``)). 13C NMR (50 MHz, DMSO): δ 160.18 (C-3), 157.61 (C-5), 139.41 (C-1``), 135.38 (C-2`), 135.26 (C-3`), 131.68 (C-4`), 128.37 (C-3``, 5``), 123.97 (C-4``), 122.58 (C-5`), 118.28 (C-2``, 6``).
N3-(4-chlorophenyl)-1-[(2,5-dichloro-3-thienyl)sulfonyl]-1,2,4-triazole-3,5-diamine 11926330
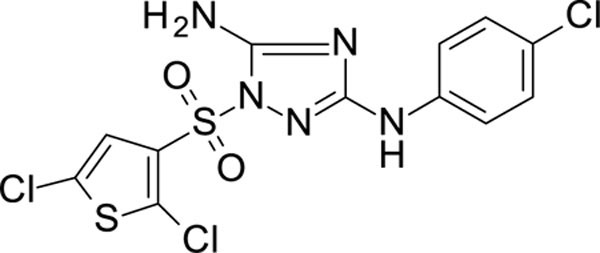
White solid in yield 34%. HRMS: calc. for C12H8Cl3N5O2S2 [M]+ 424.7145, found 424.7144. 1H NMR (200 MHz, DMSO) δ 9.47 (s, 1H, NH), 7.53 (s, 1H, HC(4`), 7.50 (d, J = 8.8 Hz, 2H, HC(3``, 5``)), 7.42 (s, 2H, NH2), 7.25 (d, J = 8.7 Hz, 2H, HC(2``, 6``)). 13C NMR (50 MHz, DMSO) δ 159.69 (C-3), 156.96 (C-5), 139.47 (C-1``), 133.53 (C-3`), 131.86 (C-2`), 128.34 (C-3``, 5``), 127.28 (C-5`), 126.43 (C-4`), 123.88 (C-4``), 118.21 (C-2``, 6``).
4-[[5-amino-1-(6-chloroimidazo[2,1-b]thiazol-5-yl)sulfonyl-1,2,4-triazol-3-yl]amino]-benzonitrile 11926331
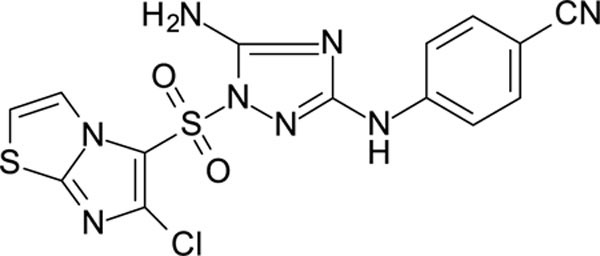
White solid in yield 49%. HRMS: calc. for C14H9ClN8O2S2 [M]+ 420.8587, found 420.8588. 1H NMR (200 MHz, DMSO): δ 9.86 (brs, 1H, NH), 8.23 (d, J = 4.4 Hz, 1H, HC(3`)), 7.78 (d, J = 4.4 Hz, 1H, HC(2`)), 7.65–7.50 (m, 4H, HC(2``,3``,5``,6``)), 7.49 (brs, 2H, NH2). 13C NMR (50 MHz, DMSO) δ 159.27 (C-3), 156.75 (C-5), 151.69 (C-7à), 144.41 (C-4``), 139.07 (C-6`), 132.83 (C-2``, C-6``), 120.85 (C-5`), 119.33 (C-3`), 117.67 (C-3``, C-5``), 116.69 (CN-C1``), 115.18 (C-2`), 101.63 (C-1``).
5-[[5-amino-3-(2,4,6-trichloroanilino)-1,2,4-triazol-1-yl]sulfonyl]naphthalene-2-carbonitrile 12026108
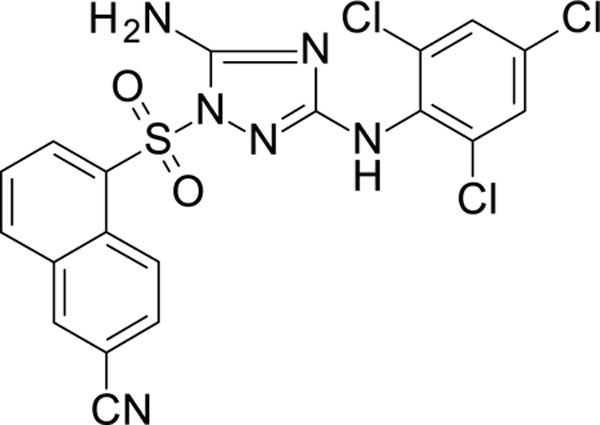
White solid in yield 54%. HRMS: calc. for C19H11Cl3N6O2S [M]+ 493.7540, found 493.7542. 1H NMR (200 MHz, DMSO): δ 8.81 (s, 1H, NH), 8.77 (d, J = 8.10 Hz, 1H, HC(4`) 8.54 (s, 1H, HC(1`)), 8.49 (d, J = 7.9 Hz, 1H, HC(8`)), 7.90 (m, 2H, HC(7`,3`), 7.55 (s, 2H, HC(3``, 5``)), 7.45 (s, 2H, NH2).
13C NMR (126 MHz, DMSO): δ 160.49 (C-3), 157.76 (C-5), 136.56 (C-1``), 135.09 (C-5`), 134.65 (C-4à), 133.98 (C-3``, C-5``), 133.31 (C-8à), 132.72 (C-2``, 6``), 131.10 (C-8`), 128.51 (C-4``), 128.19 (C-3`), 128.03 (C-3``,5``), 128.19 (C-4``), 127.72 (C-6`), 126.44 (C-7`), 126.05 (C-4`), 118.21 (CN), 109.83 (C-1`).
3-[[5-amino-3-(4-cyanoanilino)-1,2,4-triazol-1-yl]sulfonyl]benzonitrile 12026109
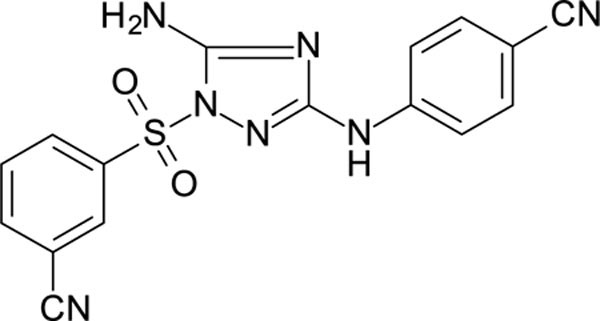
White solid in yield 56%. HRMS: calc. for C16H11N7O2S [M]+ 365.3705, found 365.3706. 1H NMR (200 MHz, DMSO): δ 9.88 (brs, 1H, NH), 8.48 (brs, 1H, HC(2`)), 8.25 (d, J = 7.7 Hz, 2H, HC(4`), HC(6`)), 7.90 (t, J = 7.7 Hz, 1H, HC(5`)), 7.72–7.56 (m, 4H, HC(2``, 3``, 5``, 6``), 7.53 (s, 2H, NH2). 13C NMR (50 MHz, DMSO): δ 159.33 (C-3), 157.24 (C-5), 144.63 (C-1``), 138.41 (C-6`), 137.01 (C-3`), 133.07 (C-3``, 5``), 131.74 (C-2`), 131.25 (C-4`), 131.04 (C-5`), 119.43 (CN-C1`), 116.91 (CN-C1``), 116.82 (C-2``, 6``), 113.03 (C-1`), 101.72 (C-4``).
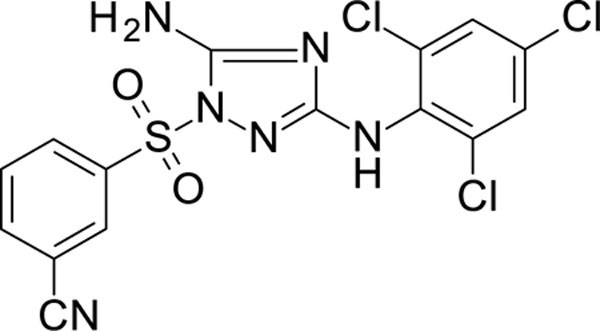
3-[[5-amino-3-(2,4,6-trichloroanilino)-1,2,4-triazol-1-yl]sulfonyl]benzonitrile 12026111
White solid in yield 76%. HRMS: calc. for C15H9Cl3N6O2S [M]+ 443.6953, found 443.6954. 1H NMR (200 MHz, DMSO): δ 8.70 (brs, 1H, NH), 8.26 (t, J = 2.1 Hz, 1H, HC(2`)), 8.24 (d, J = 2.1 Hz, 1H, HC(6`), 8.13 (dt, J = 8.2, 1.5 Hz, 1H, HC(3`)), 7.88 (t, J = 8.1 Hz, 1H, HC(5`)), 7.62 (s, 2H, HC(3``, 5``)), 7.41 (brs, 2H, NH2). 13C NMR (50 MHz, DMSO): δ 161.63 (C-3), 158.75 (C-5), 137.93 (C-1``), 136.89 (С-6`), 134.71 (C-2`), 133.95 (C-3`), 131.58 (C-4`), 131.32 (C-5`), 130.94 (C-2``, 6``)), 128.15 (C-3``, 5``, 4``), 116.89 (CN), 112.77 (C-1`).
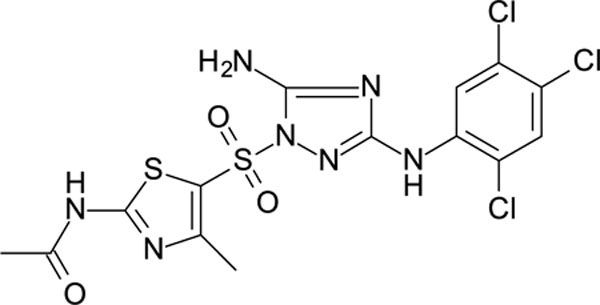
N-[5-[[5-amino-3-(2,4,5-trichloroanilino)-1,2,4-triazol-1-yl]sulfonyl]-4-methyl-thiazol-2-yl]acetamide 12026113
White solid in yield 48%. HRMS: calc. for C14H12Cl3N7O3S2 [M]+ 496.7806, found 496.7808. 1H NMR (200 MHz, DMSO): δ 12.79 (brs, 1H, CONH), 8.45 (brs, 1H, NH), 8.29 (s, 1H, HC(6`)), 7.70 (s, 1H, HC(3`)), 7.45 (brs, 2H, NH2), 2.62 (s, 3H, CH3Ar), 2.18 (s, 3H, CH3CO). 13C NMR (50 MHz, DMSO): δ 169.70 (CO), 161.42 (C-2`), 158.85 (C-3), 156.99 (C-4`), 156.87 (C-5), 136.71 (C-1``), 130.01 (C-3``), 129.92 (C-5``), 122.97 (C-4``), 120.79 (C-2``), 120.13 (C-6``), 116.97 (C-5`), 22.21 (CH3CO), 16.67 (CH3-C-4`).
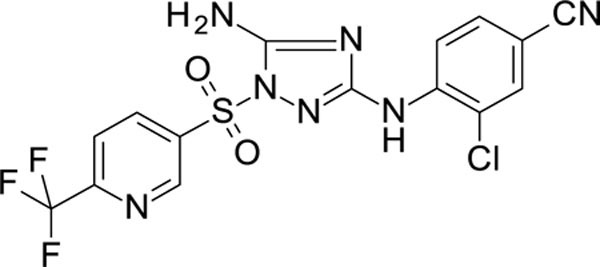
4-[[5-amino-1-[[6-(trifluoromethyl)-3-pyridyl]sulfonyl]-1,2,4-triazol-3-yl]amino]-3-chloro-benzonitrile 12026116
White solid in yield 41%. HRMS: calc. for C15H9ClF3N7O2S [M]+ 443.7918, found 443.7918. 1H NMR (200 MHz, DMSO): δ 9.33 (d, J = 2.3 Hz, 1H, HC(2`)), 8.73 (s, 1H, NH), 8.68 (dd, J = 8.4, 2.5 Hz, 1H, HC(4`)), 8.25 (d, J = 8.4 Hz, 1H, HC(5`)), 8.16 (d, J = 8.8 Hz, 1H, HC(5``)), 7.92 (d, J =2.0 Hz, 1H, HC(2``)), 7.75 (dd, J = 8.8, 2.0 Hz, 2H, HC(6``)), 7.70 (s, 2H, NH 2 ). 13C NMR (50 MHz, DMSO): δ 159.18 (C-3), 157.21 (C-5), 151.19 (q, J = 74 Hz, C(6`), 148.17 (C-2`), 140.83 (C-4``), 138.42 (C-4`), 135.56 (C-3`), 132.77 (C-2``), 131.89 (C-6``), 122.06 (C-5`), 121.8 (q, J = 260 Hz, CF3), 121.34 (C-3``), 119.25 (C-5``), 117.82 (CN), 103.81 (C-1``).
5-[[5-amino-3-[4-[(E)-2-cyanovinyl]anilino]-1,2,4-triazol-1-yl]sulfonyl]naphthalene-2-carbonitrile 12026118
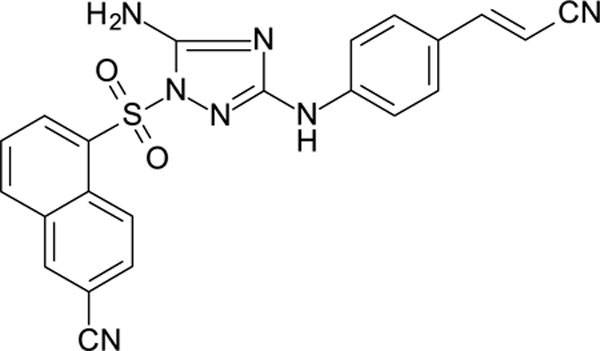
White solid in yield 62%. HRMS: calc. for C22H15N7O2S [M]+ 420.8587, found 420.8587. 1H NMR (200 MHz, DMSO): δ 9.57 (brs, 1H, NH), 9.09 (d, J = 9.0 Hz, 1H, HC(4`)), 8.78 (d, J = 1.7 Hz, 1H, HC(1`)), 8.63 (dd, J = 7.5, 1.2 Hz, 1H, HC(6`)), 8.50 (d, J = 8.2 Hz, 1H, HC(8`)), 8.11 (dd, J = 9.0, 1.8 Hz, 1H, HC(3`)), 7.93 (t, J = 7.9 Hz, 1H, HC(7`)), 7.55 (s, 2H, NH2), 7.50 (d, J = 8.7 Hz, 2H, HC(2``, 6``)), 7.48 (d, J = 16.5 Hz, 1H, (HC=)Ph), 7.40 (d, J = 8.7 Hz, 2H, HC(3``, 5``)), 6.20 (d, J = 16.5 Hz, 1H, (HC=)CN). 13C NMR (50 MHz, DMSO): δ 158.76 (C-3), 156.39 (C-5), 150.20 (PhC=), 143.01 (C-1``), 136.94 (C-5`), 135.41 (C-1`), 133.62 (C-4à), 132.78 (C-8à), 132.16 (C-3`), 129.22 (C-8`), 128.71 (C-3``, C-5``), 128.00 (C-4``), 126.64 (C-6`), 126.16 (C-7`), 125.83 (C-4`), 119.33 (CN-C2`), 118.16 (CN-C=), 116.42 (C-2``, C-6``), 110.06 (C-1`), 92.68 (=CCN).
4-[[5-amino-1-[(2-chloro-5-quinolyl)sulfonyl]-1,2,4-triazol-3-yl]amino]-3-chloro-benzonitrile 12026122
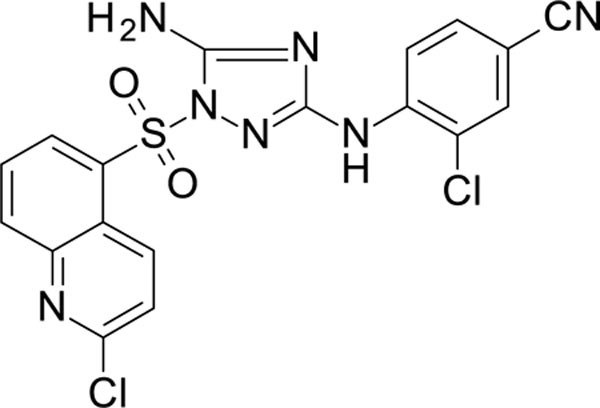
White solid in yield 33%. HRMS: calc. for C18H11Cl2N7O2S [M]+ 460.2973, found 460.2972. 1H NMR (200 MHz, DMSO): δ 8.63 (d, J = 8.4 Hz, 1H, HC(6`)), 8.62 (d, J = 8.9 Hz, 1H, HC(8`)), 8.50 (brs, 1H, NH), 8.49 (d, J = 8.7 Hz, 1H, HC(3`)), 7.91 (t, J = 7.8, 1H, HC(7`)), 7.84 (d, J = 8.4, 1H, HC(4`), 7.81 (d, J = 2.0 Hz, 1H, HC(2``)), 7.78 (d, J = 8.7 Hz, 1H, HC(5``)), 7.58 (dd, J = 8.7, 2.0 Hz, 1H, HC(6``)), 7.44 (brs, 2H, NH2). 13C NMR (50 MHz, DMSO): δ 157.85 (C-3), 157.66 (C-5), 151.69 (C-2`), 142.16 (C-8à), 141.23 (C-5`), 140.75 (C-4``), 135.92 (C-4`), 134.24 (C-7`), 132.68 (C-2``), 132.41 (C-6`), 131.62 (C-6``), 127.32 (C-4à), 126.38 (C-8`), 124.22 (C-3`), 120.72 (C-3``), 118.27 (C-5``), 117.91 (CN), 103.06 (C-1``).
4-[[5-amino-1-(6-chloroimidazo[2,1-b]thiazol-5-yl)sulfonyl-1,2,4-triazol-3-yl]amino]-2-chloro-benzonitrile 12026123
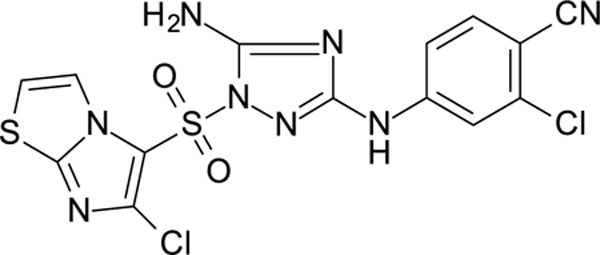
White solid in yield 76%. HRMS: calc. for C14H8Cl2N8O2S2 [M]+ 455.3034, found 455.3034. 1H NMR (200 MHz, DMSO): δ 10.16 (brs, 1H, NH), 8.21 (d, J = 4.4 Hz, 1H, HC(3`)), 7.85–7.69 (m, 3H, HC(5``), HC(2``), HC(2`)), 7.62 (brs, 2H, NH2), 7.40 (dd, J = 8.7, 2.1 Hz, 1H, HC(6``)). 13C NMR (50 MHz, DMSO): δ 158.88 (C-3), 156.84 (C-5), 151.90 (C-7à), 145.53 (C-4``), 139.41 (C-6`), 136.04 (C-2``), 134.74 (C-6``), 120.58 (C-5`), 118.12 (CN-C1``), 116.58 (C-3``), 116.43 (C-3`), 115.52 (C-2`), 115.00 (C-5``), 101.99 (C-1``).
4-[[5-amino-1-(2,4-dimethylthiazol-5-yl)sulfonyl-1,2,4-triazol-3-yl]amino]-2-chloro-benzonitrile 12026124
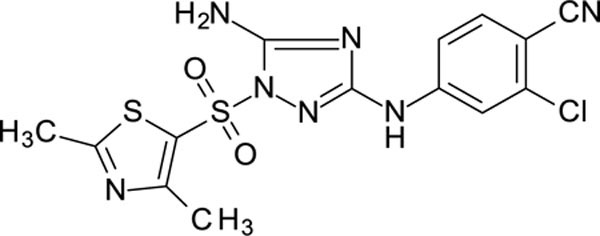
White solid in yield 79%. HRMS: calc. for C14H12ClN7O2S2 [M]+ 409.8758, found 409.8758. 1H NMR (200 MHz, DMSO): δ 10.18 (s, 1H, NH), 7.89 (d, J = 2.0 Hz, 1H, HC(3``)), 7.76 (d, J = 8.7 Hz, 1H, HC(6``), 7.54 (brs, 2H, NH2), 7.46 (dd, J = 8.7, 2.2 Hz, 1H, HC(5``), 2.67 (s, 6H, 2CH3). 13C NMR (50 MHz, DMSO): δ 172.16 (C-2`), 159.27 (C-3), 158.90 (C-4`), 157.00 (C-5), 145.69 (C-4``), 136.09 (C-2``), 134.71 (C-6``), 124.61 (C-5`), 116.49 (C-3``, CN-C1``), 115.52 (C-5``), 102.02 (C-1``), 19.18 (CH3-C2`), 16.55 (CH3-C3`).
4-[[5-amino-1-(2-chloro-4-cyano-phenyl)sulfonyl-1,2,4-triazol-3-yl]amino]-3-chloro-benzonitrile 12126046
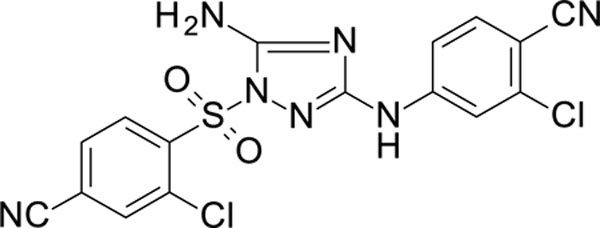
White solid in yield 45%. HRMS: calc. for C16H9Cl2N7O2S [M]+ 434.2600, found 434.2602. 1H NMR (200 MHz, DMSO): δ 10.16 (brs, 1H, NH), 8.38 (d, J = 1.6 Hz, 1H, HC(3’)), 8.36 (d, J = 8.4 Hz, 1H, HC(5’)), 8.16 (dd, J = 8.34, 1.6 Hz, 1H, HC(6’)), 7.70 (d, J = 8.8 Hz, 1H, HC(5``)), 7.66 (d, J = 2.2 Hz, 1H, HC(3``), 7.59 (brs, 2H, NH2), 7.35 (dd, J = 8.7, 2.1 Hz, 1H, HC(6``)). 13 C NMR (50 MHz, DMSO): δ 158.42 (C-3), 157.03 (C-5), 145.71 (C-4``), 138.14 (C-1`), 135.98 (C-2``), 135.71 (C-2`), 134.70 (C-6`), 132.63 (C-5`), 132.34 (C-2``3`), 132.03 (C-6``), 118.46 (C-4`C-5``), 116.49 (CN-C1``), 116.36 (C-3``), 115.94 (CN-C4`), 115.39 (C-5``), 101.91 (C-1``).
N-[5-[[5-amino-3-[4-(cyanomethyl)anilino]-1,2,4-triazol-1-yl]sulfonyl]-4-methyl-thiazol-2-yl]acetamide 12126052
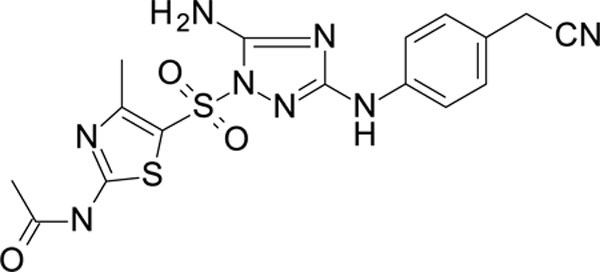
White solid in yield 19%. HRMS: calc. for C16H16N8O3S2 [M]+ 432.4824, found 432.4823. 1H NMR (200 MHz, DMSO): δ 12.74 (s, 1H, CONH), 9.31 (s, 1H, NH), 7.51 (d, J = 8.5 Hz, 2H, HC(3``, 5``)), 7.33 (brs, 2H, NH2), 7.20 (d, J = 8.3 Hz, 2H, HC(2``, 6``)), 3.89 (s, 2H, CH2), 2.61 (s, 3H, CH3Ar), 2.17 (s, 3H, CH3CO). 13C NMR (50 MHz, DMSO): δ 169.67 (CO), 161.12 (C-2`), 159.72 (C-3), 157.06 (C-5), 156.42 (C-3`), 140.05 (C-4``), 128.22 (C-3``, 5``), 122.37 (C-1``), 119.21 (CN), 117.15 (C-1`), 116.91 (C-2``, 6``), 22.22 (MeCO), 21.64 (СH2), 16.70 (CH3-C-4`).
4-[[5-amino-1-(6-chloroimidazo[2,1-b]thiazol-5-yl)sulfonyl-1,2,4-triazol-3-yl]amino]-phthalonitrile 12126053
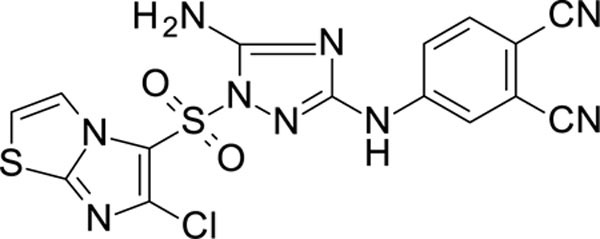
White solid in yield 67%. HRMS: calc. for C15H8ClN9O2S2 [M]+ 445.8682, found 445.8683. 1H NMR (200 MHz, DMSO): δ 10.37 (s, 1H, NH), 8.24 (d, J = 4.5 Hz, 1H, HC(3`)), 8.03 (d, J = 2.2 Hz, 1H, HC(3``), 7.91 (d, J = 8.2 Hz, 1H, HC(6``), 7.80–7.69 (m, 2H, HC(2`), HC(5``)), 7.62 (brs, 2H, NH2). 13C NMR (50 MHz, DMSO): δ 158.66 (C-3), 156.69 (C-5), 151.87 (C-7à), 144.78 (C-4``), 139.50 (C-6`), 134.62 (C-6``), 120.53 (C-3``), 120.42 (C-5``), 120.34 (C-5`), 118.00 (C-3`), 116.33 (C-2`), 115.88 (CN), 115.24 (CN), 114.9 (C-2``), 104.05 (C-1``).
5-[[5-amino-3-[4-(cyanomethyl)anilino]-1,2,4-triazol-1-yl]sulfonyl]naphthalene-2-carbonitrile 12126054
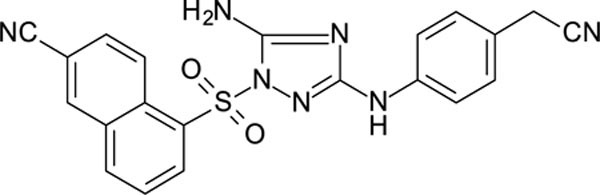
White solid in yield 57%. HRMS: calc. for C21H15N7O2S [M]+ 429.4558, found 429.4558. 1H NMR (200 MHz, DMSO): δ 9.18 (s, 1H, NH), 9.12 (d, J = 9.1 Hz, 1H, HC(4`)), 8.73 (d, J = 1.7 Hz, 1H, HC(1`)), 8.61 (d, J = 7.5 Hz, 1H, HC(6`)), 8.47 (d, J = 8.2 Hz, 1H, HC(8`)), 8.05 (dd, J = 8.9, 1.8 Hz, 1H, HC(3`)), 7.91 (t, J = 7.8 Hz ,1H, HC(7`)), 7.44 (s, 2H, NH2), 7.37 (d, J = 8.6 Hz, 2H, HC(2``), HC(6``), 7.18 (d, J = 8.4 Hz, 2H, HC(3``), HC(5``)), 3.87 (s, 2H, CH2CN). 13C NMR (50 MHz, DMSO): δ 159.09 (C-3), 156.30 (C-5), 139.86 (C-1``), 136.71 (C-5`), 135.20 (C-1`), 133.44 (C-4à), 132.77 (C-6`), 132.37 (C-8à), 129.30 (C-3`), 128.62 (C-8`), 128.24 (C-3``, 5``), 126.46 (C-7`), 126.34 (C-4`), 122.39 (C-4``), 119.19 (CN-CH2), 118.06 (CN-C(2`)), 116.88 (C-2``, 6``), 110.03 (C-2`), 21.70 (CH2).
4-[[5-amino-1-[(6-cyano-1-naphthyl)sulfonyl]-1,2,4-triazol-3-yl]amino]phthalonitrile 12126055
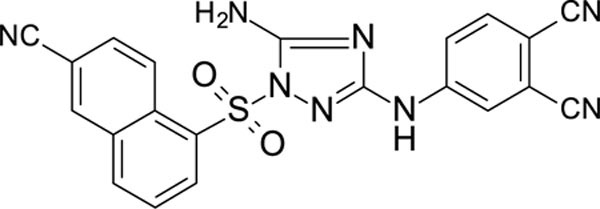
White solid in yield 51%. Mp 287–8 oC. HRMS: calc. for C21H12N8O2S [M]+ 440.4387, found 440.4387. Mp = 88.7 °C. 1H NMR (200 MHz, DMSO): δ 10.27 (s, 1H, NH), 9.06 (d, J = 9.1 Hz, 1H, HC(8`)), 8.78 (d, J = 1.7 Hz, 1H, HC(5`)), 8.66 (d, J = 7.5 Hz, 1H, HC(2`)), 8.52 (d, J = 8.2 Hz, 1H, HC(4`), 8.10 (dd, J = 9.1, 1.7 Hz, 1H, HC(7`)), 7.98–7.86 (m, 2H, HC(3`), HC(6``)), 7.88 (d, J = 2.4 Hz, 1H, HC(3``)), 7.75 (dd, J = 8.8, 2.3 Hz, 1H, HC(5``)), 7.66 (brs, 2H, NH2). 13C NMR (50 MHz, DMSO): δ 158.00 (C-3), 156.33 (C-5), 144.88 (C-4``), 137.17 (C-1`), 135.47 (C-6``), 134.84 (C-5`), 133.79 (C-8à), 132.90 (C-4à), 132.10 (C-2`), 129.25 (C-7`), 129.01 (C-4`), 126.60 (C-3`), 125.81 (C-3``), 120.24 (C-5``), 120.22 (С-8`), 118.05 (CN-C6`), 116.45 (CN-C1``), 116.07 (CN-C2``), 115.27 (C-2``), 110.29 (C-6`), 104.00 (C-1``).
4-[[5-amino-1-(2-methylthiazol-4-yl)sulfonyl-1,2,4-triazol-3-yl]amino]benzonitrile 12126056
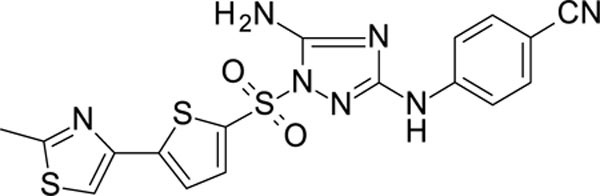
White solid in yield 37%. HRMS: calc. for C17H13N7O2S3 [M]+ 443.5291, found 443.5291. 1H NMR (200 MHz, DMSO): δ 9.90 (s, 1H, NH), 8.10 (s, 1H, HC(5`)), 7.75–7.57 (m, 4H, HC(2``,3``,5``,6``)), 7.49 (brs, 2H, NH2), 2.68 (s, 3H, CH3). 13C NMR (50 MHz, DMSO): δ 166.88 (С-2`), 159.39 (C-3), 157.45 (C-5), 145.96 (C-2`), 144.66 (C-4``), 135.92 (C-4`), 133.01 (C-2``,6``), 123.83 (C-5`), 119.42 (CN), 116.76 (C-3``, 5``), 101.60 (C-1``), 18.55 (CH3).
4-[[5-amino-1-[[6-(cyanomethyl)-1-naphthyl]sulfonyl]-1,2,4-triazol-3-yl]amino]benzonitrile 12126058
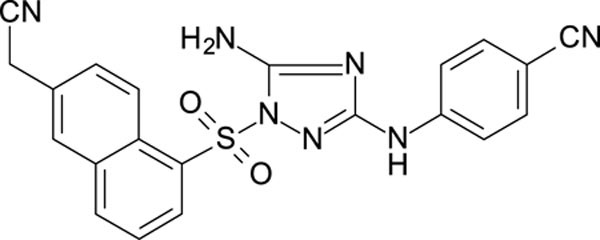
White solid in yield 47%. HRMS: calc. for C21H15N7O2S [M]+ 429.4558, found 429.4556. 1H NMR (500 MHz, DMSO): δ 9.79 (s, 1H, NH), 8.97 (d, J = 8.9 Hz, 1H, HC(8`)), 8.49 (d, J = 4.4 Hz, 1H, HC(2`)), 8.41 (d, J = 9.7 Hz, 1H, HC(4`)), 8.08 (s, 1H, HC(5`)), 7.80 (d, J = 8.9, 1H, HC(7`), 7.78 (t, J = 7.7 Hz, 1H, HC(3`)), 7.66 (d, J = 8.8 Hz, 2H, HC(2``, 6``)), 7.56 (s, 2H, NH2), 7.52 (d, J = 8.7 Hz, 2H, HC(3``, 5``)), 4.27 (s, 2H, CH2). 13C NMR (126 MHz, DMSO): δ 158.36 (C-3), 156.40 (C-5), 144.55 (C-4``), 136.31 (C-1`), 133.07 (C-2``, 6``), 132.92 (C-8à), 132.02 (C-4à), 131.42 (C-2`), 131.02 (C-4`), 129.99 (C-6`), 127.73 (C-5`), 125.38 (C-3`), 125.01 (C-7`), 123.34 (C-8`), 119.49 (CN(CH2)), 118.56 (CNAr), 116.50 (C-3``, 5``), 101.50 (С-1``), 23.32 (CH2).
4-[[5-amino-1-[[7-(cyanomethyl)-1-naphthyl]sulfonyl]-1,2,4-triazol-3-yl]amino]-2-chloro-benzonitrile 12126059
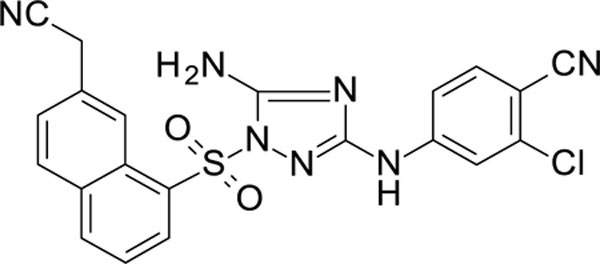
White solid in yield 45%. HRMS: calc. for C21H14ClN7O2S [M]+ 463.9005, found 463.9004. 1H NMR (500 MHz, DMSO) δ 10.07 (s, 1H, NH), 8.81 (s, 1H, HC(8`)), 8.49 (d, J = 7.7 Hz, 1H, HC(2`)), 8.43 (d, J = 8.0 Hz, 1H, HC(4`)), 8.19 (d, J = 8.4 Hz, 1H, HC(5`)), 7.80 (t, 1H, J = 7.81 Hz, HC(3`)), 7.73( d, 1H, J = 8.7 Hz, HC(6`)), 7.68 (dd, 1H, J = 1.4, 8.5 Hz, HC(6``)), 7.66 (s, 2H, NH2), 7.60 (d, J = 2.0 Hz, 1H, HC(3``)), 7.49 (dd, J = 8.5, 2.0 Hz, 1H, HC(5``)), 4.27 (s, 2H, CH2). 13C NMR (126 MHz, DMSO): δ 157.96 (C-3), 156.41 (C-5), 145.70 (C-4``), 136.42 (C-2``), 135.83 (C-1`), 134.98 (C-6``), 132.92 (C-8à), 132.18 (C-4à), 131.67 (C-2`), 131.29 (C-7`), 130.12 (C-5`), 127.60 (C-6`), 127.55 (C-4`), 125.00 (C-3`), 123.05 (C-8`), 118.49 (CN-CH2), 116.63 (CN-C1``), 116.30 (C-5``), 115.14 (C-3``), 101.78 (C-1``), 23.29 (CH2).
4-[[5-amino-1-[[6-(cyanomethyl)-1-naphthyl]sulfonyl]-1,2,4-triazol-3-yl]amino]-2-chloro-benzonitrile 12126060
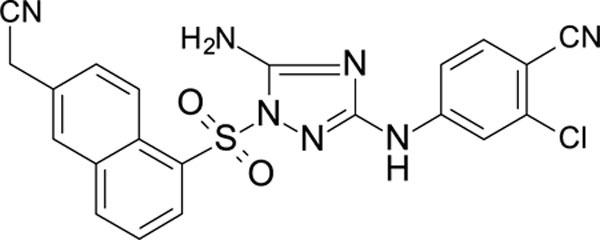
White solid in yield 23%. HRMS: calc. for C21H14ClN7O2S [M]+ 463.9005, found 463.9006. 1H NMR (500 MHz, DMSO) δ 10.04 (s, 1H, NH), 8.80 (s, 1H, HC(8`)), 8.48 (dd, J = 7.5, 1.2 Hz, 1H, HC(2`)), 8.42 (d, J = 8.2 Hz, 1H, HC(4`)), 8.18 (d, J = 8.6 Hz, 1H, HC(5`)), 7.79 (t, J = 8.0 Hz, 1H, HC(3`)), 7.76 (d, J = 8.6 Hz, 1H, HC(6``)), 7.63 (s, 2H, NH2), 7.60 (d, J = 2.2 Hz, 1H, HC(3``)), 7.48 (dd, J = 8.7, 2.2 Hz, 1H, HC(5``)), 4.26(s, 2H, CH2).
13C NMR (126 MHz, DMSO): δ 157.99 (C-3), 156.45 (C-5), 145.72 (C-4``), 135.85 (C-1``), 135.00 (C-6``), 132.93 (C-8à), 132.21 (C-4à), 131.31 (C-7`), and 130.14 (C-5`), and 127.61 (C-6`), 127.57 (C-4`), 125.02 (C-3`), 123.05* (C-8`), 118.63* and 118.50 (CN-CH2), 116.65 (CN-C1``), 116.33 (C-5``), 115.30 (C-3``), 101.82 (C-1``), 23.31 (CH2).
4-[[5-amino-1-(5-chloro-1,3-dimethyl-pyrazol-4-yl)sulfonyl-1,2,4-triazol-3-yl]amino]-2-chloro-benzonitrile 12126061
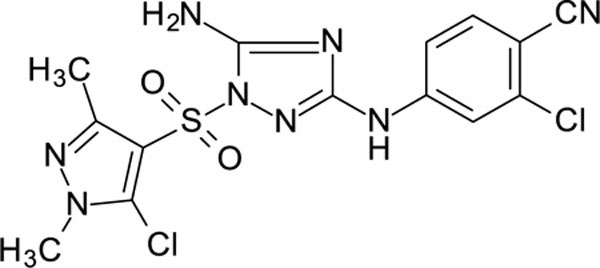
White solid in yield 59%. HRMS: calc. for C14H12Cl2N8O2S [M]+ 429.4558, found 429.4559. 1H NMR (200 MHz, DMSO): δ 10.13 (brs, 1H, NH), 7.92 (d, J = 2.4 Hz, 1H, HC(3``)), 7.74 (d, J = 8.7 Hz, 1H, HC(6``)), 7.41 (dd, J = 2.4, 8.7 Hz, 1H, HC(3``)), 7.39 (brs, 2H, NH2), 3.79 (s, 3H, NCH3), 2.43 (s, 3H, CH3-C(3`)). 13C NMR (50 MHz, DMSO): δ 158.21 (C-3), 156.27 (C-5), 148.81 (C-3`), 145.78 (C-4``), 136.07 (C-2``), 134.56 (C-6``), 130.89 (C-5`), 116.52 (C-3``), 116.31 (CN), 115.45 (C-5``), 111.85 (C-4`)), 101.75 (C-1``), 36.74 (NCH3), 13.36 (CH3-C3`).
2-[4-[[5-amino-1-[[6-(cyanomethyl)-2-naphthyl]sulfonyl]-1,2,4-triazol-3-yl]amino]phenyl]-acetonitrile 12126062
White solid in yield 35%. HRMS: calc. for C22H17N7O2S [M]+ 429.4558, found 429.4558. 1H NMR (500 MHz, DMSO): δ 9.20 (brs, 1H, NH), 8.96 (d, J = 8.9 Hz, 1H, HC(8`)), 8.45 (d, J = 4.4 Hz, 1H, HC(2`)), 8.39 (d, J = 9.7 Hz, 1H, HC(4`)), 8.07 (s, 1H, HC(5`)), 7.77 (d, J = 8.9, 1H, HC(7`), 7.75 (t, J = 7.7 Hz, 1H, HC(3`)), 7.45 (brs, 2H, NH2), 7.37 (d, J = 8.8 Hz, 2H, HC(2``, 6``)), 7.18 (d, J = 8.7 Hz, 2H, HC(3``, 5``)), 4.26 (brs, 2H, H2C-C(6`)), 3.87 (brs, 2H, H2C-C(1``)). 13C NMR (126 MHz, DMSO): δ 159.01 (C-3), 156.47 (C-5), 139.97 (C-4``), 136.11 (C-1`), 133.80 (C-8à), 132.89 (C-2`), 130.64 (C-4à), 129.89 (C-4`), 128.29 (C-2``, 6``), 127.68 (C-6`), 126.99 (C-3`), 125.31 (C-5`), 124.95 (C-7`), 123.52 (C-8`), 119.46 (CN(CH2-C(1``))), 118.68 (CN(CH2-C(2`)), 116.79 (C-3``, 5``), 122.43 (С-1``), 22.35 (CH2-C(6`)), 21.63 (CH2C-1``).
4-[[5-amino-1-(5-chloro-3-methyl-benzothiophen-2-yl)sulfonyl-1,2,4-triazol-3-yl]amino]benzonitrile 12126063
White solid in yield 53%. HRMS: calc. for C18H13ClN6O2S2 [M]+ 444.9198, found 444.9198. 1H NMR (200 MHz, DMSO): δ 10.19 (brs, 1H, NH), 8.15 (d, J = 9.0 Hz, 1H, HC(7`)), 8.13 (s, 1H, HC(4`)), 7.87 (d, J = 2.4 Hz, 1H, HC(3``)), 7.77 (d, J = 8.7 Hz, 1H, HC(6``)), 7.66 (s, 2H, NH2), 7.64 (d, J = 2.2, 9 HC(6`)), 7.41 (dd, J = 2.4, 8.7 Hz, 1H, HC(3``)), 2.78 (s, 3H, CH3-C(3`)). 13C NMR (50 MHz, DMSO): δ 158.90 (C-3), 157.12 (C-5), 145.63 (C-4``), 141.26 (C-3a), 139.77 (C-5`), 138.11 (C-7`), 136.13 (C-3`), 134.87 (C-2``), 131.76 (C-7a), 130.95 (C-6``), 128.85 (C-5`), 124.94 (C-6`), 124.31 (C-4`), 116.66 (CN), 116.42 (C-3``), 115.52 (C-5``), 101.93 (C-1``), 12.76 (CH3-C3`).
4-[[5-amino-1-[[7-[(E)-2-cyanovinyl]-1-naphthyl]sulfonyl]-1,2,4-triazol-3-yl]amino]-2-chloro-benzonitrile 12126064
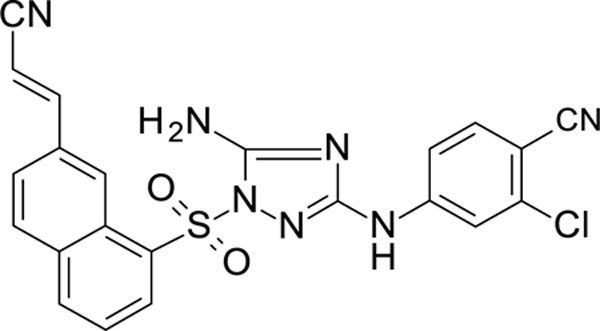
White solid in yield 67%. HRMS: calc. for C22H14ClN7O2S [M]+ 463.9005, found 463.9004. 1H NMR (200 MHz, DMSO): δ 10.05 (s, 1H, NH), 8.89 (d, J = 1.6 Hz, 1H, HC(8`)), 8.54 (d, J = 7.6 Hz, 1H, HC(2`)), 8.42 (d, J = 8.2 Hz, 1H, HC(4`)), 8.20 (d, J = 8.7 Hz, 1H, HC(5`)), 8.01 (dd, J = 8.7, 1.6 Hz, 1H, HC(6`)), 7.85 (d, J = 16.7 Hz, 1H, CH=(Ar)), 7.78 (s, 1H), 7.74 (s, 2H, NH2), 7.63 (d, J = 2.1 Hz, 1H, HC(3``)), 7.45 (dd, J = 8.7, 2.2 Hz, 1H, HC(4``)), 6.64 (d, J = 16.7 Hz, 1H, HC=(CN)). 13C NMR (126 MHz, DMSO): δ 158.03 (С-3), 156.43 (С-5), 149.66 (ArC=), 145.59 (C-4``), 136.30 (C-2``), 135.87 (C-7`), 134.89 (C-6``), 134.60 (C-1`), 133.76 (C-4`), 132.21 (C-5`), 131.94 (C-4à), 130.17 (C-2`), 127.38 (C-8à), 126.14 (C-3`), 125.03 (C-6`), 124.37 (C-8`), 118.21 (CN(C=)), 116.52 (CN-C4``), 116.26 (C-3``), 115.25 (C-5``), 101.87 (C-1``), 99.56 (CN(C=)).
4-[[5-amino-1-[[6-[(E)-2-cyanovinyl]-1-naphthyl]sulfonyl]-1,2,4-triazol-3-yl]amino]-2-chloro-benzonitrile 12126065
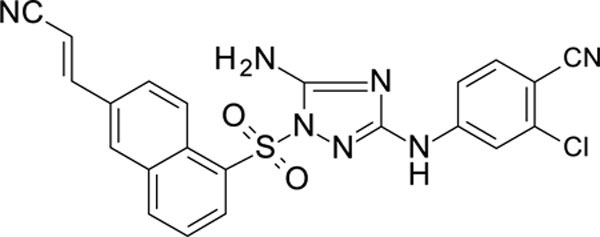
White solid in yield 24%. Mp 279–80 oC. HRMS: calc. for C22H14ClN7O2S [M]+ 463.9005, found 463.9003. Mp = 88.7 °C. 1H NMR (200 MHz, DMSO): δ 10.06 (brs, 1H, NH), 8.88 (d, J = 9.1 Hz, 1H, HC(8`)), 8.52 (d, J = 7.5 Hz, 1H, HC(2`)), 8.42 (d, J = 8.1 Hz, 1H, HC(4`)), 8.34 (d, J = 1.7 Hz, 1H, HC(5`)), 8.10 (dd, J = 9.2, 1.8 Hz, 1H, HC(7`)), 7.84 (d, J = 16.6 Hz, 1H, HC=(CN)), 7.67 (d, J = 2.1 Hz, 1H, HC(3``)), 7.65 (brs, 2H, NH2), 7.38 (dd, J = 8.7, 2.1 Hz, 1H, HC(4``)), 6.69 (d, J = 16.6 Hz, 1H, HC=(CN)). 13C NMR (50 MHz, DMSO) δ 157.99 (С-3), 156.33 (С-5), 149.25 (ArC=), 145.65 (C-4``), 137.25 (C-2``), 136.04 (C-7`), 134.86 (C-6``), 133.62 (C-1`), 132.59 (C-4`), 132.17 (C-5`), 131.83 (C-4à), 130.17 (C-2`), 128.56 (C-8à), 125.98 (C-3`), 125.86 (C-6`), 124.89 (C-8`), 118.46 (CN(C=)), 116.70 (CN-C4``), 116.19 (C-3``), 115.27 (C-5``), 101.78 (C-1``), 99.05 (CN(C=)).
(E)-3-[5-[[5-amino-3-[4-(cyanomethyl)anilino]-1,2,4-triazol-1-yl]sulfonyl]-2-naphthyl]prop-2-enenitrile 12126066
White solid in yield 38%. Mp 275–6 oC. HRMS: calc. for C23H17N7O2S [M]+ 455.4931, found 455.4931. Mp = 88.7 °C. 1H NMR (500 MHz, DMSO): δ 9.20 (s, 1H, NH), 8.94 (d, J = 9.1 Hz, 1H, HC(4`)), 8.48 (dd, J = 7.6, 1.2 Hz, 1H, HC(6`)), 8.36 (d, J = 8.2 Hz, 1H, HC(8`)), 8.29 (d, J = 1.8 Hz, 1H, HC(1`)), 8.09 (dd, J = 9.2, 1.9 Hz, 1H, HC(3`)), 7.83 – 7.78 (m, 2H, HC(7`), HC=(Ar)), 7.48 (s, 2H, NH2), 7.34 (d, J = 8.6 Hz, 2H, HC(2``, 6``)), 7.16 (d, J = 8.7 Hz, 2H, HC(3``, 5``)), 6.68 (d, J = 16.7 Hz, 1H, HC=(CN)), 3.88 (s, 2H, CH2).
13C NMR (126 MHz, DMSO) δ 156.38 (С-5), 149.20 (ArC=), 139.90 (С-1``), 136.88 (C-5`), 134.60 (C-1`), 133.57 (C-4à), 132.50 (C-1`), 132.17 (C-6`), 131.90 (C-8à), 129.96 (C-3`), 128.69 and 128.29 (C-3``, C-5``), 126.14 (C-7`), 125.76 (C-4`), 122.46*(C-4``), 116.86 (C-2``, C-6``), 99.01 (CN(C=)), 21.66 (СH2).
Cells and virus culture.
TZM-bl cells 100, HIV-1 IIIB virus 101–103, HIV-1 IIIB (A17 variant) virus 104, and the NNRTI resistant mutants (p5485, pNLGRINFQ, and p7324–1, all based on strain NL4–3) 105 were obtained from the NIH HIV Reagent Program. H9 [derivative of HuT 78] cells (ATCC® HTB176™) and 293T cells (ATCC® CRL-3216™) were obtained from the American Type Culture Collection. The IIIB and A17 viruses were grown in H9 cells to high titer, which was quantified using a HIV p24 (high sensitivity) AlphaLISA Detection Kit (PerkinElmer, Waltham, MA). The p5485, pNLGRINFQ, and p7324–1 virus plasmids were transfected into 293T cells using FuGene6 (Promega, Madison, WI) and then grown to high titer in H9 cells. The viruses were concentrated using Lenti-X Concentrator as needed (Takara Bio USA, Inc., Mountain View, CA). Infections of TZM-bl cells with these viruses was facilitated with 15 µg/mL DEAE-dextran (Sigma-Aldrich).
Inhibition and toxicity assays.
Compounds were serially diluted starting with 10 mM stocks in DMSO. DMSO concentrations were kept the same for all dilutions used within an assay (either 0.005% or 0.05% depending on highest concentration of compound used). Duplicate 96-well plates were set up with 25 µL compound dilution, 25 µL virus, and 50 µL TZM-bl cells (2x105/mL) with DEAE-dextran and incubated at 37°C 5% CO2 for 48 hr. All compounds were tested at minimum in triplicate twice, over multiple days, with single wells of EFV dilutions on each plate as positive control. All plates had 3–5 control wells without compound, 1 control well with no cells, and 1 control well with a highly toxic level of DMSO (9%). Inhibition assays were set up in clear tissue-culture treated plates with lysates moved to black assay plates or were set up in black tissue culture-treated plates. Medium was removed and the cells were lysed with Glo Lysis Buffer (Promega). Luciferase Assay System reagent (Promega) was added to cells and relative light units (RLU) was measured using an EnSpire instrument (PerkinElmer). Toxicity assays were set up in clear tissue culture-treated plates. CellTiter-Blue Cell Viability Assay reagent (Promega) was added to the wells after 46 hr incubation, followed by 2 hr incubation. Sodium lauryl sulfate was added to 1.5% to stop the reaction and the plates were measured for fluorescence (560nmEx/590nmEm) using an EnSpire instrument.
Chemicals and reagents
DEAE-dextran (Sigma-Aldrich) Cell titer Glo kit and Luciferase Assay System reagent are from Promega (Madison, WI).
NNRTI assay methods.
Inhibition of HIV-I RT was assessed two different methods:
The fluorescent method was done using an EnzChek® reverse transcriptase assay kit purchased from Molecular Probes (Eugene, OR) with a modified manufacture’s protocol. In short, compounds were serially diluted starting with 10 mM stocks in DMSO. DMSO concentrations were kept the same for all dilutions used within an assay (0.1%). The final compound concentrations ranged from 10,000 nM – 0.1 nM with six, 10-fold serial dilutions. 96-well plates were set up with 20 µL reaction mixture (poly(A) ribonucleotide template/ oligo d(T)16 primers), 4 µL diluted RT purchased from Calbiochem® (San Diego, CA), and 1 µL of compound or 0.1% DMSO control. The reaction was incubated at room temperature for 30 min followed by the addition of 125µL of PicoGreen dsDNA quantitation reagent. This was incubated for 2–5 minutes then RTase activity was quantified based on the formation of dsDNA (excitation 480 nm, emission 520 nm) using a Spectramax ID5 plate reader from Molecular Devices (San Jose, CA). A minimum of 4 positive controls and blanks were included for each plate with a minimum of 6 replicates per compound.
An additional HIV-1 RT colormetric assay was done using a kit from Roche (Indianapolis-Marion County, Indiana) using the manufacture’s protocol (500 pM final RT reaction amount). In short, compounds were serially diluted starting with 10 mM stocks in DMSO. DMSO concentrations were kept the same for all dilutions used within an assay (0.05%). Final compound concentrations ranged from 5,000 nM – 0.05 nM with 10-fold serial dilutions. 96-well plates were set up with 20 µL compound dilution, 20 µL RT, and 20 µL template/nucleotides and incubated at 37°C 5% CO2 for 1 hr in a reaction plate. 50 µL of each reaction was then transferred to a streptavidin conjugated 96-well plate and an ELISA assay was run based on manufacture’s protocol. All compounds were tested in sextuplicate, with single wells of EFV dilutions on each plate. All plates had 8 control wells without compound, 2 control wells with no RT, and 2 control blank wells.
In Vitro ADME/Tox assays
In vitro ADME studies, excluding HERG binding, were performed by BioDuro (San Diego, CA). Studies were done using standard methods, with full protocols described in the Supplementary Methods.
in vitro neurotoxicity studies
Primary cultures of mouse cortex and hippocampus.
All animal and cell culture work was performed in accordance with IACUC animal welfare guidelines and was approved by the University of North Carolina-Chapel Hill Institutional Animal Care and Use Committee. Timed gestational embryonic day 16 (E16) pregnant female CD1 mice (Charles Rivers) were anesthetized with the isoflurane drop method until breathing and heart stopped. A thoracotomy was then performed, the uterus removed, briefly rinsed ice cold 70% ethanol, and rinsed twice in ice cold, sterile HEPES-buffered Hank’s balanced salt solution (HBSS). The brain was dissected from each fetus, extensively washed, and cleaned of dura-arachnoid membrane and visible vessels. The cortex/hippocampus was dissected from each brain, minced, and transferred to a 15 ml tube containing 5 ml HBSS + 2.4 U/ml dispase + 2 U/ml DNase I and incubated for 25–30 min at 36º C. Tissue was triturated and pieces allowed to settle for 2 min. The suspended cells were transferred to a 50 ml culture tube containing 25 ml of Neurobasal Plus medium with added B27 Plus supplement, Glutamax, 5% fetal bovine serum and 20 µg/ml gentamicin. After several rounds of trituration in 2–3 ml calcium-magnesium free HBSS, the dissociated cells were seeded at a density of 20,000 cells/cm2 in 48 well plates coated with poly-D-lysine (0.1 mg.ml at 36o C, overnight) or on poly-D-lysine-treated 18 mm round coverslips. After 24 hours, cultures were transferred to Neurobasal Plus medium with added B27 Plus supplement and Glutamax. The resulting cultures were >95% neurons at day 4 after seeding.
Neuronal cell toxicity of primary mouse cultures.
Methods were described previously 48, but in short: compounds were added to primary cultures of mouse neurons at 20 days in vitro. A 10X stock dilution series of each drug was made up in artificial cerebrospinal fluid to be compatible with the medium but lacking protein supplements. A range of final concentrations from 0.1 to 10,000 nM were tested. 25 µl of each dilution was added to each well of a 48 well plate containing 225 µl of Neurobasal Plus medium with B27 Plus supplement. After 48 hr the cells were fixed in methanol:acetic acid (97:3) and stained for microtubule associated protein-2 (MAP-2). Neuron number and morphology was then quantified with the aid of Metamorph image analysis software. Details regarding the calculate of the 50% toxic concentration (TC50) are detailed in the Supplementary Methods.
Calcium Imaging.
The direct effects of the antiretroviral compounds on calcium homeostasis were tested on primary mouse neurons cultured on coverslips as described previously 48. In short, neurons at 14–18 days in vitro were loaded with the calcium indicator, Fluo-4 AM (2 µM, Molecular Probes, Inc., Eugene, OR) in aCSF (aCSF: NaCl 137 mM, KCl 5.0 mM, CaCl2 2.3 mM, MgCl2 1.3 mM, glucose 20 mM). After 30 minutes of dye loading, the coverslip was transferred to a specialized stage for imaging. Cells were maintained in aCSF and time-lapse digital images were automatically captured on an Olympus IX71 microscope using Metamorph Software. Images were captured with a 40 msec exposure every 6 seconds for 6 minutes to assess acute effects and every minute for 40 minutes to assess delayed effects. Three pre-stimulation measurements were taken to establish basal levels of fluorescence at the beginning of each experiment. After collection of the third image, the antiretroviral compound or vehicle diluted from the stock vial into aCSF was added to the neurons at the indicated final concentration. Baseline fluorescence was subtracted to correct for cell-to-cell differences in dye loading and intrinsic fluorescence. Changes in fluorescence at each time point were averaged across all cells from at least triplicate runs to provide an indication of the “typical” response. In some cases, individual cell response patterns are shown where the average masked important cell-specific profiles. All compounds were tested at 1 µM.
In vivo Studies
All in vivo pharmacokinetics and toxicity work was conducted under protocols approved by the Federal Research Center Fundamentals of Biotechnology RAS (protocol #112–2020) according to the institution’s guidelines for animal use, the state industry standards GOST 33215–2014 and GOST 33216–2014 (in harmonization to the European Directive 2010/63/ЕС). All animal work was approved by the Bioethics Committee at Research Center of Biotechnology RAS (Protocol N◦103/4, July 12th, 2022) and was carried out according to the corresponding guidelines for animal use.
Pharmacokinetics.
The pharmacokinetics of compound 12126065 was studied in Balb/C male mice aged 5 to 6 weeks (Central Research Institute of Tuberculosis, Academy of Medical Sciences (Moscow, Russia)). 24 animals with an average body weight of 20 g were selected for the study. Before the experiment the animals were kept on standard vivarium dry pelleted feed. All the test animals had free access to water but were deprived of food for 1 hour before administration of the compound. This regimen was continued for another hour after the administration. 12126065 (250 mg/kg of body weight) was administered as 0.5 ml of suspension in 1% CMC, 0.05% Tween 80 by intragastric intubation. The animals were euthanized by decapitation for blood and brain sampling. Blood, brain and liver samples (3 animals per time point) were taken at 0.5, 1, 2, 3, 5, 8 and 24 h after administration for pharmacokinetic evaluation. Blood was collected in heparinized tubes and centrifuged at 3500 RPM. Plasma was separated from formed elements and immediately frozen at -20°C in freezer. This storage continued before transferring of plasma for analysis. Mice brains were immediately frozen at -120 °C. Details for tissue-specific separation are given in the supplementary methods.
HPLC-MS quantitative analysis: UPLC-MS analysis was performed using an Impact II QqTOF high-resolution HRMS:-spectrometer (Bruker Daltonik, Germany) equipped with Apollo II ESI ion source (Bruker Daltonik, Germany) coupled to Elute UPLC (Bruker Daltonik, Germany) on Intensity solo C18–2 1.8 µm 2.1 × 100 mm reverse phase column (Bruker Daltonik, Germany) with the following conditions: gradient elution at 0.3 mL/min from 5% to 95% B in 8 min (A: 0.1% formic acid in water, B: 0.1% formic acid in acetonitrile), column at 40 °C, 10 µL injection volume, ion source in positive mode, HV capillary at 4.5 kV, spray gas – nitrogen at 2.0 bar, dry gas – nitrogen at 6 L/min 220°C, full spectra scan range m/z 50–1500 at 3 Hz scan rate, automatic internal calibration with sodium trifluoroacetate solution. Spectra were processed with Compass DataAnalysis 5.1 (Bruker Daltonik, Germany). The target compound peak area was measured by automatic integration of EIC chromatogram (m/z 476.0691±0.01). Standards were run in a blood plasma matrix and these standards were used for both blood plasma and brain quantification of compound 12126065. Pharmacokinetic parameters were calculated using Noncompartmental Pharmacokinetics Analysis (NCA) with open-source software 106 which uses standard calculation methods.
In vivo Acute Toxicity Study.
12126065 (2000 mg/kg) was administered to Balb/C male mice, aged 5 to 6 weeks (Central Research Institute of Tuberculosis, Academy of Medical Sciences (Moscow, Russia)), intragastrically. Before the experiment, 1.5 – 2 hours prior to injecting the product, mice were deprived of food and water. Animals weighing 19–21 g only were chosen for these tests. Just before administration the solutions were prepared, and 0.5 ml/20 g of the animals’ weight was administered intragastrically. The control animals received 1% starch gel intragastrically in the amount of 0.5 ml. The experimental and control groups included 5 mice each. The condition of the animals was observed for 14 days.
In vivo Subacute Toxicity Study.
Two doses of compound 12126065 were chosen for the intragastric, subacute toxicity experiments: 25 and 250 mg/kg. Following the administration of 12126065 to male, Balb/C mice (Central Research Institute of Tuberculosis, Academy of Medical Sciences (Moscow, Russia)) daily for 30 days qd, various biological activities/functions were recorded. Each experimental and control group included 17 animals with initial weights of 20.9 ± 0.8 g. The tested product was diluted with 1% starch gel with final suspensions of 2.5 and 25 mg/ml. The volume given to the animals equalled 0.2 ml per 20 g of the animal’s weight. The mice in the control group received 1% starch gel in the amount of 0.2 ml per 20 g of the animal’s weight. During the experiment, changes in the animals’ weight were registered, their coat and mucus membranes condition, and excretion patterns were also recorded.
Following the final injection, the animals’ blood was taken for analysis. Two hours prior to blood sampling, the animals were deprived of food with free access to water. The serum was prepared from the blood using standard practices (incubation at room temperature - 30 min, centrifugation - at 3500 rpm). The blood serum was then analysed for the content of total protein, urea, creatinine, aspartate aminotransferase, alanine aminotransferase and alkaline phosphatase activity. Measurements were performed with the help of a standard agent kit and biochemical analyser BAYER EXPRESS PLUS (semi-automatic, manufactured in 2004 by Bayer Health Care, USA).
While studying the pattern of the peripheral blood, the content of hemoglobin and the level of hematocrit, the amount of red and white blood cells, reticulocytes, and differential blood count were assessed.
24 hours following the last administration, the level of the orientation motor activity of the mice was estimated. This was done by registering the estimated number of rearings (upright component) within a 3-minute timeframe. For this purpose, one animal at a time was placed into a non-transparent cylinder with diameter of 160 mm, which was lighted above with an electric bulb of 60W and placed 1 m above the floor prior to recording.
Following experimental completion, the animals were euthanized, a macroscopic examination of internal organs was performed, followed by a morphometric analysis to determine the absolute and relative weight of internal organs. For the histologic examination, parts of organs were taken, and were preserved in 10% formaldehyde solution. The sections of organs were prepared by common methods with paraffin-embedding and hematoxylin and eosin stain 107.
Docking Methods.
We performed structure-based drug discovery by docking molecule designs in the numerous HIV RT protein structures. Compounds were docked into the HIV I RT wild-type (PDB: 4G1Q) and K103N/Y181C double mutant (PDB: 4RW4) using Discovery Studio (Biovia, San Diego, CA) LibDock (rigid docking). The docking sphere was chosen based on the position of the crystalized ligands of RPVand JLJ494 for wild-type and the K103N/Y181C double mutant, respectively. The docking protocols were all performed with the default settings.
t-SNE visualization.
t-SNE 108 embeds data into a lower-dimensional space. 1024 ECFP6 fingerprints were generated for all compounds. The ECFP6 fingerprints were then embedded into a 2-dimensional vector using t-SNE. All t-SNE values were generated using the scikit-learn library in python with default hyperparameters (n_components = 2, perplexity = 30, early exaggeration = 12.0, learning rate = 200, n_iter = 1000).
Supplementary Material
Acknowledgments
BioDuro are kindly acknowledged for their efforts on this project generating in vitro ADME data. The following reagents were obtained through the NIH HIV Reagent Program, Division of AIDS, NIAID, NIH: Human Immunodeficiency Virus-1 IIIB, ARP-398, contributed by Dr. Robert Gallo; Human Immunodeficiency Virus-1 IIIB (A17 Variant), ARP-1413, contributed by Dr. Emilio Emini; TZM-bl Cells, ARP-8129, contributed by Dr. John C. Kappes and Dr. Xiaoyun; HIV-1 Infectious Molecular Clone (p5485), ARP-12229, contributed by Dr. Robert Shafer; Human Immunodeficiency Virus 1 (HIV-1), Strain pNLGRINFQ, ARP-6202, contributed by Dr. Tomozumi Imamichi and Dr. H. Clifford Lane; Human Immunodeficiency Virus 1 (HIV-1), Strain NL4–3 1392 Infectious Molecular Clone (p7324–1), ARP-7396, contributed by Dr. Robert W. Shafer. Dr. Mohamed Nasr is thanked for assistance with obtaining the NIAID ChemDB HIV, Opportunistic Infection and Tuberculosis Therapeutics Database. Biovia is kindly acknowledged for providing Discovery Studio for the docking studies. Dr. Alfredo Garzino-Demo is acknowledged for early discussions and preliminary work on this project.
Grant information
Research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health under Award Number 1R01NS102164–01. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We also kindly acknowledge NIH funding: R44GM122196–02A1 from NIGMS and 1R43ES031038–01 from NIEHS (PI – Sean Ekins). “Research reported in this publication was supported by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number R43ES031038. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.”
This work was supported by the University of North Carolina at Chapel Hill Center for AIDS Research (P30 AI050410).
Abbreviations
- ADME
Absorption, distribution, metabolism, excretion
- BBB
blood brain barrier
- CNS
central nervous system
- cART
combination antiretroviral therapy
- HAART
Highly active antiretroviral therapy
- HIV
human immunodeficiency virus
- HAND
HIV associated neurocognitive disorders
- NNRTI
non-nucleoside reverse transcriptase inhibitors
- RT
reverse transcriptase
Footnotes
Competing interests:
S.E. is owner, T.R.L. is an employee at Collaborations Pharmaceuticals, Inc. All other authors have no conflicts. V.M., O.R., T.R.L. and S.E. are co-inventors on a submitted patent relating to this work.
Ancillary Information
Contents of the supplemental Information include Figures S1-S17, Tables S1-S3, PDBs of the docked compounds in wild-type (4G1Q) and A17 mutant (4RW4) RT, inhibition data of all tested compounds with corresponding SMILES, and Supplementary text and methods.
References
- 1.WHO. HIV/AIDS https://www.who.int/news-room/fact-sheets/detail/hiv-aids (accessed Mar 29, 2023).
- 2.Anon. Unaids. Global hiv & aids statistics—2020 fact sheet https://www.unaids.org/en/resources/fact-sheet. (accessed Mar 29, 2023).
- 3.UNICEF. Global and regional trends https://data.unicef.org/topic/hivaids/global-regional-trends/ (accessed Mar 29, 2023).
- 4.CDC. Statistics overview https://www.cdc.gov/hiv/statistics/overview/index.html (accessed Mar 29, 2023).
- 5.Murphy EL; Collier AC; Kalish LA; Assmann SF; Para MF; Flanigan TP; Kumar PN; Mintz L; Wallach FR; Nemo GJ; Viral Activation Transfusion Study, I. Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced hiv disease. Ann Intern Med 2001, 135, 17–26. [DOI] [PubMed] [Google Scholar]
- 6.HHS Panel on Antiretroviral Guidelines for Adults and Adolescents—A Working Group of the Office of AIDS Research Advisory Council (OARAC). Guidelines for the use of antiretroviral agents in adults and adolescents with HIVS https://www.ncbi.nlm.nih.gov/books/NBK586306/ (accessed Mar 29, 2023).
- 7.Shiau S; Abrams EJ; Arpadi SM; Kuhn L Early antiretroviral therapy in hiv-infected infants: Can it lead to hiv remission? Lancet HIV 2018, 5, e250–e258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Violari A; Cotton MF; Gibb DM; Babiker AG; Steyn J; Madhi SA; Jean-Philippe P; McIntyre JA; Team CS Early antiretroviral therapy and mortality among hiv-infected infants. N Engl J Med 2008, 359, 2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Esber A; Polyak C; Kiweewa F; Maswai J; Owuoth J; Maganga L; Adamu Y; Hickey PW; Ake JA; Crowell TA Persistent low-level viremia predicts subsequent virologic failure: Is it time to change the third 90? Clin Infect Dis 2019, 69, 805–812. [DOI] [PubMed] [Google Scholar]
- 10.Taiwo B; Gallien S; Aga E; Ribaudo H; Haubrich R; Kuritzkes DR; Eron JJ Jr. Antiretroviral drug resistance in hiv-1-infected patients experiencing persistent low-level viremia during first-line therapy. J Infect Dis 2011, 204, 515–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taramasso L; Magnasco L; Bruzzone B; Caligiuri P; Bozzi G; Mora S; Balletto E; Tatarelli P; Giacomini M; Di Biagio A How relevant is the hiv low level viremia and how is its management changing in the era of modern art? A large cohort analysis. J Clin Virol 2020, 123, 104255. [DOI] [PubMed] [Google Scholar]
- 12.Zhang T; Ding H; An M; Wang X; Tian W; Zhao B; Han X Factors associated with high-risk low-level viremia leading to virologic failure: 16-year retrospective study of a chinese antiretroviral therapy cohort. BMC Infect Dis 2020, 20, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ryscavage P; Kelly S; Li JZ; Harrigan PR; Taiwo B Significance and clinical management of persistent low-level viremia and very-low-level viremia in hiv-1-infected patients. Antimicrob Agents Chemother 2014, 58, 3585–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joya C; Won SH; Schofield C; Lalani T; Maves RC; Kronmann K; Deiss R; Okulicz J; Agan BK; Ganesan A Persistent low-level viremia while on antiretroviral therapy is an independent risk factor for virologic failure. Clin Infect Dis 2019, 69, 2145–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laprise C; de Pokomandy A; Baril JG; Dufresne S; Trottier H Virologic failure following persistent low-level viremia in a cohort of hiv-positive patients: Results from 12 years of observation. Clin Infect Dis 2013, 57, 1489–1496. [DOI] [PubMed] [Google Scholar]
- 16.Tagnouokam-Ngoupo PA; Penda IC; Tchatchueng Mbougua JB; Tetang Ndiang S; Yuya Septoh F; Kenne A; Ngalle JE; Jakpou S; Ateba Ndongo F; Warszawski J; Faye A; Tejiokem MC; Group AN-PS Virological failure and antiretroviral resistance among hiv-infected children after five years follow-up in the anrs 12225-pediacam cohort in cameroon. PLoS One 2021, 16, e0248642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao SW; Liu ZH; Wu TS; Ku SW; Tsai CL; Shie SS; Huang PY; Wu YM; Hsiao YH; Chen NY Prevalence of drug resistance mutations in hiv-infected individuals with low-level viraemia under combination antiretroviral therapy: An observational study in a tertiary hospital in northern taiwan, 2017–19. J Antimicrob Chemother 2021, 76, 722–728. [DOI] [PubMed] [Google Scholar]
- 18.Mziray SR; Kumburu HH; Assey HB; Sonda TB; Mahande MJ; Msuya SE; Kiwelu IE Patterns of acquired hiv-1 drug resistance mutations and predictors of virological failure in moshi, northern tanzania. PLoS One 2020, 15, e0232649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flexner C; Saag M The antiretroviral drug pipeline: Prospects and implications for future treatment research. Curr Opin HIV AIDS 2013, 8, 572–578. [DOI] [PubMed] [Google Scholar]
- 20.Tan IL; McArthur JC Hiv-associated neurological disorders: A guide to pharmacotherapy. CNS Drugs 2012, 26, 123–134. [DOI] [PubMed] [Google Scholar]
- 21.Sanmarti M; Ibanez L; Huertas S; Badenes D; Dalmau D; Slevin M; Krupinski J; Popa-Wagner A; Jaen A Hiv-associated neurocognitive disorders. J Mol Psychiatry 2014, 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bertrand L; Nair M; Toborek M Solving the blood-brain barrier challenge for the effective treatment of hiv replication in the central nervous system. Curr Pharm Des 2016. [DOI] [PMC free article] [PubMed]
- 23.Cross HM; Combrinck MI; Joska JA Hiv-associated neurocognitive disorders: Antiretroviral regimen, central nervous system penetration effectiveness, and cognitive outcomes. S Afr Med J 2013, 103, 758–762. [DOI] [PubMed] [Google Scholar]
- 24.Ellis RJ; Letendre S; Vaida F; Haubrich R; Heaton RK; Sacktor N; Clifford DB; Best BM; May S; Umlauf A; Cherner M; Sanders C; Ballard C; Simpson DM; Jay C; McCutchan JA Randomized trial of central nervous system-targeted antiretrovirals for hiv-associated neurocognitive disorder. Clin Infect Dis 2014, 58, 1015–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Yacovo MS; Molto J; Ferrer E; Curran A; Else L; Gisslen M; Clotet B; Tiraboschi JM; Niubo J; Vila A; Zetterberg H; Back D; Podzamczer D Antiviral activity and csf concentrations of 600/100 mg of darunavir/ritonavir once daily in hiv-1 patients with plasma viral suppression. J Antimicrob Chemother 2015, 70, 1513–1516. [DOI] [PubMed] [Google Scholar]
- 26.Van den Hof M; Blokhuis C; Cohen S; Scherpbier HJ; Wit F; Pistorius MCM; Kootstra NA; Teunissen CE; Mathot RAA; Pajkrt D Cns penetration of art in hiv-infected children. J Antimicrob Chemother 2018, 73, 484–489. [DOI] [PubMed] [Google Scholar]
- 27.Bertrand L; Meroth F; Tournebize M; Leda AR; Sun E; Toborek M Targeting the hiv-infected brain to improve ischemic stroke outcome. Nat Commun 2019, 10, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apostolova N; Funes HA; Blas-Garcia A; Galindo MJ; Alvarez A; Esplugues JV Efavirenz and the cns: What we already know and questions that need to be answered. J Antimicrob Chemother 2015, 70, 2693–2708. [DOI] [PubMed] [Google Scholar]
- 29.Decloedt EH; Rosenkranz B; Maartens G; Joska J Central nervous system penetration of antiretroviral drugs: Pharmacokinetic, pharmacodynamic and pharmacogenomic considerations. Clin Pharmacokinet 2015, 54, 581–598. [DOI] [PubMed] [Google Scholar]
- 30.Shah A; Gangwani MR; Chaudhari NS; Glazyrin A; Bhat HK; Kumar A Neurotoxicity in the post-haart era: Caution for the antiretroviral therapeutics. Neurotox Res 2016. [DOI] [PMC free article] [PubMed]
- 31.Abers MS; Shandera WX; Kass JS Neurological and psychiatric adverse effects of antiretroviral drugs. CNS Drugs 2014, 28, 131–145. [DOI] [PubMed] [Google Scholar]
- 32.Jin J; Grimmig B; Izzo J; Brown LA; Hudson C; Smith AJ; Tan J; Bickford PC; Giunta B Hiv non-nucleoside reverse transcriptase inhibitor efavirenz reduces neural stem cell proliferation in vitro and in vivo. Cell Transplant 2016. [DOI] [PMC free article] [PubMed]
- 33.Kranick SM; Nath A Neurologic complications of hiv-1 infection and its treatment in the era of antiretroviral therapy. Continuum (Minneap Minn) 2012, 18, 1319–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Funes HA; Blas-Garcia A; Esplugues JV; Apostolova N Efavirenz alters mitochondrial respiratory function in cultured neuron and glial cell lines. J Antimicrob Chemother 2015, 70, 2249–2254. [DOI] [PubMed] [Google Scholar]
- 35.Apostolova N; Funes HA; Blas-Garcia A; Alegre F; Polo M; Esplugues JV Involvement of nitric oxide in the mitochondrial action of efavirenz: A differential effect on neurons and glial cells. J Infect Dis 2015, 211, 1953–1958. [DOI] [PubMed] [Google Scholar]
- 36.Polo M; Alegre F; Funes HA; Blas-Garcia A; Victor VM; Esplugues JV; Apostolova N Mitochondrial (dys)function - a factor underlying the variability of efavirenz-induced hepatotoxicity? Br J Pharmacol 2015, 172, 1713–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sluis-Cremer N The emerging profile of cross-resistance among the nonnucleoside hiv-1 reverse transcriptase inhibitors. Viruses 2014, 6, 2960–2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melikian GL; Rhee SY; Varghese V; Porter D; White K; Taylor J; Towner W; Troia P; Burack J; Dejesus E; Robbins GK; Razzeca K; Kagan R; Liu TF; Fessel WJ; Israelski D; Shafer RW Non-nucleoside reverse transcriptase inhibitor (nnrti) cross-resistance: Implications for preclinical evaluation of novel nnrtis and clinical genotypic resistance testing. J Antimicrob Chemother 2014, 69, 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanangamudi M; Kurup S; Namasivayam V Non-nucleoside reverse transcriptase inhibitors (nnrtis): A brief overview of clinically approved drugs and combination regimens. Curr Opin Pharmacol 2020, 54, 179–187. [DOI] [PubMed] [Google Scholar]
- 40.Kerr SG; Anderson KS Pre-steady-state kinetic characterization of wild type and 3’-azido-3’-deoxythymidine (azt) resistant human immunodeficiency virus type 1 reverse transcriptase: Implication of rna directed DNA polymerization in the mechanism of azt resistance. Biochemistry 1997, 36, 14064–14070. [DOI] [PubMed] [Google Scholar]
- 41.Usach I; Melis V; Peris JE Non-nucleoside reverse transcriptase inhibitors: A review on pharmacokinetics, pharmacodynamics, safety and tolerability. J Int AIDS Soc 2013, 16, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Svarovskaia ES; Cheslock SR; Zhang WH; Hu WS; Pathak VK Retroviral mutation rates and reverse transcriptase fidelity. Front Biosci 2003, 8, d117–134. [DOI] [PubMed] [Google Scholar]
- 43.Hernandez-Gil J; Ferrer S; Cabedo N; Lopez-Gresa MP; Castineiras A; Lloret F Two copper complexes from two novel naphthalene-sulfonyl-triazole ligands: Different nuclearity and different DNA binding and cleavage capabilities. J Inorg Biochem 2013, 125, 50–63. [DOI] [PubMed] [Google Scholar]
- 44.Lin R; Connolly PJ; Huang S; Wetter SK; Lu Y; Murray WV; Emanuel SL; Gruninger RH; Fuentes-Pesquera AR; Rugg CA; Middleton SA; Jolliffe LK 1-acyl-1h-[1,2,4]triazole-3,5-diamine analogues as novel and potent anticancer cyclin-dependent kinase inhibitors: Synthesis and evaluation of biological activities. J Med Chem 2005, 48, 4208–4211. [DOI] [PubMed] [Google Scholar]
- 45.Kok BP; Ghimire S; Kim W; Chatterjee S; Johns T; Kitamura S; Eberhardt J; Ogasawara D; Xu J; Sukiasyan A; Kim SM; Godio C; Bittencourt JM; Cameron M; Galmozzi A; Forli S; Wolan DW; Cravatt BF; Boger DL; Saez E Discovery of small-molecule enzyme activators by activity-based protein profiling. Nat Chem Biol 2020, 16, 997–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang C; Dahl KD; Leung A; Chan SY; Hsueh AJ Serum bioactive follicle-stimulating hormone in men with idiopathic azoospermia and oligospermia. J Clin Endocrinol Metab 1987, 65, 629–633. [DOI] [PubMed] [Google Scholar]
- 47.Zorn KM; Lane TR; Russo DP; Clark AM; Makarov V; Ekins S Multiple machine learning comparisons of hiv cell-based and reverse transcriptase data sets. Mol Pharm 2019, 16, 1620–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson K; Liner J; Meeker RB Antiretroviral neurotoxicity. J Neurovirol 2012, 18, 388–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Das K; Bauman JD; Clark AD Jr.; Frenkel YV; Lewi PJ; Shatkin AJ; Hughes SH; Arnold E High-resolution structures of hiv-1 reverse transcriptase/tmc278 complexes: Strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci U S A 2008, 105, 1466–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wainberg MA The need for development of new hiv-1 reverse transcriptase and integrase inhibitors in the aftermath of antiviral drug resistance. Scientifica (Cairo) 2012, 2012, 238278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hermans LE; Moorhouse M; Carmona S; Grobbee DE; Hofstra LM; Richman DD; Tempelman HA; Venter WDF; Wensing AMJ Effect of hiv-1 low-level viraemia during antiretroviral therapy on treatment outcomes in who-guided south african treatment programmes: A multicentre cohort study. Lancet Infect Dis 2018, 18, 188–197. [DOI] [PubMed] [Google Scholar]
- 52.Bernal E; Gomez JM; Jarrin I; Cano A; Munoz A; Alcaraz A; Imaz A; Iribarren JA; Rivero M; Arazo P; Gutierrez F; Co RISSG Low-level viremia is associated with clinical progression in hiv-infected patients receiving antiretroviral treatment. J Acquir Immune Defic Syndr 2018, 78, 329–337. [DOI] [PubMed] [Google Scholar]
- 53.Chong P; Sebahar P; Youngman M; Garrido D; Zhang H; Stewart EL; Nolte RT; Wang L; Ferris RG; Edelstein M; Weaver K; Mathis A; Peat A Rational design of potent non-nucleoside inhibitors of hiv-1 reverse transcriptase. J Med Chem 2012, 55, 10601–10609. [DOI] [PubMed] [Google Scholar]
- 54.Cote B; Burch JD; Asante-Appiah E; Bayly C; Bedard L; Blouin M; Campeau LC; Cauchon E; Chan M; Chefson A; Coulombe N; Cromlish W; Debnath S; Deschenes D; Dupont-Gaudet K; Falgueyret JP; Forget R; Gagne S; Gauvreau D; Girardin M; Guiral S; Langlois E; Li CS; Nguyen N; Papp R; Plamondon S; Roy A; Roy S; Seliniotakis R; St-Onge M; Ouellet S; Tawa P; Truchon JF; Vacca J; Wrona M; Yan Y; Ducharme Y Discovery of mk-1439, an orally bioavailable non-nucleoside reverse transcriptase inhibitor potent against a wide range of resistant mutant hiv viruses. Bioorg Med Chem Lett 2014, 24, 917–922. [DOI] [PubMed] [Google Scholar]
- 55.Lai MT; Feng M; Falgueyret JP; Tawa P; Witmer M; DiStefano D; Li Y; Burch J; Sachs N; Lu M; Cauchon E; Campeau LC; Grobler J; Yan Y; Ducharme Y; Cote B; Asante-Appiah E; Hazuda DJ; Miller MD In vitro characterization of mk-1439, a novel hiv-1 nonnucleoside reverse transcriptase inhibitor. Antimicrob Agents Chemother 2014, 58, 1652–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hwang C; Lai MT; Hazuda D Rational design of doravirine: From bench to patients. ACS Infect Dis 2020, 6, 64–73. [DOI] [PubMed] [Google Scholar]
- 57.Xu Z; Ba M; Zhou H; Cao Y; Tang C; Yang Y; He R; Liang Y; Zhang X; Li Z; Zhu L; Guo Y; Guo C 2,4,5-trisubstituted thiazole derivatives: A novel and potent class of non-nucleoside inhibitors of wild type and mutant hiv-1 reverse transcriptase. Eur J Med Chem 2014, 85, 27–42. [DOI] [PubMed] [Google Scholar]
- 58.Liu Z; Chen W; Zhan P; De Clercq E; Pannecouque C; Liu X Design, synthesis and anti-hiv evaluation of novel diarylnicotinamide derivatives (danas) targeting the entrance channel of the nnrti binding pocket through structure-guided molecular hybridization. Eur J Med Chem 2014, 87, 52–62. [DOI] [PubMed] [Google Scholar]
- 59.Mislak AC; Frey KM; Bollini M; Jorgensen WL; Anderson KS A mechanistic and structural investigation of modified derivatives of the diaryltriazine class of nnrtis targeting hiv-1 reverse transcriptase. Biochim Biophys Acta 2014, 1840, 2203–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X; Meng Q; Qiu L; Zhan P; Liu H; De Clercq E; Pannecouque C; Liu X Design, synthesis, and anti-hiv evaluation of novel triazine derivatives targeting the entrance channel of the nnrti binding pocket. Chem Biol Drug Des 2015, 86, 122–128. [DOI] [PubMed] [Google Scholar]
- 61.Wan ZY; Yao J; Tao Y; Mao TQ; Wang XL; Lu YP; Wang HF; Yin H; Wu Y; Chen FE; De Clercq E; Daelemans D; Pannecouque C Discovery of piperidin-4-yl-aminopyrimidine derivatives as potent non-nucleoside hiv-1 reverse transcriptase inhibitors. Eur J Med Chem 2015, 97, 1–9. [DOI] [PubMed] [Google Scholar]
- 62.Lee WG; Frey KM; Gallardo-Macias R; Spasov KA; Chan AH; Anderson KS; Jorgensen WL Discovery and crystallography of bicyclic arylaminoazines as potent inhibitors of hiv-1 reverse transcriptase. Bioorg Med Chem Lett 2015, 25, 4824–4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dousson C; Alexandre FR; Amador A; Bonaric S; Bot S; Caillet C; Convard T; da Costa D; Lioure MP; Roland A; Rosinovsky E; Maldonado S; Parsy C; Trochet C; Storer R; Stewart A; Wang J; Mayes BA; Musiu C; Poddesu B; Vargiu L; Liuzzi M; Moussa A; Jakubik J; Hubbard L; Seifer M; Standring D Discovery of the aryl-phospho-indole idx899, a highly potent anti-hiv non-nucleoside reverse transcriptase inhibitor. J Med Chem 2016, 59, 1891–1898. [DOI] [PubMed] [Google Scholar]
- 64.Lu X; Li X; Yang J; Huang B; Kang D; Zhao F; Zhou Z; De Clercq E; Daelemans D; Pannecouque C; Zhan P; Liu X Arylazolyl(azinyl)thioacetanilides. Part 20: Discovery of novel purinylthioacetanilides derivatives as potent hiv-1 nnrtis via a structure-based bioisosterism approach. Bioorg Med Chem 2016, 24, 4424–4433. [DOI] [PubMed] [Google Scholar]
- 65.Tian Y; Liu Z; Liu J; Huang B; Kang D; Zhang H; De Clercq E; Daelemans D; Pannecouque C; Lee KH; Chen CH; Zhan P; Liu X Targeting the entrance channel of nnibp: Discovery of diarylnicotinamide 1,4-disubstituted 1,2,3-triazoles as novel hiv-1 nnrtis with high potency against wild-type and e138k mutant virus. Eur J Med Chem 2018, 151, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Han S; Sang Y; Wu Y; Tao Y; Pannecouque C; De Clercq E; Zhuang C; Chen FE Molecular hybridization-inspired optimization of diarylbenzopyrimidines as hiv-1 nonnucleoside reverse transcriptase inhibitors with improved activity against k103n and e138k mutants and pharmacokinetic profiles. ACS Infect Dis 2020, 6, 787–801. [DOI] [PubMed] [Google Scholar]
- 67.Zhou Z; Liu T; Kang D; Huo Z; Wu G; Daelemans D; De Clercq E; Pannecouque C; Zhan P; Liu X Discovery of novel diarylpyrimidines as potent hiv-1 nnrtis by investigating the chemical space of a less explored “hydrophobic channel”. Org Biomol Chem 2018, 16, 1014–1028. [DOI] [PubMed] [Google Scholar]
- 68.Zhou Z; Liu T; Wu G; Kang D; Fu Z; Wang Z; De Clercq E; Pannecouque C; Zhan P; Liu X Targeting the hydrophobic channel of nnibp: Discovery of novel 1,2,3-triazole-derived diarylpyrimidines as novel hiv-1 nnrtis with high potency against wild-type and k103n mutant virus. Org Biomol Chem 2019, 17, 3202–3217. [DOI] [PubMed] [Google Scholar]
- 69.Huang B; Liu X; Tian Y; Kang D; Zhou Z; Daelemans D; De Clercq E; Pannecouque C; Zhan P; Liu X First discovery of a potential carbonate prodrug of nnrti drug candidate rdea427 with submicromolar inhibitory activity against hiv-1 k103n/y181c double mutant strain. Bioorg Med Chem Lett 2018, 28, 1348–1351. [DOI] [PubMed] [Google Scholar]
- 70.Lu X; Li X; Yang J; Huang B; Kang D; Zhao F; Zhou Z; De Clercq E; Daelemans D; Pannecouque C; Zhan P; Liu X Arylazolyl(azinyl)thioacetanilides. Part 20: Discovery of novel purinylthioacetanilides derivatives as potent hiv-1 nnrtis via a structure-based bioisosterism approach. Bioorg Med Chem 2016, 24, 4424–4433. [DOI] [PubMed] [Google Scholar]
- 71.Xu C; Xin Y; Chen M; Ba M; Guo Q; Zhu C; Guo Y; Shi J Discovery, synthesis, and optimization of an n-alkoxy indolylacetamide against hiv-1 carrying nnrti-resistant mutations from the isatis indigotica root. Eur J Med Chem 2020, 189, 112071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu X; Chen P; Li X; Ba M; Jiao X; Guo Y; Xie P Design, synthesis and biological evaluation of substituted (+)-sg-1 derivatives as novel anti-hiv agents. Bioorg Med Chem Lett 2018, 28, 1699–1703. [DOI] [PubMed] [Google Scholar]
- 73.Kang D; Ruiz FX; Sun Y; Feng D; Jing L; Wang Z; Zhang T; Gao S; Sun L; De Clercq E; Pannecouque C; Arnold E; Zhan P; Liu X 2,4,5-trisubstituted pyrimidines as potent hiv-1 nnrtis: Rational design, synthesis, activity evaluation, and crystallographic studies. J Med Chem 2021, 64, 4239–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen X; Ding L; Tao Y; Pannecouque C; De Clercq E; Zhuang C; Chen FE Bioisosterism-based design and enantiomeric profiling of chiral hydroxyl-substituted biphenyl-diarylpyrimidine nonnucleoside hiv-1 reverse transcriptase inhibitors. Eur J Med Chem 2020, 202, 112549. [DOI] [PubMed] [Google Scholar]
- 75.Sang Y; Han S; Pannecouque C; De Clercq E; Zhuang C; Chen F Conformational restriction design of thiophene-biphenyl-dapy hiv-1 non-nucleoside reverse transcriptase inhibitors. Eur J Med Chem 2019, 182, 111603. [DOI] [PubMed] [Google Scholar]
- 76.Lei Y; Han S; Yang Y; Pannecouque C; De Clercq E; Zhuang C; Chen FE Design of biphenyl-substituted diarylpyrimidines with a cyanomethyl linker as hiv-1 nnrtis via a molecular hybridization strategy. Molecules 2020, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Z; Kang D; Chen M; Wu G; Feng D; Zhao T; Zhou Z; Huo Z; Jing L; Zuo X; Daelemans D; De Clercq E; Pannecouque C; Zhan P; Liu X Design, synthesis, and antiviral evaluation of novel hydrazone-substituted thiophene[3,2-d]pyrimidine derivatives as potent human immunodeficiency virus-1 inhibitors. Chem Biol Drug Des 2018, 92, 2009–2021. [DOI] [PubMed] [Google Scholar]
- 78.Sun Y; Kang D; Da F; Zhang T; Li P; Zhang B; De Clercq E; Pannecouque C; Zhan P; Liu X Identification of novel potent hiv-1 inhibitors by exploiting the tolerant regions of the nnrtis binding pocket. Eur J Med Chem 2021, 214, 113204. [DOI] [PubMed] [Google Scholar]
- 79.Feng D; Zuo X; Jing L; Chen CH; Olotu FA; Lin H; Soliman M; De Clercq E; Pannecouque C; Lee KH; Kang D; Liu X; Zhan P Design, synthesis, and evaluation of “dual-site”-binding diarylpyrimidines targeting both nnibp and the nnrti adjacent site of the hiv-1 reverse transcriptase. Eur J Med Chem 2021, 211, 113063. [DOI] [PubMed] [Google Scholar]
- 80.Kang D; Wang Z; Chen M; Feng D; Wu G; Zhou Z; Jing L; Zuo X; Jiang X; Daelemans D; De Clercq E; Pannecouque C; Zhan P; Liu X Discovery of potent hiv-1 non-nucleoside reverse transcriptase inhibitors by exploring the structure-activity relationship of solvent-exposed regions i. Chem Biol Drug Des 2019, 93, 430–437. [DOI] [PubMed] [Google Scholar]
- 81.Jiang X; Huang B; Olotu FA; Li J; Kang D; Wang Z; De Clercq E; Soliman MES; Pannecouque C; Liu X; Zhan P Exploiting the tolerant region i of the non-nucleoside reverse transcriptase inhibitor (nnrti) binding pocket. Part 2: Discovery of diarylpyrimidine derivatives as potent hiv-1 nnrtis with high fsp(3) values and favorable drug-like properties. Eur J Med Chem 2021, 213, 113051. [DOI] [PubMed] [Google Scholar]
- 82.Huang B; Chen W; Zhao T; Li Z; Jiang X; Ginex T; Vilchez D; Luque FJ; Kang D; Gao P; Zhang J; Tian Y; Daelemans D; De Clercq E; Pannecouque C; Zhan P; Liu X Exploiting the tolerant region i of the non-nucleoside reverse transcriptase inhibitor (nnrti) binding pocket: Discovery of potent diarylpyrimidine-typed hiv-1 nnrtis against wild-type and e138k mutant virus with significantly improved water solubility and favorable safety profiles. J Med Chem 2019, 62, 2083–2098. [DOI] [PubMed] [Google Scholar]
- 83.Kang D; Wang Z; Zhang H; Wu G; Zhao T; Zhou Z; Huo Z; Huang B; Feng D; Ding X; Zhang J; Zuo X; Jing L; Luo W; Guma S; Daelemans D; Clercq E; Pannecouque C; Zhan P; Liu X Further exploring solvent-exposed tolerant regions of allosteric binding pocket for novel hiv-1 nnrtis discovery. ACS Med Chem Lett 2018, 9, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhao T; Meng Q; Kang D; Ji J; De Clercq E; Pannecouque C; Liu X; Zhan P Discovery of novel indolylarylsulfones as potent hiv-1 nnrtis via structure-guided scaffold morphing. Eur J Med Chem 2019, 182, 111619. [DOI] [PubMed] [Google Scholar]
- 85.Nalli M; Armijos Rivera JI; Masci D; Coluccia A; Badia R; Riveira-Munoz E; Brambilla A; Cinquina E; Turriziani O; Falasca F; Catalano M; Limatola C; Este JA; Maga G; Silvestri R; Crespan E; La Regina G New indolylarylsulfone non-nucleoside reverse transcriptase inhibitors show low nanomolar inhibition of single and double hiv-1 mutant strains. Eur J Med Chem 2020, 208, 112696. [DOI] [PubMed] [Google Scholar]
- 86.Kang D; Zhang H; Wang Z; Zhao T; Ginex T; Luque FJ; Yang Y; Wu G; Feng D; Wei F; Zhang J; De Clercq E; Pannecouque C; Chen CH; Lee KH; Murugan NA; Steitz TA; Zhan P; Liu X Identification of dihydrofuro[3,4- d]pyrimidine derivatives as novel hiv-1 non-nucleoside reverse transcriptase inhibitors with promising antiviral activities and desirable physicochemical properties. J Med Chem 2019, 62, 1484–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kang D; Feng D; Jing L; Sun Y; Wei F; Jiang X; Wu G; De Clercq E; Pannecouque C; Zhan P; Liu X In situ click chemistry-based rapid discovery of novel hiv-1 nnrtis by exploiting the hydrophobic channel and tolerant regions of nnibp. Eur J Med Chem 2020, 193, 112237. [DOI] [PubMed] [Google Scholar]
- 88.Buemi MR; Gitto R; Ielo L; Pannecouque C; De Luca L Inhibition of hiv-1 rt activity by a new series of 3-(1,3,4-thiadiazol-2-yl)thiazolidin-4-one derivatives. Bioorg Med Chem 2020, 28, 115431. [DOI] [PubMed] [Google Scholar]
- 89.Smith SJ; Pauly GT; Hewlett K; Schneider JP; Hughes SH Structure-based non-nucleoside inhibitor design: Developing inhibitors that are effective against resistant mutants. Chem Biol Drug Des 2021, 97, 4–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu Y; Tang C; Rui R; Yang L; Ding W; Wang J; Li Y; Lai CC; Wang Y; Luo R; Xiao W; Zhang H; Zheng Y; He Y Synthesis and biological evaluation of a series of 2-(((5-akly/aryl-1h-pyrazol-3-yl)methyl)thio)-5-alkyl-6-(cyclohexylmethyl)-pyrimi din-4(3h)-ones as potential hiv-1 inhibitors. Acta Pharm Sin B 2020, 10, 512–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang Z; Kang D; Feng D; Cherukupalli S; Jiang X; Fu Z; De Clercq E; Pannecouque C; Liu X; Zhan P Targeting dual tolerant regions of binding pocket: Discovery of novel morpholine-substituted diarylpyrimidines as potent hiv-1 nnrtis with significantly improved water solubility. Eur J Med Chem 2020, 206, 112811. [DOI] [PubMed] [Google Scholar]
- 92.Zhou RL; Ju Z; Pannecouque C; Clercq E; Wang S; Chen FE Structure-guided design of novel hept analogs with enhanced potency and safety: From isopropyl-hepts to cyclopropyl-hepts. Eur J Med Chem 2023, 246, 114939. [DOI] [PubMed] [Google Scholar]
- 93.Zhao LM; Wang S; Pannecouque C; De Clercq E; Piao HR; Chen FE Discovery of novel biphenyl-substituted pyridone derivatives as potent non-nucleoside reverse transcriptase inhibitors with promising oral bioavailability. Eur J Med Chem 2022, 240, 114581. [DOI] [PubMed] [Google Scholar]
- 94.Gao S; Cheng Y; Song S; Song L; Zhao F; Xu S; Kang D; Sun L; Gao P; De Clercq E; Pannecouque C; Liu X; Zhan P Chemical space exploration around indolylarylsulfone scaffold led to a novel class of highly active hiv-1 nnrtis with spiro structural features. Eur J Med Chem 2022, 238, 114471. [DOI] [PubMed] [Google Scholar]
- 95.Das K; Lewi PJ; Hughes SH; Arnold E Crystallography and the design of anti-aids drugs: Conformational flexibility and positional adaptability are important in the design of non-nucleoside hiv-1 reverse transcriptase inhibitors. Prog Biophys Mol Biol 2005, 88, 209–231. [DOI] [PubMed] [Google Scholar]
- 96.Das K; Clark AD Jr.; Lewi PJ; Heeres J; De Jonge MR; Koymans LM; Vinkers HM; Daeyaert F; Ludovici DW; Kukla MJ; De Corte B; Kavash RW; Ho CY; Ye H; Lichtenstein MA; Andries K; Pauwels R; De Bethune MP; Boyer PL; Clark P; Hughes SH; Janssen PA; Arnold E Roles of conformational and positional adaptability in structure-based design of tmc125-r165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant hiv-1 variants. J Med Chem 2004, 47, 2550–2560. [DOI] [PubMed] [Google Scholar]
- 97.Letendre SL; Mills A; Hagins D; Swindells S; Felizarta F; Devente J; Bettacchi C; Lou Y; Ford S; Sutton K; Shaik JS; Crauwels H; D’Amico R; Patel P Pharmacokinetics and antiviral activity of cabotegravir and rilpivirine in cerebrospinal fluid following long-acting injectable administration in hiv-infected adults. J Antimicrob Chemother 2020, 75, 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Janssen Research and Development. Nonclinical introduction https://www.pmda.go.jp/drugs/2022/P20220610001/800155000_30400AMX00197_H100_1.pdf (accessed Mar 29, 2023).
- 99.Endsley JJ; Huante MB; Naqvi KF; Gelman BB; Endsley MA Advancing our understanding of hiv co-infections and neurological disease using the humanized mouse. Retrovirology 2021, 18, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wei X; Decker JM; Liu H; Zhang Z; Arani RB; Kilby JM; Saag MS; Wu X; Shaw GM; Kappes JC Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (t-20) monotherapy. Antimicrob Agents Chemother 2002, 46, 1896–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Popovic M; Read-Connole E; Gallo RC T4 positive human neoplastic cell lines susceptible to and permissive for htlv-iii. Lancet 1984, 2, 1472–1473. [DOI] [PubMed] [Google Scholar]
- 102.Popovic M; Sarngadharan MG; Read E; Gallo RC Detection, isolation, and continuous production of cytopathic retroviruses (htlv-iii) from patients with aids and pre-aids. Science 1984, 224, 497–500. [DOI] [PubMed] [Google Scholar]
- 103.Ratner L; Haseltine W; Patarca R; Livak KJ; Starcich B; Josephs SF; Doran ER; Rafalski JA; Whitehorn EA; Baumeister K; et al. Complete nucleotide sequence of the aids virus, htlv-iii. Nature 1985, 313, 277–284. [DOI] [PubMed] [Google Scholar]
- 104.Nunberg JH; Schleif WA; Boots EJ; O’Brien JA; Quintero JC; Hoffman JM; Emini EA; Goldman ME Viral resistance to human immunodeficiency virus type 1-specific pyridinone reverse transcriptase inhibitors. J Virol 1991, 65, 4887–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Balamane M; Varghese V; Melikian GL; Fessel WJ; Katzenstein DA; Shafer RW Panel of prototypical recombinant infectious molecular clones resistant to nevirapine, efavirenz, etravirine, and rilpivirine. Antimicrob Agents Chemother 2012, 56, 4522–4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu XH Noncompartmental pharmacokinetics analysis https://dash.gallery/dash-pk-calc/ (accessed Mar 29, 2023).
- 107.Romeys B Microscopic technique Foreign Literature Publishing House: Moscow, 1953. [Google Scholar]
- 108.van der Maaten L; Hinton G Visualizing data using t-SNE. J Machine Learning Research 2008, 9, 2579–2605. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


