ABSTRACT
To adapt and proliferate, bacteria must sense and respond to the ever-changing environment. Transmembrane transcription regulators (TTRs) are a family of one-component transcription regulators that respond to extracellular information and influence gene expression from the cytoplasmic membrane. How TTRs function to modulate expression of their target genes while localized to the cytoplasmic membrane remains poorly understood. In part, this is due to a lack of knowledge regarding the prevalence of TTRs among prokaryotes. Here, we show that TTRs are highly diverse and prevalent throughout bacteria and archaea. Our work demonstrates that TTRs are more common than previously appreciated and are enriched within specific bacterial and archaeal phyla and that many TTRs have unique transmembrane region properties that can facilitate association with detergent-resistant membranes.
IMPORTANCE One-component signal transduction systems are the major class of signal transduction systems among bacteria and are commonly cytoplasmic. TTRs are a group of unique one-component signal transduction systems that influence transcription from the cytoplasmic membrane. TTRs have been implicated in a wide array of biological pathways critical for both pathogens and human commensal organisms but were considered to be rare. Here, we demonstrate that TTRs are in fact highly diverse and broadly distributed in bacteria and archaea. Our findings suggest that transcription factors can access the chromosome and influence transcription from the membrane in both archaea and bacteria. This study challenges thus the commonly held notion that signal transduction systems require a cytoplasmic transcription factor and highlights the importance of the cytoplasmic membrane in directly influencing signal transduction.
KEYWORDS: cell membranes, signal transduction, transcription factors
INTRODUCTION
Signal transduction is the process through which microorganisms regulate their cellular programs according to their extracellular environment. Microorganisms are known to transduce information from outside the cell to the cytoplasm via two-component and one-component signal transduction systems (1, 2). Two-component signal transduction cascades are typically composed of a membrane localized sensor histidine kinase that, when stimulated, transfers a phosphate to a soluble response regulator, resulting in a cellular response (1–3). In contrast, one-component signal transduction systems are composed of a single protein that both directly detects a stimulus and modulates a cellular response (1–3). The vast majority of signal transduction systems in bacteria are one-component systems with most harboring DNA-binding domains and thereby controlling gene expression (2). A majority of one-component regulators are predicted to be localized within the cytoplasm, presumably to have unimpeded access to their DNA target(s) (2). Nonetheless, there are known examples of one-component regulators that are localized to the cytoplasmic membrane (Table 1). Here, we define these one-component regulators and those with similar features (i.e., the presence of a predicted transmembrane [TM] region and a predicted DNA-binding domain) as TM transcription regulators (TTRs). Functional TTRs are found in bacteria and archaea, but not in eukaryotes, due to the separation of the cytoplasmic membrane and their genomes by the nucleus (2, 4, 5). Within archaea, TTRs are known to regulate motility and pilin gene expression in response to dangerous temperatures and nutrient-limiting conditions (6, 7). TTRs are better studied in bacteria, where they have been shown to regulate bile salt resistance (8, 9), toxin production (10, 11), antibiotic resistance (12, 13), acid resistance (14–16), natural competence (17), pilin/fimbria expression (18–24), type 3 secretion systems (25, 26), biofilm formation (27, 28), and metabolism (29–31), and have been implicated in modulation of the human immune system (32) (see Table 1 for additional details).
TABLE 1.
Characterized TTRs, along with their known cellular responses and associated proteins
| TTR | Organism(s) | Cellular response(s) | Associated protein | Reference(s) |
|---|---|---|---|---|
| ToxR | Vibrio spp., Photobacterium spp. | Bile salt resistance, cationic antimicrobial peptides, pressure response, biofilm formation, and virulence factor expression | ToxS | 8, 9, 11, 19, 27, 77–88 |
| TcpP | Vibrio cholerae and Vibrio fischeri | Virulence factor (toxT expression), motility, chemotaxis, and reduction of extracellular polysaccharides | TcpH | 18, 89–92 |
| CadC | Vibrio spp., Escherichia spp., Salmonella spp., Yersinia spp. | Acid resistance | LysP | 14, 93–102 |
| TfoS | Vibrio spp. | Natural competence | Na | 17, 103 |
| VtrA/VttrA | Vibrio spp. | Type 3 secretion systems | VtrC | 25, 26, 72, 104, 105 |
| VtrB/VttrB | Vibrio spp., Salmonella spp. | Type 3 secretion systems | Na | 25, 26, 106 |
| MarT | Salmonella spp., Yersinia ruckeri | Fibronectin binding | –a | 107 – 110 |
| GvrA | Escherichia coli | Promotes expression of LEE in response to bicarbonate | Na | 111, 112 |
| YqeI | Escherichia coli | Serum resistance, flagellum synthesis, and host cell adhesion | YqeJ | 113 |
| PsaE | Yersinia pestis | Fimbria expression | PsaF | 20 – 22 |
| MyfE | Yersinia enterocolitica | Fimbria expression | MyfF | 23, 114, 115 |
| PypB | Yersinia enterocolitica and Yersinia ruckeri | Flp type IVb pilin expression | –a | 116 |
| BcrR | Enterococcus spp., Lactobacillus spp. | Bacitracin resistance | Na | 12, 13, 117, 118 |
| BreG | Lactobacillus spp., Enterococcus spp. | Bacteriocin synthesis | Na | 119, 120 |
| AguR | Enterococcus spp. | Acid tolerance | Na | 121 – 124 |
| LP_2991 | Enterococcus spp., Lactobacillus spp. | Immune modulation | Na | 32, 125 |
| HcrR | Lactobacillus plantarum | Hydroxycinnamic acid metabolism | Na | 29, 126 |
| MmsR | Lactobacillus bifermentans | Isobutyryl-CoA metabolism | Na | 30 |
| MtbS | Staphylococcus spp., Enterococcus spp., Lactobacillus spp. | Virulence factors, phosphate transport, tRNAs, etc. | Na | 127 |
| NanR | Staphylococcus spp. | Sialic acid metabolism | Na | 31 |
| WmpR | Pseudomonas tunicata | Type IV pilin, pigmentation, iron uptake, amino acid metabolism, biofilm formation, and antifouling | Na | 28, 128 |
| ArnR | Sulfolobus acidocaldarius | Motility and pilin expression | Na | 6, 7 |
| Rsp | Neisseria gonorrhoeae | Pilin expression | Na | 34 |
–, There are possible TcpH/ToxS-like genes that are uncharacterized immediately upstream or downstream of the indicated TTR.
Localization to the cytoplasmic membrane has been shown to be critical for some TTRs to influence expression of their target genes (33). TTRs are counterintuitive as their subcellular localization reduces their diffusion and thereby their ability to bind target promoter(s). However, there is a possibility that TTRs evolved from two-component systems. It is known that in Pseudomonas aeruginosa, a two-component system consisting of PilS, the membrane localized histidine kinase, and PilR, the response regulator, regulates activity of RpoN (24). The Neisseria gonorrhoeae genome encodes a chimeric protein called Rsp, which includes the membrane localized receptor of PilS at its N terminus and the PilR DNA-binding domain at its C termini and represses pilA expression (34). Bacteroides thetaiotaomicron, a constituent of the human microbiota, contains 32 hybrid histidine kinases, each with a DNA-binding domain within its genome (35). To gain a deeper understanding of TTRs, we performed a large-scale genomic analysis of TTRs across archaeal and bacterial species.
RESULTS
Transmembrane transcription regulators are prevalent in bacteria and archaea and display lineage-specific expansion.
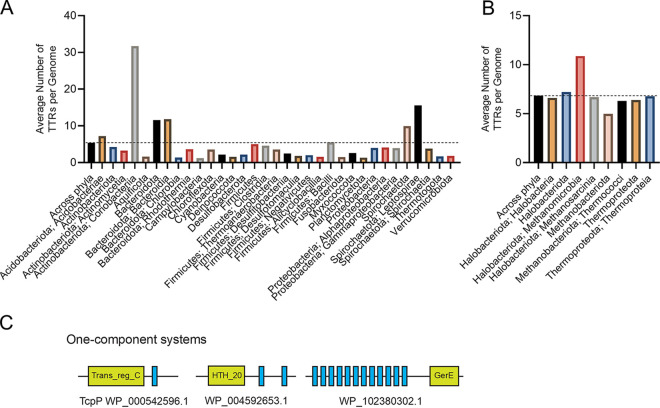
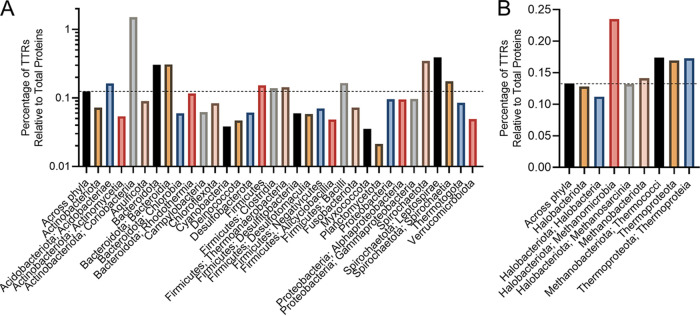
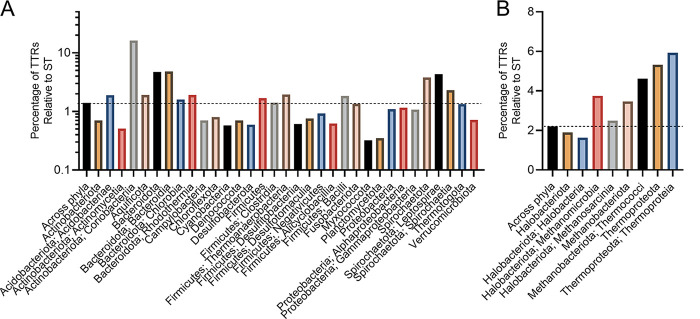
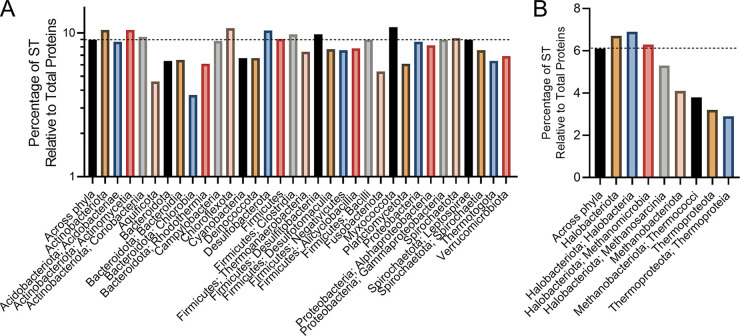
To gain a deeper understanding of the prevalence and distribution of TTRs in prokaryotes, we mined the genomes of 10,933 bacterial and 404 archaeal species for genes that encoded (i) a DNA-binding domain and (ii) at least one transmembrane region. We found that 9,306 bacterial and 367 archaeal species encoded at least one TTR (see File S1 in the supplemental material). In total, we identified 50,302 TTRs (48,918 bacterial and 1,384 archaeal) across 9,673 genomes (see File S1). On average, bacterial genomes contain 5 TTRs (±7) and archaeal genomes contain 4 TTRs (±2) containing one or more TM regions (Fig. 1; see also File S1). Species within the Coriobacteriia, Bacteroidota, Bacteroidia, Spirochaetota, Leptospirae, Acidobacteriae, and Methanomicrobia (here and throughout the manuscript: bacterial and archaeal taxonomy is according to Genome Taxonomy Database [36]), on average, contain more TTRs per genome compared to other prokaryotes (Fig. 1A and B; see also File S1). However, the number of TTRs per genome varies dramatically among phyla, with some species encoding only 1 TTR and others encoding up to 158 TTRs (e.g., Raoultibacter timonensis) (see File S1). Given that the genome size also varies dramatically between species (see File S1), we next explored whether the number of TTRs per genome was due to differences in genome size. To address this question, we normalized the number of TTRs to the total number of protein-coding sequences per genome. Across all species, we found that on average 0.15% and 0.13% of bacterial and archaeal coding sequences were composed of TTRs (Fig. 2). This normalization still indicated that species within the Coriobacteriia, Bacteroidota, Bacteroidia, Spirochaetota, Leptospirae, and Methanomicrobia were enriched with TTRs in their genomes. In addition, the Acidobacteriae, Clostridia, Thermoanaerobacteria, Bacilli, Methanobacteriota, and Thermoproteota have a slightly higher abundances of TTRs compared to other bacteria and archaea (Fig. 2). Since TTRs are a distinct type of signal transduction system, we next analyzed what portion of known signal transduction systems within both the bacterial and the archaeal domains were composed of TTRs. Across all species, we found that, on average, 1.5 to 2% of all signal transduction genes were TTRs across bacterial and archaeal species, respectively (Fig. 3). However, these data demonstrate that a high percentage of signal transduction genes are comprised of TTRs within the Coriobacteriia, Bacteroidota, Bacteroidia, Spirochaetota, Leptospirae, Methanomicrobia, Methanobacteriota, Thermococci, Thermoproteota, and Thermoproteia (Fig. 3). In addition, the Acidobacteriae, Aquificota, Chlorobia, Rhodothermia, Firmicutes, Thermoanaerobacteria, Bacilli, and Methanosarcina have a slightly higher percentages of TTRs relative to signal transduction genes (Fig. 3). Given these data, we considered the possibility that the overall abundance of signal transduction genes within bacterial and archaeal phyla and classes could skew our results. To test this, we compared the total numbers of signal transduction genes to the total numbers of protein-coding genes within each bacterial and archaeal species; these values were, on average, 9% for bacterial species and 6% for archaeal species (Fig. 4). Interestingly, we did not observe an increased ratio of signal transduction genes to total protein-coding sequences within the Coriobacteriia, Bacteroidota, Bacteroidia, Spirochaetota, Leptospirae, Methanomicrobia, Methanobacteriota, Thermococci, Thermoproteota, and Thermoproteia (Fig. 4). Taken together, these data indicate that the aforementioned bacterial and archaeal clades are indeed enriched with TTRs.
FIG 1.
(A and B) Abundance of TTRs among bacterial (A) and archaeal (B) genomes. The dashed line indicates the average number of TTRs per genome across all phyla. Bacterial and archaeal phylum are listed below their respective groups, with the class following a semicolon. To view the supporting data, see File S7 in the supplemental material. (C) Typical membrane topology and domain composition of one-component TTRs identified in this study.
FIG 2.
(A and B) Ratio of TTRs to all protein-coding genes among bacterial (A) and archaeal (B) genomes. The dashed line indicates the average ratio of TTRs to protein-coding genes across all phyla. Bacterial and archaeal phyla are listed below their respective groups, with the class following a semicolon. To view the supporting data, see File S7 in the supplemental material.
FIG 3.
(A and B) Abundance of TTRs relative to signal transduction genes among bacterial (A) and archaeal (B) genomes. The dashed line indicates the average ratio of TTRs to signal transduction genes across all phyla. Bacterial and archaeal phyla are listed below their respective groups, with class following a semicolon. To view the supporting data, see File S7 in the supplemental material.
FIG 4.
(A and B) Ratio of non-TTR signal transduction genes to total protein-coding genes among bacterial (A) and archaeal (B) genomes. The dashed line indicates the average ratio of signal transduction genes to protein-coding genes across all phyla. Bacterial and archaeal phylum are listed below their respective groups, with the class following a semicolon. To view the supporting data, see File S7 in the supplemental material.
Transmembrane transcription regulators employ a few common DNA-binding domains.
Over a hundred different DNA-binding domains were found across the identified TTRs and only a small number (19 DNA-binding domains) were not found among TTRs (see Table S1A and B in the supplemental material). However, DNA-binding domains of approximately 91% of all TTRs are represented by 11 most common domains, with the helix-turn-helix DNA-binding domain being the most predominant across all TTRs (Fig. 5A; see also Table S1A). A similar distribution of DNA-binding domains has also been reported for other one-component systems (2).
FIG 5.
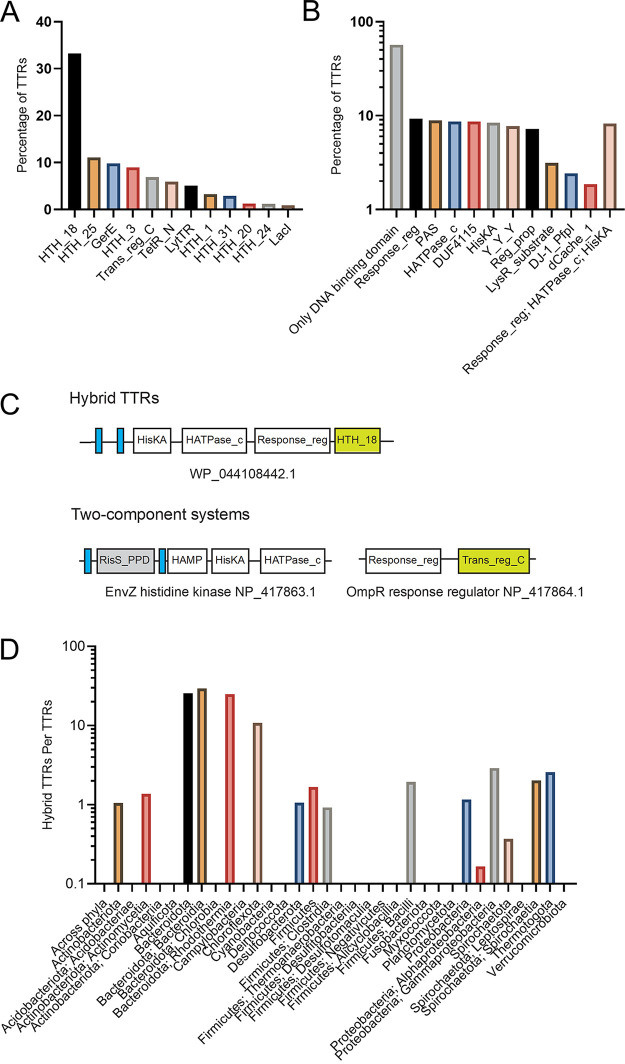
Domain composition of TTRs. (A) Most common DNA-binding domains among bacterial and archaeal TTRs. (B) Most abundant non-DNA-binding domains among bacterial and archaeal TTRs. (C) Typical membrane topology and domain composition of hybrid TTRs and two-component systems. (D) Distribution and ratio of hybrid TTR to TTR across bacterial phyla.
Hybrid TTRs are unique to bacteria.
Of the domains identified within TTRs, the most common non-DNA-binding domain is the response regulator domain (found in 9.2% of TTRs), which is a part of two-component signal transduction systems (Fig. 5B; see also Table S2) (37, 38). Response regulators catalyze the transfer of a phosphate from a histidine kinase donor and have intrinsic dephosphorylation activity (39). Response regulators are multidomain proteins typically containing a N-terminal receiver domain and a C-terminal effector domain that is commonly a DNA-binding domain (39). Phosphorylation of the receiver domain stabilizes a conformation that allows for activity of the effector domain (39). In addition, among the top five most common non-DNA-binding domains in TTRs are the HATPase_c (a histidine kinase catalytic domain [found in 8.6% of TTRs]), HisKA (a histidine kinase dimerization domain [found in 8.4% of TTRs]), and the Y_Y_Y domains (an extracellular domain found in two-component systems [found in 7.7% of TTRs]) (Fig. 5B; see also Table S2). These domains are typical for two-component signal transduction pathways (37, 38, 40, 41). In fact, approximately 8.2% of all TTRs identified by our analysis contain one or more core domain of two-component systems (response regulator, HATPase_c, and HisKA domains), which we refer to as hybrid TTRs (Fig. 5B and C; see also Table S2). Hybrid TTRs appear to be a bacterial phenomenon, since we found no archaeal TTRs containing a response_reg domain and very few containing a HisKA domain (17 TTRs) or HATPase_c domain (1 TTR).
Bacteroides thetaiotaomicron was reported to contain 32 hybrid histidine kinases with DNA-binding domains (i.e., hybrid TTRs) (35). Our data indicate that a majority of the TTRs within the Bacteroides genus are hybrid TTRs (approximately 72%) (see File S3). Furthermore, in addition to the Bacteroides genus, many other species within Bacteroidota, Bacteroidia, Rhodothermia, and Chloroflexota contain a high fraction of hybrid TTRs (Fig. 5D). Our data indicate that hybrid TTRs may have evolved from canonical two-component regulatory systems, similar to hybrid two-component regulatory systems, that evolved from canonical two-component systems (42). Our data suggest that hybrid TTRs are the product of recent evolutionary events since they are not conserved at the genus, class, or phylum level.
Single pass transmembrane regulators are the most prevalent TTRs.
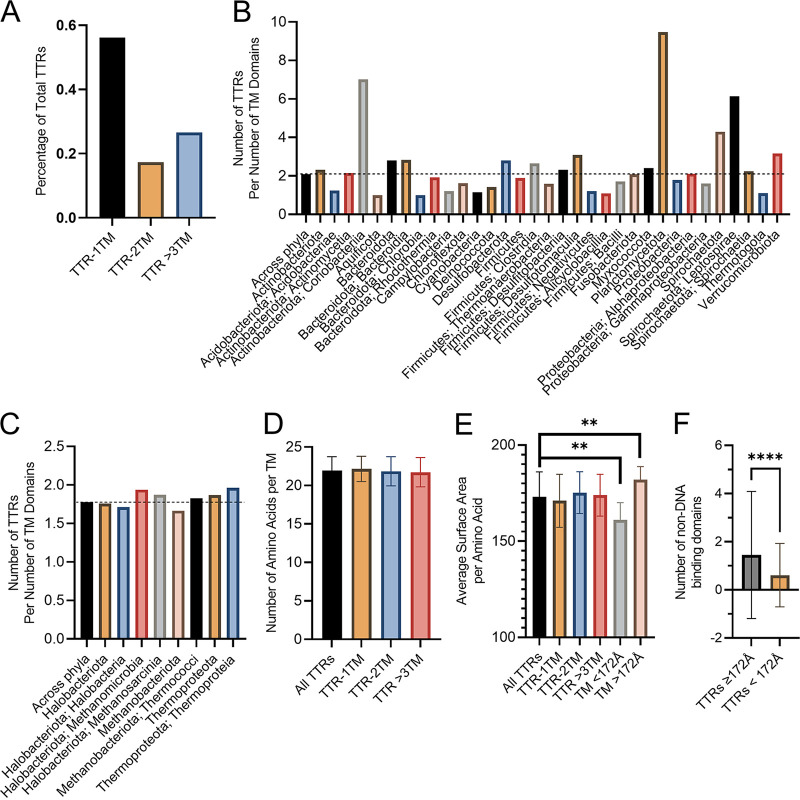
We found that approximately 56% of TTRs are single-pass transmembrane (TM) proteins, while TTRs containing two or more TM regions comprise 17 and 27% of TTRs, respectively (Fig. 6A; see also Files S4 to S6). We next questioned whether TM content of TTRs was homogenous across bacterial and archaeal phyla. Since some TTRs contain up to 22 TM regions, the average number of TM regions per TTRs, approximately 2 TM regions, is elevated within bacterial and archaeal phyla despite that the majority of TTRs are single-pass TM proteins (Fig. 6B and C; see also Files S4 to S6). The average number of TM regions per TTRs is consistent across archaeal phyla and classes while there is greater diversity among bacterial phyla. In bacteria, Coriobacteriia, Planctomycetota, Leptospirae, and Verrucomicrobiota contain TTRs with three or more TM domains on average (Fig. 6B and C).
FIG 6.
TM region analysis of TTRs. (A) Number of TM regions across all bacterial and archaeal TTRs. (B) Average number of TM regions per TTR within the bacterial phyla. (C) Average number of TM regions per TTR within the archaeal phyla. (D) Average TM region length for TTRs across all bacterial and archaeal species. (E) Average surface area of TTR TM region among bacterial and archaeal species. (F) Average number of non-DNA-binding domains per TTR based on the average surface area of the TM region. Error bars represent the standard deviations. **, P < 0.01; ****, P < 0.0001. A two-tailed unpaired Student t test was used to determine statistical significance.
TTRs have a propensity to associate with liquid-ordered membrane domains.
We hypothesized that TTRs may respond to their local membrane environment which can be influenced by extracellular conditions. A common phenomenon across bacteria and eukaryotes is the formation of small (i.e., nanometer) lipid regions, referred to here as lipid rafts (43–45). Lipid rafts and non-lipid-raft membrane regions have been shown to influence various cellular processes, including signal transduction, in part due to their ability to promote interaction between membrane localized proteins (46–55). Association with lipid raft and non-lipid-raft membrane regions is determined by the properties of the transmembrane region with length and overall surface area of the transmembrane helix being the most critical factors (56). Given that lipid rafts were found in bacteria, we asked a question whether TTR TM regions supported their association with lipid raft or non-lipid-raft membrane regions. Since the overall length and surface area of transmembrane helices controls the association of membrane proteins within lipid raft membrane regions, we calculated the overall length and surface area of all predicted transmembrane regions for TTRs analyzed here (see Files S4 to S6). Overall, we found that the TM region length and surface area remain similar across single TM or multi-TM region TTRs (Fig. 6D and E; see also Files S4 to S6). We found that a substantial number of TTR TM regions (approximately 43%) have a surface area equal to or below 172 Å per amino acid (Fig. 6E; see also Files S4 to S6). Prior studies have demonstrated that TcpP, a TTR that positively modulates virulence in Vibrio cholerae, increases its association with detergent-resistant membranes (i.e., liquid-ordered membrane domains) in the presence of ɑ-linolenic acid, a dietary fatty acid (57). The surface area of the TcpP transmembrane region is 172 Å per amino acid. These data suggest that many TTRs might have the capacity to associate with liquid-ordered membrane domains. Furthermore, this suggests that TTRs might be directly modulated by the cytoplasmic membrane. We reasoned that if the cytoplasmic membrane is a major signal for TTRs with low TM surface area that they would also contain fewer sensory domains (i.e., non-DNA-binding domains). Indeed, we found that TTRs with a surface area equal to or below 172 Å per amino acid contained fewer sensory domains compared to TTRs with a higher TM surface area (Fig. 6F).
DISCUSSION
Our analysis revealed that TTRs are widespread in bacteria and archaea and in some genomes, for example Raoultibacter timonensis, they are a major type of transcription regulators (41% of all DNA-binding transcription factors). We found that TTRs are abundant in species from the gut microbiota. For example, species within the Eggerthella genus (the Coriobacteriales order), contain a high number of TTRs per genome (~117 TTRs) (see File S1 in the supplemental material). The Eggerthellaceae family are common members of mammalian gastrointestinal tracts (58–62). Members of the Bacteroidia, commonly associated within the human gastrointestinal tract, were also found to contain a high number of TTRs and hybrid TTRs within their genomes (Fig. 1–4). There is evidence that hybrid TTRs within Bacteroides genus function to sense and respond to disaccharides (63). It remains to be seen whether TTRs within the Coriobacteriales and Bacteroidota contribute to their ability to colonize mammalian gastrointestinal environments, but our data have revealed this correlation.
The advantage of TTRs versus classical one- and two-component systems is not obvious: why bring the response regulator and DNA-binding domain to the cytoplasmic membrane? One possibility is that the membrane itself serves as a signal to further fine tune these signal transduction pathways. It is generally recognized that the membrane environment is not a homogenous. For example, a vast majority of integral membrane proteins are heterogeneously distributed in Bacillus subtilis cells, indicating that their diffusion within the cytoplasmic membrane is limited (64, 65).
Both bacterial and eukaryotic cells support the formation of small-lipid-raft membrane regions (43–45). Generally speaking, lipid raft and non-lipid-raft membrane regions differ by their overall fluidity and thickness, with lipid rafts having a lower fluidity and increased thickness, as a consequence of the phospholipid species that occupy these membrane environments (45, 46, 66–69). These membrane regions have been shown to influence many signaling pathways in eukaryotic cells, in particular T cells (46–55). There is also evidence that dietary polyunsaturated fatty acids can influence formation and stability of lipid ordered membrane domains thereby influencing signal transduction (48, 49). Here, we demonstrate that a large fraction of TTRs (approximately 43%) can associate with lipid raft membrane regions. These data support our hypothesis that the membrane environment may serve to influence TTR function; however, additional studies are required to further test this hypothesis.
In addition to the potential influence of the cytoplasmic membrane, it is also feasible that localization of TTRs to the cytoplasmic membrane facilitates tighter regulatory controls on TTR regulons. That is, not only do TTRs require a signal to impact gene expression, but that the target promoter sequence(s) must be in close proximity to the cytoplasmic membrane. This would imply that the structure of the chromosome (which is influenced by nucleoid associated proteins, transcription, translation, DNA supercoiling, and condensins) also contributes to TTR gene regulation. This would indicate that TTRs could contribute to regulation of complex lifestyles among microorganisms. Within the Alphaproteobacteria, a family of LytTR response regulators, not exclusively TTRs, were identified (70). Curiously, within Caulobacter cresentus all instances of the LytTR response regulator domain contain one or more TM regions (70). This could explain the high degree of variation of TTRs across the bacterial and archaeal domains.
Some TTRs (such as ToxR, VtrA, VtrrA, PsaE, MyfE, TcpP, and YqeI) have an associated protein that contributes to TTR mediated signal transduction by inhibiting proteolysis of the TTR or by stimulating heterodimer formation (Table 1). These TTRs are also referred to as cocomponent systems (71). Curiously, many of these associated proteins contain a lipocalin-like domain, which has been shown to be important for VtrC binding to bile, but generally are known to bind to small hydrophobic molecules (71, 72). Rather than responding to the cytoplasmic membrane itself, localization of TTRs to the cytoplasmic membrane may facilitate function of cocomponent systems.
Our data suggest that TTRs play a much larger role in signal transduction than previously thought, implying that transcription initiation also occurs at the membrane-DNA interface across bacterial and archaeal species. However, compared to the number of TTRs identified by our analysis the number of experimentally validated TTRs is extremely low indicating that a large fraction of TTRs function remains to be understood (Table 1). In support of this, a substantial fraction of TTRs identified here (~57%) contains only a DNA-binding domain and no additional recognizable domains of known function (Fig. 5B). Thus, this work provides new insights into the prevalence and distribution of TTRs within bacteria and archaea reveals unique features of these regulators.
MATERIALS AND METHODS
Identification and transmembrane domain analysis of TTRs within the MiST database.
TTRs for a representative set of genomes were collected from the MiST database by running a custom Python script on the local computer cluster (73). The script, which is available in the GitHub repository (https://github.com/bioliners/TTRs), sends requests through the database API and processes the results. For each genome, all DNA-binding signal transduction proteins that contain transmembrane regions were retrieved (see File S2 in the supplemental material). The Pfam Profile hidden Markov models of DNA-binding domains listed on the help page of the MiST database were used to identify DNA-binding domains in the proteins. All DNA-binding domains identified in TTRs and those that were checked but not identified are listed in Table S1. TM regions of the protein sequences were identified by running TMHMM, domains were verified using TREND and Pfam profile hidden Markov models (74–76). Sequences corresponding to transmembrane regions were extracted using a custom Python script. The average length, number of amino acids, and surface area for each TTR transmembrane region were calculated from data in Files S4 to S6 using a Python script. Taxonomy information for the genomes was retrieved from GTDB database (36; see File S1 for data). All used scripts are available in a GitHub repository (https://github.com/bioliners/TTRs).
Calculating statistical parameters of TTRs.
The total number of all signal transduction (ST) proteins encoded in each analyzed genome was obtained from MiST database (73). The genome size and the number of all encoded proteins were obtained from the metadata tables of GTDB (36; see Files S1 and S7 in the supplemental material). These data were cross-referenced with the number of encoded TTRs in each genome using Excel. For statistical analysis, we considered phyla or classes that contained at least 10 sequenced genomes. By running Shapiro-Wilk test and exploring quantile-quantile plots, we have established that TTR counts are not normally distributed. Therefore, we used two nonparametric tests, Spearman and Kendall correlation tests, to explore correlations. Prior to performing correlation tests, we subtracted TTR counts from the overall counts of ST proteins and ST protein counts from the sum of all proteins encoded in each genome. This was done to remove the contribution of correlation of TTR and ST proteins with themselves. Statistical analysis was carried out in the R environment.
Data availability.
The data presented here are available from the corresponding authors upon request.
ACKNOWLEDGMENTS
This study was supported by the National Institutes of Health grant R35GM131760 to I.B.Z. and a Rudolph Hugh Endowment at Michigan State University (V.J.D.). L.M.D. was a trainee of the Institutional Pharmacological Sciences Training Program at MSU (2T32 GM092715).
Study design—L.M.D., V.M.G., I.B.Z., and V.J.D.; bioinformatics—L.M.D., V.M.G., and J.X.; drafting of the manuscript—L.M.D. and V.M.G.; and critical revision of the manuscript—V.M.G., L.M.D., I.B.Z., and V.J.D.
We declare there are no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Igor B. Zhulin, Email: jouline.1@osu.edu.
Victor J. DiRita, Email: diritavi@msu.edu.
Beile Gao, South China Sea Institute of Oceanology.
REFERENCES
- 1.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu Rev Biochem 69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 2.Ulrich LE, Koonin EV, Zhulin IB. 2005. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol 13:52–56. doi: 10.1016/j.tim.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schaller GE, Shiu SH, Armitage JP. 2011. Two-component systems and their co-option for eukaryotic signal transduction. Curr Biol 21:R320–R330. doi: 10.1016/j.cub.2011.02.045. [DOI] [PubMed] [Google Scholar]
- 4.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. 2003. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA 100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeBose-Boyd RA, Brown MS, Li WP, Nohturfft A, Goldstein JL, Espenshade PJ. 1999. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell 99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- 6.Bischof LF, Haurat MF, Albers S-V. 2019. Two membrane-bound transcription factors regulate expression of various type IV pilus surface structures in Sulfolobus acidocaldarius. PeerJ 7:e6459. doi: 10.7717/peerj.6459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lassak K, Peeters E, Wróbel S, Albers S-V. 2013. The one-component system ArnR: a membrane-bound activator of the crenarchaeal archaellum. Mol Microbiol 88:125–139. doi: 10.1111/mmi.12173. [DOI] [PubMed] [Google Scholar]
- 8.Ante VM, Bina XR, Howard MF, Sayeed S, Taylor DL, Bina JE. 2015. Vibrio cholerae leuO transcription is positively regulated by ToxR and contributes to bile resistance. J Bacteriol 197:3499–3510. doi: 10.1128/JB.00419-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Provenzano D, Schuhmacher DA, Barker JL, Klose KE. 2000. The virulence regulatory protein ToxR mediates enhanced bile resistance in Vibrio cholerae and other pathogenic Vibrio species. Infect Immun 68:1491–1497. doi: 10.1128/IAI.68.3.1491-1497.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller KA, Hamilton E, Dziejman M. 2012. The Vibrio cholerae trh gene is coordinately regulated in vitro with type III secretion system genes by VttRA/VttRB but does not contribute to Caco2-BBE cell cytotoxicity. Infect Immun 80:4444–4455. doi: 10.1128/IAI.00832-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung DT, Mekalanos JJ. 2005. Bile acids induce cholera toxin expression in Vibrio cholerae in a ToxT-independent manner. Proc Natl Acad Sci USA 102:3028–3033. doi: 10.1073/pnas.0409559102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manson JM, Keis S, Smith JM, Cook GM. 2004. Acquired bacitracin resistance in Enterococcus faecalis is mediated by an ABC transporter and a novel regulatory protein, BcrR. Antimicrob Agents Chemother 48:3743–3748. doi: 10.1128/AAC.48.10.3743-3748.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gauntlett JC, Gebhard S, Keis S, Manson JM, Pos KM, Cook GM. 2008. Molecular analysis of BcrR, a membrane-bound bacitracin sensor and DNA-binding protein from Enterococcus faecalis. J Biol Chem 283:8591–8600. doi: 10.1074/jbc.M709503200. [DOI] [PubMed] [Google Scholar]
- 14.Kuper C, Jung K. 2005. CadC-mediated activation of the cadBA promoter in Escherichia coli. J Mol Microbiol Biotechnol 10:26–39. doi: 10.1159/000090346. [DOI] [PubMed] [Google Scholar]
- 15.Gu D, Wang K, Lu T, Li L, Jiao X. 2021. Vibrio parahaemolyticus CadC regulates acid tolerance response to enhance bacterial motility and cytotoxicity. J Fish Dis 44:1155–1168. doi: 10.1111/jfd.13376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rhee JE, Kim K-S, Choi SH. 2008. Activation of the Vibrio vulnificus cadBA operon by leucine-responsive regulatory protein is mediated by CadC. J Microbiol Biotechnol 18:1755–1761. doi: 10.4014/jmb.0800.121. [DOI] [PubMed] [Google Scholar]
- 17.Dalia AB, Lazinski DW, Camilli A. 2014. Identification of a membrane-bound transcriptional regulator that links chitin and natural competence in Vibrio cholerae. mBio 5:e01028-13. doi: 10.1128/mBio.01028-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Häse CC, Mekalanos JJ. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc Natl Acad Sci USA 95:730–734. doi: 10.1073/pnas.95.2.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goss TJ, Morgan SJ, French EL, Krukonis ES. 2013. ToxR recognizes a direct repeat element in the toxT, ompU, ompT, and ctxA promoters of Vibrio cholerae to regulate transcription. Infect Immun 81:884–895. doi: 10.1128/IAI.00889-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quinn JD, Weening EH, Miner TA, Miller VL. 2019. Temperature control of psaA expression by PsaE and PsaF in Yersinia pestis. J Bacteriol 201:e00217-19. doi: 10.1128/JB.00217-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Quinn JD, Weening EH, Miller VL. 2021. PsaF is a membrane-localized pH sensor that regulates psaA expression in Yersinia pestis. J Bacteriol 203:e00165-21. doi: 10.1128/JB.00165-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Y, Isberg RR. 1997. Transcriptional regulation of the Yersinia pseudotuberculosis pH6 antigen adhesin by two envelope-associated components. Mol Microbiol 24:499–510. doi: 10.1046/j.1365-2958.1997.3511719.x. [DOI] [PubMed] [Google Scholar]
- 23.Iriarte M, Vanooteghem JC, Delor I, Díaz R, Knutton S, Cornelis GR. 1993. The Myf fibrillae of Yersinia enterocolitica. Mol Microbiol 9:507–520. doi: 10.1111/j.1365-2958.1993.tb01712.x. [DOI] [PubMed] [Google Scholar]
- 24.Hobbs M, Collie ES, Free PD, Livingston SP, Mattick JS. 1993. PilS and PilR, a two-component transcriptional regulatory system controlling expression of type 4 fimbriae in Pseudomonas aeruginosa. Mol Microbiol 7:669–682. doi: 10.1111/j.1365-2958.1993.tb01158.x. [DOI] [PubMed] [Google Scholar]
- 25.Kodama T, Gotoh K, Hiyoshi H, Morita M, Izutsu K, Akeda Y, Park K-S, Cantarelli VV, Dryselius R, Iida T, Honda T. 2010. Two regulators of Vibrio parahaemolyticus play important roles in enterotoxicity by controlling the expression of genes in the Vp-PAI region. PLoS One 5:e8678. doi: 10.1371/journal.pone.0008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alam A, Tam V, Hamilton E, Dziejman M. 2010. vttRA and vttRB encode ToxR family proteins that mediate bile-induced expression of type three secretion system genes in a non-O1/non-O139 Vibrio cholerae strain. Infect Immun 78:2554–2570. doi: 10.1128/IAI.01073-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen L, Qiu Y, Tang H, Hu LF, Yang WH, Zhu XJ, Huang XX, Wang T, Zhang YQ. 2018. ToxR is required for biofilm formation and motility of Vibrio parahaemolyticus. Biomed Environ Sci 31:848–850. doi: 10.3967/bes2018.112. [DOI] [PubMed] [Google Scholar]
- 28.Stelzer S, Egan S, Larsen MR, Bartlett DH, Kjelleberg S. 2006. Unravelling the role of the ToxR-like transcriptional regulator WmpR in the marine antifouling bacterium Pseudoalteromonas tunicata. Microbiology (Reading) 152:1385–1394. doi: 10.1099/mic.0.28740-0. [DOI] [PubMed] [Google Scholar]
- 29.Santamaría L, Reverón I, López de Felipe F, de Las Rivas B, Muñoz R. 2018. Unravelling the reduction pathway as an alternative metabolic route to hydroxycinnamate decarboxylation in Lactobacillus plantarum. Appl Environ Microbiol 84:e01123-18. doi: 10.1128/AEM.01123-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steele MI, Lorenz D, Hatter K, Park A, Sokatch JR. 1992. Characterization of the mmsAB operon of Pseudomonas aeruginosa PAO encoding methylmalonate-semialdehyde dehydrogenase and 3-hydroxyisobutyrate dehydrogenase. J Biol Chem 267:13585–13592. doi: 10.1016/S0021-9258(18)42252-1. [DOI] [PubMed] [Google Scholar]
- 31.Olson ME, King JM, Yahr TL, Horswill AR. 2013. Sialic acid catabolism in Staphylococcus aureus. J Bacteriol 195:1779–1788. doi: 10.1128/JB.02294-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meijerink M, van Hemert S, Taverne N, Wels M, de Vos P, Bron PA, Savelkoul HF, van Bilsen J, Kleerebezem M, Wells JM. 2010. Identification of genetic loci in Lactobacillus plantarum that modulate the immune response of dendritic cells using comparative genome hybridization. PLoS One 5:e10632. doi: 10.1371/journal.pone.0010632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crawford JA, Krukonis ES, DiRita VJ. 2003. Membrane localization of the ToxR winged-helix domain is required for TcpP-mediated virulence gene activation in Vibrio cholerae. Mol Microbiol 47:1459–1473. doi: 10.1046/j.1365-2958.2003.03398.x. [DOI] [PubMed] [Google Scholar]
- 34.Carrick CS, Fyfe JA, Davies JK. 2000. The genome of Neisseria gonorrhoeae retains the remnants of a two-component regulatory system that once controlled piliation. FEMS Microbiol Lett 186:197–201. doi: 10.1111/j.1574-6968.2000.tb09104.x. [DOI] [PubMed] [Google Scholar]
- 35.Xu J, Chiang HC, Bjursell MK, Gordon JI. 2004. Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends Microbiol 12:21–28. doi: 10.1016/j.tim.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 36.Parks DH, Chuvochina M, Rinke C, Mussig AJ, Chaumeil P-A, Hugenholtz P. 2022. GTDB: an ongoing census of bacterial and archaeal diversity through a phylogenetically consistent, rank normalized and complete genome-based taxonomy. Nucleic Acids Res 50:D785–D794. doi: 10.1093/nar/gkab776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoch JA. 2000. Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 38.Lowe EC, Baslé A, Czjzek M, Firbank SJ, Bolam DN. 2012. A scissor blade-like closing mechanism implicated in transmembrane signaling in a Bacteroides hybrid two-component system. Proc Natl Acad Sci USA 109:7298–7303. doi: 10.1073/pnas.1200479109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao R, Mack TR, Stock AM. 2007. Bacterial response regulators: versatile regulatory strategies from common domains. Trends Biochem Sci 32:225–234. doi: 10.1016/j.tibs.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wolanin PM, Thomason PA, Stock JB. 2002. Histidine protein kinases: key signal transducers outside the animal kingdom. Genome Biol 3:REVIEWS3013. doi: 10.1186/gb-2002-3-10-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dutta R, Inouye M. 2000. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci 25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 42.Zhang W, Shi L. 2005. Distribution and evolution of multiple-step phosphorelay in prokaryotes: lateral domain recruitment involved in the formation of hybrid-type histidine kinases. Microbiology (Reading) 151:2159–2173. doi: 10.1099/mic.0.27987-0. [DOI] [PubMed] [Google Scholar]
- 43.Bramkamp M, Lopez D. 2015. Exploring the existence of lipid rafts in bacteria. Microbiol Mol Biol Rev 79:81–100. doi: 10.1128/MMBR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toledo A, Huang Z, Coleman JL, London E, Benach JL. 2018. Lipid rafts can form in the inner and outer membranes of Borrelia burgdorferi and have different properties and associated proteins. Mol Microbiol 108:63–76. doi: 10.1111/mmi.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sezgin E, Levental I, Mayor S, Eggeling C. 2017. The mystery of membrane organization: composition, regulation and roles of lipid rafts. Nat Rev Mol Cell Biol 18:361–374. doi: 10.1038/nrm.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pike LJ. 2006. Rafts defined: a report on the keystone symposium on lipid rafts and cell function. J Lipid Res 47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 47.Simons K, Toomre D. 2000. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1:31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 48.Kim W, Fan Y-Y, Barhoumi R, Smith R, McMurray DN, Chapkin RS. 2008. n-3 polyunsaturated fatty acids suppress the localization and activation of signaling proteins at the immunological synapse in murine CD4+ T cells by affecting lipid raft formation. J Immunol 181:6236–6243. doi: 10.4049/jimmunol.181.9.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chapkin RS, Wang N, Fan Y-Y, Lupton JR, Prior IA. 2008. Docosahexaenoic acid alters the size and distribution of cell surface microdomains. Biochim Biophys Acta 1778:466–471. doi: 10.1016/j.bbamem.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lorent JH, Diaz-Rohrer B, Lin X, Spring K, Gorfe AA, Levental KR, Levental I. 2017. Structural determinants and functional consequences of protein affinity for membrane rafts. Nat Commun 8:1219. doi: 10.1038/s41467-017-01328-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schuck S, Honsho M, Ekroos K, Shevchenko A, Simons K. 2003. Resistance of cell membranes to different detergents. Proc Natl Acad Sci USA 100:5795–5800. doi: 10.1073/pnas.0631579100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pike LJ. 2004. Lipid rafts: heterogeneity on the high seas. Biochem J 378:281–292. doi: 10.1042/BJ20031672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hanzal-Bayer MF, Hancock JF. 2007. Lipid rafts and membrane traffic. FEBS Lett 581:2098–2104. doi: 10.1016/j.febslet.2007.03.019. [DOI] [PubMed] [Google Scholar]
- 54.Jury EC, Flores-Borja F, Kabouridis PS. 2007. Lipid rafts in T-cell signaling and disease. Semin Cell Dev Biol 18:608–615. doi: 10.1016/j.semcdb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brassard P, Larbi A, Grenier A, Frisch F, Fortin C, Carpentier AC, Fülöp T. 2007. Modulation of T-cell signaling by non-esterified fatty acids. Prostaglandins Leukot Essent Fatty Acids 77:337–343. doi: 10.1016/j.plefa.2007.10.025. [DOI] [PubMed] [Google Scholar]
- 56.Lorent J, Diaz-Rohrer BB, Lin X, Gorfe A, Levental KR, Levental I. 2018. Structural determinants and functional consequences of protein association with membrane domains. Biophysical J 114:380a. doi: 10.1016/j.bpj.2017.11.2103. [DOI] [Google Scholar]
- 57.Demey LM, Sinha R, DiRita VJ. 2022. An essential host dietary fatty acid promotes TcpH inhibition of TcpP proteolysis enabling virulence gene expression in Vibrio cholerae. bioRxiv. https://www.biorxiv.org/content/10.1101/2022.04.28.489952v1. [DOI] [PMC free article] [PubMed]
- 58.Danylec N, Göbl A, Stoll DA, Hetzer B, Kulling SE, Huch M. 2018. Rubneribacter badeniensis gen. nov., sp. nov. and Enteroscipio rubneri gen. nov., sp. nov., new members of the Eggerthellaceae isolated from human feces. Int J Syst Evol Microbiol 68:1533–1540. doi: 10.1099/ijsem.0.002705. [DOI] [PubMed] [Google Scholar]
- 59.Beltrán D, Romo-Vaquero M, Espín JC, Tomás-Barberán FA, Selma MV. 2018. Ellagibacter isourolithinifaciens gen. nov., sp. nov., a new member of the family Eggerthellaceae, isolated from human gut. Int J Syst Evol Microbiol 68:1707–1712. doi: 10.1099/ijsem.0.002735. [DOI] [PubMed] [Google Scholar]
- 60.Clavel T, Charrier C, Braune A, Wenning M, Blaut M, Haller D. 2009. Isolation of bacteria from the ileal mucosa of TNFΔARE mice and description of Enterorhabdus mucosicola gen. nov., sp. nov. Int J Syst Evol Microbiol 59:1805–1812. doi: 10.1099/ijs.0.003087-0. [DOI] [PubMed] [Google Scholar]
- 61.Clavel T, Duck W, Charrier C, Wenning M, Elson C, Haller D. 2010. Enterorhabdus caecimuris sp. nov., a member of the family Coriobacteriaceae isolated from a mouse model of spontaneous colitis, and emended description of the genus Enterorhabdus. Int J Syst Evol Microbiol 60:1527–1531. doi: 10.1099/ijs.0.015016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, Bresciani A, Martínez I, Just S, Ziegler C, Brugiroux S, Garzetti D, Wenning M, Bui TPN, Wang J, Hugenholtz F, Plugge CM, Peterson DA, Hornef MW, Baines JF, Smidt H, Walter J, Kristiansen K, Nielsen HB, Haller D, Overmann J, Stecher B, Clavel T. 2016. The mouse intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol 1:16131. doi: 10.1038/nmicrobiol.2016.131. [DOI] [PubMed] [Google Scholar]
- 63.Sonnenburg ED, Sonnenburg JL, Manchester JK, Hansen EE, Chiang HC, Gordon JI. 2006. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc Natl Acad Sci USA 103:8834–8839. doi: 10.1073/pnas.0603249103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramadurai S, Holt A, Krasnikov V, van den Bogaart G, Killian JA, Poolman B. 2009. Lateral diffusion of membrane proteins. J Am Chem Soc 131:12650–12656. doi: 10.1021/ja902853g. [DOI] [PubMed] [Google Scholar]
- 65.Lucena D, Mauri M, Schmidt F, Eckhardt B, Graumann PL. 2018. Microdomain formation is a general property of bacterial membrane proteins and induces heterogeneity of diffusion patterns. BMC Biol 16:97. doi: 10.1186/s12915-018-0561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Nicolau DV, Jr, Burrage K, Parton RG, Hancock JF. 2006. Identifying optimal lipid raft characteristics required to promote nanoscale protein-protein interactions on the plasma membrane. Mol Cell Biol 26:313–323. doi: 10.1128/MCB.26.1.313-323.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Simons K, Vaz WLC. 2004. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct 33:269–295. doi: 10.1146/annurev.biophys.32.110601.141803. [DOI] [PubMed] [Google Scholar]
- 68.Yu J, Fischman DA, Steck TL. 1973. Selective solubilization of proteins and phospholipids from red blood cell membranes by nonionic detergents. J Supramol Struct 1:233–248. doi: 10.1002/jss.400010308. [DOI] [PubMed] [Google Scholar]
- 69.Simons K, Ikonen E. 1997. Functional rafts in cell membranes. Nature 387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 70.Nikolskaya AN, Galperin MY. 2002. A novel type of conserved DNA-binding domain in the transcriptional regulators of the AlgR/AgrA/LytR family. Nucleic Acids Res 30:2453–2459. doi: 10.1093/nar/30.11.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kinch LN, Cong Q, Jaishankar J, Orth K. 2022. Co-component signal transduction systems: fast-evolving virulence regulation cassettes discovered in enteric bacteria. Proc Natl Acad Sci USA 119:e2203176119. doi: 10.1073/pnas.2203176119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li P, Rivera-Cancel G, Kinch LN, Salomon D, Tomchick DR, Grishin NV, Orth K. 2016. Bile salt receptor complex activates a pathogenic type III secretion system. Elife 5. doi: 10.7554/eLife.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gumerov VM, Ortega DR, Adebali O, Ulrich LE, Zhulin IB. 2020. MiST 3.0: an updated microbial signal transduction database with an emphasis on chemosensory systems. Nucleic Acids Res 48:D459–D464. doi: 10.1093/nar/gkz988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gumerov VM, Zhulin IB. 2020. TREND: a platform for exploring protein function in prokaryotes based on phylogenetic, domain architecture and gene neighborhood analyses. Nucleic Acids Res 48:W72–W76. doi: 10.1093/nar/gkaa243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 76.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, Tosatto SCE, Paladin L, Raj S, Richardson LJ, Finn RD, Bateman A. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49:D412–D419. doi: 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mathur J, Waldor MK. 2004. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect Immun 72:3577–3583. doi: 10.1128/IAI.72.6.3577-3583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Welch TJ, Bartlett DH. 1998. Identification of a regulatory protein required for pressure-responsive gene expression in the deep-sea bacterium Photobacterium species strain SS9. Mol Microbiol 27:977–985. doi: 10.1046/j.1365-2958.1998.00742.x. [DOI] [PubMed] [Google Scholar]
- 79.Miller VL, Taylor RK, Mekalanos JJ. 1987. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. cell 48:271–279. doi: 10.1016/0092-8674(87)90430-2. [DOI] [PubMed] [Google Scholar]
- 80.DiRita VJ, Mekalanos JJ. 1991. Periplasmic interaction between two membrane regulatory proteins, ToxR and ToxS, results in signal transduction and transcriptional activation. Cell 64:29–37. doi: 10.1016/0092-8674(91)90206-e. [DOI] [PubMed] [Google Scholar]
- 81.Bina XR, Howard MF, Ante VM, Bina JE. 2016. Vibrio cholerae LeuO links the ToxR regulon to expression of lipid A remodeling genes. Infect Immun 84:3161–3171. doi: 10.1128/IAI.00445-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mey AR, Craig SA, Payne SM. 2012. Effects of amino acid supplementation on porin expression and ToxR levels in Vibrio cholerae. Infect Immun 80:518–528. doi: 10.1128/IAI.05851-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Krukonis ES, DiRita VJ. 2003. DNA binding and ToxR responsiveness by the wing domain of TcpP, an activator of virulence gene expression in Vibrio cholerae. Mol Cell 12:157–165. doi: 10.1016/s1097-2765(03)00222-3. [DOI] [PubMed] [Google Scholar]
- 84.DiRita VJ, Parsot C, Jander G, Mekalanos JJ. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc Natl Acad Sci USA 88:5403–5407. doi: 10.1073/pnas.88.12.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Higgins DE, DiRita VJ. 1994. Transcriptional control of toxT, a regulatory gene in the ToxR regulon of Vibrio cholerae. Mol Microbiol 14:17–29. doi: 10.1111/j.1365-2958.1994.tb01263.x. [DOI] [PubMed] [Google Scholar]
- 86.Krukonis ES, Yu RR, Dirita VJ. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol Microbiol 38:67–84. doi: 10.1046/j.1365-2958.2000.02111.x. [DOI] [PubMed] [Google Scholar]
- 87.Provenzano D, Lauriano CM, Klose KE. 2001. Characterization of the role of the ToxR-modulated outer membrane porins OmpU and OmpT in Vibrio cholerae virulence. J Bacteriol 183:3652–3662. doi: 10.1128/JB.183.12.3652-3662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kazi MI, Conrado AR, Mey AR, Payne SM, Davies BW. 2016. ToxR antagonizes H-NS regulation of horizontally acquired genes to drive host colonization. PLoS Pathog 12:e1005570. doi: 10.1371/journal.ppat.1005570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Teoh WP, Matson JS, DiRita VJ. 2015. Regulated intramembrane proteolysis of the virulence activator TcpP in Vibrio cholerae is initiated by the tail-specific protease (Tsp). Mol Microbiol 97:822–831. doi: 10.1111/mmi.13069. [DOI] [PubMed] [Google Scholar]
- 90.Matson JS, DiRita VJ. 2005. Degradation of the membrane-localized virulence activator TcpP by the YaeL protease in Vibrio cholerae. Proc Natl Acad Sci USA 102:16403–16408. doi: 10.1073/pnas.0505818102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Thompson LR, Nikolakakis K, Pan S, Reed J, Knight R, Ruby EG. 2017. Transcriptional characterization of Vibrio fischeri during colonization of juvenile Euprymna scolopes. Environ Microbiol 19:1845–1856. doi: 10.1111/1462-2920.13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Beck NA, Krukonis ES, DiRita VJ. 2004. TcpH influences virulence gene expression in Vibrio cholerae by inhibiting degradation of the transcription activator TcpP. J Bacteriol 186:8309–8316. doi: 10.1128/JB.186.24.8309-8316.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ante VM, Bina XR, Bina JE. 2015. The LysR-type regulator LeuO regulates the acid tolerance response in Vibrio cholerae. Microbiology (Reading) 161:2434–2443. doi: 10.1099/mic.0.000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rhee JE, Jeong HG, Lee JH, Choi SH. 2006. AphB influences acid tolerance of Vibrio vulnificus by activating expression of the positive regulator CadC. J Bacteriol 188:6490–6497. doi: 10.1128/JB.00533-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Merrell DS, Camilli A. 2000. Regulation of Vibrio cholerae genes required for acid tolerance by a member of the “ToxR-like” family of transcriptional regulators. J Bacteriol 182:5342–5350. doi: 10.1128/JB.182.19.5342-5350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Neely MN, Dell CL, Olson ER. 1994. Roles of LysP and CadC in mediating the lysine requirement for acid induction of the Escherichia coli cad operon. J Bacteriol 176:3278–3285. doi: 10.1128/jb.176.11.3278-3285.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Casalino M, Prosseda G, Barbagallo M, Iacobino A, Ceccarini P, Latella MC, Nicoletti M, Colonna B. 2010. Interference of the CadC regulator in the arginine-dependent acid resistance system of Shigella and enteroinvasive Escherichia coli. Int J Med Microbiol 300:289–295. doi: 10.1016/j.ijmm.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 98.Lee YH, Kim BH, Kim JH, Yoon WS, Bang SH, Park YK. 2007. CadC has a global translational effect during acid adaptation in Salmonella enterica serovar Typhimurium. J Bacteriol 189:2417–2425. doi: 10.1128/JB.01277-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lee YH, Kim JH. 2017. Direct interaction between the transcription factors CadC and OmpR involved in the acid stress response of Salmonella enterica. J Microbiol 55:966–972. doi: 10.1007/s12275-017-7410-7. [DOI] [PubMed] [Google Scholar]
- 100.Hsieh P-F, Lin H-H, Lin T-L, Wang J-T. 2010. CadC regulates cad and tdc operons in response to gastrointestinal stresses and enhances intestinal colonization of Klebsiella pneumoniae. J Infect Dis 202:52–64. doi: 10.1086/653079. [DOI] [PubMed] [Google Scholar]
- 101.Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, Roe JH. 2006. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutase and nickel transport in Streptomyces coelicolor. Mol Microbiol 59:1848–1858. doi: 10.1111/j.1365-2958.2006.05065.x. [DOI] [PubMed] [Google Scholar]
- 102.Dell CL, Neely MN, Olson ER. 1994. Altered pH lysine signaling mutants of cadC, a gene encoding a membrane-bound transcriptional activator of the Escherichia coli cadBA operon. Mol Microbiol 14:7–16. doi: 10.1111/j.1365-2958.1994.tb01262.x. [DOI] [PubMed] [Google Scholar]
- 103.Debnath A, Mizuno T, Miyoshi SI. 2020. Regulation of chitin-dependent growth and natural competence in Vibrio parahaemolyticus. Microorganisms 8:1303. doi: 10.3390/microorganisms8091303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Okada R, Matsuda S, Iida T. 2017. Vibrio parahaemolyticus VtrA is a membrane-bound regulator and is activated via oligomerization. PLoS One 12:e0187846. doi: 10.1371/journal.pone.0187846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rivera-Cancel G, Orth K. 2017. Biochemical basis for activation of virulence genes by bile salts in Vibrio parahaemolyticus. Gut Microbes 8:366–373. doi: 10.1080/19490976.2017.1287655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller KA, Sofia MK, Weaver JWA, Seward CH, Dziejman M. 2016. Regulation by ToxR-like proteins converges on vttRB expression to control type 3 secretion system-dependent Caco2-BBE cytotoxicity in Vibrio cholerae. J Bacteriol 198:1675–1682. doi: 10.1128/JB.00130-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tükel C, Akçelik M, de Jong MF, Simsek O, Tsolis RM, Bäumler AJ. 2007. MarT activates expression of the MisL autotransporter protein of Salmonella enterica serotype Typhimurium. J Bacteriol 189:3922–3926. doi: 10.1128/JB.01746-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cascales D, Guijarro JA, García-Torrico AI, Méndez J. 2017. Comparative genome analysis reveals important genetic differences among serotype O1 and serotype O2 strains of Y. ruckeri and provides insights into host adaptation and virulence. Microbiology Open 6:e00460. doi: 10.1002/mbo3.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Emmerth M, Goebel W, Miller SI, Hueck CJ. 1999. Genomic subtraction identifies Salmonella Typhimurium prophages, F-related plasmid sequences, and a novel fimbrial operon, stf, which are absent in Salmonella Typhi. J Bacteriol 181:5652–5661. doi: 10.1128/JB.181.18.5652-5661.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Blanc-Potard AB, Solomon F, Kayser J, Groisman EA. 1999. The SPI-3 pathogenicity island of Salmonella enterica. J Bacteriol 181:998–1004. doi: 10.1128/JB.181.3.998-1004.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morgan JK, Carroll RK, Harro CM, Vendura KW, Shaw LN, Riordan JT. 2016. Global regulator of virulence A (GrvA) coordinates expression of discrete pathogenic mechanisms in enterohemorrhagic Escherichia coli through interactions with GadW-GadE. J Bacteriol 198:394–409. doi: 10.1128/JB.00556-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morgan JK, Vendura KW, Stevens SM, Riordan JT. 2013. RcsB determines the locus of enterocyte effacement (LEE) expression and adherence phenotype of Escherichia coli O157: H7 spinach outbreak strain TW14359 and coordinates bicarbonate-dependent LEE activation with repression of motility. Microbiology (Reading) 159:2342–2353. doi: 10.1099/mic.0.070201-0. [DOI] [PubMed] [Google Scholar]
- 113.Xue M, Xiao Y, Fu D, Raheem MA, Shao Y, Song X, Tu J, Xue T, Qi K. 2020. Transcriptional regulator YqeI, locating at ETT2 locus, affects the pathogenicity of avian pathogenic Escherichia coli. Animals 10:1658. doi: 10.3390/ani10091658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Levine MM, Ristaino P, Marley G, Smyth C, Knutton S, Boedeker E, Black R, Young C, Clements ML, Cheney C. 1984. Coli surface antigens 1 and 3 of colonization factor antigen II-positive enterotoxigenic Escherichia coli: morphology, purification, and immune responses in humans. Infect Immun 44:409–420. doi: 10.1128/iai.44.2.409-420.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Iriarte M, Cornelis GR. 1995. MyfF, an element of the network regulating the synthesis of fibrillae in Yersinia enterocolitica. J Bacteriol 177:738–744. doi: 10.1128/jb.177.3.738-744.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Liu T, Wang K-Y, Wang J, Chen D-F, Huang X-L, Ouyang P, Geng Y, He Y, Zhou Y, Min J. 2016. Genome sequence of the fish pathogen Yersinia ruckeri SC09 provides insights into niche adaptation and pathogenic mechanism. Int J Mol Sci 17:557. doi: 10.3390/ijms17040557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Charlebois A, Jalbert L-A, Harel J, Masson L, Archambault M. 2012. Characterization of genes encoding for acquired bacitracin resistance in Clostridium perfringens. PLoS One 7:e44449. doi: 10.1371/journal.pone.0044449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Darnell RL, Nakatani Y, Knottenbelt MK, Gebhard S, Cook GM. 2019. Functional characterization of BcrR: a one-component transmembrane signal transduction system for bacitracin resistance. Microbiology (Reading) 165:475–487. doi: 10.1099/mic.0.000781. [DOI] [PubMed] [Google Scholar]
- 119.Noda M, Miyauchi R, Danshiitsoodol N, Matoba Y, Kumagai T, Sugiyama M. 2018. Expression of genes involved in bacteriocin production and self-resistance in Lactobacillus brevis 174A is mediated by two regulatory proteins. Appl Environ Microbiol 84:e02707-17. doi: 10.1128/AEM.02707-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Noda M, Miyauchi R, Danshiitsoodol N, Higashikawa F, Kumagai T, Matoba Y, Sugiyama M. 2015. Characterization and mutational analysis of a two-polypeptide bacteriocin produced by citrus Iyo-derived Lactobacillus brevis 174A. Biol Pharm Bull 38:1902–1909. doi: 10.1248/bpb.b15-00505. [DOI] [PubMed] [Google Scholar]
- 121.Suárez C, Espariz M, Blancato VS, Magni C. 2013. Expression of the agmatine deiminase pathway in Enterococcus faecalis is activated by the AguR regulator and repressed by CcpA and PTS(Man) systems. PLoS One 8:e76170. doi: 10.1371/journal.pone.0076170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Liu Y, Zeng L, Burne RA. 2009. AguR is required for induction of the Streptococcus mutans agmatine deiminase system by low pH and agmatine. Appl Environ Microbiol 75:2629–2637. doi: 10.1128/AEM.02145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Del Rio B, Linares D, Ladero V, Redruello B, Fernandez M, Martin MC, Alvarez MA. 2016. Putrescine biosynthesis in Lactococcus lactis is transcriptionally activated at acidic pH and counteracts acidification of the cytosol. Int J Food Microbiol 236:83–89. doi: 10.1016/j.ijfoodmicro.2016.07.021. [DOI] [PubMed] [Google Scholar]
- 124.Linares DM, Del Rio B, Redruello B, Ladero V, Martin MC, de Jong A, Kuipers OP, Fernandez M, Alvarez MA. 2015. AguR, a transmembrane transcription activator of the putrescine biosynthesis operon in Lactococcus lactis, acts in response to the agmatine concentration. Appl Environ Microbiol 81:6145–6157. doi: 10.1128/AEM.00959-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Hemert S, Meijerink M, Molenaar D, Bron PA, de Vos P, Kleerebezem M, Wells JM, Marco ML. 2010. Identification of Lactobacillus plantarum genes modulating the cytokine response of human peripheral blood mononuclear cells. BMC Microbiol 10:293. doi: 10.1186/1471-2180-10-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Gaur G, Oh J-H, Filannino P, Gobbetti M, van Pijkeren J-P, Gänzle MG. 2020. Genetic determinants of hydroxycinnamic acid metabolism in heterofermentative lactobacilli. Appl Environ Microbiol 86:02461-19. doi: 10.1128/AEM.02461-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Yeo W-S, Anokwute C, Marcadis P, Levitan M, Ahmed M, Bae Y, Kim K, Kostrominova T, Liu Q, Bae T. 2020. A membrane-bound transcription factor is proteolytically regulated by the AAA+ protease FtsH in Staphylococcus aureus. J Bacteriol 202:00019-20. doi: 10.1128/JB.00019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Egan S, James S, Kjelleberg S. 2002. Identification and characterization of a putative transcriptional regulator controlling the expression of fouling inhibitors in Pseudoalteromonas tunicata. Appl Environ Microbiol 68:372–378. doi: 10.1128/AEM.68.1.372-378.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00266-23-s0001.xlsx, XLSX file, 0.8 MB (859.1KB, xlsx)
Supplemental material. Download spectrum.00266-23-s0002.xlsx, XLSX file, 1.9 MB (1.9MB, xlsx)
Supplemental material. Download spectrum.00266-23-s0003.xlsx, XLSX file, 0.2 MB (245.4KB, xlsx)
Supplemental material. Download spectrum.00266-23-s0004.xlsx, XLSX file, 0.03 MB (27.5KB, xlsx)
Supplemental material. Download spectrum.00266-23-s0005.xlsx, XLSX file, 0.03 MB (30.7KB, xlsx)
Supplemental material. Download spectrum.00266-23-s0006.xlsx, XLSX file, 1.1 MB (1.1MB, xlsx)
Supplemental material. Download spectrum.00266-23-s0007.xlsx, XLSX file, 1.7 MB (1.7MB, xlsx)
Supplemental material. Download spectrum.00266-23-s0008.xlsx, XLSX file, 3.7 MB (3.7MB, xlsx)
Supplemental material. Download spectrum.00266-23-s0009.xlsx, XLSX file, 0.02 MB (20.6KB, xlsx)
Data Availability Statement
The data presented here are available from the corresponding authors upon request.