ABSTRACT
Toxigenic Vibrio cholerae serogroup O1 is the etiologic agent of the disease cholera, and strains of this serogroup are responsible for pandemics. A few other serogroups have been found to carry cholera toxin genes—most notably, O139, O75, and O141—and public health surveillance in the United States is focused on these four serogroups. A toxigenic isolate was recovered from a case of vibriosis from Texas in 2008. This isolate did not agglutinate with any of the four different serogroups’ antisera (O1, O139, O75, or O141) routinely used in phenotypic testing and did not display a rough phenotype. We investigated several hypotheses that might explain the recovery of this potential nonagglutinating (NAG) strain using whole-genome sequencing analysis and phylogenetic methods. The NAG strain formed a monophyletic cluster with O141 strains in a whole-genome phylogeny. Furthermore, a phylogeny of ctxAB and tcpA sequences revealed that the sequences from the NAG strain also formed a monophyletic cluster with toxigenic U.S. Gulf Coast (USGC) strains (O1, O75, and O141) that were recovered from vibriosis cases associated with exposures to Gulf Coast waters. A comparison of the NAG whole-genome sequence showed that the O-antigen-determining region of the NAG strain was closely related to those of O141 strains, and specific mutations were likely responsible for the inability to agglutinate. This work shows the utility of whole-genome sequence analysis tools for characterization of an atypical clinical isolate of V. cholerae originating from a USGC state.
IMPORTANCE Clinical cases of vibriosis are on the rise due to climate events and ocean warming (1, 2), and increased surveillance of toxigenic Vibrio cholerae strains is now more crucial than ever. While traditional phenotyping using antisera against O1 and O139 is useful for monitoring currently circulating strains with pandemic or epidemic potential, reagents are limited for non-O1/non-O139 strains. With the increased use of next-generation sequencing technologies, analysis of less well-characterized strains and O-antigen regions is possible. The framework for advanced molecular analysis of O-antigen-determining regions presented herein will be useful in the absence of reagents for serotyping. Furthermore, molecular analyses based on whole-genome sequence data and using phylogenetic methods will help characterize both historical and novel strains of clinical importance. Closely monitoring emerging mutations and trends will improve our understanding of the epidemic potential of Vibrio cholerae to anticipate and rapidly respond to future public health emergencies.
KEYWORDS: Vibrio cholerae, NAG, nonagglutinating Vibrio strain, USGC Vibrio cholerae, toxigenic Vibrio surveillance, MAG
INTRODUCTION
Vibrio cholerae is a genetically diverse marine bacterium that is often spread through contaminated food and water. V. cholerae causes gastrointestinal and extraintestinal (wound or ear) infections in humans and, in some cases, septicemia (1–3). Over 200 serogroups of V. cholerae have been described, but O1 and O139 are of considerable concern because of their pandemic and epidemic potential. The current circulating pandemic strain belongs to serogroup O1 (biotype El Tor) and has been attributed to global outbreaks since 1961 (4, 5), while epidemics of O139 have arisen from the Bay of Bengal beginning in 1992 (6). Serogroup O139 was initially discovered based on its inability to agglutinate with O1 antisera (7) and was subsequently shown to contain differences in O-antigen biosynthesis regions (8). Furthermore, serogroup O139 was determined to have emerged from the O1 El Tor biotype as a result of serogroup conversion and genetic exchange of O-antigen DNA (4, 8–11). Because serogroup O139 is recently descended from O1 El Tor, it harbors the genetic backbone of seventh pandemic O1 El Tor strains, including many of the same virulence genes, such as the cholera toxin genes (12).
Pandemic O1 and O139 strains contain major virulence genes on mobile genetic elements, including cholera toxin genes such as ctxAB carried on the cholera toxin prophage (CTXϕ), which is integrated into one or both chromosomes of V. cholerae, and toxin coregulated pilus (TCP), which serves as the receptor for CTXϕ and is encoded by tcp genes located on the Vibrio pathogenicity island (VPI) (13). Therefore, detection of serogroups O1 and O139 and toxin genes has been the focus of surveillance testing in laboratories across the world. Phenotypic testing for serogroups O1 and O139 is widely used due to the accessibility of commercially available antisera, while PCR tests are used for toxin gene detection (14, 15). Characterization of serogroups O1 and O139 based on conserved genetic marker genes (wbeN and wbfR, respectively) in the O-antigen-determining regions (wbe and wbf gene clusters) have made it possible for surveillance testing to consolidate important serogroup and toxin gene detection into a single molecular workflow (11, 14–19).
An important consideration for developing a comprehensive surveillance system for vibriosis is the inclusion of other serogroups that can also carry virulence genes, such as O75 and O141 (4, 20–22). Antisera for phenotypic testing are not commercially available for non-O1/non-O139 serogroups and public sequence data, and known genetic markers are limited for strains like O75 and O141 (18, 19, 22). However, the increased use of next-generation sequencing (NGS) technologies over the last 2 decades has led to detailed characterizations of non-O1/non-O139 strains, including O141 (22). Therefore, data from NGS can be used to develop analytical frameworks for other less well-characterized serogroups and where access to antisera for phenotypic classifications is limited.
The CDC monitors vibriosis through the nationwide Cholera and Other Vibrio Illness Surveillance (COVIS) system (https://www.cdc.gov/vibrio/surveillance.html), a passive surveillance system first launched in 1989 by the U.S. Gulf Coast (USGC) states (Alabama, Florida, Louisiana, and Texas) that includes reporting from all 50 states and the U.S. Food and Drug Administration. Epidemiological surveillance is complemented by laboratory analysis at the National Vibrio Reference Laboratory at the CDC, which includes species identification of V. cholerae isolates sent from states using WGS, toxin profiling, and phenotype testing of four serogroups (O1, O139, O75, and O141). Over the years, we have observed patterns in strains endogenous to the USGC, which include serogroups O1, O75, and O141, that are of public health concern due to shared virulence characteristics with pandemic strains of V. cholerae (3, 20, 21, 23–25). Multiple studies have reported V. cholerae cases from O141 and O75 strains that have caused cholera-like outbreaks in the United States (3, 20–22) and attributed them to environmental reservoirs where these strains are endemic (23). The emergence of atypical and nonagglutinating non-O1/non-O139 strains from environmental reservoirs in South Africa has also been described (26). Therefore, characterization of unusual and atypical strains that cause sporadic cases of severe clinical illness is useful for elucidating potential evolutionary changes or factors that have limited the epidemic spread of non-O1/non-O139 serogroups.
In 2008, an isolate (3528-08) was received at the CDC from Texas that was positive for cholera toxin upon routine surveillance testing. There was no history of travel or consumption of seafood, but the patient reported recreational swimming. The isolate did not agglutinate in any of the four routinely tested antisera (O1, O139, O75, or O141) and did not display a rough phenotype (i.e., agglutination was not observed in sterile water). We investigated several hypotheses that might explain the recovery of this potential nonagglutinating (NAG) strain. First, the strain might represent one of the four known toxigenic serogroups previously seen in the USGC that lost the ability to produce O antigen; second, the strain could represent a resident serogroup that recently acquired CTXϕ from one of the resident toxigenic strains; the strain represents the introduction of a toxigenic serogroup not normally included in CDC surveillance (i.e., the introduction of an emerging or novel toxigenic serogroup). We obtained whole-genome sequences (WGS) and employed a variety of bioinformatic techniques to investigate the origin of the potential NAG isolate by examining the phylogenetic relationship to other cholera toxin-positive USGC strains, compared the nucleotide sequences of the major virulence genes (ctxAB and tcpA), and compared the genetic sequence of the O-antigen-determining region in the potential NAG isolate to the genetic sequences of these regions in other cholera toxin-positive reference strains. We found that the strain likely represents an emerging O141 strain that has lost the ability to produce functional O141 O antigen. Taken together, our analysis demonstrates the utility of WGS and phylogenetic methods for the characterization of important toxin- and O-antigen-determining regions of V. cholerae.
RESULTS
Initial characterization of the Vibrio cholerae NAG strain.
A clinical isolate (3528-08) was confirmed as Vibrio cholerae in the National Vibrio Reference Laboratory in the Enteric Diseases Laboratory Branch at the CDC. The isolate was positive for the cholera toxin and the toxin coregulated pilus targets by PCR analysis, but the isolate did not agglutinate with routinely tested antisera for serogroups O1, O139, O75, and O141, which can result in clinical illness. It was also determined that the strain was not rough and did not autoagglutinate in sterile saline.
Phylogenetic analysis of the NAG strain.
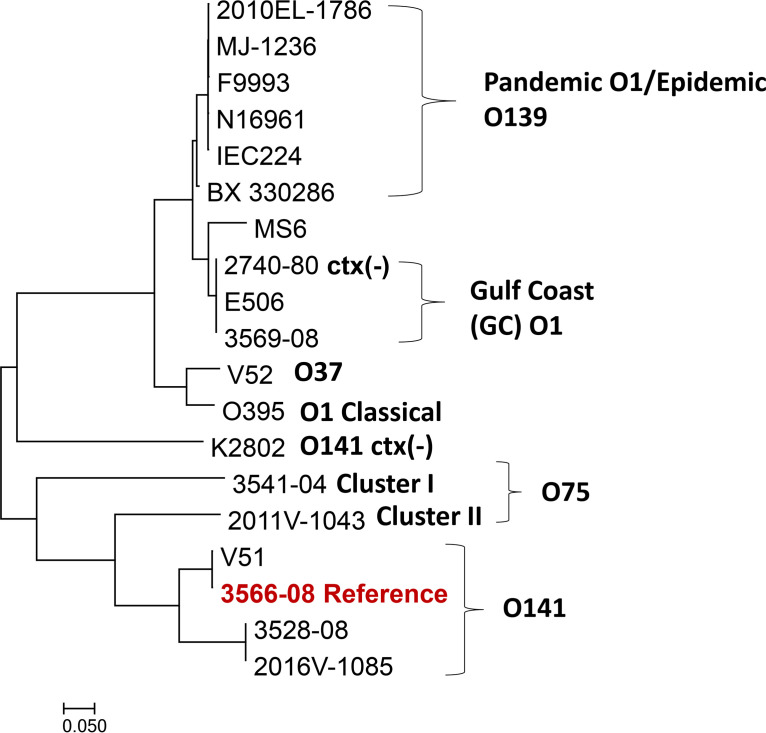
To investigate whether this potential nonagglutinating (NAG) strain (3528-08) might represent a known toxigenic serogroup, a core genome single nucleotide polymorphism (SNP) phylogeny was inferred (Fig. 1), along with 18 representative whole-genome sequences from relevant V. cholerae serogroups with publicly available data (Table 1; see Table S1 in the supplemental material). The potential NAG strain formed a well-defined monophyletic cluster with the toxigenic (ctx+) O141 strains V51, 2016V-1085, and 3566-08 (which was used as the reference for the phylogeny) (Fig. 1). The potential NAG strain clustered next to the O141 strain 2016V-1085, which was isolated in New Mexico in 2016 (Fig. 1). O1 and O139 strains with epidemic or pandemic potential formed a distinct cluster. Nontoxigenic V. cholerae strains formed separate phylogenetic groups, except the V. cholerae O1 USGC strain 2740-80, which formed a monophyletic cluster with toxigenic USGC O1 strains. Toxigenic O75 strains formed two clusters, denoted cluster I (3541-04) and cluster II (2011V-1043), basal to toxigenic O141 strains. Based on this core genome SNP phylogeny, the potential NAG strain was most closely related to O141 reference strains.
FIG 1.
Core genome SNP phylogeny of toxigenic ctx (+) Vibrio cholerae serogroup O1, O139, O37, O75, and O141 strains and the NAG strain, generated using Parsnp version 1.5.2. 3566-08 served as the reference genome to build this phylogeny. Nontoxigenic ctx (−) variants of O141 and O1 were also included for comparison.
TABLE 1.
List of strains and sequences included in the study
| Species | Strain | Serogroup | GenBank assembly accession no.a | Strain included in phylogeny? |
||
|---|---|---|---|---|---|---|
| Core genome SNP | ctxAB | tcpA | ||||
| Vibrio cholerae | 2010EL-1786 | O1 | GCA_000166455.2 | Yes | Yes | Yes |
| 2016V-1085 | O141 | GCA_021308775.1 | Yes | Yes | Yes | |
| 3528-08 | NAG | GCA_009762895.1 | Yes | Yes | Yes | |
| 3566-08 | O141 | GCA_009763105.1 | Yes | Yes | Yes | |
| 3569-08 | O1 | GCA_009762985.1 | Yes | Yes | Yes | |
| BX 330286 | O1 | GCA_000174335.1 | Yes | Yes | No | |
| 2011V-1043 | O75 | GCA_009764135.1 | Yes | Yes | Yes | |
| MJ1236 | O1 | GCA_000022585.1 | Yes | No | No | |
| N16961 | O1 | GCA_900205735.1 | Yes | Yes | No | |
| O395 | O1 | GCA_000021625.1 | Yes | Yes | Yes | |
| V51 | O141 | GCA_000152465.2 | Yes | Yes | Yes | |
| V52 | O37 | GCA_001857545.1 | Yes | Yes | Yes | |
| 3541-04 | O75 | GCA_009763045.1 | Yes | Yes | Yes | |
| E506 | O1 | GCA_001887475.1 | Yes | Yes | No | |
| F9993 | O139 | GCA_009763825.1 | Yes | No | No | |
| IEC224 | O1 | GCA_000250855.1 | Yes | No | No | |
| MS6 | O1 | GCA_000829215.1 | Yes | No | No | |
| K2802 | O141 | GCA_009763985.1 | Yes | No | No | |
| 2470-80 | O1 | GCA_001683415.1 | Yes | No | No | |
| 365-96b,c | O27 | NA | No | Yes | Yes | |
| ATCC 25872 (280 NAG) | O37 | GCA_018336415.1 | No | No | Yes | |
| CIRS101 | O1 | GCA_000175695.1 | No | Yes | No | |
| Vibrio mimicus | M1567 (523-80)d | NA | NA | No | No | Yes |
| 06-2455 | NA | GCA_009764005.1 | No | No | Yes | |
| 2011V-1073 | NA | GCA_009665195.1 | No | No | Yes | |
NA, not available.
The GenBank accession number for the 365-96 ctxAB sequence is AF390572.1.
The GenBank accession number for the 365-96 tcpA sequence is AF390571.1.
The GenBank accession number for the M1567 (523-80) tcpA sequence is FJ209007.1.
Characterization of the NAG virulence genes.
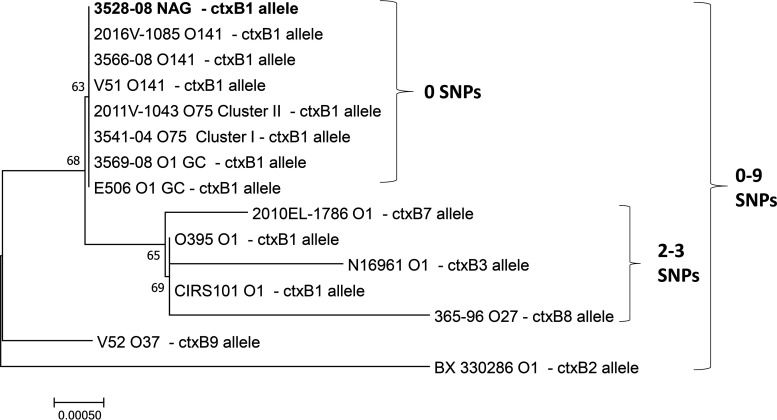
To infer the evolutionary history of the ctxAB locus of the potential NAG strain, a neighbor-joining phylogeny with a total of 15 ctx+ strains was generated (Fig. 2). The potential NAG strain had a ctxAB sequence similar to those of USGC serogroups representing O1, O75, and O141 strains, and it had the same ctxAB allele, as confirmed by SNP distances (Fig. 2). The ctxAB sequence of the potential NAG and USGC serogroups (O1, O75, and O141) was distinct from those of other ctxAB alleles, as evidenced by a separate monophyletic cluster in the ctxAB phylogeny. Additional clusters of different ctxAB alleles from toxigenic O1, O27, and O37 strains were between 0 and 9 SNPs apart from those of the potential NAG strain and USGC strains. The ctxAB sequence for O139 strain F9993 was not included in the phylogeny, as the ctxAB allele is identical to that of O1 El Tor strain N16961.
FIG 2.
Neighbor-joining phylogeny of ctxAB sequences (1,148 bp) from toxigenic (ctx+) strains of Vibrio cholerae: the NAG strain and strains from O1, O75, O141, and other toxigenic serogroups. The phylogeny was generated using MEGA7 with the Kimura 2-parameter method and 500 replications of the interior branch test. Vibrio cholerae O139 strain F9993 was not included (its ctxAB sequence is identical to that of Vibrio cholerae O1 strain N16961). ctxB alleles and SNP ranges are displayed (e.g., number of differences). Bootstrap values greater than 50 are shown.
To further characterize the ctxAB allele of the potential NAG strain, individual nucleotide sequences of the ctxA and ctxB subunits were analyzed. An alignment of the potential NAG ctxAB sequence compared to other V. cholerae ctxAB sequences showed that the representative USGC serogroup strains (O1, O75, and O141) and the potential NAG strain share a ctxA allele with O37 strains (Fig. S1). This was also corroborated by a BLAST search of the ctxA sequence of the potential NAG against the National Center for Biotechnology Information (NCBI) nucleotide databases, which revealed that the USGC ctxA allele was shared with O37 strains, in addition to another species of Vibrio, Vibrio mimicus (Fig. S1). However, distinct SNPs observed downstream in the ctxB gene distinguish the potential NAG and USGC serogroups from O37 strains, as well as pandemic serogroup O1 strains (Fig. S1). In addition, the ctxB of USGC serogroups and the potential NAG strain were unrelated to the ctxB of V. mimicus. Furthermore, ctxB genotyping of conserved SNPs revealed that the potential NAG strain contained the ctxB1 allele (Fig. S1; Fig. 2), which is also found in toxigenic USGC strains and pandemic strains of O1 (4, 12). Consistent with the previous report of sequence similarity of ctxAB among O141 strains (22), this phylogeny and nucleotide alignment show that the potential NAG strain ctxAB sequence is closely related to those of other toxigenic V. cholerae strains from the USGC, despite the diversity in times of collection and differences in O-antigen serogroups.
To characterize additional important virulence factors associated with V. cholerae, the tcpA gene sequence of the potential NAG strain was compared with those of other toxigenic V. cholerae strains (serogroups O1, O27, and O37), USGC strains (O1, O75, and O141), and V. mimicus (Fig. 3). A maximum likelihood phylogeny showed that the NAG strain tcpA is most similar to the those of the O141 strain 2016V-1085 and V. mimicus strains M1567, 2011V-1073, and 06-2455; these five strains cluster basal to O141 strains (3566-08 and V51) and the O75 cluster I strain 3541-04 (Fig. 3). The remaining USGC strains cluster further away in the tcpA phylogeny: the O1 USGC strain 3569-08 clusters with the pandemic O1 El Tor strain 2010EL-1786, while the O75 cluster II USGC strain 2011V-1043 groups with the classic O1 strain O395 and O37 strains ATCC 25872 and V52 (Fig. 3). Taken together, these findings indicate that two important virulence genes, ctxAB and tcpA, carried by the potential NAG strain are highly related to those of other toxigenic USGC V. cholerae strains (serogroups O1, O75, and O141).
FIG 3.

Maximum likelihood phylogeny of tcpA sequences (675 bp) from the Vibrio cholerae NAG strain and strains from O1, O75, O141, and other representative serogroups. The phylogenetic tree was generated using MEGA7 with the general time-reversible model and 500 bootstrap replications. Vibrio mimicus was also included for comparison. Bootstrap values greater than 50 are shown.
Characterization of the O-antigen-determining region.
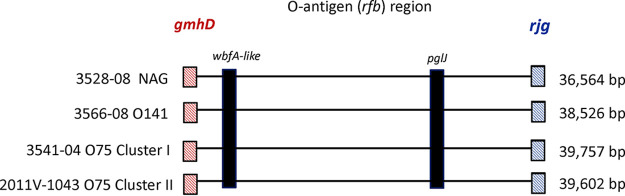
To evaluate if the potential NAG strain shared any antigenic sequence homology with serogroups commonly found in the USGC, the O-antigen region (rfb) was compared to reference O-antigen regions, including those of serotype O1, O139, O75, and O141 strains. The O-antigen region lengths for the representative reference strains 2010EL-1786 (O1), 3569-08 (O1 USGC), F9993 (O139), 3541-04 (O75 cluster I), 2011V-1043 (O75 cluster II), and 3566-08 (O141) and the NAG strain 3528-08 ranged between 24 and 40 kb (24,020, 24,032, 38,075, 39,757, 39,602, 38,526, and 36,564 bp, respectively) (Fig. 4). The O-antigen regions of the NAG strain, the O75 cluster I and II strains (3541-04 and 2011V-1043), and the O141 strain (3566-08) were genetically similar based on alignments of these regions. The 36,564-bp O-antigen region of the NAG strain showed the highest similarity to the O-antigen region of O141 reference strain 3566-08, with 99.98% average nucleotide identity (ANI) and 94.9% aligned bases, followed by the O75 strains 3541-04 (ANI, 99.37%; 73.43% aligned bases) and 2011V-1043 (ANI, 98.3%; 68% aligned bases) and the O139 strain F9993 (ANI, 98.2%; 42.37% aligned bases). The NAG O-antigen region shared a 305-bp region with O1 strains 2010EL-1786 and 3569-08, with 97.38% ANI and 1.27% aligned bases.
FIG 4.
Evidence of mutations in the NAG strain O antigen. The illustration shows the approximate locations of genes with mutations in the NAG strain 3528-08 O-antigen region—an unknown gene that is wbfA-like and the pglJ gene—as well as those in O141 reference strain 3566-08 and O75 cluster I and II reference strains 3541-04 and 2011V-1043. The location of the O-antigen junction genes gmhD and rjg are labeled in red and blue, respectively. The O-antigen region lengths are shown on the right. While the O75 sequences for these genes were not examined in detail, the wbfA-like gene and pglJ gene are also present in O75, as shown.
Genome alignments of the O antigens of the potential NAG strain, O75 cluster I and II strains (3541-04 and 2011V-1043), and the O141 strain (3566-08) also indicated genetic similarity, as depicted in a Mauve alignment with annotations (Fig. S2). While considerable homology across the O-antigen-determining region exists between the potential NAG strain and the O141 strain 3566-08, two notable regions of homology were identified across all four strains, including O75 cluster I strain 3541-04 and O75 cluster II strain 2011V-1043 (Fig. 4; Fig. S2). Specifically, an unknown gene predicted to be a hypothetical protein and the pglJ gene within the O-antigen rfb region share sequence homology across all four strains (Fig. 4; Fig. S2). These genes were located on chromosome 1 of the potential NAG strain at positions 444027 to 445544 and 421452 to 420355, respectively (GenBank accession number CP046736.1; Fig. S2). Furthermore, mutations were discovered within these two genes based on alignments between the NAG strain O-antigen DNA sequence and the O141 reference strain (3566-08) O-antigen DNA sequence (Fig. S3, Fig. S4). Specifically, a nucleotide insertion at position 964 in the unknown gene resulted in a proline amino acid substitution (Fig. S3). Furthermore, a 483-bp insertion at the 3′ end of the gene was confirmed to have homology with the wbfA domain, previously described as an O139 O-antigen gene with an unknown function (11, 16, 17). To further investigate the potential function of the insertion, the NAG hypothetical protein was subjected to a BLAST search with a full-length annotated wbfA coding sequence from O139 strain MO45 at NCBI (AB012956.1). These two sequences were 99.47% identical and had the same length (1,518 bp), suggesting that this gene may be a homolog of wbfA that was inserted into the NAG strain. Two mutations (nucleotide substitutions) were also observed in pglJ, which encodes a glycosyltransferase responsible for linking sugars to the O chain (27; Fig. S4). These mutations were T to C nonsynonymous transition mutations at positions 1031 (leucine amino acid replaced with to serine) and 1085 (isoleucine amino acid replaced with to threonine). Taken together, this series of mutations in the O-antigen-determining region of this NAG strain might be responsible for the loss of agglutination.
DISCUSSION
Here, we characterize an emerging, nonagglutinating (NAG) isolate (3528-08), which is likely related to the known toxigenic V. cholerae serogroup O141 from the USGC. Due to mutations in the O-antigen-determining region, this strain does not produce O141 O antigen. Multiple lines of evidence support this hypothesis. First, the NAG strain formed a monophyletic cluster with toxigenic O141 strains in a core genome SNP phylogeny and was most similar to an O141 strain from 2016 (2016V-1085). Second, the NAG strain formed a monophyletic cluster with USGC serogroups O1, O75, and O141 in a phylogeny generated from ctxAB gene sequences. Furthermore, the NAG strain contained the USGC ctxAB allele, which was distinct from other ctxAB alleles of pandemic O1, O37, and O27 strains. The NAG strain also shared the same ctxA and ctxB alleles as the USGC strains. Third, the NAG strain clustered with a single O141 strain (2016V-1085) in a phylogeny of tcpA, as in the core genome SNP phylogeny. It should be noted that while the NAG strain and 2016V-1085 were closely related, the genomes were not identical (ANI, 99.90%; 96.06% aligned bases and 55 SNPs different). The tcpA sequences of the NAG strain and a single O141 strain (2016V-1085) were similar to the tcpA of V. mimicus, suggesting that recombination and horizontal gene transfer with another Vibrio strain might have occurred. Fourth, the NAG strain’s O-antigen sequence was determined to be structurally and genetically similar to the reference O-antigen sequence of the toxigenic O141 strain 3566-08 based on gene structure or synteny, as well as nucleotide composition. Multiple mutations were found that could affect the ability of the NAG strain to agglutinate with O141 antisera, including mutations in the pglJ gene (a glycosyltransferase) (27), which might affect glycosylation and thus its ability to react with antisera, and a 483-bp insertion, which was identified as the wbfA domain from V. cholerae O139.
Potential limitations of this analysis include the limited lots or manufacturers of antisera: agglutination was only evaluated using O1, O139, O75, and O141 antisera, leaving the potential that this atypical strain was one of the other V. cholerae serogroups. Additionally, long-read sequencing data were not available for a closely related strain of the NAG, which could have also served as a reference for the O-antigen region (O141 strain 2016V-1085). Because of this, the O-antigen region assembly was not complete in 2016V-1085; however, O-antigen regions were identified by performing a command line nucleotide BLAST search against the complete reference O-antigen region in O141 strain 3566-08. The O-antigen segments identified in O141 strain 2016V-1085 (37,546 bp total) were less related to the NAG than the O141 reference O-antigen region from 3566-08 (87.04% aligned bases and 99.99% identical, compared to 94.90% aligned bases and 99.98% identical, respectively), supporting the use of the O141 3566-08 genome as a reference for comparing the O-antigen region in the NAG strain.
Multiple lines of evidence support the placement of this strain in serogroup O141, based on sequence-based phylogenies, characterization of virulence genes, and characterization of the O-antigen regions; in each of these analyses, the NAG strain closely resembled O141. Serogroup conversion due to the recombination of DNA in the O-antigen region has been reported in V. cholerae (10, 11). Therefore, here, we hypothesize that mutations and recombination events might be responsible for the emergence of this nonagglutinating (NAG) isolate (3528-08), which most likely represents the toxigenic O141 lineage of V. cholerae from the U.S. Gulf Coast. While the evolutionary changes in pandemic and epidemic potential V. cholerae strains have been well documented in areas where cholera is endemic, continued surveillance of strains—especially in regions such as the USGC—remains important for monitoring the potential emergence of novel pathogenic strains. Further characterization of both toxigenic and nontoxigenic serogroups of V. cholerae is also of public health importance, as some clinical opportunistic cases are attributed to nontoxigenic V. cholerae and other Vibrio species—especially in those who are immunocompromised or have underlying risk factors, such as liver disease, cancer, diabetes, HIV, or thalassemia.
MATERIALS AND METHODS
Genome acquisition.
The genome sequence for the V. cholerae NAG strain 3528-08 and representative whole-genome sequences (WGS) for the relevant V. cholerae serogroups (namely, O1, O139, O75, and O141) were downloaded from GenBank or sequenced and assembled in-house as described below (Table 1).
An additional O141 strain (2016V-1085) that agglutinated with O141 antisera was included in this analysis. Strain 2016V-1085 was grown at 37°C overnight on Trypticase soy agar supplemented with 2% sheep blood, and genomic DNA was extracted in-house using the Wizard genomic DNA purification kit (Promega, Madison, WI). Genomic DNA was sheared to a mean size of 600 bp using an LE220 focused ultrasonicator (Covaris Inc., Woburn, MA). DNA fragments were cleaned using AMPure beads (Beckman Coulter Inc., Indianapolis, IN) and used to prepare dual-indexed sequencing libraries using NEBNext Ultra DNA library prep reagents (New England Biolabs Inc., Ipswich, MA) and barcoding indices synthesized in the CDC Biotechnology Core Facility. The libraries were analyzed for size and concentration, normalized, pooled, and denatured for loading onto flow cells for cluster generation. WGS was performed on the HiSeq platform using 2 × 251-bp chemistry (Illumina, San Diego, CA) (see “Data availability” and Table 1).
Genome assembly, annotation, and WGS phylogeny.
The WGS for 2016V-1085 was assembled using SPAdes version 3.14.0 with the option –careful; short contigs (<500 bp) were filtered from the genome using the CG-Pipeline script run_assembly_filterContigs.pl (28, 29). The remaining genomes were previously downloaded from GenBank (NCBI). Genome sequences were annotated using Prokka version 1.14.5 (30). A core genome SNP phylogeny was constructed from representative genome sequences using Parsnp version 1.5.2 from the Harvest suite and visualized using MEGA7 (31, 32) (Table 1).
Virulence gene characterization.
Representative sequences for cholera toxin genes (ctxAB), as well as sequences for the A subunit of the toxin coregulated pilus (tcpA) gene, were obtained from GenBank or from the WGS using Prokka annotations (Table 1). The ctxAB and tcpA sequences were aligned using MEGA7 and the MUSCLE algorithm; an alignment was exported for ctxAB and visualized using Microsoft Word (32). A phylogeny of ctxAB was generated using MEGA7 (32); the evolutionary history was inferred using the neighbor-joining method, and the Kimura 2-parameter method was used for estimating evolutionary distances. The robustness of the tree was tested with 500 replications of the interior branch test. Single nucleotide polymorphism (SNP) distances were calculated for ctxAB sequences using the number of differences option in MEGA7 (32). A phylogeny of tcpA was also generated using MEGA7; the evolutionary history was inferred using the maximum likelihood method based on the general time-reversible model with 500 bootstrap replications (32).
O-antigen characterization.
The O-antigen regions for the NAG strain and the four main reference serogroups (O1, O139, O75, and O141) were extracted from representative genome sequences by first locating the left junction gene gmhD (GenBank accession number X90547.1) and the right junction gene rjg (AF090685.1) with BLASTn using NCBI BLAST+ version 2.9.0 (33); the regions were then extracted using the custom script extractSequence.pl (https://github.com/lskatz/lskScripts). For genome assemblies with incomplete O-antigen regions (e.g., 2016V-1085), BLASTn was used to identify the contigs with O-antigen sequences using a complete reference O-antigen region from O141 strain 3566-08, and the resulting sequences were extracted using coordinates identified in the BLAST analysis.
The NAG O-antigen region was then compared to the extracted reference O-antigen regions for the four serogroups using dnadiff from the MUMmer version 3.23 package, and the resulting out.report and out.snps files were analyzed to interpret the relatedness of the O antigens using the average nucleotide identity (ANI) and to look for mutations in the O-antigen-determining region (34). Mauve version 2.4.0 was used to align the genome sequences with annotations to visualize the O-antigen regions and features (35). Mauve was also used to verify that the O-antigen regions being visualized were flanked by gmhD and rjg, which are O-antigen junction genes that define the region (18, 19, 35). Select O-antigen gene sequences with predicted mutations or significant SNPs were aligned using MEGA7 and the MUSCLE algorithm; the alignments were exported and visualized using Microsoft Word (32).
Data availability.
The whole-genome sequence data and relevant gene sequence data used to conduct this research are publicly available at NCBI under the accession numbers provided in Table 1. The 2016V-1085 WGS is available under BioProject accession number PRJNA266293.
ACKNOWLEDGMENTS
The Centers for Disease Control and Prevention provided support for this project. The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
We thank the CDC Biotechnology Core Facility for their technical assistance.
Footnotes
Supplemental material is available online only.
Contributor Information
Christine C. Lee, Email: clee13@cdc.gov.
Paul M. Luethy, University of Maryland School of Medicine
REFERENCES
- 1.Baker-Austin C, Oliver JD, Alam M, Ali A, Waldor MK, Qadri F, Martinez-Urtaza J. 2018. Vibrio spp. infections. Nat Rev Dis Primers 4:8. doi: 10.1038/s41572-018-0005-8. [DOI] [PubMed] [Google Scholar]
- 2.Trinanes J, Martinez-Urtaza J. 2021. Future scenarios of risk of Vibrio infections in a warming planet: a global mapping study. Lancet Planet Health 5:e426–e435. doi: 10.1016/S2542-5196(21)00169-8. [DOI] [PubMed] [Google Scholar]
- 3.Crump JA, Bopp CA, Greene KD, Kubota KA, Middendorf RL, Wells JG, Mintz ED. 2003. Toxigenic Vibrio cholerae serogroup O141-associated cholera-like diarrhea and bloodstream infection in the United States. J Infect Dis 187:866–868. doi: 10.1086/368330. [DOI] [PubMed] [Google Scholar]
- 4.Safa A, Nair GB, Kong RY. 2010. Evolution of new variants of Vibrio cholerae O1. Trends Microbiol 18:46–54. doi: 10.1016/j.tim.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 5.Hu D, Liu B, Feng L, Ding P, Guo X, Wang M, Cao B, Reeves PR, Wang L. 2016. Origins of the current seventh cholera pandemic. Proc Natl Acad Sci USA 113:E7730–E7739. doi: 10.1073/pnas.1608732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cholera Working Group. 1993. Large epidemic of cholera-like disease in Bangladesh caused by Vibrio cholerae O139 synonym Bengal. Lancet 342:387–390. doi: 10.1016/0140-6736(93)92811-7. [DOI] [PubMed] [Google Scholar]
- 7.Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, Takeda Y. 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 8.Faruque SM, Sack DA, Sack RB, Colwell RR, Takeda Y, Nair GB. 2003. Emergence and evolution of Vibrio cholerae O139. Proc Natl Acad Sci USA 100:1304–1309. doi: 10.1073/pnas.0337468100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Comstock LE, Johnson JA, Michalski JM, Morris JG, Jr, Kaper JB. 1996. Cloning and sequence of a region encoding a surface polysaccharide of Vibrio cholerae O139 and characterization of the insertion site in the chromosome of Vibrio cholerae O1. Mol Microbiol 19:815–826. doi: 10.1046/j.1365-2958.1996.407928.x. [DOI] [PubMed] [Google Scholar]
- 10.Li M, Shimada T, Morris JG, Jr, Sulakvelidze A, Sozhamannan S. 2002. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect Immun 70:2441–2453. doi: 10.1128/IAI.70.5.2441-2453.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blokesch M, Schoolnik GK. 2007. Serogroup conversion of Vibrio cholerae in aquatic reservoirs. PLoS Pathog 3:e81. doi: 10.1371/journal.ppat.0030081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramamurthy T, Mutreja A, Weill FX, Das B, Ghosh A, Nair GB. 2019. Corrigendum: revisiting the global epidemiology of cholera in conjunction with the genomics of Vibrio cholerae. Front Public Health 7:237. doi: 10.3389/fpubh.2019.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farmer JJ, Janda JM, Brenner FW, Cameron DN, Birkhead KM. 2005. Genus Vibrio, p 494–546. In Brenner DJ, Krieg NR, Staley JT (ed), Bergey’s manual of systematic bacteriology, 2nd ed. Springer, New York, NY. [Google Scholar]
- 14.Huang J, Zhu Y, Wen H, Zhang J, Huang S, Niu J, Li Q. 2009. Quadruplex real-time PCR assay for detection and identification of Vibrio cholerae O1 and O139 strains and determination of their toxigenic potential. Appl Environ Microbiol 75:6981–6985. doi: 10.1128/AEM.00517-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keasler SP, Hall RH. 1993. Detecting and biotyping Vibrio cholera O1 with multiplex polymerase chain reaction. Lancet 341:1661. doi: 10.1016/0140-6736(93)90792-f. [DOI] [PubMed] [Google Scholar]
- 16.Dumontier S, Berche P. 1998. Vibrio cholerae O22 might be a putative source of exogenous DNA resulting in the emergence of the new strain of Vibrio cholerae O139. FEMS Microbiol Lett 164:91–98. doi: 10.1111/j.1574-6968.1998.tb13072.x. [DOI] [PubMed] [Google Scholar]
- 17.Stroeher UH, Jedani KE, Manning PA. 1998. Genetic organization of the regions associated with surface polysaccharide synthesis in Vibrio cholerae O1, O139 and Vibrio anguillarum O1 and O2: a review. Gene 223:269–282. doi: 10.1016/s0378-1119(98)00407-7. [DOI] [PubMed] [Google Scholar]
- 18.Ali A. 2008. Genetics of O-antigens, capsules and the rugose variant of Vibrio cholerae: genomics and molecular biology, p 101–122. In Faruque SM, Nair GB (ed), Vibrio cholerae. Caister Academic Press, Poole, UK. [Google Scholar]
- 19.Aydanian A, Tang L, Morris JG, Johnson JA, Stine OC. 2011. Genetic diversity of O-antigen biosynthesis regions in Vibrio cholerae. Appl Environ Microbiol 77:2247–2253. doi: 10.1128/AEM.01663-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dalsgaard A, Serichantalergs O, Forslund A, Lin W, Mekalanos J, Mintz E, Shimada T, Wells JG. 2001. Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J Clin Microbiol 39:4086–4092. doi: 10.1128/JCM.39.11.4086-4092.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haley BJ, Choi SY, Grim CJ, Onifade TJ, Cinar HN, Tall BD, Taviani E, Hasan NA, Abdullah AH, Carter L, Sahu SN, Kothary MH, Chen A, Baker R, Hutchinson R, Blackmore C, Cebula TA, Huq A, Colwell RR. 2014. Genomic and phenotypic characterization of Vibrio cholerae non-O1 isolates from a US Gulf Coast cholera outbreak. PLoS One 9:e86264. doi: 10.1371/journal.pone.0086264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hounmanou YMG, Sit B, Fakoya B, Waldor MK, Dalsgaard A. 2022. Genomic and phenotypic insights for toxigenic clinical Vibrio cholerae O141. Emerg Infect Dis 28:617–624. doi: 10.3201/eid2803.210715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blake PA. 1994. Endemic cholera in Australia and the United States: chapter 20. In Wachsmuth IK, Blake PA, Olsvik Ø (ed), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington, DC. [Google Scholar]
- 24.Tobin-D'Angelo M, Smith AR, Bulens SN, Thomas S, Hodel M, Izumiya H, Arakawa E, Morita M, Watanabe H, Marin C, Parsons MB, Greene K, Cooper K, Haydel D, Bopp C, Yu P, Mintz E. 2008. Severe diarrhea caused by cholera toxin-producing Vibrio cholerae serogroup O75 infections acquired in the southeastern United States. Clin Infect Dis 47:1035–1040. doi: 10.1086/591973. [DOI] [PubMed] [Google Scholar]
- 25.Crowe SJ, Newton AE, Gould LH, Parsons MB, Stroika S, Bopp CA, Freeman M, Greene K, Mahon BE. 2016. Vibriosis, not cholera: toxigenic Vibrio cholerae non-O1, non-O139 infections in the United States, 1984–2014. Epidemiol Infect 144:3335–3341. doi: 10.1017/S0950268816001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Igere BE, Okoh AI, Nwodo UU. 2022. Atypical and dual biotypes variant of virulent SA-NAG-Vibrio cholerae: an evidence of emerging/evolving patho-significant strain in municipal domestic water sources. Ann Microbiol 72:3. doi: 10.1186/s13213-021-01661-5. [DOI] [Google Scholar]
- 27.Szymanski CM, Wren BW. 2005. Protein glycosylation in bacterial mucosal pathogens. Nat Rev Microbiol 3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- 28.Kislyuk AO, Katz LS, Agrawal S, Hagen MS, Conley AB, Jayaraman P, Nelakuditi V, Humphrey JC, Sammons SA, Govil D, Mair RD, Tatti KM, Tondella ML, Harcourt BH, Mayer LW, Jordan IK. 2010. A computational genomics pipeline for prokaryotic sequencing projects. Bioinformatics 26:1819–1826. doi: 10.1093/bioinformatics/btq284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 31.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darling AC, Mau B, Blattner FR, Perna NT. 2004. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00182-23-s0001.pdf, PDF file, 0.6 MB (566.6KB, pdf)
Data Availability Statement
The whole-genome sequence data and relevant gene sequence data used to conduct this research are publicly available at NCBI under the accession numbers provided in Table 1. The 2016V-1085 WGS is available under BioProject accession number PRJNA266293.