ABSTRACT
Bleaching is one of the most relevant factors implicated in the integrity of coral reef ecosystems, with the increasing frequency and intensity of damaging events representing a serious threat to reef biodiversity. Here, we analyzed changes in coral-associated bacteria from three types of non-bleached and bleached scleractinian corals (Acropora digitifera, Galaxea fascicularis, and Porites pukoensis) in Hainan Luhuitou peninsula coastal areas. The community structure of symbiotic bacteria differed significantly among the three apparently healthy corals. The bleached corals had higher bacterial alpha diversity and some specific bacteria genera, including Ruegeria, Methyloceanibacter, Filomicrobium, Halioglobus, Rubripirellula, Rhodopirellula, Silicimonas, Blastopirellula, Sva0996 marine group, Woeseia, and unclassified_c_Gammaproteobacteria, were consistently increased in bleached groups. Network analysis revealed significantly different degrees of modularity between bleached and non-bleached groups at the bacterial genus level, and a higher proportion of links was dominated by positive co-occurrences. Functional prediction analysis illustrated that coral-associated bacteria remained relatively consistent in the bleached and non-bleached groups. Structure equation modeling revealed that the bacterial community diversity and function were directly influenced by host and environment factors. These findings suggested that coral-associated bacterial responses to bleaching occur in a host-dependent manner, informing novel strategies for restoring coral and aiding adaption to bleaching stress.
IMPORTANCE Accumulating evidence indicates that coral-associated bacteria play an important role in the health of holobionts. However, the variability of the symbiotic bacterial community structure among coral species with different coral health statuses remains largely unknown. Here, we investigated three apparent non-bleached (healthy) and bleached coral species (sampled in situ), involving related symbiotic bacterial profiles, including composition, alpha diversity, network relationship, and potential function. Structural equation modeling analysis was used to analyze the relationship between coral status and abiotic and biotic factors. The bacterial community structure of different groups was shown to exhibit host-specific traits. Both host and environmental impacts had primary effects on coral-associated microbial communities. Future studies are needed to identify the mechanisms that mediate divergent microbial consortia.
KEYWORDS: coral health status, bleaching, bacterial communities, network relationship, host-dependent profile
INTRODUCTION
The coral holobiont harbors a diverse assemblage of microorganisms (1). Taking the symbiotic microalgae as an example, impressive progress has revealed endosymbiotic dinoflagellates functional roles and photosynthate mechanisms, which transfer the majority of the coral’s energy requirements (2, 3), promoting calcification rates and coral growth, and in return, the hosts supply an acidic microenvironment and inorganic nutrients for the symbiotic dinoflagellates (4–6). Meanwhile, microbial symbionts, such as bacteria, facilitate host nutrient cycling and support more protective gene functions (7). Symbiotic partnerships, which provide niches for a wide range of diverse coral reef organisms, are a primary source of structure and nutrition in oceans.
The health of the coral holobiont hinges on multiple external and internal factors. The former includes temperature changes (8), eutrophication (9, 10), ocean acidification (11), and other anthropogenic environmental stresses (such as plastic pollution and overfishing) (12). All of these factors can destabilize the symbiosis between the host and their symbionts, leading to massive mortality and bleaching events (13). The latter emphasizes that the potential contributions of the mutualistic symbiosis among coral symbionts, members of the host, dinoflagellate symbiont Symbiodinum, and microorganisms are essential to host-symbiont homeostasis and integrity (14). However, the disruption of the symbiotic relationship can lead to the destruction of coral tissue and the escape of zooxanthellae symbionts from coral hosts, resulting in bleaching (15, 16). Moreover, extensive research into coral-associated bacteria has increased attention (17–20). Coral-associated bacteria play a role in coral health (21, 22). Different putative or opportunistic pathogens have been identified and are shown to cause coral disease and bleaching (18, 23–25). Independent experimental studies have also defined coral bleaching as a bacterial dysbiosis within the coral holobiont (26).
Nonetheless, regarding potential adaptation to stress (27), studies have indicated that the bleaching tolerance of some corals explains their ability to resist coral bleaching and substantially increases their survival (28, 29). Bleaching tolerance is a location-and species-specific trait, which is associated with the composition of the bacterial community (29, 30). There is also increasing evidence to suggest that the microbiome can regulate host resistance to thermal-induced bleaching (31, 32). Nonetheless, comparatively little is known about the structural and functional characteristics of symbiotic bacterial communities with different coral health statuses. Comparative studies on shifts in the microbial community in individuals of different coral species with non-bleached and bleached are also lacking.
Subsequently, to comprehensively address how coral bacteria differ by coral type and health status, we collected non-bleached and bleached coral colony fragments from one reef site (Hainan Luhuitou) to ensure the same level of environmental stress. We hypothesized that the bacterial community structure would differ among the different coral types and that bleached individuals would also exhibit obvious changes. To test this, three apparent non-bleached (healthy) and bleached coral species (Acropora digitifera, Galaxea fascicularis, and Porites pukoensis) were sampled in situ, and the related symbiotic bacterial profiles were investigated using the 16S rRNA gene pyrosequencing, including the composition, biodiversity, network relationship, as well as the potential function. This work aims to provide a basis for understanding the ecological relationships between corals and their symbiotic communities under bleached and non-bleached. Meanwhile, the knowledge gained from this study may pave the way for new therapeutic approaches and bioremediation techniques to combat coral bleaching.
RESULTS
Overall taxonomic characteristics and alpha diversity.
In this study, we applied high-throughput 16S rRNA gene sequencing methods to simultaneously assess the diversity of prokaryotic microbial communities from the two phenotypes (non-bleached and bleached) of three species of stony corals and seawater. In total, 2,563,874 16S rRNA sequences were recorded after quality control and sequence filtering, which were clustered into 13,357 operational taxonomic units (OTUs) at 97% similarity level, respectively. Before alpha and beta analysis and to equalize sequencing depth, OTU tables were rarefied to a minimum number per sample (22,282 quality sequences per bacterial sample).
In the taxonomically assigned OTUs from the prokaryotic data set, the Good’s coverage values ranged from 99.28% to 99.99%, revealing that sequencing depth was sufficient to capture the majority of the bacterial community (see Table S1 in the supplemental material). Principle seawater had higher alpha diversity for bacteria than coral samples. The Shannon and Chao1 indices, respectively, represent the diversity and richness of communities, and the OTU richness (Chao 1) of seawater was highest across all samples. In the coral samples, bleached coral species were significantly higher in richness and evenness (Chao 1) compared to non-bleached, except for A. digitifera (Fig. 1A). The same difference was observed regarding Shannon indices with increased diversity in bleached A. digitifera, G. fascicularis, and P. pukoensis (Fig. 1B; Table S1). This shows that bleached G. fascicularis and P. pukoensis samples possessed diversity that was more approximate to seawater samples (Fig. 1B). A Student’s t test also showed differences in alpha diversity between the three coral species under non-bleached and bleached conditions (shown in Table S2).
FIG 1.
Compassion of alpha diversity indices based on 16S rRNA genes sequencing (A, B). Asterisks indicate significant differences (*, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001) based on the Student's t test. N, non-bleached corals; B, bleached corals.
Prokaryotic community compositions and features.
The taxonomically assigned OTUs from all samples involved 384 archaeal OTUs and 11,655 bacterial OTUs after the removal of unclassified at the domain, belonged to 66 phyla, 183 classes, 456 orders, 790 families, 1,644 genera, and 3,283 species. The relative abundance at the phylum level presented in Fig. 2A revealed that the dominant phylum was Proteobacteria (near 12.9 to 74.8%) in the coral and seawater groups but not in the non-bleached P. pukoensis (Bacteroidota was the most dominant phylum at 52.7% average relative abundance). After Proteobacteria and Bacteroidota were the predominant phylums, followed by Firmicutes, Cyanobacteria, and Actinobacteriota. Among them, Fimicutes showed a relative abundance ranging from 8.3% to 19.7% among the coral species and an abundance of only 0.7% in seawater. Unlike in Fimicutes, Cyanobateria showed a different distribution pattern in coral and seawater. Cyanobateria was dominant (26.2%) in seawater with only 0.3% to 8.4% in coral samples. Actinobacteriota did not exhibit significant differences in the relative abundance in all tested samples, which ranged from 2.4 to 9.4%.
FIG 2.
Composition of microbial prokaryotes present in three non-bleached (N) and bleached (B) coral species and seawater. Bar plots show the taxonomic classification of OTU in each sample at the phyla (A), class (B), order (C), and genus (D) levels (other: relative abundance <2%).
At the class level (Fig. 2B), there were remarkable differences between the seawater and coral samples. While the former was dominated by Cyanobacteria (26.1%) and Alphaproteobacteria (19.4%), the latter was dominated by Gammaproteobacteria (16.7%) and Bacteriodia (17.1%). The most obvious change in bleached A. digitifera was increased Bacilli and Actinobacteria. The dominant class also changed markedly in G. fascicularis and P. pukoensis. For G. fascicularis, the dominant class in non-bleached coral was Gammaproteobacteria (23.8%) and Alphaproteobacteria (25.0%) in bleached coral. Furthermore, additional obvious changes in bleached coral were a reduction in Bacilli and Actinobacteria. For P. pukoensis, the Chlorobia and Bacteroidiav were decreased by more than 3-fold and Alphaproteobacteria increased almost 6-fold compared with the non-bleached samples.
At the order level, the differences between species were further amplified. The relative abundance of the three analyzed coral species fluctuated by coral type (Fig. 2C). The dominant order-species in the healthy A. digitifera, G. fascicularis, and P. pukoensis were Rickettsiales, Chloroplast, and Chlorobiales, respectively. In bleached conditions, these members were reduced in relative abundance including Burkholderiales and Chloroplast (Fig. 2C). Some enriched species were also increased in bleached coral, including Lachnospirales and Enterobacterales in A. digitifera, Rhodobacterales, Rhizobiales, and Peptostreptococcales-Tissierellales in G. fascicularis, as well as Rhodobacterales, Rhizobiales, and Flavobacteriales in P. pukoensis. These were upregulated 3.5 to 19.8-fold. Moreover, at the genera level prokaryotic microbiome composition was further observed to be specific in each non-bleached and bleached coral species (Fig. 2D). Analyzing the relative abundance of genus bar plots, which demonstrated a common trend, the proportion of the most dominant genus (such as norank_f_Mitochondria, norank_f_norank_o_Chloroplast, Prosthecochloris) in bleached coral decreased compared to non-bleached corals.
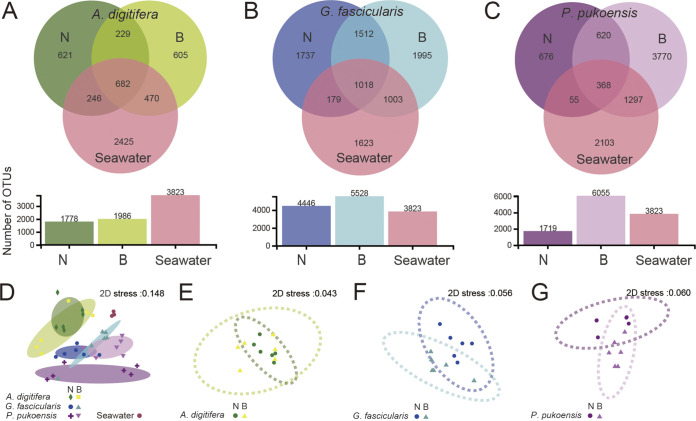
Unique and shared OTUs at different conditions in each coral species and seawater are shown in the Venn diagrams in Fig. 3. OTU numbers found uniquely in non-bleached and bleached P. pukoensis were 676 OTUs and 3,770 OTUs (Fig. 3C), respectively. The number of shared OTUs between was markedly higher in bleached corals and seawater than in non-bleached corals among the three corals. Based on this, we speculated that the symbiotic bacteria of bleached coral were more likely to be affected by the surrounding seawater, showing stronger convergence. In contrast, the healthy corals were more capable of shaping their own bacterial community composition, showing stronger heterogeneity.
FIG 3.
(A to C) Venn diagram for the common and unique number of OTUs among non-bleached and bleached corals and seawater among A. digitifera (A), G. fascicularis (B), and P. pukoensis (C). Prokaryotic community structure and relative dispersion of the non-bleached and bleached coral species and seawater samples, analyzed with nonmetric multidimensional scaling (NMDS) plots using Bray-Curtis dissimilarity. (D) Three coral species (non-bleached and bleached) and seawater. (E to G) A. digitifera, G. fascicularis, and P. pukoensis in non-bleached and bleached model, respectively. N, non-bleached; B, bleached.
Bacterial beta diversity was quantified via nonmetric multidimensional scaling analysis (NMDS) analysis based on Bray-Cutis dissimilarities, which further revealed the community separation among coral and seawater OTUs. The bacterial communities of coral formed distinct clusters with significant differences being found at the OTU level (ANOMIS test) (Fig. 3D). The bleached G. fascicularis (Fig. 3F; stress: 0.056; P value = 0.023) and P. pukoensis (Fig. 3G; stress: 0.060; P value = 0.015) bacterial communities demonstrated a significant separation from their healthy communities, apart from the A. digitifera samples in Fig. 3E (stress: 0.043; P value = 0.353). The bacteria structure of G. fascicularis and P. pukoensis was significantly different in non-bleached and bleached samples. This may be because bleaching facilitates high biodiversity of bacteria due to the dramatic migration from surrounding seawater bacteria. This is one source of opportunistic and potentially pathogenic bacteria that cause coral disease and bleaching. Unlike G. fascicularis and P. pukoensis, the difference in the diversity of A. digitifera was not obvious, indicating that its symbiotic bacteria have a certain buffer or tolerance when facing stress.
Specific bacteria between corals.
Despite the relatively low bacterial abundance at the genus level, this was considered the primary reason for the bacterial shifts in the phenotypic status of the two corals. As shown in Fig. 4, 14 genera were significantly higher in bleached samples [unclassified_f_Rhodobacteraceae (P = 0.023), unclassified_c_Alphaproteobacteria (P = 0.002), Ruegeria (P = 0.008), Methyloceanibacter (P = 0.004), Filomicrobium (P = 0.002), Halioglobus (P = 0.013), Rubripirellula (P = 0.008), Rhodopirellula (P = 0.010), Silicimonas (P = 0.299), Blastopirellula (P = 0.015), Sva0996 marine group (P = 0.002), Woeseia (P = 0.0005), Pir4 lineage (P = 0.001), and unclassified_c_Gammaproteobacteria (P = 0.001)] compared with non-bleached. These results indicated that those genera may be associated with bleached coral and bacterial dysbiosis.
FIG 4.
Distribution of the relative abundance of the specific bacteria between corals.
The response species also have a strong host correlation. Taxa such as norank_Mitochondria (P = 0.0001) and Halodesulfovibrio (P = 0.047) were particularly enriched in A. digitifera, and Prosthecochloris (P = 0.036) was highly enriched in P. pukoensis. In addition, norank_Chlroplast (P = 0.023) and Ralstonia (P = 0.028) were observed higher in A. digitifera and G. fascicularis. The relative abundances of these genera in bleached coral samples were significantly lower than in non-bleached samples, which may suggest their vital function in non-bleached corals. Moreover, the shifts of the individual bacterial genera were dependent not only on their coral species but also on host conditions.
Bacterial profiles also varied from coral species to species. Probiotics and opportunistic and infectious pathogens play an important role in affecting coral health by impacting the functioning of the coral holobiont to suit the prevailing environmental conditions (18, 33). Here, we observed that the relative abundance of some probiotics (Erythrobacter and Ruegeria) increased in the bleached corals. In comparison, the relative abundance of other probiotics (Halomonas, Endozoicomonas) and pathogens (Vibrio) varied by sample.
Interactions between non-bleached and bleached coral bacteria.
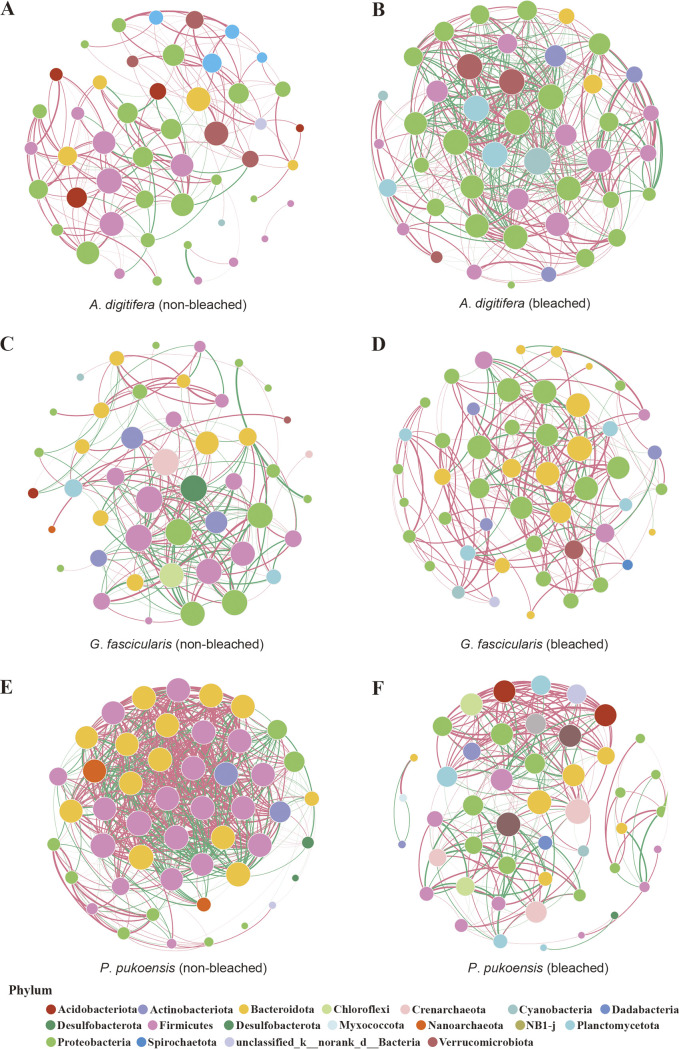
To investigate how interactions with bacterial genera and the complex patterns of interrelationships shifted with bleaching, the topological properties of networks were calculated to reconstruct a genus co-occurrence network in each coral species. The bacteria from each non-bleached and bleached coral species and their individual networks are presented in Fig. 5. In the resulting networks, each node was a bacterial genus with green edges indicating a positive connection (co-occurrent) and pink edges denoting a negative connection (mutual exclusion). The network from bleached A. digitifera (45 nodes and 444 edges) was more complex than non-bleached (48 nodes and 189 edges). In contrast, the bacterial network of the bleached P. pukoensis group (48 nodes and 632 edges) had more edges than the non-bleached group (24 nodes and 258 edges). There was no change between non-bleached (46 nodes and 217 edges) and bleached G. fascicularis (49 nodes and 202 edges).
FIG 5.
Co-occurrence network of coral bacterial communities at the genus level between two conditions. (A, C, E) Non-bleached. (B, D, F) Bleached. The size of the nodes is proportional to the average relative abundance of the genera. Edge thickness linking two nodes is proportional to the absolute value of the Spearman’s correlation coefficients. Node colors are used to differentiate between phyla. Green edges indicate the co-occurrence of connected nodes, pink edges denote mutual exclusivity, and the width of edges reflects the strength of interaction.
Networks for the six groups were constructed with a frequency of >50%. According to the calculated properties, the coral-associated bacteria had higher positive edge correlations among all non-bleached and bleached samples. Stable coexistence in a community represents high modularity (34). We found that community networks decreased in modularity as the negative connection increased (Table S3). The ratio of negative edges in the non-bleached A. digitifera network was approximately 1/3 (positive edges = 149; negative edges = 40) with a modularity of 0.435, whereas the ratio of negative edges in the bleached group was 1/2 (positive edges = 233; negative edges = 211) with a modularity of 0.228. The ratio of negative connections of G. fascicularis and P. pukoensis were both 1/3 in the bleached groups with a higher modularity (modularity index was 0.491 and 0.410, respectively) compared to the non-bleached groups (the ratio of negative edges were both 1/2 and modularity index was 0.302 and 0.102, respectively), indicating that bacterial communities with high modularity exist with more positive connections, which are dominant in more stable networks, rather than in negative associations between taxa.
In bleached samples, Proteobacteria was the dominant phylum with the most connections with other phylum and was increasingly common compared to non-bleached coral networks, especially in A. digitifera (Fig. 5A and B) and G. fascicularis (Fig. 5C and D) groups. In non-bleached P. pukoensis (Fig. 5E), Firmicutes, and Bacteroidota were the most dominant hubs with a high level of centrality degrees, abundance of these two hubs prevented the bleached P. pukoensis group (Fig. 5F) from forming a modularized community network (modularity index = 0.410; Table S3). Similarly, compared with non-bleached G. fascicularis, the modularity index of 0.491 in the bleached group, suggested that the network had a modular structure. Conversely, the lowest network density of 0.168 was observed in non-bleached A. digitifera, which had a higher modularity index (modularity index = 0.435; Table S3) than the bleached group. The co-occurrence network showed that bacteria in different corals were significantly different depending on host species and status, they were more modular, and they were dominated by positive co-occurrences.
Functional profiling of bacterial communities in corals.
To study the functional alternative of bacterial communities in the non-bleached and bleached coral species, the PICRUSt2 bioinformatics tool was used to predict the functional potential and profiles of the coral-associated bacteria based on 16S amplicon sequencing profiles. A level 3 of Kyoto Encyclopedia of Gene and Genomes (KEGG) categories was obtained between the coral samples as shown in Fig. S1. Different patterns in the frequencies of these categories by two conditions of coral species were identified, exhibiting a high frequency of metabolic pathways, biosynthesis of secondary metabolites, microbial metabolism in diverse environments, amino acid biosynthesis, carbon metabolism, and nucleotide metabolism (pyrimidine metabolism). However, other categories such as energy metabolism (methane metabolism, carbon fixation pathway in prokaryotes, and sulfur metabolism), amino acid metabolism (arginine and proline metabolism and arginine biosynthesis lysine degradation), metabolism of cofactors and vitamins (pantothenate and CoA biosynthesis, nicotinate, and nicotinamide metabolism) were less frequent in all groups. G. fascicularis and P. pukoensis had a more similar trend involved in the relative frequencies of most categories and were increased in the bleached groups except for A. digitifera samples.
Linkage of abiotic and biotic to coral health status.
Structural equation modeling (SEM) was performed to test the relationships or directional influences among coral, microbiome, and environment (Fig. 6). Our results showed that coral status was directly influenced by seawater physicochemical parameters (salinity, temperature, Fe3+, silicate, ammonium, nitrite, nitrate, phosphate, pH, and ChI a) (P < 0.001) and the predictor microbial phenotypes (P < 0.001) (Table S4). In addition, the host had a strong direct negative effect on microbial diversity (P < 0.001) and a strong direct positive effect on microbial community function (P < 0.001), which induced several effects (Fig. 6; Table S5). These results suggested that the host was the dominant determinant of the progress of microbial diversity and function.
FIG 6.

Structure equation modeling showing the relative influence between seawater abiotic and biotic coral associated with microbial factors on coral status. Response variables are represented as solid and predictor variables are represented as bold-dashed boxes. Red and blue arrows indicate positive and negative relationships, respectively. Proportion of variance explained (R2) is represented by orange numbers for each response variable. Significant paths imply a causal influence of different variables with arrow widths proportional to the degree of influence, as shown in the bottom right. Arrow numbers represent the standardized path strength.
DISCUSSION
Coral ecosystems are facing a dramatic increase in the frequency and intensity of stressors (35), such as heat waves (36, 37), EI Nino-Southern oscillation, and La Nina events (28, 38–42). Moreover, outbreaks of crown-of-thorns starfish, typhoons, diseases, and strong solar radiation also can cause coral bleaching (43, 44). The ways in which coral holobionts respond to bleaching events are often complex and spatiotemporally heterogeneous because of different stress tolerances of coral species, namely, induced coral acclimation and extrinsic environmental factors.
In this study, the bacterial communities associated with three bleached and non-bleached coral species, A. digitifera, G. fascicularis, and P. pukoensis, were characterized. From the overall results, we found that different coral species possessed distinct bacterial communities through NMDS plots and composition analysis, suggesting that coral taxonomy is the major determinant for the symbiotic bacterial structure and composition variations. As shown previously, the top bacterial phyla in all the coral samples were Proteobacteria, Cyanobacteria, Actinobacteriota, and Firmicutes (44). Despite relatively stable coral-bacteria associations at the phylum level, the composition of the three target coral species had different diversities and relative abundance when analyzed at lower taxonomic levels (such as class and order levels), further suggesting that coral bacteria are diverse and host specific.
There are three main reasons for the host dependence of coral symbiotic bacteria. First, sensitivity to environmental pressures induces bleaching. Different coral species displayed variant degrees of bleaching and disease susceptibility as reported in several studies (45–47). There is mounting evidence to suggest that Porites and Galaxea were relatively resistant to heat tolerance and undergo lower mortality and bleaching rates compared to Acropora (40, 48, 49). In this study, we observed a higher number of OTUs in P. pukoensis and G. fascicularis compared to A. digitifera, suggesting that P. pukoensis and G. fascicularis exhibit higher bleaching thresholds. The host symbiont can obtain or lose high-species variability, depending on the species’ own capacity to resist stressor intensity, symbiotic partnership, and energy metabolism level (48, 49). Differences in the bacteria community structure of coral species are also related to the host’s physiological characteristics of the test species. Among the three corals, A. digitifera and G. fascicularis have a larger specific surface area, whereas P. pukoensis has a relatively smaller specific surface area. Therefore, to some extent, the bacteria of P. pukoensis had different characteristics than the other two coral species in this study.
The second reason may be lifestyle. Fast-growing branching coral taxa, such as Acropora, are normally highly susceptible to thermal stress (50, 51). The contrasting response of bleaching susceptibility also has been observed in other studies (50, 52, 53). Taking Acropora as an example, its individual life history traits are possible factors shaping its rapid adaptation potential (50). Most autotrophic coral holobionts with low heterotrophic capacity (A. digitifera) reflect a specialized strategy at low microbiome flexibility for microbiome adaptation to environmental change. Accordingly, A. digitifera harbors a highly flexible microbiome, which is thought to contribute to coral holobiont plasticity and adaptation.
The third possible reason is the ecological adaptation strategy. Changes in bacterial community structure tend to show the potential mechanism with metabolic flexibility and functional redundancy (54). The fluctuations of A. digitifera and P. pukoensis bacteria composition influence the host health outcome. These findings suggest that coral species exhibit different degrees of flexibility in holobiont structure and composition.
Normal coral bacteria play an important role in maintaining the host health. Thus, it is important to understand the dynamics of the coral-associated bacterial community structures in healthy and bleached corals. An increase in bacterial diversity often accompanies the holobiont response to stress as a result of emerging opportunistic taxa that are otherwise absent or suppressed (55). Here, the order of alpha diversity in bleached samples was G. fascicularis, P. pukoensis, and A. digitifera. Venn diagrams showed that the majority of OTUs were recorded in bleached groups, which have a large percentage of OTUs shared with environmental samples (seawater) compared to non-bleached groups. A possible reason may be that the symbiont provides an entry niche for opportunistic taxa from seawater that can be able to colonize bleached coral hosts during bleaching events. Non-bleached coral communities can escape thermal bleaching in the same biotope and might benefit from their stable immune systems (56). Under this situation, alpha diversity was increased in the bleached samples. However, our results differ from those obtained by Pollock et al. (57) who conclude that bleached corals had half the number of healthy coral OTUs. This may be because coral immune capacity may be suppressed in bleached coral holobionts with specific microorganism recruitment from the environment, which affects a coral’s susceptibility to disease (57).
Rhodobacteraceae, Flavobacteriaceae, and Synechococcaeae were significantly increased in bleached coral samples and were the dominant taxa in seawater. Based on their thermal sensitivity, these genera have the potential as biomarkers of high temperature in coral ecosystems (58). Meanwhile, shifts in coral-associated bacterial communities may contribute more to the resilience and survival of coral holobionts to environmental disturbances (59, 60). The beneficial or probiotic bacteria of corals, such as Erythrobacter, Endozoicomonas, and Ruegeria, occupied a relatively higher proportion in bleached coral and seem to be a defensive mechanism, whereas pathogens (for example, Vibrio) were significantly increased in the bleached G. fascicularis group (61). In contrast, the relative abundance of Vibrio in A. digitifera and P. pukoensis had lower abundance and there was no significant difference between the bleached and non-bleached corals. One possible explanation could be that A. digitifera and P. pukoensis have stronger resistance/resilience than G. fascicularis, which reduced colonization by the opportunistic Vibrio from surrounding water or sediments. Previous reports have also shown that a lower abundance of Vibrio was found in bleached Diploria strigosa and Siderastrea sidereal, both Caribbean coral species, while a higher abundance was observed in bleached Pavona duerdeni and Porites lutea of the Pacific coral reef (62). Similarly, consistent with our study, Krishnaswamy et al. (63) did not observe significant pathogens like Vibrio in the diseased Porites lutea and suggested that “unique species owning a unique microbial composition” contribute to it (63). Changes in the abundance of Vibrio were shown to vary across both coral species and geographic locations. In contrast, Endozoicomonas remained stable even under the conditions of coral bleaching and nonbleaching proportionally among A. digitifera and P. pukoensis, except for G. fascicularis. Most studies reported that the relative abundance of Endozoicomonas was reduced with stress, disease, and bleached corals (64). As Ritchie (65) initially discovered, potential pathogens are inhibited by antibiotics secreted by beneficial bacteria associated with corals. Bacterial consortia in corals were manipulated with the increasing of probiotic bacteria, which can help to partially mitigate coral bleaching and alleviate pathogens (32) and benefit coral heat resistance (66).
The topological properties of networks have been increasingly used to define potential bacterial interconnections and community stability (67, 68). Co-occurrence analyses show that non-bleached and bleached corals had a higher proportion of positive than negative correlations. Among the bacterial community networks, except the A. digitifera, both bleached P. pukoensis and bleached G. fascicularis groups had evident modular architecture, indicating that the bacterial groups showed a reduced influence to pressure. The higher modularity in healthy corals suggests that their bacteria may have more stable synergistic interactions that allow them to respond to external perturbations more efficiently (69). However, the relatively low complexity of the network is regarded as remarkably vulnerable to environmental interference (70). Proteobacteria, Bacteroidetes, and Firmicutes are the most prevalent taxa among coral associated with diverse bacterial assemblages (71), which were shown to have a strong correlation with other taxa in this study. Among these three taxa, the hub Proteobacteria were increased in the bleached group, suggesting that they play an important role in constructed microbiota interaction networks when coral holobionts become bleached. Taken together, the complexity of the co-occurrence networks of bacteria in different status was significantly different, indicating that the efficiency of the co-occurrence network hinge on host-holobiont-specific responses to disturbances.
To study the potential roles of microbiota in non-bleached and bleached coral groups, we performed PICRUSt pathway analysis. The microbiota was more involved in basic metabolic pathways, including biosynthesis of secondary metabolites, microbial metabolism in diverse environments, biosynthesis of amino acids, and carbon metabolism. KEGG analysis results showed that bacterial consortia between non-bleached and bleached corals had no obvious differences. Similar results have shown that bacterial metabiotic functions remain conserved after suffering from bleaching events (72, 73). However, opposite results have been found in coral-associated microbiomes that undergo eutrophication stress (74). Structural changes and functional varieties may have different sensitivities to stress. In fact, Krishnaswamy et al. (63) and Pogoreutz et al. (75) speculated that the functional redundancy of coral microorganisms contributes to the limited differences between the healthy and diseased groups. Based on this, we propose that the changed microbial communities associated with the relatively stable functions of coral are explained by the decoupling effect of taxonomy and function (76). This suggests a possible evolutionary strategy that allows bacteria to undergo microevolution to adapt to stressors in their specific host, maintaining stable balance and well-being for the bacterial communities, which might provide stable situations and opportunities for bleached coral recovery. The potential association between bacteria-environment-coral was established by using the SEM analysis. Coral host and seawater physicochemical parameters had a relatively significant impact on bacterial profiles (community and function). Bacterial phenotypes drive a negative influence on bleached corals and a positive influence on non-bleached corals.
Conclusion.
The current study characterized the bacterial composition of three coral species under non-bleached and bleached conditions. The different coral species maintained relatively conserved bacterial populations, indicating that coral symbiotic bacteria have certain host specificity. However, the community structure for a given host can vary by health status. Higher alpha biodiversity was observed in bleached coral samples, and some indicator bacteria acted as potential biomarkers in bleached individuals. Additionally, the bleached and non-bleached groups had significantly different network modularity profiles. After multiparameter coupling analysis by SEM, the results (Fig. 6) revealed that the coral host has the main effect on microbial diversity and function. This study improves our understanding of the bacterial composition of different coral species exhibiting host dependence and how these symbionts contribute to the coral holobiont’s environmental adaptability. However, it should be noted that the current research is only based on limited samples taken at one time point. In the future, we need to conduct tracking and collect a larger number of sequential samples in the natural environment. In addition, the metabolic potential of this work is based on functional prediction. Metagenomic analysis is needed to clarify the exact functional mechanism of coral symbiotic bacteria.
MATERIALS AND METHODS
Sampling and processing.
Three types of corals (Acropora digitifera, Galaxea fascicularis, and Porites pukoensis) fragments (approximately 5 to 15 cm each) and seawater were collected on April 4, 2021, in the South China Sea off the coast of Luhuitou, Sanya, Hainan (18°12’N, 109°28’E) (Fig. 7), when the seawater temperature was 24.49 ± 0.29°C at salinity of 32.2 ± 0.2‰, pH of 8.16 ± 0.03. The three coral species were identified based on their characteristic morphological features via an online website http://www.coralsoftheworld.org/page/home/. From this, fragments were collected from each colony of both visually non-bleached and bleached tissue using sterilized scissors and forceps, separately into 100-mL sterile plastic bottles. Surrounding seawater was filtered (3 L) onto 0.2-μm MCE membrane filters (47-mm diameter; Millipore, Billerica, USA) in duplicate. Each filter (n = 3) was placed into aseptic centrifugal tubes (50 mL) with a screw cap. All samples were immediately frozen in a solid carbon dioxide and were transported to the laboratory, at which point they were store at −80°C until required.
FIG 7.
(A) Map of the South China Sea study site. (B) Corals and seawater were sampled at one coral reef site off Luhuitou peninsula coastal, which is marked with a circle. (C) The site is located approximately 200 m northeast of Luhuitou (image of sampling site). (D) In situ monitoring of coral bleaching through fixed undersea observation equipment at the Hainan Island. (E) Representative real-time captured image of bleached corallite.
DNA isolation and sequencing of microbial symbionts.
Per coral samples (6 × non-bleached A. digitifera, 6 × bleached A. digitifera, 6 × non-bleached G. fascicularis, 6 × bleached G. fascicularis, 6 × non-bleached P. pukoensis, and 6 × bleached P. pukoensis), a fragment of 1 g was weighed and was immediately ground to a fine powder using liquid nitrogen and a mortar and pestle. DNA was extracted from 36 coral samples and 3 surrounding seawater samples using the Dneasy PowerSoil Pro kit (Qiagen, Hilden, Germany) according to manufacturer’s protocols. DNA integrity was verified through 1% (vol/vol) agarose gel electrophoresis, DNA concentration was examined by using Qubit 2.0. DNA purity was measured using Nanodrop (optical density at 260 nm/optical density at 280 nm ratio) and stored at −20°C before use.
In total 39 DNA samples were sent to Majorbio Bio-Parm Technology Co., Ltd. (Shanghai, China) on solid carbon dioxide for next-generation shotgun sequencing. The V4 hypervariable region of bacterial and archaeal 16S rRNA gene was amplified with primers 515FomdF (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806RmodR (5′-GGACTACNVGGGTWTCTAAT-3′) (77, 78). PCR amplification profile was performed using the following program: initial denaturation of 15 min at 94°C, 27 cycles of denaturation at 95°C for 40 s, annealing at 55°C for 40 s, and amplification at 72°C for 40 s, a final elongation step of 10 min at 72°C. All 16S PCR products were purified by E.Z.N.A. Gel Extraction kit (Omega, USA) and sent for sequencing (Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China) on the Illumina Hiseq2500 platform.
Read processing, assembly, and data analysis.
All the obtained raw reads were merged, trimmed, quality filtered, and clustered into OTUs at the ≥97% similarity level using Uparse pipeline (79) (version 7.0.1090), which were aligned and compared with the SILVA database v138 with a bootstrap confidence cutoff of 70%. To reduce the nonsystematic variation from amplification bias and sequencing errors, the sequences were rarefied to normalization to the minimum number of reads per sample before calculating the respective alpha diversity (Shannon indices) and richness (Chao1 indices) by Student’s t test; the Good’s coverage was calculated by mothur (80) (version 1.30.2) to evaluate OTU similarity level (97%). Venn diagram and bar plot analysis were conducted using the “vegan” package in R (version 3.3.1) for Linux to visualize shifts in the percentage of community abundances on different levels among different samples. NMDS using based on the Bray-Curtis dissimilarity index of the beta diversity of bacterial community among coral and seawater samples was potted in QIIME (version 1.9.1) software.
Network feature, functional profile, and statistical analysis.
To describe the topological properties of the co-occurrence patterns among microbiome between non-bleached and bleached coral samples, the opened source platform Gephi (version 0.9.2) was used to visualize entity relationship with the network. We selected bacterial genera with a frequency of ≥50% with the Spearman coefficients absolute values ≥0.5 and false discovery rate corrected P value of ≤0.05. Each node represents one genus, and its color indicates one phylum. Edges make significant correlations (negative and positive relationships) among nodes. The clustering coefficient, closeness values, modularity, and eigenvectors were calculated to identify the topological properties of six different co-occurrence networks.
Analysis of variance and Tukey’s honestly significant difference post hoc analyses identified differentially expressed probiotic microbes between non-bleached and bleached coral samples. The relative abundance of bacterial genera among the six groups (coral samples) was statistically analyzed using the Kruskal-Wallis H test followed by the Scheffé’s post hoc test.
To assess the potential functional profiles of the microbiome between the non-bleached and bleached coral samples, PICRUSt2 was used to predict metagenomes from 16S data in the bacterial microbiome. A heatmap of the imputed relative abundances of KEGG pathways in each coral sample predicted by the KEGG catalog (81). In addition, an SEM was analyzed via the online website https://spssau.com (82) to identify the relationship among seawater physicochemical parameters, microbial diversity, predictor factors (microbial community function and phenotype), and coral status. As the abiotic and biotic drivers drive the coral symbiont’s health, the priori model was set as follows: (1) the host, microbial diversity, and community function are driven by abiotic factors; (2) the biotic factors are driving the host status; and (3) the host drives the predictor factors.
Data availability.
All sequencing runs and data filtering were conducted at Majorbio, Inc. (Shanghai, China), and sequence data utilized for this project were deposited in the NCBI Short Read Archive database and can be accessed with the BioProject (PRJNA922724).
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (NSCF; 42106096), S&T Projects of Shenzhen Science and Technology Innovation Committee (KCXFZ20211020165547011, RCJC20200714114433069, and JCYJ20200109142818589), and Hainan Key Research and Development Program (ZDYF2021SHFZ059), China Postdoctoral Science Foundation (2021M691804), as well as the Cross Research and Innovation Funding of Tsinghua Shenzhen International Graduates School (SIGS; JC2022004).
Footnotes
Supplemental material is available online only.
Contributor Information
Guofu Chen, Email: chenguofu@hitwh.edu.cn.
Jin Zhou, Email: zhou.jin@sz.tsinghua.edu.cn.
Jianjun Wang, Nanjing Institute of Geography and Limnology Chinese Academy of Sciences.
REFERENCES
- 1.Rohwer F, Seguritan V, Azam F, Knowlton N. 2002. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser 243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- 2.Falkowski P, Dubinsky Z, Muscatine L, Porter J. 1984. Light and the bioenergetics of a symbiotic coral. Bioscience 34:705–709. doi: 10.2307/1309663. [DOI] [Google Scholar]
- 3.Muscatine L, McCloskey LR, Marian RE. 1981. Estimating the daily contribution of carbon from zooxanthellae to coral animal respiration. Limnol Oceanogr 26:601–611. doi: 10.4319/lo.1981.26.4.0601. [DOI] [Google Scholar]
- 4.Burriesci MR, Raab TK, Pringle JR. 2012. Evidence that glucose is the major transferred metabolite in dinoflagellate-cnidarian symbiosis. J Exp Biol 215:3467–3477. doi: 10.1242/jeb.070946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rahav O, Dubinsky Z, Achituv Y, Falkowski P. 1989. Ammonium metabolism in the zooxanthellate coral, Stylophora pistillata. Proc R Soc Ser B Biol Sci 236:325–337. doi: 10.1098/rspb.1989.0026. [DOI] [Google Scholar]
- 6.Barott KL, Venn AA, Perez SO, Tambutte S, Tresguerres M. 2015. Coral host cells acidify symbiotic algal microenvironment to promote photosynthesis. Proc Natl Acad Sci USA 112:607–612. doi: 10.1073/pnas.1413483112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lima LFO, Alker AT, Papudeshi B, Morris MM, Edwards RA, de Putron SJ, Dinsdale EA. 2022. Coral and seawater metagenomes reveal key microbial functions to coral health and ecosystem functioning shaped at reef scale. Microb Ecol doi: 10.1007/s00248-022-02094-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leggat W, Seneca F, Wasmund K, Ukani L, Yellowlees D, Ainsworth TD. 2011. Differential responses of the coral host and their algal symbiont to thermal stress. PLoS One 6:e26687. doi: 10.1371/journal.pone.0026687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malone TC, Newton A. 2020. The globalization of cultural eutrophication in the coastal ocean: causes and consequences. Front Mar Sci 7:1–30. doi: 10.3389/fmars.2020.00670.32802822 [DOI] [Google Scholar]
- 10.Vega-Thurber RL, Burkepile DE, Fuchs C, Shantz AA, McMinds R, Zaneveld JR. 2014. Chronic nutrient enrichment increases prevalence and severity of coral disease and bleaching. Glob Chang Biol 20:544–554. doi: 10.1111/gcb.12450. [DOI] [PubMed] [Google Scholar]
- 11.Kaniewska P, Chan CK, Kline D, Ling EY, Rosic N, Edwards D, Hoegh-Guldberg O, Dove S. 2015. Transcriptomic changes in coral holobionts provide insights into physiological challenges of future climate and ocean change. PLoS One 10:e0139223. doi: 10.1371/journal.pone.0139223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ying L, Sinutok S, Pramneechote P, Aiyarak P, Ralph PJ, Chotikarn P. 2021. Physiological responses of Pocillopora acuta and Porites lutea under plastic and fishing net stress. Front Mar Sci 8:712214. doi: 10.3389/fmars.2021.712214. [DOI] [Google Scholar]
- 13.Faizal A, Amri K, Rani C, Nessa MN, Jompa J. 2020. Dynamic model; the effects of eutrophication and sedimentation on the degradation of coral reefs in Spermonde Archipelago, Indonesia. IOP Conf Ser Earth Environ Sci 564:e012084. doi: 10.1088/1755-1315/564/1/012084. [DOI] [Google Scholar]
- 14.Ziegler M, Seneca FO, Yum LK, Palumbi SR, Voolstra CR. 2017. Bacterial community dynamics are linked to patterns of coral heat tolerance. Nat Commun 8:14213. doi: 10.1038/ncomms14213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jokiel PL, Coles SL. 1977. Effects of temperature on the mortality and growth of Hawaiian reef corals. Mar Biol 43:201–208. doi: 10.1007/BF00402312. [DOI] [Google Scholar]
- 16.Gates RD, Baghdasarian G, Muscatine L. 1992. Temperature stress causes host cell detachment in symbiotic cnidarians: implications for coral bleaching. Biol Bull 182:324–332. doi: 10.2307/1542252. [DOI] [PubMed] [Google Scholar]
- 17.Meyer JL, Gunasekera SP, Scott RM, Paul VJ, Teplitski M. 2016. Microbiome shifts and the inhibition of quorum sensing by Black Band Disease cyanobacteria. ISME J 10:1204–1216. doi: 10.1038/ismej.2015.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munn CB. 2015. The role of Vibrios in diseases of corals. Microbiol Spectr 3:1–12. doi: 10.1128/microbiolspec.VE-0006-2014. [DOI] [PubMed] [Google Scholar]
- 19.Thatcher C, Hoj L, Bourne DG. 2022. Probiotics for coral aquaculture: challenges and considerations. Curr Opin Biotechnol 73:380–386. doi: 10.1016/j.copbio.2021.09.009. [DOI] [PubMed] [Google Scholar]
- 20.Ravindran C, Raveendran HP, Irudayarajan L. 2022. Ciliated protozoan occurrence and association in the pathogenesis of coral disease. Microb Pathog 162:105211. doi: 10.1016/j.micpath.2021.105211. [DOI] [PubMed] [Google Scholar]
- 21.Tout J, Jeffries TC, Webster NS, Stocker R, Ralph PJ, Seymour JR. 2014. Variability in microbial community composition and function between different niches within a coral reef. Microb Ecol 67:540–552. doi: 10.1007/s00248-013-0362-5. [DOI] [PubMed] [Google Scholar]
- 22.Rosenberg E, Koren O, Reshef L, Efrony R, Zilber-Rosenberg I. 2007. The role of microorganisms in coral health, disease and evolution. Nat Rev Microbiol 5:355–362. doi: 10.1038/nrmicro1635. [DOI] [PubMed] [Google Scholar]
- 23.Voss JD, Mills DK, Myers JL, Remily ER, Richardson LL. 2007. Black band disease microbial community variation on corals in three regions of the wider Caribbean. Microb Ecol 54:730–739. doi: 10.1007/s00248-007-9234-1. [DOI] [PubMed] [Google Scholar]
- 24.Séré MG, Tortosa P, Chabanet P, Quod JP, Sweet MJ, Schleyer MH. 2015. Identification of a bacterial pathogen associated with Porites white patch syndrome in the Western Indian Ocean. Mol Ecol 24:4570–4581. doi: 10.1111/mec.13326. [DOI] [PubMed] [Google Scholar]
- 25.Sweet MJ, Croquer A, Bythell JC. 2014. Experimental antibiotic treatment identifies potential pathogens of white band disease in the endangered Caribbean coral Acropora cervicornis. Proc Biol Sci 281:20140094. doi: 10.1098/rspb.2014.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boilard A, Dube CE, Gruet C, Merciere A, Hernandez-Agreda A, Derome N. 2020. Defining coral bleaching as a microbial dysbiosis within the coral holobiont. Microorganisms 8:1682. doi: 10.3390/microorganisms8111682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurihara H, Watanabe A, Tsugi A, Mimura I, Hongo C, Kawai T, Reimer JD, Kimoto K, Gouezo M, Golbuu Y. 2021. Potential local adaptation of corals at acidified and warmed Nikko Bay, Palau. Sci Rep 11:11192. doi: 10.1038/s41598-021-90614-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes TP, Anderson KD, Connolly SR, Heron SF, Kerry JT, Lough JM, Baird AH, Baum JK, Berumen ML, Bridge TC, Claar DC, Eakin CM, Gilmour JP, Graham NAJ, Harrison H, Hobbs JPA, Hoey AS, Hoogenboom M, Lowe RJ, McCulloch MT, Pandolfi JM, Pratchett M, Schoepf V, Torda G, Wilson SK. 2018. Spatial and temporal patterns of mass bleaching of corals in the Anthropocene. Science 359:80–83. doi: 10.1126/science.aan8048. [DOI] [PubMed] [Google Scholar]
- 29.Yu XP, Yu KF, Chen B, Liao ZH, Liang JY, Yao QC, Qin ZJ, Wang H, Yu JY. 2021. Different responses of scleractinian coral Acropora pruinosa from Weizhou Island during extreme high temperature events. Coral Reefs 40:1697–1711. doi: 10.1007/s00338-021-02182-y. [DOI] [Google Scholar]
- 30.Yu X, Yu K, Huang W, Liang J, Qin Z, Chen B, Yao Q, Liao Z. 2020. Thermal acclimation increases heat tolerance of the scleractinian coral Acropora pruinosa. Sci Total Environ 733:139319. doi: 10.1016/j.scitotenv.2020.139319. [DOI] [PubMed] [Google Scholar]
- 31.Sweet MJ, Bulling MT. 2017. On the importance of the microbiome and pathobiome in coral health and disease. Front Mar Sci 4:9. doi: 10.3389/fmars.2017.00009. [DOI] [Google Scholar]
- 32.Rosado PM, Leite DCA, Duarte GAS, Chaloub RM, Jospin G, Nunes da Rocha U, Saraiva P, Dini-Andreote F, Eisen JA, Bourne DG, Peixoto RS. 2019. Marine probiotics: increasing coral resistance to bleaching through microbiome manipulation. ISME J 13:921–936. doi: 10.1038/s41396-018-0323-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peixoto RS, Sweet M, Villela HDM, Cardoso P, Thomas T, Voolstra CR, Høj L, Bourne DG. 2021. Coral probiotics: premise, promise, prospects. Annu Rev Anim Biosci 9:265–288. doi: 10.1146/annurev-animal-090120-115444. [DOI] [PubMed] [Google Scholar]
- 34.Stouffer DB, Bascompte J. 2011. Compartmentalization increases food-web persistence. Proc Natl Acad Sci USA 108:3648–3652. doi: 10.1073/pnas.1014353108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Woesik R, Kratochwill C. 2022. A global coral-bleaching database, 1980–2020. Sci Data 9:20. doi: 10.1038/s41597-022-01121-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kennedy J, Blunden J, Alvar-Beltrán J, Kappelle M. 2021. State of the global climate 2020. World Meteorological Organization, Geneva, Switzerland. [Google Scholar]
- 37.Lyu YH, Zhou ZH, Zhang YM, Chen ZQ, Deng W, Shi RG. 2022. The mass coral bleaching event of inshore corals from South China Sea witnessed in 2020: insight into the causes, process and consequence. Coral Reefs 41:1351–1364. doi: 10.1007/s00338-022-02284-1. [DOI] [Google Scholar]
- 38.Claar D, Szostek L, McDevitt-Irwin J, Schanze J, Baum J. 2018. Global patterns and impacts of El Niño events on coral reefs: a meta-analysis. PLoS One 13:e0190957. doi: 10.1371/journal.pone.0190957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun FL, Yang HQ, Wang G, Shi Q. 2020. Combination analysis of metatranscriptome and metagenome reveal the composition and functional response of coral symbionts to bleaching during an El Niño event. Front Microbiol 11:448. doi: 10.3389/fmicb.2020.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cannon SE, Aram E, Beiateuea T, Kiareti A, Peter M, Donner SD. 2021. Coral reefs in the Gilbert Islands of Kiribati: resistance, resilience, and recovery after more than a decade of multiple stressors. PLoS One 16:e0255304. doi: 10.1371/journal.pone.0255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riegl B, Glynn PW, Banks S, Keith I, Rivera F, Vera-Zambrano M, D’Angelo C, Wiedenmann J. 2019. Heat attenuation and nutrient delivery by localized upwelling avoided coral bleaching mortality in northern Galapagos during 2015/2016 ENSO. Coral Reefs 38:773–785. doi: 10.1007/s00338-019-01787-8. [DOI] [Google Scholar]
- 42.Sun F, Yang H, Zhang X, Tan F, Shi Q. 2022. Response characteristics of bacterial communities in multiple coral genera at the early stages of coral bleaching during El Niño. Ecol Indic 144:109569. doi: 10.1016/j.ecolind.2022.109569. [DOI] [Google Scholar]
- 43.Anthony KRN, Kerswell AP. 2007. Coral mortality following extreme low tides and high solar radiation. Mar Biol 151:1623–1631. doi: 10.1007/s00227-006-0573-0. [DOI] [Google Scholar]
- 44.Ohara T, Hoeksema BW, Wee HB, Reimer JD. 2021. Downslope migration of free-living corals (Scleractinia: Fungiidae) in typhoon-exposed reef habitats at Okinawa, Japan. Mar Environ Res 170:105445. doi: 10.1016/j.marenvres.2021.105445. [DOI] [PubMed] [Google Scholar]
- 45.Ernesto W, Aldo C, Isabel U. 2009. Temporal variability and impact of coral diseases and bleaching in La Parguera, Puerto Rico from 2003–2007. Caribb J Sci 45:221–246. doi: 10.18475/cjos.v45i2.a10. [DOI] [Google Scholar]
- 46.Williams L, Smith TB, Burge CA, Brandt ME. 2020. Species-specific susceptibility to white plague disease in three common Caribbean corals. Coral Reefs 39:27–31. doi: 10.1007/s00338-019-01867-9. [DOI] [Google Scholar]
- 47.Calnan JM, Smith TB, Nemeth RS, Kadison E, Blondeau J. 2008. Coral disease prevalence and host susceptibility on mid-depth and deep reefs in the United States Virgin Islands. Int J Trop Biol 56:223–234. [Google Scholar]
- 48.Madeira C, Dias M, Ferreira A, Gouveia R, Cabral H, Diniz MS, Vinagre C. 2022. Does predation exacerbate the risk of endosymbiont loss in heat stressed hermatypic corals? Molecular cues provide insights into species-specific health outcomes in a multi-stressor ocean. Front Physiol 13:801672. doi: 10.3389/fphys.2022.801672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xu HL, Feng BX, Xie MR, Ren YX, Xia JQ, Zhang Y, Wang AM, Li XB. 2020. Physiological characteristics and environment adaptability of reef-building corals at the Wuzhizhou Island of South China Sea. Front Physiol 11:390. doi: 10.3389/fphys.2020.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guest JR, Baird AH, Maynard JA, Muttaqin E, Edwards AJ, Campbell SJ, Yewdall K, Affendi YA, Chou LM. 2012. Contrasting patterns of coral bleaching susceptibility in 2010 suggest an adaptive response to thermal stress. PLoS One 7:e33353. doi: 10.1371/journal.pone.0033353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ong RH, King AJC, Kaandorp JA, Mullins BJ, Caley MJ. 2017. The effect of allometric scaling in coral thermal microenvironments. PLoS One 12:e0184214. doi: 10.1371/journal.pone.0184214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Monroe AA, Ziegler M, Roik A, Röthig T, Hardenstine RS, Emms MA, Jensen T, Voolstra CR, Berumen ML. 2018. In situ observations of coral bleaching in the central Saudi Arabian Red Sea during the 2015/2016 global coral bleaching event. PLoS One 13:e0195814. doi: 10.1371/journal.pone.0195814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pratchett MS, McCowan D, Maynard JA, Heron SF. 2013. Changes in bleaching susceptibility among corals subject to ocean warming and recurrent bleaching in Moorea, French Polynesia. PLoS One 8:e70443. doi: 10.1371/journal.pone.0070443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Avila-Jimenez ML, Burns G, He Z, Zhou J, Hodson A, Avila-Jimenez JL, Pearce D. 2020. Functional associations and resilience in microbial communities. Microorganisms 8:951. doi: 10.3390/microorganisms8060951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McDevitt-Irwin JM, Baum JK, Garren M, Vega Thurber RL. 2017. Responses of coral-associated bacterial communities to local and global stressors. Front Mar Sci 4:1–16. doi: 10.3389/fmars.2017.00262. [DOI] [Google Scholar]
- 56.Bosch TCG. 2013. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu Rev Microbiol 67:499–518. doi: 10.1146/annurev-micro-092412-155626. [DOI] [PubMed] [Google Scholar]
- 57.Pollock FJ, Lamb JB, van de Water J, Smith HA, Schaffelke B, Willis BL, Bourne DG. 2019. Reduced diversity and stability of coral-associated bacterial communities and suppressed immune function precedes disease onset in corals. R Soc Open Sci 6:190355. doi: 10.1098/rsos.190355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Laas P, Ugarelli K, Absten M, Boyer B, Briceno H, Stingl U. 2021. Composition of prokaryotic and eukaryotic microbial communities in waters around the Florida Reef Tract. Microorganisms 9:1120. doi: 10.3390/microorganisms9061120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kanisan DP, Quek ZBR, Oh RM, Afiq-Rosli L, Lee JN, Huang D, Wainwright BJ. 2023. Diversity and distribution of microbial communities associated with reef corals of the Malay Peninsula. Microb Ecol 85:37–48. doi: 10.1007/s00248-022-01958-1. [DOI] [PubMed] [Google Scholar]
- 60.Wada N, Pollock FJ, Willis BL, Ainsworth T, Mano N, Bourne DG. 2016. In situ visualization of bacterial populations in coral tissues: pitfalls and solutions. PeerJ 4:e2424. doi: 10.7717/peerj.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hou J, Xu T, Su D, Wu Y, Cheng L, Wang J, Zhou Z, Wang Y. 2018. RNA-Seq reveals extensive transcriptional response to heat stress in the stony coral Galaxea fascicularis. Front Genet 9:37. doi: 10.3389/fgene.2018.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kellogg C, Piceno Y, Tom L, DeSantis T, Gray M, Zawada D, Andersen G. 2013. Comparing bacterial community composition between healthy and white plague-like disease states in orbicella annularis using phyloChip™ G3 microarrays. PLoS One 8:e79801. doi: 10.1371/journal.pone.0079801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krishnaswamy VG, Mani K, Senthil Kumar P, Rangasamy G, Sridharan R, Rethnaraj C, Amirtha Ganesh SS, Kalidas S, Palanisamy V, Chellama NJ, Chowdula S, Parthasarathy V, Rajendran S. 2023. Prevalence of differential microbiome in healthy, diseased and nipped colonies of corals, Porites lutea in the Gulf of Kachchh, north-west coast of India. Environ Res 216:114622. doi: 10.1016/j.envres.2022.114622. [DOI] [PubMed] [Google Scholar]
- 64.Gignoux-Wolfsohn SA, Aronson FM, Vollmer SV. 2017. Complex interactions between potentially pathogenic, opportunistic, and resident bacteria emerge during infection on a reef-building coral. FEMS Microbiol Ecol 93:fix080. doi: 10.1093/femsec/fix080. [DOI] [PubMed] [Google Scholar]
- 65.Ritchie KB. 2006. Regulation of microbial populations by coral surface mucus and mucus-associated bacteria. Mar Ecol Prog Ser 322:1–14. doi: 10.3354/meps322001. [DOI] [Google Scholar]
- 66.Doering T, Wall M, Putchim L, Rattanawongwan T, Schroeder R, Hentschel U, Roik A. 2021. Towards enhancing coral heat tolerance: a “microbiome transplantation” treatment using inoculations of homogenized coral tissues. Microbiome 9:102. doi: 10.1186/s40168-021-01053-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Vries FT, Griffiths RI, Bailey M, Craig H, Girlanda M, Gweon HS, Hallin S, Kaisermann A, Keith AM, Kretzschmar M, Lemanceau P, Lumini E, Mason KE, Oliver A, Ostle N, Prosser JI, Thion C, Thomson B, Bardgett RD. 2018. Soil bacterial networks are less stable under drought than fungal networks. Nat Commun 9:3033. doi: 10.1038/s41467-018-05516-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hernandez DJ, David AS, Menges ES, Searcy CA, Afkhami ME. 2021. Environmental stress destabilizes microbial networks. ISME J 15:1722–1734. doi: 10.1038/s41396-020-00882-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu Y, Xiao G, Zhou J, Wang Y, Wang Z, Kurths J, Schellnhuber HJ. 2016. System crash as dynamics of complex networks. Proc Natl Acad Sci USA 113:11726–11731. doi: 10.1073/pnas.1612094113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Banerjee S, Walder F, Büchi L, Meyer M, Held AY, Gattinger A, Keller T, Charles R, van der Heijden MGA. 2019. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J 13:1722–1736. doi: 10.1038/s41396-019-0383-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huggett MJ, Apprill A. 2019. Coral microbiome database: integration of sequences reveals high diversity and relatedness of coral-associated microbes. Environ Microbiol Rep 11:372–385. doi: 10.1111/1758-2229.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Badhai J, Ghosh TS, Das SK. 2016. Composition and functional characterization of microbiome associated with mucus of the coral Fungia echinata collected from Andaman Sea. Front Microbiol 7:936. doi: 10.3389/fmicb.2016.00936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bulan DE, Wilantho A, Krainara P, Viyakarn V, Chavanich S, Somboonna N. 2018. Spatial and seasonal variability of reef bacterial communities in the upper gulf of Thailand. Front Mar Sci 5:441. doi: 10.3389/fmars.2018.00441. [DOI] [Google Scholar]
- 74.Degregori S, Casey JM, Barber PH. 2021. Nutrient pollution alters the gut microbiome of a territorial reef fish. Mar Pollut Bull 169:112522. doi: 10.1016/j.marpolbul.2021.112522. [DOI] [PubMed] [Google Scholar]
- 75.Pogoreutz C, Radecker N, Cardenas A, Gardes A, Wild C, Voolstra CR. 2018. Dominance of Endozoicomonas bacteria throughout coral bleaching and mortality suggests structural inflexibility of the Pocillopora verrucosa microbiome. Ecol Evol 8:2240–2252. doi: 10.1002/ece3.3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen H, Ma K, Huang Y, Yao Z, Chu C. 2021. Stable soil microbial functional structure responding to biodiversity loss based on metagenomic evidences. Front Microbiol 12:716764. doi: 10.3389/fmicb.2021.716764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sampson TR, Debelius JW, Thron T, Janssen S, Shastri GG, Ilhan ZE, Challis C, Schretter CE, Rocha S, Gradinaru V, Chesselet MF, Keshavarzian A, Shannon KM, Krajmalnik-Brown R, Wittung-Stafshede P, Knight R, Mazmanian SK. 2016. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson' disease. Cell 167:1469–1480.e12. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA, Apprill A, Knight R. 2016. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems 1:e00009. doi: 10.1128/mSystems.00009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 80.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes J, Clemente J, Burkepile D, Vega Thurber R, Knight R, Beiko R, Huttenhower C. 2013. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol 31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li QC, Wang B, Zeng YH, Cai ZH, Zhou J. 2022. The microbial mechanisms of a novel photosensitive material (treated rape pollen) in anti-biofilm process under marine environment. Int J Mol Sci 23:3837. doi: 10.3390/ijms23073837. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and Tables S1 to S5. Download spectrum.04910-22-s0001.pdf, PDF file, 0.6 MB (649KB, pdf)
Data Availability Statement
All sequencing runs and data filtering were conducted at Majorbio, Inc. (Shanghai, China), and sequence data utilized for this project were deposited in the NCBI Short Read Archive database and can be accessed with the BioProject (PRJNA922724).