ABSTRACT
Clostridioides difficile, the primary cause of nosocomial antibiotic-associated diarrhea, has a complex relationship with antibiotics. While the use of broad-spectrum antibiotics disrupts the gut microbiota and increases the risk of C. difficile infection (CDI), antibiotics are also the primary treatment for CDI. However, only a few antibiotics, including vancomycin, fidaxomicin, and rifaximin, are effective against CDI, and resistance to these antibiotics has emerged recently. In this study, we report the identification of two RT027 C. difficile clinical isolates (TGH35 and TGH64) obtained from symptomatic CDI-diagnosed patients in Tampa, Florida in 2016. These two strains showed an elevated minimum inhibitory concentration (MIC) of vancomycin (MIC = 4 μg/mL, compared to the EUCAST breakpoint of 2 μg/mL) and contained a vanRCd 343A>G mutation resulting in a Thr115Ala substitution in the VanRCd response regulator. This mutation was absent in the vancomycin-sensitive control epidemic strain RT027/R20291. TGH64 was also resistant to rifaximin (MIC ≥ 128 μg/mL) and carried the previously reported Arg505Lys and Ile548Met mutations in RpoB. Furthermore, we report on the antimicrobial resistance (AMR) and genomic characterization of additional C. difficile isolates, including RT106/TGH120, RT017/TGH33, and RT017/TGH51, obtained from the same patient sample cohort representing the highly prevalent and regionally distributed C. difficile ribotypes worldwide. Considering that the VanRCd Thr115Ala mutation was also independently reported in seven C. difficile clinical isolates from Texas and Israel in 2019, we recommend epidemiological surveillance to better understand the impact of this mutation on vancomycin resistance.
IMPORTANCE The perpetually evolving antimicrobial resistance (AMR) of C. difficile is an important contributor to its epidemiology and is a grave concern to global public health. This exacerbates the challenge of treating the infections caused by this multidrug-resistant causative organism of potentially life-threatening diarrhea. Further, the novel resistance-determining factors can be transferred between different strains and species of bacteria and cause the spread of AMR in clinical, environmental, and community settings. In this study, we have identified a mutation (vanRCd 343A>G) that causes a Thr115Ala substitution and is linked to an increased MIC of vancomycin in clinical isolates of C. difficile obtained from Florida in 2016. Understanding the mechanisms of AMR, especially those of newly evolving strains, is essential to effectively guide antibiotic stewardship policies to combat antibiotic resistance as well as to discover novel therapeutic targets.
KEYWORDS: Clostridioides difficile, antibiotic resistance, vancomycin, novel resistance-determining mechanisms, reduced susceptibility to vancomycin, single base pair mutation, van operon, vanGCd operon, vanRCd
OBSERVATION
Clostridioides difficile, which is a cause of mild to life-threatening diarrhea that was responsible for 12,800 deaths in the USA in 2017, has been declared by the CDC as one of the top five urgent antibiotic-resistant threats (1). The emergence of new strains, which are often more virulent and antibiotic-resistant, has been associated with the recent increase in the prevalence and severity of CDI (2).
As a life-threatening disease with a high incidence of recurrence and limited effective therapeutic options, the resistance of C. difficile to vancomycin, which is the first line of therapy that is recommended for CDI treatment, is a grave public health concern (2, 3). Vancomycin inhibits bacterial cell wall synthesis by binding with high affinity to the d-Ala-d-Ala C terminus of peptidoglycan precursors, which thereby prevents the addition of late precursors to the nascent peptidoglycan chain (4). A gene cluster called the “van operon” has been described to mediate vancomycin resistance in enterococci (4). The two-component regulatory system of van operons encompasses a sensor histidine kinase (VanS) and a response regulator (VanR). The vanG-type of operon contains the resistance genes VanG (d-Ala-d-Ser ligase), VanXY (a bifunctional D,d-dipeptidase/d,d-carboxypeptidase), and VanT (serine racemase). When vancomycin is sensed by the membrane-bound VanS, it undergoes the ATP-dependent autophosphorylation of a histidine residue. This phosphoryl group is then transferred to the cytoplasmic VanR, which, in turn, transcriptionally activates the expression of downstream resistance genes. VanT converts l-Ser to d-Ser and VanG ligates d-Ala and d-Ser, forming low-affinity precursors and modifying the vancomycin binding target. VanXY hydrolyzes peptidoglycan precursors that end with d-Ala residues, thereby eliminating the high-affinity binding targets of vancomycin (4). However, although a functional vanG operon-like gene cluster called “vanGCd” has been found in about 85% of C. difficile clinical isolates, it was not associated with vancomycin resistance in C. difficile (5).
We previously reported 139 C. difficile clinical isolates that were obtained from symptomatic patients who were diagnosed with CDI in Tampa, FL, USA (6). Based on broth microdilution-based screening for antimicrobial susceptibility, we selected isolates that showed a reduced susceptibility to multiple antibiotics and conducted capillary PCR ribotyping at the Dutch National Reference Laboratory at the Leiden University Medical Center (LUMC), using a standardized protocol (7). Considering that the epidemiology of C. difficile shows distinct geographical distributions, we selected five isolates to represent the most prevalent C. difficile ribotypes that are found in different geographical locations around the world, namely, RT027 (Europe and North America) (2), RT106 (USA) (8), and RT017 (Asia) (9), for further analysis in the present study.
We cultured the five selected C. difficile isolates, namely, TGH35, TGH64, TGH120, TGH33, and TGH51, in brain heart infusion (BHI) broth (Sigma) at 37°C under anerobic conditions. Following genomic DNA extraction with a kit (Qiagen) and whole-genome sequencing using paired-end libraries and an Illumina HiSeq 3000 platform, we de novo assembled reads into contigs using Qiagen CLC Genomics Workbench 11.0.1 (10) (Table S1). After ordering the contigs against the reference CD630 genome (GenBank accession: CP010905.2) using Mauve (v2.4.0) (11) and annotating them using Rapid Annotations using Subsystems Technology (RAST) (12), we compared the whole-genomes of the five isolates with three reference strain genomes, namely, CD630/RT012/Clade1, R20291/RT027/Clade2, and M68/RT017/Clade4 (GenBank accessions: CP010905.2, FN545816.1, and FN668375.1, respectively), and we visualized them using the BLAST Ring Image Generator (BRIG) (13). The housekeeping genes adk, atpA, glyA, sodA, dxr, recA, and tpi were used for web-based multilocus sequence typing (MLST) on a CGE MLST platform (14), and a phylogenetic analysis was conducted using MEGA7 (15). Prophages were predicted using the PHAge Search Tool (PHAST) (16). We searched for plasmids using CGEPlasmidFinder-2.0 (17) and manually evaluated the presence of sequences corresponding to pCD-METRO (GenBank: OM972905.1) (18). We used the comprehensive antibiotic resistance database (CARD) to search for putative AMR genes using the Resistance Gene Identifier (RGI) tool for resistome predictions, based on homology and SNP models (19). The features of the characterized genomes are illustrated in Fig. 1, Table 1, and Table S2.
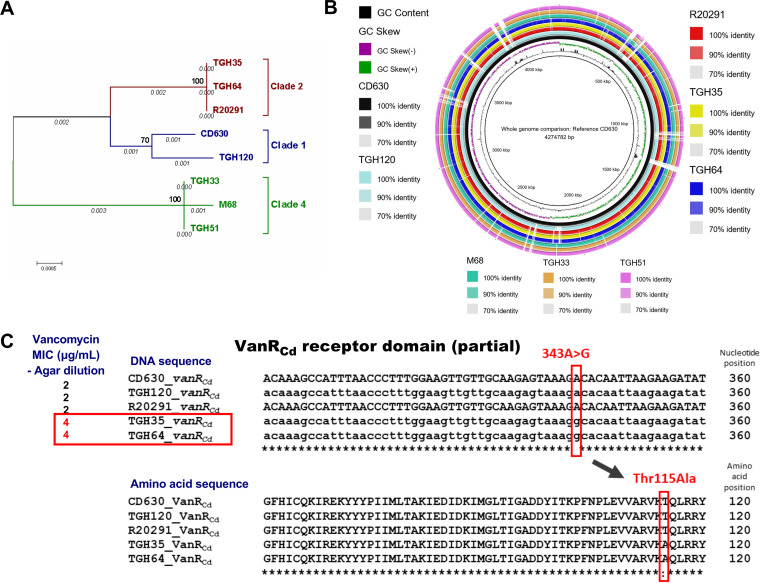
FIG 1.
Genome analysis of the five C. difficile isolates and three control strains CD630, R20291, and M68 (GenBank accessions CP010905.2, FN545816.1, and FN668375.1, respectively). (A) Phylogenetic tree generated with seven housekeeping genes, using MEGA7 software. The seven housekeeping genes were concatenated to generate a supergene alignment using MUSCLE, and a phylogenetic tree was generated using the maximum likelihood method. A general time reversible (GTR) model was used to generate the substitution model using gamma distributed with invariant sites (G+I) and 1,000 bootstrap replications. (B) BLAST ring image generator-based comparison and visualization of the whole-genomes. (C) Multiple sequence alignment (Clustal Omega) of the vanRCd gene (upper panel) and amino acid sequence (lower panel) in comparison with the vancomycin MICs obtained via the agar dilution method (left) of the novel isolates and control strains. The vanGCd operon is absent in the RT017 strains M68, TGH33, and TGH51.
TABLE 1.
Genome features, MICs by agar dilution, AMR determinants, and predicted intact prophages of the isolatesa
| Strain | Ribotype | MLST11 | Clade | MIC (μg/mL) determined by agar dilution (sensitivity criteria, source) |
AMR determinants (CARD database) |
Predicted intact prophages (PHAST) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VAN (S ≤ 2, R > 2; EUCAST ECOFFs) | MTZ (S ≤ 8, I = 16, R ≥ 32; CLSI) | FDX (S < 1, I > 1; literature) | RFX (S ≤ 0.0.004, I = 0.004 to 16, R ≥ 16; literature) | ARO term | AMR gene family | Resistance mechanism | % identity of matching region | % length of reference sequence | |||||
| TGH35 | RT027 | ST1 | 2 | 4 | 0.5 | ≤0.5 | ≤0.125 | APH(3′)-Ia | APH(3′) | Antibiotic inactivation | 100.0 | 30.63 | PHAGE_ Clostr_ phiMMP01_ NC_028883 |
| cdeA | Multidrug and toxic compound extrusion (MATE) transporter | Antibiotic efflux | 99.09 | 100.00 | |||||||||
| APH(3′)-Ia | APH(3′) | Antibiotic inactivation | 96.77 | 67.90 | |||||||||
| vanRG | Glycopeptide resistance gene cluster, vanR | Antibiotic target alteration | 77.45 | 99.15 | |||||||||
| tet(W/N/W) | Tetracycline-resistant ribosomal protection protein | Antibiotic target protection | 69.28 | 100.00 | |||||||||
| vanXYG | Glycopeptide resistance gene cluster, vanXY | Antibiotic target alteration | 58.82 | 105.51 | |||||||||
| TGH64 | RT027 | ST1 | 2 | 4 | 0.25 | ≤0.5 | ≥128 | dfrF | Trimethoprim resistant dihydrofolate reductase dfr | Antibiotic target replacement | 100.0 | 100.00 | PHAGE_ Clostr_ phiMMP01_ NC_028883 |
| cdeA | MATE transporter | Antibiotic efflux | 99.32 | 100.00 | |||||||||
| vanRG | Glycopeptide resistance gene cluster, vanR | Antibiotic target alteration | 77.45 | 99.15 | |||||||||
| vanXYG | Glycopeptide resistance gene cluster, vanXY | Antibiotic target alteration | 58.82 | 105.51 | |||||||||
| tet(W/N/W) | Tetracycline-resistant ribosomal protection protein | Antibiotic target protection | 69.28 | 100.00 | |||||||||
| ermB | Erm 23S ribosomal RNA methyltransferase | Antibiotic target alteration | 98.78 | 98.79 | |||||||||
| TGH120 | RT106 | ST42 | 1 | 2 | 0.5 | ≤0.5 | ≤0.125 | APH(3′)-Ia | APH(3′) | Antibiotic inactivation | 100.0 | 29.89 | - |
| cdeA | MATE transporter | Antibiotic efflux | 99.32 | 100.00 | |||||||||
| vanRG | Glycopeptide resistance gene cluster, vanR | Antibiotic target alteration | 77.87 | 99.15 | |||||||||
| vanXYG | Glycopeptide resistance gene cluster, vanXY | Antibiotic target alteration | 58.82 | 105.51 | |||||||||
| TGH33 | RT017 | ST37 | 4 | 2 | 0.5 | ≤0.5 | ≤0.125 | cdeA | MATE transporter | Antibiotic efflux | 98.41 | 100.00 | PHAGE_ Clostr_ phiCD506_ NC_028838 PHAGE_ Clostr_ phiMMP01_ NC_028883 |
| APH(3′)-Ia | APH(3′) | Antibiotic inactivation | 99.1 | 40.96 | |||||||||
| APH(3′)-Ia | APH(3′) | Antibiotic inactivation | 100.0 | 28.7 | |||||||||
| TGH51 | RT017 | ST37 | 4 | 1 | 0.5 | ≤0.5 | ≤0.125 | cdeA | MATE transporter | Antibiotic efflux | 98.41 | 100.00 | PHAGE_ Clostr_ phiCD506_ NC_028838 PHAGE_ Clostr_ phiCDHM19_ NC_028996 PHAGE_ Clostr_ phiCDHM19_ NC_028996 PHAGE_ Clostr_ phiMMP01_ NC_028883 |
| APH(3′)-Ia | APH(3′) | Antibiotic inactivation | 97.79 | 99.26 | |||||||||
The MICs of vancomycin via the agar dilution method for both control strains RT027/R20291 and RT012/CD630 were 2 μg/mL. The MICs of vancomycin were further determined via BHI broth microdilution: TGH35 (2 μg/mL), TGH64 (4 μg/mL), TGH120 (1 μg/mL), TGH33 (1 μg/mL), TGH51 (1 μg/mL), and R20291 (1 μg/mL). The MICs of vancomycin were also tested via Brucella blood agar dilution and E-tests at LUMC: TGH35 (1 μg/mL), TGH64 (2 μg/mL), TGH120 (0.25 μg/mL), TGH33 (0.5 μg/mL), R20291 (0.5 μg/mL). An EMBL Clustal Omega-based multiple sequence alignment was performed to explore the presence of the mutations that were previously reported to be associated with a reduced susceptibility to vancomycin in in vitro C. difficile strains. Pro108Leu in MurG was detected in all of the isolates in the present study, and G733T in rpoC was detected in none of them. MIC, minimum inhibitory concentration; AMR, antimicrobial resistance; ECOFFs, epidemiological cutoff values; VAN, vancomycin; MTZ, metronidazole; FDX, fidaxomicin; RFX, rifaximin; MLST, multi locus sequence typing; CARD, comprehensive antibiotic resistance database; ARO, Antibiotic resistance ontology. Resistance Gene Identifier criteria used by the CARD database: Strict. Detection criteria: Protein homolog model.
The antibiotic susceptibility testing (AST) of the isolates to the currently clinically used antibiotics for CDI treatment, namely, vancomycin, metronidazole, fidaxomicin, and rifaximin, was conducted using the reference standard agar dilution method, based on the Clinical and Laboratory Standards Institute (CLSI) guidelines M11-A7 (volume 27, no 2, ISBN:1-56238-626-3), using the epidemic RT027/R20291 C. difficile strain as a control with a minimum of three technical replicates in two independent experiments. While all strains were susceptible to metronidazole and fidaxomicin, high resistance to rifaximin (MIC ≥ 128 μg/mL) was detected only in TGH64. Rifaximin, which is an adjunct therapeutic for CDI, acts by binding to the β-subunit of RNA polymerase (RpoB), which thereby inhibits bacterial RNA synthesis (20). Since mutations in the rifamycin resistance-determining region (RRID) of RpoB have been previously associated with rifamycin resistance in C. difficile without imposing a fitness cost (20), we performed a EMBL Clustal Omega-based multiple sequence alignment (MSA) (21) on the rpoB genes in all isolates. We detected two rpoB SNPs in TGH64, namely, 1514G>A and 1644A>G, that result in Arg505Lys and Ile548Met, respectively. This combination of mutations has previously been associated with rifamycin resistance in C. difficile clinical isolates. Structural modeling by Dang et al. suggested that Arg505Lys results in the loss of the energetically favorable pi-stacking interactions between RRID and rifaximin, which inhibits drug binding, thereby leading to resistance (20).
Two isolates that were ribotyped as RT027, namely, TGH35 and TGH64, showed elevated vancomycin MIC (MIC = 4 μg/mL; compared to the EUCAST breakpoint of 2 μg/mL) (Table 1) values. We further assessed the vancomycin susceptibility of these strains via two other methods: the BHI broth microdilution, based on the CLSI guidelines, and Etest, which was conducted at LUMC. In agreement with the results of previous studies, both of these methods gave lower MIC values than were obtained via the reference agar dilution method for several strains (22, 23). However, with all three methods, the elevated vancomycin MIC in TGH35 and TGH64 persisted compared to the other strains.
CARD-based AMR prediction revealed two genes of the vanGCd operon, namely, vanRCd and vanXYCd, in TGH35, TGH64, and TGH120. While vanXYCd was identical in all three isolates, the vanRCd of TGH120 differed from the two RT027 strains (Table 1). Therefore, we performed a MSA of the entire vanGCd operon in these isolates with the reference strains CD630 and R20291. The vanGCd operon is absent in the TGH33, TGH51, and M68 strains. Remarkably, only one nucleotide difference was detected in the whole >6 kb vanGCd operon comparison between the five strains: 343A>G in the vanRCd gene of TGH35 and TGH64 (Fig. 1C). The resulting sense mutation Thr115Ala in the receptor domain of VanRCd likely affects the expression of the downstream resistance genes. We reported this mutation at the ASM Microbe Annual Conference in June of 2019 (24). Later, the VanRCd Thr115Ala mutation was also independently reported in seven C. difficile clinical isolates from the Texas Medical Center, USA as well as in two C. difficile clinical isolates from Israel (25). These isolates showed the constitutive expression of the vanGCd operon and elevated MICs of vancomycin, which could be reversed in the VanRCd-mutant isolates by silencing vanGCd, whereas vanGCd silencing had no effect on the MIC of the control R20291 strain. Shen et al. also used the structural homology modeling of VanRCd to propose that the Thr115Ala substitution provides better stability for its interaction with DNA, thereby enhancing the capability for the transcriptional activation of downstream genes. Since single base pair mutations under selection can quickly lead to the development of resistance, the present work highlights the need for epidemiological surveillance to monitor the prevalence of this mutation, especially in vancomycin-treated patients, to better understand its effects on the resistance to an antibiotic that is currently crucial in the treatment of C. difficile infection.
Data availability.
The whole-genome shotgun projects for the strains TGH35, TGH64, TGH120, TGH33, and TGH51 have been deposited into DDBJ/ENA/GenBank under the accession numbers JAJNGZ000000000, JAJNHA000000000, JAJNHB000000000, JAJNHC000000000, and JAJNHD000000000, respectively.
ACKNOWLEDGMENTS
We acknowledge Bastian V. H. Hornung from the Leiden University Medical Center for his advice and guidance on the genomic assembly and bioinformatic analyses. We also thank the members of the C. difficile research group at USF for their assistance and comments in this project.
This work was supported in part by the National Institutes of Health grants (R01-AI132711, R01-AI149852, and R21-AI159745) to X. Sun.
Footnotes
Supplemental material is available online only.
Contributor Information
Xingmin Sun, Email: sun5@usf.edu.
Yuan Pin Hung, Tainan Hospital, Ministry of Health and Welfare.
REFERENCES
- 1.Eubank TA, Gonzales-Luna AJ, Hurdle JG, Garey KW. 2022. Genetic mechanisms of vancomycin resistance in Clostridioides difficile: a systematic review. Antibiotics (Basel) 11:258. doi: 10.3390/antibiotics11020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He M, Miyajima F, Roberts P, Ellison L, Pickard DJ, Martin MJ, Connor TR, Harris SR, Fairley D, Bamford KB, D'Arc S, Brazier J, Brown D, Coia JE, Douce G, Gerding D, Kim HJ, Koh TH, Kato H, Senoh M, Louie T, Michell S, Butt E, Peacock SJ, Brown NM, Riley T, Songer G, Wilcox M, Pirmohamed M, Kuijper E, Hawkey P, Wren BW, Dougan G, Parkhill J, Lawley TD. 2013. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald LC, Gerding DN, Johnson S, Bakken JS, Carroll KC, Coffin SE, Dubberke ER, Garey KW, Gould CV, Kelly C, Loo V, Shaklee Sammons J, Sandora TJ, Wilcox MH. 2018. Clinical practice guidelines for Clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin Infect Dis 66:e1–e48.4. doi: 10.1093/cid/cix1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Courvalin P. 2006. Vancomycin resistance in gram-positive cocci. Clin Infect Dis 42 Suppl 1:S25–34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 5.Ammam F, Meziane-Cherif D, Mengin-Lecreulx D, Blanot D, Patin D, Boneca IG, Courvalin P, Lambert T, Candela T. 2013. The functional vanGCd cluster of Clostridium difficile does not confer vancomycin resistance. Mol Microbiol 89:612–625. doi: 10.1111/mmi.12299. [DOI] [PubMed] [Google Scholar]
- 6.Peng Z, Addisu A, Alrabaa S, Sun X. 2017. Antibiotic resistance and toxin production of Clostridium difficile isolates from the hospitalized patients in a large hospital in Florida. Front Microbiol 8:2584. doi: 10.3389/fmicb.2017.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fawley WN, Knetsch CW, MacCannell DR, Harmanus C, Du T, Mulvey MR, Paulick A, Anderson L, Kuijper EJ, Wilcox MH. 2015. Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS One 10:e0118150. doi: 10.1371/journal.pone.0118150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kociolek LK, Gerding DN, Hecht DW, Ozer EA. 2018. Comparative genomics analysis of Clostridium difficile epidemic strain DH/NAP11/106. Microbes Infect 20:245–253. doi: 10.1016/j.micinf.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imwattana K, Knight DR, Kullin B, Collins DA, Putsathit P, Kiratisin P, Riley TV. 2019. Clostridium difficile ribotype 017 – characterization, evolution and epidemiology of the dominant strain in Asia. Emerg Microbes Infect 8:796–807. doi: 10.1080/22221751.2019.1621670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.QIAGEN. 2018. CLC Genomics Workbench 11.0.1.
- 11.Rissman AI, Mau B, Biehl BS, Darling AE, Glasner JD, Perna NT. 2009. Reordering contigs of draft genomes using the Mauve aligner. Bioinformatics 25:2071–2073. doi: 10.1093/bioinformatics/btp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alikhan N-F, Petty NK, Zakour NLB, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. 2011. PHAST: a fast phage search tool. Nucleic Acids Res 39:W347–52. doi: 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boekhoud IM, Hornung BVH, Sevilla E, Harmanus C, Bos-Sanders I, Terveer EM, Bolea R, Corver J, Kuijper EJ, Smits WK. 2020. Plasmid-mediated metronidazole resistance in Clostridioides difficile. Nat Commun 11:598. doi: 10.1038/s41467-020-14382-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–d525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dang UT, Zamora I, Hevener KE, Adhikari S, Wu X, Hurdle JG. 2016. Rifamycin resistance in Clostridium difficile is generally associated with a low fitness burden. Antimicrob Agents Chemother 60:5604–5607. doi: 10.1128/AAC.01137-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastey CJ, Dale SE, Nary J, Citron D, Law JH, Roe-Carpenter DE, Chesnel L. 2017. Comparison of Clostridium difficile minimum inhibitory concentrations obtained using agar dilution vs broth microdilution methods. Anaerobe 44:73–77. doi: 10.1016/j.anaerobe.2017.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Poilane I, Cruaud P, Torlotin JC, Collignon A. 2000. Comparison of the E test to the reference agar dilution method for antibiotic susceptibility testing of Clostridium difficile. Clin Microbiol Infect 6:155–156. doi: 10.1046/j.1469-0691.2000.00034-4.x. [DOI] [PubMed] [Google Scholar]
- 24.Wickramage I, Peng Z, Harmanus C, Hornung BVH, Wang S, Kuijper EJ, Smits WK, Sun X. 2019. Genomic analysis of vancomycin-resistant Clostridioides difficile clinical isolates of the epidemic PCR ribotype RT027. ASM Microbe 2019. [Google Scholar]
- 25.Shen WJ, Deshpande A, Hevener KE, Endres BT, Garey KW, Palmer KL, Hurdle JG. 2020. Constitutive expression of the cryptic vanGCd operon promotes vancomycin resistance in Clostridioides difficile clinical isolates. J Antimicrob Chemother 75:859–867. doi: 10.1093/jac/dkz513. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.03777-22-s0001.pdf, PDF file, 0.05 MB (52.4KB, pdf)
Data Availability Statement
The whole-genome shotgun projects for the strains TGH35, TGH64, TGH120, TGH33, and TGH51 have been deposited into DDBJ/ENA/GenBank under the accession numbers JAJNGZ000000000, JAJNHA000000000, JAJNHB000000000, JAJNHC000000000, and JAJNHD000000000, respectively.