ABSTRACT
Escherichia coli isolates from inflammatory bowel disease (IBD) patients are often multidrug resistant, including to streptomycin. Streptomycin resistance (StrR) mutations can alter bacterial behavior, which may influence intestinal disease. We generated a spontaneous StrR strain of the intestinal adherent-invasive E. coli (AIEC) strain NC101. Whole-genome sequencing revealed a single missense mutation in rpsL that commonly confers StrR, rpsL-K43N. StrR NC101 exhibited a striking loss of aggregation and significantly increased motility, behaviors that can impact host-microbe interactions. Behavioral changes were associated with reduced transcription of csgA, encoding the biofilm component curli, and increased transcription of fliC, encoding flagellin. Scanning electron microscopy (SEM) detailed morphologic changes consistent with the observed alterations in multicellular behavior. Because intestinal E. coli isolates exhibit remarkable strain-specific differences, we generated spontaneous StrR mutants of 10 clinical E. coli phylotype B2 strains from patients with IBD, colorectal cancer, and urinary tract infection. Out of these 10 StrR clinical strains, two had altered colony morphology on Congo red agar (suggesting changes in extracellular products), and three had significant changes in motility. These changes were not associated with a particular rpsL mutation nor with the presence of virulence genes encoding the inflammation-associated E. coli metabolites yersiniabactin or colibactin. We conclude that common mutations in rpsL, which confer StrR, can differentially alter disease-associated phenotypes across intestinal E. coli strains. These findings highlight the heterogeneity among seemingly similar intestinal E. coli strains and reveal the need to carefully study the strain-specific effects of antibiotic resistance mutations, particularly when using these mutations during strain selection studies.
IMPORTANCE We demonstrate that StrR, commonly acquired through a single point mutation in rpsL (a gene encoding part of the 30S bacterial ribosome), strikingly alters the morphology and behavior of a key intestinal AIEC strain, NC101. These changes include remarkably diminished aggregation and significantly increased motility, traits that are linked to AIEC-defining features and disease development. Phenotypic changes were heterogeneous among other StrR clinical E. coli strains, underscoring the need to evaluate the strain-specific effects of commonly acquired antibiotic resistance mutations. This is important, as the results of studies using mutant StrR Enterobacteriaceae strains (e.g., for cloning or in vivo selection) may be confounded beyond our demonstrated effects. Long term, these findings can help researchers better distinguish the contribution of specific E. coli traits to functional changes in the microbiota. Evaluating these strain-level differences could provide insight into the diversity of IBD symptoms and lead to improved therapies for microbiota-driven intestinal disorders.
KEYWORDS: Escherichia coli, adherent-invasive, aggregation, clinical strains, motility, rpsL, streptomycin
OBSERVATION
Escherichia coli isolates from IBD patients are frequently multidrug resistant, especially to aminoglycosides like streptomycin (1). Streptomycin resistance (StrR) can be acquired through spontaneous mutations in rpsL, which encodes the 30S ribosomal protein S12 (2). StrR is frequently caused by a missense mutation in rpsL that results in the substitution of lysine with asparagine at codon 42 or 43 (3). In this study, we reveal that the acquisition of StrR induced distinct behavioral and morphological changes in intestinal adherent-invasive E. coli (AIEC) strain NC101, similar to those observed in related pathogens (4–7). However, these alterations were not consistent among 10 additional, predominantly intestinal, clinical E. coli strains from the same B2 phylogroup as NC101. Inducing StrR is a common laboratory approach used during strain selection, and our results demonstrate that mutations in rpsL may not be as neutral as some may assume. Additionally, our studies highlight the importance of examining strain-specific behavioral traits that may alter host-microbe interactions with intestinal E. coli.
The rpsL-K43N StrR mutation drives behavioral and morphologic alterations in AIEC NC101.
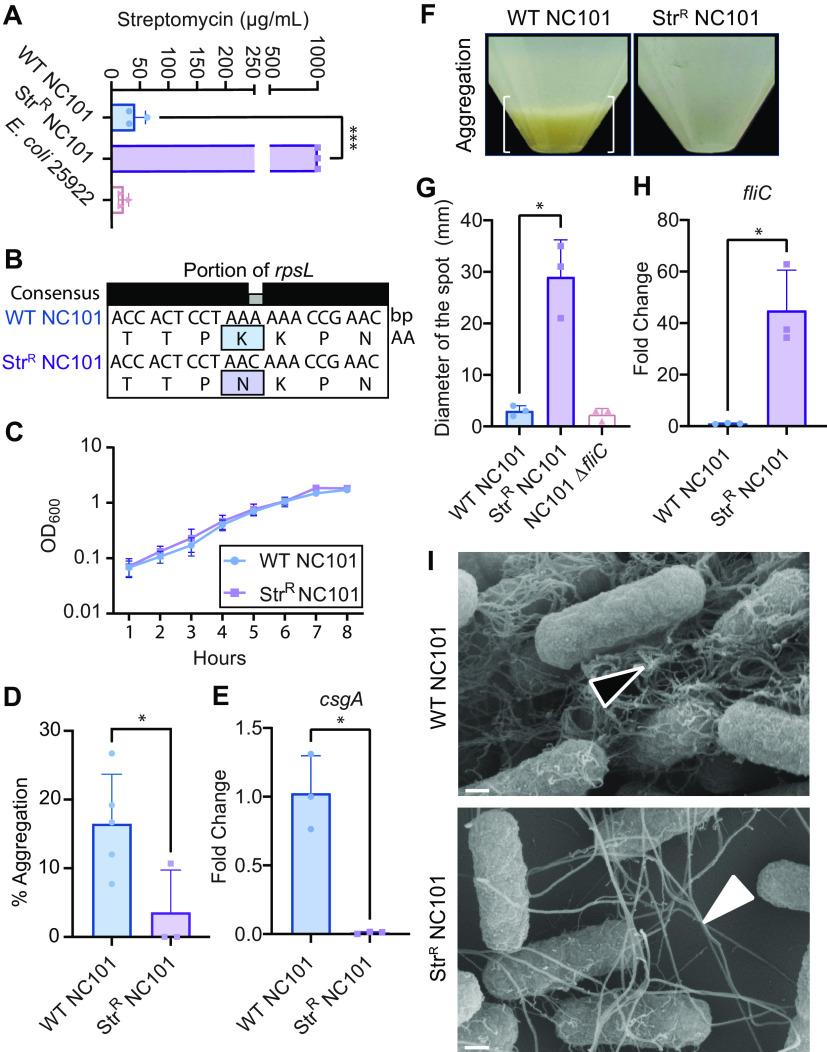
E. coli NC101 is a proinflammatory and procarcinogenic intestinal AIEC strain (6–9). To examine how StrR impacts the function of this broadly used strain, we generated a spontaneous StrR NC101 strain by passaging wild-type (WT) NC101 with increasing amounts of streptomycin and validated resistance via an MIC assay (Fig. 1A). Whole-genome sequencing revealed that StrR NC101 had only a single missense mutation in codon 43 of the rpsL gene, resulting in the substitution of lysine (K) with asparagine (N). We termed this mutation rpsL-K43N (Fig. 1B). Although there was no difference in the growth of WT and StrR NC101 cells in minimal defined medium (MM) (10) (Fig. 1C), StrR NC101 cells failed to aggregate in liquid culture, suggesting changes in extracellular product production (Fig. 1D and F).
FIG 1.
(A) MIC assays confirmed StrR. (B) rpsL gene sequencing. (C) Growth curve in MM. (D) Aggregation quantification of panel F. (E) csgA expression, normalized to uidA, expressed as the fold change to WT NC101. (F) Aggregation (white brackets). (G) Motility swarms in MM. (H) fliC expression. (I) SEM (×100,000; scale bar, 200 nm); the open triangle points to curlinacious fibers, and the closed triangle points to flagella-like products. All graphs represent the means ± standard error of the mean. *, P < 0.05; ***, P < 0.001; Welch’s t test.
As the extracellular product curli is an important component of Enterobacteriaceae biofilms, we assessed the expression of the curli biosynthesis gene csgA. Indeed, StrR NC101 expressed significantly less csgA (Fig. 1E). Because aggregation and swimming motility are often inversely activated, and altered motility can be a feature of AIEC (11), we measured the bacterial motility. As expected, StrR NC101 had significantly increased motility versus WT NC101 and the nonmotile control strain NC101 ΔfliC (Fig. 1G). The motility changes matched an upregulation of the flagellar biosynthesis gene fliC in StrR NC101 (Fig. 1H). Scanning electron microscopy (SEM) revealed changes to the cellular morphology between WT and StrR E. coli NC101, where WT cells had an abundance of fibers resembling curli and StrR NC101 predominately had flagella-appearing structures (11–13) (Fig. 1I). These data suggest that the rpsL-K43N mutation in NC101 confers StrR and, in MM, mediates a switch from the sessile to motile state for this important AIEC strain.
Acquired StrR has differential effects across clinical E. coli strains.
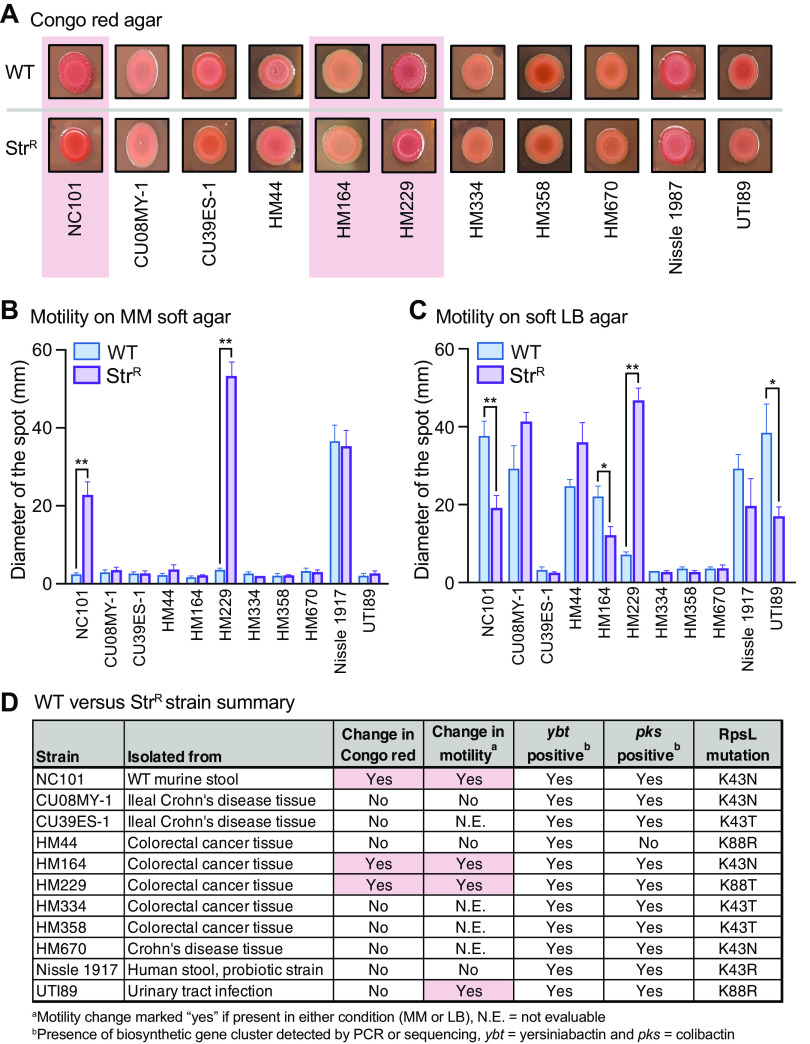
E. coli isolates exhibit remarkable strain-specific differences in genome composition, behavior, and disease-inducing potential. Thus, we generated StrR isolates of 10 clinical E. coli strains, predominantly isolated from intestines and of the same B2 phylotype as NC101. None of these strains aggregated in MM; therefore, to qualitatively determine if StrR was associated with broad changes in extracellular products, we plated the strains on Congo red agar. WT NC101 displayed a red, dry, and rough (RDAR) morphology, previously shown to be dependent upon the production of curli and cellulose (14, 15). In contrast, StrR NC101 presented as red and smooth on Congo red, supporting observations in Fig. 1. Congo red also revealed that two strains, HM164 and HM229 (16), had similar changes as NC101 upon acquiring StrR (Fig. 2A).
FIG 2.
(A) Congo red morphology; the pink boxes indicate strains with phenotypic changes. (B and C) Motility swarms on MM (B) and LB (C) (mean ± standard error of the mean; *, P < 0.05; **, P < 0.01; t test, n = 3 to 7). (D) Strain table showing the phenotypes, virulence genes, and rpsL mutations; the pink boxes indicate key changes in WT versus StrR strains.
We next performed motility assays on MM and LB soft agar, as nutrient availability can alter bacterial motility (17). Indeed, the motility of seven strains appeared different between MM and LB (Fig. 2B and C). More importantly, StrR significantly altered the motility of NC101 and three clinical strains (HM164, HM229, and UTI89), three were unaffected (CU08MY-1, HM44, and Nissle 1917), and the remaining four were nonmotile in the tested conditions (Fig. 1G and 2B to D).
Since there are several mutations in rpsL known to confer StrR, we sequenced the rpsL gene of each WT and StrR strain. Sequencing revealed various rpsL mutations (Fig. 2D), suggesting that the observed changes in Congo red morphology and motility were not driven by a particular rpsL mutation. Furthermore, these mutations were not associated with the presence of ybt or pks genes, which encode disease-modulating metabolites yersiniabactin and colibactin (6, 8) (Fig. 2D). Taken together, we conclude that the effects of StrR on E. coli morphology and behavior are heterogeneous and driven by strain-specific differences.
In summary, we reveal that the common rpsL-K43N mutation conferring StrR is associated with dramatic changes in AIEC NC101 behavior, notably the production of extracellular products and motility. Extracellular products and motility impact proinflammatory behaviors, and hypermotile lineages of E. coli can be used to distinguish Crohn’s disease patients from healthy controls (11, 15, 16, 18, 19). However, these morphological changes were inconsistent among NC101 and the 10 StrR clinical E. coli strains. These findings highlight the importance of evaluating phenotypic changes across multiple E. coli strains that inhabit the intestinal niche, rather than generalizing conclusions based on the results from one strain, particularly in studies using antibiotic resistance mutations (11, 18, 20–22).
With the broad usage of antibiotics, there is a surge of antibiotic resistance across resident intestinal bacteria (1, 23, 24). One study found that an increased proportion of Enterobacteriaceae isolates from IBD patients were ciprofloxacin and rifaximin/rifampicin resistant versus isolates from healthy individuals (1). In addition, similar to our study, changes in aggregation/motility have been associated with fluoroquinolone resistance and can enhance AIEC-defining characteristics (9, 11, 15, 25). Thus, it is important to evaluate disease-associated E. coli traits in response to these and other clinically important antibiotics. These findings may help explain the broad functional variation among genetically similar gut microbiota members and better inform treatment for IBD-related disorders.
Methods.
(i) Bacterial strains. The NC101 and NC101 StrR sequences are available at GenBank under accession numbers CP072787 and CP070227; the strains were generated as previously described (21). Strains CU39ES-1 and CU08MY-1 were from Kenneth W. Simpson at Cornell University (22); strains HM44, HM164, HM229, HM334, HM358, and HM670 were a gift from Barry Campbell at the University of Liverpool (16); and NC101 (6, 7), ATCC 25922 (26), and Nissle 1917 (26) were archived in the Arthur lab at the University of North Carolina (UNC), Chapel Hill. UTI89 was from Harry Mobley at the University of Michigan Medical School, and NC101 ΔfliC was from Melissa Ellermann at the University of South Carolina, Columbia.
(ii) Media and growth conditions. The strains were stored at −80°C with glycerol, and E. coli cells were grown at 37°C with shaking at 220 rpm in MM (containing nicotinic acid, as NC101 is an auxotroph) (10). The aggregation and Congo red assays were previously described (15). For the motility assay, MM or LB soft agar plates (0.25% [wt/vol] agar) were inoculated with 1 μL overnight culture, and the motility swarm diameters were measured after 6 h (LB) or 8 h (MM) at 37°C. For the MIC assay, streptomycin was diluted 2-fold from 1,000 μg/mL in 200 μL/well MM and inoculated with 2 μL overnight bacterial culture. After 12 to 18 h at 37°C with shaking at 220 rpm, the MIC was recorded as the lowest concentration of streptomycin required to prevent E. coli growth (optical density at 600 nm [OD600]). ATCC 25922 was a susceptible control strain.
(iii) Transcriptional analysis. Bacterial overnight cultures were back-diluted to an OD600 value of 0.05 in MM and grown at 37°C with shaking at 220 rpm for 8 h. Quantitative PCR (qPCR) was performed as previously described (26). Threshold cycle (CT) values were normalized to uidA. The primers used were as follows: uidA forward (uidA-F), 5′-TCAACAGACGCGTGGTTACAGTCT-3′; uidA reverse (uidA-R), 5′-TCCATCGCAGCGTAATGCTCTACA-3′; csgA-F, 5′-GGTAATAACAGCGGCCCGAA-3′; csgA-R, 5′-TGTCATCAGAACCTTGGCCC-3′; fliC-F, 5′-ACAACTACTGAGGATGCGGC-3′; and fliC-R, 5′-GCGGAATTTGTCGATCTGGC-3′.
(iv) SEM. SEM was performed at the Microscopy Services Lab (MSL) at UNC on bacteria pelleted from 8-h cultures in MM. The samples were imaged using a Zeiss Supra 25 FESEM device (5 kV, SE2 detector, 5 mm working distance, 30-μm aperture; Carl Zeiss SMT Inc., Peabody, MA).
(v) Targeted rpsL sequencing. A region of rpsL encoding amino acids 30 to 98 was amplified for Sanger sequencing (Genewiz/Azenta Life Sciences): F, 5′-AAACGTGGCGTATGTACTCG-3′, and R, 5′-TTGGAACGAGCCTGCTTAC-3′.
(vi) Rigor and reproducibility. All data are representative of at least three independent experiments.
ACKNOWLEDGMENTS
We thank Belgin Dogan in the Simpson Lab for critically reading the manuscript and Kristin K. White in the MSL at UNC.
We acknowledge funding from the NIH (R01DK124617 and R21AI159786) and pilot funding from UNC CGIBD (subcontract from P30DK034987) and NC TraCS (550KR252046). The MSL is partly supported by the UNC Lineberger Comprehensive Cancer Center (P30CA016086).
Contributor Information
Janelle C. Arthur, Email: janelle_arthur@med.unc.edu.
Xianqin Yang, Agriculture and Agri-Food Canada.
REFERENCES
- 1.Dogan B, Scherl E, Bosworth B, Yantiss R, Altier C, McDonough PL, Jiang Z-D, DuPont HL, Garneau P, Harel J, Rishniw M, Simpson KW. 2013. Multidrug resistance is common in Escherichia coli associated with ileal Crohn’s disease. Inflamm Bowel Dis 19:141–150. doi: 10.1002/ibd.22971. [DOI] [PubMed] [Google Scholar]
- 2.Carter AP, Clemons WM, Brodersen DE, Morgan-Warren RJ, Wimberly BT, Ramakrishnan V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407:340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 3.Paulander W, Maisnier-Patin S, Andersson DI. 2009. The fitness cost of streptomycin resistance depends on rpsL mutation, carbon source and RpoS (σS). Genetics 183:539–546. doi: 10.1534/genetics.109.106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen C, Blumentritt CA, Curtis MM, Sperandio V, Torres AG, Dudley EG. 2013. Restrictive streptomycin resistance mutations decrease the formation of attaching and effacing lesions in Escherichia coli O157:H7 strains. Antimicrob Agents Chemother 57:4260–4266. doi: 10.1128/AAC.00709-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Björkman J, Hughes D, Andersson DI. 1998. Virulence of antibiotic-resistant Salmonella typhimurium. Proc Natl Acad Sci USA 95:3949–3953. doi: 10.1073/pnas.95.7.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arthur JC, Perez-Chanona E, Mühlbauer M, Tomkovich S, Uronis JM, Fan T-J, Campbell BJ, Abujamel T, Dogan B, Rogers AB, Rhodes JM, Stintzi A, Simpson KW, Hansen JJ, Keku TO, Fodor AA, Jobin C. 2012. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338:120–123. doi: 10.1126/science.1224820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. 2005. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 128:891–906. doi: 10.1053/j.gastro.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 8.Ellermann M, Gharaibeh RZ, Fulbright L, Dogan B, Moore LN, Broberg CA, Lopez LR, Rothemich AM, Herzog JW, Rogala A, Gordon IO, Rieder F, Brouwer CR, Simpson KW, Jobin C, Sartor RB, Arthur JC. 2019. Yersiniabactin-producing adherent/invasive Escherichia coli promotes inflammation-associated fibrosis in gnotobiotic Il10−/− mice. Infect Immun 87:e00587-19. doi: 10.1128/IAI.00587-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Darfeuille-Michaud A, Neut C, Barnich N, Lederman E, Martino PD, Desreumaux P, Gambiez L, Joly B, Cortot A, Colombel J-F. 1998. Presence of adherent Escherichia coli strains in ileal mucosa of patients with Crohn’s disease. Gastroenterology 115:1405–1413. doi: 10.1016/s0016-5085(98)70019-8. [DOI] [PubMed] [Google Scholar]
- 10.Lopez LR, Barlogio CJ, Broberg CA, Wang J, Arthur JC. 2021. A nadA mutation confers nicotinic acid auxotrophy in pro-carcinogenic intestinal Escherichia coli NC101. Front Microbiol 12:670005. doi: 10.3389/fmicb.2021.670005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elhenawy W, Tsai CN, Coombes BK. 2019. Host-specific adaptive diversification of Crohn’s disease-associated adherent-invasive Escherichia coli. Cell Host Microbe 25:301–312.e5. doi: 10.1016/j.chom.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Evans ML, Chapman MR. 2014. Curli biogenesis: order out of disorder. Biochim Biophys Acta 1843:1551–1558. doi: 10.1016/j.bbamcr.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abed SE, Ibnsouda SK, Latrache H, Hamadi F. 2012. Scanning electron microscopy (SEM) and environmental SEM: suitable tools for study of adhesion stage and biofilm formation. In Kazmiruk V (ed), Scanning electron microscopy. InTechOpen, London, UK. doi: 10.5772/34990. [DOI] [Google Scholar]
- 14.Römling U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci 62:1234–1246. doi: 10.1007/s00018-005-4557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellermann M, Huh EY, Liu B, Carroll IM, Tamayo R, Sartor RB. 2015. Adherent-invasive Escherichia coli production of cellulose influences iron-induced bacterial aggregation, phagocytosis, and induction of colitis. Infect Immun 83:4068–4080. doi: 10.1128/IAI.00904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin HM, Campbell BJ, Hart CA, Mpofu C, Nayar M, Singh R, Englyst H, Williams HF, Rhodes JM. 2004. Enhanced adherence and invasion in Crohn’s disease and colon cancer. Gastroenterology 127:80–93. doi: 10.1053/j.gastro.2004.03.054. [DOI] [PubMed] [Google Scholar]
- 17.Wright KJ, Seed PC, Hultgren SJ. 2005. Uropathogenic Escherichia coli flagella aid in efficient urinary tract colonization. Infect Immun 73:7657–7668. doi: 10.1128/IAI.73.11.7657-7668.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shaler CR, Elhenawy W, Coombes BK. 2019. The unique lifestyle of Crohn’s disease-associated adherent invasive Escherichia coli. J Mol Biol 431:2970–2981. doi: 10.1016/j.jmb.2019.04.023. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Medina M, Naves P, Blanco J, Aldeguer X, Blanco JE, Blanco M, Ponte C, Soriano F, Darfeuille-Michaud A, Garcia-Gil LJ. 2009. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol 9:202–202. doi: 10.1186/1471-2180-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viladomiu M, Metz ML, Lima SF, Jin W-B, Chou L, Bank JLC, Guo C-J, Diehl GE, Simpson KW, Scherl EJ, Longman RS, JRI Live Cell Bank . 2021. Adherent-invasive E. coli metabolism of propanediol in Crohn’s disease regulates phagocytes to drive intestinal inflammation. Cell Host Microbe 29:607–619.e8. doi: 10.1016/j.chom.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Bleich RM, Zarmer S, Zhang S, Dogan B, Simpson KW, Arthur JC. 2021. Long-read sequencing to interrogate strain-level variation among adherent-invasive Escherichia coli isolated from human intestinal tissue. PLoS One 16:e0259141. doi: 10.1371/journal.pone.0259141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang S, Morgan XC, Dogan B, Martin F-P, Strickler SR, Oka A, Herzog J, Liu B, Dowd SE, Huttenhower C, Pichaud M, Dogan EI, Satsangi J, Longman R, Yantiss R, Mueller LA, Scherl E, Sartor RB, Simpson KW. 2022. Mucosal metabolites fuel the growth and virulence of E. coli linked to Crohn’s disease. JCI Insight 7:e157013. doi: 10.1172/jci.insight.157013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Becattini S, Taur Y, Pamer EG. 2016. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 22:458–478. doi: 10.1016/j.molmed.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Covington A, Pamer EG. 2017. The intestinal microbiota: antibiotics, colonization resistance, and enteric pathogens. Immunol Rev 279:90–105. doi: 10.1111/imr.12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dogan B, Zhang S, Kalla SE, Dogan EI, Guo C, Ang CR, Simpson KW. 2020. Molecular and phenotypic characterization of Escherichia coli associated with granulomatous colitis of boxer dogs. Antibiotics (Basel) 9:540. doi: 10.3390/antibiotics9090540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sadecki PW, Balboa SJ, Lopez LR, Kedziora KM, Arthur JC, Hicks LM. 2021. Evolution of polymyxin resistance regulates colibactin production in Escherichia coli. ACS Chem Biol 16:1243–1254. doi: 10.1021/acschembio.1c00322. [DOI] [PMC free article] [PubMed] [Google Scholar]