ABSTRACT
Host metabolism reprogramming is a key feature of Mycobacterium tuberculosis (Mtb) infection that enables the survival of this pathogen within phagocytic cells and modulates the immune response facilitating the spread of the tuberculosis disease. Here, we demonstrate that a previously uncharacterized secreted protein from Mtb, Rv1813c, manipulates the host metabolism by targeting mitochondria. When expressed in eukaryotic cells, the protein is delivered to the mitochondrial intermembrane space and promotes the enhancement of host ATP production by boosting the oxidative phosphorylation metabolic pathway. Furthermore, the release of cytochrome c from mitochondria, an early apoptotic event in response to short-term oxidative stress, is delayed in Rv1813c-expressing cells. This study reveals a novel class of mitochondria targeting effectors from Mtb that might participate in host cell metabolic reprogramming and apoptosis control during Mtb infections.
IMPORTANCE In this article, using a combination of techniques (bioinformatics, structural biology, and cell biology), we identified and characterized a new class of effectors present only in intracellular mycobacteria. These proteins specifically target host cell mitochondria when ectopically expressed in cells. We showed that one member of this family (Rv1813c) affects mitochondria metabolism in a way that might twist the immune response. This effector also inhibits the cytochrome c exit from mitochondria, suggesting that it might alter normal host cell apoptotic capacities, one of the first defenses of immune cells against Mtb infection.
KEYWORDS: Mycobacterium tuberculosis, apoptosis, metabolism, mitochondria
INTRODUCTION
Mycobacterium tuberculosis (Mtb) encodes secreted virulence factors contributing to its successful infection of host cells and its ability to actively replicate inside the phagosome (1, 2). After phagocytosis, Mtb blocks phagosomal maturation, escapes phagosomes, and subverts the host immune response. Several virulence factors (e.g., proteins, lipids) have been already described to mediate such mechanisms, but corruption of host cell defenses is clearly multifactorial (3). It is estimated that more than 20% of bacterial proteins have functions outside the bacterial cytoplasm and are exported to their designated locations by protein export systems (4). Identification of secreted proteins remains a challenging task (5–8). The comparison of Mtb secreted proteins reported in various proteomic studies revealed only a small number of proteins consistently identified (8). As experiments were made in various culture conditions, it is not surprising that secretion patterns differ from one experiment to another. Furthermore, the host cell environment also plays an important role in defining the secretion pattern, as recently revealed by studies focusing on the identification of secreted proteins during infection (9, 10). To get a broader view on the Mtb secretome, we used multidisciplinary approaches including bioinformatics, structural and biochemical techniques, and cellular biology analyses. We identified putative Mtb secreted proteins using protein primary sequence analysis combined with structure modeling. Among the selected targets, we studied the protein coded by the rv1813c gene, which is present only in mycobacterial pathogens. The Rv1813c protein has been used as vaccine adjuvant and displays immunogenicity properties (11). Rv1813c expression was reported to be MprA- and DosR-regulated, and the Mtb ΔRv1813c mutant was attenuated in the low-dose aerosol model of murine tuberculosis (12).
In this article, we describe molecular and functional analyses of this protein. We showed that Rv1813c defines a new class of effectors, with an original fold, addressed to mitochondria. Mitochondrion plays critical functions not only supplying cells with energy but also contributing to several cellular mechanisms, including cell cycle, apoptosis, and signaling pathways. Metabolism modulation dictates macrophage function and subsequent Mtb infection progression. Here, we demonstrate that Rv1813c affects some mitochondrial metabolic functions and cellular responses to oxidative stress. These results suggest that Rv1813c might play regulatory roles in the metabolic and apoptotic responses occurring in Mtb-infected macrophages.
RESULTS
Bioinformatic analysis of Mtb genome for identification of secreted proteins.
Mtb possesses three different secretion systems using distinct secretion determinants (structural and/or motif-based) present on transported proteins (13). To predict secreted proteins in silico, we analyzed the predicted Mtb H37Rv proteome using an in-house protein analysis toolkit (PAT) (14). First, the SignalP version 4.1 and PredTAT software packages were used to predict the presence of known signal peptides and/or structural features necessary for secretion. In addition, transmembrane segments were inferred using either Uniprot annotations or the TmHMM prediction software. The number of predicted transmembrane segments and the position of the last transmembrane segment were also manually examined to identify signals potentially missed by the other methods. To search for potential type VII secretion system (T7SS)-mediated secreted proteins, we first performed helix structure prediction of each protein using Psipred (15) and then searched for the YXXX(D/E) motif in between the characteristic two helices (16). These data were compared with various proteomic data and model databases (ModBase, Interpro, and GO). Among the proteins identified here as potentially secreted, we studied Rv1813c, a 143-amino-acid protein (14.9 kDa) with a predicted folded domain of unknown function.
Rv1813c protein sequence features and secretion.
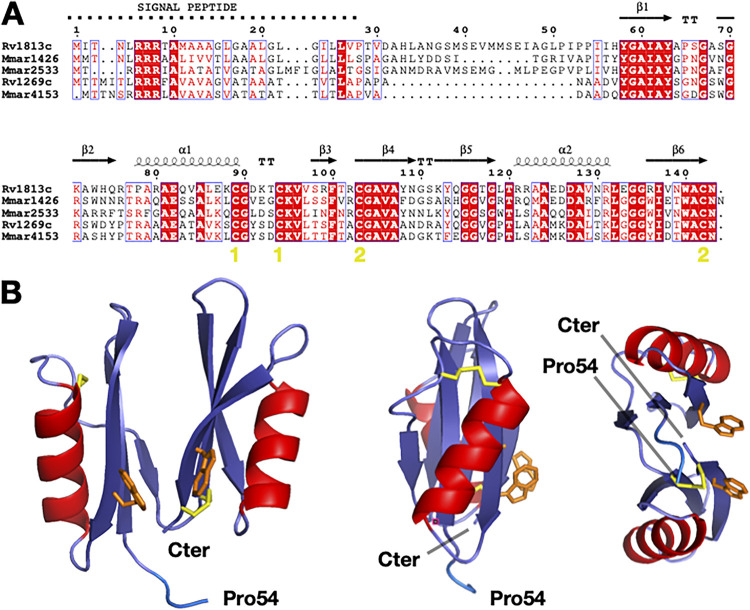
Primary sequence analysis of the Rv1813c protein unambiguously identified a potential signal sequence (residues 1 to 28) with an upstream arginine repeat (residues 6 to 8), indicating that the protein could be exported by the Tat export system (Fig. 1A). Consistent with its genuine export signal, Rv1813c has been identified in one culture filtrate proteome (8) but surprisingly is absent in three others published secretomes (5–7). Conversely, Western blot analysis of Mtb culture filtrates using a specific antibody here confirmed that Rv1813c is secreted during active growth in culture medium (Fig. S1). Rv1813c homologous proteins are mostly found in Actinobacteria (Mycobacterium, Nocardia, and Streptomyces genera). In addition to Mtb, the protein is present in various mycobacteria, including Mycobacterium marinum (Mmar), Mycobacterium avium, Mycobacterium ulcerans, and Mycobacterium abscessus. Multiple paralogs exist within the same bacteria. For instance, Mtb possesses only one ortholog (Rv1269c), whereas Mmar harbors three paralogs (MMAR_1426, MMAR_2533, and MMAR_4153). Remarkably, Rv1269c has been detected in all culture filtrates proteomes published so far (5–7). Therefore, the secretion in culture medium might be a common feature of Rv1813c homologous proteins. The sequence similarity between these various proteins is high (between 45% and 70%), with a lower sequence identity for the N-terminal part (residues 28 to 54 for Rv1813c) of the protein (Fig. 1A). Four cysteine residues are present and conserved. The last four amino acid residues (140WACN143) composed a strictly conserved motif in all Rv1813c homologous proteins that includes one of the conserved cysteines. The fold-recognition and modeling server @TOME2 previously used in many studies for protein function identification even at low sequence identity (17) failed to identify any close or distant Rv1813c structural homologs.
FIG 1.
Rv1813c defines a new protein family. (A) M. tuberculosis and M. marinum Rv1813c homolog sequence alignments. The secondary structure of Rv1813c is reported above the alignment. The yellow numbers indicate the cysteines engaged in disulfide bridges. (B) Rv1813c structure determined by multidimensional NMR. Three cartoon representations of the structure. Only four residues of the N-terminal unfolded part of the protein (residues 28 to 57) are represented. The cysteine residues, all engaged in disulfide bridges, are represented in yellow, while the two solvent-exposed tryptophan amino acids are represented in orange.
Rv1813c defines a new protein family and a unique protein fold.
The Rv1813c-coding sequence without the first codons corresponding to the protein signal peptide (residues 1 to 27) was cloned into an Escherichia coli expression vector. The protein was overexpressed as inclusion bodies, purified, and refolded. The purified protein was used for multidimensional nuclear magnetic resonance (NMR) experiments. Preliminary examination of the [1H,15N] heteronuclear single quantum coherence (HSQC) spectrum revealed that 30 residues were unfolded (Fig. S2). A full multidimensional NMR study led to the protein three-dimensional structure resolution (Fig. 1B; Table S1). Structure resolution demonstrated that residues 28 to 57 were unfolded and that the protein possessed a 86-residue folded domain at its carboxyl terminus. This domain is composed of two duplicate lobes facing each other, and each lobe comprises a series of three β-strands with a hydrophobic surface and an α-helix (β/β/α/β). The four conserved cysteines are engaged in two disulfide bonds located in different parts of each lobe. The proteins from the family defined by the Rv1813c sequence (Pfam domain: DUF4189) contain a conserved WACN motif (Trp-Ala-Cys-Asn residues) with the cysteine engaged in a disulfide bond linking strands β6 and β4, while its tryptophan is solvent-exposed, as well as the second tryptophan (Trp140) in Rv1813c. In addition to protein folding and stability properties, these solvent-accessible tryptophan residues might be functionally important for Rv1813c, contributing either to an hypothetic Rv1813c active site or to interactions with other ligands as classically described for solvent-exposed residues. The overall structure defines a previously undescribed fold, as both Dali (18) and FATCAT (19) servers failed to detect any structural homologs. Consequently, sequence and structure comparison analysis did not bring any indication on the potential biological function of the Rv1813c protein family.
Rv1813c is addressed to mitochondria in Dictyostelium cells.
To assess the function of Rv1813c in host cells, we first used the ameba Dictyostelium discoideum. This professional phagocyte is amenable to biochemical, cell biological, and genetic approaches and has proven to be an advantageous host cell model to analyze the virulence of several pathogenic bacteria (20, 21). Furthermore, the intracellular replication of Mmar has been extensively studied in D. discoideum and shows similarity to Mtb replication in macrophages (22), indicating that comparable molecular mechanisms are at play in infected D. discoideum and mammalian cells. We first analyzed the intracellular localization of Rv1813c when overexpressed in D. discoideum (ectopic expression). Although protein expression levels might differ from what is encountered during Mtb infection, ectopic expression allows the advantageous analysis of individual secreted mycobacterial proteins without potential sources of complexity brought by other bacteria effectors. Hence, this unphysiological expression of Rv1813c serves as a first step toward the biological characterization of this protein and might give some hints on its potential function(s) during Mtb infections.
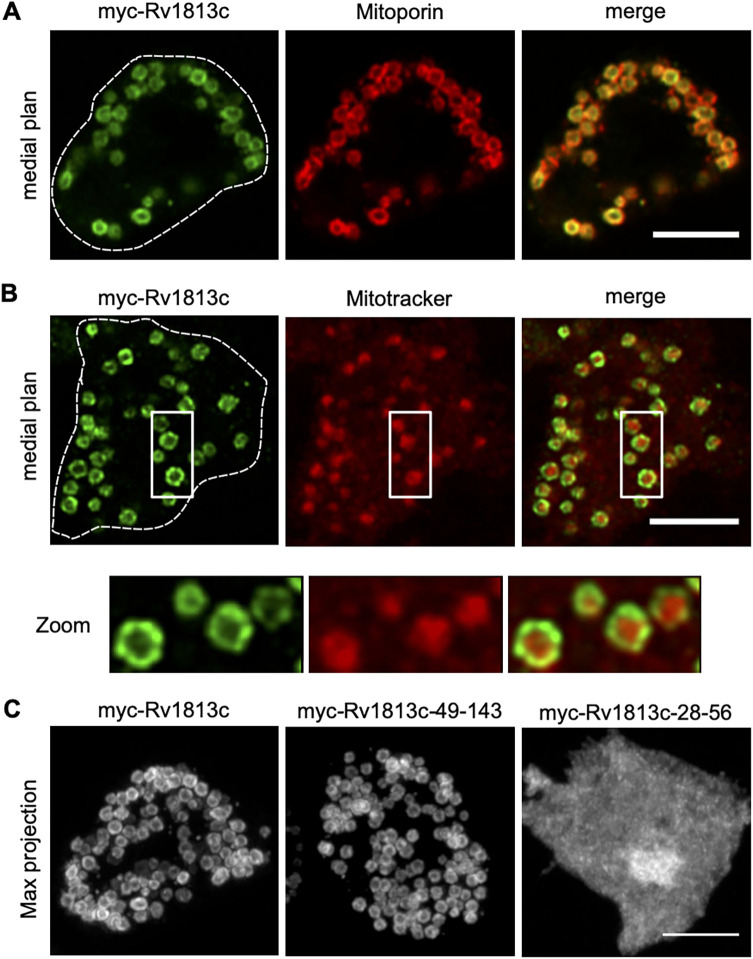
Rv1813c deleted of its predicted signal peptide (first 27 amino acid residues) was tagged with a myc epitope at its N terminus (myc-Rv1813c_P28-N143, hereafter referred to as myc-Rv1813c, 12.3 kDa) and stably expressed in D. discoideum. Confocal microscopy analysis revealed colocalization in ring-like structures of myc-Rv1813c coinciding with a mitochondrial outer membrane protein, Mitoporin (23) (Fig. 2A). Mitochondrial targeting was also observed in cells expressing Rv1813c tagged at the C terminus (Rv1813c-myc) but was lost when Rv1813c was fused to green fluorescent protein (GFP) (Fig. S3A). This specific targeting was independent of the added myc tag as staining with an anti-Rv1813c polyclonal antibody of untagged Rv1813c showed identical results (Fig. S3B). Mitoporin staining patterns were similarly observed in both parental (Ax2) and Rv1813c transfected cells (Fig. S3B and C) excluding gross mitochondrial morphological defects induced by Rv1813c expression in D. discoideum. In cells labeled with MitoTracker deep red, a specific dye accumulating inside mitochondria, myc-Rv1813c surrounded labeled mitochondria and was mostly excluded from internal structures (Fig. 2B). This result suggested that Rv1813c might be attached either to the internal or the cytosolic sides of mitochondrial outer membranes. Interestingly, deletion of the unfolded N-terminal region of Rv1813c (deletion of residues 28 to 48; myc-Rv1813c_49-143) had no effect on Rv1813c localization, whereas Rv1813c deprived of the folded region (deletion of residues 57 to 143; myc-Rv1813c_28-56) was not transported to mitochondria (Fig. 2C). Thus, the Rv1813c folded domain, which does not contain any known mitochondrial targeting signals, was sufficient to specifically direct this protein to mitochondrial outer membranes, whereas the unfolded N-terminal region appeared to be dispensable for Rv1813c targeting to mitochondria.
FIG 2.
Mitochondrial localization of Rv1813c in Dictyostelium. Dictyostelium cells expressing the indicated constructs were fixed, processed for immunofluorescence, and analyzed by confocal microscopy (Airyscan). (A) Colocalization of myc-Rv1813c (detected with a rabbit polyclonal to Rv1813c) and mitochondrial Mitoporin in ring-shaped structures. (B) Labeling of mitochondria of the indicated cells lines with MitoTracker deep red dye. Close-ups of mitochondria are shown in the insets. (C) Maximum fluorescence intensity projection of Z confocal sections of cells expressing the full-length protein (myc-Rv1813c), the sole structured domain (myc-Rv1813c_49-143), and the unfolded domain (myc-Rv1813c_28-56) labeled with an anti-myc antibody. Cell contours are indicated by dotted lines. Bar, 5 μm.
Rv1813c homologs are addressed to mitochondria in Dictyostelium cells.
Intracellular localization studies were next extended to members of the Rv1813c family in Mtb and Mmar in the amoba. All these proteins were detected in mitochondria; however, some Rv1813c-like proteins affected mitochondria morphology. Whereas the overexpression of Rv1813c Mmar orthologs (MMA_1426 and MMA_2533) did not induce any apparent morphological defects in mitochondria, cells expressing Rv1269c or its Mmar ortholog MMA_4153 displayed mitochondria with aberrant shapes and sizes (Fig. S3D). In addition to mitochondria, MMA_4153 also localized to the cytosol. Taken together, these results indicated that mitochondrial targeting in D. discoideum is a characteristic feature of the Rv1813c family, and for some members, this localization leads to defective mitochondrial morphology.
Rv1813c resides in the mitochondrial intermembrane space.
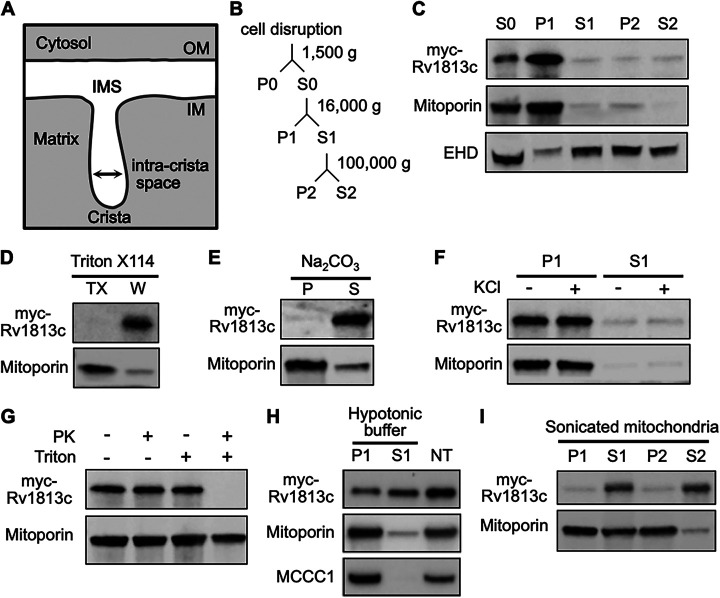
Mitochondria are composed of two membranes, the outer and inner membranes, separated by an intermembrane space (IMS) (Fig. 3A). To determine more precisely the localization of Rv1813c within these submitochondrial compartments in D. discoideum, we next applied a biochemical approach. First, mitochondria-enriched fractions (hereafter referred to as mitochondria) were obtained by subcellular fractionation (see scheme Fig. 3B). As expected, Rv1813c was recovered from the mitochondrial fraction confirmed by Mitoporin enrichment (Fig. 3C). Next, Triton X-114 phase-partitioning experiments revealed that Rv1813c is not an integral membrane protein, in agreement with the absence of any predicted transmembrane domains (Fig. 3D) and its exclusion from the Mtb cell wall (Fig. S1). Consistently, Rv1813c was extracted from mitochondrial membranes by sodium carbonate treatment, a characteristic feature of peripheral membrane proteins (Fig. 3E). Since Rv1813c was not released from mitochondria by high-salt washes (Fig. 3F) and was protected from proteinase K digestion of intact mitochondria (Fig. 3G), we concluded that Rv1813c resides inside mitochondria. In addition, Rv1813c was partially released from mitochondria upon the specific rupture of mitochondrial outer membranes in hypotonic buffer or by sonication, indicating that Rv1813c accumulates into the mitochondrial IMS, where it could be weakly attached to the internal or external sides of mitochondrial outer or inner membranes, respectively (Fig. 3H and I).
FIG 3.
Biochemical analysis of Rv1813c mitochondrial localization in Dictyostelium. (A) Schematic ultrastructure of a single crista in mitochondria. (B) Fractionation scheme of differential centrifugation steps used to purify the Rv1813c enriched fraction from Dictyostelium cells. (C) Fractions were analyzed by immunoblotting with antibodies to Mitoporin (mitochondria), EHD (endocytic vacuoles), and myc tag (myc-Rv1813c). (D) The mitochondrial fraction was fractionated by Triton X-114 extraction. The separated Triton X-114 (TX) and aqueous (W) phases were analyzed as above. Rv1813c is not extracted by Triton X-114, indicating no insertion inside membranes. (E) Mitochondria were incubated in sodium carbonate for 30 min. Rv1813c is mainly detected in the supernatant fraction (S) after centrifugation at 100,000 × g of treated mitochondria, a characteristic of soluble and/or membrane peripheral proteins. (F) For high-salt protein extraction, mitochondria were incubated in buffer ± 200 mM KCl for 30 min and centrifuged at 16,000 × g for 10 min. (G) Intact or Triton X-100-treated mitochondria were subjected to proteinase K digestion for 30 min and analyzed by immunoblotting. Mitoporin was only partially digested after the Triton X-100/PK treatment, whereas Rv1813c was fully degraded since mitochondrial membranes were mainly permeabilized rather than solubilized in our experimental conditions. (H) Mitochondria swelling was induced by hypotonic buffer incubation for 30 min. Released proteins (S1) from broken outer membranes (P1) were recovered by centrifugation at 16,000 × g for 10 min and analyzed by Western blotting to detect myc-Rv1813c, Mitoporin (mitochondrial outer membrane), and mitochondrial 3-methylcrotonyl-CoA carboxylase α (MCCC1; mitochondrial matrix). Nontreated (NT) mitochondria incubated for 30 min in mitochondria isolation buffer A served as release specificity control. (I) Mitochondria were sonicated for 30 s to disrupt membranes and centrifuged at 16,000 × g for 10 min. The supernatant S1 mainly containing ruptured membranes was further centrifuged at 100,000 × g to separate membranes (P2) from soluble material (S2). Fractions were analyzed by Western blotting with the indicated antibodies. IM, inner membrane; IMS, intermembrane space; OM, outer membrane; P, pellet; S, supernatant. EHD, C-terminal Eps15-homology-domain protein; PK, Proteinase K.
Rv1813c and orthologous proteins are addressed to mitochondria in mammalian cells.
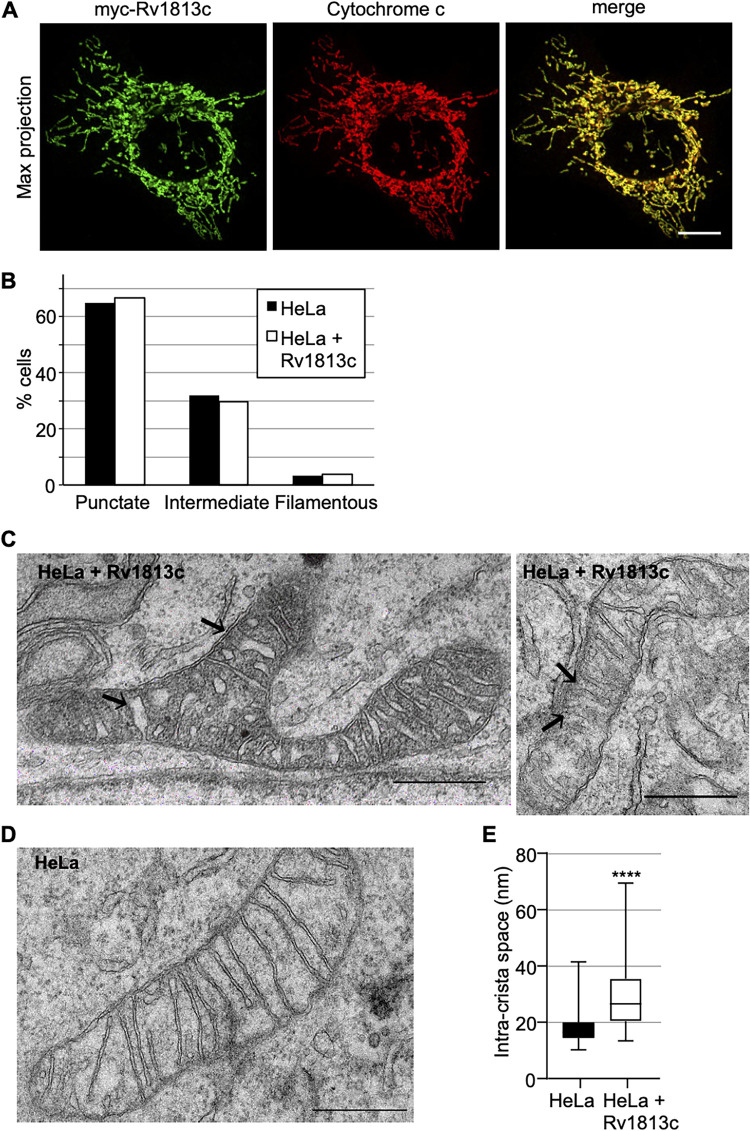
We next extended the analysis to mammalian cells. Native and myc-tagged Rv1813c were transiently expressed in HeLa cells, and their intracellular localization was determined by confocal microscopy. As observed in Dictyostelium, Rv1813c was efficiently targeted to mitochondria in HeLa cells (Fig. 4A; Fig. S4) without any detectable effects on the mitochondrial morphology (Fig. 4B). MMA_1436 and MMA_2533, two Mmar orthologs of Rv1813c, also localized to mitochondria. However, in contrast to Dictyostelium cells, Rv1269c remained in the cytosol similarly to MMA_4153, the Mmar ortholog of Rv1269c, which also showed a faint mitochondrial localization (Fig. S4). Although it might denote an intrinsic feature, we cannot rule out that the folding of these proteins might not proceed appropriately in mammalian cells, impeding their efficient mitochondrial localization.
FIG 4.
Targeting of Rv1813c to mitochondria in mammalian cells. (A) Confocal microscopy analysis of HeLa cells transiently expressing myc-Rv1813. The cells were fixed 48 h post-transfection, processed for immunofluorescence with antibodies to RV1813c and cytochrome c, and analyzed by Airyscan microscopy. Bar, 10 μm. (B) Quantitative analysis of the mitochondria morphology observed in HeLa cells transiently transfected with pCI-myc-Rv1813 or empty vector as control. Mitochondria morphology was manually identified by confocal microscopy and classified in 100 cells of one representative experiment from three independent analyses. (C, D) Representative mitoch7ondria ultrastructure determined by transmission electronic microscopy of magnetic cell sorting (MACS)-enriched HeLa cells transiently transfected with pMACS-4-IRES-II Rv1813c (C) or vector alone (D). Black arrows indicate some enlarged intracristae spaces in Rv1813c-expressing cells. Bars, 500 nm. (E) Box plot of intracristae spaces measurements for the indicated cell lines (100 random measurements each; ****, P < 0.0001 in Student’s test). The bar inside the box indicates the median value. The bottom and top edges of the box indicate the 25th and 75th percentiles, respectively. The whiskers extend to the most extreme data points not considered outliers.
Whereas the overall morphology of mitochondria was preserved upon Rv1813c ectopic expression, transmission electronic microscopy (TEM) revealed some ultrastructural modifications. Hence, Rv1813c expressed in HeLa cells contained mitochondria with either normal or electron-dense matrix, and the intracristae space appeared significantly enlarged compared to parental HeLa cells (Fig. 4C and D). This altered mitochondrial morphology is reminiscent to what is observed in Mtb-infected macrophages (24). Since cristae membranes are enriched in resident proteins involved in oxidative phosphorylation, this particular ultrastructure might lead to several mitochondrial energetic/metabolism consequences.
Rv1813c overexpression enhances cell metabolism and mitochondrial ROS production.
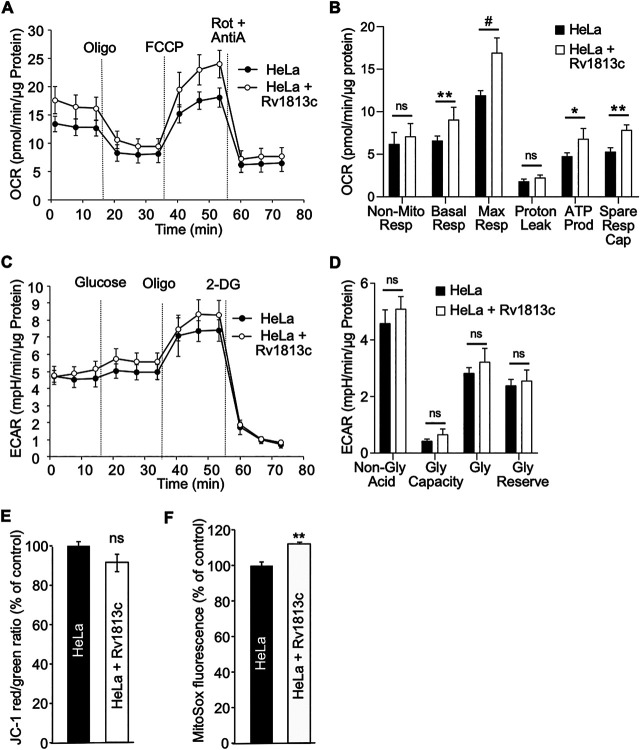
Next, we investigated whether changes in the mitochondrial morphology might cause energy metabolism disorders. Oxidative phosphorylation (OXPHOS) and glycolysis were simultaneously analyzed in intact cells by making use of an extracellular flux analyzer (XF, Agilent Seahorse). In this assay, mitochondrial respiratory characteristics are evaluated by recording the oxygen consumption rate (OCR) upon sequential chemical perturbation of selected mitochondrial functions (as detailed in the legend to Fig. 5). In Rv1813c transfected HeLa cells, basal respiration, ATP-linked respiration, maximal respiratory capacity, and reserve capacity were significantly increased compared to parental HeLa cells (Fig. 5A and B). Glycolysis was also assayed using a glycolysis stress test (Agilent Technologies) and measurements of extracellular acidification rates (ECARs) in incubation media. This assay revealed similar glycolytic profiles in control and Rv1813c-expressing HeLa cells (Fig. 5C and D).
FIG 5.
Functional consequences of Rv1813c mitochondrial localization. (A, B) Analysis of cell respiratory functions. HeLa cells were transiently transfected with pMACS-4-IRES-II Rv1813c or vector alone and enriched to >94% of expressing cells. Cell respiratory profiles (oxygen consumption rate [OCR]) (A) and respiratory parameters (B) were obtained using an extracellular flux analyzer (Seahorse XF analyzer) and the mitochondrial respiration test. After reaching basal respiration, the cells were subjected to 1 μM oligomycin to inhibit the ATP synthase and measure the mitochondrial ATP-linked OCR; followed by 1 μM cyanide-4-(trifluoromethoxy)phenylhydrazone (FCCP) to uncouple mitochondrial respiration and maximize OCR; and finally 1 μM antimycin A (AntiA) and 100 nM rotenone (Rot) to inhibit complex III and I in the electron transfer chain (ETC), respectively, and shut down respiration. (B) The analyzed respiratory parameters are nonmitochondrial respiration (Non-Mito Resp), basal respiration (Basal Resp), maximal respiration (Max Resp), proton leak, ATP production (ATP Prod), and spare respiratory capacity (Spare Resp Cap). (C, D) Analysis of glycolytic functions. Extracellular acidification (ECAR) profiles (C) and glycolytic parameters (D) of the same MACS-enriched transfected cells were determined simultaneously to OCR analysis using the glycolysis stress test and the XF analyzer. After reaching nonglycolytic acidification, 10 mM glucose was added, followed by 1 μM oligomycin (Oligo) to inhibit the ATP synthase and induce maximal glycolysis. Finally, 100 mM 2-deoxyglucose (2-DG) was added to shut down glycolysis. This last injection resulted in a decreased ECAR, confirming that the recorded ECAR was only due to glycolysis. In panel D, the analyzed glycolitic parameters are nonglycolitic acidification (Non-Gly Acid), glycolytic capacity (Gly Capacity), glycolysis (Gly), and glycolytic reserve (Gly Reserve). The values are means ± SD according to Student’s t test relative to HeLa cells. #, P < 0.000001; **, P < 0.0005; *, P < 0.005; ns, not significant. (E, F) HeLa cells were transiently transfected with pCI-myc-Rv1813c or empty vector as control and analyzed 48 h later by flow cytometry. (E) Flow cytometry analysis of the indicated HeLa cell lines stained with JC-1 to monitor mitochondrial membrane potential. JC-1 Red/Green ratio were calculated and expressed as the percentage of this ratio in HeLa cells. The values are means ± SEM of three independent experiments. ns, not significantly different Student’s t test. (F) Flow cytometry analysis of MitoSox-stained HeLa cell lines. MitoSox fluorescence was expressed as the percentage of fluorescence in HeLa cells. The values are means ± SEM of three independent experiments. **, P ≤ 0.01 Student’s t test.
Then we examined mitochondrial membrane potential (ΔΨM). The accumulation of the JC-1 dye in mitochondria depends on their ΔΨM ; thus, altered mitochondrial functions would result in concomitant reduced ΔΨM and JC-1 staining. This assay revealed that expression of Rv1813c in HeLa cells had no effect on ΔΨM in resting cells (Fig. 5E). However, these cells showed a slight but significant increase in mitochondrial ROS production (Fig. 5F). Taken together, these results indicate that Rv1813c expression improves mitochondrial respiratory capacities without altering glycolytic functions, driving cells into an energy-activated state. This higher mitochondrial respiration is associated with a moderately increased mitochondrial free radical formation without changes in the mitochondrial membrane potential.
Oxidative stress-induced translocation of cytochrome c is delayed in Rv1813c-expressing cells.
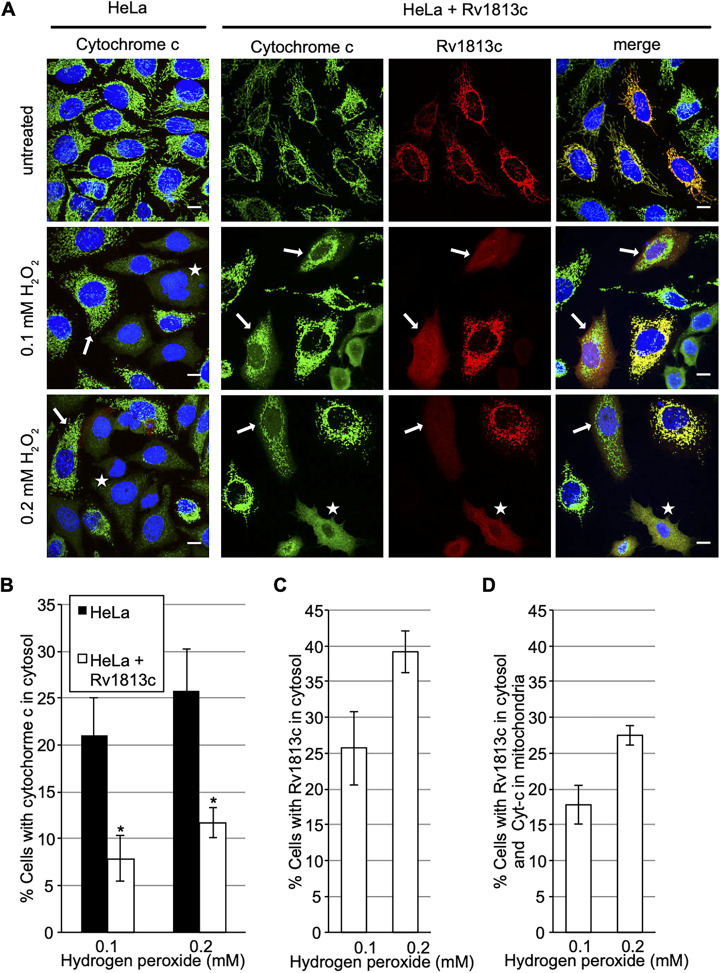
We next assessed whether these mitochondrial alterations might alter the ability of Rv1813c-expressing HeLa cells to cope with oxidative stress, a situation encountered during Mtb infection. We choose to monitor the release of cytochrome c (Cyt-c) from mitochondria into the cytosol in response to hydrogen peroxide, an early event in apoptotic cell death (25). Hence, the cells were incubated with hydrogen peroxide for 3 h, and the localization of Cyt-c and Rv1813c was analyzed by confocal microscopy. As expected, Cyt-c showed diffuse cytosolic staining in 21% of parental HeLa cells upon addition of 0.1 mM hydrogen peroxide (Fig. 6A and B). Rv1813c release from mitochondria was also observed in cells overexpressing Rv1813c in response to hydrogen peroxide treatments (Fig. 6A and C). In contrast, Cyt-c release from mitochondria into the cytosol was reduced in Rv1813c-expressing cells, with only 7.9% of cells displaying cytosolic Cyt-c staining upon oxidative stress conditions (Fig. 6A and B). Note that cells with cytosolic Cyt-c always showed a strict concomitant Rv1813c cytosolic localization; thus, we did not detect any cells harboring cytosolic Cyt-c and mitochondrial Rv1813c localizations together. Strikingly, Rv1813c release from mitochondria was more frequently observed than Cyt-c translocation leading to another cell population with Rv1813c in the cytosol but Cyt-c still in mitochondria (Fig. 6D). We conclude that the massive exit of Rv1813c from mitochondria in response to oxidative stress might delay the normal stress-induced Cyt-c translocation.
FIG 6.
Analysis of cytochrome c (Cyt-c) and Rv1813c release from mitochondria upon oxidative stress. (A) Confocal microscopy analysis of HeLa cells transiently expressing myc-Rv1813. At 48 h post-transfection, the cells were treated with 0.1 or 0.2 mM hydrogen peroxide for 3 h, fixed, processed for immunofluorescence with anti-cytochrome c (green) and anti-Rv1813c (red) antibodies, and observed by confocal microscopy. The nuclei were stained with Hoechst (blue). White arrows and white stars indicate cells with Cyt-c in mitochondria and cytosol, respectively. Bar, 10 μm. (B to D) Quantification of cells with Cyt-c in cytosol (B), with Rv1813c in cytosol (C), and Rv1813c in cytosol but Cyt-c in mitochondria (D) upon incubation with hydrogen peroxide for 3 h. The values are means ± SEM of three independent experiments, with 100 cells analyzed for each condition. *, P < 0.05 Student’s t test.
DISCUSSION
Several intracellular pathogens (i.e., Rickettsia, Legionella, and Salmonella) disrupt mitochondrial functions during infection mainly due to indirect effects (26, 27) but more rarely by direct mitochondrial targeting of bacterial effectors having adverse functions (28). For instance, the EspF effector from enteropathogenic E. coli is addressed to mitochondria via a mitochondrial import signal and promotes caspase-mediated apoptosis in intestinal epithelial cells (29). Furthermore, the MitF protein from Legionella pneumophilia was reported to alter mitochondria fission dynamics and promote a Warburg-like phenotype in macrophages (30). Using bioinformatics screening and culture filtrate analysis, we have identified Rv1813c from Mtb as a secreted protein. The protein serves a role of vaccine adjuvant (31) and is highly immunogenic (11). The corresponding gene is nonessential for Mtb growth; however, its deletion impairs Mtb virulence in a low-dose murine model (12). To our knowledge, no functional studies have been reported so far; thus, the molecular basis of this attenuation is still unknown. Interestingly, Rv1813c expression is regulated by MprA and DosR proteins (12). DosR is a transcriptional regulator induced by host intracellular stimuli, such as nitric oxide (NO), carbon monoxide (CO), and hypoxia (12), while MrpA responds to environmental stress and residence within the macrophage (32, 33) and is required during infection (34). Accordingly, reference transcriptomes have revealed that Rv1813c is overexpressed (×2 and ×4 after 24 and 48 h postinfection, respectively) in activated infected macrophages (35), and in the BALB/c mouse model (×4, ×5, and ×6 after 7, 14, and 21 days postinfection, respectively) (36).
In this study, we established that Rv1813c belongs to a new protein family constitutively secreted by Mtb in culture medium and specifically addressed to mitochondria, where it accumulates the IMS when ectopically expressed in host cells (Fig. S1; Fig. 2 and 4). We solved the structure of Rv1813c that included a new protein fold with no similarity with any structures solved to date (Fig. 1). Interestingly, this small 9-kDa conserved folded domain was sufficient to specifically address the protein into mitochondria (Fig. 2C). Furthermore, we demonstrated that this localization subsequently enhances OXPHOS and inhibits cytochrome c exit upon oxidative stress in Rv1813c-expressing cells. These Rv1813c-dependent phenotypes might be connected to important host defense mechanisms against Mtb infections and will have to be further addressed in Mtb-infected cells.
Would the increased host cell mitochondrial ATP production induced by a Mtb effector protein bring any benefits to the bacteria for its intracellular replication or for avoiding normal host defense mechanisms? The activation of macrophages in response to Mtb infection is known to induce their polarization toward the toward an M1 profile (37). This important step is achieved by metabolic reprogramming after NF-κB pathway activation either by pathogen-associated molecular patterns (PAMPs) or interferon-γ (IFN-γ). NF-κB promotes the expression of the inducible nitric-oxide synthase (iNOS) and subsequent nitric oxide (NO) release. In addition to bactericidal activity, NO directly inactivates the electron transfer chain (ETC) proteins, triggering a complex series of events, mainly dependent on the production of reactive oxygen species (ROS) and a change in the metabolite balance (i.e., NAD/NADP ratio). When the Krebs cycle is consequently blocked, citrate accumulates enhancing glycolysis and lipid biosynthesis. In addition, succinate also accumulates, leading to hypoxia-inducible factor-1 (HIF-1α) stabilization, which completes this metabolic switch, similarly to the Warburg effect observed in tumors (38). HIF-1α not only promotes the expression of enzymes involved in glycolytic ATP production but also induces expression pattern, leading to synthesis of important immune effectors, including inflammatory cytokines and chemokines under normoxic conditions (39).
Very few studies have assessed the precise change into the level of ATP produced in Mtb-infected cells (40), and they have led to contradictory results. Recently, bioenergetic analyses have been performed in infected macrophages. Hence, XF experiments and metabolites analysis have revealed a decrease of cell energetic flux through glycolysis and the tricarboxylic acid (TCA) cycle in infected macrophages. Consequently, the total level of ATP produced in Mtb-infected cells 5 and 24 h postinfection is reduced (41). These observations confirm previous results, indicating that the glycolytic flux was reduced in macrophages infected with virulent H37rv bacteria, a possible hallmark of a bacterial effector-induced incomplete or delayed metabolic shift (42).
In contrast, other studies have suggested that maintaining host cell ATP production is beneficial for Mtb in order to avoid ROS production and apoptosis. For instance, a much higher ATP/ADP ration was observed in H37Rv-infected cells compared to cells infected with avirulent H37Ra (24, 43). In agreement, an elevated ATP/ADP ratio was also correlated to lower apoptosis rates observed in H37Rv-infected cells (24, 44). Taken together, these data indicate that maintaining high ATP production might be beneficial to delay a deleterious full metabolic shift and/or apoptosis of the host cell. Consistent with this hypothesis, secretion of Rv1813c could participate in maintaining a higher ATP production within mitochondria during Mtb infection.
Mitochondrial proton leak generated from the ETC is the major source of mitochondrial ROS. Excessive ROS amounts result in multiple effects, including cytochrome c translocation followed by caspase-dependent apoptosis, as well as inflammasome activation (24). Accordingly, the slight increase of ROS observed in resting Rv1813c-expressing cells (Fig. 5F) might be due to ETC and/or ATP synthase-boosted functions raising the ATP production in these cells. Interestingly, the artificial increase of ROS by exogenous hydrogen peroxide did not readily induce the release of Cyt-c from mitochondria in Rv1813c-expressing cells compared to parental HeLa cells. The release of cytochrome c from mitochondria is an early event in apoptotic cell death and an early defense mechanism in infected Mtb macrophages (45). Thus, Rv1813c-based inhibition of this process might bring some advantages for Mtb. The molecular mechanism responsible for this inhibition will be further addressed. In summary, this study provided a detailed analysis of morphological and functional consequences of Rv1813c ectopical expression in cells and paved the way for future studies on how this secreted protein could influence metabolic and apoptotic responses in Mtb-infected macrophages.
MATERIALS AND METHODS
Purification of recombinant 6His-Rv1813c28-143 in E. coli.
E. coli BL21(DE3) strains containing pET::rv181328–143 vector were used to inoculate 1 liter of LB medium supplemented with ampicillin (100 μg/mL), and the resulting cultures were incubated at 37°C with shaking until A600 reached ~0.5. Then, a final concentration of 1 mM isopropyl 1-thio-β-d-galactopyranoside was added, and growth was continued for 3 h at 37°C. The cells were harvested by centrifugation, and the resulting cell pellet was resuspended in buffer A (50 mM Tris-HCl, pH 8.5, 150 mM NaCl, 2 mM dithiothreitol [DTT]). The cells were then lysed by sonication, and cell debris and insoluble materials were separated by centrifugation. The pellet was then resuspended in buffer B (buffer A + 8 M urea). After centrifugation the supernatant was loaded into a HiTrap IMAC HP column (Amersham Biosciences) and equilibrated in buffer B and 4% of buffer C (buffer B supplemented with 300 mM imidazole). The column was washed with successive applications of buffer B (approximately 30 mL in total) to remove all the impurities, and then buffer C was increased over 20 mL to 100%. Fractions containing the Rv1813c proteins were identified by SDS-PAGE, then pooled, and concentrated using a 5 K cutoff concentrator to a 2 mg/mL concentration. The protein was dialyzed against buffer A overnight at 4°C. The refolded protein was very unstable until removal of the 6His tag using 3C protease (4 h of digestion at 4°C). The protein was then loaded to a Superdex 75 26/60 (Amersham biosciences) size-exclusion column, equilibrated in buffer (20 mM sodium phosphate, pH 6.2, 150 mM NaCl). Again, fractions containing the protein were identified by SDS-PAGE, then pooled, and stored at −20°C until required. This protocol was carried out for all the nonlabeled constructs of Rv1813c, as well as for 15N- and 15N -13C-labeled constructs, except that the cultures were grown in a minimum medium containing 15NH4Cl and 15NH4Cl/13C6-glucose as the sole nitrogen and carbon sources.
Solution structure of Rv1813c (residues 28 to 143).
All NMR experiments were generally carried out at 25°C on a Bruker Avance III 700 (1H-15N double resonance experiments) or Avance III 500 (1H-13C-15N triple-resonance experiments) spectrometer equipped with a 5-mm z-gradient TCI cryoprobe, using the standard pulse sequences. NMR samples consist of approximately 0.9 mM 15N- or 15N,13C-labeled protein dissolved in 25 mM sodium citrate, 150 mM NaCl (pH 5.6) with 10% D2O for the lock. 1H chemical shifts were directly referenced to the methyl resonance of dextran sulfate sodium (DSS), while 13C and 15N chemical shifts were referenced indirectly to the absolute 15N/1H or 13C/1H frequency ratios. All NMR spectra were processed and analyzed with GIFA. Backbone and Cβ resonance assignments were made using standard HNCA, HNCACB, CBCA(CO)NH, HNCO, and HN(CA)CO experiments performed on the 15N,13C-labeled Rv1813c28-143 sample. NOE cross-peaks identified on 3D [1H,15N] nuclear Overhauser effect spectroscopy (NOESY)-HSQC (mixing time, 120 ms) were assigned through automated NMR structure calculations with CYANA 2.1, whereas NOE on 3D [1H,13C] NOESY-HSQC were assigned manually. Backbone φ and ψ torsion angle constraints were obtained from a database search procedure on the basis of backbone (15N, HN, 13C′, 13Cα, Hα, 13Cβ) chemical shifts using the program TALOS+ (46). Hydrogen bond restraints were derived using standard criteria on the basis of the amide 1H/2H exchange experiments and NOE data. When identified, the hydrogen bond was enforced using the following restraints: ranges of 1.8 to 2.0 Å for d(N-H,O) and 2.7 to 3.0 Å for d(N,O). This established the final list of restraints, from which values redundant with the covalent geometry have been eliminated. The 30 best structures (based on the final target penalty function values) were minimized with CNS 1.2 according the RECOORD procedure (47) and analyzed with PROCHECK (48). The root mean square deviations (RMSDs) were calculated with MOLMOL (49). All statistics are given in Table S1. The chemical shift table was deposited in the BMRB data bank (accession number 7NHZ).
Antibodies.
The following primary antibodies were used in this study: mouse anti-myc (Invitrogen, catalog no. 13-2500, 1:200 for immunofluorescence, 1:500 for immunoblot), mouse anti-cytochrome c (clone 6H2.B4, BD PharMingen, 1:500 for immunofluorescence), mouse anti-Dictyostelium Mitoporin (catalog no. 70-100-1; 1:2,000 for immunofluorescence and immunoblot) (23), rabbit anti-Rv1813c raised using recombinant Rv1813c (ProteoGenix SAS, Schiltigheim, France) (1:2,000 for immunofluorescence, 1:5,000 for immunoblot), rabbit anti-Grp75 (D13H4, XP catalog no. 3593, Cell Signaling, 1:100 for immunofluorescence), and rabbit anti-EHD (50) (1:4,000 for immunoblot). The secondary antibodies used for immunoblotting were horseradish peroxidase (HRP)-conjugated donkey anti-mouse IgG (H+L) (catalog no. 715-035-151) and HRP-conjugated donkey anti-rabbit IgG (H+L) (catalog no. 715-035-152) (Jackson ImmunoResearch). MCCC1 (mitochondrial matrix) was revealed by staining with HRP-conjugated streptavidin as previously described (51). The secondary antibodies used for immunofluorescence were Alexa Fluor 568-conjugated goat anti-mouse IgG (H+L) (catalog no. A11031), Alexa Fluor 594-conjugated donkey anti-rabbit IgG (H+L) (catalog no. A21207), Alexa Fluor 488-conjugated goat anti-rabbit IgG (H+L) (catalog no. A11029), and Alexa Fluor 488-conjugated donkey anti-rabbit IgG (H+L) (catalog no. A21206) (Thermo Fisher Scientific, Illkirsh, France). All secondary antibodies were used at 1:500 for immunofluorescence. Prolong Golf Antifade and Hoechst 33342 (catalog no. 62249) were purchased from Molecular Probes (Thermo Fisher Scientific, Illkirsh, France).
Preparation of M. tuberculosis culture.
M. tuberculosis was grown in Middlebrook 7H9 liquid medium supplemented with 10% (vol/vol) albumin-dextrose complex (ADC), 0.2% (vol/vol) glycerol, and 0.1% Tween 80 (wt/vol), at 37°C in a roller incubator. Bacterial growth was followed by measurement of absorbance at 580 nm using a spectrophotometer or by CFU counting on 7H10 agar.
Mycobacterial cell fractionation.
Mycobacteria cell fractionation was done as described elsewhere (52). Briefly, the cells were lysed in a buffer that contained 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 20 mM KCl, 10 mM MgCl2. Bacterial culture was homogenized with a Minilys homogenizer (Bertin Instruments) using glass beads. A cocktail of proteinase/phosphatase inhibitors (Roche, UK) were used in all buffers. Lysates were centrifuged for 1 h at 27,000 × g, and the pellets were washed in a carbonate buffer (pH 11) and used as cell wall material. The supernatant was centrifuged again for 4 h at 100,000 × g. The supernatant from this step was used as cytoplasmic fraction, and the pellets (membrane fractions) were washed once in carbonate buffer, pH 11, and twice in Tris-buffered saline (TBS) buffer. Proteins from cellular fractions were separated on SDS-PAGE. The purity of fractions was confirmed by the detection of diagnostic proteins as described below.
Protein electrophoresis and Western blot.
The proteins were separated on 4% to 20% gradient SERVA gels and transferred onto a nitrocellulose membrane using a Trans-Blot Turbo transfer system (Bio-Rad) according to the manufacturer’s instructions. SignalFire Elite ECL reagent (Cell Signaling, UK) was used to visualize proteins on a C-DiGit chemiluminescent blot scanner (LI-COR Biosciences), according to the manufacturer’s instructions. All the secondary antibodies were from Cell Signaling, UK. Diagnostic proteins were used for all the cellular fractions: GlnA (membrane protein), GarA (secreted and cytoplasmic protein), RpfB (membrane and cell wall protein). and FtsZ (cytoplasmic protein).
Cell culture and transfection conditions.
D. discoideum strain Ax2 was grown at 22°C in HL5c medium supplemented with 18 g/liter maltose (Formedium, Norfolk, UK). For ectopic expression in Dictyostelium, Rv1813c family coding sequences with Dictyostelium optimized codons (Integrated DNA Technologies, Inc., Coralville, IA) were cloned into pDXA-3C-myc (53). The plasmids were linearized by ScaI and transfected by electroporation as described (54). The clones were selected in 5 μg/mL G418.
HeLa (ATCC CRM-CCL-2) and HEK-293T (ATCC CRL-3216) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) and high glucose containing 5% and 10% heat-inactivated fetal bovine serum, respectively, and supplemented with GlutaMAX (Gibco Life Technologies), penicillin (100 units/mL), and streptomycin (100 μg/mL). Transfections of HeLa cells were performed using JetPEI transfection reagent (PolyPlus-Transfection, Ozyme, Saint Quentin, France), according to the manufacturer. Cells plated 1 day before transfection were incubated with JetPEI-DNA complexes (N/P = 5), and after 5 h, the medium was changed. All assays were performed at 48 h post-transfection.
For confocal microscopy analysis, HeLa cells were seeded on glass coverslips coated with 0.001% poly-l-lysine (catalog no. P4707, Sigma). For localization, Rv1813c family coding sequences with human optimized codons were cloned into the mammalian expression vector pCI (a kind gift from Solange Desagher, IGMM, Montpellier, France). Cells on glass coverslips were transfected on a 24-well culture plate and analyzed 48 h later. For mitochondrial membrane potential, mitochondrial ROS, and oxidative stress studies, cells were transfected on 6-well culture plates. After 24 h, resuspended cells were pooled and plated either on glass coverslips for confocal microscopy or on 6-well culture plates at a density of 2 to 3 × 105 cells/well for fluorescence-activated cell sorter (FACS) analysis. For extracellular flux analysis, HeLa cells seeded into five 100-mm tissue culture dishes were transfected with Rv1813c DNA cloned into pMACS 4-IRESII vector (Miltenyi Biotec, France), allowing Rv1813c coexpression with a truncated CD4 surface marker. After 24 h, EDTA-resuspended cells were pooled, and CD4-positive cells were selected through magnetic cell sorting (MACS) as described below.
Mitochondria isolation and biochemical treatments.
Mitochondria were isolated as described (55). Briefly, Dictyostelium cells were washed in ice-cold buffer A (20 mM HEPES, pH 7, 1 mM EDTA, 250 mM sucrose, proteinase inhibitors), resuspended at a cell density of 3 × 108 cells/mL, and broken with a ball bearing homogenizer (8.02-mm bore, 8.002-mm ball; 20 strokes). Unbroken cells were removed by low-speed centrifugation (5 min, 1,500 × g). The supernatant was next centrifuged for 15 min at 16,000 × g. The pellet was resuspended in buffer A, and the centrifugation was repeated to yield the enriched mitochondria fraction. For further subcellular fractionation, this fraction was further centrifuged at 100,000 × g for 1 h. Triton X-114 phase fractionation was performed as described (56). Briefly, mitochondria were incubated for 20 min at 4°C in 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 1% Triton X-114. The samples were loaded on a 6% sucrose cushion, incubated at 30°C for 3 min for condensation, and centrifuged at 300 × g for 3 min at room temperature. Supernatants were adjusted to 1% Triton X-114, and the procedure was repeated. The detergent and aqueous phases were analyzed by Western blotting.
For carbonate extraction of integral membrane proteins, mitochondria were incubated for 30 min at 4°C in 0.1 M Na2CO3, pH 11.5, and centrifuged for 30 min at 100,000 × g as previously described (57). The pellets were resuspended in buffer A. The proteins in resuspended pellets and supernatants were precipitated with 15% TCA and resuspended in SDS-PAGE loading buffer. Integral membrane proteins were recovered in the pellet, while soluble and peripheral proteins were present in the supernatant. For high-salt washes, intact mitochondria were incubated in 10 mM Tris-HCl, pH 7.3, 250 mM sucrose, 200 mM KCl and incubated for 30 min at 4°C. Mitochondria were then centrifuged for 10 min at 16,000 × g. The pellets and supernatants were treated as above. For proteinase K digestions of mitochondrial peripheral membrane proteins, mitochondria in 20 mM HEPES, pH 7, 250 mM sucrose, 100 mM KCl, 2 mM MgCl2, 1 mM KH2PO4 were incubated with 100 μg/mL proteinase K for 30 min at 4°C ± 1% Triton X-100. Samples were then treated with TCA for protein precipitation. To break selectively mitochondrial outer membranes, mitochondria were resuspended in hypotonic buffer (2 mM HEPES, pH 7, 5 mM KCl, proteinase inhibitors) for 30 min at room temperature. After centrifugation at 16,000 × g for 10 min, the pellets and supernatants were treated with TCA as above.
Immunocytochemistry.
Dictyostelium cells were applied on glass coverslips for 3 h and then fixed with 4% paraformaldehyde for 30 min, washed, and permeabilized for 2 min in −20°C methanol. The cells were incubated with the indicated antibodies for 1 h, washed, and then stained with the appropriate fluorescent secondary antibodies for 30 min. After three washes, the coverslips were mounted in Mowiol. Mammalian cells were cultured on glass coverslips and fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min. The cells were washed in TBS (25 mM Tris, pH 7.4, 150 mM NaCl) for 10 min. After permeabilization with 0.2% Triton X-100 in TBS for 4 min, nonspecific binding was blocked with 0.2% gelatin from cold water fish skin (Sigma-Aldrich, France) in TBS for 30 min. The cells were incubated with primary antibodies in blocking buffer for 1 h and were then washed three times with 0.008% Triton X-100 in TBS for 10 min. The cells were incubated for 30 min with Alexa Fluor-labeled secondary antibodies in blocking buffer. After rinsing in washing buffer, the cell nuclei were stained with 1 μg/mL Hoechst in TBS for 5 min. Finally, the coverslips were mounted with Prolong Gold Antifade (catalog no. P36934, Thermo Fisher Scientific). The slides were examined under a Leica TCS SPE confocal microscope equipped with a 40×/1.15 or 63×/1.33 ACS APO oil-immersion objective or a Zeiss LSM880 AiryScan confocal microscope equipped with a 40×/1.4 or 63×/1.4 Oil Plan-apochromat DIC objective. Fluorescence images were adjusted for brightness, contrast, and color balance using ImageJ software.
Flow cytometry analysis of JC-1 and MitoSox-stained cells.
For MitoSox red staining of HeLa cells, 2.5 × 105 cells resuspended in CPBS buffer (PBS, 2.67 mM KCl, 0.5 mM MgCl2, 0.7 mM CaCl2, and 0.1% glucose) were incubated in 5 μM MitoSox red. After 20 min at 37°C with shaking, the cells were washed twice in CPBS buffer before FACS analysis. JC-1 staining of HeLa cells was made according to the manufacturer’s recommendations. Briefly, cells cultured in 6-well culture plates (2.5 × 105/well) were incubated at 37°C in culture medium supplemented with 2 μM JC1. After 30 min, the cells were washed, resuspended in PBS, and directly analyzed by flow cytometry. As positive control of JC-1 staining, 5 μM carbonyl cyanide m-chlorophenyl hydrazone (CCCP) was added to cells during JC-1 cell incubation.
MACS enrichment of CD4-Rv1813c transfected cells.
MACS enrichment of transfected cells was done with MACSelect transfected cell selection kit from Miltenyi Biotec, according to the supplier. Briefly, HeLa cells were transfected with empty pMACS4-IRESII (as control) or pMACS4-IRESII-Rv1813c plasmids allowing expression of truncated CD4 cell surface marker alone or in combination with Rv1813c, respectively. After 24 h, ~107 cells were washed, dissociated in ice-cold PBS containing 5 mM EDTA, centrifuged at 200 × g for 10 min at 4°C, and resuspended in 320 μL ice-cold degassed PBS supplemented with 0.5% bovine serum albumin and 5 mM EDTA (PBE). Magnetic labeling of the transfected cells was achieved by incubating cells with 80 μL of anti-CD4 coupled MACSelect microbeads on ice for 15 min. The volume was adjusted to 2 mL with PBE, and the cells were subjected to magnetic separation using a LS column (Miltenyi Biotec) and a MACS separator. After three washes with 3 mL of PBE, the cells were flushed out with 5 mL of PBE. To increase the purity of the magnetically labeled fraction, magnetic separation was repeated once on a second LS column. After the final wash, the cells were flushed out with 5 mL of cell culture medium, counted, and seeded at a density of 1.85 × 104 cells/well on XF96 cell culture microplates (Seahorse, Agilent Technologies, France) previously coated with 0.1 mg/mL poly-d-lysine (catalog no. P7280, Sigma) or on glass coverslips to evaluate the level of MACS enrichment of transfected cells by immunofluorescence. The cells were incubated at 37°C and analyzed 24 h later using the Seahorse XF96 extracellular flux analyzer or by confocal microscopy.
Extracellular flux analysis.
The cells plated the day before on XF96 cell culture microplates were washed with prewarmed cell culture medium 5 h before analysis to eliminate dead cells. Extracellular flux analysis was performed using a Seahorse XF extracellular flux analyzer, allowing simultaneous measurement of the oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). Mitochondrial respiration and glycolytic function of the cells were measured using a Cell Mito stress test kit (catalog no. 103015-100) and a cell glycolysis stress test kit (catalog no. 103020-100), respectively (Agilent Technologies, France). The cells were incubated in Seahorse XF DMEM, pH 7.4 (catalog no. 103575-100, Agilent) supplemented with 1 mM sodium pyruvate, 2 mM glutamine, and 10 mM glucose (Cell Mito stress test kit) or without glucose (cell glycolysis stress test kit) in a 37°C incubator without CO2 for 1 h prior to the assay. After calibration and three initial measurements at baseline, different perturbing chemicals corresponding to each kit were sequentially injected, and three successive measurements were taken after each injection.
Transmission electron microscopy.
MACS-enriched cells on glass coverslips were successively fixed with 2.5% gluteraldehyde in 0.1 M cacodylate buffer pH 7.4, washed with cacodylate buffer, postfixed in 1% osmium tetroxide in cacodylate buffer, washed with distilled water, and finally incubated in 1% uranyl acetate. Dehydration was performed through acetonitrile series. The samples were impregnated first in epon 118: acetonitrile 50:50 and then in 100% epon. After overnight polymerization at 60°C, coverslips were detached by thermal shock with liquid nitrogen. Polymerization was then prolonged for 48 h at 60°C. Ultrathin sections of 70 nm were cut with a Leica UC7 ultramicrotome (Leica Microsystems) and counterstained with lead citrate and uranyl acetate prepared in ethanol. The sections were observed in a Jeol 1200 EXII transmission electron microscope. All chemicals were from Electron Microscopy Sciences (USA), and the solvents were from Sigma. The images were processed using Fiji software.
ACKNOWLEDGMENTS
Flow cytometry and microscopy analyses of uninfected cells were performed at the Montpellier RIO imaging facility of the University of Montpellier, member of the national infrastructure France-BioImaging, supported by the French National Research Agency under program ANR-10-INBS-04, “Investments for the Future.” The Centre de Biochimie Structurale acknowledges support from the French Infrastructure for Integrated Structural Biology (FRISBI) with grant ANR-10-INSB-05-01.
The following reagents were obtained through BEI Resources, NIAID, National Institutes of Health: monoclonal anti-M. tuberculosis GlnA (Gene Rv2220), clone IT-58 (CBA5) (produced in vitro), NR-44103, and polyclonal anti-M. tuberculosis FtsZ (Gene Rv2150c) (antiserum rabbit).
M.M., A.d.V., P.B., Y.-M.B., O.T., T.A., L.H., J.G., C.G., G.V.M., F.L., and M.C.-G. performed the experiments; M.M., G.M., C.G., O.N., F.L., and M.C.-G. analyzed the data; F.L. and M.C.-G. conceived this study. All authors contributed to manuscript writing.
We declare no conflict of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
François Letourneur, Email: francois.letourneur@inserm.fr.
Martin Cohen-Gonsaud, Email: martin@cbs.cnrs.fr.
Eric Cascales, Centre National de la Recherche Scientifique, Aix-Marseille Université.
REFERENCES
- 1.Hmama Z, Peña-Díaz S, Joseph S, Av-Gay Y. 2015. Immunoevasion and immunosuppression of the macrophage by Mycobacterium tuberculosis. Immunological Rev 264:220–232. doi: 10.1111/imr.12268. [DOI] [PubMed] [Google Scholar]
- 2.Winden VJC, Van Houben ENG, Braunstein M. 2019. Protein export into and across the atypical diderm cell envelope of mycobacteria. Microbiol Spectr 7. doi: 10.1128/microbiolspec.GPP3-0043-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicholson KR, Mousseau CB, Champion MM, Champion PA. 2021. The genetic proteome: using genetics to inform the proteome of mycobacterial pathogens. PLoS Pathogens 17:e1009124. doi: 10.1371/journal.ppat.1009124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kostakioti M, Newman CL, Thanassi DG, Stathopoulos C. 2005. Mechanisms of protein export across the bacterial outer membrane. J Bacteriol 187:4306–4314. doi: 10.1128/JB.187.13.4306-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Målen H, Berven FS, Fladmark KE, Wiker HG. 2007. Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7:1702–1718. doi: 10.1002/pmic.200600853. [DOI] [PubMed] [Google Scholar]
- 6.de Souza GA, Leversen NA, Målen H, Wiker HG. 2011. Bacterial proteins with cleaved or uncleaved signal peptides of the general secretory pathway. J Proteomics 75:502–510. doi: 10.1016/j.jprot.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 7.Albrethsen J, Agner J, Piersma SR, Højrup P, Pham TV, Weldingh K, Jimenez CR, Andersen P, Rosenkrands I. 2013. Proteomic profiling of Mycobacterium tuberculosis identifies nutrient-starvation-responsive toxin-antitoxin systems. Mol Cell Proteomics 12:1180–1191. doi: 10.1074/mcp.M112.018846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tucci P, Portela M, Chetto CR, González-Sapienza G, Marín M. 2020. Integrative proteomic and glycoproteomic profiling of Mycobacterium tuberculosis culture filtrate. PLoS One 15:e0221837. doi: 10.1371/journal.pone.0221837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perkowski EF, Zulauf KE, Weerakoon D, Hayden JD, Ioerger TR, Oreper D, Gomez SM, Sacchettini JC, Braunstein M. 2017. The EXIT strategy: an approach for identifying bacterial proteins exported during host infection. mBio 8:e00333-17. doi: 10.1128/mBio.00333-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Penn BH, Netter Z, Johnson JR, Von Dollen J, Jang GM, Johnson T, Ohol YM, Maher C, Bell SL, Geiger K, Golovkine G, Du X, Choi A, Parry T, Mohapatra BC, Storck MD, Band H, Chen C, Jäger S, Shales M, Portnoy DA, Hernandez R, Coscoy L, Cox JS, Krogan NJ. 2018. An Mtb-human protein-protein interaction map identifies a switch between host antiviral and antibacterial responses. Mol Cell 71:637–648.e5. doi: 10.1016/j.molcel.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang Y, Zhang X, Bai X, Yang Y, Gong W, Wang T, Ling Y, Zhang J, Wang L, Wang J, Li G, Chen Y, Chen X, Wu X. 2019. Immunogenicity and therapeutic effects of latency-associated genes in a Mycobacterium tuberculosis reactivation mouse model. Hum Gene Ther Methods 30:60–69. doi: 10.1089/hgtb.2018.211. [DOI] [PubMed] [Google Scholar]
- 12.Bretl DJ, He H, Demetriadou C, White MJ, Penoske RM, Salzman NH, Zahrt TC. 2012. MprA and DosR coregulate a Mycobacterium tuberculosis virulence operon encoding Rv1813c and Rv1812c. Infect Immun 80:3018–3033. doi: 10.1128/IAI.00520-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feltcher ME, Sullivan JT, Braunstein M. 2010. Protein export systems of Mycobacterium tuberculosis: novel targets for drug development? Future Microbiol 5:1581–1597. doi: 10.2217/fmb.10.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gracy J, Chiche L. 2005. PAT: a protein analysis toolkit for integrated biocomputing on the web. Nucleic Acids Res 33:W65–W71. doi: 10.1093/nar/gki455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McGuffin LJ, Bryson K, Jones DT. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 16.Daleke MH, Ummels R, Bawono P, Heringa J, Vandenbroucke-Grauls CMJE, Luirink J, Bitter W. 2012. General secretion signal for the mycobacterial type VII secretion pathway. Proc Natl Acad Sci USA 109:11342–11347. doi: 10.1073/pnas.1119453109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turapov O, Waddell SJ, Burke B, Glenn S, Sarybaeva AA, Tudo G, Labesse G, Young DI, Young M, Andrew PW, Butcher PD, Cohen-Gonsaud M, Mukamolova GV. 2014. Oleoyl coenzyme A regulates interaction of transcriptional regulator RaaS (Rv1219c) with DNA in mycobacteria. J Biol Chem 289:25241–25249. doi: 10.1074/jbc.M114.577338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm L, Rosenström P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye Y, Godzik A. 2004. Database searching by flexible protein structure alignment. Protein Sci 13:1841–1850. doi: 10.1110/ps.03602304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Steinert M. 2011. Pathogen-host interactions in Dictyostelium, Legionella, Mycobacterium and other pathogens. Semin Cell Dev Biol 22:70–76. doi: 10.1016/j.semcdb.2010.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Müller-Taubenberger A, Kortholt A, Eichinger L. 2013. Simple system–substantial share: the use of Dictyostelium in cell biology and molecular medicine. Eur J Cell Biol 92:45–53. doi: 10.1016/j.ejcb.2012.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Cardenal-Muñoz E, Barisch C, Lefrançois LH, López-Jiménez AT, Soldati T. 2017. When dicty met myco, a (not so) romantic story about one amoeba and its intracellular pathogen. Front Cell Infect Microbiol 7:529. doi: 10.3389/fcimb.2017.00529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Troll H, Malchow D, Müller-Taubenberger A, Humbel B, Lottspeich F, Ecke M, Gerisch G, Schmid A, Benz R. 1992. Purification, functional characterization, and CDNA sequencing of mitochondrial porin from Dictyostelium discoideum. J Biol Chem 267:21072–21079. doi: 10.1016/S0021-9258(19)36799-7. [DOI] [PubMed] [Google Scholar]
- 24.Jamwal S, Midha MK, Verma HN, Basu A, Rao KVS, Manivel V. 2013. Characterizing virulence-specific perturbations in the mitochondrial function of macrophages infected with Mycobacterium tuberculosis. Sci Rep 3:1328. doi: 10.1038/srep01328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stridh H, Kimland M, Jones DP, Orrenius S, Hampton MB. 1998. Cytochrome c release and caspase activation in hydrogen peroxide- and tributyltin-induced apoptosis. FEBS Lett 429:351–355. doi: 10.1016/s0014-5793(98)00630-9. [DOI] [PubMed] [Google Scholar]
- 26.Spier A, Stavru F, Cossart P. 2019. Interaction between intracellular bacterial pathogens and host cell mitochondria. Microbiol Spectr 7:3–13. doi: 10.1128/microbiolspec.BAI-0016-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garaude J. 2019. Reprogramming of mitochondrial metabolism by innate immunity. Curr Opin Immunol 56:17–23. doi: 10.1016/j.coi.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 28.Hicks SW, Galán JE. 2013. Exploitation of eukaryotic subcellular targeting mechanisms by bacterial effectors. Nat Rev Microbiol 11:316–326. doi: 10.1038/nrmicro3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hua Y, Yan K, Wan C. 2018. Clever cooperation: interactions between EspF and Host proteins. Front Microbiol 9:2831. doi: 10.3389/fmicb.2018.02831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Escoll P, Song O-R, Viana F, Steiner B, Lagache T, Olivo-Marin J-C, Impens F, Brodin P, Hilbi H, Buchrieser C. 2017. Legionella pneumophila modulates mitochondrial dynamics to trigger metabolic repurposing of infected macrophages. Cell Host Microbe 22:302–316.e7. doi: 10.1016/j.chom.2017.07.020. [DOI] [PubMed] [Google Scholar]
- 31.Bertholet S, Ireton GC, Kahn M, Guderian J, Mohamath R, Stride N, Laughlin EM, Baldwin SL, Vedvick TS, Coler RN, Reed SG. 2008. Identification of human T cell antigens for the development of vaccines against Mycobacterium tuberculosis. J Immunol 181:7948–7957. doi: 10.4049/jimmunol.181.11.7948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haydel SE, Clark-Curtiss JE. 2004. Global expression analysis of two-component system regulator genes during Mycobacterium tuberculosis growth in human macrophages. FEMS Microbiol Lett 236:341–347. doi: 10.1016/j.femsle.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Pan F, Ye T, Sun P, Gui S, Liang B, Li L, Zheng D, Wang J, Hesketh RL, Yang L, Zheng C. 2020. Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology 295:715–721. doi: 10.1148/radiol.2020200370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zahrt TC, Deretic V. 2001. Mycobacterium tuberculosis signal transduction system required for persistent infections. Proc Natl Acad Sci USA 98:12706–12711. doi: 10.1073/pnas.221272198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schnappinger D, Schoolnik GK, Ehrt S. 2006. Expression profiling of host pathogen interactions: how Mycobacterium tuberculosis and the macrophage adapt to one another. Microbes Infect 8:1132–1140. doi: 10.1016/j.micinf.2005.10.027. [DOI] [PubMed] [Google Scholar]
- 36.Talaat AM, Lyons R, Howard ST, Johnston SA. 2004. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci USA 101:4602–4607. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi L, Jiang Q, Bushkin Y, Subbian S, Tyagi S. 2019. Biphasic dynamics of macrophage immunometabolism during Mycobacterium tuberculosis infection. mBio 10:e02550-18. doi: 10.1128/mBio.02550-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi L, Eugenin EA, Subbian S. 2016. Immunometabolism in tuberculosis. Front Immunol 7:150. doi: 10.3389/fimmu.2016.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson JL, Mayr HK, Weichhart T. 2019. Metabolic programming of macrophages: implications in the pathogenesis of granulomatous disease. Front Immunol 10:2265. doi: 10.3389/fimmu.2019.02265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohareer K, Medikonda J, Vadankula GR, Banerjee S. 2020. Mycobacterial control of host mitochondria: bioenergetic and metabolic changes shaping cell fate and infection outcome. Front Cell Infect Microbiol 10:457. doi: 10.3389/fcimb.2020.00457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cumming BM, Addicott KW, Adamson JH, Steyn AJ. 2018. Mycobacterium tuberculosis induces decelerated bioenergetic metabolism in human macrophages. eLife 7:e39169. doi: 10.7554/eLife.39169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simeone R, Bobard A, Lippmann J, Bitter W, Majlessi L, Brosch R, Enninga J. 2012. Phagosomal rupture by Mycobacterium tuberculosis results in toxicity and host cell death. PLoS Pathog 8:e1002507. doi: 10.1371/journal.ppat.1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jamwal SV, Mehrotra P, Singh A, Siddiqui Z, Basu A, Rao KVS. 2016. Mycobacterial escape from macrophage phagosomes to the cytoplasm represents an alternate adaptation mechanism. Sci Rep 6:23089. doi: 10.1038/srep23089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mehrotra P, Jamwal SV, Saquib N, Sinha N, Siddiqui Z, Manivel V, Chatterjee S, Rao KVS. 2014. Pathogenicity of Mycobacterium tuberculosis is expressed by regulating metabolic thresholds of the host macrophage. PLoS Pathog 10:e1004265. doi: 10.1371/journal.ppat.1004265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, Wang X. 1997. Cytochrome c and DATP-dependent formation of Apaf-1/Caspase-9 complex initiates an apoptotic protease cascade. Cell 91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 46.Shen Y, Bax A. 2015. Protein structural information derived from NMR chemical shift with the neural network program TALOS-N. Methods Mol Biol 1260:17–32. doi: 10.1007/978-1-4939-2239-0_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CA, Nabuurs SB, Güntert P, Livny M, Markley JL, Nilges M, Ulrich EL, Kaptein R, Bonvin AM. 2005. RECOORD: a recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins 59(4):662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski RA, Moss DS, Thornton JM. 1993. Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 49.Koradi R, Billeter M, Wüthrich K. 1996. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 50.Dias M, Blanc C, Thazar-Poulot N, Ben Larbi S, Cosson P, Letourneur F. 2013. Dictyostelium ACAP-A is an ArfGAP involved in cytokinesis, cell migration and actin cytoskeleton dynamics. J Cell Sci 126(Pt 3):756–766. doi: 10.1242/jcs.113951. [DOI] [PubMed] [Google Scholar]
- 51.Davidson AJ, King JS, Insall RH. 2013. The use of streptavidin conjugates as immunoblot loading controls and mitochondrial markers for use with Dictyostelium discoideum. Biotechniques 55:39–41. doi: 10.2144/000114054. [DOI] [PubMed] [Google Scholar]
- 52.Turapov O, Forti F, Kadhim B, Ghisotti D, Sassine J, Straatman-Iwanowska A, Bottrill AR, Moynihan PJ, Wallis R, Barthe P, Cohen-Gonsaud M, Ajuh P, Vollmer W, Mukamolova GV. 2018. Two Faces of CwlM, an Essential PknB Substrate, in Mycobacterium tuberculosis. Cell Rep 25:57–67.e5. doi: 10.1016/j.celrep.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manstein DJ, Schuster HP, Morandini P, Hunt DM. 1995. Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene 162:129–134. doi: 10.1016/0378-1119(95)00351-6. [DOI] [PubMed] [Google Scholar]
- 54.Cornillon S, Pech E, Benghezal M, Ravanel K, Gaynor E, Letourneur F, Brückert F, Cosson P. 2000. Phg1p is a nine-transmembrane protein superfamily member involved in dictyostelium adhesion and phagocytosis. J Biol Chem 275:34287–34292. doi: 10.1074/jbc.M006725200. [DOI] [PubMed] [Google Scholar]
- 55.Aubry L, Klein G. 2006. Purification techniques of subcellular compartments for analytical and preparative purposes. Methods Mol Biol 346:171–185. doi: 10.1385/1-59745-144-4:171. [DOI] [PubMed] [Google Scholar]
- 56.Bordier C. 1981. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem 256:1604–1607. [PubMed] [Google Scholar]
- 57.Fujiki Y, Hubbard AL, Fowler S, Lazarow PB. 1982. Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J Cell Biol 93:97–102. doi: 10.1083/jcb.93.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental legends. Download spectrum.01066-23-s0001.pdf, PDF file, 0.10 MB (100.9KB, pdf)
Fig. S1. Download spectrum.01066-23-s0002.tif, TIF file, 7.8 MB (7.8MB, tif)
Fig. S2. Download spectrum.01066-23-s0003.tif, TIF file, 4.4 MB (4.4MB, tif)
Fig. S3. Download spectrum.01066-23-s0004.tif, TIF file, 6.7 MB (6.7MB, tif)
Fig. S4. Download spectrum.01066-23-s0005.tif, TIF file, 6.7 MB (6.7MB, tif)
Table S1. Download spectrum.01066-23-s0006.docx, DOCX file, 0.02 MB (16.4KB, docx)