ABSTRACT
Aspergillus fumigatus is a ubiquitous environmental mold that can cause severe disease in immunocompromised patients and chronic disease in individuals with underlying lung conditions. Triazoles are the most widely used class of antifungal drugs to treat A. fumigatus infections, but their use in the clinic is threatened by the emergence of triazole-resistant isolates worldwide, reinforcing the need for a better understanding of resistance mechanisms. The predominant mechanisms of A. fumigatus triazole resistance involve mutations affecting the promoter region or coding sequence of the target enzyme of the triazoles, Cyp51A. However, triazole-resistant isolates without cyp51A-associated mutations are frequently identified. In this study, we investigate a pan-triazole-resistant clinical isolate, DI15-105, that simultaneously carries the mutations hapEP88L and hmg1F262del, with no mutations in cyp51A. Using a Cas9-mediated gene-editing system, hapEP88L and hmg1F262del mutations were reverted in DI15-105. Here, we show that the combination of these mutations accounts for pan-triazole resistance in DI15-105. To our knowledge, DI15-105 is the first clinical isolate reported to simultaneously carry mutations in hapE and hmg1 and only the second with the hapEP88L mutation.
IMPORTANCE Triazole resistance is an important cause of treatment failure and high mortality rates for A. fumigatus human infections. Although Cyp51A-associated mutations are frequently identified as the cause of A. fumigatus triazole resistance, they do not explain the resistance phenotypes for several isolates. In this study, we demonstrate that hapE and hmg1 mutations additively contribute to pan-triazole resistance in an A. fumigatus clinical isolate lacking cyp51-associated mutations. Our results exemplify the importance of and the need for a better understanding of cyp51A-independent triazole resistance mechanisms.
KEYWORDS: Aspergillus fumigatus, triazole drug resistance, hmg1, hapE, CCAAT box binding complex
INTRODUCTION
Aspergillus fumigatus is a ubiquitous environmental mold, spores of which are commonly inhaled by humans (1). While innocuous in immunocompetent individuals, A. fumigatus can cause life-threatening disease when the host immune system is compromised, especially in patients undergoing hematopoietic stem cell or solid organ transplantation (2–5). A. fumigatus infections can also manifest as chronic disease in individuals with certain underlying lung conditions, such as asthma, lung cavitary disease (e.g., tuberculosis or sarcoidosis), cystic fibrosis, or chronic obstructive pulmonary disease (6–8). It is estimated that over 200,000 life-threatening Aspergillus infections occur every year, with mortality rates as high as 20 to 90% (9–12).
Mold-active triazoles are first-line therapy against A. fumigatus infections (13). Triazole antifungals inhibit the A. fumigatus sterol demethylases Cyp51A and Cyp51B (14), causing an impairment in ergosterol biosynthesis and an accumulation of toxic sterols that culminates in fungicidal activity (15–18). Currently, four triazole drugs have utility for the management of Aspergillus infections, i.e., voriconazole (VRC), itraconazole (ITRA), posaconazole (POS), and isavuconazole (ISA) (13). Unfortunately, an alarming number of triazole-resistant clinical and environmental A. fumigatus isolates have been reported worldwide, encouraging the search for novel therapeutic strategies and reinforcing the need for a better understanding of antifungal resistance mechanisms (19, 20).
The predominant mechanism of triazole resistance in A. fumigatus is genetic mutation leading to changes in the amino acid sequence and/or expression levels of one of the two target enzymes, Cyp51A (19, 20). Tandem repeats (TRs) in the promoter of the cyp51A gene coupled with single point mutations in the coding sequence, such as TR34/L98H or TR46/Y121F/T289A, have been widely reported in clinical and environmental isolates. Such TRs in the promoter lead to upregulation of cyp51A expression, which in turn increases the amount of triazole necessary for enzyme inhibition (21, 22). Moreover, amino acid substitutions in hot spot residues, such as G54 and M220, as well as those occurring with TRs in the promoter, are thought to cause conformational changes that reduce the binding affinity of the triazole drugs with the target enzyme (19, 20, 23, 24).
While cyp51A-associated mutations are the most common and best-characterized mechanism of triazole drug resistance, non-cyp51A triazole-resistant clinical isolates have been increasingly reported, with prevalence rates varying from 15 to 60% depending on geographic region (25–28). However, cyp51A-indepent triazole drug resistance remains poorly understood and underinvestigated. One of the potential causes of triazole drug resistance is the overexpression of ATP binding cassette (ABC) and major facilitator superfamily (MFS) multidrug transporters (29–31), which is often observed among triazole-resistant A. fumigatus isolates. Overexpression of these transporters is thought to cause a decrease in intracellular drug concentrations, therefore decreasing the amount of drug available to inhibit sterol demethylases (29–31).
Mutations in hmg1, which encodes the 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase Hmg1, have also recently emerged as a genetic determinant of triazole drug resistance in A. fumigatus (26). Hmg1 initiates ergosterol biosynthesis by catalyzing the reduction of HMG-CoA into mevalonate and has a conserved motif known as the sterol-sensing domain (SSD), which is predicted to participate in the regulation of HMG-CoA reductase activity (32–34). Although the clinical prevalence of hmg1 mutations has not been assessed in a systematic manner, such mutations have been identified in over 150 isolates thus far reported in the literature, many of which are resistant to triazole drugs and lack cyp51A-associated mutations (25–28, 35–44). The most frequent Hmg1 amino acid substitutions associated with triazole drug resistance occur in residues putatively located in the Hmg1 SSD, such as S269, S305, G307, and I412, with several studies showing that some of these mutations, namely, F262del, S305P, I412S, S269F, and E306K, impart decreased susceptibility to multiple triazoles when genetically introduced into a susceptible laboratory strain (25, 26, 36, 40). The mechanism by which hmg1 mutations alter triazole resistance remains to be defined; however, it is speculated that amino acid substitutions in the SSD impair its ability to sense sterols and signal for Hmg1 degradation (25, 26, 28).
Another recently identified mechanism of triazole drug resistance involves mutations in the hapE gene, which encodes one of the three subunits of the CCAAT box binding complex (CBC), a transcription factor that has been shown to repress the expression of several ergosterol biosynthesis genes, including cyp51A (45, 46). It has been suggested that the hapEP88L mutation decreases the binding affinity of the CBC for the promoter region of cyp51A (47), impairing its ability to repress gene expression, which culminates in overexpression of cyp51A. Thus far, hapE mutations seem to be rare occurrences, having been reported for only 3 isolates (26, 46, 48).
In this study, we investigated a pan-triazole-resistant clinical isolate, DI15-105, that simultaneously carries the mutations hapEP88L and hmg1F262del and lacks mutations in cyp51A. Our goal was to determine the impact of these mutations individually and in combination on the triazole resistance phenotype of DI15-105. Using a Cas9-mediated gene-editing system, hapEP88L and hmg1F262del mutations were reverted in DI15-105. Our results demonstrated that, together, these mutations account for the high levels of pan-triazole resistance in DI15-105.
RESULTS
Correction of the hapEP88L and hmg1F262del mutations recovers DI15-105 susceptibility to triazole drugs.
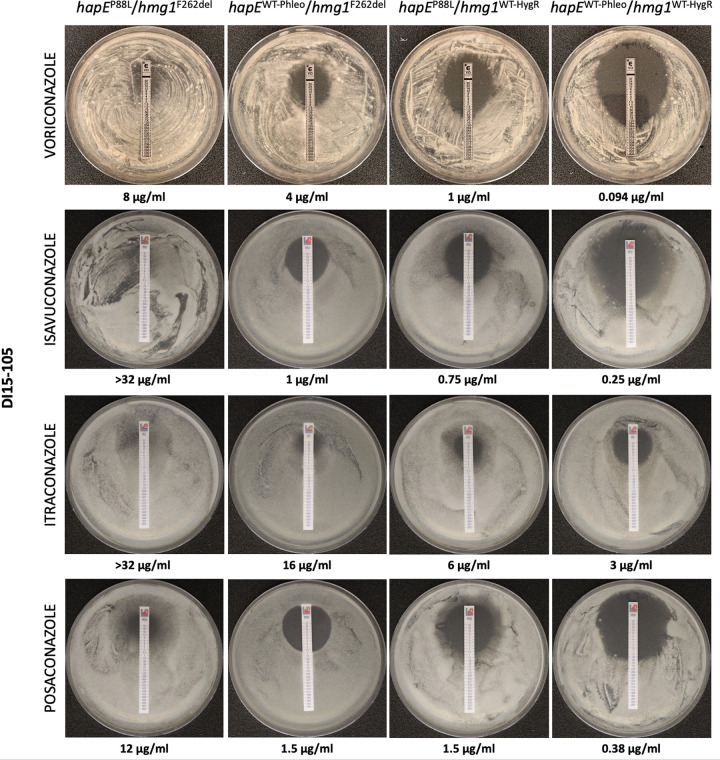
The hapEP88L and hmg1F262del mutations have been individually observed in triazole-resistant clinical isolates and independently shown to influence triazole susceptibility (25, 26, 46). Using in vitro-assembled Cas9 ribonucleoproteins (RNPs), allele swaps were performed and successfully replaced the mutated versions of hapE and hmg1 (previously performed [26]) with their respective wild-type (WT) versions (derived from AF293) in the clinical isolate DI15-105, as confirmed by PCR screening (see Fig. S1 and S2 in the supplemental material) and Sanger sequencing (data not shown). Using gradient diffusion test strips (Fig. 1), triazole susceptibility was determined and revealed that correction of either hapE or hmg1 gene sequences decreased DI15-105 MICs by 2-fold and 5-fold, respectively. MIC values decreased even further, ≥10-fold, when hapE and hmg1 gene sequences were both corrected.
FIG 1.
Antifungal susceptibility of A. fumigatus DI15-105 clinical isolates after correction of hapEP88L and hmg1F262del mutations, using gradient diffusion test strips. Conidia were inoculated in RPMI 1640 agar with VRC, ISA, ITRA, or POS gradient diffusion test strips and incubated at 35°C for 48 h. MICs were determined as the concentration at which a zone of growth clearance intercepted the drug strip; MIC values are indicated at the bottom of each image.
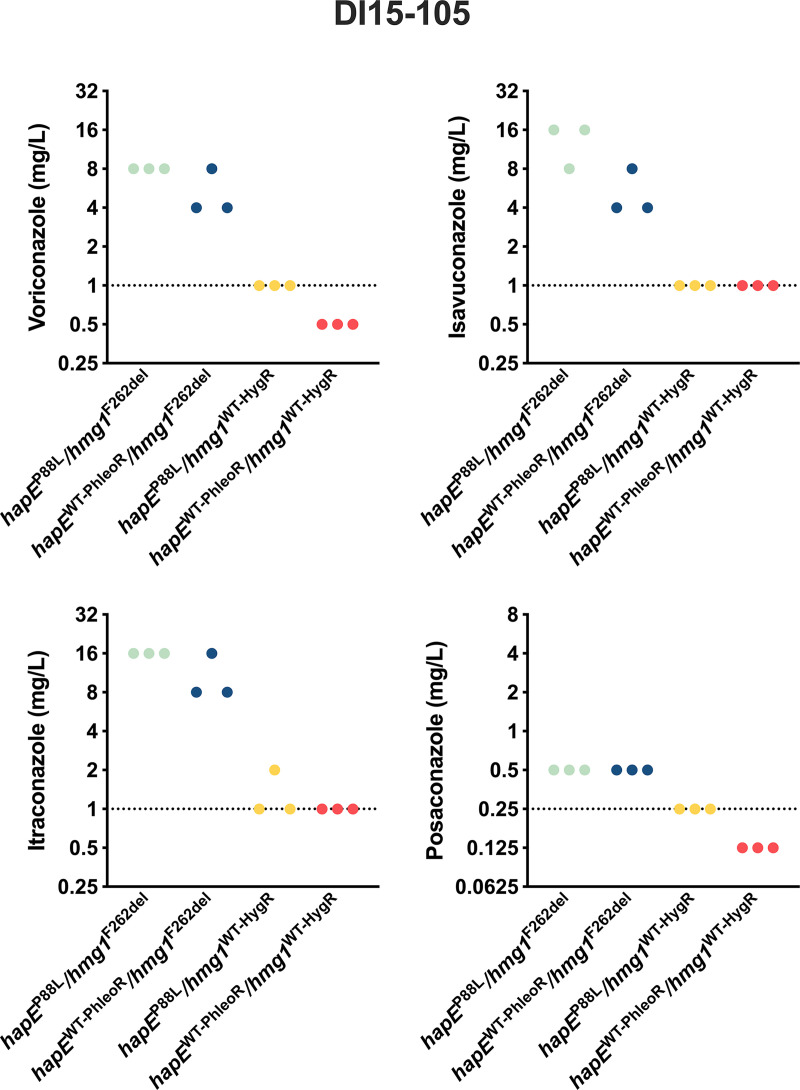
Triazole susceptibility of DI15-105 and derived strains was also assessed by broth microdilution assays according to the Clinical and Laboratory Standards Institute (CLSI) methodology (49) (Fig. 2). Correction of the hapE mutation in DI15-105_hapEWT-PhleoR/hmg1F262del decreased the MICs for VRC, ISA, and ITRA by 2-fold, while correction of the hmg1 mutation in DI15-105_hapEP88L/hmg1WT-HygR decreased the MICs for all triazoles (VRC and ISA, 8-fold; ITRA, 16-fold; POS, 2-fold) (Fig. 2). Correction of both hapE and hmg1 mutations in DI15-105_hapEWT-PhleoR/hmg1WT-HygR fully restored triazole susceptibility of DI15-105 as established by the CLSI epidemiological cutoff values, reducing the MIC levels by 4-fold (POS) to 16-fold (VRC, ISA, and ITRA).
FIG 2.
Antifungal susceptibility of A. fumigatus DI15-105 clinical isolates after correction of hapEP88L and hmg1F262del mutations, using triazole broth microdilution assays. Triazole MICs were determined according to CLSI methodology. VRC, ISA, ITRA, and POS MICs of triplicates are represented, and dotted lines indicate CLSI epidemiological cutoff values for each triazole drug.
Correction of the hapEP88L and hmg1F262del mutations in DI15-105 reduces cyp51A expression levels.
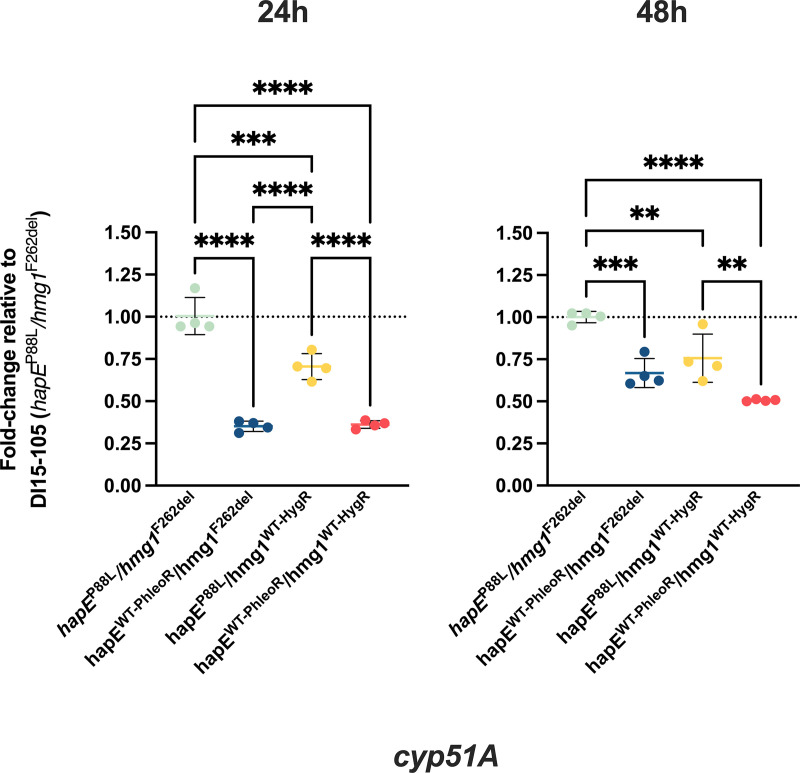
It has been suggested that the hapEP88L mutation reduces the affinity of CBC for the promoter region of cyp51A, thus impairing its ability to repress gene expression. This ultimately leads to an increase in cyp51A expression, resulting in increased MICs for triazole drugs. It is not clear by what mechanism hmg1 mutations, particularly hmg1F262del, increase triazole resistance, and previous studies have not reported changes in cyp51A expression. Since we observed an additive effect of these two mutations contributing to triazole resistance, we assessed expression levels of cyp51A upon correction of hapEP88L and hmg1F262del mutations. As expected, correction of the hapE mutation in both DI15-105_hapEWT-PhleoR/hmg1F262del and DI15-105_hapEWT-PhleoR/hmg1WT-HygR significantly decreased expression of cyp51A after 24 h (~2.8-fold and ~2.7-fold, respectively) and 48 h (~1.5-fold and ~2-fold, respectively) of fungal growth (Fig. 3). Correction of hmg1 also caused a less prominent but statistically significant decrease in cyp51A expression at 24 h (~1.4-fold) and 48 h (~1.3-fold) of fungal growth.
FIG 3.
Expression levels of cyp51A in DI15-105 isolates after correction of hapEP88L and hmg1F262del mutations. The cyp51A expression levels of hmg1 and hapE correction strains were assessed using RT-qPCR. RPMI 1640 medium was inoculated with A. fumigatus conidia at a concentration of 2 × 104 conidia/mL and incubated at 35°C and 250 rpm for 24 h or 48 h. Mycelia were harvested and subjected to RNA extraction. cDNA was synthesized, and qPCR was performed using tubA, which encodes β-tubulin A, as the housekeeping gene for data normalization. Changes in gene expression among isolates were calculated using the 2−ΔΔCT method. Experiments were performed using four biological replicates. Statistical analysis was performed using one-way ANOVA with Tukey’s multiple-comparison test. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Correction of the hapEP88L mutation improves DI15-105 radial growth and induces hypersensitivity to oxidative stress.
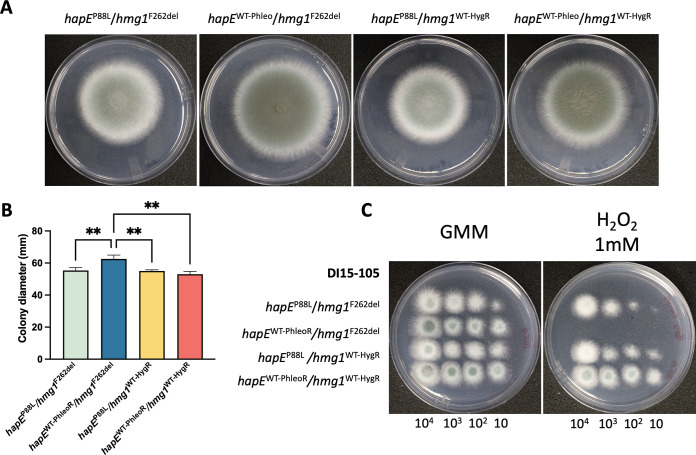
In some studies, the hapEP88L mutation has been associated with decreased fungal growth (45, 47). Therefore, we analyzed colony macromorphology (Fig. 4A) and radial growth (Fig. 4B) of the DI15-105 isolate and derived strains. Macromorphological analysis revealed that the strain with a hapE single correction (DI15-105_hapEWT-PhleoR/hmg1F262del) presented significantly enhanced radial growth, which was not observed in the double correction strain DI15-105_hapEWT-PhleoR/hmg1WT-HygR. We also observed that the hapE correction strains DI15-105_hapEWT-PhleoR/hmg1F262del and DI15-105_hapEWT-PhleoR/hmg1WT-HygR presented a darker green color than hapEP88L strains, suggesting greater conidium production; however, quantification of conidia did not show significant differences in the amounts of conidia per colony area among the isolates (Fig. S3).
FIG 4.
Phenotypic characterization of DI15-105 isolates after correction of hapEP88L and hmg1F262del mutations. (A and B) Macromorphology (A) and radial growth (B) were assessed after point inoculation of 104 A. fumigatus conidia/5 μL in GMM agar and incubation for 96 h at 37°C. (C) Fungal growth under oxidative stress was analyzed through spot dilution assays in GMM agar supplemented with 2 mM H2O2 using 5-μL suspensions containing 104, 103, 102, or 10 conidia/5 μL, followed by incubation at 37°C for 48 h. Experiments were performed using three biological replicates. Statistical analysis was performed using one-way ANOVA with Tukey’s multiple-comparison test. **, P < 0.01.
In order to evaluate the impact of hmg1 and hapE mutations on the DI15-105 stress response, strains were grown in agar medium supplemented with several stressors, namely, H2O2 (oxidative stress) (Fig. 4C), iron (depletion and excess amounts) (see Fig. S4A), calcofluor white and Congo red (cell wall stress) (see Fig. S4B), and sorbitol (osmotic stress) (see Fig. S4B). Only correction of the hapEP88L mutation in DI15-105_hapEWT-PhleoR/hmg1F262del rendered the isolate hypersensitive to oxidative stress, which once again was not observed when the hmg1 mutation was also corrected (Fig. 4; also see Fig. S5).
DISCUSSION
The emergence of triazole drug resistance represents an important challenge in the therapy of A. fumigatus infections (50). Genetic alterations in the cyp51A gene coding sequence or promoter region are the most frequently identified and best-characterized cause of A. fumigatus clinical triazole resistance. However, resistance to triazoles is not always explained by genetic alterations in cyp51A (25, 26, 28, 35, 46, 48, 51). In an earlier study, whole-genome sequencing of A. fumigatus clinical isolates collected in the United States revealed a clinical isolate, DI15-105, that had high levels of resistance to mold-active triazoles and no mutations in cyp51A (26). Instead, this isolate was shown to carry the hmg1F262del and hapEP88L mutations. Both mutations were previously reported separately in A. fumigatus clinical isolates and shown to directly contribute to triazole drug resistance (26, 46). Here, our goal was to determine the impact of these mutations individually and in combination on the triazole resistance phenotype of DI15-105.
The hmg1F262del mutation is predicted to alter a residue in the SSD of Hmg1, a conserved motif shown to be essential for the negative regulation of sterol biosynthesis in other eukaryotes, including Schizosaccharomyces pombe (32, 33, 52). We previously observed that restoration of the hmg1 WT sequence in DI15-105 restored triazole susceptibility to all mold-active triazoles (26). The same hmg1F262del mutation introduced in a susceptible A. fumigatus laboratory strain induced increases of at least 4-fold in the MICs for all triazoles, accumulation of ergosterol precursors, and significant increases in total cellular ergosterol levels (26). In a similar way, amino acid substitutions in other residues of the Hmg1 SSD have also been identified, and they are often associated with triazole-resistant isolates (25–27, 35, 36). Some of these mutations, namely, S305P, I412S, S269F, and E306K, have been shown to impart decreased susceptibility to multiple triazoles when genetically inserted in a triazole-susceptible laboratory strain (25, 26, 36). It is important to note that many of the hmg1 mutations so far reported in the literature occur in isolates that also carry cyp51A mutations that are known to drive triazole drug resistance (25–28, 36). However, it was demonstrated that, in some cases, the presence of the hmg1 mutation led to atypical and increased triazole MIC levels in the isolates (26, 27). For instance, genetic reversion of the cyp51A mutations TR34/L98H, G138C and M263I to the WT sequence did not recover triazole susceptibility in different clinical isolates (26). Those isolates also carried the hmg1 mutations E105K, Y250H, and I412S, respectively; the latter was previously shown to contribute directly to triazole resistance (26). Additionally, in a clinical isolate carrying the cyp51AM220I amino acid substitution, reversion of the hmg1S305P mutation reduced the VRC MIC while maintaining ITRA and POS MIC values (36). These data support the evidence that hmg1 mutations are important determinants of clinical triazole resistance. However, the mechanism by which Hmg1 SSD mutations lead to triazole resistance remains to be fully elucidated, and it is speculated that such mutations might affect the negative regulation of HMG-CoA reductase activity (25, 26).
The hapEP88L mutation present in DI15-105 was previously reported in a single clinical isolate that did not also carry mutations in cyp51A (46). The hapE gene encodes one of the three subunits of the CBC, a highly conserved transcription factor that has been shown to repress expression of several ergosterol biosynthesis genes, such as cyp51A, cyp51B, erg7B, and erg13B (45). It has been demonstrated that the hapEP88L mutation decreases the binding affinity of the CBC for the promoter region of cyp51A, impairing its ability to repress gene expression, which culminates in overexpression of cyp51A and thus increased tolerance to triazoles (47). Our results show that the single correction of the hapEP88L mutation in DI15-105_hapEWT-PhleoR/hmg1F262del leads to significant decreases in triazole MICs. Recently, a triazole-resistant clinical isolate without cyp51A mutations but carrying a HapE splicing site mutation (c.154-1G>A) was identified; it presented higher cyp51A expression levels than did an isogenic and triazole-susceptible isolate that had been collected previously from the same patient (48). However, the direct effect of this hapE splicing site mutation on triazole resistance has not yet been characterized. Interestingly, in both reports, hapE mutations in clinical isolates were thought to have been acquired in the patient after prolonged therapy with triazoles (47, 48). To our knowledge, DI15-105 is only the second isolate that has been reported to carry the hapEP88L mutation (26). These examples reinforce the clinical importance of hapE mutations for triazole resistance.
In our study, we observed that the hmg1F262del mutation has a major impact on the DI15-105 triazole resistance phenotype, since the reversion of this mutation generated a greater decrease in triazole MICs than did the correction of the hapEP88L mutation, in both strip and broth microdilution assays. In addition, correction of both hmg1F262del and hapEP88L mutations in DI15-105_hapEWT-PhleoR/hmg1WT-HygR led to a greater reduction in triazole MICs, demonstrating the additive effects of the two mutations on triazole resistance. In order to elucidate how the two mutations enhanced triazole resistance, we evaluated whether the increased susceptibility of the correction strains to triazole drugs was a consequence of cyp51A expression downregulation. Reverse transcription-quantitative PCR (RT-qPCR) results showed a statistically significant but slight decrease in cyp51A expression in all correction strains, although the decrease in cyp51A expression levels was more prominent in the DI15-105_hapEWT-PhleoR/hmg1F262del and DI15-105_hapEWT-PhleoR/hmg1WT-HygR strains, in which the hapE mutation was corrected. Given that the single correction of the hapEP88L mutation did not reduce triazole MICs to the extent observed with the reconstitution of both hapE and hmg1 mutations, we think that cyp51A expression is not the only mechanism operating in DI15-105. Further investigation is needed to demonstrate how the presence of the hmg1F262del and hapEP88L mutations together leads to increased MIC values in DI15-105.
In order to identify other ways in which hmg1F262del and hapEP88L mutations affect A. fumigatus, we analyzed fungal growth and responses to oxidative, iron, osmotic, and cell wall stress in DI15-105 and derived strains. While no significant changes were observed in the DI15-105_hapEP88L/hmg1WT-HygR and DI15-105_hapEWT-PhleoR/hmg1WT-HygR strains, surprisingly a slight improvement in radial growth was detected in the strain in which only the hapEP88L mutation was corrected. Although it was not observed in DI15-105_hapEWT-PhleoR/hmg1WT-HygR, this recovery of radial growth observed after single correction of hapE is consistent with previous reports that showed that the hapEP88L mutation, as well as the gene deletion of other subunits of the CBC, generate impairment of fungal growth (45, 47). Moreover, while no unique phenotypes were displayed when strains were grown under iron, osmotic, or cell wall stress conditions, single correction of the hapEP88L mutation in DI15-105_hapEWT-PhleoR/hmg1F262del resulted in hypersensitivity to H2O2. The fact that these phenotypes appear only in the strain in which single correction of hapE was performed is intriguing and suggests that, in DI15-105_hapEWT-PhleoR/hmg1F262del, recovery of hapE ability to regulate gene expression favors fungal growth under basal conditions while also disturbing the strain’s ability to respond to oxidative stress when the hmg1F262del mutation is present. Further investigation is required for clarification of this phenomenon.
In summary, we report for the first time that, in a clinical isolate with no cyp51A-associated mutations, hmg1F262del and hapEP88L mutations additively contribute to pan-triazole resistance. Because cyp51A expression levels do not seem to entirely explain the phenotypes observed in this study, the mechanism by which the two mutations lead to A. fumigatus triazole resistance remains unclear. This study is another example of the clinical importance of non-cyp51A-associated triazole resistance and reinforces the need for a better understanding of the mechanisms that drive triazole resistance in A. fumigatus.
MATERIALS AND METHODS
Isolates and growth conditions.
The pan-triazole-resistant clinical isolate DI15-105 was kindly donated by Nathan P. Wiederhold at the Fungus Testing Laboratory at the University of Texas Health Science Center at San Antonio. In a previous study, whole-genome sequencing revealed that this isolate presented no mutations in cyp51A but carried the hapEP88L and hmg1F262del mutations (26). In the same study, a DI15-105-derived strain (DI15-105_ hapEP88L/hmg1WT-HygR) in which the hmg1F262del mutation was reconstituted with a WT gene sequence was created, and it is also used here.
Fresh conidia from each isolate were obtained after harvesting of 3-day-old glucose minimal medium (GMM) agar cultures at 37°C. The concentrations of conidia in water stocks were then determined using a hemocytometer, and suspensions were kept at 4°C or used to prepare 50% glycerol stocks. Radial growth was determined by point inoculation of GMM plates with a 5-μL drop of a conidial suspension of 2 × 106 conidia/mL, and colony diameters were measured after incubation at 37°C for 96 h. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test with GraphPad Prism 9 software.
Stress analysis was performed through spot dilution (104, 103, 102, or 10 conidia per 5 μL suspension) on GMM agar plates supplemented with H2O2 (2 mM), FeSO4 (0, 30 μM, or 5 mM), calcofluor white (80 μg/mL), Congo red (40 μg/mL), or sorbitol (1.2 M) and incubation at 37°C for 48 h or 72 h.
Gene-editing strategy and A. fumigatus transformation mediated by CRISPR/Cas9.
In order to correct the hapEP88L and hmg1F262del mutations in the clinical isolate DI15-105, allele swaps mediated by a Cas9 system were performed, as described previously (26, 53). For reference, all CRISPR RNA (crRNA) sequences and primers (Integrated DNA Technologies) used to build strains are described in Table S1 in the supplemental material, and a detailed scheme of the gene-editing strategy is depicted in Fig. S1 and S2. Repair templates were constructed using WT hapE (Afu6g05300) and hmg1 (Afu2g03700) gene sequences obtained from AF293 genomic DNA. For hapE gene manipulation, the WT gene open reading frame (ORF) was cloned into the pAGRP plasmid (54), which contains a phleomycin resistance cassette (PhleoR), using the restriction enzymes NotI and PacI. After confirmation of successful insertion of hapE by diagnostic digestion and Sanger sequencing (data not shown), the plasmid (pAGRP-hapE) was used as a template for amplification of the repair template, with primers that incorporated 35-bp microhomology regions with the hapE ORF at both the 5′ and 3′ ends.
The DI15-105_ hapEP88L/hmg1WT-HygR strain (26) was created previously, as follows. For hmg1 gene manipulation, a two-component repair template that consisted of a split hmg1 allele and a hygromycin resistance marker (HygR) was used. A WT hmg1 gene sequence, including the ORF and around 50 bp upstream and 500 bp downstream, was PCR amplified using a 3′ primer that inserted 80 bases with homology to the 3′ end of the hygromycin B resistance gene ORF. The HygR cassette was PCR amplified from the pUCGH plasmid (55) using a 3′ primer that introduced approximately 70 bases with homology to the downstream region of hmg1.
In order to replace the whole hmg1 or hapE native ORF with a sequence of interest, two separate guide RNAs (gRNA) were selected to generate double-stranded DNA breaks at the 5′ and 3′ regions of the target gene ORF. gRNA duplexes were built using equimolar concentrations of a gene-specific 5′ or 3′ crRNA (see Table S1) and a universal transactivating crRNA (tracrRNA), and Cas9 RNP complexes were then assembled in vitro by mixing both 5′ and 3′ gRNA duplexes with the Cas9 enzyme, according to the manufacturer’s instructions. The transformation was carried out by mixing Cas9 RNPs with protoplasts generated from DI15-105 mycelia through enzymatic digestion and 2 to 5 μg of the desired repair template, as described in detail elsewhere (53). Transformation reactions were carried out on sorbitol minimal medium (SMM) agar plates (1.5% agar), and protoplasts were allowed to recover overnight at room temperature. For hmg1 transformations, SMM top agar (0.75% agar) containing 450 μg/mL hygromycin was added to the plates and incubated at 37°C for 3 to 5 days. For hapE transformations, SMM top agar with 375 μg/mL phleomycin was added to the plates and incubated until the next day at room temperature, followed by incubation at 30°C for 3 to 5 days. Single colonies were then subcultured in GMM supplemented with 150 μg/mL hygromycin or 125 μg/mL phleomycin, genomic DNA was extracted, and mutants were confirmed by multiple screening PCRs (see Fig. S1 and S2) and Sanger sequencing (data not shown).
The genetic manipulations performed in this study generated strain DI15-105_hapEWT-PhleoR/hmg1F262del, in which the mutated hapE allele was replaced with the WT gene sequence, and strain DI15-105_hapEWT-PhleoR/hmg1WT-HygR, in which both mutated hapE and hmg1 alleles were replaced with the respective WT gene sequences. The previously constructed DI15-105_hapEP88L/hmg1WT-HygR strain (26) was used as a background to generate the double correction strain DI15-105_hapEWT-PhleoR/hmg1WT-HygR.
Antifungal susceptibility testing.
Antifungal susceptibility to all clinically available mold-active triazoles was determined using gradient diffusion test strips and broth microdilution assays. VRC, ISA, ITRA, and POS strips were applied in the center of RPMI 1640 agar plates that had been inoculated with 500 μL of a suspension of 2 × 106 conidia/mL. The plates were incubated for 48 h at 35°C, and the MIC was defined where a zone of growth inhibition intercepted the strip reading scale. Triazole broth microdilution assays were performed according to CLSI M38-A2 methodology with modifications (49). Briefly, 2-fold serial dilutions of triazole drugs were distributed in U-bottomed 96-well plates, followed by a suspension of A. fumigatus conidia with a final concentration of 2 × 104 conidia/mL. The final drug concentrations tested varied from 0.03125 to 32 μg/mL. The plates were incubated for 48 h at 35°C, and MICs were determined.
Determination of cyp51A expression levels.
The cyp51A expression levels of DI15-105 and derived strains were assessed using RT-qPCR. Briefly, RPMI 1640 medium was inoculated with A. fumigatus conidia at a concentration of 2 × 104 conidia/mL and incubated at 35°C and 250 rpm for 24 h or 48 h. Cultures supernatants were discarded, and mycelia were harvested, washed in sterile distilled water, placed in screw-cap tubes, and immediately frozen in liquid N2. RNA extraction was performed using the RiboPure-Yeast kit (Invitrogen) according to the manufacturer’s instructions, followed by an extra purification step with isopropanol. RNA concentrations were determined using a NanoDrop spectrophotometer, and 0.5 μg of RNA was used for cDNA synthesis with the RevertAid first-strand cDNA synthesis kit (Thermo Fisher Scientific). qPCR was performed using the SsoAdvanced universal SYBR green supermix (Bio-Rad) with 1 μL of a 1:50 cDNA dilution as the template. The tubA gene, which encodes β-tubulin A, was selected as the housekeeping gene for data normalization, and changes in gene expression among isolates were calculated using the 2−ΔΔCT method with the average ΔCT of DI15-105 as the control. Primers are listed in Table S1 in the supplemental material. Experiments were performed using four biological replicates, each analyzed in technical duplicates. Statistical analysis was performed using one-way ANOVA followed by Tukey’s multiple-comparison test with GraphPad Prism 9 software.
ACKNOWLEDGMENTS
This work was supported by NIH grant R01 AI143197 to P.D.R. and J.R.F.
A.C.O.S. performed experiments, interpreted results, and wrote the manuscript. W.G. performed experiments. N.P.W. provided critical input and feedback. J.M.R. interpreted results, provided critical feedback, and assisted in writing the manuscript. J.R.F. and P.D.R. interpreted results, provided critical feedback, assisted in writing the manuscript, and supervised the study.
Footnotes
Supplemental material is available online only.
Contributor Information
Jarrod R. Fortwendel, Email: jfortwen@uthsc.edu.
P. David Rogers, Email: dave.rogers@stjude.org.
Teresa R. O'Meara, University of Michigan
REFERENCES
- 1.Latge JP, Chamilos G. 2019. Aspergillus fumigatus and aspergillosis in 2019. Clin Microbiol Rev 33:e00140-18. doi: 10.1128/CMR.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denning DW. 1998. Invasive aspergillosis. Clin Infect Dis 26:781–803. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 3.Dandachi D, Wilson Dib R, Fernández-Cruz A, Jiang Y, Chaftari A-M, Hachem R, Raad I. 2018. Invasive pulmonary aspergillosis in patients with solid tumours: risk factors and predictors of clinical outcomes. Ann Med 50:713–720. doi: 10.1080/07853890.2018.1518581. [DOI] [PubMed] [Google Scholar]
- 4.Lien M-Y, Chou C-H, Lin C-C, Bai L-Y, Chiu C-F, Yeh S-P, Ho M-W. 2018. Epidemiology and risk factors for invasive fungal infections during induction chemotherapy for newly diagnosed acute myeloid leukemia: a retrospective cohort study. PLoS One 13:e0197851. doi: 10.1371/journal.pone.0197851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kriegl L, Boyer J, Egger M, Hoenigl M. 2022. Antifungal stewardship in solid organ transplantation. Transpl Infect Dis 24:e13855. doi: 10.1111/tid.13855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith NL, Denning DW. 2011. Underlying conditions in chronic pulmonary aspergillosis including simple aspergilloma. Eur Respir J 37:865–872. doi: 10.1183/09031936.00054810. [DOI] [PubMed] [Google Scholar]
- 7.Denning DW, Pleuvry A, Cole DC. 2013. Global burden of allergic bronchopulmonary aspergillosis with asthma and its complication chronic pulmonary aspergillosis in adults. Med Mycol 51:361–370. doi: 10.3109/13693786.2012.738312. [DOI] [PubMed] [Google Scholar]
- 8.Hammond EE, McDonald CS, Vestbo J, Denning DW. 2020. The global impact of Aspergillus infection on COPD. BMC Pulm Med 20:241. doi: 10.1186/s12890-020-01259-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown GD, Denning DW, Gow NAR, Levitz SM, Netea MG, White TC. 2012. Hidden killers: human fungal infections. Sci Transl Med 4:165rv13. doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 10.Maertens JA, Raad II, Marr KA, Patterson TF, Kontoyiannis DP, Cornely OA, Bow EJ, Rahav G, Neofytos D, Aoun M, Baddley JW, Giladi M, Heinz WJ, Herbrecht R, Hope W, Karthaus M, Lee D-G, Lortholary O, Morrison VA, Oren I, Selleslag D, Shoham S, Thompson GR, Lee M, Maher RM, Schmitt-Hoffmann A-H, Zeiher B, Ullmann AJ. 2016. Isavuconazole versus voriconazole for primary treatment of invasive mould disease caused by Aspergillus and other filamentous fungi (SECURE): a phase 3, randomised-controlled, non-inferiority trial. Lancet 387:760–769. doi: 10.1016/S0140-6736(15)01159-9. [DOI] [PubMed] [Google Scholar]
- 11.van Paassen J, Russcher A, In 't Veld-van Wingerden AW, Verweij PE, Kuijper EJ. 2016. Emerging aspergillosis by azole-resistant Aspergillus fumigatus at an intensive care unit in the Netherlands, 2010 to 2013. Euro Surveill 21:30300. doi: 10.2807/1560-7917.ES.2016.21.30.30300. [DOI] [PubMed] [Google Scholar]
- 12.Koehler P, Hamprecht A, Bader O, Bekeredjian-Ding I, Buchheidt D, Doelken G, Elias J, Haase G, Hahn-Ast C, Karthaus M, Kekulé A, Keller P, Kiehl M, Krause SW, Krämer C, Neumann S, Rohde H, La Rosée P, Ruhnke M, Schafhausen P, Schalk E, Schulz K, Schwartz S, Silling G, Staib P, Ullmann A, Vergoulidou M, Weber T, Cornely OA, Vehreschild MJGT. 2017. Epidemiology of invasive aspergillosis and azole resistance in patients with acute leukaemia: the SEPIA Study. Int J Antimicrob Agents 49:218–223. doi: 10.1016/j.ijantimicag.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 13.Patterson TF, Thompson GR, Denning DW, Fishman JA, Hadley S, Herbrecht R, Kontoyiannis DP, Marr KA, Morrison VA, Nguyen MH, Segal BH, Steinbach WJ, Stevens DA, Walsh TJ, Wingard JR, Young J-AH, Bennett JE. 2016. Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin Infect Dis 63:e1–e60. doi: 10.1093/cid/ciw326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warrilow AGS, Melo N, Martel CM, Parker JE, Nes WD, Kelly SL, Kelly DE. 2010. Expression, purification, and characterization of Aspergillus fumigatus sterol 14-α demethylase (CYP51) isoenzymes A and B. Antimicrob Agents Chemother 54:4225–4234. doi: 10.1128/AAC.00316-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly SL, Arnoldi A, Kelly DE. 1993. Molecular genetic analysis of azole antifungal mode of action. Biochem Soc Trans 21:1034–1038. doi: 10.1042/bst0211034. [DOI] [PubMed] [Google Scholar]
- 16.Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Kelly DE. 1995. Mode of action and resistance to azole antifungals associated with the formation of 14α-methylergosta-8,24(28)-dien-3β,6α-diol. Biochem Biophys Res Commun 207:910–915. doi: 10.1006/bbrc.1995.1272. [DOI] [PubMed] [Google Scholar]
- 17.Vanden Bossche H, Marichal P, Le Jeune L, Coene MC, Gorrens J, Cools W. 1993. Effects of itraconazole on cytochrome P-450-dependent sterol 14α-demethylation and reduction of 3-ketosteroids in Cryptococcus neoformans. Antimicrob Agents Chemother 37:2101–2105. doi: 10.1128/AAC.37.10.2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marichal P, Vanden Bossche H. 1995. Mechanisms of resistance to azole antifungals. Acta Biochim Pol 42:509–516. doi: 10.18388/abp.1995_4904. [DOI] [PubMed] [Google Scholar]
- 19.Lestrade PPA, Meis JF, Melchers WJG, Verweij PE. 2019. Triazole resistance in Aspergillus fumigatus: recent insights and challenges for patient management. Clin Microbiol Infect 25:799–806. doi: 10.1016/j.cmi.2018.11.027. [DOI] [PubMed] [Google Scholar]
- 20.Rybak JM, Fortwendel JR, Rogers PD. 2019. Emerging threat of triazole-resistant Aspergillus fumigatus. J Antimicrob Chemother 74:835–842. doi: 10.1093/jac/dky517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellado E, Garcia-Effron G, Alcázar-Fuoli L, Melchers WJG, Verweij PE, Cuenca-Estrella M, Rodríguez-Tudela JL. 2007. A new Aspergillus fumigatus resistance mechanism conferring in vitro cross-resistance to azole antifungals involves a combination of cyp51A alterations. Antimicrob Agents Chemother 51:1897–1904. doi: 10.1128/AAC.01092-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van der Linden JWM, Camps SMT, Kampinga GA, Arends JPA, Debets-Ossenkopp YJ, Haas PJA, Rijnders BJA, Kuijper EJ, van Tiel FH, Varga J, Karawajczyk A, Zoll J, Melchers WJG, Verweij PE. 2013. Aspergillosis due to voriconazole highly resistant Aspergillus fumigatus and recovery of genetically related resistant isolates from domiciles. Clin Infect Dis 57:513–520. doi: 10.1093/cid/cit320. [DOI] [PubMed] [Google Scholar]
- 23.Nywening AV, Rybak JM, Rogers PD, Fortwendel JR. 2020. Mechanisms of triazole resistance in Aspergillus fumigatus. Environ Microbiol 22:4934–4952. doi: 10.1111/1462-2920.15274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snelders E, Karawajczyk A, Schaftenaar G, Verweij PE, Melchers WJG. 2010. Azole resistance profile of amino acid changes in Aspergillus fumigatus CYP51A based on protein homology modeling. Antimicrob Agents Chemother 54:2425–2430. doi: 10.1128/AAC.01599-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hagiwara D, Arai T, Takahashi H, Kusuya Y, Watanabe A, Kamei K. 2018. Non-cyp51A azole-resistant Aspergillus fumigatus isolates with mutation in HMG-CoA reductase. Emerg Infect Dis 24:1889–1897. doi: 10.3201/eid2410.180730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rybak JM, Ge W, Wiederhold NP, Parker JE, Kelly SL, Rogers PD, Fortwendel JR. 2019. Mutations in hmg1, challenging the paradigm of clinical triazole resistance in Aspergillus fumigatus. mBio 10:e00437-19. doi: 10.1128/mBio.00437-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Resendiz-Sharpe A, Hokken MWJ, Mercier T, Merckx R, Verhagen K, Dewitte L, Melchers WJG, Verweij PE, Maertens J, Lagrou K. 2020. Hmg1 gene mutation prevalence in triazole-resistant Aspergillus fumigatus clinical isolates. J Fungi (Basel) 6:227. doi: 10.3390/jof6040227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gonzalez-Jimenez I, Lucio J, Roldan A, Alcazar-Fuoli L, Mellado E. 2021. Are point mutations in HMG-CoA reductases (Hmg1 and Hmg2) a step towards azole resistance in Aspergillus fumigatus? Molecules 26:5975. doi: 10.3390/molecules26195975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajendran R, Mowat E, McCulloch E, Lappin DF, Jones B, Lang S, Majithiya JB, Warn P, Williams C, Ramage G. 2011. Azole resistance of Aspergillus fumigatus biofilms is partly associated with efflux pump activity. Antimicrob Agents Chemother 55:2092–2097. doi: 10.1128/AAC.01189-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Slaven JW, Anderson MJ, Sanglard D, Dixon GK, Bille J, Roberts IS, Denning DW. 2002. Increased expression of a novel Aspergillus fumigatus ABC transporter gene, atrF, in the presence of itraconazole in an itraconazole resistant clinical isolate. Fungal Genet Biol 36:199–206. doi: 10.1016/s1087-1845(02)00016-6. [DOI] [PubMed] [Google Scholar]
- 31.Fraczek MG, Bromley M, Buied A, Moore CB, Rajendran R, Rautemaa R, Ramage G, Denning DW, Bowyer P. 2013. The cdr1B efflux transporter is associated with non-cyp51a-mediated itraconazole resistance in Aspergillus fumigatus. J Antimicrob Chemother 68:1486–1496. doi: 10.1093/jac/dkt075. [DOI] [PubMed] [Google Scholar]
- 32.Theesfeld CL, Pourmand D, Davis T, Garza RM, Hampton RY. 2011. The sterol-sensing domain (SSD) directly mediates signal-regulated endoplasmic reticulum-associated degradation (ERAD) of 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase isozyme Hmg2. J Biol Chem 286:26298–26307. doi: 10.1074/jbc.M111.244798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Burg JS, Espenshade PJ. 2011. Regulation of HMG-CoA reductase in mammals and yeast. Prog Lipid Res 50:403–410. doi: 10.1016/j.plipres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcazar-Fuoli L, Mellado E, Garcia-Effron G, Lopez JF, Grimalt JO, Cuenca-Estrella JM, Rodriguez-Tudela JL. 2008. Ergosterol biosynthesis pathway in Aspergillus fumigatus. Steroids 73:339–347. doi: 10.1016/j.steroids.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 35.Sharma C, Nelson-Sathi S, Singh A, Radhakrishna Pillai M, Chowdhary A. 2019. Genomic perspective of triazole resistance in clinical and environmental Aspergillus fumigatus isolates without cyp51A mutations. Fungal Genet Biol 132:103265. doi: 10.1016/j.fgb.2019.103265. [DOI] [PubMed] [Google Scholar]
- 36.Arai T, Umeyama T, Majima H, Inukai T, Watanabe A, Miyazaki Y, Kamei K. 2021. Hmg1 mutations in Aspergillus fumigatus and their contribution to triazole susceptibility. Med Mycol 59:980–984. doi: 10.1093/mmy/myab026. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, van den Heuvel J, Debets AJM, Verweij PE, Melchers WJG, Zwaan BJ, Schoustra SE. 2017. Evolution of cross-resistance to medical triazoles in Aspergillus fumigatus through selection pressure of environmental fungicides. Proc Biol Sci 284:20170635. doi: 10.1098/rspb.2017.0635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gonzalez-Jimenez I, Lucio J, Amich J, Cuesta I, Sanchez Arroyo R, Alcazar-Fuoli L, Mellado E. 2020. A Cyp51B mutation contributes to azole resistance in Aspergillus fumigatus. J Fungi (Basel) 6:315. doi: 10.3390/jof6040315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakano Y, Tashiro M, Urano R, Kikuchi M, Ito N, Moriya E, Shirahige T, Mishima M, Takazono T, Miyazaki T, Izumikawa K. 2020. Characteristics of azole-resistant Aspergillus fumigatus attached to agricultural products imported to Japan. J Infect Chemother 26:1021–1025. doi: 10.1016/j.jiac.2020.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Handelman M, Morogovsky A, Liu W, Ben-Ami R, Osherov N. 2021. Triazole-resistant Aspergillus fumigatus in an Israeli patient with chronic cavitary pulmonary aspergillosis due to a novel E306K substitution in Hmg1. Antimicrob Agents Chemother 65:e01089-21. doi: 10.1128/AAC.01089-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang T, Yang X, Li R, Yang E, Wang Q, Osherov N, Chen W, Wan Z, Liu W. 2021. Emergence of W272C substitution in Hmg1 in a triazole-resistant isolate of Aspergillus fumigatus from a Chinese patient with chronic cavitary pulmonary aspergillosis. Antimicrob Agents Chemother 65:e00263-21. doi: 10.1128/AAC.00263-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takazono T, Ito Y, Tashiro M, Nakano Y, Hirayama T, Hosogaya N, Saijo T, Yamamoto K, Imamura Y, Miyazaki T, Yanagihara K, Kohno S, Mukae H, Izumikawa K. 2021. Transition of triazole-resistant Aspergillus fumigatus isolates in a Japanese tertiary hospital and subsequent genetic analysis. J Infect Chemother 27:537–539. doi: 10.1016/j.jiac.2020.11.027. [DOI] [PubMed] [Google Scholar]
- 43.Takeda K, Suzuki J, Watanabe A, Arai T, Koiwa T, Shinfuku K, Narumoto O, Kawashima M, Fukami T, Tamura A, Nagai H, Matsui H, Kamei K. 2021. High detection rate of azole-resistant Aspergillus fumigatus after treatment with azole antifungal drugs among patients with chronic pulmonary aspergillosis in a single hospital setting with low azole resistance. Med Mycol 59:327–334. doi: 10.1093/mmy/myaa052. [DOI] [PubMed] [Google Scholar]
- 44.Toyotome T, Onishi K, Sato M, Kusuya Y, Hagiwara D, Watanabe A, Takahashi H. 2021. Identification of novel mutations contributing to azole tolerance of Aspergillus fumigatus through in vitro exposure to tebuconazole. Antimicrob Agents Chemother 65:e02657-20. doi: 10.1128/AAC.02657-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gsaller F, Hortschansky P, Furukawa T, Carr PD, Rash B, Capilla J, Müller C, Bracher F, Bowyer P, Haas H, Brakhage AA, Bromley MJ. 2016. Sterol biosynthesis and azole tolerance is governed by the opposing actions of SrbA and the CCAAT binding complex. PLoS Pathog 12:e1005775. doi: 10.1371/journal.ppat.1005775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camps SMT, Dutilh BE, Arendrup MC, Rijs AJMM, Snelders E, Huynen MA, Verweij PE, Melchers WJG. 2012. Discovery of a HapE mutation that causes azole resistance in Aspergillus fumigatus through whole genome sequencing and sexual crossing. PLoS One 7:e50034. doi: 10.1371/journal.pone.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hortschansky P, Misslinger M, Mörl J, Gsaller F, Bromley MJ, Brakhage AA, Groll M, Haas H, Huber EM. 2020. Structural basis of HapEP88L-linked antifungal triazole resistance in Aspergillus fumigatus. Life Sci Alliance 3:e202000729. doi: 10.26508/lsa.202000729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ito Y, Takazono T, Koga S, Nakano Y, Ashizawa N, Hirayama T, Tashiro M, Saijo T, Yamamoto K, Imamura Y, Miyazaki T, Yanagihara K, Izumikawa K, Mukae H. 2021. Clinical and experimental phenotype of azole-resistant Aspergillus fumigatus with a HapE splice site mutation: a case report. BMC Infect Dis 21:573. doi: 10.1186/s12879-021-06279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clinical and Laboratory Standards Institute. 2008. Reference method for broth dilution antifungal susceptibility testing of filamentous fungi; approved standard, 2nd ed. CLSI document M38-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 50.Verweij PE, Chowdhary A, Melchers WJG, Meis JF. 2016. Azole resistance in Aspergillus fumigatus: can we retain the clinical use of mold-active antifungal azoles? Clin Infect Dis 62:362–368. doi: 10.1093/cid/civ885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bueid A, Howard SJ, Moore CB, Richardson MD, Harrison E, Bowyer P, Denning DW. 2010. Azole antifungal resistance in Aspergillus fumigatus: 2008 and 2009. J Antimicrob Chemother 65:2116–2118. doi: 10.1093/jac/dkq279. [DOI] [PubMed] [Google Scholar]
- 52.Burg JS, Powell DW, Chai R, Hughes AL, Link AJ, Espenshade PJ. 2008. Insig regulates HMG-CoA reductase by controlling enzyme phosphorylation in fission yeast. Cell Metab 8:522–531. doi: 10.1016/j.cmet.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Al Abdallah Q, Ge W, Fortwendel JR. 2017. A simple and universal system for gene manipulation in Aspergillus fumigatus: in vitro-assembled Cas9-guide RNA ribonucleoproteins coupled with microhomology repair templates. mSphere 2:e00446-17. doi: 10.1128/mSphere.00446-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fortwendel JR, Juvvadi PR, Rogg LE, Asfaw YG, Burns KA, Randell SH, Steinbach WJ. 2012. Plasma membrane localization is required for RasA-mediated polarized morphogenesis and virulence of Aspergillus fumigatus. Eukaryot Cell 11:966–977. doi: 10.1128/EC.00091-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langfelder K, Philippe B, Jahn B, Latgé JP, Brakhage AA. 2001. Differential expression of the Aspergillus fumigatus pksP gene detected in vitro and in vivo with green fluorescent protein. Infect Immun 69:6411–6418. doi: 10.1128/IAI.69.10.6411-6418.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.05188-22-s0001.pdf, PDF file, 3.0 MB (3MB, pdf)