Abstract
We determined the complete nucleotide sequence of the mitochondrial genome of an angiosperm, sugar beet (Beta vulgaris cv TK81-O). The 368 799 bp genome contains 29 protein, five rRNA and 25 tRNA genes, most of which are also shared by the mitochondrial genome of Arabidopsis thaliana, the only other completely sequenced angiosperm mitochondrial genome. However, four genes identified here (namely rps13, trnF-GAA, ccb577 and trnC2-GCA) are missing in Arabidopsis mitochondria. In addition, four genes found in Arabidopsis (ccb228, rpl2, rpl16 and trnY2-GUA) are entirely absent in sugar beet or present only in severely truncated form. Introns, duplicated sequences, additional reading frames and inserted foreign sequences (chloroplast, nuclear and plasmid DNA sequences) contribute significantly to the overall size of the sugar beet mitochondrial genome. Nevertheless, 55.6% of the genome has no obvious features of information. We identified a novel tRNACys gene (trnC2-GCA) which shows no sequence homology with any tRNACys genes reported so far in higher plants. Intriguingly, this tRNA gene is actually transcribed into a mature tRNA, whereas the native tRNACys gene (trnC1-GCA) is most likely a pseudogene.
INTRODUCTION
Over the past decade much effort has been directed towards determining the entire genomic sequence of the mitochondrial DNAs (mtDNAs) from a wide array of organisms (1). However, the studied organisms are taxonomically highly biased; most of the complete mtDNA sequences published are from animals, protists and ascomycete fungi, whereas the available sequences for plants are limited to the liverwort Marchantia polymorpha (2) and the angiosperm Arabidopsis thaliana (3).
Despite the fact that mitochondria have essentially the same metabolic function in all eukaryotes which contain them, higher plant mtDNAs exhibit a number of unique structural features (e.g. large size, rapid evolutionary rearrangements and chloroplast DNA insertions) and specific modes of expression (e.g. cis- and trans-splicing, RNA editing and universal genetic code usage) (4). Moreover, they encode significantly more genes than do their fungal and animal counterparts. Ribosomal protein genes as well as the genes involved in cytochrome c biogenesis comprise a significant fraction of such additional genes identified so far in plant mitochondria (4). The complete sequence of the Arabidopsis mtDNA reveals approximately 85 open reading frames (ORFs) of unknown function, some of which may encode plant-specific mitochondrial proteins (3).
It is also interesting to note that the study of different plant lineages has demonstrated the recent evolutionary transfer of certain mitochondrial genes to the nucleus, and therefore the gene content of mitochondria may vary in different species (4). The determination of the complete mtDNA sequence of another angiosperm would thus allow us to examine the entire coding potential of plant mitochondrial genomes and to ascertain whether or not the plant mitochondrial gene content and organization have been conserved during flowering plant evolution. In addition, it should greatly facilitate the identification of new genes and provide insights into specific mechanisms underlying mitochondrial genome divergence.
In this article we describe the complete sequence of the mitochondrial genome of sugar beet (Beta vulgaris L.). We found an intriguing diversity of gene complements between sugar beet and Arabidopsis mitochondrial genomes, although most of the genes reported in Arabidopsis are shared by sugar beet. Of particular interest is the occurrence in sugar beet mtDNA of a gene for tRNACys which shows no significant similarity to the plant mitochondrial, chloroplast or nuclear tRNACys genes identified to date. This novel gene is actually transcribed in sugar beet mitochondria whereas the standard (native) tRNACys gene seems not to be functional.
MATERIALS AND METHODS
Sequencing procedures
The clone bank covering the entire mitochondrial genome of normal fertile sugar beet (cv TK81-O) was previously constructed using λ-DASH (Stratagene) and cosmid pHC79 (Boehringer) (5). We selected three cosmid clones for sequencing the rrn26-carrying repeats and 21 λ clones for sequencing the remaining region of the genome. mtDNA in each λ clone was amplified by long PCR using an LA-PCR kit (Takara Shuzo) and then physically sheared. The resultant fragments were blunt-ended using T4 DNA polymerase and Klenow fragment and subcloned into pBluescript (Stratagene). The purified cosmid DNA was sonicated and used for subcloning. Shotgun sequencing was continued until a minimum of 4-fold sequence coverage was achieved for each clone. The DNA sequencer used was a Li-COR 4200L (Li-COR). The PCR products obtained by amplification of the sugar beet mtDNA were used for gap filling and confirmation of the sequences. Sequence alignments were generated using Sequencher 3.0 software (Gene Codes). A database search was done with the BLAST network service (http://www.dna.affrc.go.jp/htdocs/Blast/blast2.html ) and a tRNA gene search with the tRNAscan-SE service (http://www.genetics.wustl.edu/eddy/tRNAscan-SE/ ) (6). The nucleotide sequence reported in this paper has been deposited in the DDBJ/GenBank/EMBL databases under accession nos AP000396 and AP000397.
RNA isolation and hybridization
Mitochondria were prepared from green leaves as described (7) and were used for RNA isolation (8). Aliquots of total mtRNA (5 µg) were electrophoresed through 1.4% agarose gels containing 0.66 M formaldehyde. Gels were blotted onto Hybond N+ membranes (Amersham) and hybridized with a 32P-labeled probe.
RESULTS AND DISCUSSION
Conserved genes
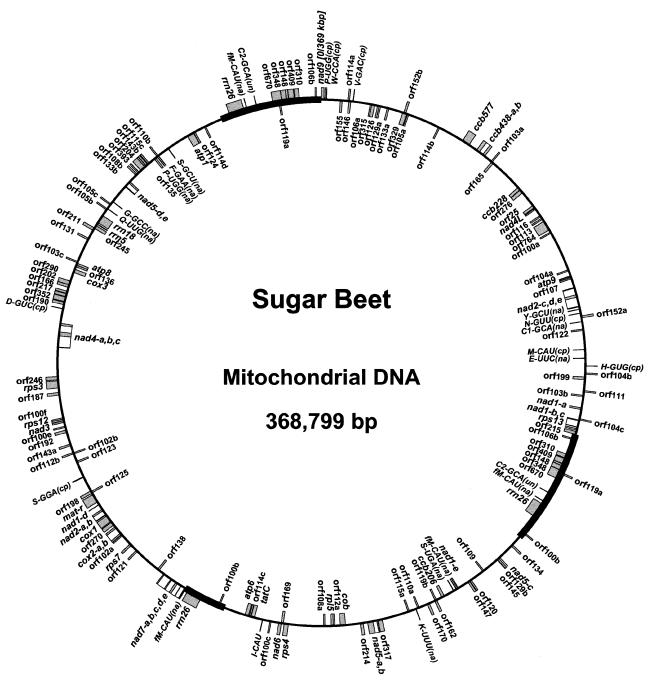
Our strategy for analysis of the sugar beet mitochondrial genome was based upon assembling the sequences of partially overlapping clones from λ and cosmid libraries and of long PCR products which have been assigned on the physical map of the genome (5). The entire genome size of the sugar beet mitochondrion is 368 799 bp, with an overall G+C content of 43.9%. These figures are comparable to those of Arabidopsis (366 924 bp in size and a G+C content of 44.8%) (3). Homology searches to match the entire genomic sequence with known sequences in the public DNA database detected 29 protein coding genes, five rRNA genes and 25 tRNA genes, which altogether account for 11.3% of the genome (Table 1). The positions of these genes in the sugar beet mitochondrial genome are depicted in Figure 1. Although most of the genes identified here are also found in Arabidopsis mitochondria, a few differences are noted in the gene content between sugar beet and Arabidopsis. As shown in Table 1, the rps13, ccb577, trnF-GAA and trnC2-GCA (see below) genes are present in sugar beet mitochondria but not in the mtDNA of Arabidopsis. Conversely, sugar beet mitochondria apparently lack four genes (ccb228, rpl2, rpl16 and trnY2-GUA) which are encoded by Arabidopsis mtDNA.
Table 1. Gene content of sugar beet (Bv) mtDNA compared to Arabidopsis (At).
| Gene (map co-ordinate in kb) | Bv | At | Gene (map co-ordinate in kb) | Bv | At |
|---|---|---|---|---|---|
| Complex I | Ribosomal proteins | ||||
| nad1* (102, 105, 152, 243) | + | + | rpl2 | – | + |
| nad2* (75, 240) | + | + | rpl5 (181) | + | + |
| nad3 (263) | + | + | rpl16 | – | + |
| nad4 (285) | + | + | rps3 (271) | + | + |
| nad4L (55) | + | + | rps4 (191) | + | + |
| nad5* (141, 171, 319) | + | + | rps7 (231) | + | + |
| nad6 (193) | + | + | rps12 (263) | + | + |
| nad7 (216) | + | + | rps13 (107) | + | – |
| nad9 (0) | + | + | Ribosomal RNAs | ||
| Complex II | rrn5 (308) | + | + | ||
| sdh4 | – | ψ? | rrn18 (309) | + | + |
| Complex III | rrn26 (125, 212, 348) | +a | + | ||
| cob (179) | + | + | Intronic ORF (in nad1-i4) | ||
| Complex IV | mat-r (244) | + | + | ||
| cox1 (238) | + | + | Sec-independent membrane targeting and translocation system | ||
| cox2 (234) | + | + | tatC (199) | + | + |
| cox3 (297) | + | + | Other ORF | ||
| Complex V | orf25 (54) | + | + | ||
| atp1 (337) | + | + | tRNAs | ||
| atp6 (201) | + | + | ‘Native’ | ||
| atp8 (299) | + | + | trnC1-GCA (83) | ψ | + |
| atp9 (72) | + | + | trnE-UUC (91) | + | + |
| Cytochrome c biogenesis | trnF-GAA (329) | + | – | ||
| ccb206 (156) | + | + | trnG-GCC (315) | + | + |
| ccb228 (49) | ψ | + | trnI-CAU (197) | + | + |
| ccb438 (37) | + | + | trnK-UUU (163) | + | + |
| ccb577 (33) | + | ψ? | trnfM-CAU (125, 153, 216, 352) | +b | + |
| trnP-UGG (329) | + | + | |||
| trnQ-UUG (313) | + | + | |||
| trnS-GCU (332) | + | + | |||
| trnS-UGA (154) | + | + | |||
| trnY1-GUA (80) | + | + | |||
| ‘Chloroplast–like’ | |||||
| trnD-GUC (289) | + | + | |||
| trnH-GUG (93) | + | + | |||
| trnN-GUU (81) | + | + | |||
| trnM-CAU (89) | + | + | |||
| trnP-UGG (1) | ψ | ψ | |||
| trnS-GGA (250) | + | + | |||
| trnV-GAC (7) | ψ | – | |||
| trnW-CCA (1) | + | + | |||
| Converted from trnF-GAA | |||||
| trnY2-GUA | – | + | |||
| Origin unknown | |||||
| trnC2-GCA (123, 354) | +c | – |
*, trans-splicing; +, present; ψ, pseudogene; ψ?, likely pseudogene; –, absent; a, three copies; b, four copies; c, two copies.
Figure 1.
Gene map of the sugar beet mitochondrial genome. Protein, rRNA and tRNA coding genes and ORFs longer than 100 amino acids are indicated by gray boxes. cis-splicing introns are shown by thin lines. Orientations are clockwise for the genes shown outside the circle and counterclockwise for the genes shown inside the circle. Abbreviations for the three classes of tRNA genes are: na, native (genuine); cp, chloroplast-like; un, origin unknown. Three-copy repeated sequences which are active in recombination are indicated by bold lines. Map coordinate 0 has been arbitrarily assigned to the first nucleotide of the nad9 ORF (top of the figure) and numbering proceeds clockwise.
An alignment of all known mitochondrial RNA editing sites (see for example 9) with the homologous nucleotide sequence of sugar beet mtDNA allowed us to predict at least 370 C→U editing sites in 28 gene/ORF transcripts of sugar beet. This number of editing events in sugar beet is somewhat fewer than in Arabidopsis (441 editings). We also experimentally verified the RNA editing-mediated creation of initiation (nad1, nad4L and atp6) and termination codons (atp6 and atp9), which is not documented in the corresponding Arabidopsis transcripts (10,11; T.Kubo et al., unpublished data).
Repeated sequences
We previously reported that the sugar beet rrn26 gene is found exclusively within a three-copy recombining-repeat family (the rrn26 repeat) (5). The present study indicates that the common sequence unit shared by all three rrn26 repeat copies is 6222 bp long and contains trnfM-CAU as well as rrn26 (Fig. 1). As can be seen in Figure 1, in two of the three repeat copies sequence identity continues a further 16 517 bp beyond the downstream junction (93 bp downstream from trnfM-CAU) of the common sequence unit. The same relationship holds between two of the three upstream rrn26 repeat flanks, which share an additional 3528 bp sequence beyond the upstream junction of the common sequence unit (Fig. 1). We found perfect identity in the base sequence over the entire length of the repeated sequence elements, suggesting that direct comparison of the repeat copies and correction of any mutational changes that appear in any one of the copies are ongoing processes.
The sugar beet mtDNA also contains 72 short repeated DNA segments, the sizes of which vary from 50 to 626 bp. These short repeats seem not to be active in frequent recombinations and they can be divided into 35 unrelated families. The repeated sequence elements cover 10.3% of the genome.
Promiscuous sequences
It is well documented that plant mtDNAs contain many sequences of foreign origin. Chloroplast DNA (cpDNA) sequences provided the first such example (12) and sequences of nuclear origin are also found in plant mitochondrial genomes (13). Our analysis indicates that cpDNA-derived sequences comprise 2.1% of the mitochondrial genome of sugar beet. The cpDNA insertions vary in size from 25 to 3366 bp. The largest fragment bears rps7, the last two exons of rps12 and trnV-GAC. Comparison of this 3.4 kb sequence to the corresponding sequences of sugar beet (accession no. AB032426) and tobacco cpDNAs (Z00044) revealed that the integrated sequence in question is closer to the sugar beet cpDNA than to tobacco cpDNA (details to be published elsewhere). This result supports the suggestion that cpDNA insertion took place after the divergence of sugar beet and tobacco. The cpDNA insertions include six intact and most likely functional tRNA genes which are also shared by Arabidopsis mtDNA (Table 1). However, most of the remaining cpDNA-like sequences are not detected in Arabidopsis.
We also found nuclear-like sequences covering 3.3% of the mitochondrial genome of sugar beet. We identified 21 retrotransposon-like sequences with lengths of between 25 and 2827 bp, only two of which exhibit similarity to the mitochondrial retrotransposon sequences of Arabidopsis. The most conspicuous example is an orf84–orf100–orf78–orf764 cluster which seems to be related to the gypsy-type retrotransposon. This cluster hybridized to sugar beet nuclear DNA (data not shown) but no corresponding sequence was observed in Arabidopsis mitochondria. Another noteworthy sequence of nuclear origin is a 168 bp segment with homology to (R)-mandelonitrile lyase (P = 8 × 10–21).
Introns
As in other higher plants, the sugar beet mitochondrial rRNA and tRNA genes lack introns. Nonetheless, sugar beet mtDNA has at least 20 introns in seven protein coding genes, all of which are categorized as group II introns based on their distinctive secondary structures. The sum of all intron lengths is ~25 800 bp, representing 7.0% of the genome. A total of six trans-splicing introns are found in the nad1, nad2 and nad5 genes and 14 cis-splicing introns are found in the seven protein coding genes (cox2, nad1, nad2, nad4, nad5, nad7 and ccb438). Interestingly, we found sugar beet rps3 to completely lack introns. This contrasts with the situation reported in other plant species (3,14,15), where the rps3 coding region is interrupted by a single intron. We also noted that the sugar beet nad4 gene does not contain the second intron found in several higher plants, including both monocots and dicots (16; N.Itchoda, T.Kubo and T.Mikami, manuscript in preparation).
Residual sequences
We searched for ORFs longer than 60 codons that begin with a methionine codon and end with a classical termination codon. A total of 559 ORFs were identified (93 ORFs are longer than 100 codons in size), none of which, however, exhibited apparent homology to any reported genes (or to any of the Arabidopsis ORFs; 3) or carried significant protein motifs. It thus seems unlikely that these ORFs code for additional important protein information. Of the sugar beet mitochondrial genome sequence 3.6% is homologous to mitochondrial plasmid DNAs. These sequences include segments of DNA and RNA polymerase-related ORFs (accession no. Y10854). We also found sequences with homology to the 5′- and 3′-untranslated regions and the intergenic sequences of mitochondrial genes in other plant species; they do not exceed 6.8% of the sugar beet genome. The major portion (55.6%) of the genome has no similarity to any nucleotide or protein sequences in the public databases.
tRNA genes
Twenty-five tRNA genes were identified by homology to their respective counterparts in Arabidopsis and other higher plants and by their predicted cloverleaf structures, which define unambiguous anticodons (Table 1). As shown in Figure 1, trnfM-CAU and trnC2-GCA are present in four and two copies, respectively, whereas the other tRNA genes occur singly. We previously reported that chloroplast-like trnP-UGG is not transcribed in sugar beet mitochondria (17). The same appears to hold true for chloroplast-like trnV-GAC (data not shown). As there are no mitochondrially encoded tRNA species for five amino acids (alanine, arginine, leucine, threonine and valine) in sugar beet, tRNA import from the cytosol must be invoked as the mechanism for making up the deficit.
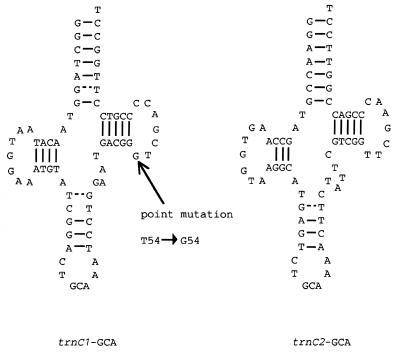
Our analysis also indicates the presence of two trnC-GCA genes; the first one is a native trnC-GCA (designated trnC1-GCA) and the other is a novel one (trnC2-GCA). When trnC1-GCA was folded into a cloverleaf secondary structure, the invariant T54 was found to be replaced by a G (Fig. 2). T54 is believed to pair with A58 to yield the familiar L-shaped tertiary structure. Hence, a G54-A58 mismatch may have a deleterious effect on expression of the trnC1-GCA gene (see below). On the other hand, the tRNA encoded by trnC2-GCA retains the invariant and semi-invariant nucleotides characteristic of standard tRNA (18; Fig. 2). Surprisingly, the best match between sugar beet trnC2-GCA and sequences in the public databases is with the trnC-GCA from a Gram-negative bacterium, Helicobacter pylori (accession no. AE000604), although it is too soon to say whether the homology (76%) is of significance. It should also be noted that the trnC2-GCA-specific probe failed to hybridize with cpDNA and nuclear DNA from sugar beet (data not shown).
Figure 2.
Cloverleaf structure deduced from the trnC1-GCA and trnC2-GCA gene sequences in sugar beet mitochondria. A mutation (T54→G54) in trnC1-GCA is indicated by an arrow.
The native trnC is likely a pseudogene whereas the novel trnC is expressed
Northern blot analysis was carried out to test for the presence in sugar beet mitochondria of RNAs corresponding to the trnC1-GCA and trnC2-GCA sequences, respectively. tRNA-sized molecules (~80 bp) were detected by hybridization with a trnC2-GCA-specific probe, confirming transcription of this gene (Fig. 3). In addition, a weaker signal (7.0 kb) indicated the presence of larger precursor molecules in the analyzed RNA fraction (Fig. 3). A probe that contains the the 5′-flanking region of the upstream trnfM-CAU gene (Fig. 1) hybridized with the 7.0 kb transcript (data not shown), an observation that is consistent with the co-transcription of trnfM-CAU and trnC2-GCA. In contrast, a trnC1-GCA-specific probe hybridized only to an RNA of 3.4 kb, indicating that the trnC1-GCA transcript remains linked to a transcript that includes the downstream trnN-GUU and trnY1-GUA genes (Fig. 1). This was confirmed by hybridization experiments with the trnN-GUU and trnY1-GUA probes, which identified a 3.4 kb transcript as well as tRNA-sized molecules (Fig. 3). These results lead us to suppose that although the trnC1-GCA sequence is transcribed, it is rarely or never processed into a mature molecule. Since no other sequence encoding tRNACys is found in the sugar beet mitochondrial genome, it seems reasonable to conclude that the trnC2-GCA identified here is the only functional tRNACys gene in the sugar beet mitochondrial genome.
Figure 3.
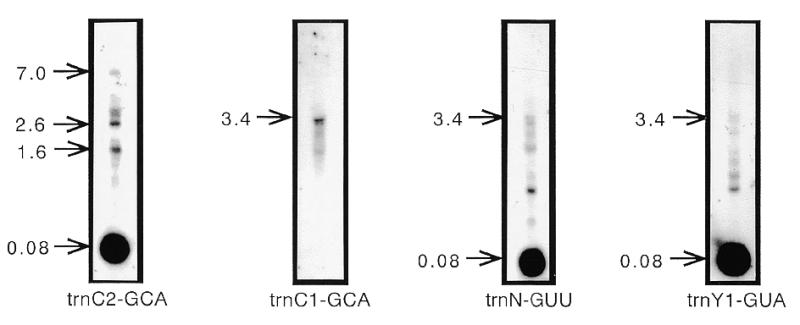
Northern blot analysis to detect and determine the relative sizes of RNA transcripts of the two sugar beet tRNACys genes. Hybridization probes were trnC2-GCA-, trnC1-GCA-, trnN-GUU- and trnY1-GUA-specific sequences whose positions are given in Figure 1. The sizes of the transcripts are shown in kilobases.
CONCLUSIONS
We found that duplicated sequences, introns, additional genes and inserted cpDNA sequences contribute significantly to the overall size of the mitochondrial genome in sugar beet. It was also noted that a large fraction (55.6%) of the sugar beet mitochondrial genomic sequences has no recognizable function and shows no apparent similarity to any sequence in the present databases. Furthermore, when the sugar beet mtDNA sequences are compared to those of Arabidopsis, the sequences shared by the two plant species total 78 057 bp, representing 21.2% of the sugar beet genome. Provided that many of the enigmatic sequences in question are of nuclear origin, one can infer that many DNA transfer events have occurred separately in the two evolutionary lineages leading to sugar beet and Arabidopsis, respectively.
A further notable finding is that the sugar beet mtDNA contains a novel tRNACys gene (trnC2-GCA) which shows no sequence homology to either the native or chloroplast-like tRNACys genes identified so far in plant mitochondria. The trnC2-GCA is most likely the only functional tRNACys gene in the sugar beet mitochondrial genome, although it remains to be seen whether the nuclear DNA-encoded tRNACys species is imported from the cytoplasm into mitochondria in sugar beet. We have recently cloned a sugar beet cDNA encoding cysteinyl-tRNA synthase. Interestingly, this protein has been found to be imported into mitochondria (T.Kubo et al., unpublished data). Whether the enzyme efficiently recognizes and aminoacylates the trnC2-GCA transcript is currently under investigation. Preliminary PCR surveys of 21 diverse genera of angiosperms indicate that the trnC2-GCA homolog is present only in members of Caryophyllales (T.Kubo et al., unpublished data). This raises the possibility that Caryophyllales acquired the trnC2-GCA sequence by horizontal transfer events from a distantly related organism. Most recently, Cho et al. (19) have described the horizontal transfer of a group I intron from a fungal donor to the mitochondrial genome of vascular plants. An alternative possibility that should be considered is that trnC2-GCA resulted from extensive genomic rearrangements, as exemplified in the trnY-GUA converted from trnF-GAA in Arabidopsis mitochondria (20) or chimeric ORFs responsible for cytoplasmic male sterility in various plants (21). Further studies are necessary to fully understand the origin and evolution of trnC2-GCA.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank Drs S. Tabata, T. Kaneko, Y. Nakamura and K. Akashi for valuable discussions and suggestions, Dr T. Tsudzuki for figure drawing and Drs Y. Kishima and Y. Nishita for technical advice and continuous encouragement. This work was done in part at the Research Center for Molecular Genetics, Hokkaido University, and was supported in part by Grants in Aid for Scientific Research from the Ministry of Education, Science and Culture, Japan, and a grant from the Program for Promotion of Basic Research Activities for Innovative Biosciences, Japan. A.E. is the recipient of a Japanese Government (Monbusho) Scholarship.
DDBJ/EMBL/GenBank accession nos AP000396, AP000397
REFERENCES
- 1.Gray M.W., Lang,B.F., Cedergren,R., Golding,G.B., Lemieux,C., Sankoff,D., Turmel,M., Brossard,N., Delage,E., Littlejohn,T.G. et al. (1998) Nucleic Acids Res., 26, 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oda K., Yamato,K., Ohta,E., Nakamura,Y., Takemura,M., Nozato,N., Akashi,K., Kanegae,T., Ogura,Y., Kohchi,T. and Ohyama,K. (1992) J. Mol. Biol., 223, 1–7. [DOI] [PubMed] [Google Scholar]
- 3.Unseld M., Marienfeld,J.R., Brandt,P. and Brennicke,A. (1997) Nature Genet., 15, 57–61. [DOI] [PubMed] [Google Scholar]
- 4.Schuster W. and Brennicke,A. (1994) Annu. Rev. Plant Physiol. Plant Mol. Biol., 45, 61–78. [Google Scholar]
- 5.Kubo T., Satoh,Y., Muro,T., Kinoshita,T. and Mikami,T. (1995) Curr. Genet., 28, 235–241. [DOI] [PubMed] [Google Scholar]
- 6.Lowe T. and Eddy,S.R. (1997) Nucleic Acids Res., 25, 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mikami T., Kishima,Y., Sugiura,M. and Kinoshita,T. (1985) Theor. Appl. Genet., 71, 166–171. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P. and Sacchi,N. (1987) Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 9.Giege P. and Brennicke,A. (1999) Proc. Natl Acad. Sci. USA, 96, 15324–15329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kubo T., Yamamoto,M.P. and Mikami,T. (2000) Theor. Appl. Genet., 100, 214–220. [Google Scholar]
- 11.Onodera Y., Yamamoto,M.P., Kubo,T. and Mikami,T. (1999) J. Plant Physiol., 155, 656–660. [Google Scholar]
- 12.Schuster W. and Brennicke,A. (1988) Plant Sci., 54, 1–10. [Google Scholar]
- 13.Knoop V., Unseld,M., Marienfeld,J., Brandt,P., Sunkel,S., Ullrich,H. and Brennicke,A. (1996) Genetics, 142, 579–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hunt M.D. and Newton,K.J. (1991) EMBO J., 10, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuster W. and Brennicke,A. (1991) Nucleic Acids Res., 19, 6923–6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gass D.A., Makaroff,C.A. and Palmer,J.D. (1992) Curr. Genet., 21, 423–430. [DOI] [PubMed] [Google Scholar]
- 17.Kubo T., Yanai,Y., Kinoshita,T. and Mikami,T. (1995) Curr. Genet., 27, 285–289. [DOI] [PubMed] [Google Scholar]
- 18.McClain W.H. (1993) J. Mol. Biol., 234, 257–280. [DOI] [PubMed] [Google Scholar]
- 19.Cho Y., Qiu,Y.-L. and Palmer,J.D. (1998) Proc. Natl Acad. Sci. USA, 95, 14244–14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen H.C., Wintz,H., Weil,J.H. and Pillay,T.N. (1989) Nucleic Acids Res., 17, 2613–2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levings C.S. (1993) Plant Cell, 5, 1285–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]