ABSTRACT
Corynebacteria of the diphtheriae species complex (CdSC) can cause diphtheria in humans and have been reported from companion animals. We aimed to describe animal infection cases caused by CdSC isolates. A total of 18,308 animals (dogs, cats, horses, and small mammals) with rhinitis, dermatitis, nonhealing wounds, and otitis were sampled in metropolitan France (August 2019 to August 2021). Data on symptoms, age, breed, and the administrative region of origin were collected. Cultured bacteria were analyzed for tox gene presence, production of the diphtheria toxin, and antimicrobial susceptibility and were genotyped by multilocus sequence typing. Corynebacterium ulcerans was identified in 51 cases, 24 of which were toxigenic. Rhinitis was the most frequent presentation (18/51). Eleven cases (6 cats, 4 dogs, and 1 rat) were monoinfections. Large-breed dogs, especially German shepherds (9 of 28 dogs; P < 0.00001), were overrepresented. C. ulcerans isolates were susceptible to all tested antibiotics. tox-positive C. diphtheriae was identified in 2 horses. Last, 11 infections cases (9 dogs and 2 cats; mostly chronic otitis and 2 sores) had tox-negative C. rouxii, a recently defined species. C. rouxii and C. diphtheriae isolates were susceptible to most antibiotics tested, and almost all of these infections were polymicrobial. Monoinfections with C. ulcerans point toward a primary pathogenic potential to animals. C. ulcerans represents an important zoonotic risk, and C. rouxii may represent a novel zoonotic agent. This case series provides novel clinical and microbiological data on CdSC infections and underlines the need for management of animals and their human contacts.
IMPORTANCE We report on the occurrence and clinical and microbiological characteristics of infections caused by members of the CdSC in companion animals. This is the first study based on the systematic analysis of a very large animal cohort (18,308 samples), which provides data on the frequency of CdSC isolates in various types of clinical samples from animals. Awareness of this zoonotic bacterial group remains low among veterinarians and veterinary laboratories, among which it is often considered commensal in animals. We suggest that in the case of CdSC detection in animals, the veterinary laboratories should be encouraged to send the samples to a reference laboratory for analysis of the presence of the tox gene. This work is relevant to the development of guidelines in the case of CdSC infections in animals and underlines their public health relevance given the zoonotic transmission risk.
KEYWORDS: Corynebacterium, diphtheria, C. ulcerans, C. diphtheriae, C. rouxii, emerging zoonosis, pet animal, case series
INTRODUCTION
Diphtheria is a potentially fatal infection in humans, caused mostly by toxigenic Corynebacterium diphtheriae isolates, which carry the tox gene coding for diphtheria toxin. This bacterial species is phylogenetically related to 5 other Corynebacterium species (C. ulcerans, C. pseudotuberculosis, C. rouxii, C. belfantii, and C. silvaticum) and together with these is grouped into the C. diphtheriae species complex (CdSC). C. ulcerans (1) can be isolated from humans and animals and is being increasingly reported (2–5). C. pseudotuberculosis causes caseous lymphadenitis in small ruminants and edematous skin disease in buffaloes. Although this species is considered potentially toxigenic, only isolates from buffaloes in Egypt were reported to produce diphtheria toxin, and there is no evidence for toxigenicity of recent isolates from caseous lymphadenitis (6–9). Two novel species, C. belfantii and C. rouxii, were recently described (10, 11). Isolates of these species were previously identified as C. diphtheriae, are mostly tox negative, and are of biovar Belfanti (starch and nitrate negative). Last, C. silvaticum was recently described from wild boars; all isolates of this species carry a disrupted tox gene, impairing their capacity to produce diphtheria toxin (12). Hence, although all members of the CdSC may potentially harbor the tox gene, toxigenic strains are frequently encountered only in C. diphtheriae and C. ulcerans. In C. diphtheriae, this gene is carried on a temperate phage that has integrated into the chromosome of a large variety of sublineages (13, 14). In C. ulcerans, in addition to a lysogenic phage, the tox gene can be carried on a pathogenicity island (15). Strains that carry the tox gene generally produce the diphtheria toxin in vitro, but a small fraction of them (~10 to 15%) do not, due to disruptions of the tox gene; these are called nontoxigenic, tox-bearing (NTTB) strains and are observed both for C. diphtheriae and for C. ulcerans (and for all C. silvaticum strains).
Although classically defined as a respiratory infection caused by toxigenic C. diphtheriae, diphtheria is sometimes defined more broadly as any infection (respiratory, cutaneous, or other) potentially leading to manifestations due to the production of the diphtheria toxin by species of the CdSC (https://www.ecdc.europa.eu/en/diphtheria). Following recent taxonomic updates, diphtheria may be defined more broadly as any infection caused by any isolate of the CdSC, irrespective of toxigenic status (16).
The typical clinical expressions of diphtheria in human are as follows: (i) classical respiratory diphtheria with pseudomembranous angina that can provoke a deadly obstruction of upper airways, usually associated with fever and enlarged anterior cervical lymph nodes and edema (“bull neck” appearance); (ii) cutaneous manifestations, with “rolled edge” ulcers usually observed on the limbs and that can be covered by a grayish pseudomembrane; and (iii) toxigenic complications such as polyneuropathy or myocarditis (17, 18). Seroprevalence studies in humans demonstrated that up to 82% of the population has titers of anti-diphtheria toxin antibodies below the limit of protection and that the proportion of nonprotected persons increases with age (19, 20).
Before 1999, no cases of C. ulcerans were reported in France (21), but between 2002 and 2013, 28 cases of toxigenic C. ulcerans were reported and between January 2018 and August 2019, 11 further human clinical cases were described (22); most of the cases were indigenous, and a similar pattern of increased reporting of C. ulcerans was found for the United Kingdom and Germany (2, 3, 5, 23).
While the transmission of C. diphtheriae is essentially interhuman, C. ulcerans is a zoonotic pathogen (2, 24). No transmission of C. ulcerans among humans was reported since its description in 1995; however, the possibility of person-to-person transmission cannot be totally excluded (25). Animals from which C. ulcerans is isolated can be asymptomatic carriers but can also present clinical symptoms, such as ulcerative dermatitis and chronic rhinitis (26–29). Horses can also carry C. ulcerans and sometimes show signs of respiratory diphtheria (30).
Similarly, C. rouxii may also be zoonotic: no interhuman transmission has been reported yet, and in addition to human cases, it has been so far identified in dogs, cats, and a fox (10, 31–33). To our knowledge, strains of C. belfantii have been isolated only from human respiratory samples.
Despite previous reports of animal CdSC infections (for examples, see references 2, 26, 29, and 34), no large case series of such infections, including precise bacterial identification and genotyping, have been reported. Here, we present a case series of CdSC infections in dogs, cats, rabbits, rats, and horses in France over 2 years and report their clinical and microbiological characteristics.
RESULTS
Isolation of members of the C. diphtheriae species complex in animals in France.
Between August 2019 and August 2021, 18,308 sick animals (dogs, cats, horses, and small mammals) were sampled from veterinary clinics from across metropolitan France. There were 2,732 nasal swabs, 4,224 cutaneous/wound/abscess swabs, and 11,352 auricular swabs (Table 1). A total of 64 consecutive, nonduplicated isolates belonging to the CdSC were identified from these samples (Table 2). Of these, 26 were carriers of the tox gene (coding for the diphtheria toxin), including 24 C. ulcerans and 2 C. diphtheriae isolates. Of the 38 nontoxigenic CdSC isolates, 27 belonged to C. ulcerans and the 11 remaining ones were C. rouxii. No nontoxigenic C. diphtheriae, C. pseudotuberculosis, or C. belfantii isolates were detected.
TABLE 1.
Overview of samples screened and sources of isolates of corynebacteria of the diphtheriae species complex (CdSC)
| Clinical source | No. (%) of samples screened | No. of C. ulcerans isolates |
No. of C. rouxii isolates (all tox negative) | No. of C. diphtheriae isolates tox positive | Total no. (%) of CdSC isolates | |
|---|---|---|---|---|---|---|
| tox positive | tox negative | |||||
| Dog | ||||||
| Ear swab | 9,439 (52) | 3 | 6 | 8 | 0 | 17 (0.2) |
| Respiratory tract sample | 1,037 (6) | 1 | 3 | 0 | 0 | 4 (0.4) |
| Wound/skin swab | 2,834 (15) | 7 | 8 | 1 | 0 | 16 (0.6) |
| Cat | ||||||
| Ear swab | 1,758 (10) | 1 | 2 | 1 | 0 | 4 (0.2) |
| Respiratory tract sample | 1,191 (7) | 6 | 4 | 0 | 0 | 10 (0.8) |
| Wound/skin swab | 1,027 (6) | 2 | 3 | 1 | 0 | 6 (0.6) |
| 1 (cystitis) | ||||||
| Horse | ||||||
| Ear swab | 5 (0.03) | 0 | 0 | 0 | 0 | 0 (0) |
| Respiratory sample | 111 (0.6) | 0 | 0 | 0 | 0 | 0 (0) |
| Wound/skin swab | 121 (0.7) | 0 | 0 | 0 | 2 (dermatitis/conjunctivitis) | 2 (1.6) |
| Rat | ||||||
| Ear swab | 6 (0.03) | 0 | 0 | 0 | 0 | 0 (0) |
| Respiratory sample | 20 (0.1) | 3 | 0 | 0 | 0 | 3 (15) |
| Wound/skin swab | 10 (0.05) | 0 | 0 | 0 | 0 | 0 (0) |
| Rabbit | ||||||
| Ear swab | 116 (0.6) | 0 | 0 | 0 | 0 | 0 (0) |
| Respiratory sample | 289 (1.6) | 1 | 0 | 0 | 0 | 1 (0.3) |
| Wound/skin swab | 127 (0.7) | 0 | 0 | 0 | 0 | 0 (0) |
| Others (goat, sheep, exotics, etc.) | ||||||
| Ear swab | 28 (0.2) | 0 | 0 | 0 | 0 | 0 (0) |
| Respiratory sample | 84 (0.5) | 0 | 0 | 0 | 0 | 0 (0) |
| Wound/skin swab | 105 (0.6) | 0 | 0 | 0 | 0 | 0 (0) |
| Total | 18,308 (100) | 24 | 27 | 11 | 2 | 64 (0.3) |
TABLE 2.
Strain characteristicsa
| Patient no. | French reference center ID of isolate | Animal species | Age (yrs) | Breed | Sex | French administrative departmentd | Monoinfection | Symptoms | Species | Toxin gene | Elek test | Sequence type | Antibiotic susceptibility test result for indicated group and drugb |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Aminoglycosides, gentamicin | Beta-lactams, amoxicillin | Macrolides Erythromycin | Spiramycin | Azithromycin | Tetracyclines, tetracycline and doxycycline | Folate pathway inhibitors, trimethoprim-sulfamethoxazole | |||||||||||||
| 1 | FRC0804 | Cat | 11 | Domestic shorthair | F | 57 | No | Chronic rhinitis | C. ulcerans | Positive | Positive | ST514 | S | S | S | S | S | S | S |
| 3 | FRC0847 | Cat | 7 | Domestic shorthair | F | 26 | Yes | Chronic rhinitis | C. ulcerans | Positive | Weakly Positive | ST325 | S | S | S | S | S | S | S |
| 5 | FRC0886 | Rat | 0.9 | ND | ND | 40 | No | Chronic rhinitis | C. ulcerans | Positive | Positive | ST332 | I | S | S | S | NA | S | S |
| 6 | FRC0895 | Rat | 1 | ND | M | 40 | No | Chronic rhinitis | C. ulcerans | Positive | NA | NA (contaminated) | S | S | S | NA | NA | S | S |
| 7 | FRC0897 | Cat | ND | Domestic shorthair | M | 41 | Yes | Chronic rhinitis + FIV | C. ulcerans | Positive | Positive | ST757 | S | S | S | S | S | S | S |
| 8 | FRC0916 | Dog | 6 | Mastiff | M | 78 | No | Sore/abscess | C. ulcerans | Positive | Positive | ST325 | S | S | S | S | S | S | S |
| 9 | FRC0922 | Cat | 1 | Domestic shorthair | F | 64 | No | Dermatitis | C. ulcerans | Positive | Positive | ST699 | S | S | S | S | S | S | S |
| 10 | FRC0929 | Dog | 3 | Basset Bleu De Gascogne | F | 71 | Yes | Sore | C. ulcerans | Positive | Positive | ST328 | S | S | S | S | S | S | S |
| 11 | FRC1009 | Dog | 3 | French bulldog | F | 17 | Yes | Chronic otitis | C. ulcerans | Positive | Positive | ST331 | S | S | S | S | S | S | S |
| 12 | FRC1027 | Cat | 8 | Domestic shorthair | M | 60 | No | Chronic otitis | C. ulcerans | Positive | Weakly Positive | ST325 | S | S | S | S | S | S | S |
| 13 | FRC1031 | Dog | 5 | German shepherd | M | 16 | No | Chronic otitis | C. ulcerans | Positive | Positive | ST331 | S | S | S | S | S | S | S |
| 14 | FRC1067 | Dog | 2 | Belgian shepherd | M | 62 | Yes | Abscess | C. ulcerans | Positive | Positive | ST331 | S | S | S | S | S | S | S |
| 15 | FRC1084 | Dog | 9 | Xc Lhasa Apso | F | 57 | No | Dermatitis | C. ulcerans | Positive | Positive | ST328 | I | S | S | S | S | S | S |
| 16 | FRC1089 | Dog | 7 | Chow chow | F | 06 | No | Chronic otitis | C. ulcerans | Positive | Weakly Positive | ST325 | I | S | S | S | S | S | S |
| 17 | FRC1102 | Cat | 13 | Domestic shorthair | M | 13 | No | Chronic rhinitis | C. ulcerans | Positive | Positive | ST325 | S | S | S | S | S | S | S |
| 18 | FRC1111 | Cat | 8 | Domestic shorthair | M | 77 | No | Sore | C. ulcerans | Positive | Positive | ST337 | S | S | S | S | S | S | S |
| 19 | FRC1115 | Dog | 8 | Small Bernese hound | M | 07 | No | Dermatitis | C. ulcerans | Positive | Positive | ST690 | I | S | S | S | S | S | S |
| 20 | FRC1123 | Dog | 7 | Jack Russell terrier | M | 42 | No | Sore | C. ulcerans | Positive | Negative | ST331 | I | S | S | S | S | S | S |
| 21 | FRC1127 | Rabbit | 0.8 | ND | F | 60 | No | Chronic rhinitis | C. ulcerans | Positive | Negative | ST358 | S | S | S | S | S | S | S |
| 22 | FRC1128 | Dog | 1 | Cane Corso | F | 12 | Yes | Chronic rhinitis | C. ulcerans | Positive | Positive | ST325 | S | S | S | S | S | S | S |
| 23 | FRC1137 | Rat | 1 | ND | M | 25 | Yes | Chronic rhinitis | C. ulcerans | Positive | Positive | ST332 | S | S | S | S | S | S | S |
| 24 | FRC1148 | Cat | 15 | Domestic shorthair | F | 78 | No | Chronic rhinitis | C. ulcerans | Positive | Negative | ST331 | I | S | S | S | S | S | S |
| 25 | FRC0855 | Cat | 5 | Domestic shorthair | F | 95 | Yes | Chronic rhinitis | C. ulcerans | Negative | NR | ST325 | S | S | S | S | S | S | S |
| 26 | FRC0856 | Dog | ND | German shepherd | ND | ND | No | Chronic otitis | C. ulcerans | Negative | NR | ST720 | I | S | S | S | S | S | S |
| 28 | FRC0915 | Cat | 1 | Maine Coon | F | 46 | No | Chronic rhinitis | C. ulcerans | Negative | NR | ST339 | S | S | S | S | S | S | S |
| 29 | FRC0921 | Dog | 5 | French mastiff | M | 57 | No | Dermatitis | C. ulcerans | Negative | NR | ST699 | S | S | S | S | S | S | S |
| 30 | FRC0971 | Dog | ND | Griffon | M | 45 | No | Chronic otitis | C. ulcerans | Negative | NR | ST339 | S | S | S | S | S | S | S |
| 31 | FRC0973 | Dog | 9 | Beauceron | F | 47 | No | Chronic otitis | C. ulcerans | Negative | NR | ST760 | S | S | S | S | S | S | S |
| 32 | FRC0981 | Dog | 4 | German shepherd | M | 77 | No | Chronic rhinitis | C. ulcerans | Negative | NR | ST339 | S | S | S | S | S | S | S |
| 34 | FRC0990 | Dog | 6 | German shepherd | M | 69 | No | Chronic rhinitis | C. ulcerans | Negative | NR | ST339 | S | S | S | S | S | S | S |
| 36 | FRC1028 | Dog | 0.3 | Great Dane | F | 77 | No | Dermatitis | C. ulcerans | Negative | NR | ST690 | S | S | S | S | S | S | S |
| 37 | FRC1032 | Cat | 11 | ND | F | 35 | Yes | Cystitis | C. ulcerans | Negative | NR | ST690 | S | S | S | S | S | S | S |
| 38 | FRC1047 | Cat | 17 | Domestic shorthair | F | 64 | No | Sore | C. ulcerans | Negative | NR | ST428 | S | S | S | S | S | S | S |
| 39 | FRC1054 | Cat | 7 | Domestic shorthair | M | 92 | No | Sore | C. ulcerans | Negative | NR | ST339 | S | S | R | R | R | S | S |
| 41 | FRC1110 | Dog | 6 | Cocker spaniel | M | 71 | No | Chronic otitis | C. ulcerans | Negative | NR | ST339 | S | S | S | S | S | S | S |
| 42 | FRC1114 | Cat | 13 | Birman | F | 16 | Yes | Chronic rhinitis | C. ulcerans | Negative | NR | ST325 | I | S | S | S | S | S | S |
| 43 | FRC1134 | Cat | 6 | Porcelaine | F | 45 | No | Chronic rhinitis | C. ulcerans | Negative | NR | ST339 | S | S | S | S | S | S | S |
| 44 | FRC1146 | Dog | 5 | Porcelaine | F | 38 | No | Sore | C. ulcerans | Negative | NR | ST339 | I | S | S | S | S | S | S |
| 45 | FRC1153 | Dog | 4 | Great Dane | F | 59 | No | Sore | C. ulcerans | Negative | NR | ST339 | R | S | S | S | S | S | S |
| 46 | FRC1164 | Dog | 6 | Great Dane | M | 57 | No | Dermatitis | C. ulcerans | Negative | NR | ST327 | S | S | S | S | S | S | S |
| 47 | FRC1165 | Cat | 7 | Domestic shorthair | F | 18 | No | Sore | C. ulcerans | Negative | NR | ST325 | S | S | S | S | S | S | S |
| 48 | FRC1170 | Dog | 9 | German shepherd | M | 13 | No | Chronic rhinitis | C. ulcerans | Negative | NR | ST339 | S | S | S | S | S | S | S |
| 49 | FRC1194 | Dog | 9 | German shepherd | M | 70 | No | Chronic otitis | C. ulcerans | Negative | NR | ST339 | S | S | S | S | S | S | S |
| 50 | FRC1193 | Cat | 7 | Rex Devon | M | 67 | No | Chronic otitis | C. ulcerans | Negative | NR | ST325 | S | S | S | I | S | S | S |
| 51 | FRC0848 | Cat | 13 | Domestic shorthair | M | 17 | No | Chronic otitis | C. ulcerans | Negative | NR | ST720 | S | S | S | S | S | S | S |
| 52 | FRC1017 | Horse | 30 | ND | F | 33 | No | Conjunctivitis | C. diphtheriae | Positive | Positive | ST669 | S | S | S | S | S | S | S |
| 53 | FRC1117 | Horse | 8 | ND | M | 40 | No | Dermatitis | C. diphtheriae | Positive | Positive | ST59 | S | S | S | S | S | S | S |
| 54 | FRC0802 | Dog | 9 | Drahthaar | M | 37 | No | Sore | C. rouxii | Negative | NR | ST694 | I | S | S | S | S | S | S |
| 55 | FRC0810 | Dog | 9 | ND | F | 84 | No | Chronic otitis | C. rouxii | Negative | NR | NA (rpoB truncated) | I | S | S | S | S | S | S |
| 56 | FRC0845 | Dog | ND | Labrador | ND | ND | No | Chronic otitis | C. rouxii | Negative | NR | ST714 | S | S | S | S | S | S | S |
| 57 | FRC0835 | Cat | ND | Domestic shorthair | ND | ND | Yes (+ Malassezia) | Chronic otitis | C. rouxii | Negative | NR | ST713 | S | S | S | S | S | S | S |
| 58 | FRC0926 | Cat | 1 | Domestic shorthair | M | 47 | No | Sore | C. rouxii | Negative | NR | ST537 | I | S | S | S | S | S | S |
| 59 | FRC0976 | Dog | 12 | German shepherd | F | 47 | No | Chronic otitis | C. rouxii | Negative | NR | ST762 | S | S | S | S | S | S | S |
| 60 | FRC1088 | Dog | 6 | American Staffordshire bull terrier | M | 84 | No | Chronic otitis | C. rouxii | Negative | NR | ST537 | I | S | S | S | S | S | S |
| 61 | FRC1100 | Dog | 13 | X Beagle | F | 17 | No | Chronic otitis | C. rouxii | Negative | NR | ST780 | S | S | S | S | S | S | S |
| 62 | FRC1150 | Dog | 7 | X Griffon | F | 64 | No | Chronic otitis | C. rouxii | Negative | NR | ST537 | S | S | S | S | S | S | S |
| 63 | FRC1159 | Dog | 9 | Labrador | M | 33 | No | Chronic otitis | C. rouxii | Negative | NR | ST537 | S | S | S | S | S | S | S |
| 64 | FRC1172 | Dog | 3 | Griffon | M | 65 | No | Chronic otitis | C. rouxii | Negative | NR | ST537 | S | S | S | S | S | S | S |
ID, identifier; F, female; M, male; FIV, feline immunodeficiency virus; ND, not documented; NR, not relevant; NA, not available; S, susceptible; I, intermediate; R, resistant.
breakpoints: gentamicin, S ≥ 18 and R < 16; amoxicillin, S ≥ 23 and R < 16; erythromycin, S ≥ 22 and R < 17; spiramycin, S ≥ 24 and R < 19; azithromycin, S ≥ 22 and R < 17; tetracycline and doxycycline, S ≥19 and R < 17; trimethoprim-sulfamethoxazole, S ≥ 16 and R < 10.
X, mixed breed.
The production of the diphtheria toxin was assessed for tox gene-bearing isolates. Both toxin gene-carrying C. diphtheriae strains were positive in the Elek test. Among the 24 tox-positive C. ulcerans isolates, one strain (FRC0895) was not available for testing, and 20 of the 23 tested isolates (87.0%) had a positive result by the Elek test, including 3 isolates with a weakly positive Elek test result. These three isolates belonged to a single genetic subtype, sequence type 325 (ST325). Three tox-positive C. ulcerans isolates (2 of ST331 and 1 of ST358) were negative by the Elek test, hence corresponding to nontoxigenic, tox gene-bearing (NTTB) strains.
Of the 64 infections with CdSC isolates, 37 were in dogs, 21 in cats, 3 in rats, and 2 in horses and 1 was in a rabbit (Tables 1 and 2). The ages of dogs and cats ranged from 1 year to 13 years. There was no statistically significant difference in prevalence of CdSC isolates among the different age groups. The most frequent breed among the C. ulcerans dog cases was the German shepherd (9 of 28 dogs of this breed). Dogs belonging to this breed were strongly associated with infections caused by C. ulcerans: there were 556 German shepherds out of 13,310 dogs (P < 0.00001).
Antimicrobial susceptibility of CdSC isolates.
C. ulcerans isolates were susceptible to most antibiotics (Table 2). Spiramycin was tested in 50 isolates, following the publication of Abbott et al. in 2020 (29); all were susceptible, except 1 resistant and 1 susceptible at a higher concentration (previously “intermediate”). Azithromycin was tested in 49 samples, and only 1 isolate (C. ulcerans FRC1054) was resistant; this isolate was also resistant to erythromycin and spiramycin. Nine C. ulcerans isolates tested intermediate for gentamicin and 1 was resistant, whereas the 42 other ones were susceptible. The C. ulcerans isolates were also susceptible to erythromycin in all but one case and were all susceptible to tetracyclines and trimethoprim-sulfamethoxazole.
The two C. diphtheriae isolates were susceptible to all antibiotics tested, including clindamycin. The C. rouxii isolates were susceptible to amoxicillin, tetracyclines, trimethoprim-sulfamethoxazole, erythromycin, spiramycin, azithromycin, and clindamycin, but only 7 isolates were susceptible to gentamicin, the 4 others being intermediate.
Genotyping of isolates using MLST.
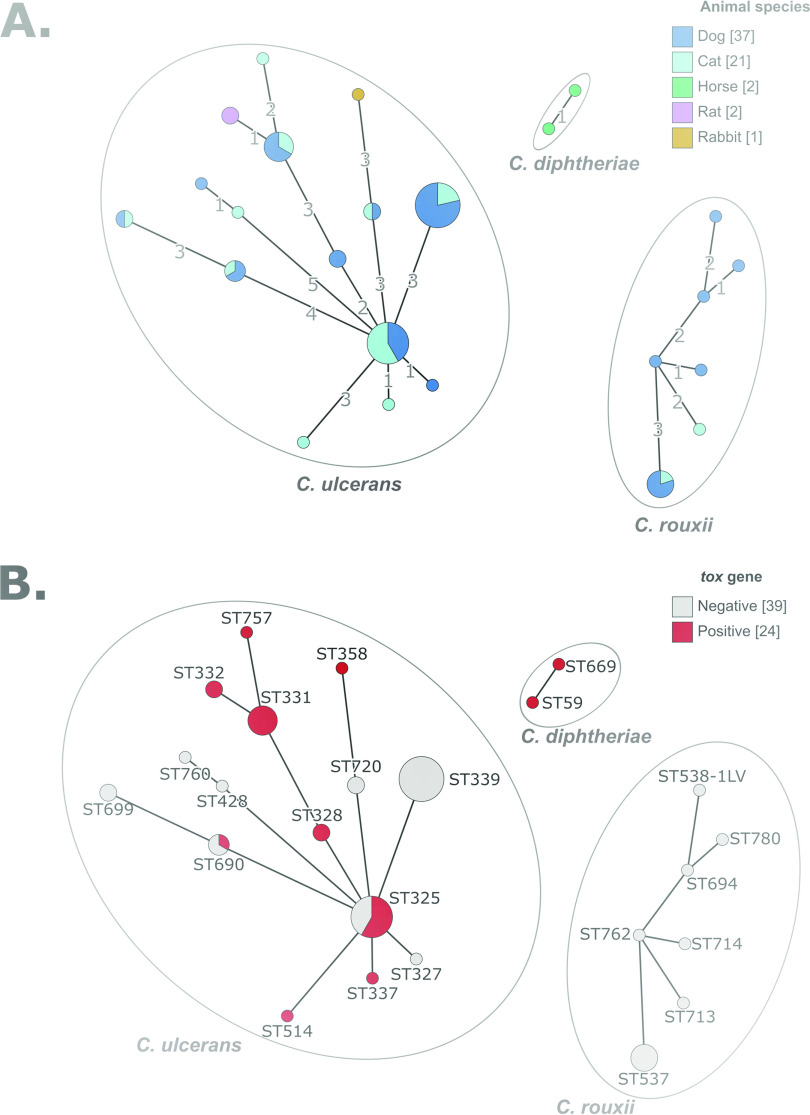
Multilocus sequence typing (MLST) analysis showed a wide diversity of C. ulcerans isolates, with 15 distinct STs (Fig. 1 and Table 2). C. rouxii was also genetically heterogeneous, with 7 STs, and the two C. diphtheriae isolates belonged to ST59 and ST699 (Fig. 1). Six C. ulcerans STs and one C. rouxii ST comprised more than one isolate. For these STs, geographical provenance and animal source were heterogeneous, indicating spread across localities and host species (see Fig. S1 in the supplemental material). Two C. ulcerans STs (ST325 and ST690) comprised both tox-positive and tox-negative isolates (Fig. 1), implying that the loss or gain of the tox gene occurred within these lineages. We noted that C. rouxii isolates originated mainly from southwestern France (Fig. 2 and Fig. S1).
FIG 1.
MLST diversity of CdSC isolates from pets. (A) Minimum spanning tree of 7-gene MLST profiles, colored in function of animal host species. (B) Same as panel A, colored by diphtheria toxin gene presence. The graphs were obtained using the GrapeTree tool, which is plugged onto the BIGSdb platform (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_diphtheria_isolates&page=plugin&name=GrapeTree).
FIG 2.
Geographic distribution of CdSC infection cases in pets. The 51 Corynebacterium ulcerans, 2 C. diphtheriae, and 9 C. rouxii are displayed within their administrative department of origin, with a species-specific symbol. tox gene-bearing strains are indicated in red and non-tox gene-bearing strains in gray. For two cases, location was not available, and they are therefore not represented here.
Clinical characteristics of toxigenic C. ulcerans isolates.
Eleven of the 24 toxigenic C. ulcerans isolates were isolated from nasal swabs. They were taken from 6 cats with chronic rhinitis and 3 rats, 1 dog, and 1 rabbit. Two of the rats were bought from the same pet shop, and they were both coinfected with Staphylococcus aureus. Euthanasia was chosen because of the zoonotic potential. Four other nasal swabs also harbored microorganisms other than C. ulcerans, including Pasteurella multocida, Escherichia coli, Bacteroides spp., Stenotrophomonas maltophilia, and Staphylococcus pseudointermedius.
The second most frequent isolation source was skin (9 of 24 cases) and included wounds, pyoderma, and abscesses. Seven of these C. ulcerans were sampled from dogs and two from cats. In 4 of the 9 skin cases, C. ulcerans was the only microorganism recovered from the samples, while in 5 cases there was a coinfection; organisms encountered were Pasteurella multocida, Pasteurella canis, Proteus mirabilis, Staphylococcus aureus, Staphylococcus pseudointermedius, Streptococcus canis, and Pseudomonas aeruginosa.
Last, there were four ear infections by toxigenic C. ulcerans. These were observed in three dogs and one cat. In 3 cases, otitis was also associated with at least another microorganism (Proteus mirabilis, Pseudomonas aeruginosa, Streptococcus canis, or Streptococcus dysgalactiae).
Three of the 24 animals infected by a toxigenic C. ulcerans strain had a bacteriological follow-up examination. Two cats, patients 3 and 4 (Table 2), had rhinitis with C. ulcerans in pure culture and tested positive again at control screenings 1 month and 18 days after the initial visit, respectively. Patient 3 had been treated for 10 days with amoxicillin-clavulanate, without success. No further control was performed. In patient 4, a 4-week cure of cefovecin did not eliminate the toxigenic C. ulcerans. However, a 1-month treatment with amoxicillin-clavulanate, to which the isolate was susceptible, was followed by a negative result for C. ulcerans on the control sample 4 months later.
One rabbit (patient 21) with C. ulcerans and Pasteurella multocida coinfection was still positive for C. ulcerans and P. multocida despite treatment with marbofloxacin for 7 days and then trimethoprim-sulfamethoxazole for 10 more days. No further control was performed.
Clinical characteristics of cases with nontoxigenic C. ulcerans isolates.
The 27 nontoxigenic C. ulcerans isolates were retrieved from 6 cases of dermatitis (6 dogs), 7 cases of rhinitis (4 cats and 3 dogs), 8 cases of otitis (6 dogs and 2 cats), 1 case of cystitis (cat), and 5 cases of nonhealing purulent wounds (3 cats and 2 dogs). In total, there were 10 cats and 17 dogs. Three monoinfections were observed and concerned two cats with chronic rhinitis and one cat with cystitis.
The other infections were polymicrobial. Regarding the otitis cases, the polyinfections consisted of coinfection with Streptococcus canis (in 3 cases), Pseudomonas aeruginosa (n = 2), Proteus mirabilis (n = 2), Staphylococcus pseudointermedius (n = 2), and Klebsiella pneumoniae (n = 1). The polyinfections in the cases of rhinitis were coinfections with Staphylococcus intermedius (n = 2), Pasteurella multocida and Bordetella bronchiseptica (n = 1), Klebsiella oxytoca (n = 1), and Staphylococcus aureus (n = 1). The dermatitis polyinfections were coinfections with S. pseudointermedius (n = 5), P. mirabilis (n = 3), and Pasteurella canis (n = 1). Last, polyinfections of sores comprised Escherichia coli (n = 2), S. aureus/pseudointermedius (1 case each), Pasteurella dagmatis/multocida (1 case each), and Streptococcus dysgalactiae (n = 1).
There was only one follow-up available (patient 25). It was for a 5-year-old cat with chronic rhinitis and a C. ulcerans monoinfection. This patient remained positive for nontoxigenic C. ulcerans for 6 months. In between, the cat was treated with doxycycline to which the strain tested susceptible in the 2 antibiotic susceptibility tests before and after treatment.
Clinical characteristics of cases with C. diphtheriae and C. rouxii isolates.
The two cases of C. diphtheriae were toxigenic and consisted of conjunctivitis and pastern dermatitis in two horses. Both cases were polymicrobial infections, in which S. dysgalactiae (conjunctivitis) and S. aureus and Enterobacter cloacae (pastern dermatitis) were also found.
C. rouxii infections were observed in 9 dogs and 2 cats and were all nontoxigenic. They consisted of 9 otitis cases (8 dogs and 1 cat) and two nonhealing wounds (one dog and one cat). C. rouxii was identified retrospectively by MLST, as this species was only recognized as a separate species during the study period (in 2020), and as matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) still identifies C. rouxii as C. diphtheriae, based on the MALDI Biotyper Compass database version 4.1.
Almost all C. rouxii infections were polymicrobial infections. There was only one monobacterial infection (ear infection of a cat), though Malassezia was also identified (Table 2). The most frequent (n = 5) coinfecting bacterium was P. aeruginosa, as expected in cases of otitis. Other coinfecting bacteria were P. mirabilis (n = 3), Staphylococcus sp. (n = 2), S. canis (n = 2), Corynebacterium amycolatum (n = 1), Citrobacter koseri (n = 1), and anaerobic bacteria (n = 1). The age of the infected animals ranged from 1 year to 13 years, with no statistically significant difference between age groups.
DISCUSSION
We report on the occurrence and clinical and microbiological characteristics of infections caused in companion animals by corynebacteria of the diphtheriae species complex (CdSC) in France. Although C. ulcerans was the most frequent species, C. rouxii and, more rarely, C. diphtheriae were also found.
All isolates initially identified as C. diphtheriae from dogs and cats were in fact C. rouxii, as identified by MLST gene sequences. This important observation suggests that this novel species has been overlooked, and it in fact appears more prevalent than C. diphtheriae itself in animal infections. As all C. rouxii infections, mostly of ears, were coinfections with other bacteria or the yeast Malassezia, this species may represent a commensal in dogs and cats. So far, all C. rouxii isolates are tox negative, with the exception of isolates from two cats from the United States (31), but in these two isolates, the tox gene was disrupted and therefore the toxin was not produced. Previously, C. rouxii has been isolated from cutaneous infections, vasculitis, and peritonitis in humans and a purulent orbital cellulitis in a dog (10). Our study corresponds to the largest group of infections with C. rouxii reported so far. We suggest that C. rouxii may represent a novel zoonotic pathogen, as pets may clearly serve as a reservoir for these human infections. However, so far, no case of transmission of C. rouxii between humans and animals has been documented. C. rouxii was more frequently isolated in southwestern France (Fig. 2) and was genetically diverse (Fig. 1 and Fig. S1). Although the sampling is still limited, this may reflect the existence of local conditions that favor the infections of dogs or cats by C. rouxii in this part of France.
Toxigenic C. diphtheriae was isolated from 2 horses. Mixed wound infections in horses have been previously described (34, 35), and colonization with C. diphtheriae was reported in 6.9% of slaughter horses in Romania (36). The pathogenic potential of C. diphtheriae in horses is questionable given its report from polymicrobial infections or asymptomatic carriage. Given the well-established human-to-human transmission of C. diphtheriae, and possible asymptomatic carriage, the possibility should be considered that the detection of this pathogen in horses corresponds to reverse zoonosis.
C. ulcerans is now well established as a zoonotic member of the CdSC. Whereas no human-to-human transmission was reported, human cases have often been associated with animal contacts, and in several cases the genetic fingerprinting of isolates supported an epidemiological link between the animal and human isolates, strongly establishing the zoonotic character of C. ulcerans (24, 26, 37). The emergence of C. ulcerans has been noted in France, Germany, and the United Kingdom (3–5, 38, 39).
This is the first study based on the systematic analysis of a very large animal cohort (18,308 samples), which provides data on the frequency of C. ulcerans in various types of clinical samples from animals. Abbott and colleagues (29) found 7 C. ulcerans isolates among 804 nasal samples (0.87%; 3 samples from 668 dogs and 4 samples from 64 cats), whereas Katsukawa et al. (28) found 44 C. ulcerans isolates in 583 pharyngeal samples of dogs (7.5%). In this study, we found 51 C. ulcerans isolates, including 24 tox-positive ones, in 18,308 clinical samples (0.27%), mostly in nasal swabs (11 tox-positive and 7 tox-negative samples).
Given the wide geographic distribution of cases (Fig. 2 and Fig. S1), the reporting of C. ulcerans in this study does not seem affected by a bias caused by local events of transmission. In addition, the genetic diversity of C. ulcerans isolates indicates that the reporting of this pathogen cannot be attributed to the clonal spread of a single emerging strain of C. ulcerans. Whether the increase in cases observed over the two last decades is due to changes in diagnostic practice (i.e., MALDI-TOF mass spectrometry) or increasing awareness of the necessity to report and test for the presence of the diphtheria toxin, rather than a real epidemiological phenomenon of emergence, remains unclear. Before 2019, C. ulcerans isolates of animal origin were sent only sporadically to the national reference laboratory. Awareness of this zoonotic bacterium remains low among veterinarians and veterinary laboratories, among which it is often considered a commensal of animals.
C. ulcerans is widely distributed, having been reported from multiple animal species (2). Whether pet animals represent a natural reservoir or (more probably) are themselves contaminated by C. ulcerans from other animal species or sources is an important question. Toxigenic C. ulcerans infections were reported for wildlife carnivores such as foxes, otters, and owls (40) and insectivores (hedgehogs and Japanese shrew owls) (41), whereas nontoxigenic C. ulcerans infections seem to be more frequent in omnivores (wild boars) and herbivores (roe deer) (38). Only wild animals with major symptoms are diagnosed, while the asymptomatic carriership is rarely investigated (42). Transmission by predatory hunting was supported by a study showing high serum diphtheria antitoxin titers in hunting dogs (43). A reservoir of symptomatic or asymptomatic carriers in small wild mammals such as herbivores, lagomorphs, or rodents, which may be prey to dogs and cats, should be further investigated. Here, we report 4 cases of toxigenic C. ulcerans in rats or rabbits. Although the cases described are from symptomatic animals, an asymptomatic carrier state of C. ulcerans in dogs, cats, horses, rodents, and wildlife animals has been described in other studies (24, 37, 41, 43, 44). Future studies should investigate the presence of CdSC in healthy animals to better understand and control its transmission to pets.
There is evidence for a pathogenic potential of C. ulcerans in animals (2), and experimental intranasal and intravenous infection studies in mice showed the pathogenic capacity of C. ulcerans, irrespective of toxigenicity (45, 46). In this study, C. ulcerans was often isolated from nasal swabs of cats with chronic rhinitis. While acute feline upper respiratory infections are mostly caused by viruses (feline herpesvirus 1 and feline calicivirus) (47) and are most of the time self-limiting, chronic rhinitis is more concerning. In general, bacteriological examinations performed on nasal swabs are often not useful because of the presence of commensal flora and only a few bacteria are considered primary pathogens (47). Toxigenic C. ulcerans may be implicated in the clinical manifestation of rhinitis. Importantly, in 7 cases of rhinitis, C. ulcerans was the only infective agent retrieved from the nasal swabs, a fact that strongly suggests a primary pathogenic nature of C. ulcerans in cats, consistent with a previous study (29). A limitation of the present study is that it was performed retrospectively; hence, the final diagnosis as well as the complete clinical picture was not available.
The pathogenicity of nontoxigenic strains is not well understood. Here, a few monoinfections by nontoxigenic C. ulcerans (2 cases of rhinitis and 1 cystitis) are reported. The infections could be favored by other virulence factors of C. ulcerans, such as phospholipase D (2).
Large dog breeds were disproportionally affected by C. ulcerans, and 32% of them were German shepherds. This interesting observation might be explained by a closer contact with small mammals, as large dogs are generally kept outdoors. But it could also reflect a breed predisposition, as German shepherd dogs are more susceptible to pyoderma (48) and other bacterial infections, for example, by Ehrlichia canis (49). Six German shepherds in this study presented with pyoderma or otitis. Further studies will be needed to investigate a possible dog breed predisposition to C. ulcerans.
In at least 3 cases of toxigenic C. ulcerans and 1 case of nontoxigenic C. ulcerans (patient numbers 3, 4, 21, and 25), the infection persisted, despite antibiotic susceptibility, as reported in previous works (26, 29, 37). One possibility is that chronic infections might involve some intracellular bacteria (50), which would be more accessible to macrolides, which can reach higher intracellular concentrations than beta-lactams. Systemic infections by C. diphtheriae suggest that this pathogen is not only able to attach to host epithelial cells but also able to gain access to deeper tissues and to live intracellularly (51). Guidelines for Corynebacterium in humans recommend the use of beta-lactams or macrolides. Yet, while chronic human carriers can be treated with azithromycin or rifampicin, the use of the latter is forbidden in the veterinary field.
Recent data suggest the use of spiramycin in cases of animal infection by C. ulcerans. A 10-day course of the combination of spiramycin and metronidazole was successful in clearing C. ulcerans from a dog, and a 6-day course of the same antibiotic combination was successful in cats (18, 29). As spiramycin only exists in a combination with metronidazole, and as metronidazole has side effects (neurotoxicity and effect on the microbiota), azithromycin seems to be a better choice for treatment.
The efficiency of the antibiotic treatment should be verified ideally by 2 samples, taken 5 to 7 days following completion of the antibiotic course. The macrolide agent (azithromycin rather than a spiramycin-metronidazole combination) and the optimal treatment duration should be investigated by future studies.
This work is relevant to the development of veterinarian guidelines in the case of CdSC infections in animals. Currently, in France no recommendations are established around the detection of toxigenic CdSC in animals, either for animal or their human contacts. From a public health perspective, the zoonotic potential of toxigenic C. diphtheriae or C. ulcerans in chronic skin infections, nonhealing wounds, rhinitis, and otitis justifies investigating these body locations for such pathogens, and these samples should not be considered to reveal only commensal organisms. We suggest that in the case of CdSC detection in animals, the veterinary laboratories should be encouraged to send the samples to a reference laboratory for analysis of the presence of the tox gene. This would be more in line with the fact that in the case of diphtheria due to toxigenic C. ulcerans in a human, recommendations exist in France that contact animals should be sampled and treated.
Toxigenic corynebacteria in animals are not mandatory to notify, and the costs of treatment, and clearance and sampling of contact animals, clearly represent a limitation for the implementation of follow-up investigations and control measures. Asymptomatic human carriers of toxigenic strains are treated with the same antibiotic regimen as symptomatic cases, with clearance swabs to ensure eradication. From an epidemiological viewpoint, it would be coherent to follow the same practice for animals, and this should be discussed between veterinary and human health institutions.
MATERIALS AND METHODS
Inclusions and microbiological characterization.
A total of 18,308 samples originating from sick animals (dogs, cats, horses, and small mammals) from metropolitan France were analyzed during the period from August 2019 to August 2021 (24 months) (Table 1). All samples from which isolates were identified as members of the CdSC were included in the present study. Although C. pseudotuberculosis was sometimes recorded, this species was excluded because the shipping and reception process of these samples is known to be biased (samples of livestock are generally sent to state laboratories and only occasionally to private laboratories).
The geographical location of the cases was recorded using the corresponding French administrative department (in metropolitan France, there are 96 geographic and administrative divisions called “départements”). Clinical data (age, sex, and clinical symptoms) were collected using information communicated on the order form.
Sterile swabs with liquid Amies culture medium were used to sample the affected animals and were sent to the laboratory under cooled conditions. There, swabs were plated on solid culture media (Columbia agar supplemented with 5% sheep blood and colistin plus nalidixic acid [CNA]) under an extractor hood. Solid culture media were incubated at 35°C under a 5% enriched CO2 atmosphere for 24 to 48 h. Isolates belonging to the Corynebacterium diphtheriae species complex form dry colonies of a dark yellow color on CNA culture medium. The identity of the suspected colonies was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS; Bruker Daltonics, Germany). Note that this method identifies C. diphtheriae and C. ulcerans but does not discriminate well other members of the CdSC from these two species.
Antibiotic susceptibility testing was performed by disk diffusion on Mueller-Hinton culture medium supplemented with 5% blood following the instructions of the French AFNOR standard NF U47-107 (https://www.boutique.afnor.org/fr-fr/norme/nf-u47107/methodes-danalyse-en-sante-animale-guide-de-realisation-des-antibiogrammes-/fa170310/40286). Diameters of the inhibition zone were read and interpreted using software tool SIRWeb (i2A, France). As there are no veterinary breakpoints, the 2013 guidelines from the human CASFM (https://www.sfm-microbiologie.org/wp-content/uploads/2020/07/CASFM_2013.pdf) were applied. The antibiotic susceptibility testing included antibiotics commonly used in veterinary clinics, including beta-lactams (amoxicillin), tetracyclines (doxycycline and tetracycline), aminoglycosides (gentamicin), macrolides (erythromycin, azithromycin, and spiramycin), and others (trimethoprim-sulfamethoxazole). Recent data suggest that C. ulcerans is less susceptible to clindamycin (52). Therefore, clindamycin was tested only for C. diphtheriae and C. rouxii. As there are no breakpoints for veterinary fluoroquinolones and cefovecin, these antibiotics were not tested.
All CdSC isolates were sent to the National Reference Center for Corynebacteria of the diphtheriae complex for confirmation of the identification and for the detection of the diphtheria toxin (tox) gene by real-time PCR (53). This multiplex assay consists of amplifying a fragment of the rpoB gene with primer sets specific for either (i) the species C. diphtheriae, C. belfantii, or C. rouxii or (ii) the species C. ulcerans and C. pseudotuberculosis; in addition, a fragment of the tox gene is detected. The production of the diphtheria toxin was assessed using the modified Elek test (54).
MLST.
Isolates were retrieved from −80°C storage and plated on tryptose-casein soy agar for 24 to 48 h. A small amount of bacterial colony biomass was resuspended in a lysis solution (20 mM Tris-HCl [pH 8], 2 mM EDTA, 1.2% Triton X-100, and lysozyme [20 mg/mL]) and incubated at 37°C for 1 h, and DNA was extracted with the DNeasy blood and tissue kit (Qiagen, Courtaboeuf, France) according to the manufacturer’s instructions. Multilocus sequence typing (MLST) was performed as previously described (13, 55); alleles and profiles were defined using the BIGSdb-Pasteur platform (https://bigsdb.pasteur.fr/diphtheria).
Statistical analyses.
Dogs and cats were divided into 3 age groups: under 2 years, 2 to 8 years, and older than 8 years. A few dogs could not be attributed to a group because the information was not available. Pearson’s chi-squared statistic was used to compare the different characteristics for the categorical variables. Statistical tests were performed in SPSS Statistics software version 25 (IBM, New York, NY).
Ethics statement.
Animals were sampled for diagnostic purposes. No ethics approval was requested for this retrospective study.
Data availability.
MLST sequences and profiles can be accessed at https://bigsdb.pasteur.fr/diphtheria.
Supplementary Material
ACKNOWLEDGMENTS
We thank Stéphanie Gilles from the bacteriology department of Cerba Vet for her support and Annick Carmi-Leroy, Annie Landier and Nathalie Armatys (Institut Pasteur) for technical help.
This work was supported financially by the French Government’s Investissement d’Avenir program Laboratoire d’Excellence “Integrative Biology of Emerging Infectious Diseases” (ANR-10-LABX-62-IBEID). This work used computational and storage services provided by the IT department at Institut Pasteur. The National Reference Center for Corynebacteria of the diphtheriae complex is supported by Institut Pasteur and Santé publique France (Public Health France). Initial microbiological analyses and logistics for referring isolates to the National Reference Laboratory were supported financially by Cerba Vet. M.H. was supported financially by the Ph.D. grant “Codes4strains” from the European Joint Program One Health, which has received funding from the European Union’s Horizon 2020 Research and Innovation Program under grant agreement no. 773830.
We declare that 2 authors (K.M. and G.R.) are employees of Cerba Vet, which performs diagnostic testing on a commercial basis.
K.M. and S.B. conceived, designed, and coordinated the study. G.R. and E.B. performed the microbiological cultures of the isolates and their biochemical and molecular characterizations. M.H. analyzed the sequence data and created the figures. G.A. and K.M. reviewed the clinical source data of the isolates. K.M. and S.B. wrote the initial version of the manuscript. All authors provided input to, read, and approved the final manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Kristina Museux, Email: kristina.museux@cerbavet.com.
Sylvain Brisse, Email: sylvain.brisse@pasteur.fr.
Florence Claude Doucet-Populaire, University Paris-Saclay, AP-HP Hôpital Antoine Béclère, Service de Microbiologie, Institute for Integrative Biology of the Cell (I2BC), CEA, CNRS.
REFERENCES
- 1.Riegel P, Ruimy R, de Briel D, Prévost G, Jehl F, Christen R, Monteil H. 1995. Taxonomy of Corynebacterium diphtheriae and related taxa, with recognition of Corynebacterium ulcerans sp. nov. nom. rev. FEMS Microbiol Lett 126:271–276. doi: 10.1111/j.1574-6968.1995.tb07429.x. [DOI] [PubMed] [Google Scholar]
- 2.Hacker E, Antunes CA, Mattos-Guaraldi AL, Burkovski A, Tauch A. 2016. Corynebacterium ulcerans, an emerging human pathogen. Future Microbiol 11:1191–1208. doi: 10.2217/fmb-2016-0085. [DOI] [PubMed] [Google Scholar]
- 3.Berger A, Teutsch B, Heinzinger S, Sing A. 2018. Corynebacterium ulcerans—ein Emerging Pathogen? https://edoc.rki.de/handle/176904/3003.
- 4.Haut Conseil de la santé publique. 2019. Avis relatif aux recommandations d’utilisation des antitoxines diphtériques en situation de pénurie. https://www.hcsp.fr/explore.cgi/avisrapportsdomaine?clefr=756.
- 5.Wagner KS, White JM, Crowcroft NS, De Martin S, Mann G, Efstratiou A. 2010. Diphtheria in the United Kingdom, 1986–2008: the increasing role of Corynebacterium ulcerans. Epidemiol Infect 138:1519–1530. doi: 10.1017/S0950268810001895. [DOI] [PubMed] [Google Scholar]
- 6.Schlicher J, Schmitt S, Stevens MJA, Stephan R, Ghielmetti G. 2021. Molecular characterization of Corynebacterium pseudotuberculosis isolated over a 15-year period in Switzerland. Vet Sci 8:151. doi: 10.3390/vetsci8080151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Selim SA. 2001. Oedematous skin disease of buffalo in Egypt. J Vet Med B Infect Dis Vet Public Health 48:241–258. doi: 10.1046/j.1439-0450.2001.00451.x. [DOI] [PubMed] [Google Scholar]
- 8.Selim SA, Mohamed FH, Hessain AM, Moussa IM. 2016. Immunological characterization of diphtheria toxin recovered from Corynebacterium pseudotuberculosis. Saudi J Biol Sci 23:282–287. doi: 10.1016/j.sjbs.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Viana MVC, Figueiredo H, Ramos R, Guimarães LC, Pereira FL, Dorella FA, Selim SAK, Salaheldean M, Silva A, Wattam AR, Azevedo V. 2017. Comparative genomic analysis between Corynebacterium pseudotuberculosis strains isolated from buffalo. PLoS One 12:e0176347. doi: 10.1371/journal.pone.0176347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Badell E, Hennart M, Rodrigues C, Passet V, Dazas M, Panunzi L, Bouchez V, Carmi-Leroy A, Toubiana J, Brisse S. 2020. Corynebacterium rouxii sp. nov., a novel member of the diphtheriae species complex. Res Microbiol 171:122–127. doi: 10.1016/j.resmic.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Dazas M, Badell E, Carmi-Leroy A, Criscuolo A, Brisse S. 2018. Taxonomic status of Corynebacterium diphtheriae biovar Belfanti and proposal of Corynebacterium belfantii sp. nov. Int J Syst Evol Microbiol 68:3826–3831. doi: 10.1099/ijsem.0.003069. [DOI] [PubMed] [Google Scholar]
- 12.Dangel A, Berger A, Rau J, Eisenberg T, Kämpfer P, Margos G, Contzen M, Busse H-J, Konrad R, Peters M, Sting R, Sing A. 2020. Corynebacterium silvaticum sp. nov., a unique group of NTTB corynebacteria in wild boar and roe deer. Int J Syst Evol Microbiol 70:3614–3624. [DOI] [PubMed] [Google Scholar]
- 13.Guglielmini J, Hennart M, Badell E, Toubiana J, Criscuolo A, Brisse S. 2021. Genomic epidemiology and strain taxonomy of Corynebacterium diphtheriae. J Clin Microbiol 59:e01581-21. doi: 10.1128/JCM.01581-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hennart M, Panunzi LG, Rodrigues C, Gaday Q, Baines SL, Barros-Pinkelnig M, Carmi-Leroy A, Dazas M, Wehenkel AM, Didelot X, Toubiana J, Badell E, Brisse S. 2020. Population genomics and antimicrobial resistance in Corynebacterium diphtheriae. Genome Med 12:107. doi: 10.1186/s13073-020-00805-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dangel A, Berger A, Konrad R, Sing A. 2019. NGS-based phylogeny of diphtheria-related pathogenicity factors in different Corynebacterium spp. implies species-specific virulence transmission. BMC Microbiol 19:28. doi: 10.1186/s12866-019-1402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haut Conseil de la santé publique. 2021. Avis relatif à la conduite à tenir autour d’un cas de diphtérie (compléments à l’avis de 2011). https://www.hcsp.fr/explore.cgi/avisrapportsdomaine?clefr=1130.
- 17.Sharma NC, Efstratiou A, Mokrousov I, Mutreja A, Das B, Ramamurthy T. 2019. Diphtheria. 1. Nat Rev Dis Primers 5:81. doi: 10.1038/s41572-019-0131-y. [DOI] [PubMed] [Google Scholar]
- 18.UK Health Security Agency. 2022. Public health control and management of diphtheria in England: 2022 guidelines. https://www.gov.uk/government/publications/diphtheria-public-health-control-and-management-in-england-and-wales.
- 19.Berbers G, van Gageldonk P, Kassteele JVD, Wiedermann U, Desombere I, Dalby T, Toubiana J, Tsiodras S, Ferencz IP, Mullan K, Griskevicius A, Kolupajeva T, Vestrheim DF, Palminha P, Popovici O, Wehlin L, Kastrin T, Maďarová L, Campbell H, Ködmön C, Bacci S, Barkoff A-M, He Q, Serosurveillance Study Team . 2021. Circulation of pertussis and poor protection against diphtheria among middle-aged adults in 18 European countries. Nat Commun 12:2871. doi: 10.1038/s41467-021-23114-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Launay O, Toneatti C, Bernède C, Njamkepo E, Petitprez K, Leblond A, Larnaudie S, Goujon C, Ungeheuer M-N, Ajana F, Raccurt C, Beytout J, Chidiac C, Bouhour D, Guillemot D, Guiso N. 2009. Antibodies to tetanus, diphtheria and pertussis among healthy adults vaccinated according to the French vaccination recommendations. Hum Vaccin 5:341–346. doi: 10.4161/hv.5.5.7575. [DOI] [PubMed] [Google Scholar]
- 21.Hommez J, Devriese LA, Vaneechoutte M, Riegel P, Butaye P, Haesebrouck F. 1999. Identification of nonlipophilic corynebacteria isolated from dairy cows with mastitis. J Clin Microbiol 37:954–957. doi: 10.1128/JCM.37.4.954-957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Reference Center for diphtheria in France. 2021. Activity report, 2019–2020. https://www.pasteur.fr/fr/sante-publique/CNR/les-cnr/corynebacteries-du-complexe-diphteriae/rapports-d-activite.
- 23.European Centre for Disease Prevention and Control. 2021. Diphtheria. Annual epidemiological report for 2018. ECDC, Stockholm, Sweden. [Google Scholar]
- 24.Othieno R, Mark K, Etherson M, Foster G, Murray S, Kalima P, Fry NK, Cameron C, Strachan J. 2019. A case of cutaneous toxigenic Corynebacterium ulcerans likely acquired from a domestic dog. Access Microbiol 1:e000025. doi: 10.1099/acmi.0.000025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konrad R, Hörmansdorfer S, Sing A. 2015. Possible human-to-human transmission of toxigenic Corynebacterium ulcerans. Clin Microbiol Infect 21:768–771. doi: 10.1016/j.cmi.2015.05.021. [DOI] [PubMed] [Google Scholar]
- 26.Lartigue M-F, Monnet X, Le Flèche A, Grimont PAD, Benet J-J, Durrbach A, Fabre M, Nordmann P. 2005. Corynebacterium ulcerans in an immunocompromised patient with diphtheria and her dog. J Clin Microbiol 43:999–1001. doi: 10.1128/JCM.43.2.999-1001.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carfora V, Scarampella F, Iurescia M, Donati V, Stravino F, Lorenzetti S, Menichini E, Franco A, Caprioli A, Battisti A. 2018. Non-toxigenic Corynebacterium ulcerans sequence types 325 and 339 isolated from two dogs with ulcerative lesions in Italy. J Vet Diagn Invest 30:447–450. doi: 10.1177/1040638718764786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katsukawa C, Komiya T, Yamagishi H, Ishii A, Nishino S, Nagahama S, Iwaki M, Yamamoto A, Takahashi M. 2012. Prevalence of Corynebacterium ulcerans in dogs in Osaka, Japan. J Med Microbiol 61:266–273. doi: 10.1099/jmm.0.034868-0. [DOI] [PubMed] [Google Scholar]
- 29.Abbott Y, Efstratiou A, Brennan G, Hallanan S, Leggett B, Leonard FC, Markey BK, Tuite C, Fry NK. 2020. Toxigenic Corynebacterium ulcerans associated with upper respiratory infections in cats and dogs. J Small Anim Pract 61:554–560. doi: 10.1111/jsap.13185. [DOI] [PubMed] [Google Scholar]
- 30.Zendri F, Isgren CM, Sinovich M, Richards-Rios P, Hopkins KL, Russell K, Groves N, Litt D, Fry NK, Timofte D. 2021. Case report: Toxigenic Corynebacterium ulcerans diphtheria-like infection in a horse in the United Kingdom. Front Vet Sci 8:650238. doi: 10.3389/fvets.2021.650238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hall AJ, Cassiday PK, Bernard KA, Bolt F, Steigerwalt AG, Bixler D, Pawloski LC, Whitney AM, Iwaki M, Baldwin A, Dowson CG, Komiya T, Takahashi M, Hinrikson HP, Tondella ML. 2010. Novel Corynebacterium diphtheriae in domestic cats. Emerg Infect Dis 16:688–691. doi: 10.3201/eid1604.091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlez K, Eisenberg T, Rau J, Dubielzig S, Kornmayer M, Wolf G, Berger A, Dangel A, Hoffmann C, Ewers C, Sing A. 2021. Corynebacterium rouxii, a recently described member of the C. diphtheriae group isolated from three dogs with ulcerative skin lesions. Antonie Van Leeuwenhoek 114:1361–1371. doi: 10.1007/s10482-021-01605-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sing A, Konrad R, Meinel DM, Mauder N, Schwabe I, Sting R. 2016. Corynebacterium diphtheriae in a free-roaming red fox: case report and historical review on diphtheria in animals. Infection 44:441–445. doi: 10.1007/s15010-015-0846-y. [DOI] [PubMed] [Google Scholar]
- 34.Leggett BA, De Zoysa A, Abbott YE, Leonard N, Markey B, Efstratiou A. 2010. Toxigenic Corynebacterium diphtheriae isolated from a wound in a horse. Vet Rec 166:656–657. doi: 10.1136/vr.b4846. [DOI] [PubMed] [Google Scholar]
- 35.Henricson B, Segarra M, Garvin J, Burns J, Jenkins S, Kim C, Popovic T, Golaz A, Akey B. 2000. Toxigenic Corynebacterium diphtheriae associated with an equine wound infection. J Vet Diagn Invest 12:253–257. doi: 10.1177/104063870001200309. [DOI] [PubMed] [Google Scholar]
- 36.Stănică E, Maximesco P, Stoian C, Pop A, Oprişan R, Potorac E. 1968. Level of antitoxic immunity and carriage of diphtheria bacilli in horses under the present circumstances of diminution of morbidity of diphtheria. Arch Roum Pathol Exp Microbiol 27:555–562. [PubMed] [Google Scholar]
- 37.Vandentorren S, Guiso N, Badell E, Boisrenoult P, Micaelo M, Troché G, Lecouls P, Moquet MJ, Patey O, Belchior E. 2014. Toxigenic Corynebacterium ulcerans in a fatal human case and her feline contacts, France, March 2014. Euro Surveill 19:20910. doi: 10.2807/1560-7917.es2014.19.38.20910. [DOI] [PubMed] [Google Scholar]
- 38.Berger A, Dangel A, Peters M, Mühldorfer K, Braune S, Eisenberg T, Szentiks CA, Rau J, Konrad R, Hörmansdorfer S, Ackermann N, Sing A. 2019. Tox-positive Corynebacterium ulcerans in hedgehogs, Germany. Emerg Microbes Infect 8:211–217. doi: 10.1080/22221751.2018.1562312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bonmarin I, Guiso N, Le Flèche-Matéos A, Patey O, Grimont Patrick AD, Levy-Bruhl D. 2009. Diphtheria: a zoonotic disease in France? Vaccine 27:4196–4200. doi: 10.1016/j.vaccine.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 40.Foster G, Patterson T, Howie F, Simpson V, Davison N, Efstratiou A, Lai S. 2002. Corynebacterium ulcerans in free-ranging otters. Vet Rec 150:524. [PubMed] [Google Scholar]
- 41.Katsukawa C, Umeda K, Inamori I, Kosono Y, Tanigawa T, Komiya T, Iwaki M, Yamamoto A, Nakatsu S. 2016. Toxigenic Corynebacterium ulcerans isolated from a wild bird (ural owl) and its feed (shrew-moles): comparison of molecular types with human isolates. BMC Res Notes 9:181. doi: 10.1186/s13104-016-1979-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Berger A, Dangel A, Schober T, Schmidbauer B, Konrad R, Marosevic D, Schubert S, Hörmansdorfer S, Ackermann N, Hübner J, Sing A. 2019. Whole genome sequencing suggests transmission of Corynebacterium diphtheriae-caused cutaneous diphtheria in two siblings, Germany, 2018. Euro Surveill 24:1800683. doi: 10.2807/1560-7917.ES.2019.24.2.1800683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katsukawa C, Komiya T, Umeda K, Goto M, Yanai T, Takahashi M, Yamamoto A, Iwaki M. 2016. Toxigenic Corynebacterium ulcerans isolated from a hunting dog and its diphtheria toxin antibody titer. Microbiol Immunol 60:177–186. doi: 10.1111/1348-0421.12364. [DOI] [PubMed] [Google Scholar]
- 44.Dias AASO, Silva FC, Pereira GA, Souza MC, Camello TCF, Damasceno JALD, Pacheco LGC, Miyoshi A, Azevedo VA, Hirata R, Bôas MHSV, Mattos-Guaraldi AL. 2010. Corynebacterium ulcerans isolated from an asymptomatic dog kept in an animal shelter in the metropolitan area of Rio de Janeiro, Brazil. Vector Borne Zoonotic Dis 10:743–748. doi: 10.1089/vbz.2009.0132. [DOI] [PubMed] [Google Scholar]
- 45.Dias AAOS, Silva FC, Jr, Santos LS, Ribeiro-Carvalho MM, Sabbadini PS, Santos CS, Filardy AA, Myioshi A, Azevedo VA, Hirata R, Villas-Bôas MHS, Mattos-Guaraldi AL. 2011. Strain-dependent arthritogenic potential of the zoonotic pathogen Corynebacterium ulcerans. Vet Microbiol 153:323–331. doi: 10.1016/j.vetmic.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Mochizuki Y, Saeki H, Iwaki M, Takagi H, Shibayama K, Amao H, Yamamoto A. 2016. A novel experimental platform for toxigenic and non-toxigenic Corynebacterium ulcerans infection in mice. Pathog Dis 74:ftv109. doi: 10.1093/femspd/ftv109. [DOI] [PubMed] [Google Scholar]
- 47.Lappin MR, Blondeau J, Boothe D, Breitschwerdt EB, Guardabassi L, Lloyd DH, Papich MG, Rankin SC, Sykes JE, Turnidge J, Weese JS. 2017. Antimicrobial use guidelines for treatment of respiratory tract disease in dogs and cats: Antimicrobial Guidelines Working Group of the International Society for Companion Animal Infectious Diseases. J Vet Intern Med 31:279–294. doi: 10.1111/jvim.14627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosser EJ. 2006. German shepherd dog pyoderma. Vet Clin North Am Small Anim Pract 36:203–211, viii. doi: 10.1016/j.cvsm.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 49.Nyindo M, Huxsoll DL, Ristic M, Kakoma I, Brown JL, Carson CA, Stephenson EH. 1980. Cell-mediated and humoral immune responses of German shepherd dogs and beagles to experimental infection with Ehrlichia canis. Am J Vet Res 41:250–254. [PubMed] [Google Scholar]
- 50.Moore LSP, Leslie A, Meltzer M, Sandison A, Efstratiou A, Sriskandan S. 2015. Corynebacterium ulcerans cutaneous diphtheria. Lancet Infect Dis 15:1100–1107. doi: 10.1016/S1473-3099(15)00225-X. [DOI] [PubMed] [Google Scholar]
- 51.Ott L, Höller M, Gerlach RG, Hensel M, Rheinlaender J, Schäffer TE, Burkovski A. 2010. Corynebacterium diphtheriae invasion-associated protein (DIP1281) is involved in cell surface organization, adhesion and internalization in epithelial cells. BMC Microbiol 10:2. doi: 10.1186/1471-2180-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marosevic DV, Berger A, Kahlmeter G, Payer SK, Hörmansdorfer S, Sing A. 2020. Antimicrobial susceptibility of Corynebacterium diphtheriae and Corynebacterium ulcerans in Germany 2011–17. J Antimicrob Chemother 75:2885–2893. doi: 10.1093/jac/dkaa280. [DOI] [PubMed] [Google Scholar]
- 53.Badell E, Guillot S, Tulliez M, Pascal M, Panunzi LG, Rose S, Litt D, Fry NK, Brisse S. 2019. Improved quadruplex real-time PCR assay for the diagnosis of diphtheria. J Med Microbiol 68:1455–1465. doi: 10.1099/jmm.0.001070. [DOI] [PubMed] [Google Scholar]
- 54.Engler KH, Glushkevich T, Mazurova IK, George RC, Efstratiou A. 1997. A modified Elek test for detection of toxigenic corynebacteria in the diagnostic laboratory. J Clin Microbiol 35:495–498. doi: 10.1128/jcm.35.2.495-498.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bolt F, Cassiday P, Tondella ML, Dezoysa A, Efstratiou A, Sing A, Zasada A, Bernard K, Guiso N, Badell E, Rosso ML, Baldwin A, Dowson C. 2010. Multilocus sequence typing identifies evidence for recombination and two distinct lineages of Corynebacterium diphtheriae. J Clin Microbiol 48:4177–4185. doi: 10.1128/JCM.00274-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.00006-23-s0001.pdf, PDF file, 0.4 MB (407.5KB, pdf)
Data Availability Statement
MLST sequences and profiles can be accessed at https://bigsdb.pasteur.fr/diphtheria.