ABSTRACT
Fosfomycin (FOS) has been recently reintroduced into clinical practice, but its effectiveness against multidrug-resistant (MDR) Enterobacterales is reduced due to the emergence of FOS resistance. The copresence of carbapenemases and FOS resistance could drastically limit antibiotic treatment. The aims of this study were (i) to investigate fosfomycin susceptibility profiles among carbapenem-resistant Enterobacterales (CRE) in the Czech Republic, (ii) to characterize the genetic environment of fosA genes among the collection, and (iii) to evaluate the presence of amino acid mutations in proteins involved in FOS resistance mechanisms. During the period from December 2018 to February 2022, 293 CRE isolates were collected from different hospitals in the Czech Republic. FOS MICs were assessed by the agar dilution method (ADM), FosA and FosC2 production was detected by the sodium phosphonoformate (PPF) test, and the presence of fosA-like genes was confirmed by PCR. Whole-genome sequencing was conducted with an Illumina NovaSeq 6000 system on selected strains, and the effect of point mutations in the FOS pathway was predicted using PROVEAN. Of these strains, 29% showed low susceptibility to fosfomycin (MIC, ≥16 μg/mL) by ADM. An NDM-producing Escherichia coli sequence type 648 (ST648) strain harbored a fosA10 gene on an IncK plasmid, while a VIM-producing Citrobacter freundii ST673 strain harbored a new fosA7 variant, designated fosA7.9. Analysis of mutations in the FOS pathway revealed several deleterious mutations occurring in GlpT, UhpT, UhpC, CyaA, and GlpR. Results regarding single substitutions in amino acid sequences highlighted a relationship between ST and specific mutations and an enhanced predisposition for certain STs to develop resistance. This study highlights the occurrence of several FOS resistance mechanisms in different clones spreading in the Czech Republic.
IMPORTANCE Antimicrobial resistance (AMR) currently represents a concern for human health, and the reintroduction of antibiotics such as fosfomycin into clinical practice can provide further option in treatment of multidrug-resistant (MDR) bacterial infections. However, there is a global increase of fosfomycin-resistant bacteria, reducing its effectiveness. Considering this increase, it is crucial to monitor the spread of fosfomycin resistance in MDR bacteria in clinical settings and to investigate the resistance mechanism at the molecular level. Our study reports a large variety of fosfomycin resistance mechanisms among carbapenemase-producing Enterobacterales (CRE) in the Czech Republic. Our study summarizes the main achievements of our research on the use of molecular technologies, such as next-generation sequencing (NGS), to describe the heterogeneous mechanisms that reduce fosfomycin effectiveness in CRE. The results suggest that a program for widespread monitoring of fosfomycin resistance and epidemiology fosfomycin-resistant organisms can aide timely implementation of countermeasures to maintain the effectiveness of fosfomycin.
KEYWORDS: Enterobacterales, carbapenemase producers, fosfomycin, drug-resistance bacteria, WGS
INTRODUCTION
Fosfomycin (FOS) is a phosphoric acid derivate, active against both Gram-negative and Gram-positive bacteria. It was discovered in 1969, and it gained renewed clinical interest in the last 20 years as a good candidate in the treatment of multidrug-resistant (MDR) bacterial infections (1). According to the Food and Drug Administration (FDA) and the European Medicine Agency (EMA), FOS is approved for oral use in uncomplicated lower urinary tract infections (UTI) and for systemic use in complicated UTI and bacterial meningitis (https://www.accessdata.fda.gov/drugsatfda_docs/label/2008/050717s005lbl.pdf). FOS binds to UDP-N-acetylglucosamine enolpyruvyl transferase (MurA), which interferes with the early stages of peptidoglycan production (2). FOS uptake inside bacterial cell depends on two transport systems: the glycerol-3-phosphate transporter (GlpT) and the glucose-6-phosphate transporter (UhpT). The expression of GlpT and UhpT is glycerol-3 (G3P) and glucose-6-phosphate (G6P) dependent and requires the presence of cyclic AMP (cAMP). Moreover, UhpT expression is further controlled by the UhpABC system (2). Inactivation of UhpABC impairs expression levels of UhpT and affects FOS uptake into the cytosol (3–8).
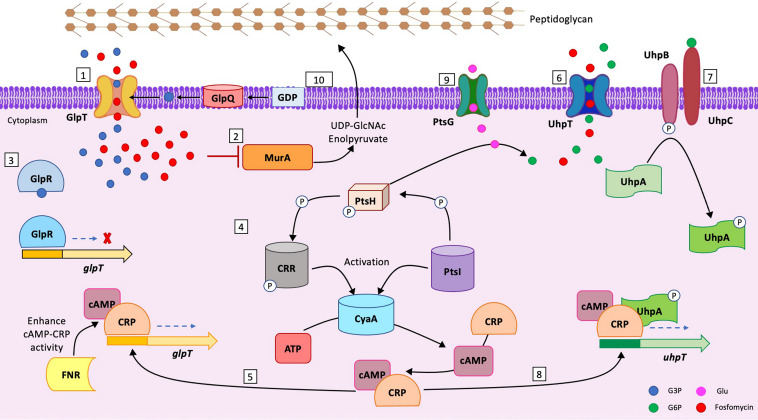
The levels of cAMP are directly related to the adenylate cyclase CyaA and to the phosphotransferase PtsI. The occurrence of mutations in both CyaA and PtsI alters cAMP intracellular levels and, consequently, FOS uptake (9) (Fig. 1).
FIG 1.
FOS uptake and pathway inside the bacterial cell. (Step 1) GlpT transports G3P and fosfomycin inside the bacterial cell. GlpT expression is mediated by G3P. (Step 2) Inside the cytoplasm, FOS binds to MurA and interferes with the formation of UDP-GlcNAc enolpyruvate, the first stage of peptidoglycan biosynthesis. (Step 3) GlpR acts as a repressor for GlpT expression. In the cytoplasm, G3P attaches to GlpR, blocking its binding to glpT promoter. (Step 4) PtsI transfers a P group from PEP (2-phosphoenolpyruvate) to PtsH. The P group is then transferred to CRR by PtsH. CRR-P and PtsI activate CyaA (4). CyaA is an adenylate cyclase that converts the ATP to cAMP. cAMP binds to the CRP, and the cAMP-CRP complex promotes expression of both GlpT and UhpT (5). (Step 5) cAMP-CRP complex promotes GlpT expression, binding to glpT promoter. The activity of the cAMP-cAMP receptor protein (CRP) complex is enhanced by FNR (6). (Step 6) UhpT promotes the entry of G6P and FOS into the cell. The presence of G6P enhances the expression levels of UhpT. (Step 7) The UhpABC system promotes the expression of UhpT. UhpC binds extracellular G6P, and through UhpB, a phosphate group is transferred to UhpA (UhpA-P). (Step 8) UhpA-P is the activate form of UhpA and, together with cAMP-CRP complex, starts UhpT transcription, binding to the uhpT promoter. (Step 9) PtsG promotes the entry of glucose into the cell (7). (Step 10) GlpQ is a periplasmic glycerophosphodiester phosphodiesterase that converts periplasmic glycerophosphodiesters to G3P (8).
Several mutations can take place at different steps of the above-mentioned pathway, affecting FOS uptake and leading to different extents of FOS resistance (2, 10, 11). Alteration of GlpT/UhpT activity and mutations in MurA are considered the primary FOS resistance mechanism, followed by impairment in UhpA, CyaA, and PtsI activity (9) (Fig. 1).
In recent years, a great deal of attention has been given to FOS-modifying enzymes, such as FosA. FosA is a metalloenzyme that catalyzes the opening of the FOS epoxide ring, leading to its inactivation (12). As of December 2022, 10 fosA variants have been reported on both chromosomes and plasmids in members of the Enterobacterales. fosA-like genes are chromosomally distributed in Providencia spp. and Serratia marcescens (fosA), in Enterobacter spp. (fosA2), in Salmonella species (fosA7), and in Klebsiella pneumoniae (fosA5 and fosA6) (12). In contrast, plasmid-mediated dissemination is recognized for fosA3, fosA4, fosA8, fosA9, and fosA10 among Escherichia coli and K. pneumoniae strains (13–16). Epidemiologically, FosA3 is the most widespread variant, with endemic cases reported from both veterinary and clinical settings in China (13, 17–19); FosA7 is predominantly found in isolates from veterinary settings (20, 21). FosA5 has been reported from clinical settings in China (22, 23) and from food in India (24), while FosA10 has been detected only in chicken meat from China (16). Few cases from food and clinical samples reported the occurrence of FosA8 (25, 26), while only one clinical case involving FosA9 has been described from a clinical case in the Netherlands (27). The co-occurrence of FosA enzymes and extended-spectrum β-lactamases (ESBLs), including carbapenemases, in Enterobacterales has been already reported in the literature, limiting the use of FOS in clinical practice (28, 29). Co-occurrence of FosA enzymes and carbapenemases has been mainly described for K. pneumoniae (17, 28, 30), with sporadic occurrences in E. coli (31–33) and two reports of occurrences in Citrobacter freundii (34).
Escherichia coli is an intestinal commensal of warmed-blood animals that has the ability to cause diseases such as extraintestinal infections (13). Citrobacter spp. are low-risk pathogens but, as reported in the literature, can act as a silent reservoir for relevant resistance genes, especially C. freundii (35). The occurrence of FosA enzymes has been documented worldwide, but large epidemiological investigations of their occurrence in E. coli and in Citrobacter spp. are lacking, especially among European countries. The Czech Republic has an active and broad surveillance program for carbapenem-resistant Enterobacterales (CRE) among clinical settings (36, 37), but no epidemiological investigation of FOS resistance among these strains has been conducted recently. Moreover, as reported in previous studies, the Czech Republic has an increasing number of cases of disease caused by carbapenemase-producing E. coli and C. freundii, and the occurrence of fosfomycin resistance mechanisms in such isolates represents a concerning public health issue (35–37).
The aim of our study was to characterize the epidemiology of FOS resistance in the Czech Republic clinical setting in E. coli and Citrobacter carbapenemase producers. Moreover, an additional aim was to characterize the resistance mechanisms of FOS resistance through the detection of fosA-like genes and through the detection of specific point mutations in the associated transporters and regulators involved in uptake.
RESULTS
All 293 Enterobacterales strains were carbapenemase producers: 132/293 produced NDM-type enzymes (111 [84%] E. coli and 21 [16%] Citrobacter isolates), 106/293 produced OXA-48-type (76 [72%] E. coli and 30 [28%] Citrobacter isolates), 50/293 produced KPC-type enzymes (33 [66%] E. coli and 17 [34%] Citrobacter isolates), and 5/293 produced VIM-type enzymes (3 [60%] E. coli and 2 [40%] Citrobacter isolates). The agar dilution method (ADM) showed that 71% (208/293) of the isolates retained susceptibility to FOS (MIC ≤ 8 μg/mL) and the remaining 29% (85/293) showed low-susceptibility/resistance profiles. In detail, 41 of 85 (38 [93%] E. coli and 3 [7%] C. freundii isolates) had FOS MICs of 16 μg/mL, 18 of 85 (16 [89%] E. coli and 2 [11%] C. freundii isolates) had FOS MICs of 32 μg/mL, and 26 of 85 (16 [62%] E. coli and 10 [38%] C. freundii isolates) had FOS MICs of ≥64 μg/mL. The sodium phosphonoformate (PPF) test was performed on strains with FOS MICs of ≥64 μg/mL, and only two of 26 FOS-resistant strains were FosA/FosC2 enzyme producers.
PCR investigations of the two strains detected fosA10 in an NDM-producing strain of E. coli (ECO49406) and a fosA7 gene in a VIM-producing C. freundii strain (CFR50714). fosA10 was successfully transferred by conjugation, while attempts to transfer fosA7 through conjugation and transformation failed.
fosA10 in NDM-producing E. coli.
Whole-genome sequencing (WGS) revealed that ECO49406 belonged to sequence type 648 (ST648)/unknown sequence type (Oxford scheme/Pasteur scheme), serotype O102:H6, and the CH type FumC4/FimH27 (fimbrial adhesion gene fimH with allele 27 and fumarate hydratase class II gene fumC with allele 4). The resistome of ECO49406 consisted of β-lactam (blaNDM-5), fosfomycin (fosA10), aminoglycoside (aadA2 and aadA5), folate pathway antagonist (dfrA12, dfrA17, and sul-1), tetracycline [tet(B)], macrolide [mph(A)], and quaternary ammonium compound (qacE) resistance genes. Moreover, ECO49406 carried a multireplicon IncFIB/FII and an IncK plasmid. Additionally, the virulome consisted of air, chuA, eilA, fyuA, gad, irp2, iss, kpsE, kpsMII_K5, lpfA, ompT, terC, traT, and yfcV (Table 1).
TABLE 1.
WGS data for FOS-susceptible and -resistant strains studieda
| Strain | Origin | Location | Species | FOS MIC (μg/mL) and category | FosA variant | Serotype | FumC/FimH | Plasmid | ST (Oxford/Pasteur) | Virulence genes | Other resistance genes | Accession no. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ECO49406 | Rectal swab | Prague | E. coli | >128, R | 10 | O_:H_ | 4/27 | IncK, IncFIB (pB171), IncFII | ST648/unknown | air, chuA, eilA, fyuA, gad, irp2, iss, kpsE, kpsMII_K5, lpfA, ompT, terC, traT, yfcV | aadA2, aadA5, sul-1, dfrA12, dfrA17, tet(B), mph(A), blaNDM-5, qacE | JAPEQT000000000 |
| ECO52246 | Urine | Nymburk | E. coli | >128, R | ND | O_:H_ | 40/30 | IncFII(K), IncR | ST131/43 | chuA, fyuA, gad, iha, irp2, iss, kpsE, ompT, sat, sitA, terC, traT, usp, yfcV | aph(6)-Id, aac(6′)-Ib3, aph(3′)-Ib, aac(6′)-Ib-cr, tet(A), mph(A), qnrB1, sul-1, sul-2, dfrA14, ARR-3, blaOXA-1, blaTEM-1B, blaKPC-3, qacE, catB3 | JAPJVB000000000 |
| ECO52259 | Rectal swab | Jičín | E. coli | >128, R | ND | O_:H_ | 40/30 | IncFII(K), IncR | ST131/43 | chuA, fyuA, gad, iha, irp2, iss, kpsE, ompT, sat, sitA, terC, traT, usp, yfcV | aph(6)-Id, aac(6′)-Ib3, aph(3′)-Ib, aac(6′)-Ib-cr, tet(A), mph(A), qnrB1, sul-1, sul-2, dfrA14, ARR-3, blaOXA-1, blaTEM-1B, blaKPC-3, qacE, catB3 | JAPJVC000000000 |
| ECO52250b | Urine | Jičín | E. coli | 4, S | ND | O_:H_ | 43/197 | IncR | 2558/unknown | chuA, clbB, cnf1, focC, fyuA, gad, hra, ibeA, iroN, irp2, iss, kpsE, kpsMII, mchB, mchC, mchF, mcmA, ompT, papA_F13, papC, pic, sfaD, sitA, terC, usp, vat, yfcV | aac(6′)-Ib3, aac(6′)-Ib-cr, mph(A), sul-1, ARR-3, blaKPC-2, blaOXA-1, qacE, catB3 | JAFEXB000000000 |
| ECO52846 | Decubitus | Jičín | E. coli | 128, R | ND | O_:H_ | 43/197 | IncR | 2558/unknown | chuA, clbB, cnf1, focC, fyuA, gad, hra, ibeA, iroN, irp2, iss, kpsE, kpsMII, mchB, mchC, mchF, mcmA, ompT, papA_F13, papC, pic, sfaD, sitA, terC, usp, vat, yfcV | aac(6′)-Ib3, aac(6′)-Ib-cr, mph(A), sul-1, ARR-3, blaKPC-2, blaOXA-1, qacE, catB3 | JAPJVD000000000 |
| ECO53083b | Stool | Hradec Kralove | E. coli | 8, S | ND | O_:H_ | 40/30 | IncFII(k), IncR | ST131/43 | chuA, fyuA, gad, iha, irp2, iss, kpsE, ompT, sat, sitA, terC, traT, usp, yfcV | aph(3′′)-Ib, aac(6′)-Ib3, aph(6)-Id, aac(6′)-Ib-cr, mph(A), sul-1, sul-2, dfrA14, ARR-3, blaKPC-3, blaOXA-1, qacE, catB3 | CP070587–CP070593 |
| CFR67526 | NA | NA | C. freundii | 128, R | ND | NA | NA | IncR, pKPC-CAV1321 | ST19 | NA | aac(6′)-Ib-cr, mcr-9, mph(A), sul-1, ARR-3, blaKPC-2, blaOXA-1, blaCMY-152, qacE, catB3 | JAPJVE000000000 |
| CFRC0593595 | Urine | Pilsen | C. freundii | 128, R | ND | NA | NA | IncFIB, IncFII, IncL | ST98 | NA | aac(6′)-Ib-cr, aph(6)-Id, aph(3′′)-Ib, aac(3′′)-IIa, qnrB1, sul-1, dfrA14, tet(A), tet(D), blaOXA-1, blaOXA-48, blaCMY-109, blaCTX-M-15, blaTEM-1b, catB3 | JAPJVF000000000 |
| CFR47654 | Urine | Prague | C. freundii | >128, R | ND | NA | NA | IncFIB, IncL | ST98 | NA | aac(6′)-Ib-cr, aph(6)-Id, aph(3′′)-Ib, aac(3′′)-IIa, qnrB1, sul-1, dfrA14, tet(A), tet(D), blaOXA-1, blaOXA-48, blaCMY-109, blaCTX-M-15, blaTEM-1b, catB3 | JAPJVG000000000 |
| CFR50714c | Urine | Prague | C. freundii | 64, R | ND | NA | NA | IncFII/IncN | ST673 | NA | aac(6′)-Ib3, aph(3′′)-Ib, aac(6′)-Ib-cr, aadA1, fosA7.9, qnrS1, dfrA14, sul-1, sul-2, blaCMY-78, blaVIM-1, qacE | JAJFDB000000000 |
| CFR47299b | Rectal swab | Prague | C. freundii | 128, R | ND | NA | NA | IncC, IncFII, IncHI2, IncHI2A, IncN | ST65 | NA | aph(6)-Id, aph(3′′)-Ib, aac(3)-IId, aac(6′)-Ib-cr, aac(3)-IIa, ARR-3, qnrA1, sul-1, sul-2, dfrA19, tet(B), blaKPC-2, blaCMY-48, blaTEM-1b, blaOXA-1, qacE, catB3, catA1 | JAFEWM000000000 |
| CFR47462b | Rectal swab | Prague | C. freundii | 128, R | ND | NA | NA | IncFII, IncHI2, IncHI2A, IncN, IncR |
ST65 | NA | aph(6)-Id, aph(3′′)-Ib, aac(3)-IId, aac(6′)-Ib-cr, aac(3)-IIa, ARR-3, qnrA1, sul-1, sul-2, dfrA19, tet(B), blaKPC-2, blaCMY-48, blaTEM-1b, blaOXA-1, qacE, catB3, catA1 | JAFEWN000000000 |
| CFR56415c | Rectal swab | Prague | C. freundii | 8, S | ND | NA | NA | IncHI2, IncHI2A | ST95 | NA | aadA1, ant(2′′)-Ia, aph(3′)-Ia, aac(6′)-Ib-cr, aadA2b, aac(6′)-Ib3, mcr-9, sul-1, dfrA19, blaVIM-4, blaCMY-51, qacE, cmlA1, catA2 | CP085726 –CP085728 |
| CFR51929c | Rectal swab | Prague | C. freundii | 16, S | ND | NA | NA | IncHI2, IncHI2A, IncM1 | ST95 | NA | aac(6′)-Ib-cr, aadA1, ant(2′′)-Ia, aph(3′)-Ia, aac(6′)-Ib3, aadA1, aadA2b, aac(3)-I, mcr-9, qnrA1, sul-1, dfrA19, tet(A), blaCMY-51, blaVIM-4, qacE, catA2, cmlA1 | CP059427–CP059429 |
| CFR47298 | Decubitus | Prague | C. freundii | >128, R | ND | NA | NA | IncFIB, IncL | ST98 | NA | aac(3′)-IIa, aac(6′)-Ib-cr, aph(6)-Id, aph(3′′)-Ib, qnrB1, dfrA14, sul-2, tet(A), tet(D), blaOXA-48, blaCTX-M-15, blaTEM-1B, blaOXA-1, blaCMY-109, catB3 | JARWKP000000000 |
NA, not available; ND, not detected; air, enteroaggregative immunoglobulin repeat protein; chuA, outer membrane hemin receptor; eilA, Salmonella HilA homolog; fyu, siderophore yersiniabactin receptor; gad, glutamate decarboxylase; irp2, high-molecular-weight protein 2 nonribosomal peptide synthetase; iss, increased serum survival; kpsE, capsule polysaccharide export inner membrane protein; kpsMII_K5, polysialic acid transport protein; lpfA, long polar fimbriae; ompT, outer membrane protease; terC, tellurium ion resistance protein; traT, outer membrane complement resistance; yfcV, fimbria-like protein.
Published in reference 36.
Included in reference 77.
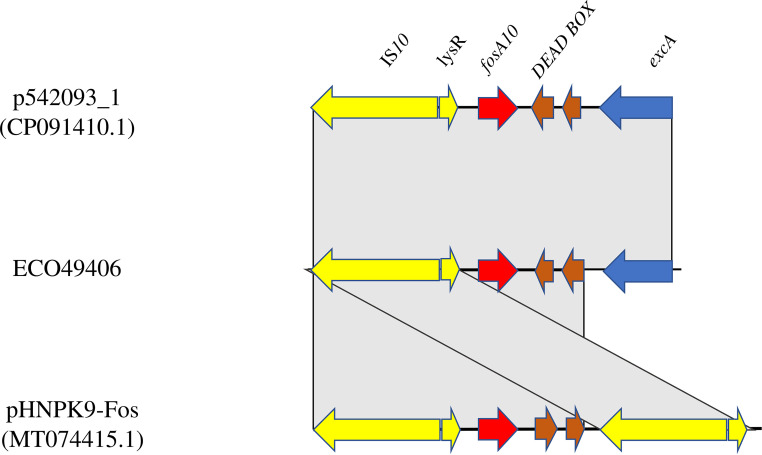
Based on short-read data and conjugation experiments, fosA10 was located on the IncK plasmid. fosA10 was in a genomic cassette of 3,835 bp consisting of excA-DEAD box-fosA10-lysR-IS10R. The BLAST results showed that the cassette shared 100% query and identity with the fosA10 cassette of the IncB/O/K/Z plasmid p542093_1 (accession no. CP091410.1) and 65% query and 100% identity with the fosA10 cassette reported in an IncFII plasmid (pHNPK9-Fos; accession no. MT074415.1) (16) collected from veterinary E. coli isolates in China. pHNPK9-fosA10 cassette (4,328 bp) differed only by (i) containing two copies of IS10 flanking fosA10 and the DEAD box and (ii) lacking excA (Fig. 2).
FIG 2.
Linear map of the fosA10 genetic environment of p542093_1, the genetic environment of the fosA10-carrying plasmid isolated from ECO49406, and the genetic environment of fosA10 harbored on an IncFII pHNPK9-Fos. Arrows show the direction of transcription of open reading frames. excA, mobile elements, fosA10, and other remaining genes are designated by blue, yellow, red, and brown, respectively. Gray shading shows homologous segments with 100% sequence identity.
fosA7.9 in VIM-producing C. freundii.
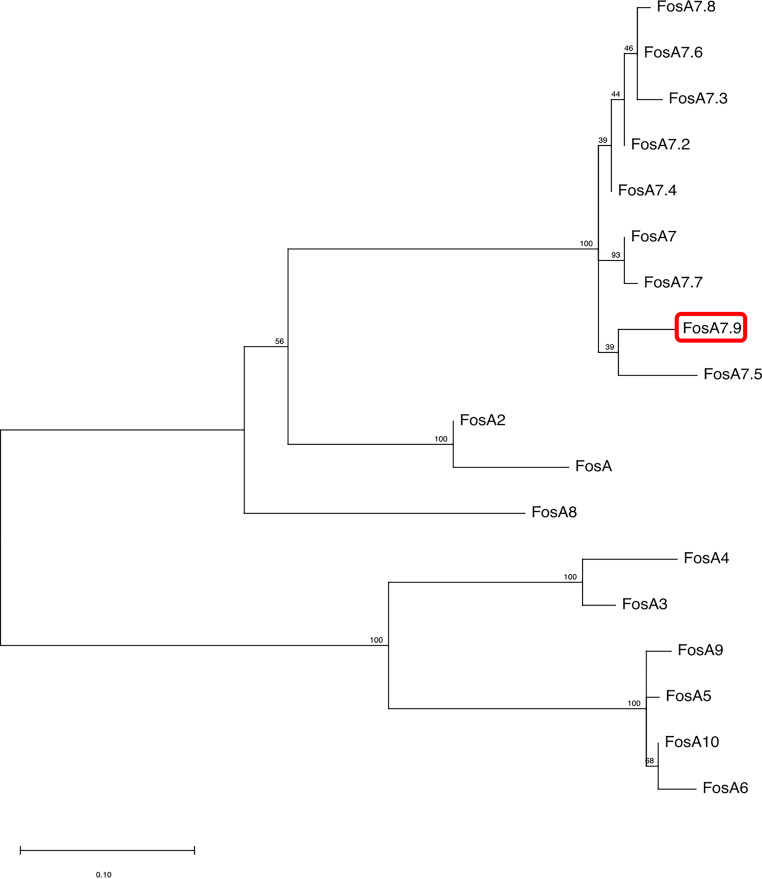
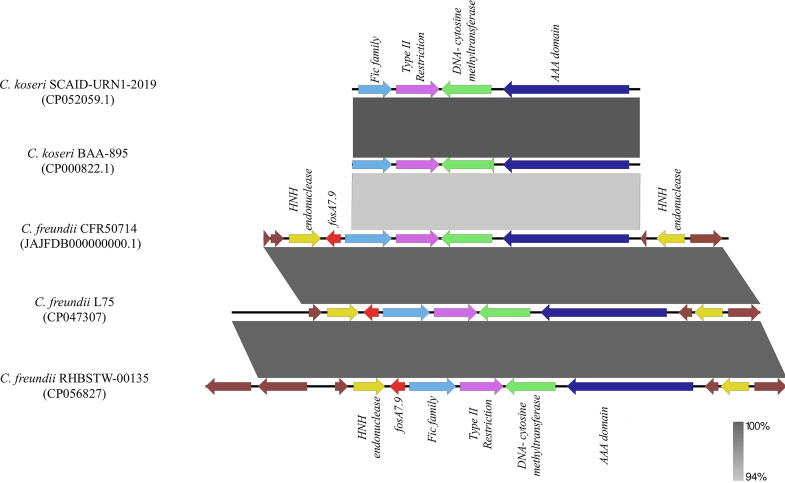
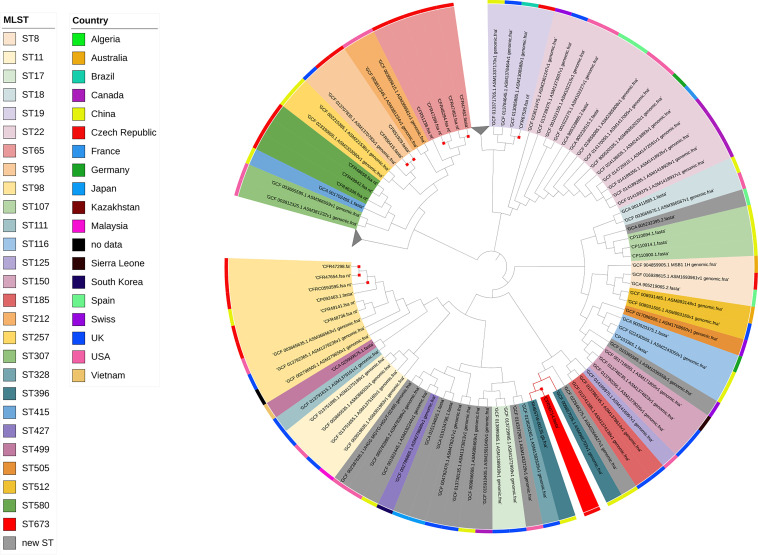
Based on WGS data, C. freundii CFR50714 belonged to ST673 and harbored antibiotic resistance genes for β-lactam (blaVIM-1 and blaCMY-78), fosfomycin (fosA7), aminoglycoside [aadA1, aac(6′)-Ib3, and aac(6′)-Ib-cr], folate pathway antagonist (dfrA14, sul-1, and sul-2), quinolone (qnrS1), macrolide [mph(A)] and quaternary ammonium compound (qacE). The fosA7 gene was 423 bp in length and encoded a 140-amino-acid protein. The fosA7 gene shared highest identity (93.38% identity and 100% query) with the fosA7.4 variant (accession no. NG_067230.1) and encoded a protein showing 95% identity with FosA7.4 (accession no. WP_023216493.1) and 93.4% with FosA7.5 (Fig. 3 and 4). fosA7.9 was inserted into a 12,065-bp cassette consisting of the following genes: the HNH endonuclease gene, fosA7.9, fic, the type II endonuclease restriction gene, the methyltransferase gene, and the HNH endonuclease gene. The BLAST results revealed that the cassette showed similarity with several strains collected worldwide: 100% query and 99.98% identity with the NDM+FosA7.9 coproducer C. freundii L75, collected in China in 2018 from a urine sample (accession no. CP047307) (38), and with the CMY-2+FosA7.9 coproducer C. freundii RHBSTW-00135, collected in 2017 from wastewater influent in the United Kingdom (accession no. CP056827) (39). Additionally, the fosA7.9 cassette shared 81% query and 94.34% identity with a chromosome cassette of Citrobacter koseri SCAID-URN1-2019 (accession no. CP052059.1) collected in Kazakhstan from a urine sample (40), and with C. koseri BAA-895, collected in Maryland from an infant with meningitis (accession no. CP000822.1). Interestingly, the cassette in C. koseri lacked the flanking HNH and fosA7.9 but had an identical fic cassette (fic-type II endonuclease restriction-methyltransferase) in the same orientation (Fig. 5). The in silico analysis and the failed attempts at conjugation suggested a chromosomal location for fosA7.9. Sequence data from strain CFR50714, C. freundii L75 (CP047307), and C. freundii RHBSTW-00135 (CP056827) were used to investigate their genomic relatedness to global isolates, to construct a single nucleotide polymorphism (SNP)-based phylogenetic tree. The three aforementioned sequences were compared against 103 genomes found in the NCBI database (Fig. 5). Strain CFR50714 clustered with C. freundii RHBSTW-00135 (ST396) and C. freundii L75 (ST396), forming a clade (Fig. 6).
FIG 3.
Alignment of amino acid sequences of the FosA7 variants: FosA7 (WP_000941934.1) (94.29% identity with FosA7.9), FosA7.2 (WP_000941935.1) (94.29% identity), FosA7.3 (WP_023231494.1) (93.57% identity), FosA7.4 (WP_023216493.1) (95%), FosA7.5 (WP_000941933.1) (93.57%), FosA7.6 (WP_061377147.1) (93.57%), FosA7.7 (WP_058653118.1) (93.57%), FosA7.8 (WP_079820715.1) (92.86%), and FosA7.9 (UYP40110.1).
FIG 4.
The evolutionary analysis and phylogenetic tree of FosA proteins found in Enterobacterales were inferred by using the maximum-likelihood method and JTT matrix-based model using MEGA 11. The tree with the highest log likelihood (−1,275.39) is shown. The percentage of trees in which the associated taxa clustered together is shown above the branches. The tree is drawn to scale. The red rectangle indicates the new FosA7.9 in C. freundii.
FIG 5.
Linear map of the fic cassette genetic environment of SCAID-URN1-2019 and BAA-895, the genetic environment of the fosA7.9 cassette harbored on the CFR50714 chromosome, L75, and RHBSTW-00135. Arrows show the direction of transcription of ORFs. The fic family, HNH endonuclease, fosA7.9, type II restriction, DNA-cytosine methyltransferase, AAA domain, and other genes are designated by light blue, yellow, red, fuchsia, green, blue and brown, respectively. Homologous segments are indicated with gray shading (light gray, 94% sequence identity; dark gray, 100% identity).
FIG 6.
Phylogenetic tree of the nine C. freundii isolates with 103 genomes downloaded from the NCBI database. Red nodes indicate the isolates harboring fosA7.9 variants. The red square shows the studied isolates. Gray triangles indicate collapsed nodes.
Fosfomycin pathway alterations.
Fifteen sequenced strains were investigated for the presence of point mutations in proteins involved in the FOS pathway. When possible, strains of the same ST but different FOS profiles were compared. Concerning E. coli strains, three of six belonged to ST131, two to ST2558, and one to ST648 (Fig. 7; also, see Fig. S1 in the supplemental material).
FIG 7.
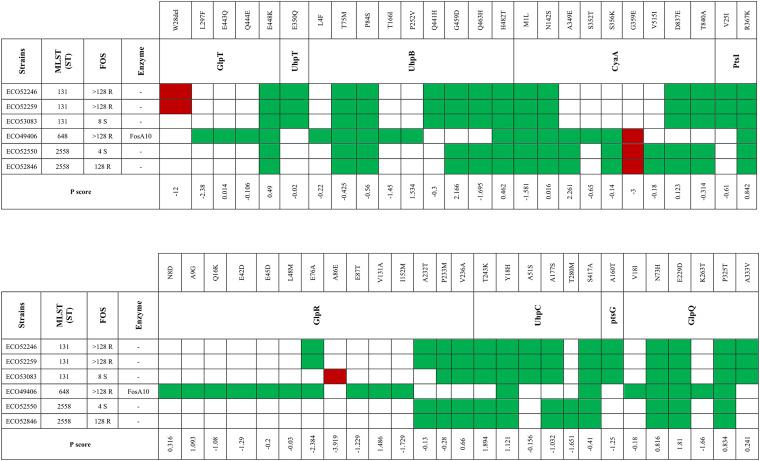
Mutations and P scores obtained by PROVEAN for selected E. coli strains. Red indicates a P score above −2.5 (categorized as deleterious); green indicates a P score below −2.5 (categorized as neutral).
All three ST131 E. coli isolates shared neutral substitutions in the amino acids of GlpT, UhpT, UhpB, CyaA, PtsI, GlpR, UhpC, PtsG, and GlpQ (Fig. 7). Additionally, two FOS-resistant (FOSr) strains (strains ECO52246 and ECO52259) out of three ST131 strains showed a deleterious W28del (PROVEAN [P] score, −12,042) of GlpT. On the other hand, the remaining FOS-susceptible (FOSs) ST131 strain (ECO53083) carried a deleterious substitution (P score, −3.919) in GlpR (A86E) (Fig. 7).
Two E. coli ST2558 strains (FOSr strain ECO528469 and FOSs strain ECO52550) shared the same neutral substitutions in GlpT, UhpB, CyaA, PtsI, GlpR, UhpC, and GlpQ. Moreover, both ST2558 strains showed a deleterious substitution (P score, −3.077) in CyaA (G359E).
The FosA10-producing strain ECO49406 exhibited a wide range of single amino acid substitutions, categorized as neutral, in GlpT, UhpB, CyaA, PtsI, and GlpR. ECO49406 carried the deleterious mutation G359E (P score, −3.077) in CyaA (Fig. 7). No alteration in the target MurA, the transporter UhpT, or the regulators PtsH, UhpA, UhpC, FNR (fumarate and nitrate reduction regulatory protein), and CRR were detected in any of the E. coli strains studied. These results imply a link between ST and mutations in the FOS pathway.
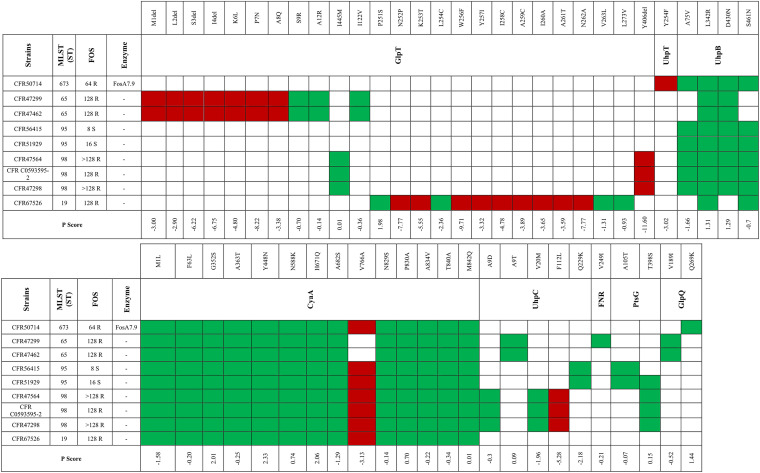
Regarding C. freundii isolates, three of nine belonged to ST98, two to ST95, two to ST65, one to ST673, and one to ST19. Analysis of the FOS pathway highlighted a similar link between ST and certain amino acid substitutions (Fig. 8). Two FOSr ST65 strains (CFR47299 and CFR47462) shared the same neutral substitutions in GlpT, UhpB, CyaA, UhpC, and GlpQ. Additionally, both isolates missed the first 4 amino acids (aa) (MLSI) and had the deleterious substitutions K6L, P7N, and A8Q (P scores, −4.8, −8.2, and −3.4, respectively) in GlpT. Two FOSs ST95 strains (CFR56415 and CFR51929) accumulated identical neutral substitutions in UhpB, CyaA, UhpC, and PtsG but shared the deleterious mutation V766A in CyaA (P score, −3.13). The same deleterious change in CyaA occurred in strains CFR47298 and CFR50714, belonging to ST98 and ST673, respectively. In contrast, strain CFR47298 had deleterious deletions in GlpT (Y406del; P score, −11.6) and in UhpC (F112L; P score, −5.28). Strain CFR67526, the only ST19 strain, shared the same reported neutral alteration in CyaA. Interestingly, the glpT sequence had an insertion of 25 nucleotides (nt) at position 788, leading to a 1,384-nt instead of a 1,359-nt gene. The frameshift mutation affects the amino acids from position 251 to 263aa, which could possibly affect the activity of the transporter (Fig. 8). No alterations in MurA and in the regulators UhpA, PtsH, PtsI, GlpR, CRP, and CRR were detected in any of the C. freundii strains studied. These results together show that the occurrence of mutations at different levels can decrease FOS susceptibility.
FIG 8.
Mutations and P scores obtained by PROVEAN for selected C. freundii strains. Red indicates a P score above −2.5 (categorized as deleterious); green indicates a P score below −2.5 (categorized as neutral).
DISCUSSION
Fosfomycin has regained importance in clinical practice and has offered an alternative first-line option against MDR bacterial infections. The pathway of fosfomycin inside bacterial cells depends mainly on GlpT and UhpT activity (2). Acquiring mutations in GlpT and UhpT can impair their transport activity, decreasing FOS uptake into the bacterial cell and hence FOS effectiveness (41–43). However, modifications in these proteins have a high fitness cost, leading to the predominance of FOS-susceptible strains (44).
In the current study, all sequenced E. coli strains had amino acid substitutions in GlpT. The substitutions E448K, Q444E, and E443Q, categorized as neutral by PROVEAN, were reported previously by Takahata et al. and Sorlozano-Puerto et al. and are recognized as not impacting GlpT functionality (3, 9). L297F has been categorized as neutral (P score, −2.375) and was reported previously by Sorlozano-Puerto and colleagues (9); however, the impact on GlpT function has not been investigated yet. Two FOSr E. coli ST131 strains (ECO52246 and ECO52259) contained W28del, classified as deleterious (P score, −12.042). We speculate that there is a possible impact of W28del on GlpT activity, leading to a FOSr profile in E. coli ST131 strains. E. coli ST131 is a hypervirulent and pandemic clone (45) associated with the global spread of ESBLs such as CTX-M-15, KPC-like, and NDM-like enzymes (46–48). The acquisition of additional antimicrobial resistance traits in such successful clones can impact the clinical outcome of infections by such isolates and reduce antibiotic availability.
UhpB is a component of the UhpABC system and is a membrane-associated protein kinase that autophosphorylates and subsequently transfers its phosphate group to UhpA, activating it. CyaA is an adenylate cyclase and catalyzes the formation of cAMP from ATP. UhpB and CyaA exhibited a consistent number of alterations, most of them categorized as neutral. Interestingly, all sequenced E. coli isolates carried the single amino acid substitution M1L (P score, −1.581) but without a clear impact on CyaA functionality. Moreover, three E. coli strains, two FOSr (ECO49406 and ECO52846) and one FOSs (ECO52550), carried the deleterious mutation G359E (P score, −3.077). We hypothesize that this substitution could affect CyaA activity but without leading to a FOSr event at a high level. We hypothesize that in the presence of G359E CyaA, other mechanisms can be involved, leading to the FOSr profiles in these E. coli strains.
GlpR is a repressor of GlpT that binds to G3P, which is essential for glpT transcription. In the literature, studies concerning the eventual effect of GlpR on FOS MIC have not been reported yet. Here, we report the deleterious mutation A86E in GlpR. This alteration occurred in a FOSs E. coli ST131 strain (MIC = 8 [susceptible {S}]), and we speculated that there is a possible effect on GlpR activity, leading to the increased FOS MIC. Our findings highlight the possible role that GlpR plays in FOS susceptibility in E. coli strains. Further investigations are needed to understand the nature of the role of GlpR in altering FOS MIC.
In the literature, there are no reports that evaluate modifications in FOS pathway among Citrobacter spp.
Here, we describe the first genetic analysis of mutations detected in FOSs and FOSr C. freundii isolates and their effect on FOS susceptibility. In our study, several amino acid substitutions and deletions were detected in GlpT, UhpT, UhpB, CyaA, UhpC, PtsG, and GlpQ. Interestingly, two FOSr C. freundii ST65 isolates lost the first 4 aa of GlpT and carried the deleterious substitutions K6L (P score, −4.8), P7N (P score, −8.22), and A8Q (P score, −3.384). Additionally, both strains lacked the first 18 aa of CyaA. These alterations together could impair the activity of GlpT and CyaA, leading to a high level of FOSr. Among the nine C. freundii strains, we detected 30 different single amino acid substitutions in CyaA. Interestingly, all C. freundii isolates except C. freundii ST65 strains had the deleterious mutation V766A in CyaA (P score, −3.126). Regarding the UhpB regulator, all C. freundii harbored the neutral substitutions L342R and D430N and the mutations A75V (P score, −1.66) and S461N (P score, −0.7) (except for C. freundii ST65 strains). Regarding the FOSr C. freundii ST98 strain (CFR 47298), we report the deletion Y406del (P score, −11.6) in GlpT and the deleterious single substitution F112L (P score, −5.28) in UhpC, which could be implicated in FOS resistance. C. freundii ST65 and ST98 are emerging clones involved in the spread of ESBLs. Samuelsen et al. described two cases of CMY-48+OXA-10-coproducing C. freundii ST65 from clinical samples in Denmark, while Schweizer et al. described an outbreak event in Germany, caused by KPC-2-producing C. freundii ST98 isolates (49, 50). The occurrence of deleterious substitutions in proteins implicated in FOS uptake could decrease the effectiveness of FOS and its use against infections by clinically relevant clones such as carbapenemase-producing C. freundii ST98 isolates.
E. coli ST648 is an international high-risk and pathogenic clone, in both clinical and veterinary settings (51). Recently, it has been recognized as a pandemic clone, able to carry carbapenemases such as KPC-2 (52), NDM-1 (53–55), and OXA-48 (53, 55). In 2016, Yang et al. reported an E. coli ST648 strain coproducing NDM-5, CTX-M-55, MCR-1, and FosA3 from a duck in China (56). Here, we report the first case of E. coli ST648 coproducing NDM-5 and FosA10 isolated from humans in the Czech Republic. The genomic environment of fosA10 showed perfect identity with the fosA10-carrying IncI1 plasmid obtained from clinical E. coli ST227 strains in the United Kingdom and close similarity with the fosA10-carrying IncFII plasmid from veterinary E. coli ST38 collected in China (57). The global ST648 epidemiology and our findings focus attention on the ease of acquisition of MDR genes in this clone, which could drastically reduce the number of still-active antibiotics. Moreover, the findings highlight (i) the transition of fosA10 from veterinary to clinical settings, (ii) the ability of the fosA10 cassette to fit in both the IncFII and IncB/O/K/Z environments, and (iii) the ability of fosA10-carrying IncK plasmid to switch from minor to successful clones, such as ST648.
Moreover, we report the first case of a C. freundii ST673 isolate producing VIM and carrying a new FosA7 variant, named FosA7.9. The FosA7.9 gene was inserted in a well-conserved cassette, surrounded by two copies of the HNH endonuclease gene. The HNH endonuclease is a group of homing endonucleases that can act as selfish genetic elements, like transposons, breaking DNA double strands and allowing the acquisition of functional attributes to the host cell, such as antimicrobial resistance (AMR) genes (58, 59). Moreover, the genomic environment of the fosA7 cassette, flanked by HNH genes, was shared with two clinical C. freundii ST396 isolates collected in China (38). These results and the lack of any insertion elements surrounding fosA7.9 suggest a possible role for HNH endonucleases in slowly spreading new AMR traits in low-risk and silent hosts, such as C. freundii (35).
Conclusions.
These results show the emergence of FOSr among CRE from clinical settings in the Czech Republic. The 3-year study revealed a decrease in FOS susceptibility among carbapenemases-producing E. coli and Citrobacter strains. In our investigation, the decrease of FOSs could be largely attributable to impairment in GlpT and CyaA activity, significantly reducing the permeability to FOS. To our knowledge, we report the first isolation of FosA10-producing E. coli ST648 and the emergence of a new FosA7 allele, FosA7.9, in C. freundii in the Czech Republic. The emergence and the spread of both chromosome- and plasmid-mediated FOSr mechanisms in CRE could compromise the usefulness of FOS against severe and complex infections. A better knowledge of the genetic mechanisms underlying FOSr may facilitate the creation of rapid DNA-based testing for FOSr.
MATERIALS AND METHODS
Identification of bacterial isolates, susceptibility determination, and detection of enzymes.
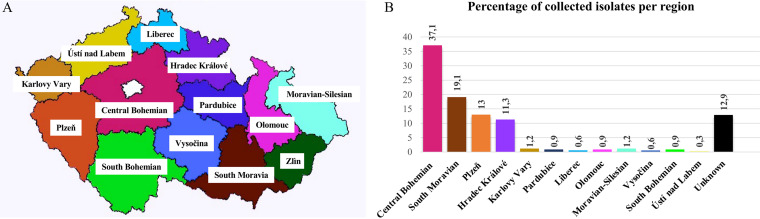
In the period from December 2018 to February 2022, 223 carbapenemase-producing E. coli isolates and 70 Citrobacter isolates (64 C. freundii, 3 Citrobacter amalonaticus, 2 Citrobacter braakii, and 1 Citrobacter youngae isolate) were collected from different health care settings in the Czech Republic and sent to University Hospital in Pilsen, Czech Republic, as part of a national surveillance effort for carbapenemase production in Enterobacterales (Fig. 9A and B). Of these, 51.1% (150/293) were collected from urine, 19% (56/293) from rectal swabs, 8% (23/293) from wounds, 8% (23/293) from sputum, 2% (6/293) from blood, and the remaining 11.9% (35/293) from different sources, including stool, pus, bronchoalveolar lavage fluid, decubitus swabs, throat swabs, and aspirates (Fig. S2).
FIG 9.
(A) Map of the Czech Republic. (B) Regional percentage of collected isolates (values expressed in percentage).
Identification of strains was confirmed by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) with MALDI Biotyper software (Bruker Daltonics, Bremen, Germany). The production of carbapenemases (metallo-β-lactamase, OXA-48, and KPC) was assessed with the double-disc synergy test with EDTA, the temocillin disc test, and the phenylboronic acid test (37). FOS MICs were evaluated using ADM and interpreted according to EUCAST clinical breakpoints (v 12.0) and guidelines. Production of FosA-like and FosC2 enzymes was detected by the PPF test (60).
Molecular investigations.
All the isolates were screened by PCR for the presence of blaKPC-like, blaNDM-like, blaVIM-like, and blaOXA-48-like genes, as reported elsewhere (36). Positive strains with PPF test were additionally screened by PCR for the presence of fosA, fosA2, fosA3, fosA4, fosA5, fosA6, fosA7, fosA10, and fosC2 (15, 16, 21, 61).
WGS and analysis.
A total of 15 strains (6 E. coli and 9 C. freundii strains) were selected as representative for genomic content and FOS pathway mutation analysis. The selection was based on FOS MICs: the selection included 11 strains (four E. coli and seven C. freundii strains) with FOS MICs of ≥64 μg/mL (resistant [R]) and four strains (two E. coli and two C. freundii strains) with FOS MICs of ≤16 μg/mL (S). The four FOS-susceptible strains were included to compare differences in FOS resistance (FOSr) and FOS susceptibility (FOSs) profiles. In detail, the selection for FOSs E. coli included FOS MICs corresponding to the FOS epidemiological cutoff (ECOFF) value and one dilution beyond the FOS ECOFF value according to EUCAST (https://mic.eucast.org/search/?search%5Bmethod%5D=mic&search%5Bantibiotic%5D=100&search%5Bspecies%5D=-1&search%5Bdisk_content%5D=-1&search%5Blimit%5D=50). For FOSs C. freundii isolates, FOS ECOFF values are not available; thus, low-level susceptibility profiles for FOS were selected (FOS MIC = 8 μg/mL and 16 μg/mL).
Genomic DNA was extracted using a NucleoSpin microbial DNA kit (Macherey-Nagel, Germany). WGS was performed on seven selected strains with the NovaSeq 6000 system with a 2 × 250 paired-end run following Nextera XT library preparation (Illumina Inc., San Diego, CA, USA). The remaining eight strains belonged to two different projects (Table 1), and WGS was previously performed with both the Illumina MiSeq platform (Illumina Inc., San Diego, CA, USA) and the Sequel I platform (Pacific Biosciences, Menlo Park, CA, USA) (36). Reads were assembled using SPAdes software (62). Assembled sequences were annotated using the RAST (Rapid Annotation using Subsystems Technology) server (63). The resistome, plasmid replicons, mobile elements, multilocus sequence types (MLST), and plasmid MLST (pMLST) were determined by uploading the assembled sequences to ResFinder 4.1 and CARD (64, 65), PlasmidFinder (66), ISfinder (67), MLST 2.0 (68), and pMLST 2.0 (66), respectively. Comparative genome alignment was done using Mauve v.2.4.0 and SnapGene (SnapGene Software). A linear map of chromosomal environments was created by using EasyFig (69) and the graphic editor Procreate (Savage Interactive, Tasmania, Australia).
Phylogenetic analysis.
Phylogenetic relationships between the selected sequenced isolates and global genomes were investigated. Phylogenetic trees were obtained using core genome, recombination, and SNPs by using parsnp v1.2, available in the harvest suite (70), and using a corresponding reference genome. Graphic illustration of the trees was build with the Interactive Tree Of Life (iTOL) (https://itol.embl.de/) (71). For the construction of the SNP-based phylogenies, 160 Escherichia coli genomes and 112 Citrobacter freundii genomes were downloaded from the NCBI assembly database, including complete and draft genomes. E. coli ECO49406 and C. freundii RHBSTW-00135 were use as respective references. The evolutionary analysis of FosA-like proteins in Enterobacterales was conducted by MEGA 11 (72), using the maximum-likelihood method and the Jones-Taylor-Thornton (JTT) matrix-based model (73).
Conjugation/transformation assay.
The conjugal transfer of fosA genes was tested in liquid medium using the E. coli A15 strain (Azdr) as a recipient. Transconjugants were selected on MacConkey agar (Scharlab, SL, Barcelona, Spain) plates containing sodium azide (100 mg/L) (Sigma-Aldrich, St. Louis, MO, USA), FOS (64 mg/L) (Sigma-Aldrich), and G6P (25 mg/L) (Roche). The presence of fosA-like genes and the plasmid content in transconjugants were further confirmed by PCR and PCR replicon typing (PBRT 2.0 kit), respectively (74). Since fosA7 was not transferable by conjugation, transformation was carried out with CFR50714; plasmid extraction was performed using a Qiagen maxi kit (Qiagen, Hilden, Germany), and competent E. coli Top10 cells were used as the recipient. Transformants were selected on Mueller-Hinton (MH) agar (Oxoid, Hampshire, UK) with 32 mg/L FOS (Sigma-Aldrich) and 25 mg/L G6P (Roche) (75).
Protein mutations.
The effects of amino acid alterations on the biological function were predicted using the online PROVEAN (Protein Variation Effect Analyzer) platform (http://provean.jcvi.org/index.php) (76). PROVEAN predicts protein sequence variations, including single or multiple amino acid substitutions, insertions, or deletions. The platform produces a delta alignment score based on the reference and variant versions of a protein query sequence with respect to sequence homologs collected from the NCBI protein database through BLAST. For each substitution, the tool provides a score (P score) categorized in three classes: (i) if the P score is equal to or below the cutoff of −2.5, the protein alteration is categorized as deleterious (potential loss of protein structure or function); (ii) if the P score is above the threshold, the alteration is marked as neutral (no alteration in the structure or function of the protein) (9). Amino acid variations in MurA, GlpT, UhpT, UhpA, UhpB, UhpC, CyaA, PtsI, PtsH (phosphocarrier protein HPr), GlpR, CRP, CRR (enzyme IIA [Glc]), PtsG (phosphotransferase system [PTS] glucose-specific EIICB component), FNR (fumarate and nitrate reduction regulatory protein), and GlpQ (glycerophosphodiester phosphodiesterase) were investigated (accession numbers are reported in Table S1).
Data availability.
The nucleotide sequence of fosA7.9 has been uploaded to GenBank under the accession number ON245013. GenBank accession numbers of the sequenced strains are presented in Table 1.
ACKNOWLEDGMENTS
The study was supported by research project grants NU20J-05-00033 and NU23J-09-00067 provided by the Czech Health Research Council and by the project National Institute of Virology and Bacteriology (Program EXCELES, ID project no. LX22NPO5103), funded by the European Union–Next Generation EU.
Contributor Information
V. Mattioni Marchetti, Email: vittoria.mattionimarche01@universitadipavia.it.
Rafael Vignoli, Instituto de Higiene.
REFERENCES
- 1.Dijkmans AC, Zacarías NVO, Burggraaf J, Mouton JW, Wilms EB, van Nieuwkoop C, Touw DJ, Stevens J, Kamerling IMC. 2017. Fosfomycin: pharmacological, clinical and future perspectives. Antibiotics 6:24. doi: 10.3390/antibiotics6040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castañeda-García A, Blázquez J, Rodríguez-Rojas A. 2013. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics (Basel) 2:217–236. doi: 10.3390/antibiotics2020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takahata S, Ida T, Hiraishi T, Sakakibara S, Maebashi K, Terada S, Muratani T, Matsumoto T, Nakahama C, Tomono K. 2010. Molecular mechanisms of fosfomycin resistance in clinical isolates of Escherichia coli. Int J Antimicrob Agents 35:333–337. doi: 10.1016/j.ijantimicag.2009.11.011. [DOI] [PubMed] [Google Scholar]
- 4.Zeng J, Hong Y, Zhao N, Liu Q, Zhu W, Xiao L, Wang W, Chen M, Hong S, Wu L, Xue Y, Wang D, Niu J, Drlica K, Zhao X. 2022. A broadly applicable, stress-mediated bacterial death pathway regulated by the phosphotransferase system (PTS) and the cAMP-Crp cascade. Proc Natl Acad Sci USA 119:e2118566119. doi: 10.1073/pnas.2118566119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimata K, Takahashi H, Inada T, Postma P, Aiba H. 1997. cAMP receptor protein-cAMP plays a crucial role in glucose-lactose diauxie by activating the major glucose transporter gene in Escherichia coli. Proc Natl Acad Sci USA 94:12914–12919. doi: 10.1073/pnas.94.24.12914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurabayashi K, Tanimoto K, Tomita H, Hirakawa H. 2017. Cooperative actions of CRP-cAMP and FNR increase the fosfomycin susceptibility of enterohaemorrhagic Escherichia coli (EHEC) by elevating the expression of glpT and uhpT under anaerobic conditions. Front Microbiol 8:426. doi: 10.3389/fmicb.2017.00426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seeto S, Notley-McRobb L, Ferenci T. 2004. The multifactorial influences of RpoS, Mlc and cAMP on ptsG expression under glucose-limited and anaerobic conditions. Res Microbiol 155:211–215. doi: 10.1016/j.resmic.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 8.Ohshima N, Yamashita S, Takahashi N, Kuroishi C, Shiro Y, Takio K. 2008. Escherichia coli cytosolic glycerophosphodiester phosphodiesterase (UgpQ) requires Mg2+, Co2+, or Mn2+ for its enzyme activity. J Bacteriol 190:1219–1223. doi: 10.1128/JB.01223-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sorlozano-Puerto A, Lopez-Machado I, Albertuz-Crespo M, Martinez-Gonzalez LJ, Gutierrez-Fernandez J. 2020. Characterization of fosfomycin and nitrofurantoin resistance mechanisms in Escherichia coli isolated in clinical urine samples. Antibiotics 9:534. doi: 10.3390/antibiotics9090534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadner RJ. 1973. Genetic control of the transport of hexose phosphates in Escherichia coli: mapping of the uhp locus. J Bacteriol 116:764–770. doi: 10.1128/jb.116.2.764-770.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsuruoka T, Yamada Y. 1975. Charactertization of spontaneous fosfomycin (phosphonomycin)-resistant cells of Escherichia coli B in vitro. J Antibiot (Tokyo) 28:906–911. doi: 10.7164/antibiotics.28.906. [DOI] [PubMed] [Google Scholar]
- 12.Zurfluh K, Treier A, Schmitt K, Stephan R. 2020. Mobile fosfomycin resistance genes in Enterobacteriaceae—an increasing threat. Microbiologyopen 9:e1135. doi: 10.1002/mbo3.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou M, Ma PP, Liu WS, Liang X, Li XY, Li YZ, Liu BT. 2021. Prevalence and antibiotic resistance characteristics of extraintestinal pathogenic Escherichia coli among healthy chickens from farms and live poultry markets in China. Animals 11:1112. doi: 10.3390/ani11041112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang L, Cao M, Hu Y, Zhang R, Xiao Y, Chen G. 2021. Prevalence and mechanisms of fosfomycin resistance among KPC-producing Klebsiella pneumoniae clinical isolates in China. Int J Antimicrob Agents 57:106226. doi: 10.1016/j.ijantimicag.2020.106226. [DOI] [PubMed] [Google Scholar]
- 15.Loras C, González-Prieto A, Pérez-Vázquez M, Bautista V, Ávila A, Campoy PS, Oteo-Iglesias J, Alós JI. 2021. Prevalence, detection and characterisation of fosfomycin-resistant Escherichia coli strains carrying fosA genes in Community of Madrid, Spain. J Glob Antimicrob Resist 25:137–141. doi: 10.1016/j.jgar.2021.02.032. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Lin Q, Zhou Q, Lv L, Wan M, Gao X, Wang C, Liu JH. 2020. Identification of fosA10, a novel plasmid-mediated fosfomycin resistance gene of Klebsiella pneumoniae origin, in Escherichia coli. Infect Drug Resist 13:1273–1279. doi: 10.2147/IDR.S251360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singkham-In U, Muhummudaree N, Chatsuwan T. 2020. fosA3 overexpression with transporter mutations mediates high-level of fosfomycin resistance and silence of fosA3 in fosfomycin-susceptible Klebsiella pneumoniae producing carbapenemase clinical isolates. PLoS One 15:e0237474. doi: 10.1371/journal.pone.0237474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang LJ, Gu XX, Zhang J, Yang L, Lu YW, Fang LX, Jiang HX. 2020. Characterization of a fosA3 carrying IncC-IncN plasmid from a multidrug-resistant ST17 Salmonella Indiana isolate. Front Microbiol 11:1582. doi: 10.3389/fmicb.2020.01582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Min C, Li J, Yu T, Hu Y, Dou Q, Zou M. 2021. Characterization of fosfomycin resistance and molecular epidemiology among carbapenem-resistant Klebsiella pneumoniae strains from two tertiary hospitals in China. BMC Microbiol 21:109. doi: 10.1186/s12866-021-02165-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rehman MA, Yin X, Persaud-Lachhman MG, Diarra MS. 2017. First detection of a fosfomycin resistance gene, fosA7, in Salmonella enterica serovar Heidelberg isolated from broiler chickens. Antimicrob Agents Chemother 61:e00410-17. doi: 10.1128/AAC.00410-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Wang Y, Wang ZY, Wu H, Mei CY, Shen PC, Pan ZM, Jiao X. 2021. Chromosomally located fosA7 in Salmonella isolates from China. Front Microbiol 12:781306. doi: 10.3389/fmicb.2021.781306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun Y, Chen W, Wang S, Cao X. 2021. Co-occurrence of fosA5, blaSHV-145 and blaOXA-48 among a Klebsiella pneumoniae high-risk ST16 from a tertiary hospital in China: focusing on the phylogeny of OXA-48 genes from global Klebsiella pneumoniae isolates. Braz J Microbiol 52:2559–2563. doi: 10.1007/s42770-021-00572-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gou JJ, Liu N, Guo LH, Xu H, Lv T, Yu X, Chen YB, Guo XB, Rao YT, Zheng BW. 2020. Carbapenem-resistant Enterobacter hormaechei ST1103 with IMP-26 carbapenemase and ESBL gene blaSHV-178. Infect Drug Resist 13:597–605. doi: 10.2147/IDR.S232514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sajeev S, Hamza M, Sivaraman GK, Ghatak S, Ojha R, Mendem SK, Murugesan D, Raisen C, Shome BR, Holmes MA. 2022. Genomic insights of beta-lactamase producing Klebsiella quasipneumoniae subsp. similipneumoniae belonging to sequence type 1699 from retail market fish, India. Arch Microbiol 204:454. doi: 10.1007/s00203-022-03071-w. [DOI] [PubMed] [Google Scholar]
- 25.Biggel M, Zurfluh K, Treier A, Nüesch-Inderbinen M, Stephan R. 2021. Characteristics of fosA-carrying plasmids in E. coli and Klebsiella spp. isolates originating from food and environmental samples. J Antimicrob Chemother 76:2004–2011. doi: 10.1093/jac/dkab119. [DOI] [PubMed] [Google Scholar]
- 26.Milner KA, Bay DC, Alexander D, Walkty A, Karlowsky JA, Mulvey MR, Sharma MK, Zhanel GG. 2020. Identification and characterization of a novel FosA7 member from fosfomycin-resistant Escherichia coli clinical isolates from Canadian hospitals. Antimicrob Agents Chemother 65:e00865-20. doi: 10.1128/AAC.00865-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ten Doesschate T, Abbott IJ, Willems RJL, Top J, Rogers MRC, Bonten MM, Paganelli FL. 2019. In vivo acquisition of fosfomycin resistance in Escherichia coli by fosA transmission from commensal flora. J Antimicrob Chemother 74:3630–3632. doi: 10.1093/jac/dkz380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hao Y, Zhao X, Zhang C, Bai Y, Song Z, Lu X, Chen R, Zhu Y, Wang Y. 2021. Clonal dissemination of clinical carbapenem-resistant Klebsiella pneumoniae isolates carrying fosA3 and blaKPC-2 coharboring plasmids in Shandong, China. Front Microbiol 12:771170. doi: 10.3389/fmicb.2021.771170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Ai W, Cao Y, Guo Y, Wu X, Wang B, Rao L, Xu Y, Zhao H, Wang X, Yu F. 2021. The co-occurrence of NDM-5, MCR-1, and FosA3-encoding plasmids contributed to the generation of extensively drug-resistant Klebsiella pneumoniae. Front Microbiol 12:811263. doi: 10.3389/fmicb.2021.811263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xiang D-R, Li J-J, Sheng Z-K, Yu H-Y, Deng M, Bi S, Hu F-S, Chen W, Xue X-W, Zhou Z-B, Doi Y, Sheng J-F, Li L-J. 2016. Complete sequence of a novel IncR-F33:A-:B- plasmid, pKP1034, harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence type 11 strain in China. Antimicrob Agents Chemother 60:1343–1348. doi: 10.1128/AAC.01488-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Q, Zhang P, Zhao D, Jiang Y, Zhao F, Wang Y, Li X, Du X, Yu Y. 2018. Emergence of tigecycline resistance in Escherichia coli co-producing MCR-1 and NDM-5 during tigecycline salvage treatment. Infect Drug Resist 11:2241–2248. doi: 10.2147/IDR.S179618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peng Z, Li X, Hu Z, Li Z, Lv Y, Lei M, Wu B, Chen H, Wang X. 2019. Characteristics of carbapenem-resistant and colistin-resistant Escherichia coli co-producing NDM-1 and MCR-1 from pig farms in China. Microorganisms 7:482. doi: 10.3390/microorganisms7110482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian X, Zheng X, Sun Y, Fang R, Zhang S, Zhang X, Lin J, Cao J, Zhou T. 2020. Molecular mechanisms and epidemiology of carbapenem-resistant Escherichia coli isolated from Chinese patients during 2002–2017. Infect Drug Resist 13:501–512. doi: 10.2147/IDR.S232010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feng J, Qiu Y, Yin Z, Chen W, Yang H, Yang W, Wang J, Gao Y, Zhou D. 2015. Coexistence of a novel KPC-2-encoding MDR plasmid and an NDM-1-encoding pNDM-HN380-like plasmid in a clinical isolate of Citrobacter freundii. J Antimicrob Chemother 70:2987–2991. doi: 10.1093/jac/dkv232. [DOI] [PubMed] [Google Scholar]
- 35.Bitar I, Caltagirone M, Villa L, Mattioni Marchetti V, Nucleo E, Sarti M, Migliavacca R, Carattoli A. 2019. Interplay among IncA and blaKPC-carrying plasmids in Citrobacter freundii. Antimicrob Agents Chemother 63:e02609-18. doi: 10.1128/AAC.02609-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kraftova L, Finianos M, Studentova V, Chudejova K, Jakubu V, Zemlickova H, Papagiannitsis CC, Bitar I, Hrabak J. 2021. Evidence of an epidemic spread of KPC-producing Enterobacterales in Czech hospitals. Sci Rep 11:15732. doi: 10.1038/s41598-021-95285-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chudejova K, Kraftova L, Mattioni Marchetti V, Hrabak J, Papagiannitsis CC, Bitar I. 2021. Genetic plurality of OXA/NDM-encoding features characterized from Enterobacterales recovered from Czech hospitals. Front Microbiol 12:641415. doi: 10.3389/fmicb.2021.641415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Lin Y, Lu L, Wang K, Yang L, Li P, Li J, Jia L, Li P, Song H. 2020. Genetic characterisation of a complex class 1 integron in an NDM-1-producing Citrobacter freundii ST396 clinical strain isolated from a urine sample. J Glob Antimicrob Resist 23:64–66. doi: 10.1016/j.jgar.2020.08.002. [DOI] [PubMed] [Google Scholar]
- 39.AbuOun M, Jones H, Stubberfield E, Gilson D, Shaw LP, Hubbard ATM, Chau KK, Sebra R, Peto TEA, Crook DW, Read DS, Gweon HS, Walker AS, Stoesser N, Smith RP, Anjum MF. The Rehab Consortium. 2021. A genomic epidemiological study shows that prevalence of antimicrobial resistance in Enterobacterales is associated with the livestock host, as well as antimicrobial usage. Microb Genom 7:e000630. doi: 10.1099/mgen.0.000630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Korotetskiy IS, Jumagaziyeva AB, Shilov SV, Kuznetsova TV, Iskakbayeva ZА, Myrzabayeva AN, Korotetskaya N, Ilin AI, Reva ON. 2020. Phenotypic and genotypic characterisation of clinical isolates of nosocomial infections. Eurasian J Appl Biotechnol doi: 10.11134/btp.1.2020.5. [DOI] [Google Scholar]
- 41.Kahan FM, Kahan JS, Cassidy PJ, Kropp H. 1974. The mechanism of action of fosfomycin (phosphonomycin). Ann N Y Acad Sci 235:364–386. doi: 10.1111/j.1749-6632.1974.tb43277.x. [DOI] [PubMed] [Google Scholar]
- 42.Hardisson C, Llaneza J. 1977. The action of fosfomycin on the growth of Pseudomonas aeruginosa. Chemotherapy 23:37–44. doi: 10.1159/000222024. [DOI] [PubMed] [Google Scholar]
- 43.Lindgren V. 1978. Mapping of a genetic locus that affects glycerol 3-phosphate transport in Bacillus subtilis. J Bacteriol 133:667–670. doi: 10.1128/jb.133.2.667-670.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nilsson AI, Berg OG, Aspevall O, Kahlmeter G, Andersson DI. 2003. Biological costs and mechanisms of fosfomycin resistance in Escherichia coli. Antimicrob Agents Chemother 47:2850–2858. doi: 10.1128/AAC.47.9.2850-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Whitmer GR, Moorthy G, Arshad M. 2019. The pandemic Escherichia coli sequence type 131 strain is acquired even in the absence of antibiotic exposure. PLoS Pathog 15:e1008162. doi: 10.1371/journal.ppat.1008162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Doi Y, Park YS, Rivera JI, Adams-Haduch JM, Hingwe A, Sordillo EM, Lewis JS, Howard WJ, Johnson LE, Polsky B, Jorgensen JH, Richter SS, Shutt KA, Paterson DL. 2013. Community-associated extended-spectrum β-lactamase-producing Escherichia coli infection in the United States. Clin Infect Dis 56:641–648. doi: 10.1093/cid/cis942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piazza A, Principe L, Comandatore F, Perini M, Meroni E, Mattioni Marchetti V, Migliavacca R, Luzzaro F. 2021. Whole-genome sequencing investigation of a large nosocomial outbreak caused by ST131 H30Rx KPC-producing Escherichia coli in Italy. Antibiotics 10:718. doi: 10.3390/antibiotics10060718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peirano G, Chen L, Nobrega D, Finn TJ, Kreiswirth BN, DeVinney R, Pitout JDD. 2022. Genomic epidemiology of global carbapenemase-producing Escherichia coli, 2015–2017. Emerg Infect Dis 28:924–931. doi: 10.3201/eid2805.212535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Samuelsen Ø, Overballe-Petersen S, Bjørnholt JV, Brisse S, Doumith M, Woodford N, Hopkins KL, Aasnæs B, Haldorsen B, Sundsfjord A. Norwegian Study Group on CPE. 2017. Molecular and epidemiological characterization of carbapenemase-producing Enterobacteriaceae in Norway, 2007 to 2014. PLoS One 12:e0187832. doi: 10.1371/journal.pone.0187832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schweizer C, Bischoff P, Bender J, Kola A, Gastmeier P, Hummel M, Klefisch FR, Schoenrath F, Frühauf A, Pfeifer Y. 2019. Plasmid-mediated transmission of KPC-2 carbapenemase in Enterobacteriaceae in critically ill patients. Front Microbiol 10:276. doi: 10.3389/fmicb.2019.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fernandes MR, Sellera FP, Moura Q, Gaspar VC, Cerdeira L, Lincopan N. 2018. International high-risk clonal lineages of CTX-M-producing Escherichia coli F-ST648 in free-roaming cats, South America. Infect Genet Evol 66:48–51. doi: 10.1016/j.meegid.2018.09.009. [DOI] [PubMed] [Google Scholar]
- 52.Sellera FP, Fernandes MR, Moura Q, Souza TA, Cerdeira L, Lincopan N. 2017. Draft genome sequence of Enterobacter cloacae ST520 harbouring blaKPC-2, blaCTX-M-15 and blaOXA-17 isolated from coastal waters of the South Atlantic Ocean. J Glob Antimicrob Resist 10:279–280. doi: 10.1016/j.jgar.2017.07.017. [DOI] [PubMed] [Google Scholar]
- 53.Solgi H, Badmasti F, Aminzadeh Z, Giske CG, Pourahmad M, Vaziri F, Havaei SA, Shahcheraghi F. 2017. Molecular characterization of intestinal carriage of carbapenem-resistant Enterobacteriaceae among inpatients at two Iranian university hospitals: first report of co-production of bla NDM-7 and bla OXA-48. Eur J Clin Microbiol Infect Dis 36:2127–2135. doi: 10.1007/s10096-017-3035-3. [DOI] [PubMed] [Google Scholar]
- 54.Qamar MU, Walsh TR, Toleman MA, Saleem S, Jahan S. 2018. First identification of clinical isolate of a novel “NDM-4” producing Escherichia coli ST405 from urine sample in Pakistan. Braz J Microbiol 49:949–950. doi: 10.1016/j.bjm.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harada S, Suzuki M, Sasaki T, Sakurai A, Inaba M, Takuya H, Wakuda M, Doi Y. 2021. Transmission of NDM-5-producing and OXA-48-producing Escherichia coli sequence type 648 by international visitors without previous medical exposure. Microbiol Spectr 9:e01827-21. doi: 10.1128/spectrum.01827-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang RS, Feng Y, Lv XY, Duan JH, Chen J, Fang LX, Xia J, Liao XP, Sun J, Liu YH. 2016. Emergence of NDM-5- and MCR-1-producing Escherichia coli clones ST648 and ST156 from a single Muscovy duck (Cairina moschata). Antimicrob Agents Chemother 60:6899–6902. doi: 10.1128/AAC.01365-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Queenan AM, Bush K. 2007. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Edgell DR. 2009. Selfish DNA: homing endonucleases find a home. Curr Biol 19:R115–117. doi: 10.1016/j.cub.2008.12.019. [DOI] [PubMed] [Google Scholar]
- 59.Li T, Chen H, Zhao J, Tao Z, Lan W, Zhao Y, Sun X. 2023. Characterization of phage vB_SalM_SPJ41 and the reduction of risk of antibiotic-resistant Salmonella enterica contamination in two ready-to-eat foods. Antibiotics (Basel) 12:364. doi: 10.3390/antibiotics12020364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakamura G, Wachino J, Sato N, Kimura K, Yamada K, Jin W, Shibayama K, Yagi T, Kawamura K, Arakawa Y. 2014. Practical agar-based disk potentiation test for detection of fosfomycin-nonsusceptible Escherichia coli clinical isolates producing glutathione S-transferases. J Clin Microbiol 52:3175–3179. doi: 10.1128/JCM.01094-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benzerara Y, Gallah S, Hommeril B, Genel N, Decré D, Rottman M, Arlet G. 2017. Emergence of plasmid-mediated fosfomycin-resistance genes among Escherichia coli isolates, France. Emerg Infect Dis 23:1564–1567. doi: 10.3201/eid2309.170560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nurk S, Bankevich A, Antipov D, Gurevich AA, Korobeynikov A, Lapidus A, Prjibelski AD, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, Clingenpeel SR, Woyke T, McLean JS, Lasken R, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J Comput Biol 20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen AV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran HK, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carattoli A, Zankari E, García-Fernández A, Voldby Larsen M, Lund O, Villa L, Møller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Pontén T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sullivan MJ, Petty NK, Beatson SA. 2011. Easyfig: a genome comparison visualizer. Bioinformatics 27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Treangen TJ, Ondov BD, Koren S, Phillippy AM. 2014. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Letunic I, Bork P. 2021. Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura K, Stecher G, Kumar S. 2021. MEGA11: Molecular Evolutionary Genetics Analysis version 11. Mol Biol Evol 38:3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 74.Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. 2005. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods 63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 75.Sambrook J, Russell DW. 2006. Preparation and transformation of competent E. coli using calcium chloride. Cold Spring Harb Protoc 2006:pdb.prot3932. doi: 10.1101/pdb.prot3932. [DOI] [PubMed] [Google Scholar]
- 76.Choi Y, Chan AP. 2015. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bitar I, Papagiannitsis CC, Kraftova L, Marchetti VM, Petinaki E, Finianos M, Chudejova K, Zemlickova H, Hrabak J. 2022. Implication of different replicons in the spread of the VIM-1-encoding integron, In110, in Enterobacterales from Czech hospitals. Front Microbiol 13:993240. doi: 10.3389/fmicb.2022.993240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequence of fosA7.9 has been uploaded to GenBank under the accession number ON245013. GenBank accession numbers of the sequenced strains are presented in Table 1.