Abstract
Spinal muscular atrophy (SMA), the leading genetic cause of infant mortality, arises from SMN protein insufficiency following SMN1 loss. Approved therapies circumvent endogenous SMN regulation and require repeated dosing or may wane. We describe genome editing of SMN2, an insufficient copy of SMN1 harboring a C6>T mutation, to permanently restore SMN protein levels and rescue SMA phenotypes. We used nucleases or base editors to modify five SMN2 regulatory regions. Base editing converted SMN2 T6>C, restoring SMN protein levels to wild-type. AAV9-mediated base editor delivery in Δ7SMA mice yielded 87% average T6>C conversion, improved motor function, and extended average lifespan, which was enhanced by one-time base editor+nusinersen co-administration (111 versus 17 days untreated). These findings demonstrate the potential of a one-time base editing treatment for SMA.
One-Sentence Summary:
Base editing enables a one-time treatment for spinal muscular atrophy (SMA) that rescues disease pathology and extends lifespan in mice.
SMA is a progressive motor neuron disease and the leading genetic cause of infant mortality(1-3). SMA is caused by homozygous loss or mutation of the essential survival motor neuron 1 (SMN1) gene. One or more copies of the nearly identical (>99.9% sequence identity) SMN2 partially compensates for the loss of SMN1(1, 4, 5). However, SMN1 and SMN2 differ by a silent C•G-to-T•A substitution at nucleotide position 6 of exon 7 (C6T) that results in exon 7 skipping in mRNA transcripts (Fig. 1A)(6, 7). The resulting truncated SMNΔ7 protein is rapidly degraded, causing SMN protein insufficiency that results in loss of motor neurons, paralysis, and death(8-10). Untreated patients with the most common form of SMA (type I) live a median of 6 months(11, 12).
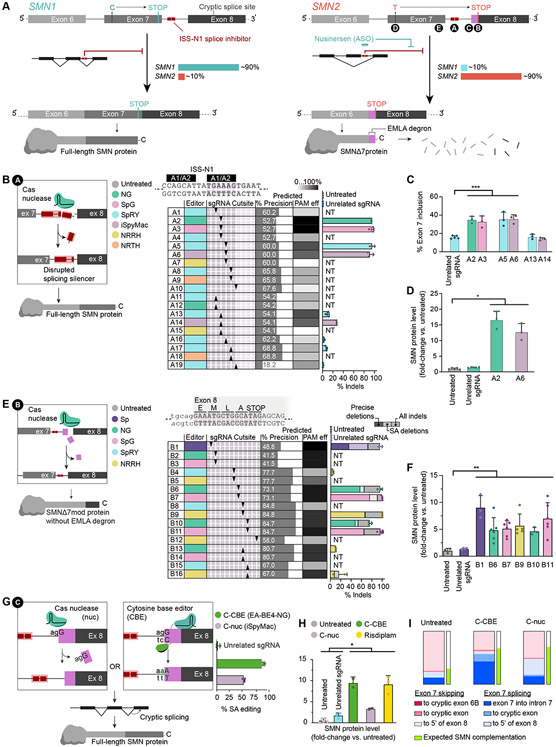
Fig. 1. Editing SMN2 regulatory regions.
(A) Genomic SMN exons 6 to 8, and SMN mRNA and protein products. (B) Nuclease editing strategy and genome editing outcomes of ISS-N1 targeting (strategy A). The table shows combinations of six nucleases, paired with ten sgRNAs complementary to the top (A1-10) or bottom strand (A11-19) identified by arrows that show the DSB site of the sgRNAs relative to the sequence above. (C) Exon 7 inclusion in SMN mRNA after editing, as indicated, measured by automated electrophoresis. (D) SMN protein levels after editing, as indicated, normalized to histone H3. (E) Nuclease editing strategy targeting and genome editing outcomes of targeting the first five codons of exon 8 (strategy B). The table shows combinations of five nucleases, paired with nine sgRNAs complementary to the top (B1-12) or bottom strand (B13-16) identified by arrows that indicate their DSB site, as above. (F) Total SMN protein levels after editing. (G) Nuclease and cytosine base editing strategies and genome editing outcomes of 3’-splice acceptor disruption at exon 8 (strategy C). (H) SMN protein levels following C-nuc and C-CBE editing or treatment with risdiplam, normalized to histone H3. i) Distribution of SMN2 transcript variants after C-nuc and C-CBE editing. Experiments are performed in Δ7SMA mESCs, NT=no treatment, *≤0.05, **≤0.01, ***≤0.005.
Upregulation of SMN protein can rescue motor function and substantially improve the prognosis of SMA patients(13-15). However, endogenous SMN protein is subject to multiple levels of regulation that differs across tissues(16-18). While SMN underexpression can fail to rescue SMN phenotypes, SMN overexpression can cause aggregation, toxicity, and tissue pathology(19-21). Three breakthrough therapeutics effectively rescue many SMA phenotypes and improve lifespan by upregulating SMN protein(22). The antisense oligonucleotide (ASO) nusinersen (Spinraza) and the small-molecule risdiplam (Evrysdi) both promote splicing inclusion of exon 7 resulting in ~2-fold upregulation of SMN levels, and have proven highly effective in the clinic(23, 24). However, SMN protein is reduced ~6.5-fold in the spinal cord of untreated SMA patients(25-27). The partial recovery of SMN protein promoted by these therapeutics may be insufficient at early timepoints and in damaged tissues, potentially underlying the limited rescue observed in some patients(28, 29). Moreover, the transient nature of these therapeutics necessitates repeated administration of costly drugs throughout patients’ lifetimes(30, 31).
AAV-mediated gene complementation of full-length SMN cDNA by onasemnogene abeparvovec-xioi (Zolgensma) leads to constitutive production of SMN in transduced cells that is not under endogenous control(32-34). In the spinal cord, Zolgensma upregulates SMN transcript levels by ~25%(35), while in other tissues such as the liver and dorsal root ganglia, gene complementation may cause SMN overexpression that under some circumstances can cause long-term toxicity(21). We do not yet know whether SMN overexpression induces toxicity in patients treated with Zolgensma, nor how long AAV-mediated expression will persist in motor neurons in patients (36, 37). As such, a therapeutic modality that restores endogenous gene expression and preserves native SMN regulation by a one-time permanent treatment may address remaining limitations of existing SMA therapies. Genome editing of SMN2, which is present in all SMA patients regardless of the nature of their SMN1 mutation, could enable a one-time treatment for SMA that restores native SMN transcript and protein levels while preserving their endogenous regulatory mechanisms.
Results
Predictable and precise nuclease editing of SMN2 ISS-N1 increases SMN protein levels
SMN protein production from SMN1 and SMN2 genes is constrained by transcriptional, transcriptomic, and post-translational regulatory sequences. We explored using Cas nucleases to create gain-of-function alleles in SMN2 regulatory sequences that upregulate SMN levels. The inclusion of exon 7, which underlies SMN protein stability, is strongly influenced by the downstream intronic splicing silencer ISS-N1 that harbors two heterogeneous nuclear ribonucleoprotein (hnRNPs) A1/A2 binding sites (Fig. 1A)(38). Deletions within, and downstream of the 3’ (3-prime) hnRNP A1/A2 binding domain improve exon 7 inclusion(38-41). We speculated that Cas9 nuclease-mediated disruption of the ISS-N1 genomic locus might increase exon 7 inclusion in SMN2 splicing and thereby increase SMN protein levels (strategy A, Fig. 1B).
We used inDelphi, a machine learning model of SpCas9 nuclease editing outcomes, to predict indel outcomes at the ISS-N1 locus that disrupt hnRNP A1/A2 binding and improve full-length SMN splicing of SMN2 (Fig. 1B)(42). InDelphi identified ten spacer sequences predicted to induce ≥4-nt deletions at ISS-N1 and loss of ≥1-nt of the 3’-hnRNP A1/A2 domain (‘predicted % precision’). We estimated editing efficiencies of these strategies based on the reported PAM compatibility of these spacer sequences with SpCas9-variant nucleases (‘predicted % PAM efficiency’)(43-46). From 19 possible nuclease editing strategies (A1-A19, defined as different combinations of genome editing agents and guide RNAs) we selected nine (A2, A3, A5, A6, A13, A14, A16, A17, A19) for experimental testing.
We co-transfected Δ7SMA mESCs—which lack endogenous Smn1, are homozygous for the full-length human SMN2 gene, carry human SMNΔ7-cDNA transgenes, and harbor a Mnx1:GFP reporter of motor neurons (SMN2+/+; SMNΔ7; Smn−/−; Mnx1:GFP)(47)—with nuclease expression plasmids that carry a blasticidin-resistance cassette and sgRNA plasmids that carry a hygromycin-resistance cassette. Both plasmids also contain Tol2 transposase sequences to enable stable transposon-mediated genomic integration and antibiotic selection. We achieved 92±5.6% average indel frequencies for the top four strategies targeting the ISS-N1 locus (A2, A3, A5, A6, Fig. 1B).
To assess whether nuclease-mediated editing of ISS-N1 improved exon 7 inclusion, we performed reverse-transcription PCR (RT-PCR) of SMN2 from exons 6 to 8, and quantified SMNΔ7 and full-length SMN products (Fig. 1C). We found that all strategies that edited ISS-N1 with high efficiency (≥85%) resulted in a significant increase in exon 7 inclusion averaging 2.2-fold relative to an unrelated sgRNA control (Welch’s two-tailed t-test p=0.01). The increase in exon 7 inclusion caused a substantial increase in SMN protein of 17-fold by A2 and 13-fold by A6 relative to untreated controls (values normalized to histone H3, Welch’s two-tailed t-test p=0.02, Fig. 1D, and fig. S1A). Collectively, these results demonstrate that disruption of the ISS-N1 genomic locus can stably increase full-length SMN splicing and protein phenotypes of SMA.
Predictable and precise genome editing of SMN2 exon 8 increases SMN protein levels
As an alternative nuclease-mediated approach to upregulate SMN levels, we disrupted post-translational regulatory sequences in SMN2 to increase SMNΔ7 protein stability. The critical difference between full-length SMN and the unstable SMNΔ7 protein is the substitution of 16 amino acids encoded by exon 7 with EMLA, a four-residue degron encoded by exon 8 (Fig. 1A)(8). Extending the coding sequence of exon 8 with five or more heterologous amino acids obscures SMNΔ7 C-terminal degradation signals. These modified SMNΔ7 (SMNΔ7mod) protein variants have increased stability and rescue survival and motor phenotypes of severe SMA mice(48). We designed strategies for Cas nuclease-mediated disruption of exon 8 to generate similar stabilized SMNΔ7mod proteins with therapeutic potential (strategy B1-B16, Supplementary Text, Fig. 1E), and observed up to 7.0-fold increase in SMN protein levels by B11 (Welch’s two-tailed t-test p=0.007, Fig. 1F, and fig. S1B).
Some exon 8 editing strategies improved SMN protein stability more than expected based on observed edited genotypes (Fig. 1, E and F). For example, precision edited genotypes were 1.9-fold higher in frequency following B9 editing than B1, yet SMNΔ7mod protein levels were greater in cells edited with B1 (9.1-fold) than B9 (5.7-fold). These data suggest that additional edited genotypes may improve SMN protein stability. Inspection of the non-precisely edited fraction of edited alleles revealed that B1 editing frequently induces indels at the exon 8 splice acceptor. Thus, we hypothesized that disrupting splicing of exon 8 improves SMN protein stability(49).
To test this hypothesis, we disrupted the canonical AG splice acceptor (SA) motif of exon 8 using either a nuclease or cytosine base editor (C-nuc or C-CBE) in Δ7SMA mESCs (Fig. 1G)(45, 50), and observed 54±2.3% indels from C-nuc and 89±2.3% cytosine base editing from C-CBE. Notably, C-nuc editing resulted in a complex mixture of indel genotypes at the intron-exon junction that resulted in deletion of additional nucleotides beyond the AG motif. Both strategies significantly increased SMN levels in Δ7SMA mESCs, similar to treatment with risdiplam (3.3-fold for C-nuc, 9.5-fold for C-CBE, 9.1-fold for risdiplam relative to untreated, Welch’s two-tailed t-test p<0.05, Fig. 1H, and fig. S1D to G), indicating that alternative splicing at exon 8 improves the stability of SMN2 gene products.
We investigated how exon 8 SA disruption affects SMN2 transcripts (Supplementary Text). C-CBE editing induced a minor increase in SMN2 mRNA that only partially explains the 9.5-fold increase in SMN levels (fig. S1H). We also observed a profound shift in SMN2 splice products (Fig. 1I). We investigated whether these alternative splice isoforms improve stability of SMN proteins, and found that transcripts including exon 7 were increased 2-fold by C-CBE (63±2.0%) and 1.6-fold by C-nuc (50±1.1%) relative to untreated cells (24±1.4%). These transcripts often retain intron 7 as in some functional transcript variants of SMN2 (ENST00000511812.5, fig. S1I). Importantly, all transcripts that include exon 7 encode full-length SMN protein and can therefore complement loss of SMN1, Thus, the substantial increase in SMN protein levels following exon 8 SA editing predominantly arises from an increase in normal full-length SMN.
Collectively, the tested SMN2 editing strategies permanently increase SMN protein levels up to 17-fold (strategy A2), 9.1-fold (strategy B1), and 9.5-fold (strategy C-CBE). As a 1.5- to 2-fold increase in SMN protein is therapeutic for SMA patients(23, 24), these strategies represent promising approaches for further studies.
Efficient and precise base editing of SMN2 splice regulatory elements
Several single-nucleotide substitutions in exon 7 strongly regulate splicing of SMN2, including the C-to-T transition at position 6 (C6T) that differentiates SMN1 (C) from SMN2 (T) genes (Fig. 1A), and T44C, G52A, and A54G at the 3’-end of exon 7(51). Using existing and newly developed BE-Hive predictive models of base-editing outcomes (Supplementary Text, fig. S2, A to E), we identified 42 strategies (combinations of base editors and guide RNAs) to modify exon 7 splicing regulatory elements (SREs, Fig. 2, A to C and fig. S2, F and G). We designed 13 spacers targeting C6T using ABE8e (strategy D1-19), or targeting C6T, T44C, G52A, and A54G using ABE8e, ABE7.10 and EA-BE4 deaminases (strategy E1-23). We paired these spacers with 12 compatible SpCas9-variants based on reported PAM preferences (‘predicted % PAM efficiency’)(43, 50, 52). We validated these strategies in Δ7SMA mESCs, and found that the BE-Hive models of SpCas9 base editors predicted edited outcomes of Cas-variant base editors with high accuracy (Cas9-NG(52), NRTH, NRRH, NRCH(44) Pearson’s r=0.810, chimeric SpyMac and iSpMac(45) Pearson’s r=0.910, Supplementary Text, Fig. 2D).
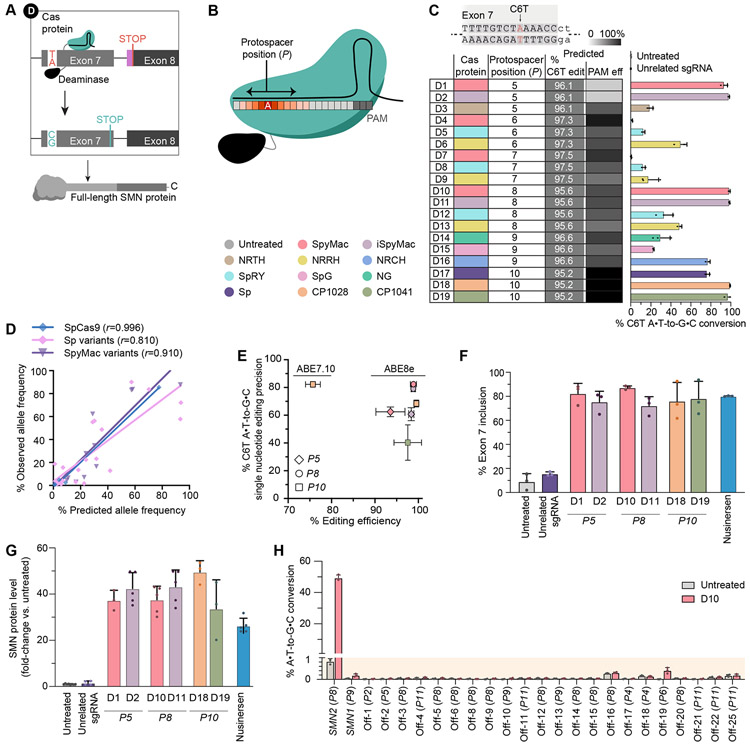
Fig. 2. Adenine base editing of SMN2 C6T.
(A) Adenine base editing of SMN2 C6T (strategy D). (B) Target nucleotide position within the protospacer (P#) for base editing. A typical base editor activity window is illustrated as a heat map. (C) The table shows ABE8e editing strategies with color-coded Cas-variant domains and their corresponding spacers. The protospacer position of the C6T target nucleotide (P#) is indicated. Graph shows genome editing outcomes in Δ7SMA mESCs. (D) Correlation of BE-Hive predicted editing outcomes with observed allele frequencies after base editing with ABE7.10 or ABE8e deaminases fused to different Cas variants. Pearson’s r is shown, 95% CI ranges are 0.9408–0.9998 for SpCas9, 0.5823–0.9201 for SpCas9 engineered and evolved variants, and 0.7557–0.9689 for SpyMac Cas variants. (E) Plot of base editing efficiency and single nucleotide editing precision of C6T by the indicated ABE and spacer combinations. (F) Exon 7 inclusion in SMN mRNA after editing by the indicated strategies, measured by automated electrophoresis. (G) SMN protein levels after editing by the indicated strategies, normalized to histone H3. (H) On-target and off-target base editing of strategy D10 in HEK293T cells. Bars show editing of the most frequently edited nucleotide at each locus, with the P# position shown in parenthesis.
Base editing of exon 7 SREs was highly efficient. At 3’-SREs, we achieved 69±5.0% T44C editing by E14, 92±4.0% G52A editing by E20, and 95±5.1% A54G editing by E23 (fig. S2, F and G). We achieved nearly complete (94%–99.5%) C6T A•T-to-G•C conversion by strategies targeting C6T at positions P5 (D1, D2), P8 (D10, D11), and P10 (D18, D19) within the protospacer (Fig. 2, A to C). The deaminase in ABE7.10 enabled up to 64±2.5% conversion of T6>C (E7, fig. S2G)(53, 54).
The frequency of edited alleles with single-nucleotide T6>C conversion alone (i.e., without any bystander edits or indels) varied substantially between the most efficient C6T editing strategies, ranging from 82±1.9% from D10 to 40±13% from D19 editing (Fig. 2E). Prior studies suggest that the coding sequence at the SMN C-terminus beyond exon 6 does not strongly affect SMN protein function and it is therefore unlikely that single-nucleotide editing precision of C6T is imperative for rescue of SMA(8, 48, 55). Maximizing the sequence similarity of modified SMN2 genes to native SMN1, however, may preserve additional regulatory interactions, including those not yet known. D10, the strategy with the highest precision and efficiency (99±0.7%), did not induce measurable indels and its induced bystander missense nucleotide substitutions (18±2.4%) have previously been shown to benefit inclusion of exon 7 by improved protein binding at the exonic splicing enhancer (fig. S2H)(56, 57). Together, these results establish efficient base editing strategies to convert SMN2 T6>C with high-fidelity and few undesired byproducts.
Base editing of SMN2 splice regulatory elements rescues SMN protein levels
Next, we asked whether base editing of exon 7 SREs results in functional rescue of cellular SMA phenotypes. The top six ABE8e editing strategies that converted C6T in >97% of alleles increased exon 7 inclusion to 78±10.2% on average, up to 9.7-fold higher than untreated cells (87±1.5% by D10 compared to 9.0±6.6% in untreated, Welch’s two-tailed t-test p<0.002, Fig. 2F). These results are on par with, or exceed, maximum exon 7 inclusion by risdiplam or nusinersen treatment of Δ7SMA mESCs (89±4.3% and 80±0.3%, respectively, Fig. 2F and fig. S1E), and resemble splicing ratios of SMN1 genes (82±7.3% in U2OS cells)(38, 39). Base editing of 3’-SREs in exon 7 also improved inclusion, averaging 60±3.2% following T44C editing by E14, 76±12% following G52A editing by E20, and 50±8.6% following A54G editing by E23 (fig. S2I). These data demonstrate that base editing of various exon 7 SREs can increase full-length SMN splice products.
Base editing of 3’-SREs increased SMN protein levels in ways that did not closely mirror observed improvements in exon 7 inclusion. We detected a 3.4-fold increase in SMN protein by E14 base editing of T44C, 23-fold increase by E20 editing of G52A, and 1.6-fold increase by E23 editing of A54G (Welch’s two-tailed t-test p=0.02), despite all three edits inducing comparable improvements in exon 7 inclusion (figs. S2I, and S3, A and B). We hypothesized that unintended bystander edits may underlie this persistent protein instability and found that the T44C and A54G editing strategies frequently ablate the nearby TAA stop codon in exon 7 (fig. S2, F and G). A failure to terminate translation in exon 7 leads to the extension of full-length SMN proteins with the EMLA degron encoded by exon 8 (Fig. 1A). Thus, imprecise editing of T44C or A54G by E14 or E23 results in the translation of unstable full-length SMN-EMLA fusions that prevent upregulation of SMN protein levels. Editing of G52A by E20 uses the EA-BE4 cytosine deaminase that does not recognize TAA as a substrate and therefore does not induce non-silent bystander changes in 99±0.1% of edited alleles, resulting in a 23-fold improvement in SMN protein levels.
Base editing of exon 7 C6T resulted in the greatest upregulation of SMN protein. The top six ABE8e editing strategies that correct C6T in >97% of alleles induced a 41-fold average increase in SMN protein levels compared to untreated controls (normalized to H3, Welch’s two-tailed t-test p<0.0002, Fig. 2G and fig. S3C), indicating complete rescue of normal SMN protein levels in Δ7SMA mESCs, which are ~40-fold reduced relative to wild-type mESCs(47). Despite inducing comparable increase in exon 7 inclusion, base editing of C6T enabled a 4.5-fold and 1.5-fold greater increase in SMN protein levels than risdiplam and nusinersen treatment of Δ7SMA mESCs (9-fold and 17-fold respectively, compared to 41-fold on average across the top six strategy D approaches, Fig. 1H, 2G, and figs. S1, D and F to G, and S3, C and E to F). Normal levels of SMN protein are essential to the function, survival, and long-term health of all species in the animal kingdom(58-61). Restoring wild-type levels of SMN protein as achieved by base editing strategy may thus best maximize the long-term health of SMA patients.
Among all genome editing strategies tested, base editing of C6T by D10 induces the greatest increase in exon 7 inclusion (87±1.5%) and best recapitulates native SMN protein levels (95% of wild-type levels, a 38-fold increased versus untreated Δ7SMA mESCs). D10 base editing is highly efficient (99±0.7%) with high on-target precision (82±0.0%). The SMN2 gene arose from a duplication of the chromosomal region containing SMN1, and shares an identical promoter and >99.9% sequence identity with SMN1, including 100% DNA conservation of its protein-coding sequence other than exon 7 C6T(1, 4, 5). We performed RT-qPCR and quantified SMN2 mRNA levels in edited cells, confirming that SMN2 mRNA abundance is not affected by D10 base editing compared to untreated Δ7SMA mESCs or following ABE8e transfection with an unrelated sgRNA (fig. S3G). Together, these data indicate that D10 editing of SMN2 faithfully reproduces the genomic sequence and function of native SMN1 alleles. Therefore, we selected strategy D10 for further study.
Off-target editing analysis of ABE8e targeting SMN2 C6T in the human genome
Some base editors can induce off-target deamination in cells, including Cas-dependent off-target DNA editing and Cas-independent off-target DNA or RNA editing(62-66). Genomic and transcriptomic off-target deamination by adenine base editors without involvement of the Cas protein component is rare, and deaminase variants that minimize these events have been reported(62, 67). We assessed the Cas-dependent genome specificity of the D10 strategy (ABE8e-SpyMac and P8 sgRNA) characterizing SpyMac Cas9 nuclease with P8 sgRNA using CIRCLE-seq(68), an unbiased and sensitive empirical in vitro off-target detection method. Potential off-target sites nominated by CIRCLE-seq can then be sequenced in-depth in base-edited human cells to provide a sensitive genome-wide analysis of off-target genome editing events induced by the D10 strategy(68, 69).
We generated purified D10 strategy ribonucleoprotein (RNP) complexes containing SpyMac nuclease and P8 sgRNA to treat human genomic DNA from HEK293T cells in vitro and analyzed rare off-target genomic cleavage events (fig. S3H). We identified 55 candidate SpyMac-dependent DNA off-target loci nominated by the CIRCLE-seq method. Next, we measured on-target and genomic off-target editing at the top 23 CIRCLE-seq-nominated loci in human cells (Supplementary Text, Fig. 2H and fig. S3I). We achieved 49±1.8% C6T on-target base editing at SMN2 in HEK293T cells and observed minimal base editing at SMN1 (0.15±0.07%), which is generally absent in SMA patients. We detected minor levels of D10 base editing at off-target site ranked 19 (0.41±0.14%), which is in an intergenic region of chromosome 15, and no evident base editing (≤0.03% over untreated cells) at the other 21 assayed potential off-target loci. These data indicate high genomic target specificity of the D10 base editing strategy for the on-target locus.
Together, these experiments did not detect any coding mutations or sequence changes of anticipated physiological significance in the human genome and support continued preclinical evaluation of the D10 strategy, including assessment of base editor off-target editing measured in various tissues that may accumulate over an extended period of time. We refer to the D10 editing strategy as the ‘ABE strategy’ hereafter.
Viral delivery of ABE enables efficient in vivo conversion of SMN2 C6T
To enable in vivo SMN2 C6T conversion in an animal model of SMA, we designed an adeno-associated virus (AAV) strategy to package ABE8e-SpyMac and the P8 sgRNA for delivery (v6 AAV-ABE8e, Supplementary Text, Fig. 3A and fig. S3J). The AAV serotype 9 (AAV9) has a well-established tropism for neurons in the CNS of a wide range of organisms, including Δ7SMA mice and human patients(70-72). In the cortex, AAV9 has been shown to almost exclusively target neurons(72), and intracerebroventricular (ICV) or systemic injection in neonates results in efficient transduction of spinal motor neurons to enable rescue of SMA disease phenotypes and lethality in both mice and humans(13, 32, 73). Thus, we selected AAV9 for delivery of our D10 ABE strategy (‘AAV9-ABE’) to Δ7SMA neonates by ICV injection to correct the SMN2 C6T target in vivo (Fig. 3B).
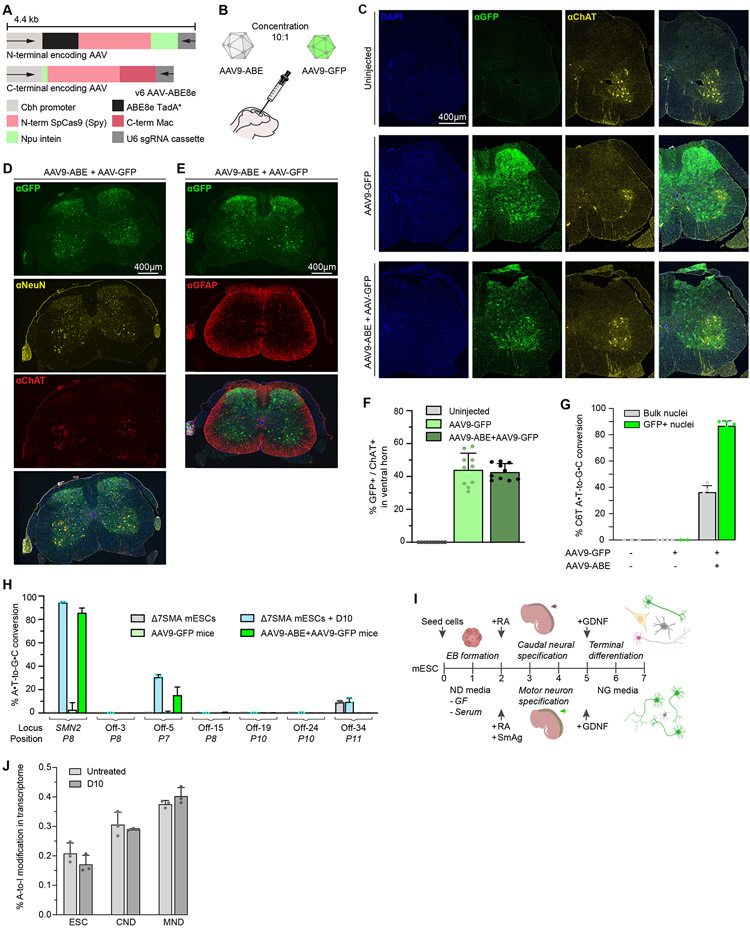
Fig. 3. Adenine base editing in Δ7SMA mice.
(A) Dual-AAV vectors encoding split-intein ABE8e-SpyMac and P8 sgRNA cassettes, v6 AAV9-ABE8e. (B) Neonatal ICV injections in Δ7SMA mice with AAV9-ABE, and AAV9-GFP as a transduction control. (C-E) Immunofluorescence images of lumbar spinal cord sections from wild-type Δ7SMA mice at 25 weeks old, ICV injected on PND0-1 with AAV9-ABE, AAV9-GFP, or uninjected as indicated. GFP indicates transduction, ChAT labels spinal motor neurons in the ventral horn, NeuN labels post-mitotic neurons, GFAP labels astrocytes, DAPI stains all nuclei. (F) Quantification of GFP and ChAT double-positive cells within the ventral horn (n=3). (G) Base editing in bulk and GFP+ flow-sorted nuclei of Δ7SMA mice treated with AAV9-ABE+AAV9-GFP (n=5), AAV9-GFP (n=4), or uninjected (n=3). (H) On-target and off-target editing following VIVO analysis of strategy D10 in Δ7SMA mESCs compared to AAV9-ABE+AAV9-GFP treatment in Δ7SMA mice. Bars show editing of the most frequently edited nucleotide at each locus, with the P# position shown in parenthesis. (I) Schematic of motor neuron differentiation (MND) and caudal-neural differentiation (CND) of Δ7SMA mESCs. (J) Whole transcriptome A-to-I RNA off-target editing analysis in Δ7SMAmESCs (n=3), and CND (n=3) and MND (n=3) differentiated cells stably expressing the D10 strategy.
We ICV injected SMA neonates with total 2.7x1013 vg/kg of the dual AAV9-ABE vectors, along with 2.7x1012 vg/kg AAV9-Cbh-eGFP-KASH (Klarsicht/ANC-1/Syne-1 homology domain, hereafter AAV9-GFP)(74) to serve as a viral transduction control. This dose is comparable to doses used for P0 ICV AAV administration of Zolgensma for rescue of Δ7SMA mice, and of other base editor AAVs that enable efficient genome editing in mice(32, 74). We observed typical transduction patterns of AAV9 in the spinal cord (Fig. 3, C to E, Supplementary Text, fig. S4A)(32, 33, 75). We quantified GFP and choline acetyl-transferase (ChAT) double-positive cells in the ventral horn of spinal cords from injected mice and observed a mean transduction efficiency of 43% in spinal motor neurons (Fig. 3F), consistent with transduction efficiencies >20% previously shown to enable significant phenotypic rescue of Δ7SMA mice following ICV injection of self-complementary AAV9-SMN (Zolgensma)(32). Transduction of spinal motor neurons using 2.97x1013 vg/kg AAV9-GFP alone was similar (median 46%) to transduction efficiencies using the ten-fold lower concentration of 2.7x1012 vg/kg, suggesting that the low-dose co-transduction of AAV9-GFP accurately represents the subset of cells transduced by AAV9-ABE.
Next, we assessed base editing in transduced cells (Supplementary Text, fig. S4B). We isolated cortical nuclei of treated animals and enriched for AAV9-transduction by sorting GFP-positive cells as previously described(74, 76). We observed 87±3.5% conversion of SMN2 C6T among transduced cells (Fig. 3G), a 2.4-fold enrichment over unsorted tissue (37%±4.7%), with high single-nucleotide precision for C6T alone (73±.2.7%) and few indels (<0.4±.1%) or bystander edits, similar to D10 editing in Δ7SMA mESCs (Fig. 2E, fig. S2H, S4C). Collectively, these data confirm that ICV injection of AAV9-ABE in Δ7SMA neonates enables efficient and precise conversion of SMN2 C6T in the CNS of treated animals with minimal undesired byproducts(56, 57, 77).
Base editing conversion of C6T effectively converts the native SMN2 gene to SMN1, thereby restoring SMN protein levels to that of wild-type cells. Current SMA drugs induce non-native SMN levels(23, 24, 32-35), and require repeated dosing or may fade over time. The permanent and precise editing of endogenous SMN2 genes that preserves native transcript levels and native regulatory mechanisms governing SMN expression thus may address shortcomings of existing SMA therapies(1, 4, 5, 21, 28, 78).
In vitro and in vivo DNA and RNA off-target analysis of ABE8e targeting SMN2 C6T
In addition to the off-target analysis in human cells described above, we also assessed the DNA and RNA specificity of the ABE strategy in mouse cells in vitro and in vivo. We performed CIRCLE-seq and validated the top 35 nominated sites in Δ7SMA mESCs (Supplementary Text, fig. S4D). We achieved 95±0.0% on-target editing at the SMN2 transgene and only observed substantial editing at off-target site 5 in an intron of the mucin 16 gene (Muc16, 31±1.9%) that is not expressed in the CNS (fig. S4E)(79). Next, we compared this analysis to off-target editing in vivo following AAV9-ABE ICV injection in Δ7SMA neonates by performing verification of in vivo off targets (VIVO)(80). We observed between 10-27% (average 15±7%) editing at off-target site 5 in intron 54 of Muc16, and between 0.1-0.9% (average 0.5±0.3%) editing at the non-coding off-target site rank 15, compared to 87±3.5% average on-target editing of SMN2 among GFP-positive cells in the CNS across five animals (Fig. 3G and H). These animals ranged from 4 to 18 weeks of age at the time of off-target analysis (26, 36, 42, 80, and 127 days old) and we observe no increase in off-target editing events over time. Thus, off-target editing outcomes observed in cell culture experiments were consistent with those observed in vivo over 18 weeks(80). The ABE strategy did not induce any detected coding mutations in either human or mouse genomes, and off-target editing in vivo was lower than in cell culture (~2-fold lower at Muc16 intron 54), likely due to lower copy number and expression levels in transduced cells in vivo or in vivo gene silencing over time(33, 36, 37).
Cas-independent RNA off-target adenine base editing in vivo is typically indistinguishable from background A-to-I conversion due to the low copy-number of ABE-expressing transgenes(33, 81). We investigated RNA off-target editing in Δ7SMA mESCs and differentiated neural lineages including motor neurons, that stably produce ABE8e from low gene copy numbers similar to those resulting from AAV9 transduction (Fig. 3I, Supplementary Text, fig. S4, F to H). Consistent with previous reports(81, 82), whole transcriptome sequencing did not reveal detected accumulation of RNA A-to-I edits over background levels of endogenous A-to-I and A-to-G changes (Fig. 3J, and fig. S4G).
Collectively, these in vitro and in vivo analyses did not reveal off-target edits of anticipated clinical or physiological significance in human or mouse cells, suggesting high target specificity of the D10 base editing approach. Continued preclinical assessment and minimization of off-target editing is important to ensure the safety of a potential base editing therapeutic for the treatment of SMA in patients.
ABE-mediated rescue of SMA pathophysiology in mice
The physiology of AAV9-ABE treated Δ7SMA mice was improved compared to untreated animals (Movies S1 and S2). We assessed the rescue of motor phenotypes by electrophysiological measurements in AAV9-ABE treated Δ7SMA mice. We measured compound muscle action potential (CMAP) amplitude and performed motor unit number estimation (MUNE) in the gastrocnemius muscle to assess loss of motor neuron functional integrity, a key feature of SMA and preclinical SMA models(83). We compared outcomes with FDA-approved therapeutics for SMA including ICV injection of Zolgensma, and daily intraperitoneal (IP) injection of risdiplam (Evrysdi) at doses that were previously demonstrated to confer a survival benefit to these mice (2.5x1013 vg/kg Zolgensma and 0.1 mg/kg risdiplam, Fig. 4A)(30, 32). MUNE were reduced by 50% in untreated Δ7SMA animals compared to heterozygous mice at postnatal day (PND) 12, and Zolgensma or 0.1 mg/kg risdiplam showed little to no improvement (50% and 75% relative to heterozygotes respectively, Kruskal-Wallis test p>0.6). In contrast, MUNE in AAV9-ABE treated SMA mice were significantly improved compared to untreated animals (Kruskal-Wallis test p<0.02) and did not significantly differ from heterozygous animals, with values averaging 91% that of heterozygotes. CMAP amplitudes were also higher for AAV9-ABE-treated mice compared to risdiplam-treated or untreated Δ7SMA mice, while CMAP amplitudes did not significantly differ between heterozygotes, Zolgensma-treated mice, and AAV9-ABE-treated animals (Kruskal-Wallis one-way ANOVA p>0.2). Thus, neonatal ICV injection of AAV9-ABE measurably rescues SMA pathophysiology of spinal motor neurons.
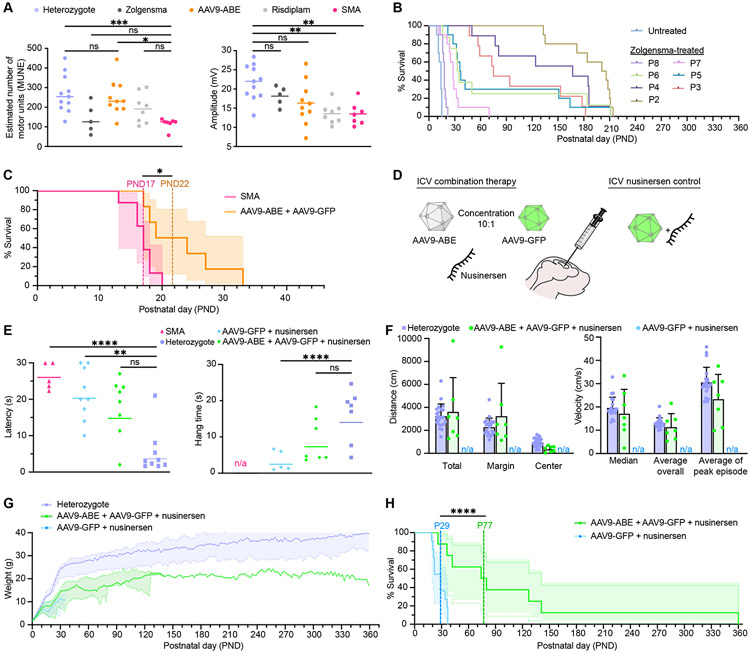
Fig. 4. AAV9-ABE mediated rescue of Δ7SMA mice.
(A) (Left) Motor unit number estimation (MUNE) and (Right) compound muscle action potential (CMAP) amplitude at PND12 in heterozygotes (n=11), and Δ7SMA mice treated with Zolgensma (n=5), AAV9-ABE (n=10), risdiplam (n=8), or uninjected (n=7). (B) Kaplan–Meier curve of Δ7SMA neonates ICV injected with Zolgensma from Robbins et al. 2014 (data extracted using PlotDigitizer). Average (av), median (md), and longest (lng) survival in days: untreated (avg-13, med-14, lng-15), PND2 (avg-187, med-204, lng-214), PND3 (avg-102, med-75, lng-182), PND4 (avg-141, med-167, lng-211), PND5 (avg-76, med-37, lng-211), PND6 (avg-73, med-34, lng-211), PND7 (avg-30, med-28, lng-70), and PND8 (avg-18, med-18, lng-22).). (C) Kaplan-Meier curve in AAV9-ABE treated (n=6) and uninjected (n=8) Δ7SMA mice. (D) Neonatal ICV co-injections with AAV9-ABE, AAV9-GFP, and nusinersen. (E) (Left) The time required for Δ7SMA mice to right themselves in the righting reflex assay at PND7. (Right) The hang time of Δ7SMA mice in the inverted screen test at PND25. (F) Analysis of voluntary movement by open field tracking at PND40. (Left) Traveled distance in cm. (Right) Velocity in cm/s. (G, H) Body weight in grams and Kaplan-Meier curve of Δ7SMA mice. Graph line shading represents (G) standard deviation or (H) 95% CI. Animals are treated as indicated. Dots represent individual animals, *≤0.02, **≤0.01, ***≤0.005, ****≤0.001.
Next, we assessed survival of ICV AAV9-ABE injected Δ7SMA mice. In SMA type I patients, therapeutic intervention can meaningfully improve disease outcomes if administered in the first several months of life(84-87), however, in Δ7SMA mice survival drops precipitously when animals receive treatment past PND6 (Fig. 4B)(88). This large difference is due in part to the highly accelerated (~150-fold greater) rate of maturation of mice compared to humans in the first month, early perinatal reduction in SMN expression that occurs in mice(89) and humans(28), and the rapid early-onset loss of motor units, which consist of spinal motor neurons and the muscle fibers that they innervate(83, 90). Restoration of SMN protein levels using inducible transgenes demonstrates that high levels of SMN are required by PND4-6 to rescue Δ7SMA mice, and delays of small numbers of days are strongly anti-correlated with survival(32, 88, 89, 91-93). In cells, complete mRNA rescue is not achieved until 7 days post D10 transfection (fig. S5A), and the time to restore SMN protein levels in vivo surpasses the extremely short therapeutic window in Δ7SMA mice.
The accumulation of SMN protein following transduction with the dual single-stranded AAV9 ABE8e vectors used in this study requires completion of (1) second-strand synthesis of each AAV9-ABE genome(94, 95), (2) transcription and translation of the split-intein ABE protein segments, (3) assembly and trans-splicing of the split ABE protein, (4) RNP assembly and base editing of SMN2, (5) transcription of full-length C6T-modified endogenous SMN2 pre-mRNA driven by its native promoter, and (6) splicing and translation of corrected SMN2 transcripts. Thus, the timing for SMN protein rescue following AAV9-ABE administration is slower than fast-acting splice-switching drugs or constitutive gene complementation from SMN cDNA encoded by a self-complementary AAV9-SMN vector such as Zolgensma(94-96). We recently demonstrated that in vivo base editing impacts protein levels by ~1-3 weeks post-administration(81).
Despite the incongruent timeline of base editing-mediated rescue for ideal rescue of Δ7SMA mice, AAV9-ABE increased the lifespan of treated animals by ~33% in two colonies in different institutions (Supplementary Text, Materials and Methods, Fig. 4C, and fig. S5, B to D). Lifespan of treated animals improved from an average of 17 days (median 17 days, maximum 20 days) to 23 days (median 22 days, maximum 33 days, Mantel-Cox test p<0.02,). As anticipated, the lifespan extension resulting from AAV9-ABE treatment is similar to that achieved by scAAV9-SMN gene therapy in post-symptomatic (>PND7) Δ7SMA mice (Fig. 4B)(32, 73, 88, 93). Collectively, these data demonstrate that postnatal conversion of SMN2 C6T by AAV9-ABE rescues SMA motor phenotypes in mice, including the number (MUNE) and output (CMAP) of functional motor units innervating muscle, and that the prolonged process of AAV9-ABE-mediated SMN restoration results in mostly post-symptomatic rescue in Δ7SMA mice that results in a significant, but limited improvement in animal lifespan.
Upregulation of SMN protein levels improves motor function and life expectancy of SMA patients and animal models if achieved prior to onset of neuromuscular pathology and symptoms(13, 32, 86-88, 93), yet even high levels of SMN protein cannot correct neuromuscular junction defects once SMA has progressed to an advanced stage and loss of motor neurons upon cell death is irreversible. We therefore sought to extend the effective therapeutic window for gene editing by transient early administration of an existing approved SMA drug to attenuate disease progression, as has previously been applied to study milder forms of SMA in mice(73, 97, 98). Since SMA patients in a gene editing clinical trial would likely be receiving an SMA drug, repeating the base editing treatment in mice receiving an existing SMA drug would also inform a potential future clinical application of this approach.
Combination therapy improves the lifespan of ABE-treated SMA mice
Transient SMA drug administration can ameliorate SMA pathology and extend survival of Δ7SMA mice. We hypothesized that attenuating disease progression using nusinersen could extend the unusually short therapeutic window of Δ7SMA mice and allow AAV9-ABE-mediated rescue to begin before extensive irreversible SMA damage occurs. The mechanism of nusinersen (binding to SMN2 pre-mRNA) is orthogonal to base editing of SMN2 genes, and co-transfection of 20 nM nusinersen did not affect base editing outcomes or inclusion of exon 7 in spliced SMN transcripts following D10 in Δ7SMA mESCs (fig. S5, A and E). We assessed whether co-administration of nusinersen can improve phenotypic rescue from AAV9-ABE treatment. A single ICV injection of nusinersen at PND0 has been shown to extend survival of Δ7SMA mice by several weeks(99), thus we co-injected a single low dose (1 μg) of nusinersen together with AAV9-ABE and AAV9-GFP in Δ7SMA neonates (Supplementary Text). As a control, we also treated Δ7SMA neonates with 1 μg nusinersen and AAV9-GFP but no base editor (Fig. 4D). We assessed motor coordination and overall muscle strength at PND7 using the righting reflex test, which measures the time needed for a mouse placed on its back to right itself (Fig. 4E). We observed significant difference between heterozygotes and nusinersen-treated or untreated Δ7SMA mice (Kruskal-Wallis test p≤0.01), but no significant difference between mice treated with combined AAV9-ABE+nusinersen compared to heterozygous littermates (Kruskal-Wallis test p>0.1).
Next, we assessed motor strength and coordination of treated and heterozygous mice using an inverted screen test, which measures how long the mice can hang inverted from a screen mesh surface. At PND25, Δ7SMA animals treated with nusinersen alone performed significantly worse than healthy heterozygous mice at inverted screen testing (Kruskal-Wallis test p=0.007, Fig. 4E). In contrast, the AAV9-ABE+nusinersen combination-treated animals showed no significant difference in the inverted screen assay from healthy heterozygous mice. Notably, half of nusinersen-only treated animals were deceased by this timepoint, and age-matched untreated Δ7SMA mice do not survive long enough for this PND25 assay.
For a more complete behavioral assessment of treated and heterozygous animals, we performed extensive multiparametric analysis of voluntary movement by open field tracking at PND40 (Fig. 4F, and fig. S5, F to J). Across 33 parameters including traveled distances, velocity, duration, and counts of various activities, the measured behaviors of AAV9-ABE+nusinersen combination-treated animals showed no significant difference with those of heterozygous mice (Mann-Whitney test p>0.5). Neither nusinersen-only treated or untreated age-matched Δ7SMA mice were available as reference for this PND40 assay due to their short lifespan.
We also assessed the effect of combination AAV9-ABE and nusinersen treatment on weight and lifespan of Δ7SMA mice. The weight of nusinersen-only and AAV9-ABE+nusinersen combination-treated Δ7SMA mice steadily increased and were indistinguishable for the first week of life, after which weight gain slowed in the nusinersen-only cohort (Fig. 4G). Combination-treated animals maintained on average 61±4.0% the weight of heterozygous animals throughout their lifespans. The nusinersen-only injection improved lifespan of Δ7SMA mice from an average of 17 days (median 17, maximum 20 days, Fig. 4C) to an average 28 days (median 29, maximum 37 days, Mantel-Cox test p=0.0001, Fig. 4H). Importantly, combination treatment of AAV9-ABE with nusinersen improved survival of Δ7SMA mice to on average of 111 days (median 77, Mantel-Cox test p=0.002), with over 60% of animals surviving beyond nusinersen-only controls, and a 10-fold increase in maximum lifespan (37 days maximum with nusinersen only, compared to 360 days maximum with AAV9-ABE). Combination AAV9-ABE+nusinersen-treated SMA mice also exhibited normal behavior and vitality well beyond the lifespan of nusinersen only-injected controls (P40, P96, and P200 in Movies S3 to S5). Collectively, these data indicate that transient extension of the very narrow therapeutic window in Δ7SMA mice can greatly improve phenotypic rescue of SMA from base editing of SMN2.
While neonatal AAV9-ABE ICV injection alone enables life extension in Δ7SMA mice that resembles >PND7 ICV injection with Zolgensma (Fig. 4B and C)(88), co-administration of 1 μg nusinersen temporarily slows disease progression and broadens the narrow therapeutic window, allowing base editing the opportunity to enable lifespan rescue that more closely resembles that of pre-symptomatic Zolgensma administration at ≤PND3 (Fig. 4H). Moreover, these data demonstrate compatibility of AAV9-ABE with nusinersen as a one-time treatment without evident adverse effects, and with apparent synergy to improve therapeutic outcomes. Such a combination therapy approach may play an important role in future clinical trial designs for one-time SMA treatments that permanently correct a genetic cause of the disease, and for clinical application in patients already receiving treatment.
Discussion
Current treatment options for SMA have revolutionized care for thousands of patients, effectively extending lifespan, preventing the loss of motor function in pre-symptomatic patients, and delaying progression in symptomatic patients by increasing full-length SMN protein levels(13, 24, 86, 87, 91, 100). However, current therapies do not restore endogenous protein levels and native regulation of SMN, which could result in pathogenic SMN insufficiency in motor neurons or potential long-term toxicity in other tissues(21, 23-28, 35). Furthermore, the transient therapies nusinersen and risdiplam require repeated dosing throughout a patient’s lifetime, and it is unclear whether Zolgensma gene complementation will persist in motor neurons(36, 37). Thus, achieving permanent and endogenously regulated rescue of SMN protein levels is an important goal of a future therapeutic for SMA patients. The optimized D10 ABE strategy developed in this work is a one-time treatment that enables permanent and precise editing of endogenous SMN2 genes while preserving native transcript levels and regulatory mechanisms that govern SMN expression(1, 4, 5, 28, 78, 101). As such, a future base editing therapeutic approach could offer substantial benefits over existing SMA therapies.
We compared 79 total nuclease and base editing strategies targeting five regions of SMN2 to induce either post-transcriptional or post-translational regulatory changes in SMN2 that upregulate SMN protein production. BE-Hive and inDelphi machine learning models enabled the design of precise editing strategies that in some cases were not obvious, and pre-selected sgRNAs for genotypic and phenotypic validation of editing outcomes. All SMA patients regardless of their SMN1 mutations must carry the SMN2 gene to complete gestation(7), and thus the genome editing strategies identified in this study have the potential to benefit all SMA patients.
While on-target Cas nuclease editing at SMN2 can be precise, DSBs can result in large deletions and chromosomal rearrangements, especially when induced simultaneously at multiple genomic loci(102). Given that SMA patients usually have multiple copies of SMN2, nuclease editing may result in unintended restructuring of the chromosome region (5q13) that harbors SMN genes(103, 104). In contrast, base editors precisely convert nucleotides without inducing DSBs(50, 105, 106), and result in greater SMN protein upregulation than the nuclease strategies in this study (up to 50-fold by base editors compared to up to 17-fold by nucleases). We therefore recommend that future gene editing therapeutic strategies for SMA use base editing rather than nucleases.
ABE strategy D10 demonstrated high on-target efficiency and specificity, with minimal Cas-dependent or Cas-independent off-target DNA or RNA editing. It is possible that extended base editor expression in cells, as can result from AAV-delivery, could result in a greater accumulation of genomic and transcriptomic off-target events. Therefore, a deeper assessment of genomic and transcriptomic off-targets and efforts to minimize off-target editing risk will be important in the preclinical development of a potential base editing therapeutic for SMA. If needed, Cas-independent editing events can be further minimized by alternative delivery strategies that shorten exposure to base editors(62), and by the use of tailored deaminases such as the V106W variant of TadA*-8e(62, 64) or TadA-8.17-m(107).
SMA has variable presentation in humans that largely correlates with the copy number of SMN2(108-112). Type I SMA patients have two SMN2 copies and present with symptoms within the first 6 months, type II patients have three copies and present with symptoms by 18 months, while type III patients have 3-4 SMN2 copies with later onset. Early intervention is paramount to achieving the best outcomes for SMA patients. The window to effectively treat type II and III patients is broader than for type I patients, who ideally receive treatment within the first few months of life and up to 18 months(13, 24, 84-87, 100). Indeed, we directly observed the critical role of differences in timing on the order of days in determining the efficacy of an AAV9-ABE treatment in Δ7SMA mice, which have an unusually short (≤6 days) therapeutic window compared to the timescale of base editing (weeks)(88). We show that the FDA-approved ASO drug nusinersen can extend the very short therapeutic window for rescue in Δ7SMA mice, allowing base editing-mediated rescue of SMN protein levels to occur to a greater extent(81). We anticipate that the broader therapeutic window in human SMA patients would provide ample opportunity for AAV9-ABE-mediated restoration of SMN protein levels to take place without the need for co-administration of a transient therapeutic. Nevertheless, our study demonstrates the compatibility of base editing with nusinersen as a combination therapy approach to treat SMA in animals, which may be valuable for future clinical applications.
The ICV-injected AAV9-ABE animals in our study exhibited mouse-specific peripheral disease phenotypes that are common in SMA mouse models including necrosis of the extremities(113, 114), while exhibiting otherwise normal behavior and vitality without displays of progressive muscle weakness. However, SMA treatment that is restricted to the CNS also reveals a late onset lethal cardiac abnormality specific to Δ7SMA mice(32, 115-117), and likely underlies the sudden late-stage fatality observed in ICV AAV9-ABE treated animals in this study. Treating both CNS and peripheral tissues may ameliorate this murine cardiac phenotype to improve lifespan of treated Δ7SMA mice compared to ICV-injected animals(115, 118). Nevertheless, peripheral restoration of SMN protein does not appear to be required to rescue SMA lethality in humans in light of patients successfully treated intrathecally with Spinraza(23, 31, 84, 91, 119).
As demonstrated in this work, dual-AAV delivery of base editors supports therapeutic levels of editing in mouse models of human disease(120, 121). After these in vivo experiments were completed, our lab developed efficient in vivo base editing using single-AAV9-ABE systems that use size-minimized AAV vector components and one of a suite of small Cas protein domains that are highly active as ABEs(81). Such single-AAV base-editing systems may simplify the development of future base editor therapeutics, and potentially minimize the required dose and potential side effects of AAV in clinical settings(122).
Materials and Methods
Cell culture
Culture of mESCs, HEK293T, and U2OS cells was performed according to previously published protocols(123). mESCs were maintained on 0.2% gelatin-coated plates feeder-free in mESC media composed of Knockout DMEM (Life Technologies) supplemented with 15% defined fetal bovine serum (FBS, HyClone), 0.1 mM nonessential amino acids (NEAA, Life Technologies), Glutamax (GM, Life Technologies), 0.55 mM 2-mercaptoethanol (b-ME, Sigma-Aldrich), 1X ESGRO LIF (Millipore), with the addition of 2i: 5 nM GSK-3 inhibitor XV (Sigma-Aldrich), and 500 nM UO126 (Sigma-Aldrich). Δ7SMA mESCs were a kind gift from Lee L. Rubin. HEK293T cells were purchased from ATCC (CRL-3216) and were maintained in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (ThermoFisher Scientific). U2OS cells were purchased from ATCC (HTB-96) and were maintained in McCoy's 5a medium (Life Technologies) supplemented with 10% fetal bovine serum (ThermoFisher Scientific). All cells were regularly tested for mycoplasma.
For genome editing experiments, cells were seeded one day prior to be ~70-80% confluent on the day of transfection and transfected with sgRNA and genome editing plasmids at a 1:1 molar ratio using Lipofectamine 3000 (ThermoFisher Scientific) in accordance with the manufacturer’s protocols. For stable integration of plasmids, cells were co-transfected with Tol2 transposase at an equimolar ratio. Cells that did not undergo antibiotic selection were cultured for 3-5 days before harvesting. For antibiotic selection, Δ7SMA mESCs were treated with 50 μg/mL hygromycin B (Life Technologies) and/or 6.67 μg/mL blasticidin as indicated, starting 24 hours after transfection. For transient selection, antibiotics were removed from the media after 48 hours. Selected cells were allowed to recover and expand prior to harvesting. All sgRNA sequences designed for this study are listed in the supplement.
For Δ7SMA mESCs nusinersen experiments, cells were transfected with 20 nM of fully 2′-O-methoxyethyl (MOE)-modified ASO (5’-TCACTTTCATAATGCTGG-3') on a phosphorothioate backbone (TriLink), using Lipofectamine 3000 (ThermoFisher Scientific). After 24 hours media was replaced every other day with fresh mESC+2i media. For splicing rescue by risdiplam, mESC media was supplemented with 0.1–1 μM of risdiplam (RG7916, Selleck Chemicals LLC) in DMSO, as indicated. Cells were harvested at the indicated timepoints.
High-throughput sequencing of genomic DNA
Sequencing library preparation was performed according to previously published protocols(50). Primers are listed in the supplement. Briefly, we isolated genomic DNA (gDNA) with the QIAamp DNA mini kit (Qiagen) and used 250-1000 ng of gDNA for individual locus editing experiments and 20 μg of gDNA for comprehensive context library samples. Sequencing libraries were amplified in two steps, first to amplify the locus of interest and second to add full-length Illumina sequencing adapters using the NEBNext Index Primer Sets 1 and 2 (New England Biolabs) or internally ordered primers with equivalent sequences. All PCRs were performed using NEBNext Ultra II Q5 Master Mix. Samples were pooled using Tape Station (Agilent) and quantified using a KAPA Library Quantification Kit (KAPA Biosystems). The pooled samples were sequenced using Illumina NextSeq or MiSeq. Alignment of fastq files and quantification of editing frequency for individual loci was performed using CRISPResso2 in batch mode(67). The editing frequency for each site was calculated as the ratio between the number of modified reads (i.e. containing nucleotide conversions or indels) and the total number of reads. Base editing characterization library analysis was performed as previously described(50).
Quantification of SMN splice products
We isolated mRNA from Δ7 mESCs with the RNeasy mini kit (Qiagen) and performed reverse transcription using SuperScript IV (ThermoFisher) according to the manufacturer’s protocols. For targeted SMN2 splice product quantitation by qPCR, high-throughput sequencing, or automated electrophoresis we performed reverse transcription with random hexamers. Inclusion of SMN2 exon 7 was quantified by automated electrophoresis using Tape Station (Agilent). For unbiased SMN2 splice product analysis by high-throughput sequencing, we performed reverse transcription using a custom oligo-dT primer with a Read 2 Illumina sequencing stub. The pooled samples were sequenced using Illumina MiSeq. All PCRs were performed using NEBNext Ultra II Q5 Master Mix, with the addition of Sybr Green for qPCR. Primers are listed in Supplementary Table 3.
Western Blot
Cells harvested for western blot were washed with ice-cold PBS and incubated at 4 °C for 30 min while rocking in RIPA lysis buffer (ThermoFisher) supplemented with 1 mM PMSF (ThermoFisher) and cOmplete EDTA-free protease inhibitor cocktail (Roche). Lysates were clarified by centrifugation at 12,000 rpm at 4 °C for 20 min. Lysates were normalized using BCA (Pierce BCA Protein Assay Kit) and combined with 4x Laemelli buffer (BioRad) and DTT (ThermoFisher) at a final concentration of 1 mM. We loaded 10 μg of reduced protein per gel lane and performed transfer with an iBlot 2 dry blotting system (ThermoFisher) using the following program: 20 V for 1 min, then 23 V for 4 min, then 25 V for 2 min for a total transfer time of 7 minutes. Blocking was performed at room temperature for 60 minutes with block buffer: 1% BSA in TBST (150 mM NaCl, 0.5% Tween-20, 50 mM Tris-Cl, pH 7.5). Membranes were then incubated in primary antibody diluted in block buffer for 2 hours at room temperature. After a washing, secondary antibodies diluted in TBST were added and incubated for 1 hour at room temperature. Membranes were washed again and imaged using a LI-COR Odyssey. Wash steps were 3x 5-minute washes in TBST. Primary antibodies used were mouse anti-human SMN (Proteintech 2C6D9), mouse anti-mouse and human SMN (Proteintech 3A8G1) and rabbit anti-histone H3 (Cell Signaling D1H2), secondary antibodies used were LI-COR IRDye 680RD goat anti-rabbit (#926–68071) and goat anti-mouse (#926–68070).
Base editor characterization library assay
For characterization of the ABE8e-SpCas9 base editor, we used mouse ESCs carrying the comprehensive context library according to previously published protocols(42, 50). Briefly, 15-cm plates with >107 initial cells were transfected with a total of 50 μg of p2T-ABE8e-SpCas9 and 30 μg of Tol2 plasmid to allow for stable genomic integration with Lipofectamine 3000 according to manufacturer protocols, and selected with 10 μg/mL blasticidin starting the day after transfection for 4 days before harvesting. We maintained an average coverage of ~ 300x per library cassette throughout. We collected gDNA from cells 5 days after transfection, after 4 days of antibiotic selection.
Cloning
Base editor plasmids were constructed by replacing deaminase and Cas-protein domains of the p2T-CMV-ABE7.10-BlastR (Addgene 152989) plasmid by USER cloning (New England Biolabs)(50). Individual sgRNAs were cloned into the SpCas9-hairpin U6 sgRNA expression plasmid (Addgene 71485) using BbsI plasmid digest and Gibson assembly (New England Biolabs). Protospacer sequences and gene-specific primers used for amplification followed by HTS are listed in Supplementary Table 1. Constructs were transformed into Mach1 chemically competent E. coli (ThermoFisher) grown on LB agar plates and liquid cultures were grown in LB broth overnight at 37 °C with 100 μg/mL ampicillin. Individual colonies were validated by Templiphi rolling circle amplification (ThermoFisher) followed by Sanger sequencing. Verified plasmids were prepared by mini, midi, or maxiprep (Qiagen).
AAV vectors were cloned by Gibson assembly (NEB) using NEB Stable Competent E. coli (High Efficiency) to insert the sgRNA sequence and C-terminal base editor half of ABE8e-SpyMac into v5 Cbh-AAV-ABE-NpuC+U6-sgRNA (Addgene 137177), and the N-terminal base editor half and a second U6-sgRNA cassette into v5 Cbh-AAV-ABE-NpuN (Addgene 137178)(74).
Neural differentiation
Differentiation of Δ7SMA mESCs was performed according to established protocols(124, 125). Briefly, Δ7SMA mESCs maintained on 0.2% gelatin-coated plates feeder-free in mESC media + 2i were plated onto irradiated mouse embryonic fibroblast (iMEF) feeders on 0.2% gelatin-coated plates in mESC media for 7 days to wean cells from 2i factors. Cells were then seeded at 106 in 10-cm tissue culture treated plates for 48 hours for priming and depletion of feeders. Media was replaced with neural differentiation (ND) media composed of 1:1 DMEM:F12 and Neurobasal media (Life Technologies) supplemented with 10% knockout serum-replacement (KOSR, Life Technologies), Glutamax (GM, Life Technologies) and 0.55 mM 2-mercaptoethanol (b-ME, Sigma-Aldrich), for one hour prior to trypsinization and seeding of 2x106 cells in 10-cm non-tissue culture treated dishes for 24 hours. Single cells and small early embryoid bodies (EBs) in suspension were collected and transferred to 10-cm tissue culture treated plates in fresh ND media for 24 hours. Small EBs that remained in suspension were collected and transferred to 10-cm tissue culture treated plates in fresh ND media with the addition of 1μM retinoic acid (RA, Sigma-Aldrich R2625) for caudal neural differentiation (CND), or with 1μM RA and 0.5 μM smoothened agonist (SmAg, Calbiochem 566660) for motor neuron differentiation (MND) for 72 hours. Large EBs were collected and split into two 10-cm tissue culture treated plates in neural growth (NG) media composed of 1:1 DMEM:F12 and Neurobasal media supplemented with GM, B27 (Life Technologies), and 10ng/mL human recombinant glial cell line-derived neurotrophic factor (GDNF, R&D Systems 212-GD-010) for 48 hours. EBs were monitored for Mnx1:GFP expression to assess motor neuron differentiation efficiency and imaged using a Zeiss inverted fluorescence microscope or collected for downstream whole transcriptome analysis.
Whole transcriptome RNA-sequencing
Library preparation, sequencing and analysis were performed by SMART-seq2 as previously described(126). Briefly, total RNA was harvested from cells using the RNeasy Mini kit (Qiagen). First, we incubated 20 ng purified total RNA with RNase inhibitor (Clontech Takara 2313B), dNTP mix (Thermo Fisher R0192), and the 3’-RT primer (5’-AAGCAGTGGTATCAACGCAGAGTAC(T30)VN-3’) at 72 °C for 3 min to anneal the RT primer. Next, we performed first-strand synthesis using the template switching oligo (TSO): (5'-AGCAGTGGTATCAACGCAGAGTACrGrG+G-3' Exiqon, Qiagen) together with RNase inhibitor, betaine (Sigma Aldrich B0300-1VL), MgCl2 (Sigma Aldrich 1028) and Maxima RNase H-minus RT (Thermo Fisher EP0751), according to the manufacturer’s protocols. We performed pre-amplification of first-strand libraries with the ISPCR primer: 5'-AAGCAGTGGTATCAACGCAGAGT-3' using KAPA HiFi HotStart (KAPA KK2601) and SYBR green (Thermo Fisher). Whole transcriptome amplification (WTA) product was washed using DNA SPRI beads (Beckman Coulter A63881) and quantified by Agilent Tapestation. We performed Tagmentation and library preparation of 0.25 ng WTA using the Nextera XT kit (Illumina) and Nextera i7 and Nextera i5 barcoding primers. Samples were pooled and washed using washed using DNA SPRI beads and quantified by Agilent Tapestation and the KAPA Universal Library Quantification kit (Roche KK4824). Libraries were run on Illumina NextSeq 550.
FASTQs were generated using bcl2fastq v2.20. Trim Galore v0.6.7 in paired-end mode with default parameters to remove low-quality bases, adapter sequences, and unpaired sequences. Trimmed reads were aligned to the GENCODE mouse reference genome M31 (GRCm39) using STAR (v2.7.10a), quantified using kallisto(127), and refined to canonical coding sequences using CCDS release 21(128). For RNA A-to-I off-target analysis, REDItools v1.3 was used to quantify the average frequency of A-to-I editing among all sequenced adenosines in each sample(129), excluding adenosines with read depth <10 or read quality score <30. The transcriptome-wide A-to-I editing frequency was calculated independently for each biological replicate as: (number of reads in which an adenosine was called as a guanosine)/(total number of reads covering all analyzed adenosines).
Purification of SpyMac Cas nuclease protein
SpyMac Cas nuclease protein was cloned into the expression plasmid pD881-SR (Atum, Cat. No. FPB-27E-269). The resulting plasmid was transformed into BL21 Star DE3 competent cells (ThermoFisher, Cat. No. C601003). Colonies were picked for overnight growth in terrific broth (TB)+25 μg/mL kanamycin at 37 °C. The next day, 2 L of pre-warmed TB were inoculated with overnight culture at a starting OD600 of 0.05. Cells were shaken at 37 °C for about 2.5 hours until the OD600 was ~1.5. Cultures were cold shocked in an ice-water slurry for 1 hour, following which L-rhamnose was added to a final concentration of 0.8% to induce. Cultures were then incubated at 18 °C with shaking for 24 hours to produce protein. Following induction, cells were pelleted and flash-frozen in liquid nitrogen and stored at −80 degrees. The next day, cells were resuspended in 30 mL cold lysis buffer (1 M NaCl, 100 mM Tris-HCl pH 7.0, 5 mM TCEP, 20% glycerol, with 5 tablets of cOmplete, EDTA-free protease inhibitor cocktail tablets (Millipore Sigma, Cat. No. 4693132001). Cells were passed three times through a homogenizer (Avestin Emulsiflex-C3) at ~18,000 psi to lyse. Cell debris was pelleted for 20 minutes using a 20,000 g centrifugation at 4 °C. Supernatant was collected and spiked with 40 mM imidazole, followed by a 1-hour incubation at 4 °C with 1 mL of Ni-NTA resin slurry (G Bioscience Cat. No. 786-940, prewashed once with lysis buffer). Protein-bound resin was washed twice with 12 mL of lysis buffer in a gravity column at 4 °C. Protein was eluted in 3 mL of elution buffer (300 mM imidazole, 500 mM NaCl, 100 mM Tris-HCl pH 7.0, 5 mM TCEP, 10% glycerol). Eluted protein was diluted in 40 mL of low-salt buffer (100 mM Tris-HCl, pH 7.0, 1 mM TCEP, 20% glycerol) just before loading into a 50 mL Akta Superloop for ion exchange purification on the Akta Pure25 FPLC. Ion exchange chromatography was conducted on a 5 mL GE Healthcare HiTrap SP HP pre-packed column (Cat. No. 17115201). After washing the column with low-salt buffer, the diluted protein was flowed through the column to bind. The column was then washed in 15 mL of low salt buffer before being subjected to an increasing gradient to a maximum of 80% high salt buffer (1 M NaCl, 100 mM Tris-HCl, pH 7.0, 5 mM TCEP, 20% glycerol) over the course of 50 mL, at a flow rate of 5 mL per minute. 1-mL fractions were collected during this ramp to high-salt buffer. Peaks were assessed by SDS-PAGE to identify fractions containing the desired protein, which were concentrated first using an Amicon Ultra 15-mL centrifugal filter (100-kDa cutoff, Cat. No. UFC910024), followed by a 0.5-mL 100-kDa cutoff Pierce concentrator (Cat. No. 88503). Concentrated protein was quantified using a BCA assay and determined to be 12.6 milligrams per milliliter (ThermoFisher, Cat. No. 23227).
CIRCLE-seq off-target editing analysis
Off-target analysis using CIRCLE-seq was performed as previously described(68, 130). Briefly, genomic DNA from HEK293T cells or NIH3T3 cells was isolated using Gentra Puregene Kit (Qiagen) according to manufacturer’s instructions. Purified genomic DNA was sheared with a Covaris S2 instrument to an average length of 300 bp. The fragmented DNA was end repaired, poly-A tailed, and ligated to an uracil-containing stem-loop adaptor using the KAPA HTP Library Preparation Kit, PCR Free (KAPA Biosystems). Adaptor ligated DNA was treated with Lambda Exonuclease (NEB) and E. coli Exonuclease I (NEB), then with USER enzyme (NEB) and T4 polynucleotide kinase (NEB). Intramolecular circularization of the DNA was performed with T4 DNA ligase (NEB) and residual linear DNA was degraded by Plasmid-Safe ATP-dependent DNase (Lucigen). In vitro cleavage reactions were performed with 250 ng of Plasmid-Safe ATP-dependent DNase-treated circularized DNA, 90 nM of SpyMac Cas9 nuclease protein, Cas9 nuclease buffer (NEB) and 90 nM of synthetic chemically modified sgRNA (Synthego), in 100 μl. Cleaved products were poly-A tailed, ligated with a hairpin adaptor (NEB), treated with USER enzyme (NEB), and amplified by PCR with barcoded universal primers NEBNext Multiplex Oligos for Illumina (NEB), using Kapa HiFi Polymerase (KAPA Biosystems). Libraries were sequenced with 150-bp paired-end reads on an Illumina MiSeq instrument. CIRCLE-seq data analyses were performed using open-source CIRCLE-seq analysis software and default recommended parameters (https://github.com/tsailabSJ/circleseq).
Husbandry of Δ7SMA mice
All experiments in animals were approved by the Institutional and Animal Care and Use Committee of the Broad Institute of MIT and Harvard and Ohio State University (OSU). Δ7SMA heterozygous mice (Smn+/−; SMN2+/+; SMNΔ7+/+) were purchased from the Jackson Laboratory (005025)(55), and maintained in the Broad Institute and OSU vivaria according to recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. Pairs of Δ7SMA heterozygotes were crossed to generate Δ7 SMA mice (Smn−/−; SMN2+/+; SMNΔ7+/+). On date of birth (PND0), pups were microtattooed on the foot pads (Aramis) with animal-grade permanent ink (Ketchum) using a sterile hypodermic needle (BD) to enable identification of individual pups. Subsequently, biopsies of ~1 mm tissue were taken from the tail using a sterile blade, lysed for genomic DNA extraction, and used for genotyping by PCR. Litter size was controlled to five pups, including 1-3 homozygous mutants, by culling and cross-fostering among same-age mice. Mice of both sexes were included in the study, although sex has been reported to not have a substantial impact on the phenotype of SMA mice (Treat-NMD SOP Code: SMA_M.2.2.003).
Electrophysiology experiments were performed at OSU. All other animal studies were performed at the Broad Institute unless indicated otherwise in the text. At the Broad Institute, the mean birthweight of heterozygous animals was 1.7±0.1 grams, and 1.5±0.1 grams for SMA pups, and any animal weighing <1.5 grams at time of birth was excluded from the study. The average weight of SMA neonates at injection on PND0 at the Broad Institute was 1.6±0.2 grams. At OSU, the mean birthweight of heterozygous animals on the day of birth was 1.3±0.1 grams and 1.2±0.1 grams for SMA pups, and any SMA, heterozygous or wild-type pup weighing ≤1.0 grams at time of birth were excluded from the study. The average weight of SMA neonates at injection on PND0 at OSU was 1.3±0.13 grams. By facility, each litter was subjected to the same exclusion criterion (Treat-NMD SOP Code: SMA_M.2.2.003). Cohort sizes were chosen based on prior experience with these animals, known to allow for determination of statistical significance. Animals were monitored daily for morbidity and mortality and weighed every other day from day of birth.
Intracerebroventricular injections
Neonatal ICV injections were performed as previously described(74, 131). Briefly, glass capillaries (Drummond 5–000-1001-X10) were pulled to a tip diameter of approximately 100 μm. High-titer qualified AAV was obtained through the Viral Vector Core at UMass Medical School and concentrated using Amicon Ultra-15 centrifugal filter units (Millipore), quantified by qPCR (AAVpro Titration Kit v.2, Clontech), and stored at 4 °C until use. For injection, a small amount of Fast Green was added to the AAV injection solution to assess ventricle targeting. The injection solution was loaded via front-filling using the included Drummond plungers. Δ7SMA pups were anesthetized by placement on ice for 2–3 minutes, until they were immobile and unresponsive to a toe pinch. Up to 4.5 μL of injection mix was injected freehand into each ventricle on PND0-2.
Immunofluorescence imaging of spinal cord sections
For immunofluorescence staining of transduced spinal motor neurons, Δ7SMA mice were perfused at 25 weeks with ice-cold PBS and ice-cold 4% PFA, the CNS was exposed, and the whole carcass was fixed overnight in 4% PFA. Whole spinal cord was isolated and fixed in 4% PFA overnight, then consecutively transferred to 10%, 20%, and 30% sucrose in three overnight incubations before embedding in OCT for long-term storage at –80°C. Embedded tissue was cryo-sectioned and stained with goat anti-Choline Acetyltransferase (Millipore AB144P), mouse anti-NeuN (EMD Millipore MAB377), mouse anti-GFAP (Sigma-Aldrich MAB3402), rabbit anti-GFP (Thermo scientific A-11122), and Alexa-Fluor secondary antibodies (Life Technologies), and imaged on an SP8 confocal microscope (Leica).
Nuclear isolation and sorting of tissues
Tissue harvest and nuclear isolation was performed as previously described(74). Briefly, deceased Δ7SMA mice were stored at −80 °C until dissection of the brain and spinal cord tissue. For isolation of the cortex and cerebella were separated from the brain postmortem using surgical scissors. Hemispheres were separated using a scalpel and the cortex was separated from underlying midbrain tissue with a curved spatula. For nuclear isolation, dissected tissue was homogenized using a glass dounce homogenizer (Sigma D8938) (20 strokes with pestle A followed by 20 strokes with pestle B) in 2 mL ice-cold EZ-PREP buffer (Sigma NUC-101). Samples were incubated for 5 minutes with an additional 2 mL EZ-PREP buffer. Nuclei were centrifuged at 500 g for 5 minutes, and the supernatant removed. For spinal cord tissue, wash steps were repeated ten times. Samples were resuspended with gentle pipetting in 4 mL ice-cold Nuclei Suspension Buffer (NSB) consisting of 100 μg/mL BSA and 3.33 μM Vybrant DyeCycle Ruby (ThermoFisher) in PBS and centrifuged at 500 g for 5 minutes. The supernatant was removed and nuclei were resuspended in 1–2 mL NSB, passed through a 35 μm strainer, and sorted into 200 μL Agencourt DNAdvance lysis buffer using a MoFlo Astrios (Beckman Coulter) at the Broad Institute flow cytometry core. All steps were performed on ice or at 4 °C. Genomic DNA was purified according to the DNAdvance (Agencourt) instructions for 200 μL volume.
Behavioral assays
Righting reflex was recorded on PND7 by placing neonates on their backs and recording the duration to right themselves with a stopwatch up to a maximum of 30 sec. For inverted screen testing, juvenile mice were subjected to the horizontal grid test for mice (Maze Engineers) on PND25 by placing the animals on a wire-mesh screen which the mice are capable of gripping, then inverting the screen over the course of 2 seconds, animal head first, over a padded surface made of bedding 4-5 cm high. The time for the animal to fall onto the bedding was recorded with a stopwatch. Each mouse was assessed with three measurements. The procedure is concluded when the animal falls onto the bedding, or if the animal exceeds 120 seconds for the measurement, in which case the screen is reverted so that the mouse is upright, and the mouse is manually removed from the screen.
Voluntary movement of adult mice is recorded on PND40 by open field testing (Omnitech Electronics). Mice were brought into the testing room under normal lighting conditions and allowed 30-60 minutes of acclimation. The animals were placed into the locomotor activity chamber with infrared beams crossing the X, Y and Z axes that plot their ambulatory and fine motor movements and rearing behavior. Recordings are analyzed using Fusion 5.1 SuperFlex software.
Electrophysiological Measurements
Compound muscle action potential (CMAP) and motor unit number estimate (MUNE) measurements were performed as previously described(132). Briefly, at PND12 the right sciatic nerve was stimulated with a pair of insulated 28-gauge monopolar needles (Teca, Oxford Instruments Medical, NY) placed in proximity to the sciatic nerve in the proximal hind limb. Recording electrodes consisted of a pair of fine ring wire electrodes (Alpine Biomed, Skovlunde, Denmark). The active recording electrode (E1) was placed distal to the knee joint over the proximal portion of the triceps surae muscle and the reference electrode (E2) over the metatarsal region of the foot. A disposable strip electrode (Carefusion, Middleton, WI) was placed on the tail to serve as the ground electrode. For CMAP, supramaximal responses were generated maintaining stimulus currents <10 mA and baseline-to-peak amplitude measurements made.
For MUNE, an incremental stimulus technique similar to a previously described procedure was used(132). Submaximal stimulation was used to obtain ten incremental responses to calculate the average single motor unit potential (SMUP) amplitude. The first increment was obtained by delivering square wave stimulations at 1 Hz at an intensity between 0.21 mA to 0.70 mA to obtain the minimal all-or-none SMUP response. If the initial response did not occur with stimulus intensity between 0.21 mA and 0.70 mA, the stimulating cathode position was adjusted either closer or farther away from the position of the sciatic nerve in the proximal thigh to decrease or increase the required stimulus intensity, respectively. This first incremental response was accepted if three duplicate responses were observed. To obtain the subsequent incremental responses the stimulation intensity was adjusted in 0.03 mA steps and incremental responses were distinguished visually in real-time to obtain nine additional increments. To be accepted, each increment was required to be: (1) observed for a total of three duplicate responses, (2) visually distinct from the prior increment, and (3) at least 25 μV larger than the prior increment. The peak-to-peak amplitude of each individual incremental response was calculated by subtracting the amplitude of the prior response. The ten incremental values were averaged to estimate average peak-to-peak single motor unit potential (SMUP) amplitude. The maximum CMAP amplitude (peak-to-peak) was divided by the average SMUP amplitude to yield the MUNE.
Statistical Analysis
Welch’s two-tailed t-tests were used to compare sequencing, splicing, mRNA levels, and immunostaining data. Error bars represent standard deviations of ≥3 independent biological replicates. Root mean squared error (RMSE) and Pearson’s r-correlation were used for correlation analysis of predicted and observed genome editing outcomes, where appropriate. Kruskal-Wallis tests were used to compare physiology measurements and behaviors of mouse cohorts under experimental conditions. Mann-Whitney tests were used to compare multiparametric measurements of voluntary behaviors of mouse cohorts. The logrank Mantel-Cox test Kaplan-Meier survival curves. All statistical tests were calculated by GraphPad Prism 9.4.1 and Microsoft Excel v16.64.
Supplementary Material
Acknowledgements:
We thank Dr. Anahita Vieira for assistance with editing, Mary O’Reilly for assistance with figures, and Alvin Hsu for assistance with computational analysis.
Funding:
US National Institutes of Health grant U01 AI142756 (D.R.L.)
US National Institutes of Health grant RM1 HG009490 (D.R.L.)
US National Institutes of Health grant R01 EB022376 (D.R.L.)
US National Institutes of Health grant R35 GM118062 (D.R.L.)
US National Institutes of Health grant P01 HL053749 (D.R.L.)
Bill and Melinda Gates Foundation (D.R.L.)
Howard Hughes Medical Institute (D.R.L.)
Friedreich’s Ataxia Accelerator grant (D.R.L.)
The Netherlands Organization for Scientific Research Rubicon Fellowship (M.A.)
US National Institutes of Health K99 Pathway to Independence Award NS119743-01A1 (M.A.)
Helen Hay Whitney Fellowship (G.A.N.)
US National Institutes of Health K99 Pathway to Independence Award HL163805 (G.A.N.)
National Science Foundation Graduate Research Fellowship (A.R., M.W.S.)
Howard Hughes Medical Institute Hanna Gray Fellowship (M.F.R.)
Harvard Chemical Biology Training Grant T32 GM095450 (K.T.Z.)
UMass Chan Internal Funding (G.G.)
US National Institute of Health grant R01HD060586 (A.H.M.B.)
Footnotes
Competing interests: M.A. and D.R.L. have filed patent applications on this work. D.R.L. is a consultant and equity owner of Beam Therapeutics, Prime Medicine, Pairwise Plants, Chroma Medicine, and Nvelop Therapeutics, companies that use or deliver genome editing or genome engineering agents. A.H.M.B is a consultant for Novartis. WDA has served as a consultant to NMD Pharma, Genentech, Catalyst Pharmaceuticals, Dyne, Avidity Biosciences, Design Therapeutics, Argenx, and Novartis and has received research funding from Novartis, Avidity Biosciences, NMD Pharma, and Biogen.
Data and materials availability: The plasmids used in this study are available through AddGene (depositor: David R. Liu). DNA sequencing files can be accessed using the NCBI SRA (SUB# PRJNA871232). All data are available in the main text or the supplementary materials.
References
- 1.Lefebvre S, Bürglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 80, 155–65 (1995). [DOI] [PubMed] [Google Scholar]
- 2.Sugarman EA, Nagan N, Zhu H, Akmaev VR, Zhou Z, Rohlfs EM, Flynn K, Hendrickson BC, Scholl T, Sirko-Osadsa DA, Allitto BA, Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of > 72 400 specimens. Eur. J. Hum. Genet 20, 27–32 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roberts DF, Chavez J, Court SDM, The genetic component in child mortality. Arch. Dis. Child 45, 33–38 (1970). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boda B, Mas C, Giudicelli C, Nepote V, Guimiot F, Levacher B, Zvara A, Santha M, LeGall I, Simonneau M, Survival motor neuron SMN1 and SMN2 gene promoters: Identical sequences and differential expression in neurons and non-neuronal cells. Eur. J. Hum. Genet 12, 729–737 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Rochette CF, Gilbert N, Simard LR, SMN gene duplication and the emergence of the SMN2 gene occurred in distinct hominids: SMN2 is unique to Homo sapiens. Hum. Genet 108, 255–266 (2001). [DOI] [PubMed] [Google Scholar]
- 6.Lorson CL, Hahnen E, Androphy EJ, Wirth B, A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl. Acad. Sci 96, 6307–6311 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AHM, McPherson JD, A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet 8, 1177–1183 (1999). [DOI] [PubMed] [Google Scholar]
- 8.Cho S, Dreyfuss G, A degron created by SMN2 exon 7 skipping is a principal contributor to spinal muscular atrophy severity. Genes Dev. 24, 438–42 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vitte J, Fassier C, Tiziano FD, Dalard C, Soave S, Roblot N, Brahe C, Saugier-Veber P, Bonnefont JP, Melki J, Refined characterization of the expression and stability of the SMN gene products. Am. J. Pathol 171, 1269–1280 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnett BG, Muñoz E, Tandon A, Kwon DY, Sumner CJ, Fischbeck KH, Regulation of SMN Protein Stability. Mol. Cell. Biol 29, 1107–1115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cobben JM, Lemmink HH, Snoeck I, Barth PA, van der Lee JH, de Visser M, Survival in SMA type I: A prospective analysis of 34 consecutive cases. Neuromuscul. Disord 18, 541–544 (2008). [DOI] [PubMed] [Google Scholar]
- 12.Kolb SJ, Coffey CS, Yankey JW, Krosschell K, Arnold WD, Rutkove SB, Swoboda KJ, Reyna SP, Sakonju A, Darras BT, Shell R, Kuntz N, Castro D, Parsons J, Connolly AM, Chiriboga CA, McDonald C, Burnette WB, Werner K, Thangarajh M, Shieh PB, Finanger E, Cudkowicz ME, McGovern MM, McNeil DE, Finkel R, Iannaccone ST, Kaye E, Kingsley A, Renusch SR, McGovern VL, Wang X, Zaworski PG, Prior TW, Burghes AHM, Bartlett A, Kissel JT, Natural history of infantile-onset spinal muscular atrophy. Ann. Neurol 82, 883–891 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendell JR, Al-Zaidy S, Shell R, Arnold WD, Rodino-Klapac LR, Prior TW, Lowes L, Alfano L, Berry K, Church K, Kissel JT, Nagendran S, L’Italien J, Sproule DM, Wells C, Cardenas JA, Heitzer MD, Kaspar A, Corcoran S, Braun L, Likhite S, Miranda C, Meyer K, Foust KD, Burghes AHM, Kaspar BK, Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med 377, 1713–1722 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Ottesen EW, ISS-N1 makes the first FDA-approved drug for spinal muscular atrophy. Transl. Neurosci 8 (2017), pp. 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoy SM, Onasemnogene Abeparvovec: First Global Approval. 79, 1255–1262 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Kernochan LE, Russo ML, Woodling NS, Huynh TN, Avila AM, Fischbeck KH, Sumner CJ, The role of histone acetylation in SMN gene expression, doi: 10.1093/hmg/ddi130. [DOI] [PubMed] [Google Scholar]
- 17.d’Ydewalle C, Ramos DM, Pyles NJ, Ng SY, Gorz M, Pilato CM, Ling K, Kong L, Ward AJ, Rubin LL, Rigo F, Bennett CF, Sumner CJ, The Antisense Transcript SMN-AS1 Regulates SMN Expression and Is a Novel Therapeutic Target for Spinal Muscular Atrophy. Neuron. 93, 66–79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo CJ, Maier VK, Davey R, Brennan J, Li G, Brothers J, Schwartz B, Gordo S, Kasper A, Okamoto TR, Johansson HE, Mandefro B, Sareen D, Bialek P, Chau BN, Bhat B, Bullough D, Barsoum J, Gene activation of SMN by selective disruption of lncRNA-mediated recruitment of PRC2 for the treatment of spinal muscular atrophy. Proc. Natl. Acad. Sci. U. S. A 114, E1509–E1518 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cucchiarini M, Madry H, Terwilliger EF, Enhanced expression of the central survival of motor neuron (SMN) protein during the pathogenesis of osteoarthritis. J. Cell. Mol. Med 18, 115–124 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blauw HM, Barnes CP, Van Vught PWJ, Van Rheenen W, Verheul M, Cuppen E, Veldink JH, Van Den Berg LH, SMN1 gene duplications are associated with sporadic ALS. Neurology. 78, 776–780 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Alstyne M, Tattoli I, Delestrée N, Recinos Y, Workman E, Shihabuddin LS, Zhang C, Mentis GZ, Pellizzoni L, Gain of toxic function by long-term AAV9-mediated SMN overexpression in the sensorimotor circuit. Nat. Neurosci, 1–11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chaytow H, Faller KME, Huang YT, Gillingwater TH, Spinal muscular atrophy: From approved therapies to future therapeutic targets for personalized medicine. Cell reports. Med 2 (2021), doi: 10.1016/J.XCRM.2021.100346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiriboga CA, Swoboda KJ, Darras BT, Iannaccone ST, Montes J, De Vivo DC, Norris DA, Bennett CF, Bishop KM, Results from a phase 1 study of nusinersen (ISIS-SMNRx) in children with spinal muscular atrophy. Neurology. 86, 890 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baranello G, Darras BT, Day JW, Deconinck N, Klein A, Masson R, Mercuri E, Rose K, El-Khairi M, Gerber M, Gorni K, Khwaja O, Kletzl H, Scalco RS, Seabrook T, Fontoura P, Servais L, Risdiplam in Type 1 Spinal Muscular Atrophy. N. Engl. J. Med 384, 915–923 (2021). [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J, Correlation between severity and SMN protein level in spinal muscular atrophy (1997). [DOI] [PubMed] [Google Scholar]
- 26.Coovert DD, Le TT, McAndrew PE, Strasswimmer J, Crawford TO, Mendell JR, Coulson SE, Androphy EJ, Prior TW, Burghes AHM, The Survival Motor Neuron Protein in Spinal Muscular Atrophy. Hum. Mol. Genet 6, 1205–1214 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Patrizi AL, Tiziano F, Zappata S, Donati MA, Neri G, Brahe C, SMN protein analysis in fibroblast, amniocyte and CVS cultures from spinal muscular atrophy patients and its relevance for diagnosis (1999). [DOI] [PubMed] [Google Scholar]
- 28.Ramos DM, d’Ydewalle C, Gabbeta V, Dakka A, Klein SK, Norris DA, Matson J, Taylor SJ, Zaworski PG, Prior TW, Snyder PJ, Valdivia D, Hatem CL, Waters I, Gupte N, Swoboda KJ, Rigo F, Frank Bennett C, Naryshkin N, Paushkin S, Crawford TO, Sumner CJ, Age-dependent SMN expression in disease-relevant tissue and implications for SMA treatment. J. Clin. Invest 129, 4817–4831 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kariya S, Obis T, Garone C, Akay T, Sera F, Iwata S, Homma S, Monani UR, Requirement of enhanced Survival Motoneuron protein imposed during neuromuscular junction maturation. J. Clin. Invest 124, 785–800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ratni H, Ebeling M, Baird J, Bendels S, Bylund J, Chen KS, Denk N, Feng Z, Green L, Guerard M, Jablonski P, Jacobsen B, Khwaja O, Kletzl H, Ko CP, Kustermann S, Marquet A, Metzger F, Mueller B, Naryshkin NA, Paushkin SV, Pinard E, Poirier A, Reutlinger M, Weetall M, Zeller A, Zhao X, Mueller L, Discovery of Risdiplam, a Selective Survival of Motor Neuron-2 (SMN2) Gene Splicing Modifier for the Treatment of Spinal Muscular Atrophy (SMA). J. Med. Chem 61, 6501–6517 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Wurster CD, Ludolph AC, Nusinersen for spinal muscular atrophy. Ther. Adv. Neurol. Disord 11 (2018), , doi: 10.1177/1756285618754459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer K, Ferraiuolo L, Schmelzer L, Braun L, McGovern V, Likhite S, Michels O, Govoni A, Fitzgerald J, Morales P, Foust KD, Mendell JR, Burghes AHM, Kaspar BK, Improving single injection CSF delivery of AAV9-mediated gene therapy for SMA: A dose-response study in mice and nonhuman primates. Mol. Ther 23, 477–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Armbruster N, Lattanzi A, Jeavons M, Van Wittenberghe L, Gjata B, Marais T, Martin S, Vignaud A, Voit T, Mavilio F, Barkats M, Buj-Bello A, Efficacy and biodistribution analysis of intracerebroventricular administration of an optimized scAAV9-SMN1 vector in a mouse model of spinal muscular atrophy. Mol. Ther. - Methods Clin. Dev 3, 16060 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bevan AK, Duque S, Foust KD, Morales PR, Braun L, Schmelzer L, Chan CM, McCrate M, Chicoine LG, Coley BD, Porensky PN, Kolb SJ, Mendell JR, Burghes AHM, Kaspar BK, Systemic gene delivery in large species for targeting spinal cord, brain, and peripheral tissues for pediatric disorders. Mol. Ther 19, 1971–1980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomsen G, Burghes AHM, Hsieh C, Do J, Chu BTT, Perry S, Barkho B, Kaufmann P, Sproule DM, Feltner DE, Chung WK, McGovern VL, Hevner RF, Conces M, Pierson CR, Scoto M, Muntoni F, Mendell JR, Foust KD, M Burghes AH, Hsieh C, Do J, T Chu BT, Perry S, Barkho B, Kaufmann P, Sproule DM, Feltner DE, Chung WK, McGovern VL, Hevner RF, Conces M, Pierson CR, Scoto M, Muntoni F, Mendell JR, Foust KD, Biodistribution of onasemnogene abeparvovec DNA, mRNA and SMN protein in human tissue. Nat. Med. 2021 2710 27, 1701–1711 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Das A, Vijayan M, Walton EM, Stafford VG, Fiflis DN, Asokan A, Epigenetic Silencing of Recombinant Adeno-associated Virus Genomes by NP220 and the HUSH Complex. J. Virol 96, e0203921 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greig JA, Breton C, Martins1 KM, Zhu Y, He Z, White J, Bell P, Wang L, Wilson JM, Loss of transgene expression limits liver gene therapy in primates. BioRxiv. 8.5.2017 (2022). [Google Scholar]
- 38.Singh NK, Singh NN, Androphy EJ, Singh RN, Splicing of a critical exon of human Survival Motor Neuron is regulated by a unique silencer element located in the last intron. Mol. Cell. Biol 26, 1333–1346 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR, Antisense Masking of an hnRNP A1/A2 Intronic Splicing Silencer Corrects SMN2 Splicing in Transgenic Mice. Am. J. Hum. Genet 82, 834–848 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li JJ, Lin X, Tang C, Lu YQ, Hu X, Zuo E, Li H, Ying W, Sun Y, Lai LL, Chen HZ, Guo XX, Zhang QJ, Wu S, Zhou C, Shen X, Wang Q, Lin MT, Ma LX, Wang N, Krainer AR, Shi L, Yang H, Chen WJ, Disruption of splicing-regulatory elements using CRISPR/Cas9 to rescue spinal muscular atrophy in human iPSCs and mice. Natl. Sci. Rev 7, 92–101 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou M, Tang S, Duan N, Xie M, Li Z, Feng M, Wu L, Hu Z, Liang D, Targeted-Deletion of a Tiny Sequence via Prime Editing to Restore SMN Expression. Int. J. Mol. Sci 23 (2022), doi: 10.3390/IJMS23147941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen* M, Arbab* M, Hsu JY, Worstell D, Culbertson SJ, Krabbe O, Cassa A, Liu DR, Gifford DK, Sherwood RI, Predictable and precise template-free CRISPR editing of pathogenic variants. Nature. 563, 646–651 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walton RT, Christie KA, Whittaker MN, Kleinstiver BP, Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science (80-. ) 368, 290–296 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller SM, Wang T, Randolph PB, Arbab M, Shen MW, Huang TP, Matuszek Z, Newby GA, Rees HA, Liu DR, Continuous evolution of SpCas9 variants compatible with non-G PAMs, doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chatterjee P, Lee J, Nip L, Koseki SRT, Tysinger E, Sontheimer EJ, Jacobson JM, Jakimo N, A Cas9 with PAM recognition for adenine dinucleotides. Nat. Commun 11, 1–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O, Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science (80-. ) 361, 1259–1262 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez-Muela N, Litterman NK, Norabuena EM, Mull JL, Galazo MJ, Sun C, Ng SY, Makhortova NR, White A, Lynes MM, Chung WK, Davidow LS, Macklis JD, Rubin LL, Single-Cell Analysis of SMN Reveals Its Broader Role in Neuromuscular Disease. Cell Rep. 18, 1484–1498 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolstencroft EC, Mattis V, Bajer AA, Young PJ, Lorson CL, A non-sequence-specific requirement for SMN protein activity: the role of aminoglycosides in inducing elevated SMN protein levels, doi: 10.1093/hmg/ddi131. [DOI] [PubMed] [Google Scholar]
- 49.Madocsai C, Lim SR, Geib T, Lam BJ, Hertel KJ, Correction of SMN2 Pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol. Ther 12, 1013–1022 (2005). [DOI] [PubMed] [Google Scholar]
- 50.Arbab M, Shen MW, Mok B, Wilson C, Matuszek Ż, Cassa CA, Liu DR, Determinants of Base Editing Outcomes from Target Library Analysis and Machine Learning. Cell. 182, 463–480.e30 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh NN, Singh RN, Androphy EJ, Modulating role of RNA structure in alternative splicing of a critical exon in the spinal muscular atrophy genes. Nucleic Acids Res. 35, 371–389 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O, Shi X, Gao L, Abudayyeh OO, Gootenberg JS, Holmes B, Zhang F, Ishiguro S, Mori H, Tanaka M, Seki M, Aburatani H, Noda T, Oura S, Ikawa M, Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science (80-. ) 361, 1259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oakes BL, Fellmann C, Rishi H, Taylor KL, Ren SM, Nadler DC, Yokoo R, Arkin AP, Doudna JA, Savage DF, CRISPR-Cas9 Circular Permutants as Programmable Scaffolds for Genome Modification. Cell. 176, 254–267.e16 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang TP, Zhao KT, Miller SM, Gaudelli NM, Oakes BL, Fellmann C, Savage DF, Liu DR, Circularly permuted and PAM-modified Cas9 variants broaden the targeting scope of base editors. Nat. Biotechnol (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Le TT, Pham LT, Butchbach MER, Zhang HL, Monani UR, Coovert DD, Gavrilina TO, Xing L, Bassell GJ, Burghes AHM, SMNΔ7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet 14, 845–857 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Singh NN, Androphy EJ, Singh RN, An extended inhibitory context causes skipping of exon 7 of SMN2 in spinal muscular atrophy. Biochem. Biophys. Res. Commun 315, 381–388 (2004). [DOI] [PubMed] [Google Scholar]
- 57.Cartegni L, Hastings ML, Calarco JA, De Stanchina E, Krainer AR, Determinants of Exon 7 Splicing in the Spinal Muscular Atrophy Genes, SMN1 and SMN2. Am. J. Hum. Genet 78, 63 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schrank B, Götz R, Gunnersen JM, Ure JM, V Toyka K, Smith AG, Sendtner M, Gotz R, Gunnersen JM, Ure JM, V Toyka K, Smith AG, Sendtner M, Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. U. S. A 94, 9920–9925 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Monani UR, Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron. 48, 885–896 (2005). [DOI] [PubMed] [Google Scholar]
- 60.Singh RN, Howell MD, Ottesen EW, Singh NN, Diverse role of survival motor neuron protein. Biochim. Biophys. Acta - Gene Regul. Mech 1860 (2017), pp. 299–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Briese M, Esmaeili B, Fraboulet S, Burt EC, Christodoulou S, Towers PR, Davies KE, Sattelle DB, Deletion of smn-1, the Caenorhabditis elegans ortholog of the spinal muscular atrophy gene, results in locomotor dysfunction and reduced lifespan. Hum. Mol. Genet 18, 97–104 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Richter MF, Zhao KT, Eton E, Lapinaite A, Newby GA, Thuronyi BW, Wilson C, Koblan LW, Zeng J, Bauer DE, Doudna JA, Liu DR, Phage-assisted evolution of an adenine base editor with improved Cas domain compatibility and activity. Nat. Biotechnol (2020), doi: 10.1038/s41587-020-0453-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doman JL, Raguram A, Newby GA, Liu DR, Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol, doi: 10.1038/s41587-020-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rees HA, Wilson C, Doman JL, Liu DR, Analysis and minimization of cellular RNA editing by DNA adenine base editors. Sci. Adv 5 (2019), doi: 10.1126/sciadv.aax5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu Y, Leete TC, Born DA, Young L, Barrera LA, Lee SJ, Rees HA, Ciaramella G, Gaudelli NM, Cytosine base editors with minimized unguided DNA and RNA off-target events and high on-target activity. Nat. Commun 11, 1–10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhou C, Sun Y, Yan R, Liu Y, Zuo E, Gu C, Han L, Wei Y, Hu X, Zeng R, Li Y, Zhou H, Guo F, Yang H, Off-target RNA mutation induced by DNA base editing and its elimination by mutagenesis. Nature. 571, 275–278 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, Cole MA, Liu DR, Joung JK, Bauer DE, Pinello L, CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat. Biotechnol 37 (2019), pp. 224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tsai SQ, Nguyen NT, Malagon-Lopez J, V Topkar V, Aryee MJ, Joung JK, CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods. 14, 607–614 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhao Y, Dai Z, Liang Y, Yin M, Ma K, He M, Ouyang H, Teng C-B, Sequence-specific inhibition of microRNA via CRISPR/CRISPRi system. Sci. Rep 4, 3943 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Foust KD, Nurre E, Montgomery CL, Hernandez A, Chan CM, Kaspar BK, Curtis M, Kaspar BK, Intravascular AAV9 preferentially targets neonatal-neurons and adult-astrocytes in CNS. Nat. Biotechnol 27, 59–65 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hammond SL, Leek AN, Richman EH, Tjalkens RB, Cellular selectivity of AAV serotypes for gene delivery in neurons and astrocytes by neonatal intracerebroventricular injection. PLoS One. 12 (2017), doi: 10.1371/journal.pone.0188830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mathiesen SN, Lock JL, Schoderboeck L, Abraham WC, Hughes SM, CNS Transduction Benefits of AAV-PHP.eB over AAV9 Are Dependent on Administration Route and Mouse Strain. Mol. Ther. Methods Clin. Dev 19, 447 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Arnold WD, McGovern VL, Sanchez B, Li J, Corlett KM, Kolb SJ, Rutkove SB, Burghes AH, The neuromuscular impact of symptomatic SMN restoration in a mouse model of spinal muscular atrophy. Neurobiol. Dis 87, 116–123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Levy JM, Yeh WH, Pendse N, Davis JR, Hennessey E, Butcher R, Koblan LW, Comander J, Liu Q, Liu DR, Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng 4, 97–110 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schuster DJ, Dykstra JA, Riedl MS, Kitto KF, Belur LR, Scott McIvor R, Elde RP, Fairbanks CA, Vulchanova L, McIvor RS, Elde RP, Fairbanks CA, Vulchanova L, Biodistribution of adeno-associated virus serotype 9 (AAV9) vector after intrathecal and intravenous delivery in mouse. Front. Neuroanat 8, 42 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swiech L, Heidenreich M, Banerjee A, Habib N, Li Y, Trombetta J, Sur M, Zhang F, In vivo interrogation of gene function in the mammalian brain using CRISPR-Cas9. Nat. Biotechnol 33, 102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cartegni L, Krainer AR, Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002 304 30, 377–384 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Groen EJN, Perenthaler E, Courtney NL, Jordan CY, Shorrock HK, Van Der Hoorn D, Huang YT, Murray LM, Viero G, Gillingwater TH, Temporal and tissue-specific variability of SMN protein levels in mouse models of spinal muscular atrophy. Hum. Mol. Genet 27, 2851–2862 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karlsson M, Zhang C, Méar L, Zhong W, Digre A, Katona B, Sjöstedt E, Butler L, Odeberg J, Dusart P, Edfors F, Oksvold P, von Feilitzen K, Zwahlen M, Arif M, Altay O, Li X, Ozcan M, Mardonoglu A, Fagerberg L, Mulder J, Luo Y, Ponten F, Uhlén M, Lindskog C, A single–cell type transcriptomics map of human tissues. Sci. Adv 7, eabh2169 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Akcakaya P, Bobbin ML, Guo JA, Malagon-Lopez J, Clement K, Garcia SP, Fellows MD, Porritt MJ, Firth MA, Carreras A, Baccega T, Seeliger F, Bjursell M, Tsai SQ, Nguyen NT, Nitsch R, Mayr LM, Pinello L, Bohlooly YM, Aryee MJ, Maresca M, Joung JK, Bohlooly-Y M, Aryee MJ, Maresca M, Joung JK, In vivo CRISPR editing with no detectable genome-wide off-target mutations. Nature. 561, 416–419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis JR, Wang X, Witte IP, Huang TP, Levy JM, Raguram A, Banskota S, Seidah NG, Musunuru K, Liu DR, Efficient in vivo base editing via single adeno-associated viruses with size-optimized genomes encoding compact adenine base editors. Nat. Biomed. Eng 6, 1272–1283 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rothgangl T, Dennis MK, Lin PJC, Oka R, Witzigmann D, Villiger L, Qi W, Hruzova M, Kissling L, Lenggenhager D, Borrelli C, Egli S, Frey N, Bakker N, Walker JA, Kadina AP, Victorov DV, Pacesa M, Kreutzer S, Kontarakis Z, Moor A, Jinek M, Weissman D, Stoffel M, van Boxtel R, Holden K, Pardi N, Thöny B, Häberle J, Tam YK, Semple SC, Schwank G, In vivo adenine base editing of PCSK9 in macaques reduces LDL cholesterol levels. Nat. Biotechnol 39, 949–957 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.David Arnold W, Porensky PN, Mcgovern VL, Iyer CC, Duque S, Li X, Meyer K, Schmelzer L, Kaspar BK, Kolb SJ, Kissel JT, Burghes AHM, Electrophysiological biomarkers in spinal muscular atrophy: proof of concept. Ann. Clin. Transl. Neurol 1, 34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tscherter A, Rüsch CT, Baumann D, Enzmann C, Hasselmann O, Jacquier D, Jung HH, Kruijshaar ME, Kuehni CE, Neuwirth C, Stettner GM, Klein A, Baumann D, Enzmann C, Hasselmann O, Jacquier D, Jung HH, Klein A, Kruijshaar ME, Kuehni CE, Lötscher N, Neuwirth C, Ramelli GP, Ripellino P, Scheidegger O, Stettner GM, Tscherter A, Wille D-A, Evaluation of real-life outcome data of patients with spinal muscular atrophy treated with nusinersen in Switzerland. Neuromuscul. Disord 0 (2022), doi: 10.1016/J.NMD.2022.02.001/ATTACHMENT/96B80B10-FB76-49C7-9B7B-571ECC277841/MMC1.DOCX. [DOI] [PubMed] [Google Scholar]
- 85.Dangouloff T, Servais L, Clinical Evidence Supporting Early Treatment Of Patients With Spinal Muscular Atrophy: Current Perspectives. Ther. Clin. Risk Manag 15, 1153 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strauss KA, Farrar MA, Muntoni F, Saito K, Mendell JR, Servais L, McMillan HJ, Finkel RS, Swoboda KJ, Kwon JM, Zaidman CM, Chiriboga CA, Iannaccone ST, Krueger JM, Parsons JA, Shieh PB, Kavanagh S, Tauscher-Wisniewski S, McGill BE, Macek TA, Onasemnogene abeparvovec for presymptomatic infants with two copies of SMN2 at risk for spinal muscular atrophy type 1: the Phase III SPR1NT trial. Nat. Med. 2022 287 28, 1381–1389 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.De Vivo DC, Bertini E, Swoboda KJ, Hwu WL, Crawford TO, Finkel RS, Kirschner J, Kuntz NL, Parsons JA, Ryan MM, Butterfield RJ, Topaloglu H, Ben-Omran T, Sansone VA, Jong YJ, Shu F, Staropoli JF, Kerr D, Sandrock AW, Stebbins C, Petrillo M, Braley G, Johnson K, Foster R, Gheuens S, Bhan I, Reyna SP, Fradette S, Farwell W, Nusinersen initiated in infants during the presymptomatic stage of spinal muscular atrophy: Interim efficacy and safety results from the Phase 2 NURTURE study. Neuromuscul. Disord 29, 842–856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Robbins KL, Glascock JJ, Osman EY, Miller MR, Lorson CL, Defining the therapeutic window in a severe animal model of spinal muscular atrophy. Hum. Mol. Genet 23, 4559–4568 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Le TT, McGovern VL, Alwine IE, Wang X, Massoni-Laporte A, Rich MM, Burghes AHM, Temporal requirement for high SMN expression in SMA mice. Hum. Mol. Genet 20, 3578–3591 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Flurkey K, Currer JM, Harrison DE, Mouse Models in Aging Research. Mouse Biomed. Res 3, 637–672 (2007). [Google Scholar]
- 91.Mercuri E, Darras BT, Chiriboga CA, Day JW, Campbell C, Connolly AM, Iannaccone ST, Kirschner J, Kuntz NL, Saito K, Shieh PB, Tulinius M, Mazzone ES, Montes J, Bishop KM, Yang Q, Foster R, Gheuens S, Bennett CF, Farwell W, Schneider E, De Vivo DC, Finkel RS, Nusinersen versus Sham Control in Later-Onset Spinal Muscular Atrophy. N. Engl. J. Med 378, 625–635 (2018). [DOI] [PubMed] [Google Scholar]
- 92.Finkel RS, Day JW, Darras BT, Kuntz NL, Connolly AM, Crawford T, Butterfield RJ, Shieh PB, Tennekoon G, Iannaccone ST, Meriggioli M, Tauscher-Wisniewski S, Shoffner J, Ogrinc FG, Kavanagh S, Kernbauer E, Whittle J, Sproule DM, Feltner DE, Mendell JR, One-Time Intrathecal (IT) Administration of AVXS-101 IT Gene-Replacement Therapy for Spinal Muscular Atrophy: Phase 1 Study (STRONG) (2493). Neurology. 94 (2020). [Google Scholar]
- 93.Lutz CM, Kariya S, Patruni S, Osborne MA, Liu D, Henderson CE, Li DK, Pellizzoni L, Rojas J, Valenzuela DM, Murphy AJ, Winberg ML, Monani UR, Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J. Clin. Invest 121, 3029–3041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Z, Ma HI, Li J, Sun L, Zhang J, Xiao X, Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther. 10, 2105–2111 (2003). [DOI] [PubMed] [Google Scholar]
- 95.Hauck B, Zhao W, High K, Xiao W, Intracellular Viral Processing, Not Single-Stranded DNA Accumulation, Is Crucial for Recombinant Adeno-Associated Virus Transduction. J. Virol 78, 13678–13686 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McCarty DM, Monahan PE, Samulski RJ, Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 8, 1248–1254 (2001). [DOI] [PubMed] [Google Scholar]
- 97.Feng Z, Ling KKY, Zhao X, Zhou C, Karp G, Welch EM, Naryshkin N, Ratni H, Chen KS, Metzger F, Paushkin S, Weetall M, Ko CP, Pharmacologically induced mouse model of adult spinal muscular atrophy to evaluate effectiveness of therapeutics after disease onset. Hum. Mol. Genet 25, 964–975 (2016). [DOI] [PubMed] [Google Scholar]
- 98.Naryshkin NA, Weetall M, Dakka A, Narasimhan J, Zhao X, Feng Z, Ling KKY, Karp GM, Qi H, Woll MG, Chen G, Zhang N, Gabbeta V, Vazirani P, Bhattacharyya A, Furia B, Risher N, Sheedy J, Kong R, Ma J, Turpoff A, Lee CS, Zhang X, Moon YC, Trifillis P, Welch EM, Colacino JM, Babiak J, Almstead NG, Peltz SW, Eng LA, Chen KS, Mull JL, Lynes MS, Rubin LL, Fontoura P, Santarelli L, Haehnke D, McCarthy KD, Schmucki R, Ebeling M, Sivaramakrishnan M, Ko CP, Paushkin SV, Ratni H, Gerlach I, Ghosh A, Metzger F, SMN2 splicing modifiers improve motor function and longevity in mice with spinal muscular atrophy. Science (80-. ) 345, 688–693 (2014). [DOI] [PubMed] [Google Scholar]
- 99.Passini MA, Bu J, Richards AM, Kinnecom C, Sardi SP, Stanek LM, Hua Y, Rigo F, Matson J, Hung G, Kaye EM, Shihabuddin LS, Krainer AR, Bennett CF, Cheng SH, Antisense Oligonucleotides Delivered to the Mouse CNS Ameliorate Symptoms of Severe Spinal Muscular Atrophy. Sci. Transl. Med 3, 72ra18–72ra18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Finkel RS, Mercuri E, Darras BT, Connolly AM, Kuntz NL, Kirschner J, Chiriboga CA, Saito K, Servais L, Tizzano E, Topaloglu H, Tulinius M, Montes J, Glanzman AM, Bishop K, Zhong ZJ, Gheuens S, Bennett CF, Schneider E, Farwell W, De Vivo DC, Nusinersen versus Sham Control in Infantile-Onset Spinal Muscular Atrophy. N. Engl. J. Med 377, 1723–1732 (2017). [DOI] [PubMed] [Google Scholar]
- 101.Wadman RI, Stam M, Jansen MD, Der Van Weegen Y, Wijngaarde CA, Harschnitz O, Sodaar P, Braun KPJ, Dooijes D, Lemmink HH, Van Den Berg LH, Van Ludo Der Pol W, A comparative study of SMN protein and mRNA in blood and fibroblasts in patients with spinal muscular atrophy and healthy controls. PLoS One. 11, e0167087–e0167087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, Zhang F, Koonin EV, Discovery and Functional Characterization of Diverse Class 2 CRISPR-Cas Systems. Mol. Cell 60, 385–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Butchbach MER, Copy number variations in the survival motor neuron genes: Implications for spinal muscular atrophy and other neurodegenerative diseases. Front. Mol. Biosci 3 (2016), , doi: 10.3389/fmolb.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Kato T, Hara S, Goto Y, Ogawa Y, Okayasu H, Kubota S, Tamano M, Terao M, Takada S, Creation of mutant mice with megabase-sized deletions containing custom-designed breakpoints by means of the CRISPR/Cas9 system. Sci. Rep 7 (2017), doi: 10.1038/S41598-017-00140-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR, Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature. 551, 464–471 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR, Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 533, 420–424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gaudelli NM, Lam DK, Rees HA, Solá-Esteves NM, Barrera LA, Born DA, Edwards A, Gehrke JM, Lee SJ, Liquori AJ, Murray R, Packer MS, Rinaldi C, Slaymaker IM, Yen J, Young LE, Ciaramella G, Directed evolution of adenine base editors with increased activity and therapeutic application. 38, 892–900 (2020). [DOI] [PubMed] [Google Scholar]
- 108.McAndrew PE, Parsons DW, Simard LR, Rochette C, Ray PN, Mendell JR, Prior TW, Burghes AHM, Identification of Proximal Spinal Muscular Atrophy Carriers and Patients by Analysis of SMNT and SMNC Gene Copy Number. Am. J. Hum. Genet 60, 1411–1422 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mailman MD, Heinz JW, Papp AC, Snyder PJ, Sedra MS, Wirth B, Burghes AHM, Prior TW, Molecular analysis of spinal muscular atrophy and modification of the phenotype by SMN2. Genet. Med 4, 20–26 (2002). [DOI] [PubMed] [Google Scholar]
- 110.Ruhno C, McGovern VL, Avenarius MR, Snyder PJ, Prior TW, Nery FC, Muhtaseb A, Roggenbuck JS, Kissel JT, Sansone VA, Siranosian JJ, Johnstone AJ, Nwe PH, Zhang RZ, Swoboda KJ, Burghes AHM, Complete sequencing of the SMN2 gene in SMA patients detects SMN gene deletion junctions and variants in SMN2 that modify the SMA phenotype. Hum. Genet. 2019 1383 138, 241–256 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Calucho M, Bernal S, Alías L, March F, Venceslá A, Rodríguez-Álvarez FJ, Aller E, Fernández RM, Borrego S, Millán JM, Hernández-Chico C, Cuscó I, Fuentes-Prior P, Tizzano EF, Correlation between SMA type and SMN2 copy number revisited: An analysis of 625 unrelated Spanish patients and a compilation of 2834 reported cases. Neuromuscul. Disord 28, 208–215 (2018). [DOI] [PubMed] [Google Scholar]
- 112.Prior TW, Krainer AR, Hua Y, Swoboda KJ, Snyder PC, Bridgeman SJ, Burghes AHM, Kissel JT, A Positive Modifier of Spinal Muscular Atrophy in the SMN2 Gene. Am. J. Hum. Genet 85, 408–413 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Besse A, Astord S, Marais T, Roda M, Giroux B, Lejeune FX, Relaix F, Smeriglio P, Barkats M, Biferi MG, AAV9-Mediated Expression of SMN Restricted to Neurons Does Not Rescue the Spinal Muscular Atrophy Phenotype in Mice. Mol. Ther 28, 1887–1901 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yu D, Lv M, Chen W, Zhong S, Zhang X, Chen L, Ma T, Tang J, Zhao J, Role of miR-155 in drug resistance of breast cancer. Tumor Biol. 36, 1395–1401 (2015). [DOI] [PubMed] [Google Scholar]
- 115.Bevan AK, Hutchinson KR, Foust KD, Braun L, McGovern VL, Schmelzer L, Ward JG, Petruska JC, Lucchesi PA, Burghes AHMM, Kaspar BK, Early heart failure in the SMNDelta7 model of spinal muscular atrophy and correction by postnatal scAAV9-SMN delivery. Hum. Mol. Genet 19, 3895–3905 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shababi M, Habibi J, Yang HT, Vale SM, Sewell WA, Lorson CL, Cardiac defects contribute to the pathology of spinal muscular atrophy models. Hum. Mol. Genet 19, 4059–4071 (2010). [DOI] [PubMed] [Google Scholar]
- 117.McGovern VL, Iyer CC, Arnold WD, Gombash SE, Zaworski PG, Blatnik AJ, Foust KD, Burghes AHMM, David Arnold W, Gombash SE, Zaworski PG, Blatnik AJ, Foust KD, Burghes AHMM, SMN expression is required in motor neurons to rescue electrophysiological deficits in the SMNΔ7 mouse model of SMA. Hum. Mol. Genet 24, 5524–5541 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hua Y, Sahashi K, Rigo F, Hung G, Horev G, Bennett CF, Krainer AR, Peripheral SMN restoration is essential for long-term rescue of a severe spinal muscular atrophy mouse model. Nature. 478, 123–126 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lipnick SL, Agniel DM, Aggarwal R, Makhortova NR, Finlayson SG, Brocato A, Palmer N, Darras BT, Kohane I, Rubin LL, Systemic nature of spinal muscular atrophy revealed by studying insurance claims. PLoS One. 14, e0213680 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Villiger L, Grisch-Chan HM, Lindsay H, Ringnalda F, Pogliano CB, Allegri G, Fingerhut R, Haberle J, Matos J, Robinson MD, Thony B, Schwank G, Häberle J, Matos J, Robinson MD, Thöny B, Schwank G, Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat Med. 24, 1519–1525 (2018). [DOI] [PubMed] [Google Scholar]
- 121.Yeh WH, Chiang H, Rees HA, Edge ASB, Liu DR, In vivo base editing of post-mitotic sensory cells. Nat. Commun 9 (2018), doi: 10.1038/s41467-018-04580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kuzmin DA, Shutova MV, Johnston NR, Smith OP, Fedorin VV, Kukushkin YS, van der Loo JCM, Johnstone EC, The clinical landscape for AAV gene therapies. Nat. Rev. Drug Discov 20, 173–174 (2021). [DOI] [PubMed] [Google Scholar]
- 123.Arbab M, Srinivasan S, Hashimoto T, Geijsen N, Sherwood RII, Cloning-free CRISPR. Stem Cell Reports. 5, 908–917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wichterle H, Lieberam I, Porter JA, Jessell TM, Directed differentiation of embryonic stem cells into motor neurons. Cell. 110, 385–397 (2002). [DOI] [PubMed] [Google Scholar]
- 125.Wichterle H, Peljto M, Differentiation of Mouse Embryonic Stem Cells to Spinal Motor Neurons. Curr. Protoc. Stem Cell Biol 5 (2008), doi: 10.1002/9780470151808.sc01h01s5. [DOI] [PubMed] [Google Scholar]
- 126.Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R, Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 127.Bray NL, Pimentel H, Melsted P, Pachter L, Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 128.Pujar S, O’Leary NA, Farrell CM, Loveland JE, Mudge JM, Wallin C, Girón CG, Diekhans M, Barnes I, Bennett R, Berry AE, Cox E, Davidson C, Goldfarb T, Gonzalez JM, Hunt T, Jackson J, Joardar V, Kay MP, Kodali VK, Martin FJ, McAndrews M, McGarvey KM, Murphy M, Rajput B, Rangwala SH, Riddick LD, Seal RL, Suner MM, Webb D, Zhu S, Aken BL, Bruford EA, Bult CJ, Frankish A, Murphy T, Pruitt KD, Consensus coding sequence (CCDS) database: A standardized set of human and mouse protein-coding regions supported by expert curation. Nucleic Acids Res. 46, D221–D228 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.DIroma MA, Ciaccia L, Pesole G, Picardi E, Elucidating the editome: Bioinformatics approaches for RNA editing detection. Brief. Bioinform 20, 436–447 (2019). [DOI] [PubMed] [Google Scholar]
- 130.Lazzarotto CR, Nguyen NT, Tang X, Malagon-Lopez J, Guo JA, Aryee MJ, Joung JK, Tsai SQ, Defining CRISPR–Cas9 genome-wide nuclease activities with CIRCLE-seq. Nat. Protoc 13, 2615–2642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Porensky PN, Mitrpant C, McGovern VL, Bevan AK, Foust KD, Kaspar BK, Wilton SD, Burghes AHM, A single administration of morpholino antisense oligomer rescues spinal muscular atrophy in mouse. Hum. Mol. Genet 21, 1625–1638 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Arnold WD, Sheth KA, Wier CG, Kissel JT, Burghes AH, Kolb SJ, Electrophysiological Motor Unit Number Estimation (MUNE) Measuring Compound Muscle Action Potential (CMAP) in Mouse Hindlimb Muscles. J. Vis. Exp, 52899 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cobb MS, Rose FF, Rindt H, Glascock JJ, Shababi M, Miller MR, Osman EY, Yen PF, Garcia ML, Martin BR, Wetz MJ, Mazzasette C, Feng Z, Ko CP, Lorson CL, Development and characterization of an SMN2-based intermediate mouse model of spinal muscular atrophy. Hum. Mol. Genet (2013), doi: 10.1093/hmg/ddt037. [DOI] [PubMed] [Google Scholar]
- 134.Farooq F, Balabanian S, Liu X, Holcik M, MacKenzie A, p38 Mitogen-activated protein kinase stabilizes SMN mRNA through RNA binding protein HuR. Hum. Mol. Genet 18, 4035–4045 (2009). [DOI] [PubMed] [Google Scholar]
- 135.Singh RN, Singh NN, "Mechanism of splicing regulation of spinal muscular atrophy genes" in Advances in Neurobiology (2018; file:///Users/marbab/Library/Application Support/Mendeley Desktop/Downloaded/Singh, Singh - Unknown - Mechanism of Splicing Regulation of Spinal Muscular Atrophy Genes.pdf), vol. 20, pp. 31–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yoshimoto S, Harahap NIF, Hamamura Y, Ar Rochmah M, Shima A, Morisada N, Shinohara M, Saito T, Saito K, Lai PS, Matsuo M, Awano H, Morioka I, Iijima K, Nishio H, Alternative splicing of a cryptic exon embedded in intron 6 of SMN1 and SMN2. Hum. Genome Var 3, 1–3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Seo J, Singh NN, Ottesen EW, Lee BM, Singh RN, A novel human-specific splice isoform alters the critical C-terminus of Survival Motor Neuron protein. Sci. Rep 6, 1–14 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ottesen EW, Luo D, Seo J, Singh NN, Singh RN, Human Survival Motor Neuron genes generate a vast repertoire of circular RNAs. Nucleic Acids Res. 47, 2884–2905 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Allen F, Crepaldi L, Alsinet C, Strong AJ, Kleshchevnikov V, De Angeli P, Páleníková P, Khodak A, Kiselev V, Kosicki M, Bassett AR, Harding H, Galanty Y, Muñoz-Martínez F, Metzakopian E, Jackson SP, Parts L, Predicting the mutations generated by repair of Cas9-induced double-strand breaks. Nat. Biotechnol 37, 64–82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kim HK, Min S, Song M, Jung S, Choi JW, Kim Y, Lee S, Yoon S, Kim H, Deep learning improves prediction of CRISPR-Cpf1 guide RNA activity. Nat. Biotechnol 36, 239–241 (2018). [DOI] [PubMed] [Google Scholar]
- 141.Kim HK, Kim Y, Lee S, Min S, Bae JY, Choi JW, Park J, Jung D, Yoon S, Kim HH, SpCas9 activity prediction by DeepSpCas9, a deep learning–based model with high generalization performance. Sci. Adv 5, eaax9249–eaax9249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, Virgin HW, Listgarten J, Root DE, Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 34, 184–191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR, Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 556, 57 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chatterjee P, Lee J, Nip L, Koseki SRTT, Tysinger E, Sontheimer EJ, Jacobson JM, Jakimo N, A Cas9 with PAM recognition for adenine dinucleotides. Nat. Commun 11, 1–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lin X, Chen H, Lu YQ, Hong S, Hu X, Gao Y, Lai LL, Li JJ, Wang Z, Ying W, Ma L, Wang N, Zuo E, Yang H, Chen WJ, Base editing-mediated splicing correction therapy for spinal muscular atrophy. Cell Res. 30 (2020), pp. 548–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lapinaite A, Knott GJ, Palumbo CM, Lin-Shiao E, Richter MF, Zhao KT, Beal PA, Liu DR, Doudna JA, DNA capture by a CRISPR-Cas9-guided adenine base editor. Science (80-. ) 369, 566–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nagaoka SI, Nakaki F, Miyauchi H, Nosaka Y, Ohta H, Yabuta Y, Kurimoto K, Hayashi K, Nakamura T, Yamamoto T, Saitou M, ZGLP1 is a determinant for the oogenic fate in mice. Science (80-. ) 367 (2020), doi: 10.1126/science.aay5947. [DOI] [PubMed] [Google Scholar]
- 148.Lee HK, Smith HE, Liu C, Willi M, Hennighausen L, Cytosine base editor 4 but not adenine base editor generates off-target mutations in mouse embryos. Commun. Biol 3, 1–6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Doman JL, Raguram A, Newby GA, Liu DR, Evaluation and minimization of Cas9-independent off-target DNA editing by cytosine base editors. Nat. Biotechnol 38, 620–628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Reichart D, Newby GA., Wakimoto H, Lun M, Gorham JM, Curran JJ, Raguram A, DeLaughter DM, Conner DA, Marsiglia JDC, Kohli S, Chmatal L, Page DC, Zabaleta N, Vandenberghe L, Liu DR, J. G. Seidman1, C. Seidman, Efficient in vivo Genome Editing Prevents Hypertrophic Cardiomyopathy in Mice. Nat. Med in press (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huang X, Guo H, Tammana S, Jung YC, Mellgren E, Bassi P, Cao Q, Tu ZJ, Kim YC, Ekker SC, Wu X, Wang SM, Zhou X, Gene transfer efficiency and genome-wide integration profiling of sleeping beauty, Tol2, and PiggyBac transposons in human primary t cells. Mol. Ther 18, 1803–1813 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Briggs JA, Li VC, Lee S, Woolf CJ, Klein A, Kirschner MW, Mouse embryonic stem cells can differentiate via multiple paths to the same state. Elife. 6, e26945 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ryu SM, Koo T, Kim K, Lim K, Baek G, Kim ST, Kim HS, Kim DE, Lee H, Chung E, Kim JS, Adenine base editing in mouse embryos and an adult mouse model of Duchenne muscular dystrophy. Nat. Biotechnol 36, 536–539 (2018). [DOI] [PubMed] [Google Scholar]
- 154.Zettler J, Schütz V, Mootz HD, The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 583, 909–914 (2009). [DOI] [PubMed] [Google Scholar]
- 155.Lim CKW, Gapinske M, Brooks AK, Woods WS, Powell JE, Zeballos C. MA, Winter J, Perez-Pinera P, Gaj T, Treatment of a Mouse Model of ALS by In Vivo Base Editing. Mol. Ther (2020), doi: 10.1016/j.ymthe.2020.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Banskota S, Raguram A, Suh S, Du SW, Davis JR, Choi EH, Wang X, Nielsen SC, Newby GA, Randolph PB, Osborn MJ, Musunuru K, Palczewski K, Liu DR, Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell. 185, 250–265.e16 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Lutz CM, Kariya S, Patruni S, Osborne MA, Liu D, Henderson CE, Li DK, Pellizzoni L, Rojas J, Valenzuela DM, Murphy AJ, Winberg ML, Monani UR, Postsymptomatic restoration of SMN rescues the disease phenotype in a mouse model of severe spinal muscular atrophy. J. Clin. Invest 121, 3029–3041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Paushkin S, Gubitz AK, Massenet S, Dreyfuss G, The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol 14, 305–312 (2002). [DOI] [PubMed] [Google Scholar]
- 159.Gubitz AK, Feng W, Dreyfuss G, The SMN complex. Exp. Cell Res 296, 51–56 (2004). [DOI] [PubMed] [Google Scholar]
- 160.Chan YB, Miguel-Aliaga I, Franks C, Thomas N, Trülzsch B, Sattelle DB, Davies KE, van den Heuvel M, Neuromuscular defects in a Drosophila survival motor neuron gene mutant. Hum. Mol. Genet 12, 1367–1376 (2003). [DOI] [PubMed] [Google Scholar]
- 161.Szunyogova E, Zhou H, Maxwell GK, Powis RA, Francesco M, Gillingwater TH, Parson SH, Survival Motor Neuron (SMN) protein is required for normal mouse liver development. Sci. Rep 6 (2016), doi: 10.1038/srep34635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Chaytow H, Huang YT, Gillingwater TH, Faller KME, The role of survival motor neuron protein (SMN) in protein homeostasis. Cell. Mol. Life Sci 75 (2018), pp. 3877–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Blatnik AJ, McGovern VL, Le TT, Iyer CC, Kaspar BK, Burghes AHM, Conditional deletion of SMN in cell culture identifies functional SMN alleles. Hum. Mol. Genet 29, 3477–3492 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sivanesan S, Howell MD, Didonato CJ, Singh RN, Antisense oligonucleotide mediated therapy of spinal muscular atrophy. Transl. Neurosci 4 (2013), pp. 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.