Abstract
Objective
Neurodegenerative conditions often manifest radiologically with the appearance of premature aging. Multiple sclerosis (MS) biomarkers related to lesion burden are well developed, but measures of neurodegeneration are less well‐developed. The appearance of premature aging quantified by machine learning applied to structural MRI assesses neurodegenerative pathology. We assess the explanatory and predictive power of “brain age” analysis on disability in MS using a large, real‐world dataset.
Methods
Brain age analysis is predicated on the over‐estimation of predicted brain age in patients with more advanced pathology. We compared the performance of three brain age algorithms in a large, longitudinal dataset (>13,000 imaging sessions from >6,000 individual MS patients). Effects of MS, MS disease course, disability, lesion burden, and DMT efficacy were assessed using linear mixed effects models.
Results
MS was associated with advanced predicted brain age cross‐sectionally and accelerated brain aging longitudinally in all techniques. While MS disease course (relapsing vs. progressive) did contribute to advanced brain age, disability was the primary correlate of advanced brain age. We found that advanced brain age at study enrollment predicted more disability accumulation longitudinally. Lastly, a more youthful appearing brain (predicted brain age less than actual age) was associated with decreased disability.
Interpretation
Brain age is a technically tractable and clinically relevant biomarker of disease pathology that correlates with and predicts increasing disability in MS. Advanced brain age predicts future disability accumulation.
Introduction
Neuroimaging is critical in the diagnosis of neurological illness, including multiple sclerosis (MS). 1 Clinicians use neuroimaging to understand disability, 2 monitor response to therapy, 3 or predict future disease severity. 4 Data reduction strategies attempt to summarize images based on features (e.g., atrophy patterns) but rely on prior anatomical or physiological assumptions which may bias outcomes. In many neurological diseases, the appearance of imaging features consistent with premature or accelerated aging (e.g., atrophy) is appreciated on radiological assessment of individual patients. Machine learning (ML) algorithms can effectively predict age in healthy people from routine neuroimaging datasets. 5 , 6 Advanced predicted “brain age” compared to chronologic age has emerged as a (non‐specific) biomarker of brain pathology. 6 This “brain age gap” has been proposed as a biomarker of MS, 7 , 8 as well as Alzheimer disease, 9 physical fitness, 10 psychosis, 11 and neurological effects of human immunodeficiency virus infection. 12 However, methodological problems related to derived quantities (e.g., brain age gap) have been identified which complicate the interpretation and application of brain age to the study of human disease. 13
Brain age analysis identifies pathological changes manifesting as increased brain age in patients with MS. 6 , 7 , 8 Machine learning assessments of brain age are driven by volume changes (i.e., atrophy) and other structural features which may be measured independently. Brain age analysis confers the advantage of an implicit normative comparison value (i.e., the chronological age of the participant) and reduces a topographically complex process (regional atrophy) into a single scalar value which simplifies statistical and clinical inference. In this study, we utilize neuroimaging data from a multi‐site, international collaboration collecting longitudinal, standardized imaging in MS patients. The Multiple Sclerosis Partners Advancing Technology and Health Solutions (MS PATHS) study is an observational study collecting clinical assessments and standardized, high‐resolution T1 and FLAIR images in MS patients as part of their standard of care. 14 , 15 Images from 6,732 individual MS patients, contributing 13,852 imaging studies, were examined using brain age analysis to understand and make predictions about MS trajectory. We implemented and compared three brain age algorithms to examine the effect of MS, MS disease course (e.g., relapsing vs. progressive), and disability on predicted brain age in cross‐sectional and longitudinal cohorts. Additionally, we examined the power of brain age to predict future disability accumulation.
Methods
The Multiple Sclerosis Partners Advancing Technology and Health Solutions (MS PATHS) dataset
Clinical and imaging data were obtained from the MS PATHS 14 network which comprises multiple sites in the United States and Europe. MS PATHS collects clinical, performance, and imaging data from a large, heterogeneous MS population as part of routine clinical care. Images from healthy control participants were also collected using identical imaging protocols. MS PATHS participants consent to share pseudo‐randomized data with the research sponsor (Biogen) and the network investigators under the supervision of local Institutional Review Boards. Written informed consent was provided. This study was approved by the IRB at Washington University in St. Louis. A total of 13,852 sessions from 6,734 participants were included in these analyses (also see Table S1).
MS PATHS data consist of clinical information including demographics, performance on standardized testing, and standardized 3D T1‐weighted and 3D T2‐FLAIR images (both 1 mm isotropic voxels). 16 These data are described in more detail below.
Clinical characterization of MS PATHS participants
Clinical measurements closest to each imaging session were associated with the imaging data and used for statistical analysis. The following clinical variables were utilized:
MS disease course
Patients characterized themselves as having one of the following courses of MS: relapsing–remitting (RR), secondary progressive (SP), primary progressive (PP), or progressive relapsing (PR). Patients could also identify as clinically isolated syndrome (CIS).
Patient Determined Disease Steps (PDDS)
The PDDS is a self‐reported disability score with a strong correlation (r = 0.78) with the Expanded Disability Status Scale. 17 Patients rate their disability from 0 to 8, with 0 corresponding to normal, 1—mild disability, 2—moderate disability, 3—gait disability, 4—early use of a cane, 5—late use of a cane, 6—bilateral support, 7—wheelchair or scooter dependent, and 8—bedridden.
Disease Modifying Therapy (DMT)
As part of MS PATHS, patients self‐report DMT use. Patients were categorized based on their DMT exposure as none, low, or high efficacy. When a patient had been on multiple DMTs, their most efficacious category was used. Low efficacy medications included glatiramer acetate, interferons, teriflunomide, fumarate class, and non‐MS‐specific immunosuppression (e.g., azathioprine). High efficacy medications included the S1P‐modulators, B‐cell depletion, natalizumab, and cladribine.
Imaging parameters
Imaging parameters have been previously described. 16 In brief, each subject and visit contribute a 3D T1‐weighted MP‐RAGE and a 3D T2‐weighted FLAIR image. Both sequences were acquired with 1 mm3 isotropic voxels the same field of view. To increase homogeneity, only 3T Siemens scanners were included. However, the model of scanner varied across sites. White matter lesion volume was derived from the MS PATHS dataset using the MSPie proprietary algorithm which has been previously validated.
Image preprocessing and brain age calculation
A growing number of brain age algorithms exist in the literature with no consensus on a superior algorithm. Thus, it is unclear if divergent findings between studies are related to algorithm choice, sample selection, sampling error, statistical analysis, or a combination of these. To address this, we implemented three different brain age algorithms which utilize different preprocessing strategies and differ with respect to their assumptions and numerical properties. The three algorithms used in this report are BrainAgeR, 18 , 19 Amyloid Negative Brain Age (ANBA), 20 and DeepBrainNet. 21 Each algorithm is discussed in detail in its respective publications and only described briefly for the present report.
BrainAgeR 18
The raw T1‐weighted MP‐RAGE was processed through a customized pipeline implemented in DARTEL 22 in SPM12 which includes bias field correction, template normalization, and smoothing and ultimately generates a gray matter, white matter, and cerebrospinal fluid segmentation. These segmented images were then nonlinearly registered to a template in MNI152 space. Volumetric estimates were subjected to Gaussian process regression using coefficients that were trained in a previous, publicly available dataset.
ANBA 20
The image preprocessing is identical to BrainAgeR. However, only gray matter segmentations are analyzed by the pretrained algorithm. The key innovation of this algorithm was training on an aging sample known to be free of Alzheimer disease.
DeepBrainNet 21
The raw T1‐weighted MP‐RAGE underwent skull stripping via HD‐BET, 23 bias field correction using the N4 tools, 24 and linear registration to the target template space using FLIRT. 25 This image was then subjected to the DeepBrainNet algorithm pretrained on the University of Pennsylvania LifeSpanCN dataset. This convolutional neural network does not depend on a priori segmentation.
To assess the predictive performance of the three different brain age algorithms, thalamic volume, cortical gray matter volume, and deep gray matter volume were measured using the MS Pie tool provided by MS PATHS.
Statistical analysis
A commonly reported statistic in the brain age literature is the “brain age gap” defined as the difference between predicted brain age and measured chronological age. Significant statistical challenges are associated with this quantity. 13 In this work, we avoid explicit calculation of the brain age gap to avoid this confound for reasons detailed in the Supplemental Methods.
All statistical analyses were performed in R v3.6.3. Cross‐sectional models were constructed as linear models. Post hoc contrasts were extracted for purposes of reporting statistical significance. Missing data were excluded, not interpolated. Predicted brain age from each algorithm separately were modeled as a function of sex, actual age, MS disease course, disability measured by PDDS, DMT category, and lesion volume. Interactions were allowed between actual age, MS‐disease course, disability, DMT category, and lesion volume. Longitudinal models were constructed using linear mixed models. To account for the longitudinal nature of the analysis, a dummy‐variable indexing time since baseline was created and this was nested within the random effect of subject. The rate of change in brain age attributable to specific variables (i.e., PDDS, MS disease course, DMT efficacy) was calculated by summing the patient specific beta values corresponding to 2nd or higher order interactions including the variable of interest and Time. In this way, the marginal increase (or decrease) in brain age accumulation related to the variable of interest was captured. Values greater than 0 indicate accelerated brain age accumulation related to the variable of interest; values less than 0 indicate relatively slowed brain age accumulation. All models included a random effect of site to account for differences in patient characteristics or scanner differences across the enrolling sites. Of note, the effect of site was small and statistical results did not vary with the inclusion vs. exclusion of this parameter.
Survival models were fit using Cox Proportional Hazard models with time to sustained disability accumulation as the outcome of interest. Sustained disability accumulation was defined as an increase of PDDS by 1 that remained increased until the end of the available data. By definition, increased PDDS at the last data sample could not represent sustained disability accumulation. Risk factors in the model included predicted brain age, actual age, MS disease course, and baseline disability measured with the PDDS. Predicted brain age was modeled two ways. The first approach was as a continuous variable. The second approach was dichotomized based on a predicted brain age gap greater than or less than the median for the group. Hazard ratios were extracted from these models, and reconstructed survival curves are shown.
Results
Brain age algorithms accurately predict actual age from structural MRI data
We first compared the ability of the candidate brain age algorithms to predict the actual age of healthy control participants. The initial imaging sessions from N = 207 controls (Table 1) were subjected to the BrainAgeR, ANBA, and DeepBrainNet brain age analysis. Each algorithm resulted in a high correlation between predicted age and actual age, but the correlation was higher in the BrainAgeR (Fig. 1A; r = 0.82) and DeepBrainNet (Fig. 1C; r = 0.80) prediction compared to ANBA (Fig. 1B; r = 0.68). Figure 1D shows the distribution of the differences between predicted age and actual age for each model with BrainAgeR and DeepBrainNet demonstrating numerical superiority in cross‐sectional analysis.
Table 1.
Demographics.
| Controls | CIS | RRMS | SPMS | PPMS | PRMS | |
|---|---|---|---|---|---|---|
| Cross‐Sectional Cohort | ||||||
| N | 207 | 813 | 3784 | 1110 | 353 | 465 |
| Age | 44.1 (13.0) | 46.0 (13.1) | 45.7 (12.1) | 49.2 (11.4) | 49.7 (12.5) | 48.2 (12.5) |
| F/M | 131/47 | 225/561 | 2804/880 | 783/295 | 238/107 | 308/144 |
| PDDS | N/A | 1.1 (1.8) | 1.4 (1.7) | 3.3 (2.1) | 3.3 (2.3) | 2.9 (2.3) |
| DMT None/Low/High Efficacy | N/A | 63/317/433 | 327/1357/2100 | 107/339/664 | 76/80/197 | 48/141/276 |
| Education (years) | 15.6 (2.8) | 15.0 (2.7) | 14.9 (2.6) | 14.4 (2.7) | 14.2 (2.9) | 13.9 (2.5) |
| Race White/Black/Asian | 137/22/7 | 581/109/11 | 2914/463/19 | 868/128/2 | 271/40/2 | 318/79/4 |
| Longitudinal Cohort | ||||||
| 2 Visits | 75 | 198 | 1127 | 301 | 100 | 142 |
| 3 Visits | 22 | 198 | 586 | 194 | 56 | 62 |
| 4 Visits | 2 | 91 | 248 | 90 | 20 | 24 |
| 5 Visits | 2 | 28 | 117 | 38 | 4 | 14 |
| 6+ Visits | 1 | 17 | 74 | 20 | 4 | 6 |
| Mean Follow‐up (years) | 0.68 (0.8) | 1.2 (1.2) | 1.1 (1.2) | 1.1 (1.3) | 1.0 (1.2) | 1.0 (1.2) |
Demographic factors for each group. Top panel shows cross‐sectional characteristics. Bottom panel shows the number of subjects with exactly M visits and the mean follow‐up time. Means (SD) are reported as applicable. Patient derived disease steps (PDDS) indicates mild disability for CIS/RRMS, and gait disability for SPMS/PPMS/PRMS. There is a large number of missing self‐reported race in these data.
Figure 1.
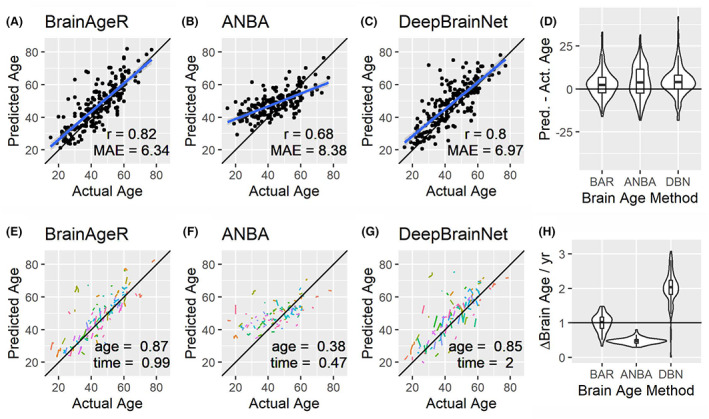
Brain age algorithms accurately predict actual age in healthy controls. The predicted age as a function of actual age is shown for the baseline visit of the healthy control group for the BrainAgeR (A), ANBA (B), and DeepBrainNet (C) algorithm. Regression line with 95% confidence interval is shown in blue. For each algorithm, the slope of the fitted line is less than 1 indicating regression towards the mean. Practically, young ages are over‐estimated and older ages are under‐estimated. The quality of the fit was assessed by correlation of predicted and actual age (r, inset) and by mean absolute error between predicted and actual age (MAE, inset). The distribution of predicted age − actual age for each method is shown (D). Absent any pathology, a difference of 0 indicates ideal performance. Thus, BrainAgeR and DeepBrainNet performed best. Longitudinal scatter plots (i.e., spaghetti plots) show each longitudinally studies subject as a line segment (E–G). Predicted age was fit to a mode including baseline age and a longitudinal nested variable time. Values close to 1 indicate ideal performance and are shown as inset. The calculated rate of change of brain age is shown in H. For healthy controls, as one calendar year passes the brain age should increase by 1. This behavior is best approximated by BrainAgeR.
We subjected the predicted brain age to a linear mixed effects model with the participants baseline actual age (age) and a dummy longitudinal variable (time) indexing years since baseline image as factors. The fitted regression βs are shown in Fig. 1E–G as insets. Beta values close to 1 indicate an additional brain age year for each year of age or year of longitudinal follow‐up. Figure 1H shows the distribution of the estimated slope across participants. BrainAgeR had the median values closest to 1 demonstrating that BrainAgeR is the algorithm with features closest to the numerically ideal behavior. Thus, for the remainder of this report, BrainAgeR results are shown in the main text and parallel results derived from ANBA and DeepBrainNet will be presented as supplemental material.
MS and MS‐related disability associate with increased brain age
We examined the effect of MS on cross‐sectionally estimated brain age of N = 6,732 participants (Table 1). We fit linear models with age, sex, disability (quantified by the PDDS), MS disease course, lesion volume, and DMT category as factors. ANOVA and individual coefficients are shown in Tables S3 and S4, respectively. Male sex was associated with greater brain age (F = 84.93, p < 10−16, Fig. S4) which has previously been observed in other imaging modalities. 26 The third order interactions of MS disease course × PDDS × lesion volume (F = 2.68, p = 0.020) and PDDS × DMT category × lesion volume (F = 3.31, p = 0.037) were observed and investigated further. Greater disability was associated with increased brain age in the RRMS (Fig. 2A), SPMS (Fig. 2B) and PPMS (Fig. 2C) groups. The effect gradient was most evident in the progressive groups where greater disability level was associated with larger over‐estimation of brain age. The effect of MS disease course was modest at PDDS = 0 (Fig. 2D), PDDS = 3 (Fig. 2E), and PDDS = 6 (Fig. 2F). The effect of lesion volume in an RRMS patient with PDDS = 0 (Fig. 2G) and PDDS = 3 (Fig. 2H) demonstrated larger overestimation of brain age with greater lesion burden. Similar results were seen in progressive MS. The effect of none vs. low vs. high efficacy DMT was modest in RRMS (Fig. 2I) but more pronounced in SPMS (Fig. 2J). Similar results were observed using the other brain age estimation algorithms (Figs. S5 and S6). Brain age was correlated with disability and increasing lesion burden. The effects of greater disability and lesion burden were larger than the effect of MS disease course.
Figure 2.
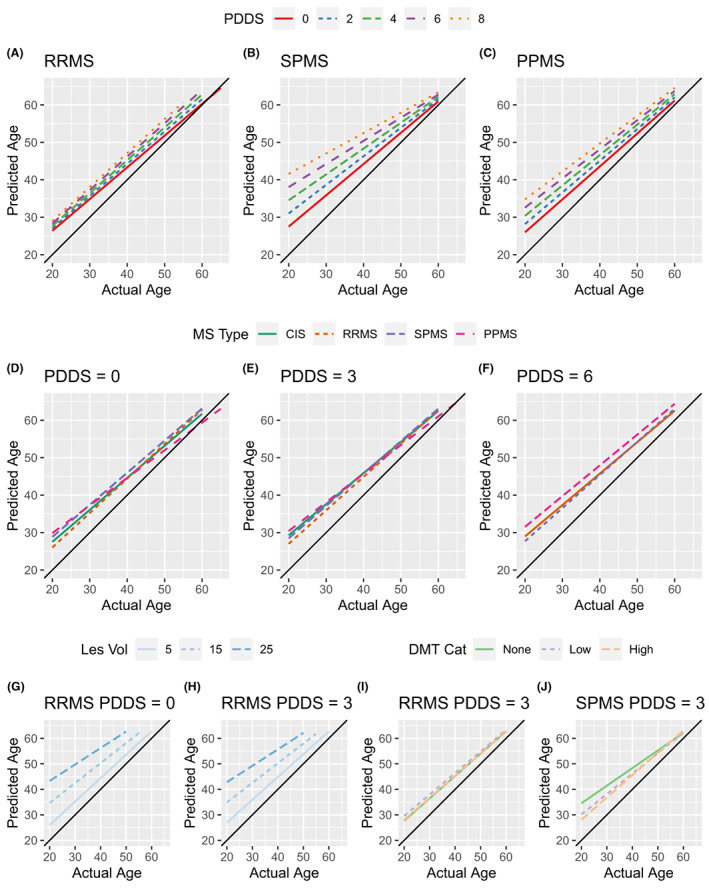
MS and MS‐related disability is associated with increased predicted brain age. Brain age was modeled as a function of actual age, sex, MS disease course, disability, lesion burden, and DMT efficacy. Statistical results are shown in Tables S3 and S4. Using these models, predicted values were generated to visualize the effects across the age span. (A) The effect of increasing disability is estimated in the RRMS group. Each increase in disability is associated with an increased estimate of brain age relative to chronological age. (B and C) The effect of disability on SPMS and PPMS in the same style as A. The effect of disability is most evident in the progressive groups. (D) To facilitate comparisons across MS disease courses, the estimates for the different MS disease courses are shown for PDDS 0. This shows that brain age is not dramatically difference in SPMS or PPMS compared to CIS and RRMS. (E and F) The effect of MS disease course in PDDS 3 and 6 in the style of D. Note, CIS is not included in the PDDS 6 plot since no patients met these criteria. The salient feature of D–F is that MS disease course is less distinct at higher disability levels. The effect of increasing lesion burden is shown for an RRMS PDDS 0 (G) and RRMS PDDS 3 (H) patient demonstrating the impact of increasing lesion burden. In RRMS PDDS 3 patients, the effect of DMT efficacy is minimal (I). In a similarly disabled SPMS patient, higher efficacy DMT is associated with decreased brain age at young ages (J). Black line represents where predicted age = actual age.
Longitudinal rate of change in predicted brain age is associated with MS‐related disability
We next examined the longitudinal behavior of predicted brain age using all available longitudinal data. Spaghetti plots are shown for a random sample of CIS (Fig. 3A), RRMS (Fig. 3B), SPMS (Fig. 3C), and PPMS (Fig. 3D) patients. For each MS disease course, predicted brain age was higher than actual age. To these data, we fit linear mixed effects models with sex, age at baseline, MS‐disease course, disability (PDDS), lesion burden, and DMT category as factors. A dummy‐variable indexing time since baseline scan was nested within the random effect of subject. ANOVA and regression tables for each brain age algorithm are shown in Tables S5 and S6. On the basis of these linear models, we estimated the marginal change in the rate of increasing brain age (ΔBrainAge/year) attributable to PDDS, MS disease course, and DMT efficacy compared to a reference of PDDS = 0, healthy controls, and no DMT, respectively. Increasing disability was associated with an increased rate of brain age accumulation (up to ~1 brain age year/calendar year) beyond what would have been measured in a non‐disabled (PDDS 0) individual (Fig. 3E). CIS and RRMS did not increase the rate of brain age accumulation whereas progressive MS was associated with more rapid brain aging (Fig. 3F). Neither low nor high efficacy DMT significantly modulated the rate of brain aging compared to no DMT (Fig. 3G). Together, these results suggest that the effect of accelerated brain aging is related to disability accumulation and progressive disease and is not modulated by DMT efficacy category.
Figure 3.
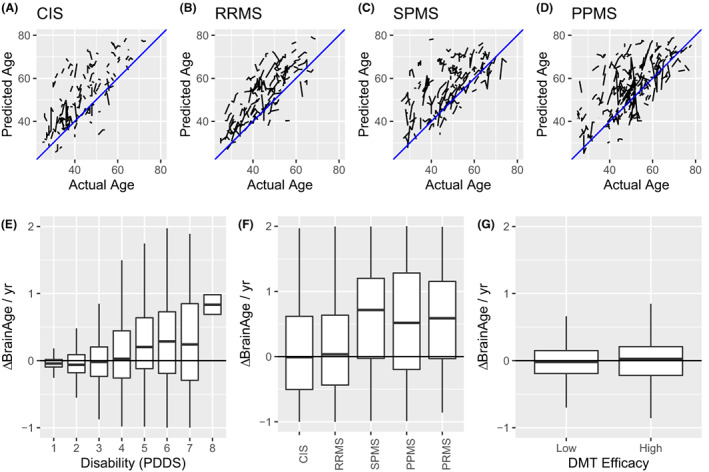
Longitudinal changes in brain age are related to disability. Longitudinally predicted brain age by BrainAgeR is shown as spaghetti plots for CIS (A), RRMS (B), SPMS (C), and PPMS (D) participants. In each case, predicted age over‐estimates actual age and single subjects demonstrate increasing predicted brain age with increasing actual age. To facilitate visualization, only a randomly selected fraction of the total data are shown for purpose of illustration; all subjects are considered statistically. Next, the longitudinal marginal rate of change in predicted brain age was calculated for each subject and expressed as change in predicted brain age (ΔBrainAge) per actual year attributable to PDDS (E), MS disease course (F), and DMT efficacy (G). Higher disability was associated with accelerated brain aging.
We next assessed the extent to which brain age provided additional statistical information compared to more conventional measures of atrophy. Identical statistical models were fit with thalamic, cortical gray matter, and deep gray matter volume as dependent variables. The variance explained (R2) was compared between these models and each of the three brain age models. In each case, the brain age models outperformed the direct volume measurement models (Table S7) demonstrating the added value of the brain age analysis procedure.
Advanced brain age predicts disability accumulation
We next examined if advanced brain age predicts future MS‐related disability accumulation. We fit Cox Proportional Hazard models with time to sustained disability (PDDS) accumulation as the outcome of interest. Brain age calculated from the subject's first imaging session was the predictor variable. Additional included factors were actual age, MS disease course, baseline disability, and DMT category. Hazard ratios and summary statistics are shown in Fig. 4A. Advanced brain age was associated with a shorter time to disability accumulation. For display purposes only, we dichotomized the difference between brain age vs. actual age into higher than the median and lower than the median (Fig. S5) and demonstrated that higher brain age gap at baseline predicts more rapid disability accumulation (Fig. 4B). Higher levels of baseline disability were also predictive of future disability accumulation (Fig. 4C) but were less predictive than brain age. MS disease course and DMT efficacy did not significantly modulate ability of brain age to predict time to disability accumulation over the span of this study. These results suggest that higher brain age as calculated by imaging is a biomarker of future disability accumulation.
Figure 4.
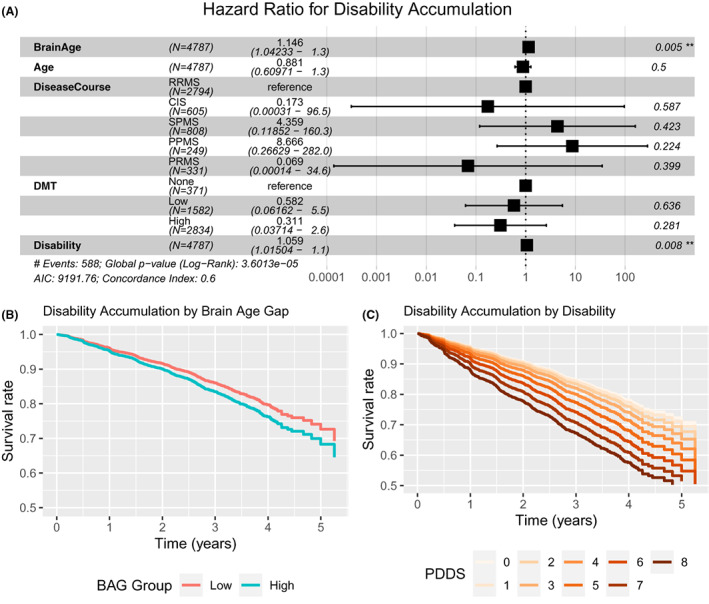
Advanced brain age predicts future disability accumulation. Cox proportional hazard models were fit with the outcome of sustained increase in PDDS and brain age, actual age, MS disease course, baseline disability (PDDS), and DMT efficacy as covariates. Brain age was indexed two ways. First, brain age was modeled as a continuous variable (A) with time units of decades. Second, two groups were defined on the basis of brain age—actual age above or below the median for the group (B). Hazard ratios and summary statistics are shown for each model separately. Survival curves are constructed from the dichotomized model which show more rapid disability accumulation in patients with large brain age gaps and higher levels of pre‐existing disability.
Demographic and clinical correlates of brain youthfulness
Most MS patients exhibited brain ages exceeding their actual chronological age. However, a subset of patients demonstrated brain ages less than their chronological age (appearing below the predicted age = actual age identity line in e.g. Fig. 2). We sought to determine what demographic or clinical factors were associated with this calculated brain youthfulness. A logistic regression was performed with sex, MS disease course, race, age of symptom onset, education level, medical comorbidities, and DMT efficacy as predictors (Table 2). The presence of brain age less than chronological age was associated with being female, being less disabled, having a higher age of symptom onset, being highly educated and free from diabetes were associated with chronologic age greater than brain age. Importantly, presence of a high efficacy DMT was associated with brain age less than chronological age.
Table 2.
Clinical variables associated with brain youthfulness.
| OR | 2.5% | 97.5% | p value | |
|---|---|---|---|---|
| Male | 0.634 | 0.547 | 0.733 | <0.001 |
| RRMS | 0.996 | 0.824 | 1.207 | 0.967 |
| SPMS | 0.987 | 0.774 | 1.260 | 0.916 |
| PPMS | 0.897 | 0.646 | 1.239 | 0.514 |
| PRMS | 0.979 | 0.722 | 1.322 | 0.892 |
| PDDS | 0.951 | 0.919 | 0.985 | 0.005 |
| Am. Indian/Alaska Nat. | 0.558 | 0.084 | 2.248 | 0.464 |
| Asian | 0.543 | 0.214 | 1.204 | 0.160 |
| Black | 0.749 | 0.618 | 0.905 | 0.003 |
| Nat. HI/Pac. Islander | 1.495 | 0.210 | 7.127 | 0.636 |
| Other | 0.684 | 0.476 | 0.960 | 0.033 |
| Age at Symptom Onset | 1.035 | 1.029 | 1.042 | <0.001 |
| Education | 1.081 | 1.055 | 1.107 | <0.001 |
| No Cardiac Disease | 1.562 | 0.228 | 31.248 | 0.695 |
| No Diabetes | 1.329 | 1.052 | 1.692 | 0.019 |
| No Dyslipidemia | 1.115 | 0.954 | 1.304 | 0.173 |
| Low Efficacy DMT | 0.935 | 0.752 | 1.165 | 0.546 |
| High Efficacy DMT | 1.263 | 1.086 | 1.404 | 0.005 |
A logistic regression was performed to determine variables associated with brain youth (brain age < actual age). MS disease course reference group was CIS; race reference group was white. Youthful brain age was associated with being female, having low disability, not having diabetes, a greater age of symptom onset, higher education, and being on a high efficacy DMT. Identifying as black was negatively associated with brain youthfulness.
Discussion
We applied machine learning‐based brain age analysis to a large, real‐world sample of images from patients with MS across the disease spectrum. We found that brain age was greater in patients with MS compared to healthy controls, particularly those with high levels of disability. Advanced brain age correlated with disability and, importantly, predicted time to further disability accumulation. Previous work by Cole and colleagues has established the power of brain age for predicting future disability in MS. 8 This work confirms and extends that work in a larger dataset and using a statistical analysis that avoids potential issues with calculations of brain age gaps. Thus, we propose that an increased predicted brain age is a parsimonious and potentially useful biomarker of pathology in MS which indicates a higher risk of accumulating disability in the future.
Brain age prediction algorithms provide robust and interpretable biomarkers
We compared the performance and results of three distinct brain age prediction algorithms 18 , 19 , 20 , 21 and examined statistical issues surrounding brain age analysis. The three selected algorithms are based on different machine learning principles, numerical assumptions, and preprocessing strategies. Importantly, these algorithms differed in input data. Algorithms which incorporated WM information (i.e., BrainAgeR and DeepBrainNet) statistically performed better than ANBA which incorporated only GM information, suggesting that pathology in both GM and WM contribute to MS‐related disability. Our results suggest that the prediction of age on the basis of structural MRI is an application well suited to machine learning and is not strongly dependent on algorithm choice. Moreover, we found that each brain age algorithm provided increased explanatory power compared to more traditional volumetric measurements. Therefore, brain age prediction and the biological conclusions drawn therefrom appear to be robust to algorithmic choice which facilitates comparison across diseases and methodologies.
The brain age gap, the difference between predicted and actual age, has a well‐established negative dependence on actual age. 27 A common approach in the brain age prediction literature is to orthogonalize the brain age gap with respect to actual age (i.e., regress out the negative slope). However, this approach inflates common measures of prediction performance and could artifactually inflate correlation with age‐dependent covariates. 13 To avoid this confound, our primary statistical analysis does not rely on the calculation of the brain age gap. Rather, age is included in the statistical model as a covariate. Our results demonstrate that brain age, handled in this way, remains a robust biomarker.
Advanced and accelerated brain aging is primarily related to disability
Previous applications of predicted brain age analyses have found increased predicted brain age in MS patients 6 , 7 and correlations with future disability accumulation. 8 Cole and colleagues 8 demonstrated a larger brain age gap in MS patients compared to controls with no difference between relapsing and progressive MS patients. The modified brain age gap correlated with disability as measured by the expanded disability status scale (EDSS) and predicted longitudinally increasing EDSS. Hogestol and colleagues 7 found a larger brain age gap in MS patients compared to controls that increased over time, but no association with clinical status. Importantly, both studies used a modified brain age gap. Kaufmann and colleagues 6 found that MS patients had increased brain age compared to controls across the brain, but with the frontal and temporal lobes relatively spared. The present study extends these previous findings by studying a larger number of MS patients and demonstrates that these findings are not specific to a single brain age algorithm. We found that advanced brain age was correlated with lesion burden but not because lesions themselves impacted brain age calculation (See Supplemental Results). The methodological innovations of the present study coupled with the large sample size demonstrate that predicted brain age is a reliable biomarker of MS‐related pathology.
MS disease course and disability level contributed independently to increasing brain age. On the basis of the longitudinal imaging results, disability measured by PDDS was a more powerful contributor to accelerated brain aging than MS clinical course. This suggests that brain age is most sensitive to damage resulting in disability and not to the inflammatory profile on which MS clinical disease courses are based (e.g., relapsing vs non‐relapsing). This result is concordant with recent work demonstrating that clinically defined disease courses capture only a small fraction of the variance in clinical outcomes. 28 Clinical MS characterizations are imprecise clinical constructs and SPMS is by definition a retrospective designation. Our results and those of Cole 8 emphasize the potential for using brain age determined by imaging as a biomarker in prognostication in MS patients beyond characterization of clinical disease course.
Advanced brain age predicts more rapid disability accumulation
We found that predicted brain age and current disability were strong prognosticators of future disability accumulation. Importantly, this analysis drew on the first image available for each subject and thus demonstrates feasibility as a useful biomarker even when the brain age determination was not calculated using the MRI at disease onset. Thus, these results provide further support for brain age as a clinically tractable biomarker of future MS disability.
Several imaging biomarkers are used in MS but typically focus on the relapsing or inflammatory component of the disease. Quantities such as new or expanding white matter lesions, 29 contrast enhancing lesions, and black hole formation 30 are widely used clinically, in research, and as secondary endpoints in clinical trials. The most common biomarker of progressive or degenerative pathology is atrophy. 31 The use of brain age is computationally straightforward and provides a single quantitative measure that can predict disability over time. 32 MS PATHS provides a large dataset which supports the utility of brain age as a potentially useful clinical tool. However, many measures in the MS PATHS dataset are subjective (e.g., PDDS). Future studies with objective measures are needed to validate this tool as useful in predicting disease course thereby informing treatment decisions. Demonstration of robust, within‐subject reliability would support future clinical applications.
Strengths and weakness of the present results
The presented results were drawn from a large, multinational observational study of MS. Strengths of the study include the harmonized imaging protocol, large sample size, and standardized evaluations. The use of three brain age algorithms that provided similar results supports the generalizability of the findings. Weaknesses include the observational nature of the data collection. The brain age algorithms themselves are generic, applying to several disease processes. More specifically, many demographic and pathologic processes result in advanced brain age making the finding of advanced brain age non‐specific. The predictive power of brain age in the present, real‐world dataset likely reflects the impacts of comorbidities on brain age and disability accumulation. Future studies with more specific MS tools may provide more precise predictions, but the present results demonstrate the utility of this approach in real‐world data. In particular, a MS‐specific brain age algorithm may benefit from incorporating both T1 and T2 information. Imaging and clinical assessment frequency were not standardized, with varied sampling frequency across sites and patients. Also, disability was self‐reported by patients using the PDDS. Nevertheless, the large numbers and robust statistical estimates indicate that predicted brain age is a strong marker of MS‐related pathology with the power to predict future disability. Lastly, we were unable to quantify DMT course and indexed patients by the most effective DMT they reported being exposed to.
Conclusion
Brain age is a non‐specific but easily estimated imaging biomarker that has demonstrated utility in predicting MS‐related disability. The results of this analysis establish that brain age is reliably and consistently estimated via a variety of techniques and can be analyzed successfully while avoiding statistically confounded procedures. We found the primary correlate of advanced brain age to be disability, both in cross‐sectional and longitudinal estimates. Finally, we expanded, in a large cohort, previous demonstrations 8 that advanced brain age predicts more rapid disability accumulation. These results support the use of brain age as a biomarker of MS‐related disability accumulation.
Conflict of Interest Statement
MRB has received fees for consultation from Genentech and EMD Serono. ZL, ML, HK, LL, WD, and JEM have no disclosures. AHC has received fees for consulting or contracted research from Biogen, Brisol Myers Squibb, EMD Soreno (Merck), Genentech, Horizon Therapeutics, Jazz Pharmaceuticals, Novartis, and TG Therapeutics. TLSB has investigator‐initiated research funding from the NIH, the Alzheimer's Association, the Foundation at Barnes‐Jewish Hospital and Avid Radiopharmaceuticals (a wholly owned subsidiary of Eli Lilly and Company). She participates as a site investigator in clinical trials sponsored by Avid Radiopharmaceuticals, Eli Lilly and Company, Biogen, Eisai, Jaansen, and Roche. She serves as an unpaid consultant to Eisai and Siemens, and is on the Speaker's Bureau for Biogen. RTN has consulted for Abata Therapeutics, Banner Life Sciences, BeiGene, Biogen, Bristol Myers Squibb, Genentech, Genzyme, Janssen, GW Therapeutics, Horizon Therapeutics, Lundbeck, NervGen, TG Therapeutics. SC has received consulting fees from Biogen, Novartis, Sanofi Genzyme, BMS and Jansen. SC has received funding for investigator‐initiated research from Biogen, BMS and the BMS foundation.
Supporting information
Appendix S1: Supporting Information
Table S4:
Table S5:
Table S6:
Acknowledgements
MS PATHS is funded by Biogen. MRB received research support from the NIH (2R25NS090978‐06; KL2TR002346). HK was supported by NIH K01MH122741. JEM received support from NSF DMS 2054199 and NIH R01AG052550. AHC was supported by the Manny & Rosalyn Rosenthal—Dr. John L. Trotter MS Center Chair in Neuroimmunology. Data are available to through Biogen or participating healthcare institutions in the MS PATHS program.
Funding Information
National Institutes of Health: 2R25NS090978‐06 : K01MH122741 : KL2TR002346 : R01AG052550 : Biogen National Science Foundation: DMS2054199
Funding Statement
This work was funded by National Institutes of Health grants 2R25NS090978‐06, K01MH122741, KL2TR002346, and R01AG052550; Biogen ; National Science Foundation grant DMS2054199.
References
- 1. Filippi M, Rocca MA, Ciccarelli O, et al. MRI criteria for the diagnosis of multiple sclerosis: MAGNIMS consensus guidelines. Lancet Neurol. 2016;15(3):292‐303. doi: 10.1016/S1474-4422(15)00393-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zivadinov R, Leist TP. Clinical‐magnetic resonance imaging correlations in multiple sclerosis. J Neuroimaging. 2005;15(4 SUPPL):10S‐21S. doi: 10.1177/1051228405283291 [DOI] [PubMed] [Google Scholar]
- 3. Filippi M, Preziosa P, Rocca MA. Magnetic resonance outcome measures in multiple sclerosis trials: time to rethink? Curr Opin Neurol. 2014;27(3):290‐299. doi: 10.1097/WCO.0000000000000095 [DOI] [PubMed] [Google Scholar]
- 4. Tousignant A, Paul Lemaître M, Doina Precup C, Arnold DL. Prediction of disease progression in multiple sclerosis patients using deep learning analysis of MRI data. Proc Mach Learn Res. 2019;102:483‐492. http://proceedings.mlr.press/v102/tousignant19a.html [Google Scholar]
- 5. Franke K, Ziegler G, Klöppel S, Gaser C. Estimating the age of healthy subjects from T1‐weighted MRI scans using kernel methods: exploring the influence of various parameters. Neuroimage. 2010;50(3):883‐892. doi: 10.1016/j.neuroimage.2010.01.005 [DOI] [PubMed] [Google Scholar]
- 6. Kaufmann T, van der Meer D, Doan NT, et al. Common brain disorders are associated with heritable patterns of apparent aging of the brain. Nat Neurosci. 2019;22(10):1617‐1623. doi: 10.1038/s41593-019-0471-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Høgestøl EA, Kaufmann T, Nygaard GO, et al. Cross‐sectional and longitudinal MRI brain scans reveal accelerated brain aging in multiple sclerosis. Front Neurol. 2019;10(APR):1‐9. doi: 10.3389/fneur.2019.00450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cole JH, Raffel J, Friede T, et al. Longitudinal assessment of multiple sclerosis with the brain‐age paradigm. Ann Neurol. 2020;88(1):93‐105. doi: 10.1002/ana.25746 [DOI] [PubMed] [Google Scholar]
- 9. Gaser C, Franke K, Klöppel S, Koutsouleris N, Sauer H. BrainAGE in mild cognitive impaired patients: predicting the conversion to Alzheimer's disease. PLoS One. 2013;8(6):e67346. doi: 10.1371/journal.pone.0067346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Steffener J, Habeck C, O'Shea D, Razlighi Q, Bherer L, Stern Y. Differences between chronological and brain age are related to education and self‐reported physical activity. Neurobiol Aging. 2016;40:138‐144. doi: 10.1016/j.neurobiolaging.2016.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hajek T, Franke K, Kolenic M, et al. Brain age in early stages of bipolar disorders or schizophrenia. Schizophr Bull. 2019;45(1):191‐198. doi: 10.1093/schbul/sbx172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cole JH, Underwood J, Caan MWA, et al. Increased brain‐predicted aging in treated HIV disease. Neurology. 2017;88(14):1349‐1357. doi: 10.1212/WNL.0000000000003790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Butler ER, Chen A, Ramadan R, et al. Pitfalls in brain age analyses. Hum Brain Mapp. 2021;42(13):4092‐4101. doi: 10.1002/hbm.25533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mowry EM, Bermel RA, Williams JR, et al. Harnessing real‐world data to inform decision‐making: multiple sclerosis partners advancing technology and health solutions (MS PATHS). Front Neurol. 2020;11:632. doi: 10.3389/fneur.2020.00632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fisher E, Kober T, Tsang A, et al. Magnetic resonance imaging (MRI) metrics in routine clinical practice: proof of concept in MS PATHS (multiple sclerosis partners advancing Technology for Health Solutions) (1356). Neurology. 2020;94(15 Supplement). [Google Scholar]
- 16. Brier MR, Snyder AZ, Tanenbaum A, et al. Quantitative signal properties from standardized MRIs correlate with multiple sclerosis disability. Ann Clin Transl Neurol. 2021;8(5):1096‐1109. doi: 10.1002/acn3.51354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kister I, Chamot E, Salter AR, Cutter GR, Bacon TE, Herbert J. Disability in multiple sclerosis: a reference for patients and clinicians. Neurology. 2013;80(11):1018‐1024. doi: 10.1212/WNL.0b013e3182872855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cole JH, Poudel RPK, Tsagkrasoulis D, et al. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. Neuroimage. 2017;163:115‐124. doi: 10.1016/j.neuroimage.2017.07.059 [DOI] [PubMed] [Google Scholar]
- 19. Cole JH, Ritchie SJ, Bastin ME, et al. Brain age predicts mortality. Mol Psychiatry. 2018;23(5):1385‐1392. doi: 10.1038/mp.2017.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ly M, Yu GZ, Karim HT, et al. Improving brain age prediction models: incorporation of amyloid status in Alzheimer's disease. Neurobiol Aging. 2020;87:44‐48. doi: 10.1016/j.neurobiolaging.2019.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bashyam VM, Erus G, Doshi J, et al. MRI signatures of brain age and disease over the lifespan based on a deep brain network and 14 468 individuals worldwide. Brain. 2020;143(7):2312‐2324. doi: 10.1093/brain/awaa160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ashburner J. A fast diffeomorphic image registration algorithm. Neuroimage. 2007;38(1):95‐113. doi: 10.1016/j.neuroimage.2007.07.007 [DOI] [PubMed] [Google Scholar]
- 23. Isensee F, Schell M, Pflueger I, et al. Automated brain extraction of multisequence MRI using artificial neural networks. Hum Brain Mapp. 2019;40(17):4952‐4964. doi: 10.1002/HBM.24750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tustison NJ, Avants BB, Cook PA, et al. N4ITK: improved N3 bias correction. IEEE Trans Med Imaging. 2010;29(6):1310‐1320. doi: 10.1109/TMI.2010.2046908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. Neuroimage. 2012;62(2):782‐790. doi: 10.1016/J.NEUROIMAGE.2011.09.015 [DOI] [PubMed] [Google Scholar]
- 26. Goyal MS, Blazey TM, Su Y, et al. Persistent metabolic youth in the aging female brain. Proc Natl Acad Sci U S A. 2019;116(8):3251‐3255. doi: 10.1073/pnas.1815917116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Smith SM, Vidaurre D, Alfaro‐Almagro F, Nichols TE, Miller KL. Estimation of brain age delta from brain imaging. Neuroimage. 2019;200(February):528‐539. doi: 10.1016/j.neuroimage.2019.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eshaghi A, Young AL, Wijeratne PA, et al. Identifying multiple sclerosis subtypes using unsupervised machine learning and MRI data. Nat Commun. 2021;12(1):1‐12. doi: 10.1038/s41467-021-22265-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Preziosa P, Pagani E, Meani A, et al. Slowly expanding lesions predict 9‐year multiple sclerosis disease progression. Neurol Neuroimmunol Neuroinflamm. 2022;9(2):e1139. doi: 10.1212/NXI.0000000000001139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sahraian MA, Radue EW, Haller S, Kappos L. Black holes in multiple sclerosis: definition, evolution, and clinical correlations. Acta Neurol Scand. 2010;122(1):1‐8. doi: 10.1111/J.1600-0404.2009.01221.X [DOI] [PubMed] [Google Scholar]
- 31. Fisher E, Lee JCC, Nakamura K, Rudick RA. Gray matter atrophy in multiple sclerosis: a longitudinal study. Ann Neurol. 2008;64(3):255‐265. doi: 10.1002/ana.21436 [DOI] [PubMed] [Google Scholar]
- 32. Wood DA, Kafiabadi S, Busaidi A, et al. Accurate brain‐age models for routine clinical MRI examinations. Neuroimage. 2022;249:118871. doi: 10.1016/J.NEUROIMAGE.2022.118871 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting Information
Table S4:
Table S5:
Table S6:


