Abstract
Synaptic modification in postnatal development is essential for the maturation of neural networks. Developmental maturation of excitatory synapses occurs at the loci of dendritic spines that are dynamically regulated by growth and pruning. Striatal spiny projection neurons (SPNs) receive excitatory input from the cerebral cortex and thalamus. SPNs of the striatonigral direct pathway (dSPNs) and SPNs of the striatopallidal indirect pathway (iSPNs) have different developmental roots and functions. The spatial and temporal dynamics of dendritic spine maturation of these two types of SPNs remain elusive. Here, we delineate the developmental trajectories of dendritic spines of dSPNs and iSPNs in the caudoputamen and nucleus accumbens (NAc). We labeled dendritic spines of SPNs by microinjecting Cre-dependent AAV-eYFP viruses into newborn Drd1-Cre or Adora2a-Cre mice, and analyzed spinogenesis at three levels, including different SPN cell types, subregions and postnatal times. In the dorsolateral striatum, spine pruning of dSPNs and iSPNs occurred at postnatal day (P)30–P50. In the dorsomedial striatum, the spine density of both dSPNs and iSPNs reached its peak between P30 and P50, and spine pruning occurred after P30 and P50, respectively, for dSPNs and iSPNs. In the NAc shell, spines of dSPNs and iSPNs were pruned after P21–P30, but no significant pruning was observed in iSPNs of lateral NAc shell. In the NAc core, the spine density of dSPNs and iSPNs reached its peak at P21 and P30, respectively, and subsequently declined. Collectively, the developmental maturation of dendritic spines in dSPNs and iSPNs follows distinct spatiotemporal trajectories in the dorsal and ventral striatum.
Keywords: basal ganglia, spine formation, spine pruning, spinogenesis, striatum, synaptogenesis
Significance Statement
The direct striatonigral and indirect striatopallidal pathways are engaged in neural circuits of basal ganglia for the regulation of movement and drug addiction. Such circuit functions rely on precise synaptic connectivity that goes through the maturation process. Excitatory synaptic connectivity can be traced by examining the development of dendritic spines. Here, we provide a comprehensive characterization of the development of dendritic spines in spiny projection neurons (SPNs) of the direct and indirect pathways from juvenile and adolescent to adult stages in the mouse brain. We found distinct cell type-specific trajectories of dendritic spines in the caudoputamen and nucleus accumbens (NAc). Our study provides a basic reference for neuropsychiatric diseases in which dysfunction of spinogenesis and synaptogenesis is targeted during development, including autism and schizophrenia.
Introduction
The basal ganglia are a group of subcortical nuclei serving a variety of neurologic functions, including motor control, reward learning, motivation, and emotion (Graybiel and Grafton, 2015; Keiflin and Janak, 2015; Kim and Hikosaka, 2015; Pessoa et al., 2019). The striatum is the main input structure of the basal ganglia. The striatum consists of two populations of projection neurons: dopamine D1 receptor-expressing striatonigral projection neurons of the direct pathway (dSPNs) and dopamine D2 receptor-expressing striatopallidal projection neurons of the indirect pathway (iSPNs; Gerfen et al., 1990; Gerfen and Surmeier, 2011). The prevailing theory suggests that activation of the direct pathway facilitates movement, whereas activation of the indirect pathway inhibits movement (Albin et al., 1989; Gerfen et al., 1990; Kravitz et al., 2010). The coordination of neuronal activity in direct and indirect pathways is thus important for movement control (Macpherson and Hikida, 2019; Arber and Costa, 2022).
The striatal complex is divided into the dorsal striatum (caudoputamen) and the ventral striatum [nucleus accumbens (NAc); Haber, 2016; Chen et al., 2020]. The caudoputamen is involved in the regulation of movement, motor learning, habits and decision-making (Mink, 2003; Gittis and Kreitzer, 2012; DeLong and Wichmann, 2015; Gunaydin and Kreitzer, 2016; Macpherson and Hikida, 2019). The nucleus accumbens is a key hub of reward circuits that are engaged in processing reward, motivation, emotion and drug addiction (Volkow et al., 2012; Russo and Nestler, 2013; Klawonn and Malenka, 2018; Nestler and Lüscher, 2019). The NAc is divided into the core and shell regions, and they have distinct neuronal connectivity that enables specific functions (West and Carelli, 2016; Li et al., 2018; Yang et al., 2018; Ma et al., 2020).
The neuronal architecture is specialized for neural processing in networks (Parekh and Ascoli, 2015). Dendritic spines are small membrane protrusions and specialized cell compartments through which neurons receive excitatory input from presynaptic axonal terminals. Dynamic changes in dendritic spine size, density and morphologic types occur not only in neuronal development but also in neuronal plasticity such as learning and memory (Segal, 2017; Runge et al., 2020). Abnormality in dendritic spine formation has frequently been found in neuropsychiatric diseases, including depression, schizophrenia and drug addiction (Roberts et al., 1996; Russo et al., 2010; Francis et al., 2017).
Synaptogenesis occurs after axonal outgrowth to innervate target cells during development. Activity-dependent modification of synaptogenesis further refines synaptic connectivity through synapse pruning (Südhof, 2018; Batool et al., 2019). Dendritic spine pruning plays an important role in the formation of mature excitatory synaptic connectivity, since the synaptic strength of neurotransmission is influenced by the density of spines/synapses (Carlisle and Kennedy, 2005; Ding et al., 2011; Grueter et al., 2012; Sala and Segal, 2014). Dendritic spines undergo plastic changes in response to environmental stimuli. The prevailing theory suggests that in the developing mammalian brains, dendritic spines initially overgrowth, followed by pruning during juvenile and adolescent stages before maturation in adulthood (Segal et al., 2000; Semple et al., 2013). Importantly, defective synaptic pruning in postnatal brains has been implicated in the pathogenesis of neuropsychiatric diseases (Hutsler and Zhang, 2010; Forrest et al., 2018; Lima Caldeira et al., 2019; Eltokhi et al., 2020), which calls for the understanding of synaptic modification during postnatal maturation. Previous studies have reported the development process of spinogenesis in the cerebellum, cerebral cortex, and hippocampus (García-López et al., 2010; Elston and Fujita, 2014). However, the developmental trajectory of spinogenesis of striatal neurons remains yet elusive.
In the present study, we investigated the developmental trajectory of dendritic spinogenesis of dSPNs and iSPNs in the caudoputamen and NAc. We found distinct trajectories of spinogenesis of dSPNs and iSPNs in subregions of the striatum during postnatal development, suggesting that synaptic wiring in striatal subregions is under different spatiotemporal control.
Materials and Methods
Animals
All animals were housed in groups in a 12/12 h light/dark cycle-specific pathogen-free room with food and water available ad libitum at the Animal Center of National Yang Ming Chiao Tung University. All experimental procedures in this study were approved by the Institutional Animal Care and Use Committee of National Yang Ming Chiao Tung University. Dopamine receptor D1 (Drd1)-Cre mice (STOCK Tg(Drd1-cre)EY262Gsat/Mmucd, RRID: MMRRC_017264-UCD) and adenosine receptor 2a (Adora2a)-Cre mice (STOCK Tg(Adora2a-cre)KG139Gsat/Mmucd, RRID: MMRRC_031168-UCD) were obtained from Mutant Mouse Resource and Research Centers supported by the National Institutes of Health. Drd1-Cre and Adora2a-Cre mice were maintained by intercrossing with wild-type C57BL/6J mice. Male mice were used in the current study, because gender-specific regulation of SPN maturation by sex hormones has previously been reported in rodents (Cao et al., 2018).
Genotyping
Genotypes of the mice were identified at postnatal day (P)0 by PCR with genomic DNA. For DNA extraction, ∼0.2 mm of tail tissue was heated and dissolved in 25 mm NaOH/0.2 mm EDTA for 10 min at 100°C. The dissolved tissue was then neutralized with an equal volume of 40 mm Tris-EDTA buffer (pH5.5) and cooled on ice. The PCR protocols were as follows: 95°C for 3 min, 31 cycles at 95°C for 30 s, 58°C for 30 s, 72°C for 45 s with a thermocycler (T3000, Biometra). The reaction was completed at 72°C for an additional 5 min and paused at 4°C. The primers used to identify the Cre sequences are 5′-ATGCTTCTGTCCGTTTGCCG-3′ and 5′-TGAGTGAACGAACCTGGTCG-3′. The expected size of the product was 316 bp.
Stereotaxic microinjections
Cre-dependent AAV9.EF1α.DIO.eYFP.WPRE.hGH (UPenn vector core; catalog #V-9-27056; lot #CS0977) viruses were diluted at 1:100 in D-PBS (CORNING-cellgro) for microinjections. The mouse pups (P0–P2) were anesthetized by hypothermia. AAV9.EF1α.DIO.eYFP.WPRE.hGH viruses (50 nl/site) were microinjected into the bilateral dorsal striatum (AP: +2.8 mm, ML: ±1.5 mm, DV: 1.5 mm) or ventral (AP: +2.8 mm, ML: ±1.0 mm, DV: 1.7 mm) striatum of Drd1-Cre or Adora2a-Cre mice.
Tissue preparation and immunohistochemistry
The brains of Drd1-Cre and Adora2a-Cre mice were harvested at P13, P21, P30, P50, and P100 by transcardial perfusion of 0.9% saline followed by 4% paraformaldehyde (PFA) in 0.1 m PBS (pH 7.4). The perfused brains were postfixed in 4% PFA/0.1 m PBS at 4°C overnight and were then cryoprotected with 30% sucrose/0.1 m PBS for 48 h. The brains were cut into 80-μm coronal sections with a vibratome (D.S.K., DTK-1000) and stored in 0.1 m phosphate buffer (PB) with 0.1% sodium azide at 4°C. Immunohistochemistry was performed as previously described (Chen et al., 2016). Briefly, after permeabilization with 0.2% Triton X-100/0.1 m PBS and removal of endogenous peroxidase with 10% methanol/3% H2O2/0.2% Triton X-100/0.1 m PBS, brain sections were blocked with 3% normal donkey serum/0.1 m PBS for 1 h at room temperature. After rinsing with 0.1 m PBS, brain sections were incubated in chicken anti-GFP primary antibody (1:1000; Abcam catalog #ab13970, RRID: AB_300798) at 4°C overnight. The next day, brain sections were incubated with the biotinylated secondary antibody donkey anti-chicken (1:500, Vector Laboratories) for 1 h, followed by the avidin-biotin-peroxidase complex (1:250, Vector catalog #PK-6100, RRID: AB_2336819) for another 1 h. The sections were then developed in 0.02% diaminobenzidine/0.08% nickel ammonium sulfate/0.003% H2O2 in 0.1 m PB.
Microscope imaging and quantification
Z-stack images of dendritic spines with 0.42 μm step size were acquired using an Olympus BX63 microscope with a 100× oil immersion objective. Neurons filled with eYFP signals were imaged. All imaged neurons were analyzed, except that the neurites of the imaged neurons were intermingled with the neurites of neighboring neurons. Images of eYFP-positive neurons were taken from the dorsal striatum at Bregma levels +1.7 to +0.14. A vertical line was drawn in the middle striatum to separate the dorsomedial and dorsolateral striatum. For the ventral striatum at Bregma +1.7 to +0.74, the core and shell of the NAc were identified by the gradient of cell density under a microscope. The medial and lateral parts of the NAc were divided by a vertical line from the middle part of the anterior commissure. The numbers of dendritic spines were manually counted in the proximal pars of the secondary dendrites and normalized to the lengths of the analyzed dendrites. Dendritic spine density was calculated as the average number of spines per 10 μm. We categorized the morphology of dendritic spines into six types according to Harris et al., with slight modifications (Harris et al., 1992). Spines were categorized as “stubby spines” if the diameter of the neck was similar to the length of the spine. Spines were categorized as “thin/filopodial spines” if the length of the spine was longer than the diameter of the neck; and the diameters of the head and neck were similar with a ratio is no more than two. Spines were categorized as “mushroom spines” if the diameter of the head was >2.5-fold of the diameter of the neck. Spines were categorized as “branched spines” and “multibranched spines” if they had two and more than two heads, respectively. The spines that could not be classified into a specific group were assigned to the category of “atypical spines.”
Statistical analysis
Statistical analysis was performed with SPSS (IBM, version 21). First, all the data were tested for normality with Shapiro–Wilk test. All data in this study were consistent with a normal distribution and we used two-way ANOVA to examine the influences of developmental times and genotypes on dendritic spine density. In the event of no interaction, one-way ANOVA followed by Tukey’s HSD post hoc tests was used to analyze the effects of developmental times in each genotype. If there was an interaction, simple main effects were performed to identify significant differences between developmental times. To analyze the temporal changes of each type of spine, one-way ANOVA followed by Tukey’s HSD post hoc tests were used for normally distributed data. For the data that are not normally distributed, Kruskal–Wallis one-way ANOVA followed by Dunn’s pairwise multiple comparisons tests or Mann–Whitney U test was used. To analyze the differences between the dorsomedial and dorsolateral striatum at each developmental time point, Student’s t tests were used. To analyze the differences among the NAc subregions at each developmental time point, one-way ANOVA followed by Tukey’s HSD post hoc tests were used. Data were presented as mean ± SEM if the data were normally distributed. Data were presented as median ± interquartile range if the data were analyzed by nonparametric analysis. Table 1 shows the details of statistical analyses.
Table 1.
Statistical analyses
| Data structure | Type of test | 95% confidence interval | ||
|---|---|---|---|---|
| Lower bound | Upper bound | |||
| Figure 1D, dSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 4.713 | 8.103 |
| P21 | Normal distribution | Two-way ANOVA | 7.393 | 10.782 |
| P30 | Normal distribution | Two-way ANOVA | 12.956 | 16.345 |
| P50 | Normal distribution | Two-way ANOVA | 9.656 | 13.046 |
| P100 | Normal distribution | Two-way ANOVA | 8.951 | 13.341 |
| Figure 1D, iSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 8.062 | 11.452 |
| P21 | Normal distribution | Two-way ANOVA | 9.312 | 12.701 |
| P30 | Normal distribution | Two-way ANOVA | 15.355 | 18.744 |
| P50 | Normal distribution | Two-way ANOVA | 11.748 | 15.137 |
| P100 | Normal distribution | Two-way ANOVA | 7.623 | 11.012 |
| Figure 1F, dSPN | ||||
| Stubby | ||||
| P13 | Normal distribution | Kruskal–Wallis test | 36.742 | 53.250 |
| P21 | Normal distribution | 38.707 | 50.243 | |
| P30 | Normal distribution | 28.789 | 43.423 | |
| P50 | Normal distribution | 27.797 | 41.213 | |
| P100 | Non-normal distribution | 31.236 | 39.342 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | One-way ANOVA | 34.391 | 45.181 |
| P21 | Normal distribution | 36.123 | 48.871 | |
| P30 | Normal distribution | 44.159 | 54.677 | |
| P50 | Normal distribution | 42.864 | 57.136 | |
| P100 | Normal distribution | 35.728 | 43.648 | |
| Mushroom | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | 4.471 | 16.927 |
| P21 | Normal distribution | 7.168 | 12.234 | |
| P30 | Normal distribution | 7.523 | 14.089 | |
| P50 | Normal distribution | 6.555 | 14.385 | |
| P100 | Normal distribution | 16.950 | 25.212 | |
| Branched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.184 | 8.220 |
| P21 | Non-normal distribution | 0.326 | 5.594 | |
| P30 | Non-normal distribution | 0.261 | 5.791 | |
| P50 | Normal distribution | 1.996 | 7.238 | |
| P100 | Non-normal distribution | 2.080 | 5.166 | |
| Multibranched | ||||
| P13 | Not applicable | Kruskal–Wallis test | Not applicable | |
| P21 | Not applicable | Not applicable | ||
| P30 | Non-normal distribution | −0.240 | 0.620 | |
| P50 | Non-normal distribution | −0.515 | 1.331 | |
| P100 | Non-normal distribution | −0.170 | 0.814 | |
| Atypical | ||||
| P13 | Not applicable | Mann–Whitney U | Not applicable | |
| P21 | Non-normal distribution | −0.467 | 1.207 | |
| P30 | Non-normal distribution | −0.574 | 1.484 | |
| P50 | Not applicable | Not applicable | ||
| P100 | Not applicable | Not applicable | ||
| Figure 1F, iSPN | ||||
| Stubby | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | 29.847 | 40.033 |
| P21 | Normal distribution | 39.921 | 47.440 | |
| P30 | Normal distribution | 38.523 | 48.398 | |
| P50 | Normal distribution | 30.236 | 40.799 | |
| P100 | Normal distribution | 33.705 | 40.828 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | One-way ANOVA | 37.272 | 51.416 |
| P21 | Normal distribution | 37.240 | 44.236 | |
| P30 | Normal distribution | 37.849 | 44.907 | |
| P50 | Normal distribution | 43.051 | 45.783 | |
| P100 | Normal distribution | 38.402 | 46.334 | |
| Mushroom | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | 6.244 | 28.854 |
| P21 | Normal distribution | 9.035 | 16.238 | |
| P30 | Normal distribution | 7.531 | 14.029 | |
| P50 | Normal distribution | 11.163 | 17.228 | |
| P100 | Normal distribution | 11.965 | 19.652 | |
| Branched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | 0.655 | 5.430 |
| P21 | Normal distribution | 1.023 | 4.459 | |
| P30 | Normal distribution | 1.844 | 6.378 | |
| P50 | Normal distribution | 3.338 | 7.656 | |
| P100 | Normal distribution | 2.466 | 4.936 | |
| Multibranched | ||||
| P13 | Not applicable | Kruskal–Wallis test | Not applicable | |
| P21 | Non-normal distribution | −0.118 | 0.525 | |
| P30 | Non-normal distribution | −0.341 | 0.882 | |
| P50 | Non-normal distribution | −0.050 | 0.572 | |
| P100 | Non-normal distribution | −0.004 | 1.459 | |
| Atypical | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.158 | 0.408 |
| P21 | Not applicable | Not applicable | ||
| P30 | Not applicable | Not applicable | ||
| P50 | Non-normal distribution | −0.142 | 0.367 | |
| P100 | Non-normal distribution | −0.162 | 0.418 | |
| Figure 2D, dSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 5.139 | 8.236 |
| P21 | Normal distribution | Two-way ANOVA | 8.373 | 11.469 |
| P30 | Normal distribution | Two-way ANOVA | 10.991 | 14.087 |
| P50 | Normal distribution | Two-way ANOVA | 9.548 | 12.645 |
| P100 | Normal distribution | Two-way ANOVA | 8.567 | 11.663 |
| Figure 2D, iSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 7.237 | 10.334 |
| P21 | Normal distribution | Two-way ANOVA | 9.057 | 12.154 |
| P30 | Normal distribution | Two-way ANOVA | 11.254 | 14.351 |
| P50 | Normal distribution | Two-way ANOVA | 13.300 | 16.396 |
| P100 | Normal distribution | Two-way ANOVA | 7.192 | 10.289 |
| Figure 2F, dSPN | ||||
| Stubby | ||||
| P13 | Normal distribution | One-way ANOVA | 33.063 | 46.911 |
| P21 | Normal distribution | 35.711 | 49.029 | |
| P30 | Normal distribution | 35.143 | 42.312 | |
| P50 | Normal distribution | 28.874 | 40.205 | |
| P100 | Normal distribution | 30.579 | 39.438 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | One-way ANOVA | 37.429 | 49.957 |
| P21 | Normal distribution | 41.409 | 54.360 | |
| P30 | Normal distribution | 41.981 | 49.121 | |
| P50 | Normal distribution | 44.564 | 53.695 | |
| P100 | Normal distribution | 39.988 | 45.904 | |
| Mushroom | ||||
| P13 | Normal distribution | Kruskal–Wallis test | 9.872 | 18.020 |
| P21 | Normal distribution | 3.334 | 9.160 | |
| P30 | Normal distribution | 8.098 | 15.757 | |
| P50 | Non-normal distribution | 7.214 | 13.084 | |
| P100 | Normal distribution | 13.042 | 22.791 | |
| Branched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.301 | 3.987 |
| P21 | Non-normal distribution | 0.249 | 5.323 | |
| P30 | Normal distribution | 1.342 | 4.814 | |
| P50 | Non-normal distribution | 3.301 | 8.600 | |
| P100 | Normal distribution | 2.381 | 5.878 | |
| Multibranched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.227 | 1.008 |
| P21 | Non-normal distribution | −0.394 | 1.819 | |
| P30 | Non-normal distribution | −0.157 | 0.731 | |
| P50 | Non-normal distribution | −0.118 | 0.580 | |
| P100 | Not applicable | Not applicable | ||
| Atypical | ||||
| P13 | Non-normal distribution | Mann–Whitney U | −0.178 | 0.460 |
| P21 | Not applicable | Not applicable | ||
| P30 | Non-normal distribution | −0.233 | 1.091 | |
| P50 | Not applicable | Not applicable | ||
| P100 | Not applicable | Not applicable | ||
| Figure 2F, iSPN | ||||
| Stubby | ||||
| P13 | Normal distribution | One-way ANOVA | 31.928 | 49.053 |
| P21 | Normal distribution | 31.945 | 42.072 | |
| P30 | Normal distribution | 36.102 | 47.205 | |
| P50 | Normal distribution | 30.633 | 41.191 | |
| P100 | Normal distribution | 32.026 | 42.913 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | One-way ANOVA | 37.451 | 51.261 |
| P21 | Normal distribution | 42.865 | 52.515 | |
| P30 | Normal distribution | 37.090 | 50.568 | |
| P50 | Normal distribution | 29.703 | 48.579 | |
| P100 | Normal distribution | 40.278 | 49.134 | |
| Mushroom | ||||
| P13 | Normal distribution | Kruskal–Wallis test | 6.817 | 17.795 |
| P21 | Non-normal distribution | 7.399 | 18.539 | |
| P30 | Normal distribution | 8.693 | 19.137 | |
| P50 | Normal distribution | 13.486 | 26.657 | |
| P100 | Non-normal distribution | 10.660 | 16.904 | |
| Branched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | 0.390 | 3.971 |
| P21 | Non-normal distribution | 0.380 | 3.682 | |
| P30 | Non-normal distribution | 0.058 | 3.146 | |
| P50 | Normal distribution | 2.054 | 7.134 | |
| P100 | Normal distribution | 2.139 | 5.769 | |
| Multibranched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.631 | 1.631 |
| P21 | Non-normal distribution | −0.187 | 0.792 | |
| P30 | Not applicable | Not applicable | ||
| P50 | Non-normal distribution | −0.178 | 0.460 | |
| P100 | Non-normal distribution | −0.112 | 0.289 | |
| Atypical | ||||
| P13 | Non-normal distribution | Mann–Whitney U | −0.210 | 0.544 |
| P21 | Not applicable | Not applicable | ||
| P30 | Not applicable | Not applicable | ||
| P50 | Non-normal distribution | −0.178 | 0.460 | |
| P100 | Not applicable | Not applicable | ||
| Figure 3D, dSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 8.769 | 11.674 |
| P21 | Normal distribution | Two-way ANOVA | 8.813 | 11.717 |
| P30 | Normal distribution | Two-way ANOVA | 10.400 | 13.305 |
| P50 | Normal distribution | Two-way ANOVA | 7.344 | 10.249 |
| P100 | Normal distribution | Two-way ANOVA | 7.008 | 9.913 |
| Figure 3D, iSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 7.340 | 10.245 |
| P21 | Normal distribution | Two-way ANOVA | 6.423 | 9.328 |
| P30 | Normal distribution | Two-way ANOVA | 7.042 | 9.947 |
| P50 | Normal distribution | Two-way ANOVA | 8.567 | 11.472 |
| P100 | Normal distribution | Two-way ANOVA | 7.165 | 10.070 |
| Figure 3F, dSPN | ||||
| Stubby | ||||
| P13 | Normal distribution | One-way ANOVA | 35.886 | 46.900 |
| P21 | Normal distribution | 37.407 | 50.410 | |
| P30 | Normal distribution | 35.555 | 46.824 | |
| P50 | Normal distribution | 29.784 | 40.883 | |
| P100 | Normal distribution | 25.271 | 33.411 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | One-way ANOVA | 36.756 | 48.498 |
| P21 | Normal distribution | 35.852 | 48.491 | |
| P30 | Normal distribution | 31.125 | 41.900 | |
| P50 | Normal distribution | 36.678 | 47.183 | |
| P100 | Normal distribution | 47.323 | 50.933 | |
| Mushroom | ||||
| P13 | Normal distribution | One-way ANOVA | 6.090 | 11.397 |
| P21 | Normal distribution | 7.172 | 16.549 | |
| P30 | Normal distribution | 13.375 | 23.500 | |
| P50 | Normal distribution | 15.079 | 24.542 | |
| P100 | Normal distribution | 14.252 | 18.435 | |
| Branched | ||||
| P13 | Normal distribution | Kruskal–Wallis test | 3.497 | 8.541 |
| P21 | Non-normal distribution | 0.039 | 2.728 | |
| P30 | Normal distribution | 1.777 | 4.880 | |
| P50 | Normal distribution | 1.181 | 4.671 | |
| P100 | Non-normal distribution | 2.544 | 6.045 | |
| Multibranched | ||||
| P13 | Normal distribution | Kruskal–Wallis test | 0.314 | 1.658 |
| P21 | Non-normal distribution | −0.048 | 1.399 | |
| P30 | Non-normal distribution | −0.163 | 0.804 | |
| P50 | Not applicable | Not applicable | ||
| P100 | Non-normal distribution | −0.152 | 1.939 | |
| Atypical | ||||
| P13 | Non-normal distribution | Mann–Whitney U | −0.294 | 0.759 |
| P21 | Not applicable | Not applicable | ||
| P30 | Non-normal distribution | −0.114 | 0.538 | |
| P50 | Not applicable | Not applicable | ||
| P100 | Not applicable | Not applicable | ||
| Figure 3F, iSPN | ||||
| Stubby | ||||
| P13 | Normal distribution | One-way ANOVA | 33.192 | 48.068 |
| P21 | Normal distribution | 34.031 | 41.462 | |
| P30 | Normal distribution | 28.308 | 42.659 | |
| P50 | Normal distribution | 36.953 | 40.510 | |
| P100 | Normal distribution | 31.264 | 38.340 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | One-way ANOVA | 31.176 | 46.244 |
| P21 | Normal distribution | 43.619 | 50.504 | |
| P30 | Normal distribution | 35.150 | 46.630 | |
| P50 | Normal distribution | 34.313 | 41.881 | |
| P100 | Normal distribution | 38.587 | 43.987 | |
| Mushroom | ||||
| P13 | Normal distribution | One-way ANOVA | 10.895 | 19.485 |
| P21 | Normal distribution | 8.370 | 15.747 | |
| P30 | Normal distribution | 11.283 | 22.671 | |
| P50 | Normal distribution | 14.385 | 22.627 | |
| P100 | Normal distribution | 18.163 | 24.865 | |
| Branched | ||||
| P13 | Normal distribution | Kruskal–Wallis test | 2.889 | 6.931 |
| P21 | Normal distribution | 1.528 | 4.401 | |
| P30 | Non-normal distribution | 1.394 | 11.906 | |
| P50 | Normal distribution | 2.690 | 5.878 | |
| P100 | Normal distribution | 0.839 | 2.958 | |
| Multibranched | ||||
| P13 | Not applicable | Mann–Whitney U | Not applicable | |
| P21 | Not applicable | Not applicable | ||
| P30 | Not applicable | Not applicable | ||
| P50 | Non-normal distribution | −0.221 | 0.983 | |
| P100 | Non-normal distribution | −0.120 | 1.117 | |
| Atypical | ||||
| P13 | Non-normal distribution | Mann–Whitney U | −0.101 | 1.222 |
| P21 | Non-normal distribution | −0.214 | 0.553 | |
| P30 | Not applicable | Not applicable | ||
| P50 | Not applicable | Not applicable | ||
| P100 | Not applicable | Not applicable | ||
| Figure 4D, dSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 7.472 | 10.088 |
| P21 | Normal distribution | Two-way ANOVA | 8.464 | 11.079 |
| P30 | Normal distribution | Two-way ANOVA | 12.942 | 15.557 |
| P50 | Normal distribution | Two-way ANOVA | 9.576 | 12.191 |
| P100 | Normal distribution | Two-way ANOVA | 9.615 | 12.231 |
| Figure 4D, iSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 8.194 | 10.809 |
| P21 | Normal distribution | Two-way ANOVA | 9.231 | 11.846 |
| P30 | Normal distribution | Two-way ANOVA | 10.682 | 13.297 |
| P50 | Normal distribution | Two-way ANOVA | 9.553 | 12.168 |
| P100 | Normal distribution | Two-way ANOVA | 8.374 | 10.989 |
| Figure 4F, dSPN | ||||
| Stubby | ||||
| P13 | Normal distribution | One-way ANOVA | 33.850 | 41.769 |
| P21 | Normal distribution | 41.638 | 46.655 | |
| P30 | Normal distribution | 37.210 | 48.537 | |
| P50 | Normal distribution | 32.869 | 46.236 | |
| P100 | Normal distribution | 27.686 | 40.016 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | One-way ANOVA | 42.065 | 48.362 |
| P21 | Normal distribution | 37.953 | 45.448 | |
| P30 | Normal distribution | 33.192 | 41.123 | |
| P50 | Normal distribution | 36.004 | 48.050 | |
| P100 | Normal distribution | 34.177 | 42.528 | |
| Mushroom | ||||
| P13 | Normal distribution | One-way ANOVA | 8.380 | 15.331 |
| P21 | Normal distribution | 8.463 | 13.480 | |
| P30 | Normal distribution | 10.926 | 22.004 | |
| P50 | Normal distribution | 11.574 | 19.644 | |
| P100 | Normal distribution | 16.809 | 29.022 | |
| Branched | ||||
| P13 | Normal distribution | One-way ANOVA | 2.906 | 6.428 |
| P21 | Normal distribution | 1.359 | 4.719 | |
| P30 | Normal distribution | 1.509 | 5.285 | |
| P50 | Normal distribution | 0.578 | 4.441 | |
| P100 | Normal distribution | 2.127 | 6.565 | |
| Multibranched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.130 | 0.940 |
| P21 | Non-normal distribution | −0.180 | 0.466 | |
| P30 | Non-normal distribution | −0.136 | 0.351 | |
| P50 | Non-normal distribution | −0.160 | 0.413 | |
| P100 | Non-normal distribution | −0.676 | 1.748 | |
| Atypical | ||||
| P13 | Not applicable | Not applicable | Not applicable | |
| P21 | Not applicable | Not applicable | ||
| P30 | Not applicable | Not applicable | ||
| P50 | Non-normal distribution | −0.221 | 0.572 | |
| P100 | Not applicable | Not applicable | ||
| Figure 4F, iSPN | ||||
| Stubby | ||||
| P13 | Normal distribution | One-way ANOVA | 39.285 | 46.048 |
| P21 | Normal distribution | 39.255 | 48.969 | |
| P30 | Normal distribution | 36.944 | 43.832 | |
| P50 | Normal distribution | 35.812 | 45.406 | |
| P100 | Normal distribution | 36.527 | 41.910 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | Kruskal–Wallis test | 35.846 | 41.869 |
| P21 | Normal distribution | 35.167 | 45.447 | |
| P30 | Normal distribution | 35.582 | 43.339 | |
| P50 | Non-normal distribution | 36.507 | 43.133 | |
| P100 | Normal distribution | 35.375 | 42.509 | |
| Mushroom | ||||
| P13 | Normal distribution | One-way ANOVA | 12.688 | 19.001 |
| P21 | Normal distribution | 10.056 | 13.477 | |
| P30 | Normal distribution | 13.213 | 18.533 | |
| P50 | Normal distribution | 13.252 | 21.245 | |
| P100 | Normal distribution | 15.118 | 22.548 | |
| Branched | ||||
| P13 | Normal distribution | One-way ANOVA | 0.855 | 4.189 |
| P21 | Normal distribution | 1.312 | 5.933 | |
| P30 | Normal distribution | 2.452 | 5.705 | |
| P50 | Normal distribution | 0.907 | 3.740 | |
| P100 | Normal distribution | 1.152 | 4.363 | |
| Multibranched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.139 | 0.359 |
| P21 | Non-normal distribution | −0.243 | 0.627 | |
| P30 | Non-normal distribution | −0.104 | 0.505 | |
| P50 | ||||
| P100 | Non-normal distribution | −0.126 | 0.623 | |
| Atypical | ||||
| P13 | Not applicable | Not applicable | Not applicable | |
| P21 | Not applicable | Not applicable | ||
| P30 | Not applicable | Not applicable | ||
| P50 | Not applicable | Not applicable | ||
| P100 | Not applicable | Not applicable | ||
| Figure 5D, dSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 6.917 | 9.796 |
| P21 | Normal distribution | Two-way ANOVA | 10.298 | 12.574 |
| P30 | Normal distribution | Two-way ANOVA | 8.920 | 11.798 |
| P50 | Normal distribution | Two-way ANOVA | 8.629 | 11.508 |
| P100 | Normal distribution | Two-way ANOVA | 7.408 | 9.758 |
| Figure 5D, iSPN | ||||
| P13 | Normal distribution | Two-way ANOVA | 7.217 | 10.096 |
| P21 | Normal distribution | Two-way ANOVA | 7.404 | 10.283 |
| P30 | Normal distribution | Two-way ANOVA | 9.421 | 12.299 |
| P50 | Normal distribution | Two-way ANOVA | 9.540 | 12.418 |
| P100 | Normal distribution | Two-way ANOVA | 7.003 | 9.353 |
| Figure 5F, dSPN | ||||
| Stubby | ||||
| P13 | Normal distribution | One-way ANOVA | 35.906 | 43.439 |
| P21 | Normal distribution | 36.294 | 47.263 | |
| P30 | Normal distribution | 27.286 | 43.385 | |
| P50 | Normal distribution | 28.076 | 40.691 | |
| P100 | Normal distribution | 29.939 | 38.538 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | One-way ANOVA | 40.338 | 51.234 |
| P21 | Normal distribution | 39.407 | 47.054 | |
| P30 | Normal distribution | 42.124 | 54.845 | |
| P50 | Normal distribution | 36.213 | 48.237 | |
| P100 | Normal distribution | 35.388 | 41.348 | |
| Mushroom | ||||
| P13 | Normal distribution | One-way ANOVA | 9.094 | 13.298 |
| P21 | Normal distribution | 7.943 | 15.476 | |
| P30 | Normal distribution | 7.968 | 16.939 | |
| P50 | Normal distribution | 13.707 | 23.974 | |
| P100 | Normal distribution | 19.883 | 27.036 | |
| Branched | ||||
| P13 | Normal distribution | Kruskal–Wallis test | 0.842 | 4.111 |
| P21 | Normal distribution | 1.277 | 3.529 | |
| P30 | Non-normal distribution | −0.936 | 5.831 | |
| P50 | Normal distribution | 1.767 | 7.091 | |
| P100 | Normal distribution | 2.020 | 4.736 | |
| Multibranched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | 0.032 | 1.144 |
| P21 | Non-normal distribution | −0.107 | 1.309 | |
| P30 | Non-normal distribution | −0.267 | 1.857 | |
| P50 | Non-normal distribution | −0.154 | 0.398 | |
| P100 | Non-normal distribution | −0.045 | 0.625 | |
| Atypical | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.150 | 0.713 |
| P21 | Non-normal distribution | −0.135 | 0.690 | |
| P30 | Non-normal distribution | −0.611 | 1.579 | |
| P50 | Not applicable | Not applicable | ||
| P100 | Non-normal distribution | −0.040 | 0.572 | |
| Figure 5F, iSPN | ||||
| Stubby | ||||
| P13 | Normal distribution | Kruskal–Wallis test | 32.329 | 40.112 |
| P21 | Non-normal distribution | 31.046 | 43.463 | |
| P30 | Normal distribution | 34.412 | 42.389 | |
| P50 | Normal distribution | 33.412 | 41.381 | |
| P100 | Normal distribution | 34.637 | 39.443 | |
| Thin/Filopodial | ||||
| P13 | Normal distribution | One-way ANOVA | 40.414 | 48.479 |
| P21 | Normal distribution | 38.564 | 48.200 | |
| P30 | Normal distribution | 34.343 | 43.229 | |
| P50 | Normal distribution | 36.824 | 42.283 | |
| P100 | Normal distribution | 34.603 | 40.100 | |
| Mushroom | ||||
| P13 | Normal distribution | One-way ANOVA | 9.046 | 16.662 |
| P21 | Normal distribution | 12.614 | 21.038 | |
| P30 | Normal distribution | 15.105 | 22.990 | |
| P50 | Normal distribution | 14.417 | 22.040 | |
| P100 | Normal distribution | 19.207 | 26.732 | |
| Branched | ||||
| P13 | Normal distribution | One-way ANOVA | 2.419 | 8.748 |
| P21 | Normal distribution | 0.658 | 4.182 | |
| P30 | Normal distribution | 2.002 | 5.530 | |
| P50 | Normal distribution | 2.462 | 5.799 | |
| P100 | Normal distribution | 1.196 | 2.832 | |
| Multibranched | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.603 | 1.978 |
| P21 | Non-normal distribution | −0.149 | 0.384 | |
| P30 | Not applicable | Not applicable | ||
| P50 | Non-normal distribution | −0.375 | 1.239 | |
| P100 | Non-normal distribution | −0.294 | 1.313 | |
| Atypical | ||||
| P13 | Non-normal distribution | Kruskal–Wallis test | −0.263 | 0.680 |
| P21 | Not applicable | Not applicable | ||
| P30 | Not applicable | Not applicable | ||
| P50 | Non-normal distribution | −0.263 | 0.680 | |
| P100 | Non-normal distribution | −0.132 | 0.362 | |
Data and materials availability
All data are available in the main text.
Results
Dendritic spines were pruned in dSPNs and iSPNs of the dorsolateral striatum after P30
We microinjected Cre-dependent AAV9-EF1α-DIO-eYFP reporter viruses into subregions of the striatum of P0–P2 striatum of Drd1a-Cre and Adora2a-Cre mice to label the dendritic spines of dSPNs and iSPNs, respectively. Microinjected brains were harvested for spine analysis at stages of early juvenile (P13), late juvenile (P21), early adolescence (P30), late adolescence (P50), and adulthood (P100).
Because the dorsolateral and dorsomedial parts of the striatum have distinct roles in the regulation of motor learning, habit formation and drug addiction (Thorn et al., 2010; Lipton et al., 2019), we investigated dendritic spinogenesis in the dorsolateral and dorsomedial striatum. In the dorsolateral striatum, the density of the spines in dSPNs increased to reach its peak at P30. Subsequently, the dendritic spines were significantly pruned between P30 and P50. At P100, the spine density returned to a level comparable to that of P21 (Fig. 1A,B,D).
Figure 1.
Development of dendritic spines in dSPNs and iSPNs of the dorsolateral striatum. A, Schematic drawings show the regions of the dorsolateral striatum that are included for analysis. B, C, Immunohistochemistry of eYFP shows the development of eYFP-labeled dendritic spines in dSPNs and iSPNs, respectively, in the dorsolateral striatum (caudoputamen) of Drd1-Cre mice (B) and Adora2a-Cre mice (C). D, Quantification of spine density. E, Quantitative analysis of morphologic profiles of spines during development. F, Dynamic changes of specific types of spines during development. * differences between ages. *p < 0.05, **p < 0.01, ***p < 0.001. # differences between genotypes. ##p < 0.01. Two-way ANOVA is used in D. One-way ANOVA followed by Tukey’s HSD post hoc tests is used in F for the data that are normally distributed. Kruskal–Wallis one-way ANOVA followed by Dunn’s pairwise multiple comparisons tests are used in F for data that are not normally distributed. Data in D and E are mean ± SEM; Thin/Filopodial spine data in F are mean ± SEM; Stubby, Mushroom, Branched, Multi-branched and Atypical spine data in F are median ± interquatile range. n = 10 cells from 2–3 mice/age. P, postnatal day. Scale bar: 10 μm.
Regarding the development of different types of dendritic spines in dSPNs, the mushroom type of spines gradually increased from P13 to P100 along with a reduction in thin/filopodial spines between P50 and P100 (Fig. 1E,F).
For iSPNs, the spine density progressively increased to reach the maximum level at P30. Spine pruning in the iSPN population subsequently occurred in P30–P100, which was longer than the pruning time window of P30–P50 in the dSPN population (Fig. 1A,C,D). However, with all spines, the percentage of each type of spine in iSPNs appeared to remain about the same level across developmental times (Fig. 1E,F).
Dendritic spines were pruned in dSPNs and iSPNs of the dorsomedial striatum after P50
In the dorsomedial striatum of dSPNs, the density of the spine reached its peak at P30, and the plateau level was maintained from P30 to P50. Subsequently, the dendritic spines underwent pruning between P50 and P100. By P100, the spine density dropped back to a level comparable to that of P21 (Fig. 2A,B,D).
Figure 2.
Development of dendritic spines in dSPNs and iSPNs of the dorsomedial striatum. A, Schematic drawings show the regions of the dorsomedial striatum that are included for analysis. B, C, Immunohistochemistry of eYFP shows the development of eYFP-labeled dendritic spines in dSPNs and iSPNs, respectively, in the dorsomedial striatum (caudoputamen) of Drd1-Cre mice (B) and Adora2a-Cre mice (C). D, Quantification of spine density. E, Quantitative analysis of morphologic profiles of spines during development. F, Dynamic changes of specific types of spines during development. * differences between ages. *p < 0.05, **p < 0.01, ***p < 0.001. # differences between genotypes. ##p < 0.01. Two-way ANOVA is used in D. One-way ANOVA followed by Tukey’s HSD post hoc tests is used in F for the data that are normally distributed. Kruskal–Wallis one-way ANOVA followed by Dunn’s pairwise multiple comparisons tests are used in F for data that are not normally distributed. Data in D and E are mean ± SEM; Stubby and Thin/Filopodial spine data in F are mean ± SEM; Mushroom, Branched, Multi-branched and Atypical spine data in F are median ± interquatile range. n = 10 cells from 2–3 mice/age. P, postnatal day. Scale bar: 10 μm.
For iSPNs, the spine density remained stable before P21 and progressively increased after P21. Compared with the peak of spines at P30 in the dSPN population, the spine density of iSPNs reached its maximum level later between P30–P50. By P100, the spine density of iSPNs fell back to a level comparable to that of P13–P21 (Fig. 2A,C,D). Interestingly, we found a higher spine density in the iSPNs at P13 and P50 compared with the dSPNs, which may result from a late-onset spine pruning of iSPNs. By P100 at adulthood, the spine density between dSPNs and iSPNs was similar.
As for temporal profiles of spine classification in the dorsomedial striatum, the proportion of mushroom spines were significantly increased in dSPNs by 1.87-fold at P100 (Fig. 2E,F). In contrast, no significant change in temporal profiles of spine classification was observed in iSPNs of the dorsomedial striatum (Fig. 2E,F).
Although the developmental trajectories of dendritic spines were different between the dorsolateral and dorsomedial SPNs, the spine densities of dSPNs and iSPNs were comparable between these two regions during postnatal development except at P30, the spiny density of iSPNs of the dorsomedial striatum was higher than that of the dorsolateral striatum (Table 2).
Table 2.
Postnatal development of dendritic spines of striatonigral (dSPNs) and striatopallidal neurons (iSPNs) in the dorsolateral and dorsomedial striatum of male mice
| Striatonigral neurons/dSPNs | Striatopallidal neurons/iSPNs | |||
|---|---|---|---|---|
| Age | DLS | DMS | DLS | DMS |
| P13 | 6.408 ± 0.650 | 6.687 ± 0.777 | 9.076 ± 0.719 | 8.786 ± 0.639 |
| P21 | 9.087 ± 0.658 | 9.921 ± 0.992 | 11.007 ± 0.659 | 10.606 ± 0.319 |
| P30 | 14.650 ± 0.917 | 12.539 ± 0.893 | 17.079 ± 1.129** | 12.803 ± 0.844 |
| P50 | 11.351 ± 1.051 | 11.351 ± 1.051 | 13.443 ± 1.169 | 14.848 ± 1.124 |
| P100 | 10.646 ± 0.744 | 10.115 ± 0.399 | 9.318 ± 0.573 | 8.741 ± 0.320 |
The numbers indicate the density of dendritic spines (mean ± SEM) on different postnatal days (P).
**p < 0.01. DLS, dorsolateral striatum; DMS, dorsomedial striatum.
Dendritic spines were preferentially pruned in dSPNs but not in iSPNs in the shell region of NAc
It has been shown that dSPN and iSPN populations in the medial and lateral parts of the NAc shell serve different functions, e.g., dSPNs and iSPNs in the NAc shell differentially regulate reward and aversion (Yang et al., 2018; Yao et al., 2021). We investigated spinogenesis in the medial and lateral parts of the NAc shell.
In the medial part of the NAc shell, the spine density of dSPNs was already high at P13, and a high level was maintained until P30. Marked spine pruning was found between P30 and P50. A trend of decreasing spine density was observed at P30–P100 (Fig. 3A,B,D). In contrast to dSPNs, no prominent spine pruning was found in iSPNs from P13 to P100, although there was a moderately increasing trend from P21 to P50 (Fig. 3A,C,D).
Figure 3.
Development of dendritic spines in dSPNs and iSPNs of the medial shell of the nucleus accumbens. A, Schematic drawings show the regions of the medial shell of the nucleus accumbens that are included for analysis. B, C, Immunohistochemistry of eYFP shows the development of eYFP-labeled dendritic spines in dSPNs and iSPNs, respectively, in the medial shell of the nucleus accumbens of Drd1-Cre mice (B) and Adora2a-Cre mice (C). D, Quantification of spine density. E, Quantitative analysis of morphologic profiles of spines during development. F, Dynamic changes of specific types of spines during development. * differences between ages. *p < 0.05, **p < 0.01, ***p < 0.001. # differences between genotypes. #p < 0.05. ##p < 0.01. Two-way ANOVA is used in D. One-way ANOVA followed by Tukey’s HSD post hoc tests is used in F for the data that are normally distributed. Kruskal–Wallis one-way ANOVA followed by Dunn’s pairwise multiple comparisons tests were used in F for data that are not normally distributed. Data in D and E are mean ± SEM; Stubby, Thin/Filopodial and Mushroom spine data in F are mean ± SEM; Branched, Multi-branched and Atypical spine data in F are median ± interquatile range. n = 10 cells from 2–3 mice/age. P, postnatal day. Scale bar: 10 μm.
In the lateral part of the NAc shell, the spine density of dSPNs reached its peak at P30, and spine pruning was observed between P30 and P50. At P100, the spine density was slightly higher than that at P13–P21 (Fig. 4A,B,D). For iSPNs, no substantial changes in the spine density were found in the lateral part of the NAc shell from P13 to P100 (Fig. 4A,C,D).
Figure 4.
Development of dendritic spines in dSPNs and iSPNs of the lateral shell of the nucleus accumbens. A, Schematic drawings show the regions of the lateral shell of the nucleus accumbens that are included for analysis. B, C, Immunohistochemistry of eYFP shows the development of eYFP-labeled dendritic spines in dSPNs and iSPNs, respectively, in the lateral shell of the nucleus accumbens of Drd1-Cre mice (B) and Adora2a-Cre mice (C). D, Quantification of spine density. E, Quantitative analysis of morphologic profiles of spines during development. F, Dynamic changes of specific types of spines during development. * differences between ages. *p < 0.05, **p < 0.01, ***p < 0.001. # differences between genotypes. #p < 0.05. Two-way ANOVA is used in D. One-way ANOVA followed by Tukey’s HSD post hoc tests is used in F for the data that are normally distributed. Kruskal–Wallis one-way ANOVA followed by Dunn’s pairwise multiple comparisons tests are used in F for data that are not normally distributed. Data in D and E are mean ± SEM; Stubby, Thin/Filopodial, Mushroom, Branched spine data of dSPN and Stubby, Mushroom, Branched spine data of iSPN in F are mean ± SEM; Multi-branched, Atypical spine data of dSPN and Thin/Filopodial, Multi-branched, Atypical spine data of iSPN in F are median ± interquatile range. n = 10 cells from 2–4 mice/age. P, postnatal day. Scale bar: 10 μm.
Regarding temporal profiles of morphologic changes of dendritic spines, progressively increasing proportions of mushroom spines were found in both dSPN and iSPN of lateral and medial parts of the NAc shell (Figs. 3E,F, 4E,F). Decreasing levels of stubby or thin/filopodial spines were also found in dSPNs of medial and lateral parts of the NAc shell and iSPNs of the medial part of the NAc shell, although there was a slight increase in thin/filopodial spines between P30 and P100 in dSPNs of medial NAc shell (Figs. 3E,F, 4E,F). These results suggest a progressive maturation of dendritic spines in dSPNs and iSPNs of NAc shell from immature to mature types during postnatal development.
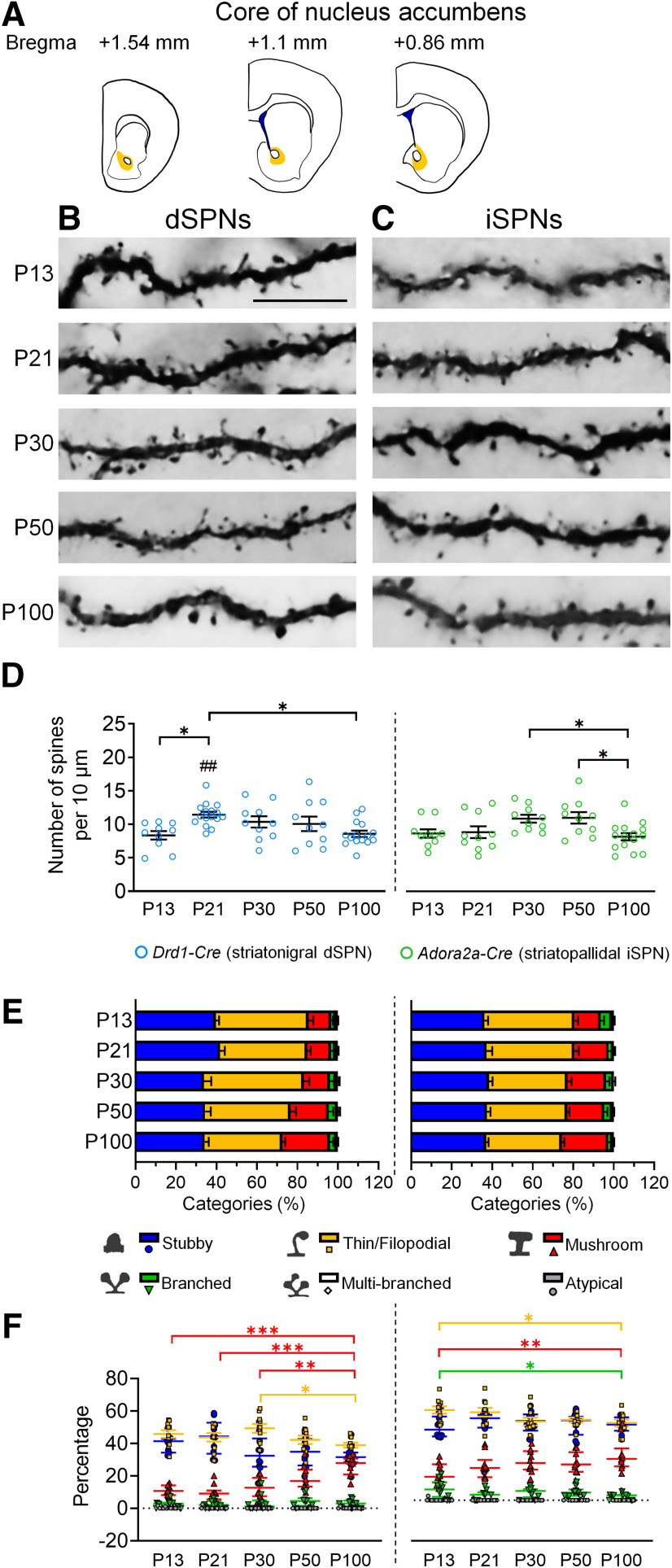
Spine pruning progressively occurred in dSPNs and iSPNs in the core region of NAc
In the NAc core, the maximum level of spine density of dSPNs was observed at P21. Subsequently, spine density decreased until P100, at which time the dendritic spines were at a level comparable to that of P13 (Fig. 5A,B,D). For iSPNs, spine density reached its peak level at P30 and P50 and was then gradually decreased from P50 to P100 (Fig. 5A,C,D).
Figure 5.
Development of dendritic spines in dSPNs and iSPNs of the core of the nucleus accumbens. A, Schematic drawings show the regions of the core of the nucleus accumbens that are included for analysis. B, C, Immunohistochemistry of eYFP shows the development of eYFP-labeled dendritic spines in dSPNs and iSPNs, respectively, in the core of the nucleus accumbens of Drd1-Cre mice (B) and Adora2a-Cre mice (C). D, Quantification of spine density. E, Quantitative analysis of morphologic profiles of spines during development. F, Dynamic changes of specific types of spines during development. * differences between ages. *p < 0.05, **p < 0.01, ***p < 0.001. # differences between genotypes. ##p < 0.01. Two-way ANOVA is used in D. One-way ANOVA followed by Tukey’s HSD post hoc tests are used in F for the data that are normally distributed. Kruskal–Wallis one-way ANOVA followed by Dunn’s pairwise multiple comparisons tests are used in F for data that are not normally distributed. Data in D and E are mean ± SEM; Stubby, Thin/Filopodial, Mushroom spine data of dSPN and Thin/Filopodial, Mushroom, Branched spine data of iSPN in F are mean ± SEM; Branched, Multi-branched, Atypical spine data of dSPN and Stubby, Multi-branched, Atypical spine data of iSPN in F are median ± interquatile range. n = 10–16 cells from 2–3 mice/age. P, postnatal day. Scale bar: 10 μm.
For morphologic analysis, thin/filopodial spines were reduced after P30, accompanied by a gradual increase of mushroom spines in dSPNs throughout postnatal development (Fig. 5E,F). As for iSPNs, a gradually increased proportion of mushroom spines was found from P13 to P100. Similar to dSPNs, the proportion of thin/filopodial spines in iSPNs was lower at P100 compared with that of P13 (Fig. 5E,F).
Among the different regions of the NAc, the spine density of dSPNs in the core of NAc core was lower than that of dSPNs in the lateral part of NAc shell at P30 and P100 (Table 3), which was consistent with the prominent pruning found in the core of NAc after P21 (Fig. 5D). Furthermore, the spine density of iSPNs in the medial part of NAc shell was lowest at P21 compared with other regions of NAc (Table 3; Fig. 3D).
Table 3.
Postnatal development of dendritic spines of striatonigral (dSPNs) and striatopallidal neurons (iSPNs) in subregions of the nucleus accumbens of male mice
| Age | NAcc | NAcsLat | NAcsMed |
|---|---|---|---|
| Striatonigral neurons/dSPNs | |||
| P13 | 8.356 ± 0.634 | 8.780 ± 0.803 | 10.222 ± 0.562 |
| P21 | 11.492 ± 0.685 | 9.772 ± 0.546 | 10.265 ± 0.712 |
| P30 | 10.359 ± 0.849# | 14.250 ± 0.596 | 11.852 ± 0.822 |
| P50 | 10.068 ± 1.087 | 10.884 ± 0.919 | 8.797 ± 0.486 |
| P100 | 8.700 ± 0.675# | 10.923 ± 0.474 | 8.461 ± 1.215# |
| Striatopallidal neurons/iSPNs | |||
| P13 | 8.657 ± 0.616 | 9.501 ± 0.557 | 8.792 ± 0.825 |
| P21 | 8.843 ± 0.878* | 10.539 ± 0.681** | 7.876 ± 0.659 |
| P30 | 10.860 ± 0.587 | 11.990 ± 0.536 | 8.495 ± 0.699 |
| P50 | 10.979 ± 0.857 | 10.861 ± 0.764 | 10.019 ± 0.613 |
| P100 | 8.137 ± 0.816 | 9.681 ± 0.575 | 8.618 ± 0.395 |
The numbers indicate the density of dendritic spines (mean ± SEM) on different postnatal days (P). # versus NAcsLat, #p < 0.05. * versus NAcsMed, *p < 0.05, **p < 0.01. NAcc: nucleus accumbens core; NAcsLat: lateral part of nucleus accumbens shell; NAcsMed: medial part of nucleus accumbens shell.
Discussion
We investigated the developmental trajectories of dendritic spines of dSPN and iSPN populations in the caudoputamen and NAc. We took advantage of the Cre/LoxP system and viral targeting strategies to label specific cell types of SPNs. With the aid of eYFP immunohistochemistry, it allows us to identify the protrusion of eYFP-positive dendritic spines of SPNs, although the resolution was limited to resolve the morphologic detail of individual spines. Our study complements previous reports and provides new information on spine formation in striatal neurons at three levels. First, we analyzed spinogenesis in subregions of the striatal complex, including the medial and lateral parts of the caudoputamen (dorsal striatum), and the core and shell regions of the NAc (ventral striatum). Second, in each striatal subregion, we provide a cell-type characterization of spinogenesis in dSPN and iSPN populations. Third, by analyzing multiple time points in postnatal stages, including early juvenile at P13, late juvenile at P21, early adolescence at P30, late adolescence at P50, and adulthood at P100, we delineated the developmental trajectories of dSPN and iSPN populations in each striatal subregion.
In general, we found that the dendritic spines of dSPNs and iSPNs progressively increased at the early stages of postnatal development, followed by prominent spine pruning beginning from adolescence in both the dorsal and ventral striatum. The developmental maturation of dendritic spines in dSPNs and iSPNs, however, follow different spatiotemporal trajectories in the dorsal and ventral striatum, implicating cell type-specific maturation of striatal circuits.
Developmental maturation of dendritic spines in the caudoputamen of striatal circuitry
Dendritic spines are presumably the loci of excitatory synapses. SPNs receive excitatory inputs from the cerebral cortex and the thalamus. Despite some discrepancies among different species, corticostriatal and thalamostriatal axonal terminals preferentially innervate, respectively, the head and shaft of spines (Smith et al., 2004; Liu et al., 2011). Previous studies have characterized spinogenesis in the dorsal striatum in early postnatal development. Immature dendritic spines of striatal neurons occur as early as P6 (Lee and Sawatari, 2011). Mature spines appear in striatal neurons at P8-P9 (Lee and Sawatari, 2011; Chen et al., 2016), followed by extensive growth of spines at P10–P28 (Tepper et al., 1998; Uryu et al., 1999; Peixoto et al., 2016; Krajeski et al., 2019). Electron microscopic study has reported synaptic pruning in the striatum from P18 to adulthood (Uryu et al., 1999). These previous studies, however, do not provide information on spine development and maturation in specific cell types in the striatum.
In the present study, we found that in the caudoputamen, the density of spines in dSPNs and iSPNs progressively increased in the early stages of the postnatal period before P30, followed by prominent spine pruning in both medial and lateral parts of the caudoputamen after P30–P50 onwards. The findings of progressive increases in SPN spines before P30 are likely to reflect the increasing innervations of SPNs by the cortex and thalamus (Krajeski et al., 2019). Consistent with these results, previous electrophysiological study with morphologic analysis shows that a gradual increase in dendritic spines occurs parallelly in dSPNs and iSPNs at P3–P28, although this study does not have spine data after P35 (Krajeski et al., 2019).
We further found a difference in the trajectory of spine pruning in dSPNs and iSPNs. For the dSPN population, the time window of spine pruning occurred after P30, which was earlier than the iSPN population in which spine pruning occurred after P50. The cellular mechanisms underlying the differential regulation of spine pruning time windows in dSPNs and iSPNs are not yet known. Distinct intrinsic electrophysiological properties and biased inhibitory inputs to dSPN and iSPN populations after the second postnatal week may be involved in the differential regulation of spinogenesis (Krajeski et al., 2019).
The developmental maturation of dendritic spines is likely to reflect the functional maturation of excitatory synapses. Previous electrophysiological and optogenetic study has demonstrated that progressively increased amplitudes of optically evoked excitatory postsynaptic currents in corticostriatal circuits are correlated with developmental increases in the ratio of AMPA/NMDA currents and dendritic spine density of SPNs in the first postnatal month period (Peixoto et al., 2016), suggesting a strong functional interaction between the developing cortex and striatum during corticostriatal circuit wiring.
Developmental maturation of dendritic spines in the nucleus accumbens of striatal circuitry
Despite that neural plasticity of dendritic spines of SPNs is well characterized in the NAc under different behavioral states, including reward learning, drug addiction and chronic social defeat-induced stress (LaPlant et al., 2010; Fox et al., 2020; Iino et al., 2020; Thompson et al., 2021), the developmental trajectory of spinogenesis in the NAc has not yet been fully characterized. Previous studies have reported that spine density is stably maintained in SPNs of the NAc core of the rat brain at P21–P70 (Bringas et al., 2013; Tendilla-Beltrán et al., 2016).
Our present study found a dynamic profile of spine development and maturation in dSPNs and iSPNs of the NAc. The spine density of dSPNs and iSPNs in the NAc core reached its peak level at P21 and P30, respectively, and subsequently decreased in postnatal periods. Microglia plays an important role in spine pruning (Paolicelli et al., 2011). Mallya et al., have reported that microglia-mediated engulfment of presynaptic terminals and postsynaptic dendritic spines is involved in the elimination of synapses in the prefrontal cortex during adolescence (Mallya et al., 2019). Interestingly, a marked increase in the proliferation of microglia occurs in the NAc during the third postnatal week (Hope et al., 2020). Given microglia-mediated phagocytic activity to prune synapses evident in the prefrontal cortex in postnatal development (Mallya et al., 2019), microglia-mediated synaptic pruning may be involved in shaping the maturation patterns of spines in dSPNs and iSPNs that we observed in the NAc. Such microglia-mediated synaptic modification conceivably may have a behavioral impact. For example, microglia and complement-mediated phagocytic activity have been shown to shape NAc development by eliminating dopamine D1R, which impacts the development of social behavior in adolescent rats (Kopec et al., 2018). Interestingly, microglia with distinct physiological characteristics emerge in different regions of basal ganglia during the second postnatal week (De Biase et al., 2017). Because microglia are implicated in the differential responses of excitatory postsynaptic currents in dSPNs and iSPNs of the dorsal striatum in adult mice (Hayes et al., 2022), it will be of interest to see whether region-specific microglia play a role in determining differential trajectories of spine maturation in SPNs of the caudoputamen and NAc.
Activity-dependent regulation of the development and maturation of spines/synapses of SPNs in striatal circuitry
Sensory inputs and learning-related experiences can induce neuroplastic changes in spine formation and elimination (Berry and Nedivi, 2017), indicating that neuronal activity plays an important role in spinogenesis/synaptogenesis. The striatum receives three major afferents from the cerebral cortex, thalamus, and ventral midbrain. SPNs integrate glutamatergic inputs through corticostriatal and thalamostriatal pathways. The activities of glutamatergic inputs are influential in spinogenesis and synaptic wiring during development. It has been proposed that the balanced activity of dSPN and iSPN pathways controls glutamatergic synaptogenesis/spinogensis via recurrent closed loops of cortico-basal ganglia-thalamus circuits (Kozorovitskiy et al., 2012). It is yet unknown whether thalamostriatal activity could affect synaptogenesis/spinogensis of SPNs. Despite that the percentages of corticostriatal and thalamostriatal synapses in dSPNs and iSPNs are varied among different studies (Doig et al., 2010; Lei et al., 2013), it has been shown that repetitive stimulations of corticostriatal and thalamostriatal pathways, respectively, increase and decrease postsynaptic depolarization in dSPNs and iSPNs (Ding et al., 2008). Given the nature of activity-dependent regulation of spine/synapse formation, it will be of great interest to look into the possibility of how corticostriatal and thalamostriatal glutamatergic inputs are integrated and as a consequence impact the trajectories of spine maturation in dSPNs and iSPNs that we observed in the present study.
Synaptic plasticity, including long-term potentiation (LTP) and long-term depression (LTD) that occurs in glutamatergic synapses of SPNs, may play a role in sculpting dendritic spines during development. In the rat striatum, LTP and LTD can occur in striatal neurons in early postnatal stages of P12–P15 (Partridge et al., 2000). However, in the mouse striatum, Maltese et al., have reported that LTP and LTD cannot be stably induced, respectively, before P24 and P28 (Maltese et al., 2018). Furthermore, the endocannabinoid receptor (CB1R)-mediated LTD can only be induced in corticostriatal synapses, but not in thalamostriatal synapses in adult brains (Wu et al., 2015). Intriguingly, the CB1R expression in corticostriatal terminals undergoes prominent downregulation after P28 in both the cortex and striatum (Van Waes et al., 2012), which coincides with the period of spine pruning in SPNs of the caudoputamen. Given the ability of LTP and LTD in shaping spine formation and elimination, it will be of interest to see whether neural plasticity mechanisms of LTP and LTD may participate in the spine/synapse maturation of dSPNs and iSPNs.
Dopamine inputs from the ventral midbrain are important for striatal development and maturation. Dopamine increases dSPN activity but decreases iSPN activity (Nishi et al., 2000), which may, in turn, regulate spinogenesis. It has been shown that chemogenetic inhibition of dSPNs and iSPNs during P8–P14, respectively, decreases and increases dendritic spines (Kozorovitskiy et al., 2012). Dopamine is known to be involved in neural plasticity of striatal LTP, LTD and spinogenesis (Martel and Galvan, 2022), and activation of D1R and D2R promotes, respectively, LTP and LTD in striatal glutamatergic synapses (Shen et al., 2008; Higley and Sabatini, 2010). Interestingly, the spine formation of SPNs can be modulated by D1R and A2aR signalings in P8–P13 (Kozorovitskiy et al., 2015). The co-activation of D1R and A2aR increases the spines of iSPNs (Kozorovitskiy et al., 2015), whereas genetic deletion of D1R decreases the spines of dSPNs and iSPNs (Suarez et al., 2020).
Intrinsic inputs from local interneurons may sculpt the trajectory of SPN spinogenesis. Late-onset maturation of local inhibitory interneurons may contribute to shaping the excitability of SPNs by increasing inhibitory inputs that could result in spine pruning (Krajeski et al., 2019). Notably, as cholinergic interneurons can modulate corticostriatal LTD (Wang et al., 2006), such cholinergic interneuron-modulated LTD activity may, in turn, regulate spinogenesis.
Dysregulation of synaptogenesis and spinogenesis in neuropsychiatric diseases
Developmental modification of dendritic spines is critical to the maturation of precise neural networks. Defective development and plasticity of dendritic spines are associated with the pathophysiology of neuropsychiatric disorders (Forrest et al., 2018). Aberrant synaptic pruning had been reported in cortical pyramidal neurons in patients with autism spectrum disorder (ASD) (Hutsler and Zhang, 2010) and animal models of ASD (Pan et al., 2010; Jiang et al., 2013; Isshiki et al., 2014). Abnormal spinogenesis occurs in striatal neurons of ASD mouse models (Peça et al., 2011; Chen et al., 2016), e.g., genetic mutation of the autism-risk gene Shank3 results in a selective decrease in spine density of iSPNs but not dSPNs (Wang et al., 2017). Furthermore, the re-introduction of Shank3 gene expression in adult striatal neurons rescues dendritic spine deficits and behavioral abnormalities in Shank3 knock-out mice (Mei et al., 2016). Excessive spine pruning and spine dysgenesis have also been reported in patients with schizophrenia (Feinberg, 1982; Roberts et al., 1996; MacDonald et al., 2017; McKinney et al., 2019; Onwordi et al., 2020), in which excessive synaptic pruning may be mediated by microglia (Sellgren et al., 2019). These clinical and preclinical studies highlight the importance of understanding synaptogenesis and spinogenesis in postnatal brains during maturation. Our current study, therefore, provides a basic reference to neurodevelopmental diseases in which spinogenesis and synaptogenesis are affected in striatal neurons.
Limitations and issues for further work
Our study is subject to limitations. We have only focused on the characterization of spine development. We did not investigate the potential role of cortical and thalamic excitatory inputs in the regulation of the development and maturation of dendritic spines in dSPNs and iSPNs. This is an interesting and important question, given the key role of neuronal activity in determining spine/synapse structure and function (Segal and Andersen, 2000; Martel and Galvan, 2022). The whole-brain mapping study has shown that the direct dSPN and indirect iSPN pathways receive overlapping and yet differential inputs from the cortex and thalamus (Wall et al., 2013; Guo et al., 2015). Network connectivity coupled with intrinsic neurochemical and molecular differences between dSPNs and iSPNs may underlie differential development and maturation of spines in the caudoputamen and NAc. The resulting precise synaptic wiring through spine/synapse maturation ensures proper circuit functions in the basal ganglia.
Despite these and other limitations, our study delineates the trajectories of spine maturation in dSPNs and iSPNs in the motor circuitry-related caudoputamen and the limbic circuitry-associated NAc of the striatal complex. Our study provides a basic reference for future studies on the maturation of striatal circuitry and neurologic diseases related to basal ganglia.
Acknowledgments
Acknowledgments: We thank the Ministry of Science and Technology, National Science and Technology Council, and the Ministry of Education in Taiwan for supporting this project.
Synthesis
Reviewing Editor: Christophe Bernard, INSERM & Institut de Neurosciences des Systèmes
Decisions are customarily a result of the Reviewing Editor and the peer reviewers coming together and discussing their recommendations until a consensus is reached. When revisions are invited, a fact-based synthesis statement explaining their decision and outlining what is needed to prepare a revision will be listed below. The following reviewer(s) agreed to reveal their identity: Kenneth Campbell.
Your reviewers and I agree that some attempt should be made in the discussion to evaluate those results in the context of overall maturation of striatal neuronal circuitry.
We also regret that you did not try to assess the striatal afferent inputs in relation to the spine development/maturation. The available tissue would be sufficient to perform such experiments. But we also understand that you may not wish to include such analysis. This is up to you.
In conclusion, if you decide not to address the previous issue, just expand the discussion as requested.
References
- Albin RL, Young AB, Penney JB (1989) The functional anatomy of basal ganglia disorders. Trends Neurosci 12:366–375. 10.1016/0166-2236(89)90074-x [DOI] [PubMed] [Google Scholar]
- Arber S, Costa RM (2022) Networking brainstem and basal ganglia circuits for movement. Nat Rev Neurosci 23:342–360. 10.1038/s41583-022-00581-w [DOI] [PubMed] [Google Scholar]
- Batool S, Raza H, Zaidi J, Riaz S, Hasan S, Syed NI (2019) Synapse formation: from cellular and molecular mechanisms to neurodevelopmental and neurodegenerative disorders. J Neurophysiol 121:1381–1397. 10.1152/jn.00833.2018 [DOI] [PubMed] [Google Scholar]
- Berry KP, Nedivi E (2017) Spine dynamics: are they all the same? Neuron 96:43–55. 10.1016/j.neuron.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringas ME, Carvajal-Flores FN, López-Ramírez TA, Atzori M, Flores G (2013) Rearrangement of the dendritic morphology in limbic regions and altered exploratory behavior in a rat model of autism spectrum disorder. Neuroscience 241:170–187. 10.1016/j.neuroscience.2013.03.030 [DOI] [PubMed] [Google Scholar]
- Cao J, Willett JA, Dorris DM, Meitzen J (2018) Sex differences in medium spiny neuron excitability and glutamatergic synaptic input: heterogeneity across striatal regions and evidence for estradiol-dependent sexual differentiation. Front Endocrinol (Lausanne) 9:173. 10.3389/fendo.2018.00173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlisle HJ, Kennedy MB (2005) Spine architecture and synaptic plasticity. Trends Neurosci 28:182–187. 10.1016/j.tins.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Chen SY, Lu KM, Ko HA, Huang TH, Hao JH, Yan YT, Chang SL, Evans SM, Liu FC (2020) Parcellation of the striatal complex into dorsal and ventral districts. Proc Natl Acad Sci U S A 117:7418–7429. 10.1073/pnas.1921007117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Kuo HY, Bornschein U, Takahashi H, Chen SY, Lu KM, Yang HY, Chen GM, Lin JR, Lee YH, Chou YC, Cheng SJ, Chien CT, Enard W, Hevers W, Pääbo S, Graybiel AM, Liu FC (2016) Foxp2 controls synaptic wiring of corticostriatal circuits and vocal communication by opposing Mef2c. Nat Neurosci 19:1513–1522. 10.1038/nn.4380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biase LM, Schuebel KE, Fusfeld ZH, Jair K, Hawes IA, Cimbro R, Zhang HY, Liu QR, Shen H, Xi ZX, Goldman D, Bonci A (2017) Local cues establish and maintain region-specific phenotypes of basal ganglia microglia. Neuron 95:341–356.e6. 10.1016/j.neuron.2017.06.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong MR, Wichmann T (2015) Basal ganglia circuits as targets for neuromodulation in Parkinson disease. JAMA Neurol 72:1354–1360. 10.1001/jamaneurol.2015.2397 [DOI] [PubMed] [Google Scholar]
- Ding J, Peterson JD, Surmeier DJ (2008) Corticostriatal and thalamostriatal synapses have distinctive properties. J Neurosci 28:6483–6492. 10.1523/JNEUROSCI.0435-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding JB, Oh WJ, Sabatini BL, Gu C (2011) Semaphorin 3E-Plexin-D1 signaling controls pathway-specific synapse formation in the striatum. Nat Neurosci 15:215–223. 10.1038/nn.3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doig NM, Moss J, Bolam JP (2010) Cortical and thalamic innervation of direct and indirect pathway medium-sized spiny neurons in mouse striatum. J Neurosci 30:14610–14618. 10.1523/JNEUROSCI.1623-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elston GN, Fujita I (2014) Pyramidal cell development: postnatal spinogenesis, dendritic growth, axon growth, and electrophysiology. Front Neuroanat 8:78. 10.3389/fnana.2014.00078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltokhi A, Janmaat IE, Genedi M, Haarman BCM, Sommer IEC (2020) Dysregulation of synaptic pruning as a possible link between intestinal microbiota dysbiosis and neuropsychiatric disorders. J Neurosci Res 98:1335–1369. 10.1002/jnr.24616 [DOI] [PubMed] [Google Scholar]
- Feinberg I (1982) Schizophrenia: caused by a fault in programmed synaptic elimination during adolescence? J Psychiatr Res 17:319–334. 10.1016/0022-3956(82)90038-3 [DOI] [PubMed] [Google Scholar]
- Forrest MP, Parnell E, Penzes P (2018) Dendritic structural plasticity and neuropsychiatric disease. Nat Rev Neurosci 19:215–234. 10.1038/nrn.2018.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox ME, Figueiredo A, Menken MS, Lobo MK (2020) Dendritic spine density is increased on nucleus accumbens D2 neurons after chronic social defeat. Sci Rep 10:12393. 10.1038/s41598-020-69339-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis TC, Chandra R, Gaynor A, Konkalmatt P, Metzbower SR, Evans B, Engeln M, Blanpied TA, Lobo MK (2017) Molecular basis of dendritic atrophy and activity in stress susceptibility. Mol Psychiatry 22:1512–1519. 10.1038/mp.2017.178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-López P, García-Marín V, Freire M (2010) Dendritic spines and development: towards a unifying model of spinogenesis—a present day review of Cajal’s histological slides and drawings. Neural Plast 2010:769207. 10.1155/2010/769207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Surmeier DJ (2011) Modulation of striatal projection systems by dopamine. Annu Rev Neurosci 34:441–466. 10.1146/annurev-neuro-061010-113641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Engber TM, Mahan LC, Susel Z, Chase TN, Monsma FJ Jr, Sibley DR (1990) D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science 250:1429–1432. 10.1126/science.2147780 [DOI] [PubMed] [Google Scholar]
- Gittis AH, Kreitzer AC (2012) Striatal microcircuitry and movement disorders. Trends Neurosci 35:557–564. 10.1016/j.tins.2012.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graybiel AM, Grafton ST (2015) The striatum: where skills and habits meet. Cold Spring Harb Perspect Biol 7:a021691. 10.1101/cshperspect.a021691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grueter BA, Rothwell PE, Malenka RC (2012) Integrating synaptic plasticity and striatal circuit function in addiction. Curr Opin Neurobiol 22:545–551. 10.1016/j.conb.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaydin LA, Kreitzer AC (2016) Cortico–basal ganglia circuit function in psychiatric disease. Annu Rev Physiol 78:327–350. 10.1146/annurev-physiol-021115-105355 [DOI] [PubMed] [Google Scholar]
- Guo Q, Wang D, He X, Feng Q, Lin R, Xu F, Fu L, Luo M (2015) Whole-brain mapping of inputs to projection neurons and cholinergic interneurons in the dorsal striatum. PLoS One 10:e0123381. 10.1371/journal.pone.0123381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber SN (2016) Corticostriatal circuitry. Dialogues Clin Neurosci 18:7–21. 10.31887/DCNS.2016.18.1/shaber [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B (1992) Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J Neurosci 12:2685–2705. 10.1523/JNEUROSCI.12-07-02685.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes LN, An K, Carloni E, Li F, Vincent E, Trippaers C, Paranjpe M, Dölen G, Goff LA, Ramos A, Kano S-i, Sawa A (2022) Prenatal immune stress blunts microglia reactivity, impairing neurocircuitry. Nature 610:327–334. 10.1038/s41586-022-05274-z [DOI] [PubMed] [Google Scholar]
- Higley MJ, Sabatini BL (2010) Competitive regulation of synaptic Ca2+ influx by D2 dopamine and A2A adenosine receptors. Nat Neurosci 13:958–966. 10.1038/nn.2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope KT, Hawes IA, Moca EN, Bonci A, De Biase LM (2020) Maturation of the microglial population varies across mesolimbic nuclei. Eur J Neurosci 52:3689–3709. 10.1111/ejn.14740 [DOI] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H (2010) Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res 1309:83–94. 10.1016/j.brainres.2009.09.120 [DOI] [PubMed] [Google Scholar]
- Iino Y, Sawada T, Yamaguchi K, Tajiri M, Ishii S, Kasai H, Yagishita S (2020) Dopamine D2 receptors in discrimination learning and spine enlargement. Nature 579:555–560. 10.1038/s41586-020-2115-1 [DOI] [PubMed] [Google Scholar]
- Isshiki M, Tanaka S, Kuriu T, Tabuchi K, Takumi T, Okabe S (2014) Enhanced synapse remodelling as a common phenotype in mouse models of autism. Nat Commun 5:4742. 10.1038/ncomms5742 [DOI] [PubMed] [Google Scholar]
- Jiang M, Ash RT, Baker SA, Suter B, Ferguson A, Park J, Rudy J, Torsky SP, Chao HT, Zoghbi HY, Smirnakis SM (2013) Dendritic arborization and spine dynamics are abnormal in the mouse model of MECP2 duplication syndrome. J Neurosci 33:19518–19533. 10.1523/JNEUROSCI.1745-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiflin R, Janak PH (2015) Dopamine prediction errors in reward learning and addiction: from theory to neural circuitry. Neuron 88:247–263. 10.1016/j.neuron.2015.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HF, Hikosaka O (2015) Parallel basal ganglia circuits for voluntary and automatic behaviour to reach rewards. Brain 138:1776–1800. 10.1093/brain/awv134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klawonn AM, Malenka RC (2018) Nucleus accumbens modulation in reward and aversion. Cold Spring Harb Symp Quant Biol 83:119–129. 10.1101/sqb.2018.83.037457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopec AM, Smith CJ, Ayre NR, Sweat SC, Bilbo SD (2018) Microglial dopamine receptor elimination defines sex-specific nucleus accumbens development and social behavior in adolescent rats. Nat Commun 9:3769. 10.1038/s41467-018-06118-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozorovitskiy Y, Saunders A, Johnson CA, Lowell BB, Sabatini BL (2012) Recurrent network activity drives striatal synaptogenesis. Nature 485:646–650. 10.1038/nature11052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozorovitskiy Y, Peixoto R, Wang W, Saunders A, Sabatini BL (2015) Neuromodulation of excitatory synaptogenesis in striatal development. Elife 4:e10111. 10.7554/eLife.10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajeski RN, Macey-Dare A, van Heusden F, Ebrahimjee F, Ellender TJ (2019) Dynamic postnatal development of the cellular and circuit properties of striatal D1 and D2 spiny projection neurons. J Physiol 597:5265–5293. 10.1113/JP278416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kravitz AV, Freeze BS, Parker PR, Kay K, Thwin MT, Deisseroth K, Kreitzer AC (2010) Regulation of parkinsonian motor behaviours by optogenetic control of basal ganglia circuitry. Nature 466:622–626. 10.1038/nature09159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, et al. (2010) Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci 13:1137–1143. 10.1038/nn.2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Sawatari A (2011) Medium spiny neurons of the neostriatal matrix exhibit specific, stereotyped changes in dendritic arborization during a critical developmental period in mice. Eur J Neurosci 34:1345–1354. 10.1111/j.1460-9568.2011.07852.x [DOI] [PubMed] [Google Scholar]
- Lei W, Deng Y, Liu B, Mu S, Guley NM, Wong T, Reiner A (2013) Confocal laser scanning microscopy and ultrastructural study of VGLUT2 thalamic input to striatal projection neurons in rats. J Comp Neurol 521:1354–1377. 10.1002/cne.23235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chen Z, Fan G, Li A, Yuan J, Xu T (2018) Cell-type-specific afferent innervation of the nucleus accumbens core and shell. Front Neuroanat 12:84. 10.3389/fnana.2018.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima Caldeira G, Peça J, Carvalho AL (2019) New insights on synaptic dysfunction in neuropsychiatric disorders. Curr Opin Neurobiol 57:62–70. 10.1016/j.conb.2019.01.004 [DOI] [PubMed] [Google Scholar]
- Lipton DM, Gonzales BJ, Citri A (2019) Dorsal striatal circuits for habits, compulsions and addictions. Front Syst Neurosci 13:28. 10.3389/fnsys.2019.00028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Ouyang L, Mu S, Zhu Y, Li K, Zhan M, Liu Z, Jia Y, Lei W (2011) The morphological characteristics of corticostriatal and thalamostriatal neurons and their intrastriatal terminals in rats. Surg Radiol Anat 33:807–817. 10.1007/s00276-011-0823-9 [DOI] [PubMed] [Google Scholar]
- Ma L, Chen W, Yu D, Han Y (2020) Brain-wide mapping of afferent inputs to accumbens nucleus core subdomains and accumbens nucleus subnuclei. Front Syst Neurosci 14:15. 10.3389/fnsys.2020.00015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald ML, Alhassan J, Newman JT, Richard M, Gu H, Kelly RM, Sampson AR, Fish KN, Penzes P, Wills ZP, Lewis DA, Sweet RA (2017) Selective loss of smaller spines in schizophrenia. Am J Psychiatry 174:586–594. 10.1176/appi.ajp.2017.16070814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson T, Hikida T (2019) Role of basal ganglia neurocircuitry in the pathology of psychiatric disorders. Psychiatry Clin Neurosci 73:289–301. 10.1111/pcn.12830 [DOI] [PubMed] [Google Scholar]
- Mallya AP, Wang HD, Lee HNR, Deutch AY (2019) Microglial pruning of synapses in the prefrontal cortex during adolescence. Cereb Cortex 29:1634–1643. 10.1093/cercor/bhy061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maltese M, Stanic J, Tassone A, Sciamanna G, Ponterio G, Vanni V, Martella G, Imbriani P, Bonsi P, Mercuri NB, Gardoni F, Pisani A (2018) Early structural and functional plasticity alterations in a susceptibility period of DYT1 dystonia mouse striatum. Elife 7:e33331. 10.7554/eLife.33331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel AC, Galvan A (2022) Connectivity of the corticostriatal and thalamostriatal systems in normal and parkinsonian states: an update. Neurobiol Dis 174:105878. 10.1016/j.nbd.2022.105878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney BC, MacDonald ML, Newman JT, Shelton MA, DeGiosio RA, Kelly RM, Fish KN, Sampson AR, Lewis DA, Sweet RA (2019) Density of small dendritic spines and microtubule-associated-protein-2 immunoreactivity in the primary auditory cortex of subjects with schizophrenia. Neuropsychopharmacology 44:1055–1061. 10.1038/s41386-019-0350-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Monteiro P, Zhou Y, Kim JA, Gao X, Fu Z, Feng G (2016) Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature 530:481–484. 10.1038/nature16971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mink JW (2003) The basal ganglia and involuntary movements: impaired inhibition of competing motor patterns. Arch Neurol 60:1365–1368. 10.1001/archneur.60.10.1365 [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Lüscher C (2019) The molecular basis of drug addiction: linking epigenetic to synaptic and circuit mechanisms. Neuron 102:48–59. 10.1016/j.neuron.2019.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi A, Bibb JA, Snyder GL, Higashi H, Nairn AC, Greengard P (2000) Amplification of dopaminergic signaling by a positive feedback loop. Proc Natl Acad Sci U S A 97:12840–12845. 10.1073/pnas.220410397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onwordi EC, Halff EF, Whitehurst T, Mansur A, Cotel MC, Wells L, Creeney H, Bonsall D, Rogdaki M, Shatalina E, Reis Marques T, Rabiner EA, Gunn RN, Natesan S, Vernon AC, Howes OD (2020) Synaptic density marker SV2A is reduced in schizophrenia patients and unaffected by antipsychotics in rats. Nat Commun 11:246. 10.1038/s41467-019-14122-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Aldridge GM, Greenough WT, Gan WB (2010) Dendritic spine instability and insensitivity to modulation by sensory experience in a mouse model of fragile X syndrome. Proc Natl Acad Sci U S A 107:17768–17773. 10.1073/pnas.1012496107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, Giustetto M, Ferreira TA, Guiducci E, Dumas L, Ragozzino D, Gross CT (2011) Synaptic pruning by microglia is necessary for normal brain development. Science 333:1456–1458. 10.1126/science.1202529 [DOI] [PubMed] [Google Scholar]
- Parekh R, Ascoli GA (2015) Quantitative investigations of axonal and dendritic arbors: development, structure, function, and pathology. Neuroscientist 21:241–254. 10.1177/1073858414540216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge JG, Tang KC, Lovinger DM (2000) Regional and postnatal heterogeneity of activity-dependent long-term changes in synaptic efficacy in the dorsal striatum. J Neurophysiol 84:1422–1429. 10.1152/jn.2000.84.3.1422 [DOI] [PubMed] [Google Scholar]
- Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G (2011) Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature 472:437–442. 10.1038/nature09965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peixoto RT, Wang W, Croney DM, Kozorovitskiy Y, Sabatini BL (2016) Early hyperactivity and precocious maturation of corticostriatal circuits in Shank3B(-/-) mice. Nat Neurosci 19:716–724. 10.1038/nn.4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessoa L, Medina L, Hof PR, Desfilis E (2019) Neural architecture of the vertebrate brain: implications for the interaction between emotion and cognition. Neurosci Biobehav Rev 107:296–312. 10.1016/j.neubiorev.2019.09.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RC, Conley R, Kung L, Peretti FJ, Chute DJ (1996) Reduced striatal spine size in schizophrenia: a postmortem ultrastructural study. Neuroreport 7:1214–1218. 10.1097/00001756-199604260-00024 [DOI] [PubMed] [Google Scholar]
- Runge K, Cardoso C, de Chevigny A (2020) Dendritic spine plasticity: function and mechanisms. Fron Synaptic Neurosci 12:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, Nestler EJ (2013) The brain reward circuitry in mood disorders. Nat Rev Neurosci 14:609–625. 10.1038/nrn3381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo SJ, Dietz DM, Dumitriu D, Morrison JH, Malenka RC, Nestler EJ (2010) The addicted synapse: mechanisms of synaptic and structural plasticity in nucleus accumbens. Trends Neurosci 33:267–276. 10.1016/j.tins.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof TC (2018) Towards an understanding of synapse formation. Neuron 100:276–293. 10.1016/j.neuron.2018.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Segal M (2014) Dendritic spines: the locus of structural and functional plasticity. Physiol Rev 94:141–188. 10.1152/physrev.00012.2013 [DOI] [PubMed] [Google Scholar]
- Segal I, Korkotian I, Murphy DD (2000) Dendritic spine formation and pruning: common cellular mechanisms? Trends Neurosci 23:53–57. 10.1016/s0166-2236(99)01499-x [DOI] [PubMed] [Google Scholar]
- Segal M (2017) Dendritic spines: morphological building blocks of memory. Neurobiol Learn Mem 138:3–9. 10.1016/j.nlm.2016.06.007 [DOI] [PubMed] [Google Scholar]
- Segal M, Andersen P (2000) Dendritic spines shaped by synaptic activity. Curr Opin Neurobiol 10:582–586. 10.1016/s0959-4388(00)00123-9 [DOI] [PubMed] [Google Scholar]
- Sellgren CM, Gracias J, Watmuff B, Biag JD, Thanos JM, Whittredge PB, Fu T, Worringer K, Brown HE, Wang J, Kaykas A, Karmacharya R, Goold CP, Sheridan SD, Perlis RH (2019) Increased synapse elimination by microglia in schizophrenia patient-derived models of synaptic pruning. Nat Neurosci 22:374–385. 10.1038/s41593-018-0334-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple BD, Blomgren K, Gimlin K, Ferriero DM, Noble-Haeusslein LJ (2013) Brain development in rodents and humans: identifying benchmarks of maturation and vulnerability to injury across species. Prog Neurobiol 106-107:1–16. 10.1016/j.pneurobio.2013.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen W, Flajolet M, Greengard P, Surmeier DJ (2008) Dichotomous dopaminergic control of striatal synaptic plasticity. Science 321:848–851. 10.1126/science.1160575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Y, Raju DV, Pare JF, Sidibe M (2004) The thalamostriatal system: a highly specific network of the basal ganglia circuitry. Trends Neurosci 27:520–527. 10.1016/j.tins.2004.07.004 [DOI] [PubMed] [Google Scholar]
- Suarez LM, Solis O, Sanz-Magro A, Alberquilla S, Moratalla R (2020) Dopamine D1 receptors regulate spines in striatal direct-pathway and indirect-pathway neurons. Mov Disord 35:1810–1821. 10.1002/mds.28174 [DOI] [PubMed] [Google Scholar]
- Tendilla-Beltrán H, Arroyo-García LE, Diaz A, Camacho-Abrego I, de la Cruz F, Rodríguez-Moreno A, Flores G (2016) The effects of amphetamine exposure on juvenile rats on the neuronal morphology of the limbic system at prepubertal, pubertal and postpubertal ages. J Chem Neuroanat 77:68–77. 10.1016/j.jchemneu.2016.05.004 [DOI] [PubMed] [Google Scholar]
- Tepper JM, Sharpe NA, Koós TZ, Trent F (1998) Postnatal development of the rat neostriatum: electrophysiological, light- and electron-microscopic studies. Dev Neurosci 20:125–145. 10.1159/000017308 [DOI] [PubMed] [Google Scholar]
- Thompson BL, Oscar-Berman M, Kaplan GB (2021) Opioid-induced structural and functional plasticity of medium-spiny neurons in the nucleus accumbens. Neurosci Biobehav Rev 120:417–430. 10.1016/j.neubiorev.2020.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn CA, Atallah H, Howe M, Graybiel AM (2010) Differential dynamics of activity changes in dorsolateral and dorsomedial striatal loops during learning. Neuron 66:781–795. 10.1016/j.neuron.2010.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uryu K, Butler AK, Chesselet MF (1999) Synaptogenesis and ultrastructural localization of the polysialylated neural cell adhesion molecule in the developing striatum. J Comp Neurol 405:216–232. [DOI] [PubMed] [Google Scholar]
- Van Waes V, Beverley JA, Siman H, Tseng KY, Steiner H (2012) CB1 cannabinoid receptor expression in the striatum: association with corticostriatal circuits and developmental regulation. Front Pharmacol 3:21. 10.3389/fphar.2012.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Fowler JS, Tomasi D (2012) Addiction circuitry in the human brain. Annu Rev Pharmacol Toxicol 52:321–336. 10.1146/annurev-pharmtox-010611-134625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, De La Parra M, Callaway EM, Kreitzer AC (2013) Differential innervation of direct- and indirect-pathway striatal projection neurons. Neuron 79:347–360. 10.1016/j.neuron.2013.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Li C, Chen Q, van der Goes MS, Hawrot J, Yao AY, Gao X, Lu C, Zang Y, Zhang Q, Lyman K, Wang D, Guo B, Wu S, Gerfen CR, Fu Z, Feng G (2017) Striatopallidal dysfunction underlies repetitive behavior in Shank3-deficient model of autism. J Clin Invest 127:1978–1990. 10.1172/JCI87997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Kai L, Day M, Ronesi J, Yin HH, Ding J, Tkatch T, Lovinger DM, Surmeier DJ (2006) Dopaminergic control of corticostriatal long-term synaptic depression in medium spiny neurons is mediated by cholinergic interneurons. Neuron 50:443–452. 10.1016/j.neuron.2006.04.010 [DOI] [PubMed] [Google Scholar]
- West EA, Carelli RM (2016) Nucleus accumbens core and shell differentially encode reward-associated cues after reinforcer devaluation. J Neurosci 36:1128–1139. 10.1523/JNEUROSCI.2976-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YW, Kim JI, Tawfik VL, Lalchandani RR, Scherrer G, Ding JB (2015) Input- and cell-type-specific endocannabinoid-dependent LTD in the striatum. Cell Rep 10:75–87. 10.1016/j.celrep.2014.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, de Jong JW, Tak Y, Peck J, Bateup HS, Lammel S (2018) Nucleus accumbens subnuclei regulate motivated behavior via direct inhibition and disinhibition of VTA dopamine subpopulations. Neuron 97:434–449.e4. 10.1016/j.neuron.2017.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y, Gao G, Liu K, Shi X, Cheng M, Xiong Y, Song S (2021) Projections from D2 neurons in different subregions of nucleus accumbens shell to ventral pallidum play distinct roles in reward and aversion. Neurosci Bull 37:623–640. 10.1007/s12264-021-00632-9 [DOI] [PMC free article] [PubMed] [Google Scholar]