Abstract
As a tool for functional genomics, a hairpin ribozyme gene library with randomized target recognition sequences was constructed in a retroviral vector. This library has the potential to target and cleave any possible RNA substrate. Mouse fibroblasts transduced with this ribozyme gene vector library were selected in a focus formation assay to isolate in vivo functional ribozymes that promote cell transformation in tissue culture. After two successive rounds of selection by focus formation assay, a transforming ribozyme (Rz007) was identified. The sequence of this ribozyme was used to identify the putative target genes responsible for the transformation. A candidate gene target for Rz007 encodes telomerase reverse transcriptase (mTERT). Both mRNA level and enzymatic activity of mTERT were down-regulated in Rz007-transformed cells. Furthermore, newly designed ribozymes, recognizing other potential ribozyme cleavage sites unique to the mTERT mRNA, also cause cell transformation, thus validating the role of mTERT in suppressing the transformation phenotype. These surprising results suggest that the commonly accepted role of telomerase in maintaining cellular immortalization is more complicated than previously thought. These studies also demonstrate the utility of this novel ‘reverse’ functional genomics approach, enabling the targeted discovery of genes, whether previously known or not, that are involved in any selectable phenotype.
INTRODUCTION
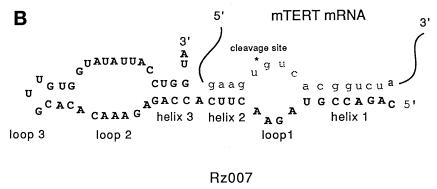
Ribozymes (Rz) are small catalytically active RNA molecules that are found in nature and cleave natural target RNAs. Ribozymes contain conserved sequences composed of catalytic and structural determinants, as well as variable regions that determine target sequence specificity. A ribozyme can be engineered with specific target recognition sequences that allow it to cleave a specific target RNA. This specifically designed ribozyme can be used to inactivate a given gene function in vivo if it is introduced into cells. Ribozyme sequences can be cloned into an expression cassette and stably introduced into cells where they function as ‘ribozyme genes’ to express the ribozymes inside the cells. The typical structure of a hairpin ribozyme is shown in Figure 1B. The target binding sequences are designated helices 1 and 2. Any GUC sequence on an RNA molecule could be a potential hairpin ribozyme target site (1).
Figure 1.
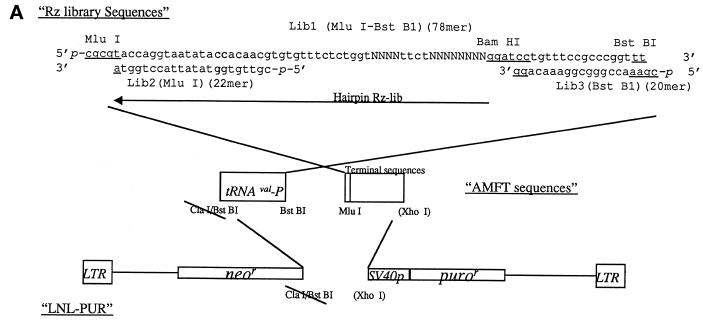
(A) Construction scheme of the retroviral vector containing a tRNAVal Pol III promoter cassette-expressing ribozyme gene library. The sequences represent random sequences of the ribozyme library. N can be any one of the four deoxyribonucleotides. (B) Structure of a hairpin ribozyme and its RNA target, using Rz007 and the putative target mTERT as examples. The target cleavage site is shown by *. GUC is required for any hairpin ribozyme target site (1). Helices 1 and 2 are formed by the target sequences of the RNA substrate and the target recognition sequences of the hairpin ribozyme flanking XGUC of the target RNA.
Ribozymes are powerful tools for studying somatic cell genetics and can be applied in at least two aspects of functional genomics and pharmaceutical drug development. Firstly, they can be used in gene target validation, which involves inactivating a specific gene target with a specific ribozyme. The resultant phenotypic change helps to elucidate gene function. Secondly, ribozymes can be used in a ‘genetic screening’ approach which identifies genes, without prior sequence information, that are involved in a specific function. In this approach, a ribozyme gene vector library with random target recognition sequences is used to transfer the ribozyme genes to recipient cells in culture such that each cell expresses a single ribozyme with a single specificity. The mRNAs of those genes responsible for a specific phenotype will be cleaved in cells containing a specific complementary ribozyme. If the cells with phenotypic changes can be isolated, the ribozymes responsible for the phenotypic alterations can also be rescued from these cells. Subsequently, a ribozyme target gene(s) can be identified based on the complementary target recognition sequences predicted from the ribozymes.
NIH 3T3 is a classic cell system for testing various transformation agents such as oncogenes and chemical carcinogens (2). It is easily transformed (requiring only ‘one hit’) and the transformation is readily assayed in tissue culture by focus formation or soft agar colony formation assay. The cells with transformed phenotype can be separated from the non-transformed cells using these assays, which makes efficient rescue of the ribozymes responsible for the transformation possible. Therefore, NIH 3T3 is an ideal cell system for testing the ribozyme gene library strategy in isolating transformation-potent ribozymes in vivo and the corresponding transformation suppressor genes that they target. This report describes using this method to isolate a transformation-inducing hairpin ribozyme and identify its gene target.
MATERIALS AND METHODS
Cells
NIH 3T3 mouse fibroblast cells were obtained from Dr Jhian (Ohio State University) (3); 293 cells, a transformed human embryonic kidney cell line, and HT1080 cells, a human fibrosarcoma cell line, were purchased from the American Type Culture Collection (ATCC, Rockville, MD) (ATCC CR-1573 and ATCC-CCL-121, respectively). The cells were cultured at 37°C in DMEM medium (Gibco BRL, Rockville, MD) containing 10% fetal calf serum under 5% CO2.
Construction of the plasmid ribozyme library
The structure of the retroviral vector, LHPM-Rz-library, is shown in Figure 1A. The two drug resistance genes, for neomycin and puromycin resistance, were inserted in the vector for selection of vector-containing cells after transduction. The human pol III tRNAVal promoter was used to drive efficient ribozyme gene expression in vivo (4). Briefly the LHPM-CNR3 vector was constructed by ligating a ClaI–XhoI fragment (containing the tRNAVal promoter and a CNR3 ribozyme sequence cassette) from the AMFT-CNR3 vector (5; Welch et al., unpublished results) into the pLNL-PUR vector in place of a XhoI–BstBI fragment, resulting in loss of the BstBI and ClaI sites in vector LHPM-CNR3. CNR3 is a ribozyme against the HCV minus strand 5′-UTR and was used as a control ribozyme in the study. A 2 kb stuffer fragment was then inserted into the vector at the BamHI and MluI sites in place of the CNR3 ribozyme to generate the cloning vector, LHPM-2kb, for inserting the ribozyme library or other ribozymes. The LHPM-2kb vector fragments were gel purified (Gel Isolation Kit; Qiagen, Valencia, CA) after BstBI and MluI digestion, recovered by ethanol precipitation and resuspended at 1 µg/ml in water. Three oligonucleotides, Lib1, -2 and -3, (Fig. 1A) were chemically synthesized, 5′-end phosphorylated and PAGE purified (Integrated DNA Technologies, Coralville, IA). These three oligonucleotides were mixed at a molar ratio of 1:20:20 (6). They were annealed together to form a partial double-stranded DNA (Fig. 1A) by boiling for 2 min then cooling slowly to room temperature. The vector fragments were ligated with the annealed oligonucleotides at a molar ratio of 1:5 at 14°C overnight (Gibco BRL). Twenty micrograms of the ligated mixtures were transformed into DH12s cells (Gibco BRL) by electroporation at 1 µg DNA/40 µl/bacteria/electroporation (GenePulser; Bio-Rad, Hercules, CA). The transformed bacteria were plated on 20 150-mm LB–ampicillin plates and incubated overnight at 37°C. In parallel, small aliquots of the transformed bacteria were serially diluted and plated to determine the total yield of transformants. The bacteria from all 20 plates were pooled and stored as aliquots with 1000 full complement library equivalents (1.7 × 107 cells/library) at –80°C. To prepare the library plasmid DNA, one vial of library bacteria was grown in 500 ml of ampicillin–LB overnight at 37°C. Sequence analysis of the individual clones of the library was performed (Retrogen, San Diego, CA).
Transduction of NIH 3T3 cells with the ribozyme gene library retroviral vector and fibroblast focus formation assay
The library vector was generated by triple transfection of 293 cells with DNA of the LHPM-Rz-library vector and two helper vectors expressing Gag-Pol (7) and VSV-G (8), using a calcium phosphate precipitation method (Gibco BRL). The supernatant was harvested, 0.2 µm filtered and titrated. The titer of the library was determined by counting G418-resistant colonies after transduction of HT1080 cells with the serially diluted vector.
The transduction of NIH 3T3 cells with the ribozyme gene vector library was performed as follows. The cells were plated in five 150-mm plates on day 1 at a density of 106 cells/plate. On day 2, four plates were transduced with library vector at an MOI of 1 (one quarter of the library was used in this case) and one plate was mock-transduced. The cells were refed on day 3 with G418-containing medium (500 µg/ml) and continuously cultured in the presence of G418 to confluence with the medium changed twice a week.
The confluent cultures were incubated under the same conditions and the medium changed twice a week after reaching confluence. After 3–4 weeks, foci started to be visible. The foci were harvested by tapping the plate gently and collecting the floating cells, since the focus cells detach easily from the dish. These cells were treated with trypsin for 5 min to disaggregate them and replated in 6-well plates. The morphology was observed at various time points.
Ribozyme rescue from the transformed cells
For viral rescue, the transformed cells were plated in 10-cm dishes on day 1. On day 2, when the culture was 50% confluent, they were co-transfected with DNAs of two helper vectors expressing Gag-Pol and VSV-G at 10 µg each. The vector supernatants were collected, filtered (0.2 µm) and used to transduce fresh 3T3 cells.
For PCR rescue, the high molecular weight DNA was extracted from the transformed cells (Blood Kit; Qiagen) and used as the template for amplification of the ribozyme sequences. The PCR reaction conditions were 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, 72°C for 20 s and one cycle of 72°C for 5 min. The purified PCR products were then digested with BamHI and MluI and the digested products were cloned into the LHPM-2kb vector with the 2 kb stuffer fragment removed. Thousands of colonies were obtained by electroporation. All the transformants were pooled and used for plasmid preparation and the plasmids were used to generate the vector for the next round of transduction as described above. Sample colonies were also picked for sequence analysis.
Construction of a vector containing a disabled 007 ribozyme, Rz007(–), and mTERT validation ribozymes
The vector with the disabled ribozyme, Rz007(–), was generated by PCR using LHPM-CNR3 as template and the two primers 007(–) (5′-gatatcggatcccagaccgtagaacttcaccagagcgucacacgttgtggtat-3′) and Rz-UR (5′-gtcgacacgcgtaccaggtaatataccac-3′). The vectors with various target validation ribozymes were generated by PCR using pLHPM-CNR3 as template and the primers Rz-UR (reverse) and the corresponding forward ribozyme primer (mTERTRz6, -7, -8, -9, -10 or -11). The purified PCR products were cloned into the LHPM-2kb vector.
Assessing the growth properties of the transformed cells
The transformed cells and non-transformed control were plated in 100-mm plates at an equal density of 5 × 105 cells/plate on day 1. They were allowed to grow for 3 days. On day 4, the cultures were observed. The number of cells was determined.
For growth in the low serum or serum-free conditions, the cells were plated in four sets in 10% serum on day 1. On day 2, one set of the cells was trypsinized and counted to establish the starting cell number. The other three sets of cultures were washed with the serum-free media and refed with medium containing 0, 1 or 10% fetal calf serum, respectively, for each set. On day 4, culture morphology was examined and the number of cells determined.
Quantitation of mTERT message
Two 150-mm plates of subconfluent transformed 3T3 cells and control cells were lysed on the plate and the mRNA extracted using a mRNA isolation kit (Ambion, Austin, TX). mTERT message was quantitated using the PE TaqMan One step RT–PCR procedure [TaqMan Gold RT-PCR Kit P/N N808-0233 (PE Applied Biosytems, Foster City, CA) and Prism 7700 Sequence Detection System (PE Applied Biosystems)]. Oligonucleotide primers and the probe were chosen from the mTERT gene sequences using Primer Express Software as follows: forward primer, mTERTf (5′-tgggtctcccctgtaccaaa-3′); reverse primer, mTERTr (5′-gcctgtaactagcggacacaga-3′); fluorescence labeled oligonucleotide probe, mTERTp (5′-ttgtgccaccacggatatctggcc-3′). Mouse glyceraldehyde 3-phosphate dehydrogenase (musgapdh) mRNA was measured as an internal control. The primers and probe for musgapdh were as follows: forward primer, mGAPf (5′-ggcaaattcaacggcacagt-3′); reverse primer, mGAPr (5′-agatggtgatgggcttccc-3′); fluorescence labeled probe, mGAPp (5′-aaggccgagaatgggaagcttgtcatc-3′).
TRAP-based ELISA assay of telomerase activity in the transformed 3T3 cells
The cells from one T-75 flask of subconfluent culture were harvested, lysed and telomerase activity measured using the telomeric repeat amplification protocol (TRAP)-based ELISA kit from Roche Biosciences (Palo Alto, CA). The experiments were performed according to the manufacturer’s recommended protocol. Briefly, cells from a 100-mm plate (nearly confluent) were harvested using 1 mM EDTA–phosphate-buffered saline (PBS) and washed once with PBS. They were counted and lysed with the lysis solution provided by the manufacturer. The lysate supernatant was used for total protein determination and TRAP PCR amplification as the source of telomerase (9). The total protein in the cell lysates was quantitated by Bradford assay (US Biochemical, Cleveland, OH). The TRAP PCR products were quantitated with the ELISA kit and the telomerase activity was normalized to total protein. The normalized telomerase activity of the functional ribozyme-transformed cells were compared with that of the control.
RESULTS
Complexity of the constructed hairpin ribozyme gene library
A randomized hairpin ribozyme library was constructed in a retroviral vector (Fig. 1A). A total of 12 random positions were introduced into the target recognition region of the library: 8 nt in helix 1 and 4 nt in helix 2. This results in a maximum complexity of the library of 1.7 × 107 (= 412) different ribozyme molecules. Efficient cloning of the ribozyme library sequences into the plasmid vector was necessary to maximize complexity of the library. A total of 5 × 107 independent transformed colonies were generated in this experiment, demonstrating that the resultant ribozyme library should have the full complexity of 1.7 × 107 with 95% confidence. The randomness of the library was also evaluated by sequencing 64 independent colonies from the original transformation. All 64 sequences were unique, suggesting randomization of the sequences. The frequencies of the four nucleotides in the random regions are: A, 29.0 ± 4.5%; T, 29.0 ± 4.5%; C, 24.2 ± 4.2%; G, 17.7 ± 3.9%. It was noted there are variations in the use of the four nucleotides from the expected frequency of 25% for each nucleotide and these variations could reduce the complexity of the library. Although the actual complexity of the constructed library was difficult to accurately assess, it is significantly lower than the maximum complexity of 1.7 × 107. Considering that the size of the average gene is between 500 bp and a few kilobases, most genes should have multiple ribozyme sites, and this library should still have at least one representative for each gene target.
Isolation of the ribozyme responsible for transforming 3T3 cells by focus formation assays
A total of 4 × 106 transduction units of vector, containing roughly one-quarter of the library complexity, were used to transduce NIH 3T3 cells. The NIH 3T3 cells used in this study are from a clone with a low background of spontaneous transformation (3). The transduction was conducted at an MOI of 1. The transduced cells were subsequently selected in medium containing G418, so that all the surviving cells would contain the vector.
The focus formation assay is a convenient method to monitor fibroblast transformation. Normal fibroblasts, like NIH 3T3, grow as a monolayer and stop dividing when the culture reaches confluence due to contact inhibition. When these cells are transformed, they lose contact inhibition and continue to grow even after the culture reaches confluence. They grow on top of each other to form foci, in contrast to the uniform monolayer formed by non-transformed cells. This morphological change is an indicator of cell transformation and provides a way to isolate transformed cells from non-transformed cells.
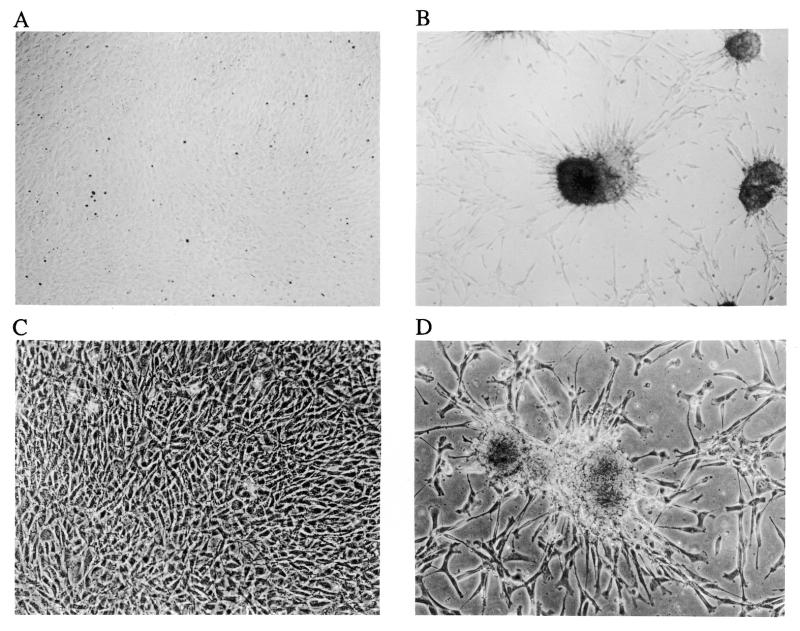
The G418-selected cells were cultured to reach confluence and the confluent cultures were maintained to allow focus formation. There were a number of foci appearing in both library- and mock-transduced cultures 3–4 weeks after the cultures reached confluence. The number of foci in the library-transduced cultures was slightly higher (2- to 5-fold) than that in the mock-transduced culture (3 foci in the one mock-transduced culture and 6, 8, 10 and 15 foci in four library-transduced cultures, respectively). To determine whether focus formation was genetically stable, the foci derived from both the mock and the ribozyme gene library-transduced cultures were harvested and re-plated. The mock vector-transduced ‘focus cells’ still grew as a typical monolayer culture, as did the original 3T3 (Fig. 2A), suggesting that they are not true focus forming cells or truly transformed cells. In contrast, the library-transduced ‘focus cells’ showed a morphologically transformed phenotype and formed many foci (Fig. 2B). These distinct properties of the foci from ribozyme library-transduced and mock-transduced cultures suggested that certain ribozymes in the ribozyme library might have promoted genetically stable cellular transformation.
Figure 2.
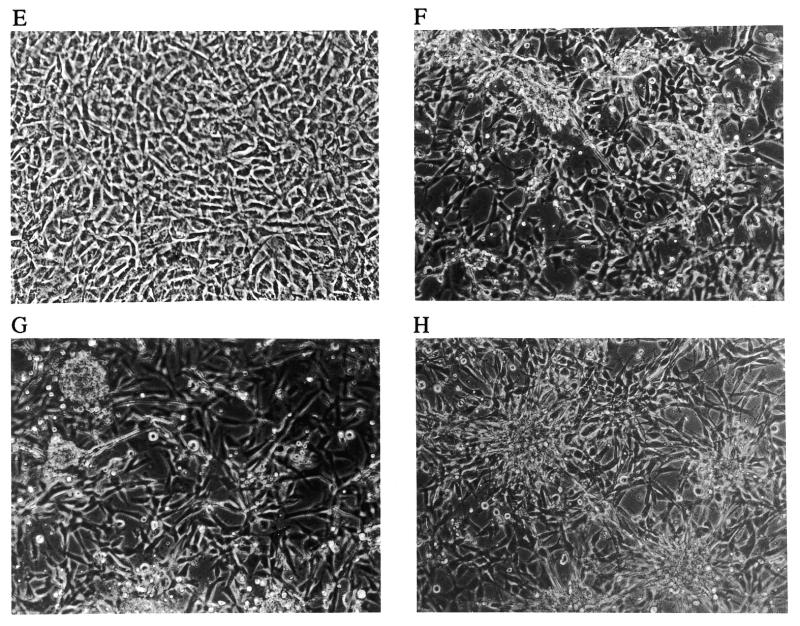
Morphology of the re-plated focus cells from the foci in the focus formation assay. The foci from the focus formation assays were harvested and re-plated into 6-well plates. The morphology was observed after they reached confluence. (A) Mock and (B) Rz library are from a focus formation experiment with the original library transduction; (C) CNR3 and (D) Rz007 are from a confirmation experiment in which Rz007 was transduced as a single ribozyme; (E) CNR3, (F) Rz007, (G) mTERTRz10 and (H) mTERTRz11 are from a validation experiment with transduction by ‘validation ribozymes’ against mTERT.
The ribozymes in the transformed cells were rescued by PCR and sequenced to determine the enrichment in the population at this point in the selection scheme. Four independent colonies were found to be unrelated. They did not appear in the 64 clones sequenced during library characterization and in the later rescues, suggesting that they might be random clones. The population does not appear to be significantly enriched for a particular gene, although more clones would need to be sequenced to confirm this. The PCR rescue colonies were pooled and used to generate a ‘PCR sub-library’. In addition, cells from the first round selection were transfected with helper plasmids to generate a ‘viral rescue sub-library’. These two rescued ‘sub-libraries’ were used in separate experiments for a second round of transduction on fresh 3T3 cells. The transduced cells were tested in the focus formation assay as before. In parallel, an irrelevant ribozyme vector, CNR3, in which the ribozyme library sequences in the library vector were replaced with the CNR3 ribozyme (see Materials and Methods), was used as a control in these experiments. There was no change in phenotype in the CNR3 control after re-plating, similar to the previously mock-transduced cultures. Re-plating of the foci derived from both ‘sub-library’-transduced cells showed strikingly transformed phenotypes (not shown), similar to re-plating of the foci from the first round selection of the original library-transduced culture. These results further confirmed the presence of the transforming ribozymes in both rescued pools.
The ribozyme sequences from the second round pooled foci were rescued by the PCR method. We analyzed six sequences from both rescue schemes. In both cases, there was enrichment of a dominant ribozyme gene sequence, Rz007. Rz007 appeared at frequencies of 3/6 and 4/6 in the two rescue schemes, respectively. Enrichment of the same ribozyme from two independent selections indicated that Rz007 was potentially responsible for the transformation.
To confirm its role in promoting transformation, Rz007, as a single ribozyme gene, was transduced into fresh 3T3 cells, using CNR3 as a control. In three independent experiments Rz007 was repeatedly shown to transform 3T3 (Fig. 2D), while the control CNR3 did not (Fig. 2C). This suggests that Rz007 alone is sufficient to cause transformation without participation of other ribozymes. We also generated a disabled Rz007, Rz007(–), in which nucleotides AAA in loop 2 of Rz007 were replaced by CGU, inactivating the ribozyme activity (5). The helices 1 and 2 structures, and thus the target binding ability, are maintained. Rz007(–) was unable to cause transformation (data not shown), which suggests that the transformation caused by Rz007 was due to ribozyme activity, not an antisense or aptamer effect.
Characterization of the growth properties of Rz007-transformed NIH 3T3 cells
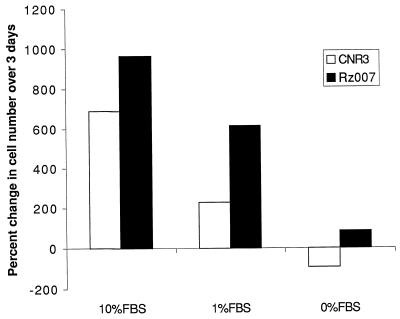
The transformed cells derived from the foci caused by Rz007 were further characterized for their phenotype. First, we compared their growth rate with that of the CNR3 control-transduced cells. We did not observe pronounced differences in the growth rate between them when the cells were grown at a relatively low density. At higher cell density, the control cells grew more slowly or stopped growing due to contact inhibition, while the Rz007-transformed cells continue to grow. When these cultures were grown in media with a low serum concentration or without serum, the difference in growth rate was even more dramatic (Fig. 3). The transformed cells in 1% serum, although growing slower than in 10%, grew much faster than non-transformed cells. In the serum-free media the non-transformed cells died quickly (within 3 days), while the transformed cell survived for more than 2 weeks. This suggests that the Rz007-transformed 3T3 cells reduced the requirement for serum growth factors for proliferation and survival, one of the hallmarks of fibroblast transformation (10,11). In addition, conditioned media from the transformed cell culture grown in the absence of serum seemed to help the non-transformed cells survive without serum (data not shown), indicating a secreted factor(s) produced by the Rz007-transformed cells. These observations are not surprising since it has been reported that some transformed fibroblasts secret PDGF-like growth factors and reduce the requirement for exogenous growth factor for growth (12,13). However, the putative secreted factor(s) in our Rz007-transformed cells has yet to be investigated.
Figure 3.
Growth properties of the Rz007-transformed NIH 3T3 cells. The conditions for this assay are described in Materials and Methods.
The mouse mTERT gene is a potential target of Rz007 based on its sequences
The potential gene target of Rz007 can be identified based on the specific sequences in helices 1 and 2 of the ribozyme hairpin structure (Fig. 1B). The ribozyme sequence was used to predict a putative target mRNA sequence 5′-gaagxgucacggtcta-3′, the ‘ribozyme sequence tag’ of Rz007, or RST007. The public sequence database was searched for potential matches with RST007 using BLAST. Several potential candidates were identified, with the best match (15/16) being the sequence 2947–2962 of the mouse gene encoding telomerase reverse transcriptase (mTERT), the catalytic subunit of telomerase. There is only one mismatch at the last nucleotide at the 3′-end of helix 1 (A in mTERT and G in RST007) (Fig. 1B). Since a hairpin ribozyme with a 7 bp helix 1 is generally active, mTERT could be a potential target for Rz007.
Down-regulation of mTERT expression in the Rz007-transformed cells
If the true target of Rz007 is mTERT mRNA, the mTERT message should be down-regulated by the ribozyme. The mRNA level of mTERT in both the Rz007-transformed cells and CNR3-transduced control cells was analyzed by the TaqMan one-step RT–PCR RNA quantitation procedure. musgapdh mRNA was measured as an internal control. The results show that the mTERT message was decreased in the transformed cells by roughly one-half compared to that in the CNR3 control (Table 1). This suggests that Rz007 reduced the mTERT mRNA level and that mTERT could be a candidate target.
Table 1. mTERT mRNA level and telomerase activity in cells transformed by Rz007.
| Ribozymes | mTERT mRNA levela (%) | Telomerase activityb (%) |
|---|---|---|
| CNR3 | 100 ± 10 | 100 ± 6 |
| Rz007 | 47 ± 13 | 16 ± 2 |
amRNA level was measured by the TaqMan RT–PCR method. It is represented by the mean of mTERT to mGAPDH ratio of duplicates and presented as a relative quantity.
bTelomerase activity was measured by TRAP-based ELISA (see text). It is represented by the mean of the activity to total protein ratio of duplicates and is presented as a relative quantity.
Next, the telomerase enzymatic activity of the Rz007-transformed cells was compared to CNR3-transduced cells by a TRAP-based ELISA assay. In this experiment there was an ~80% decrease in telomerase activity in the Rz007-transformed cells (Table 1), in agreement with down-regulation of the mRNA in these cells. The down-regulated telomerase activity associated with the transformed cells again supports a role for this target in the cell transformation.
Validation of the role of mTERT in cell transformation using hairpin ribozymes recognizing other GUC sites on the mTERT mRNA target
The knock-down of mTERT mRNA and enzymatic activity is consistent with the hypothesis that the transformed phenotype observed with Rz007 was due to a decrease in telomerase activity. However, it is possible that Rz007 targeted another gene besides mTERT that has similarity in the target recognition domain. To exclude this possibility and validate that the transformed phenotype was a direct consequence of mTERT knock-down, we generated ribozymes recognizing other GUC sites unique to mTERT. Six such ‘validation ribozymes’ (mTERTRz6, -7, -8, -9, -10 and -11) with the exact same structure as Rz007 except for their helices 1 and 2 binding areas were designed to recognize six different GUC sites on mTERT mRNA (GUC at nucleotides 424, 591, 1400, 1800, 2455 and 2534). These six ribozyme vectors were transduced into 3T3 cells in parallel with the CNR3 negative control and Rz007. Among the six ‘validation ribozymes‘, at least two of them (mTERTRz10 and -11) caused focus formation (Fig. 2G and H) just as with Rz007, while CNR3 did not. Furthermore, the cells transformed by the ‘validation ribozymes’ also showed reduced telomerase activity, similar to that of Rz007-transformed cells (Table 2). Together, these observations demonstrate that down-regulation of mTERT can result in cell transformation.
Table 2. Telomerase activity in cells transformed by ‘validation ribozymes’.
| Ribozyme | Telomerase activity (%) |
|---|---|
| CNR3 | 100 ± 8 |
| Rz007 | 58 ± 5 |
| mTERTRz10 | 51 ± 15 |
| mTERTRz11 | 63 ± 3 |
The parameter is the same as in Table 1.
DISCUSSION
A functionally active ribozyme responsible for mouse fibroblast transformation, Rz007, was successfully isolated using a ribozyme gene library strategy in combination with two rounds of selection by focus formation assay. The role of Rz007 in transformation was supported by two experimental observations: (i) Rz007 appeared dominantly in both ‘viral rescued foci’ and ‘PCR-rescued foci’; and (ii) the transforming ability of Rz007 was confirmed by introducing this single ribozyme into 3T3 cells. It is noteworthy that it usually took a few weeks for the ‘real foci’ to form, implying the involvement of multiple factors or multiple steps. The experiments suggest that the combinatorial ribozyme gene library is an effective strategy to isolate functionally active ribozymes responsible for certain phenotypes in tissue culture, provided that an efficient assay to isolate the cells associated with the phenotype is available.
Telomerase has recently become a focus of studies concerning development, cancer and aging due to its important role in controlling cell proliferation. Telomerase is a ribonucleoprotein enzyme complex consisting of RNA template components and protein components, including a specialized reverse transcriptase (TERT) (14,15). Telomerase is responsible for maintaining the physical termini of eukaryotic chromosomes. Although little is known about the mechanism of regulation, telomerase activity in cells is apparently regulated at the level of TERT mRNA.
Rz007 was shown here to transform NIH 3T3 cells by inactivating mTERT mRNA. The present observation of mTERT involvement as a suppressor of transformation, however, was somewhat unexpected from the current point of view in the field. Activation of human telomerase, or an increased level of human TERT (hTERT), has been observed in many human tumors, although some human tumor cells lack telomerase activity (16–18). Conversely, inhibition of telomerase activity causes tumor cells to undergo senescence. Telomerase activity is believed to contribute to the immortalization, and thus transformation, of human cells (19). In fact, hTERT has been widely viewed as a potential therapeutic target against cancers (20).
However, mouse telomerase and telomeres have differences from their human counterparts. Unlike human telomerase, mouse telomerase is widely expressed in adult tissues and in primary cell cultures and is not regulated following crisis. Mouse chromosomes also have significantly longer telomeres in contrast to human chromosomes (21). Experiments have shown that mouse cells can be transformed in the absence of telomerase, indicating that telomerase is not a requisite for the process of transformation (22). Furthermore, telomerase deficiency has also recently been implicated in the increased tumorigenicity seen in telomerase knockout mice (23), presumably due to effects on chromosomal stability. Our observations, in agreement with the knockout data, suggest a novel role of telomerase in suppressing fibroblast transformation. Further studies should include determining the effects of ribozyme-mediated telomerase knock-down on telomere length and chromosomal stability. The observation that telomerase can suppress cell transformation may be important to those developing drugs to inhibit telomerase for the treatment of cancer.
Statistically, since every 64 nt (43) nucleic acid length should have a GUC, it is likely that every mRNA has more than one GUC site and could be a potential target of a hairpin ribozyme library of full complement. However, the actual chance of cleaving any mRNA target using a ribozyme library in cells is likely much smaller than 100%. Many factors could affect the actual outcome from application of even a full complement library. Many ribozymes may not be expressed at a sufficient level. Some of the GUC sites of a particular mRNA may not be properly exposed to ribozymes in vivo due to cellular compartmentation, secondary structure or protection resulting from binding to proteins. In addition, some targets may tolerate a certain degree of down-regulation without being selected by a given assay system. Due to these factors, as well as the fact that only a fraction of the library was used, we did not identify other ribozymes besides Rz007 causing transformation in the experiment described. It is possible that there are additional ribozymes from the second rescue that can cause transformation. However, preliminary experiments using the five other rescued ribozymes sequenced did not transform cells and did not have reasonable matches to any known gene targets.
Functional characterization of a gene is a bottleneck in human functional genomics. However, the progress of current human genomics will ensure that the majority of human gene sequences will be available in databases in the near future. Together with more powerful search tools, it is possible to efficiently identify a gene target based on RST sequences. The described approach demonstrates the utility of using a ribozyme gene library, cell assays and bioinformatics to identify genes based on function. In the event that the appropriate candidate gene is not present in the existing databases, other experimental approaches using the RST as a molecular tag can be utilized to identify the unknown target genes from mRNAs of the target cells (Welch et al., in preparation). We believe that this approach could be an important addition to the field of functional genomics, which connects genes to their biological phenotype and identifies novel gene functions, and can be applied to explore pharmaceutical drug targets, as well as develop powerful ribozyme drugs.
Acknowledgments
ACKNOWLEDGEMENTS
The authors would like to thank Drs R. Tritz, S.-H. Xing and E. Marcuson for their suggestions and critical reading of the manuscript. This work was funded by a grant from the US Department of Energy (DEFG0398ER62624).
REFERENCES
- 1.Turner P.C. (ed.) (1997) Ribozyme Protocols. Humana Press, Totowa, NJ, pp. 1 and 171.
- 2.Shih C. and Weinberg,R.A. (1982) Cell, 29, 161–169. [DOI] [PubMed] [Google Scholar]
- 3.Xing S. (1997) Endocrinology, 137, 1512–1519. [DOI] [PubMed] [Google Scholar]
- 4.Tritz R., Leavitt,M. and Barber,J.R. (1999) In Rossi,J.J. and Couture,L. (eds), Intracellular Ribozyme Applications: Principle and Protocols. Horizon Scientific Press, Wymondham, UK, p. 115.
- 5.Welch P.J., Tritz,R., Yei,S., Leavitt,M., Yu,M. and Barber,J. (1996) Gene Ther., 3, 994–1001. [PubMed] [Google Scholar]
- 6.Cwirla S.E., Peters,E.A., Barrett,R.W. and Dower,W.J. (1990) Proc. Natl Acad. Sci. USA, 87, 6378–6382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Landar N.F. and Littman,D.R. (1992) J. Virol., 66, 5110–5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burns J.C, Friedman,T., Driever,W., Burrascano,M. and Yee,J.K. (1993) Proc. Natl Acad. Sci. USA, 90, 8033–8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim N.W. and Wu,F. (1997) Nucleic Acids Res., 25, 2595–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scher C.D., Pledger,W.J., Martin,P., Antoniades,H. and Stiles,C.D. (1998) J. Cell. Physiol., 97, 371–380. [DOI] [PubMed] [Google Scholar]
- 11.Zhan X. and Goldfarb,M. (1986) Mol. Cell. Biol., 6, 3541–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowen-Pope D.F., Vogel,A. and Ross,R. (1984) Proc. Natl Acad. Sci. USA, 81, 2396–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dicker P., Pohjanpelto,P., Pettican,P. and Rozengurt,E. (1981) J. Cell. Physiol., 97, 371–380. [Google Scholar]
- 14.Blackurn E. (1999) In The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 603–635.
- 15.Greider C.W. (1996) Annu. Rev. Biochem.,65, 337–365. [DOI] [PubMed] [Google Scholar]
- 16.Kim N.W. (1994) Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 17.Shay J.W. and Bacchetti,A. (1997) Eur. J.Cancer, 33, 787–791. [DOI] [PubMed] [Google Scholar]
- 18.Bryan T.M., Englezou,A., Gupta,J., Bacchetti,S. and Reddel,R.R. (1995) EMBO J., 14, 4240–4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahn W.C., Counter,C.M., Lundberg,A.S., Beijersbergen,R.L., Knoll,J.H.M., Meyerson,M., Brooks,M.W., Weinberg,R.A. et al. (1999) Nature, 400, 464–468. [DOI] [PubMed] [Google Scholar]
- 20.Hahn W.C., Stewart,S.A., Brooks,M.W., York,S.G., Eaton,E., Kurachi,A., Beijersbergen,R.L., Knoll,J.H.M., Meyerson,M. and Weinberg,R.A. (1999) Nature Med., 10, 1164–1170. [DOI] [PubMed] [Google Scholar]
- 21.Greenberg R.A., Allopp,R.C., Chin,L., Morin,G.B. and DePinho,R.A. (1998) Oncogene, 16, 1723–1730. [DOI] [PubMed] [Google Scholar]
- 22.Blasco M.A., Lee,H-W., Hande,M.P., Samper,E., Lanorp,P.M., DePinho,R.A. and Greider,C.W. (1997) Cell, 91, 25–34. [DOI] [PubMed] [Google Scholar]
- 23.Rudolph L.K., Chang,S., Lee,H-W., Blasco,M., Gottlieb,F.J., Greider,C. and DePinho,R.A. (1999) Cell, 96, 701–712. [DOI] [PubMed] [Google Scholar]