Abstract
Objective
Mendelian randomisation (MR) is a technique that aims to assess causal effects of exposures on disease outcomes. The paper aims to present the main assumptions that underlie MR, the statistical methods used to estimate causal effects and how to account for potential violations of the key assumptions.
Methods
We discuss the key assumptions that should be satisfied in an MR setting. We list the statistical methodologies used in two-sample MR when summary data are available to estimate causal effects (ie, Wald ratio estimator, inverse-variance weighted and maximum likelihood method) and identify/adjust for potential violations of MR assumptions (ie, MR-Egger regression and weighted Median approach). We also present statistical methods and graphical tools used to evaluate the presence of heterogeneity.
Results
We use as an illustrative example of a published two-sample MR study, investigating the causal association of body mass index with three psychiatric disorders (ie, bipolar disorder, schizophrenia and major depressive disorder). We highlight the importance of assessing the results of all available methods rather than each method alone. We also demonstrate the impact of heterogeneity in the estimation of the causal effects.
Conclusions
MR is a useful tool to assess causality of risk factors in medical research. Assessment of the key assumptions underlying MR is crucial for a valid interpretation of the results.
Keywords: Schizophrenia and psychotic disorders
Introduction
Mendelian randomisation (MR) is a technique that aims to investigate if exposure is causally contributing to a disease outcome using genetic variants as proxies for environmental exposures.1 The reason for utilising MR is to overcome many drawbacks of observational epidemiology, such as confounding and reverse causation. MR is analogous to a randomised control trial (RCT) where instead of the allocation of participants to different treatment groups, individuals are randomised by nature to carry or not carry genetic variants that may modify the risk of an exposure.1
An MR analysis is feasible using either individual-level data (one-sample) or summary data (two-sample), where the association of genetic variant(s) with the exposure and the outcome are available. For one-sample MR, the association of genetic variant(s) with the exposure and genetic variant(s) with the outcome are estimated in the same sample, while in the latter in different non-overlapping samples.2 For two-sample MR, the increasing availability and scale of summary data from genome-wide association studies (GWAS) are used to estimate the causal effect of the exposure on an outcome. By using such summary statistics, one can avoid additional complications arising from confidentiality agreements, especially when it comes to large consortia. Additionally, collaborative efforts from large GWAS have identified numerous genetic variants, explaining a high proportion of the heritability of the tested phenotypes and can be used to derive accurate and precise causal effects.3
The aim of this review is to provide an overview of two-sample MR when summary data are available, its assumptions and estimation methods. We include an illustrative example of medical relevance with a focus on the field of mental health.
Methods
MR assumptions
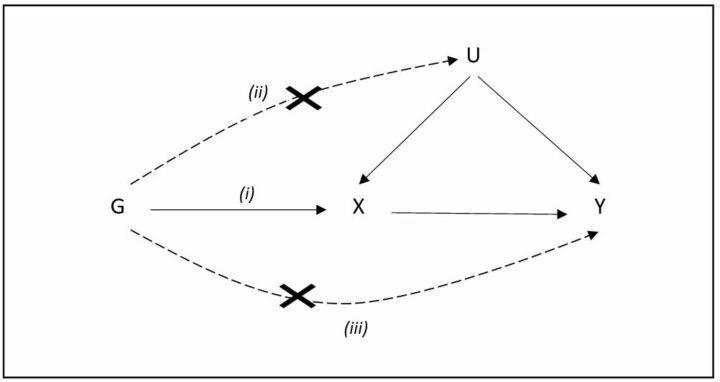
The instrumental variable method, initially introduced in econometrics and social sciences, is an approach to account for confounding and thus infer causality in observational settings.4 5 An instrumental variable is selected as to mimic the randomisation of individuals to the exposure ensuring compatibility of groups with respect to any measured or unmeasured confounders. An MR uses genetic variants associated with exposure as instruments to infer causality on an outcome. For a genetic variant to be a valid instrument, three assumptions must be satisfied, (i) the genetic variant (G) must be strongly associated with the exposure of interest X; (ii) the genetic variant is not associated with any confounder (U) of the exposure–outcome association; (iii) the genetic variant is only associated with the outcome (Y) through the exposure (figure 1).4
Figure 1.
Directed acyclic graph of Mendelian randomisation representing the assumptions (i) the genetic variant (G) must be associated with the exposure X, (ii) must not be associated with any confounder (U) of the exposure-outcome association and (iii) must be associated with the outcome (Y) only via the exposure.
The first assumption is the only assumption that can be formally tested and in practice, genetic variants that are related to a given exposure at the genome-wide significance level are used as instruments (p value <5×10−8). The second and third assumptions can be assessed by estimating the associations between the genetic variants and a large set of confounders. However, when summarised data are available (ie, two-sample MR), assessing the associations between the genetic variants and confounders is based solely on literature evidence, which could be not measured/reported.6 Overall, there is no way to prove that the second and third MR assumptions definitively hold. However, it is often possible to find empirical evidence suggesting that the instruments under consideration are invalid. In practice, we can indirectly evaluate MR assumptions by checking if there is high reproducibility of the MR causal estimates in different studies and if all available MR estimation methods yield concordant results.
Selection of instruments
Genetic variants for gene–exposure associations can be obtained either by extensive catalogues of published GWAS, such as GWAS Catalog and MR-BASE,7 8 or publicly available data of genetic consortia.
Table 1 contains a list of genetic consortia for mental health disorders. Genetic variants must be strongly associated with the exposure at a genome-wide significance level (p value <5×10−8). Genetic variants are then pruned and independent variants (not in linkage disequilibrium (LD) which is the non-random association of alleles at two or more loci in a general population) are taken forward for analysis. However, methods accounting for the correlation structure have been also proposed and can be used to increase statistical power when only a few variants for the exposure of interest are available and these explain a low proportion of the variability.9
Table 1.
Genetic consortia with publicly available data for psychiatric disorders
| Genetic consortium | Disease under study |
| PGC | Anorexia, autism, bipolar disorder, schizophrenia, OCD, PTSD, ADHD |
| CommonMind Consortium | Bipolar disorder, schizophrenia |
| IGAP | Alzheimer’s disease |
ADHD, attention deficit/hyperactivity disorder; IGAP, International Genomics of Alzheimer’s Project; OCD, obsessive compulsive disorder; PGC, Psychiatric Genomics Consortium; PTSD, post-traumatic stress disorder.
Estimation methods
MR estimation methods can be broadly grouped into two main categories depending on the number of the available instruments: (i) when a single variant is available (Wald ratio estimator) and (ii) when multiple variants are available (inverse-variance weighted [IVW] method and maximum likelihood [ML] method).
Wald ratio estimator
When a single genetic variant is available, the easiest method for calculating the causal effect of an exposure on an outcome is the Wald ratio. This can be considered as the change in the outcome resulting from a unit change in the exposure and can be calculated by the ratio of the regression coefficient of the gene–outcome association with the regression coefficient of the gene–exposure association.10 Thus, if we denote by and the estimated regression coefficients of the exposure and the outcome, respectively, on the genetic variant, then the ratio estimate can be expressed as:
A SE can be approximately estimated using the Delta method.11
IVW method
When multiple genetic variants are associated with the exposure of interest, the Wald ratio method can be extended by borrowing methodology from the field of meta-analysis. In particular, the ratio estimates of the causal effects from each genetic variant are combined employing an IVW meta-analysis framework. Thus, the IVW method is a weighted average of the causal effects derived from the genetic variants. This method is equivalent to fitting a weighted linear regression of the associations of the instruments with the outcome on the instruments with the exposure setting the intercept term to zero. Notably, this method assumes that all instruments are valid and no pleiotropic effects exist (ie, the genetic variants are not associated with multiple exposures). Thus, differences in the causal estimates as estimated by each genetic variant individually are due to sampling variability (homogeneity assumption).6 9 12
ML method
Assuming that outcome and exposure are linearly dependent and jointly normally distributed, the causal effect of exposure on the outcome can be estimated by direct maximisation of the likelihood, allowing for uncertainty in both exposure and outcome.6 9
When gene–exposure associations are precisely estimated, then the IVW and ML method give similar results. However, when considerable imprecision exists, IVW produces over-precise causal estimates, while ML results in wider and therefore appropriately-sized CIs, as ML allows for uncertainty in both gene–exposure and gene–outcome associations.9
Accounting for violation of MR assumptions
When all genetic variants satisfy the assumptions of an MR study, causal effect estimates derived from IVW and ML are unbiased. There are several methods that have been developed to identify, allow and correct for violations of assumptions when some of the selected variants are invalid instruments.
MR-Egger
In the presence of pleiotropy, one could fit a weighted linear regression of the associations of the instruments with the outcome on the instruments with the exposure, while assuming an unconstrained intercept term (unlike the IVW approach where intercept term is constrained and set to zero), resulting in the so-called MR-Egger regression method.13 14 The slope of the MR-Egger regression is a robust estimate of the causal effect accounting for potential horizontal pleiotropy (ie, when the genetic variant(s) has an effect on the outcome, independently of the exposure under study). MR-Egger requires that gene–exposure and gene–outcome associations are independent (Instrument Strength Independent on Direct Effect-InSIDE assumption) and that the variance of the association of the genetic variants with the exposure association is negligible (No Measurement Error-NOME assumption). However, the MR-Egger approach can be underpowered when few instruments are available.
Weighted Median estimator
When up to 50% of genetic variants are invalid instruments, then a causal effect can be estimated as the median of the weighted ratio estimates using as weights the reciprocal of the variance of the ratio estimate.15 The InSIDE assumption is not necessary. Violations of the second and the third assumptions are also allowed.
Heterogeneity as an indication of pleiotropy
In an MR setting, we assume that all the instruments estimate the same underlying causal effect and any discrepancies are an indication of pleiotropy. It is likely that the pleiotropic effects of individual genetic variants cancel each other out as they could be either positive or negative. However, when substantial heterogeneity is present, the estimated causal effect will be imprecisely estimated. This heterogeneity can be quantified using the Cochran’s Q statistic or the I 2 metric. A scatter plot with gene–outcome against gene–exposure associations provides a visual inspection of pleiotropy. Under the hypothesis of no heterogeneity, all plotted points must be compatible with a line passing through the origin. One could also plot the precision of the instruments against MR causal estimates and any asymmetry is an indication of potential pleiotropic effects.16
Leave one out analysis
As already discussed, IVW and MR-Egger methods are formulated as a regression of the gene–outcome associations on gene–exposure associations with an intercept term to be constrained or not to zero, respectively. As in any regression model, outlying data points could bias the estimated causal effect. Therefore, the influence of each variant can be assessed by re-estimating the causal association after excluding one genetic variant at a time and any deviances may serve as an indication of potential pleiotropic effects.14
Results
Illustrative example
Hartwig and colleagues conducted a two-sample MR study to investigate the potential causal associations of body mass index (BMI) with three psychiatric disorders (bipolar disorder, schizophrenia and major depressive disorder).17
Publicly available genetic data for BMI were retrieved from the Genetic Investigation of Anthropometric Traits (GIANT) consortium for 322 154 European ancestry individuals.18 Corresponding genetic variants for the three psychiatric disorders were extracted from the Psychiatric Genomics Consortium (PCG) (http://www.med.unc.edu/pgc/downloads). This included summary association data for a total of 7481 bipolar disorder cases/9250 controls; 34 241 schizophrenia cases/45 604 ancestry-matched controls; and 9240 major depressive disorder cases/9 519 controls.19–21 In total, 97 genetic variants single-nucleotide polymorphisms (SNPs) were identified at a genome-wide significance level (p value <5×10−8) for BMI. For schizophrenia, one SNP (ie, rs12016871) was not available. For major depressive disorder, 90 SNPs were extracted (for 28 out of these 90 SNPs proxies were chosen using the 1000 Genomes Pilot 1 and HapMap release 22 as reference panels).
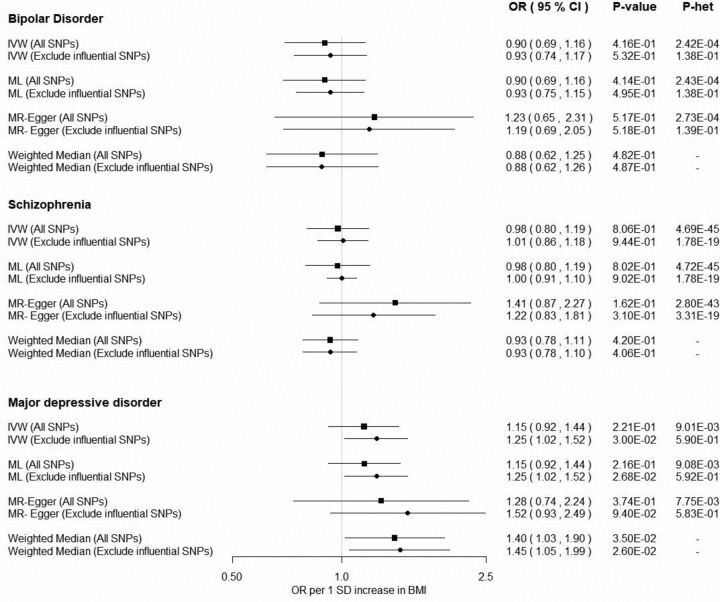
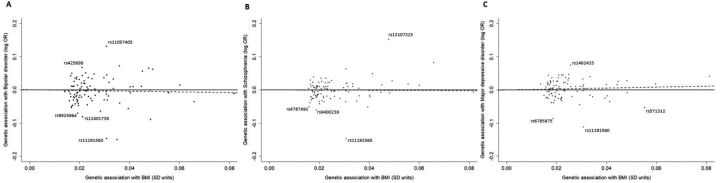
The SNP–BMI associations were calculated applying an inverse-normal transformation on BMI measurements and the MR estimates corresponded to 1 SD increase in BMI. For bipolar disorder and schizophrenia, IVW and ML methods yielded identical non-significant MR estimates (bipolar disorder OR: 0.90; 95% CI 0.69 to 1.16 and schizophrenia OR: 0.98; 95% CI 0.80 to 1.19). There was no evidence for an association of BMI with bipolar disorder and schizophrenia when using the weighted median approach (bipolar disorder OR: 0.88, 95% CI 0.62 to 1.25; and schizophrenia OR: 0.93, 95% CI 0.78 to 1.11). Notably, the MR-Egger approach yielded directionally inconsistent estimates with an OR equal to 1.23 (95% CI 0.65 to 2.31) for bipolar disorder and an OR equal to 1.41 (95% CI 0.87 to 2.27) for schizophrenia. All methods showed little evidence for association with consistent direction for the association of BMI with the major depressive disorder with some evidence of association from the weighted median approach (OR: 1.40; 95% CI 1.03 to 1.90) (figure 2). For all psychiatric disorders, the MR-Egger intercept was approximately equal to 1.00 indicating no violation of the MR assumptions due to pleiotropic effects (bipolar disorder OR: 0.99; 95% CI 0.97 to 1.01, schizophrenia OR: 0.99; 95% CI 0.98 to 1.00, MD OR: 1.00; 95% CI 0.98 to 1.01). Substantial heterogeneity was quantified by the Q-statistics for all tested outcomes (figure 2). As shown in figure 3, there were some SNPs that could be considered outliers and potentially increase statistical heterogeneity. Removing outlying variants resulted in nearly identical results for bipolar disorder. For schizophrenia, the MR estimates did not change for all methods with the exception of the MR-Egger approach, where a weaker causal association with increased precision was observed (OR: 1.22; 95% CI 0.83 to 1.81). Excluding possible pleiotropic variants in major depressive disorder, all methods yielded comparable results in direction with some evidence of association derived by the IVW method (OR: 1.25; 95% CI1.02 to 1.52).
Figure 2.
Mendelian randomisation results of BMI with psychiatric disorders. All estimates are reported per 1 SD increase of BMI. The data used in this example are obtained from a published study of Hartwig and coworkers.17 BMI, body mass index; IVW, inverse-variance weighted; ML, maximum likelihood; P-het, Cohrans’s Q p value; SNPs, single-nucleotide polymorphisms.
Figure 3.
Scatter plots of associations of the selected variants with (A) bipolar disorder, (B) schizophrenia and (C) major depressive disorder and BMI. The data used in this example are obtained from a published study of Hartwig and coworkers.17 BMI, body mass index; logOR, natural logarithm of OR.
Conclusively, only for major depressive disorder, the results of all methods were concordant and the association was even stronger after removing influential SNPs. Therefore, one can conclude that there might be a true causal effect of BMI on major depressive disorder although the statistical evidence was weak. In contrast, the discrepancy between methods in bipolar disorder and schizophrenia does not allow for any safe conclusions. Thus, the reported association of BMI with psychiatric disorders in observational studies may have been confounded.22–24
Discussion
MR aims to identify and quantify potential causal associations between exposures and outcomes of major health importance. Mostly due to the substantial increment of information available by genetic studies, relating the genetic architecture of various phenotypes and diseases, its popularity has increased. Implementing statistical methods borrowed from the field of meta-analysis, to synthesise the summary effects of multiple genetic variants to estimate causal effects, has increased its feasibility. As a result, even researchers with non-statistical backgrounds can implement an MR study effectively, by using software such as the MR-Base for this purpose.8 However, it is crucial that one keeps in mind the assumptions on which the validity of the estimated causal effect relies, especially when using summary-level data.
As discussed in this article, methods exist to assesses and adjust for different degrees of violation of the key assumptions. Both weighted median and MR-Egger can be used to estimate the causal effect of exposure on an outcome, making different assumptions about the degree of violation of the MR assumptions. MR-Egger estimates robust causal effects of the exposure on the outcome, even if all genetic variants are invalid instruments. In contrast, the weighted median requires at least half of them to be valid. Additionally, weighted median allows violation of the MR assumptions in a more general framework, while MR-Egger relaxes second and third assumptions by replacing them with weaker but still untestable assumptions (InSIDE and NOME assumptions).13 Generally, it is recommended to critically appraise all methods together. If the various methods yield results of similar magnitude, then it is more plausible that the produced results are reliable. On the other hand, if estimated causal effects are contradictory further evaluation should be considered and the results should be interpreted with caution.
In the era of -omics, MR is considered a powerful and promising technique, as it could utilise metabolomic, proteomic and DNA methylation data, to better explain the contribution of certain metabolic pathways and gene regulations in the development of diseases.25 Therefore, it is crucial for researchers to recognise that MR is a method that requires statistical and biological knowledge to make inferences that reflect more reliably the nature of complex diseases.
Footnotes
Contributors: All authors contributed to writing and commenting on this paper.
Funding: ND was supported by the IKY scholarship programme in Greece, which is co-financed by the European Union (European Social Fund - ESF) and Greek national funds through the action entitled ”Reinforcement of Postdoctoral Researchers”, in the framework of the Operational Programme ”Human Resources Development Program, Education and Lifelong Learning” of the National Strategic Reference Framework (NSRF) 2014 – 2020 .PP, NM, ES have not declared a specific grant for this research from any funding agency in the public, commercial or not-for-profit sectors.
Disclaimer: The authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Competing interests: None declared.
Ethics approval: An ethics approval was not required as we used summary data publicly available at https://www.ncbi.nlm.nih.gov/pubmed/27601421.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data sharing statement: Data are available in an open access publication https://www.ncbi.nlm.nih.gov/pubmed/27601421.
References
- 1. Smith GD, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol 2003;32:1–22. 10.1093/ije/dyg070 [DOI] [PubMed] [Google Scholar]
- 2. Lawlor DA. Commentary: two-sample Mendelian randomization: opportunities and challenges. Int J Epidemiol 2016;45:908–15. 10.1093/ije/dyw127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pierce BL, Ahsan H, Vanderweele TJ. Power and instrument strength requirements for Mendelian randomization studies using multiple genetic variants. Int J Epidemiol 2011;40:740–52. 10.1093/ije/dyq151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lawlor DA, Harbord RM, Sterne JA, et al. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 2008;27:1133–63. 10.1002/sim.3034 [DOI] [PubMed] [Google Scholar]
- 5. Zhang Z, Uddin MJ, Cheng J, et al. Instrumental variable analysis in the presence of unmeasured confounding. Ann Transl Med 2018;6:182. 10.21037/atm.2018.03.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 2013;37:658–65. 10.1002/gepi.21758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Buniello A, MacArthur JAL, Cerezo M, et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res 2019;47(D1):D1005–D1012. 10.1093/nar/gky1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hemani G, Zheng J, Elsworth B, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife 2018;7. 10.7554/eLife.34408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Burgess S, Scott RA, Timpson NJ, et al. Using published data in Mendelian randomization: a blueprint for efficient identification of causal risk factors. Eur J Epidemiol 2015;30:543–52. 10.1007/s10654-015-0011-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Burgess S, Small DS, Thompson SG. A review of instrumental variable estimators for Mendelian randomization. Stat Methods Med Res 2017;26:2333–55. 10.1177/0962280215597579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Thomas DC, Lawlor DA, Thompson JR, et al. Re: Estimation of bias in nongenetic observational studies using “Mendelian triangulation” by Bautista et al . Ann Epidemiol 2007;17:511–3. 10.1016/j.annepidem.2006.12.005 [DOI] [PubMed] [Google Scholar]
- 12. Greco M FD, Minelli C, Sheehan NA, et al. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med 2015;34:2926–40. 10.1002/sim.6522 [DOI] [PubMed] [Google Scholar]
- 13. Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 2015;44:512–25. 10.1093/ije/dyv080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol 2017;32:377–89. 10.1007/s10654-017-0255-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bowden J, Davey Smith G, Haycock PC, et al. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol 2016;40:304–14. 10.1002/gepi.21965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burgess S, Bowden J, Fall T, et al. Sensitivity analyses for robust causal inference from mendelian randomization analyses with multiple genetic variants. Epidemiology 2017;28:30–42. 10.1097/EDE.0000000000000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hartwig FP, Bowden J, Loret de Mola C, et al. Body mass index and psychiatric disorders: a Mendelian randomization study. Sci Rep 2016;6:32730. 10.1038/srep32730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206. 10.1038/nature14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–7. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011;43:977–83. 10.1038/ng.943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ripke S, Wray NR, Lewis CM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry 2013;18:497–511. 10.1038/mp.2012.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abou Abbas L, Salameh P, Nasser W, et al. Obesity and symptoms of depression among adults in selected countries of the Middle East: a systematic review and meta-analysis. Clin Obes 2015;5:2–11. 10.1111/cob.12082 [DOI] [PubMed] [Google Scholar]
- 23. Luppino FS, de Wit LM, Bouvy PF, et al. Overweight, obesity, and depression: a systematic review and meta-analysis of longitudinal studies. Arch Gen Psychiatry 2010;67:220–9. 10.1001/archgenpsychiatry.2010.2 [DOI] [PubMed] [Google Scholar]
- 24. McElroy SL, Keck PE. Obesity in bipolar disorder: an overview. Curr Psychiatry Rep 2012;14:650–8. 10.1007/s11920-012-0313-8 [DOI] [PubMed] [Google Scholar]
- 25. Burgess S, Timpson NJ, Ebrahim S, et al. Mendelian randomization: where are we now and where are we going? Int J Epidemiol 2015;44:379–88. 10.1093/ije/dyv108 [DOI] [PubMed] [Google Scholar]