IMPORTANCE:
This is a study of lipid metabolic gene expression patterns to discover precision medicine for sepsis.
OBJECTIVES:
Sepsis patients experience poor outcomes including chronic critical illness (CCI) or early death (within 14 d). We investigated lipid metabolic gene expression differences by outcome to discover therapeutic targets.
DESIGN, SETTING, AND PARTICITPANTS:
Secondary analysis of samples from prospectively enrolled sepsis patients (first 24 hr) and a zebrafish endotoxemia model for drug discovery. Patients were enrolled from the emergency department or ICU at an urban teaching hospital. Enrollment samples from sepsis patients were analyzed. Clinical data and cholesterol levels were recorded. Leukocytes were processed for RNA sequencing and reverse transcriptase polymerase chain reaction. A lipopolysaccharide zebrafish endotoxemia model was used for confirmation of human transcriptomic findings and drug discovery.
MAIN OUTCOMES AND MEASURES:
The derivation cohort included 96 patients and controls (12 early death, 13 CCI, 51 rapid recovery, and 20 controls) and the validation cohort had 52 patients (6 early death, 8 CCI, and 38 rapid recovery).
RESULTS:
The cholesterol metabolism gene 7-dehydrocholesterol reductase (DHCR7) was significantly up-regulated in both derivation and validation cohorts in poor outcome sepsis compared with rapid recovery patients and in 90-day nonsurvivors (validation only) and validated using RT-qPCR analysis. Our zebrafish sepsis model showed up-regulation of dhcr7 and several of the same lipid genes up-regulated in poor outcome human sepsis (dhcr24, sqlea, cyp51, msmo1, and ldlra) compared with controls. We then tested six lipid-based drugs in the zebrafish endotoxemia model. Of these, only the Dhcr7 inhibitor AY9944 completely rescued zebrafish from lipopolysaccharide death in a model with 100% lethality.
CONCLUSIONS:
DHCR7, an important cholesterol metabolism gene, was up-regulated in poor outcome sepsis patients warranting external validation. This pathway may serve as a potential therapeutic target to improve sepsis outcomes.
Keywords: genetics, lipids, sepsis, transcriptomics, zebrafish
KEY POINTS
Question: Can lipid metabolism gene expression patterns distinguish between poor versus favorable outcomes in sepsis and can they be used to identify drug targets for sepsis?
Findings: In this prospective cohort study with a derivation/validation design, the cholesterol metabolism gene 7-dehydrocholesterol reductase (DHCR7) was significantly up-regulated in in poor outcome sepsis compared with rapid recovery patients and in 90-day nonsurvivors. Blockade of Dhcr7 in a zebrafish endotoxemia model led to complete rescue from death in a model with 100% lethality.
Meaning: DHCR7 was up-regulated in poor outcome sepsis patients and may serve as a potential therapeutic target in sepsis.
Sepsis is life-threatening organ dysfunction caused by a dysregulated host response to infection and is the costliest reason for hospital admission world-wide (1–4). It occurs when a systemic infection results in a dysregulated immune response that leads to organ dysfunction and potentially death (3). Survivors of sepsis are frequently left with reduced quality of life physical function, and long-term survival (5–7). Our group has defined and described clinically relevant outcomes that include early death (death within the first 14 d of sepsis), chronic critical illness (CCI, ICU stay > 14 d with organ dysfunction), and rapid recovery (neither early death nor CCI). CCI patients frequently develop the persistent inflammation immunosuppression and catabolism syndrome (PICS), characterized by impaired physical function and 1-year mortality rates over 40% (7, 8).
We and others have described the protective role of lipids and lipoproteins in sepsis (9–12). High-density lipoprotein (HDL) has antioxidant and anti-inflammatory proteins (paraoxonase-1 and apolipoprotein A-I) that protect against lipid oxidation, prevent inflammatory cell activation and chemotaxis, bind and clear bacterial toxins, and down-regulate inflammatory pathways (9–17). Similarly low-density lipoprotein (LDL) may play a role in bacterial endotoxin clearance via the LDL receptor, particularly in gram negative sepsis, with proprotein convertase subtilisin/kexin type 9 playing an important regulatory role (18–21). However, dysregulated lipid metabolism occurs in sepsis leading to HDL’s conversion to dysfunctional and pro-inflammatory HDL (Dys-HDL) that may play a role in progression of organ dysfunction, and the pathogenesis of CCI, and PICS (9, 22, 23).
Recent studies have shed some light on the genetic underpinnings of lipid metabolism in sepsis. A U.K. Biobank study identified an important link between genetically determined HDL cholesterol (HDL-C) levels and decreased risk of hospitalizations for infectious disease, lower odds of outpatient antibiotic usage, and reduced risk of mortality from sepsis (24). LDL cholesterol (LDL-C) and triglyceride levels did not confer the same risk reduction (24). However, the U.K. Biobank population was of homogenous ancestry. Another study identified that a rare missense variant in the cholesteryl ester transfer protein (CETP) gene (lowers HDL-C levels) was linked with reductions in HDL-C during sepsis (25). Carriers of this risk allele had more severe organ failure and reduced 28-day survival.
Genetic studies of diverse cohorts are needed to understand the role of dysregulated lipid and lipoprotein metabolism in sepsis. This study sought to leverage a diverse prospective cohort of sepsis patients to investigate transcriptional profiles relevant to lipid metabolism in sepsis and associate these differences with relevant outcomes. The primary objective was to analyze leukocyte gene expression patterns of sepsis patients by clinical outcomes by performing both an unbiased RNA sequencing (RNA-seq) analysis and a focused analysis of relevant lipid metabolism genes (47 genes selected a priori). Results were corroborated in a zebrafish endotoxemia model, which further allowed the functional testing of genes relevant to sepsis. Zebrafish were selected as they are vertebrates that share many anatomic and physiologic similarities with humans, most aspects of the immune response, and nearly all elements of lipid and lipoprotein metabolism (26–28). These investigations may aid the identification of lipid metabolic pathways that are critical for regulating the response to sepsis and identifying new potential therapies.
METHODS
Design
We performed a secondary analysis of transcriptomic data from four prospective studies of sepsis patients enrolled between November 2016 and July 2022 from the emergency department at UF Health Jacksonville. All human studies were approved by the University of Florida Institutional Review Board (IRB-01, approved through September 18, 2025) and registered with clinicaltrials.gov (NCT02934997, NCT04576819, and NCT03405870). STrengthening the Reporting of OBservational studies in Epidemiology guidelines for observational studies were followed (29). Approval for all zebrafish work was granted by the Institutional Animal Care and Use Committee (IACUC protocol PRO00010679; expiration date March 10, 2025) at The University of Michigan (Animal Welfare Assurance Number on file with the NIH Office of Laboratory Animal Welfare is A3114-01). All animal experiments were performed in accordance with the ARRIVE 2.0 guidelines (30).
Patient Selection and Enrollment
Patients enrolled in the UF JAX Sepsis Biobank were considered eligible for inclusion after IRB approval. UF Health Jacksonville emergency department patients meeting Sepsis-3 criteria were identified prospectively by trained research coordinators or providers within 24 hours of sepsis recognition (3). Patient enrollment occurred 7 days per week between the hours of 8 am and 10 pm. Patients from three observational studies and one ongoing clinical trial (LIPid Intensive Drug Therapy for Sepsis Pilot) were included in this analysis (31, 32). Exclusion criteria were overall similar to prior studies (31, 32). Healthy controls were patients presenting to the emergency department (ED) with minor noninfectious complaints (e.g., medication refills, or similar) and with normal vital signs (excluded for fever, tachycardia, hypotension, or hypoxia). Patients were excluded if they had abdominal pain, bleeding, respiratory complaints, suspicion of viral or bacterial infection, or fever or infection in the week preceding enrollment. They were also excluded if they had received antibiotics in the preceding 2 weeks.
Data Collection
All clinical and laboratory data were reviewed and entered into a Research Electronic Data Capture database by trained research coordinators (33, 34). Prospectively collected data included demographics, place of residence, source of infection, and comorbidities. Clinical variables included triage and enrollment vital signs, Sequential Organ Failure Assessment (SOFA) score, timing of antibiotics, volume of IV fluids, vasopressor use and duration, and mechanical ventilation use and duration. Hospital length of stay (LOS), and ICU LOS were documented.
Clinical Outcomes and Adjudication
The primary outcome was one of three categories: 1) early death (within 2 wk of sepsis onset), 2) CCI (total ICU stay > 14 d with organ dysfunction or total ICU ≤ 14 d but discharged to long-term acute care, another hospital, or hospice), or 3) rapid recovery (all others) (8). Group adjudication by at least two clinician-investigators was performed for the sepsis diagnosis, primary outcomes, primary and secondary source of infection, culture positivity, and hospital disposition during sepsis adjudication meetings (35). Discrepancies were resolved by the inclusion of a third clinician-investigator. The social security death index was used to determine mortality for patients lost to follow-up. Twenty-eight ninety-day mortality was also recorded.
Blood Sampling, RNA-seq, and RT-qPCR Analysis
Blood was drawn at the time of enrollment and within 24 hours of sepsis recognition and before any clinical trial drug administration. Clinical laboratory testing included cholesterol levels, and SOFA score laboratory measures including platelets, creatinine, and total bilirubin levels. Serum total cholesterol, HDL-C, and triglyceride levels were directly measured from serum samples. LDL-C was calculated using the Friedewald formula (36). RNA-seq was performed using the Illumina NextSeq 550 system (San Diego, CA). Reverse transcriptase polymerase chain reaction (RT-qPCR) was performed using Bio-Rad (Hercules, CA) iQ SYBR Green Supermix (Cat no. 1708882). For details on RNA-seq and RT-qPCR refer to Supplemental Digital Content (http://links.lww.com/CCX/B201).
Zebrafish Experiments
Zebrafish were maintained according to protocols approved by the University of Michigan
Animal Care and Use Committee. All wild-type fish were a hybrid line generated by crossing AB and TL fish acquired from the Zebrafish International Resource Center (Eugene, OR). For details on cholesterol metabolism drug experiments, RT-qPCR, and RNA-seq analysis refer to Supplemental Digital Content (http://links.lww.com/CCX/B201).
Data Analysis
Univariate Comparisons
Presenting vital signs, cholesterol levels, demographic information, clinical features, and clinical management data across the outcome groups and by mortality (28 and 90 d) were analyzed. We calculated medians and interquartile ranges for continuous variables and counts and proportions for categorical variables. To test for differences among outcome groups, we ran the Shapiro-Wilkes test of normality for each of the continuous variables. Only age was found to be normally distributed. Age was also found to have homogeneity of variances, per Bartlett’s test, thereby meeting the requirements to use an analysis of variance procedure (37). For all other continuous variables, we used the nonparametric Kruskal-Wallis procedure. We used Fisher exact test to compare differences in categorical variables. We conducted a total of 28 tests comparing differences with the outcome group variable (Tables 1 and 2), then applied Bonferroni adjustment to proportionally correct our presented p values. Analysis and calculations were completed in R (R Statistical Software v4.1.2; R Core Team 2021; Vienna, Austria) using statistical tests from the Stats package.
TABLE 1.
Demographic Features, Presenting Vital Signs, and Presenting Lipid Levels by Outcome
| Variable | All Patients (n = 128) | Rapid Recovery (n = 89) | Chronic Critical Illness (n = 21) | Early Death (n = 18) | p |
|---|---|---|---|---|---|
| Demographic features | |||||
| Age (yr), median (interquartile range [IQR]) | 61.5 (56.0–70.0) | 60.0 (54.0–66.0) | 72.0 (65.0–78.0) | 61.5 (57.0–65.8) | 0.036a |
| Gender, n (%) | |||||
| Male | 76 (59%) | 59 (66%) | 9 (43%) | 8 (44%) | 1b |
| Female | 52 (41%) | 30 (34%) | 12 (57%) | 10 (56%) | |
| Race, n (%) | |||||
| African American | 71 (55%) | 49 (55%) | 13 (62%) | 9 (50%) | 1b |
| Caucasian | 54 (42%) | 38 (43%) | 7 (33%) | 9 (50%) | |
| Other | 3 (2%) | 2 (2%) | 1 (5%) | 0 (0%) | |
| Presenting vital signs | |||||
| Systolic blood pressure (mm Hg), median (IQR) | 112.0 (97.0–129.8) | 115.0 (103.0–129.0) | 106.0 (92.0–136.0) | 102.5 (87.5–124.5) | 1c |
| Diastolic blood pressure (mm Hg), median (IQR) | 61.0 (53.0–74.0) | 61.0 (54.0–74.0) | 65.0 (53.0–77.0) | 60.5 (44.8–69.8) | 1c |
| Heart rate (beats/min), median (IQR) | 97.0 (84.0–117.5) (1 missing) | 97.0 (86.0–116.5) (1 missing) | 88.0 (73.0–101.0) | 106.0 (90.5–121.5) | 1c |
| Respiratory rate (breaths/min), median (IQR) | 19.0 (17.0–22.5) (1 missing) | 19.0 (16.0–22.0) | 20.0 (16.0–24.0) | 22.0 (19.0–26.0) (1 missing) | 1c |
| Temperature (F), median (IQR) | 99.0 (98.1–100.5) (1 missing) | 99.1 (98.1–100.7) (1 missing) | 98.4 (97.7–99.3) | 99.3 (98.4–100.2) | 1c |
| Oxygen saturation (%), median (IQR) | 98.0 (95.0–99.0) (1 missing) | 98.0 (95.0–99.0) | 98.0 (96.0–99.3) (1 missing) | 97.0 (95.0–99.0) | 1c |
| Presenting lipid levels | |||||
| High-density lipoprotein (mg/dL), median (IQR) | 21.7 (13.1–34.6) | 21.0 (13.8–34.5) | 18.1 (10.0–30.0) | 26.0 (14.7–35.8) | 1c |
| Low-density lipoprotein (mg/dL), median (IQR) | 58.5 (29.3–84.8) (2 missing) | 67.0 (47.0–99.0) | 27.0 (21.0–61.1) | 26.5 (17.8–45.3) (2 missing) | < 0.001c |
| Triglycerides (mg/dL), median (IQR) | 99.5 (58.8–142.3) | 95.5 (47.0–131.0) | 116.0 (65.0–144.0) | 99.0 (69.8–153.0) | 1c |
| Total cholesterol (mg/dL), median (IQR) | 90.9 (74.0–122.3) | 102.0 (79.8–125.0) | 75.0 (65.0–120.0) | 76.5 (70.3–95.3) | 0.709c |
One-way analysis of variance.
Fisher exact test.
Kruskal-Wallis test.
TABLE 2.
Clinical Features and Management by Outcome
| Variable | All Patients (n = 128) | Rapid Recovery (n = 89) | Chronic Critical Illness (n = 21) | Early Death (n = 18) | p |
|---|---|---|---|---|---|
| Clinical features | |||||
| T0 Sequential Organ Failure Assessment score, median (interquartile range IQR]) | 6.0 (4.0–10.0) | 5.0 (3.0–8.0) | 11.0 (8.0–11.0) | 10.0 (6.5–13.0) | < 0.001b |
| Acute Physiology and Chronic Health Evaluation II score, median (IQR) | 17.0 (11.0–21.0) (39 missing) | 13.0 (9.0–18.0) (32 missing) | 18.0 (14.8–29.0) (5 missing) | 21.0 (17.8–25.0) (2 missing) | 0.010b |
| Vasopressor use (0/1), n (%) | 57 (45%) | 25 (28%) | 15 (71%) | 17 (94%) | < 0.001c |
| Pressor durationa (hr), median (IQR) | 41.0 (24.0–66.0) | 34.0 (17.2–41.0) | 43.0 (26.1–97.5) | 62.1 (44.0–128.8) | 0.343b |
| Diabetes (0/1), n (%) | 54 (42%) | 38 (43%) | 8 (38%) | 8 (44%) | 1c |
| Chronic obstructive pulmonary disease (0/1), n (%) | 18 (14%) | 11 (12%) | 3 (14%) | 4 (22%) | 1c |
| End-stage renal disease (0/1), n (%) | 15 (12%) | 11 (12%) | 3 (14%) | 1 (6%) | 1c |
| HIV (0/1), n (%) | 4 (3%) | 4 (4%) | 0 (0%) | 0 (0%) | 1c |
| Primary infection source, n (%) (2 missing) | |||||
| Blood without another source | 5 (4%) | 4 (4%) | 0 (0%) | 1 (6%) | |
| Endocarditis | 4 (3%) | 2 (2%) | 1 (5%) | 1 (6%) | |
| Intra-abdominal | 11 (9%) | 6 (7%) | 2 (10%) | 3 (18%) | |
| IV catheter-related bloodstream | 2 (2%) | 1 (1%) | 0 (0%) | 1 (6%) | |
| Necrotizing soft tissue | 1 (1%) | 0 (0%) | 1 (5%) | 0 (0%) | |
| Other | 3 (2%) | 3 (3%) | 0 (0%) | 0 (0%) | |
| Pulmonary | 39 (30%) | 24 (27%) | 10 (48%) | 5 (28%) | |
| Skin/soft tissue | 17 (13%) | 14 (16%) | 2 (10%) | 1 (6%) | |
| Surgical site | 1 (1%) | 1 (1 %) | 0 (0%) | 0 (0%) | |
| Surgical thoracic | 1 (1%) | 0 (0%) | 1 (5%) | 0 (0%) | |
| Unknown | 1 (1%) | 1 (1%) | 0 (0%) | 0 (0%) | |
| Urinary tract | 42 (33%) | 33 (37%) | 4 (19%) | 5 (28%) | |
| Treatment variables | |||||
| Time to antibiotics from triage (min), median (IQR) | 120.0 (76.5–179.0) (1 missing) | 122.0 (77.0–180.0) | 109.0 (67.0–156.0) | 134.0 (90.0–190.0) (1 missing) | 1b |
| Fluids volume in first 6 hr from triage (mL), median (IQR) | 2,000 (1,000–3,137) | 2,000 (1,000–3,000) | 3,000 (1,500–3,330) | 2,500 (1,000–3,500) | 1b |
| Mechanical ventilation use (0/1), n (%) | 46 (36%) | 16 (18%) | 15 (71%) | 15 (83%) | < 0.001c |
| Mechanical ventilation duration (d), median (IQR) (2 missing) | 2.9 (0.6–9.5) (2 missing) | 2.3 (0.7–3.2) | 7.8 (1.2–19.4) (1 missing) | 4.5 (0.5–9.6) (1 missing) | 1b |
| Length of hospital stay (d), median (IQR) | 8.0 (4.9–13.4) | 7.1 (5.0–12.9) | 19.5 (12.0–27.9) | 6.3 (1.0–9.0) | 0.044b |
| ICU (0/1), n (%) | 90 (70%) | 52 (58%) | 20 (95%) | 18 (100%) | < 0.001c |
| Length of ICU staya (d), median (IQR) | 5.0 (3.0–11.0) | 4.0 (2.0–5.3) | 17.0 (8.0–29.0) | 7.5 (3.3–10.8) | < 0.001b |
Statistical test and comparisons were applied, respectively, for 57 patients on vasopressors and 90 patients in ICU.
Kruskal-Wallis test.
Fisher exact test.
Differential Expression Data Analysis
For data alignment, gene counts were obtained by aligning reads to the hg38 genome (GRCh38.p11) using STAR (38) (v.2.7.9a) and featureCounts (39) (v.2.0.3). We had two steps of analysis for the differential expression analysis: derivation and validation. We ensured a similar distribution of clinical outcomes across derivation and validation sets to detect differential expression patterns by outcome. To simplify the differential expression analysis, we combined early death and CCI patients into a “poor outcomes” group and compared them to rapid recovery patients who had more favorable outcomes. In a similar manner, we also performed a differential expression analysis by 90-day mortality. Twenty healthy control samples were analyzed with the sepsis samples in the derivation set to compare gene expression patterns between the broader cohort of sepsis patients to healthy controls. The same differentially expressed genes detection protocol was used for both the derivation and validation steps of analysis. We included samples from two duplicate patients (both included in the validation set) enrolled in the study during two different sepsis episodes, over 1 year apart. Data were analyzed with and without these two additional patient encounters; their inclusion did not change the significant differentially expressed genes and so these encounters were included in the final results. In brief, the differential expression analysis was performed using DESeq2 (40) in R (R Statistical Software v4.1.2; R Core Team 2021). Gene counts were modeled with a negative binomial generalized linear model and adjusted for batch effects. Wald tests were conducted for pairwise comparisons. We identified genes with adjusted p values (i.e., p values after false discovery rate correction) less than 0.05 as the differentially expressed genes. We focused our analysis on a set of 47 prespecified lipid metabolism genes (Supplemental Digital Content—Table 1, http://links.lww.com/CCX/B201).
RESULTS
The analysis included 128 sepsis patient encounters and 20 healthy controls. The derivation cohort included 96 patients and controls (12 early death, 13 CCI, 51 rapid recovery, and 20 controls) and the validation cohort had 52 patients (6 early death, 8 CCI, and 38 rapid recovery). For sepsis patients, presenting vital signs were similar in outcomes. Distribution of comorbidities across the outcome groups was similar (Table 1). Initial LDL-C levels were significantly lower for patients with early death or CCI compared with rapid recovery patients. Total cholesterol, HDL-C, and triglyceride levels were not statistically significantly different between groups. CCI patients were significantly older (median 72 yr) than early death (median 61.5 yr) or rapid recovery (median 60 yr). Median SOFA and Acute Physiology And Chronic Health Evaluation (APACHE) II scores were significantly higher for CCI (11, 18, respectively) and early death (10, 21, respectively) compared with rapid recovery (5, 13, respectively) patients. There was a higher proportion of septic shock patients in the early death and CCI groups compared with rapid recovery. The most common source of infection was pulmonary (27%), urinary tract (25%), and multiple sources of infection (17%). There were no significant differences inpatient management characteristics (Table 2).
For the differential expression analysis, the derivation cohort had 96 single-end sequencing samples, including 12 early death, 13 CCI, 51 rapid recoveries, and 20 healthy control patient samples. The validation cohort had 58 paired-end sequencing samples of sepsis patients, including eight early death, 12 CCI, and 38 rapid recoveries. Patients included in the derivation cohort had a similar age, gender, and race distribution compared with patients in the validation set. With the exception of triglycerides, presenting cholesterol and lipid levels were similar between derivation and validation cohorts. They also had similar APACHE II and SOFA scores, proportions of shock patients, and clinical management (Supplemental Digital Content, Tables 2 and 3, http://links.lww.com/CCX/B201).
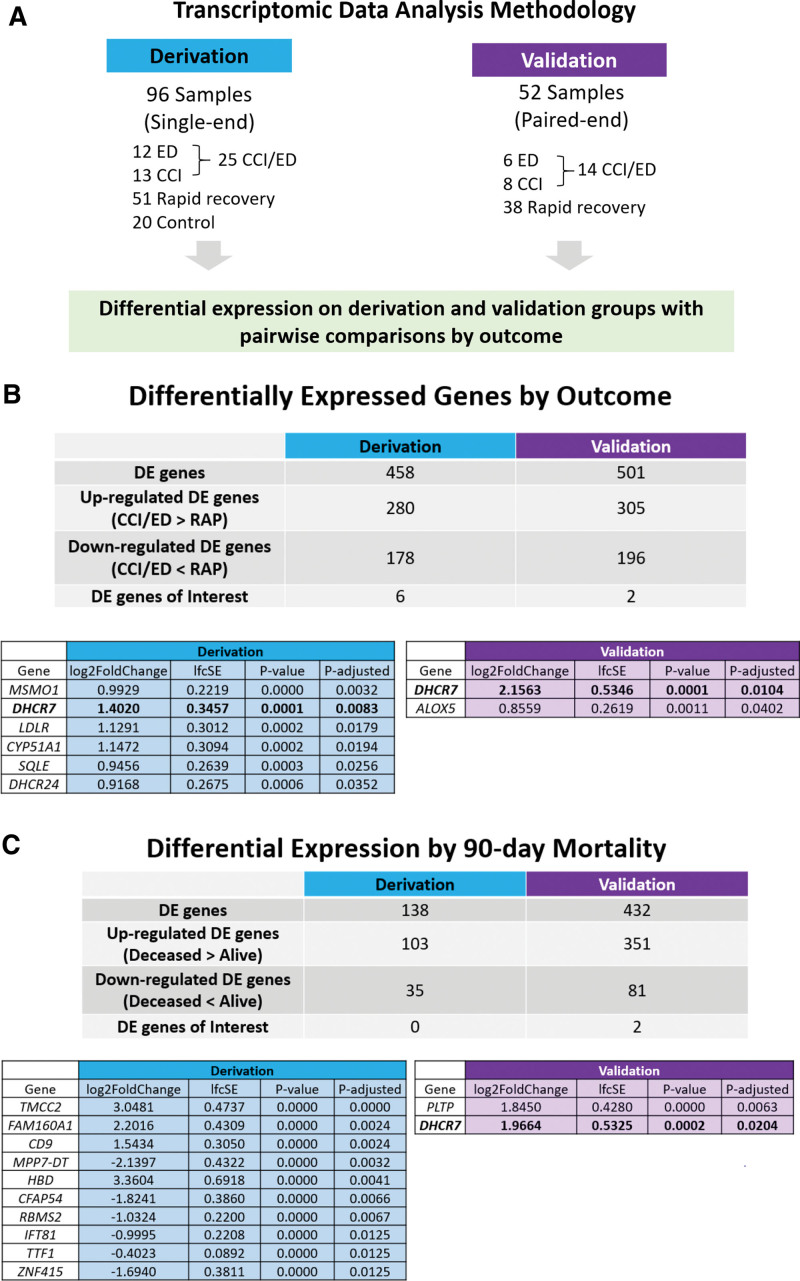
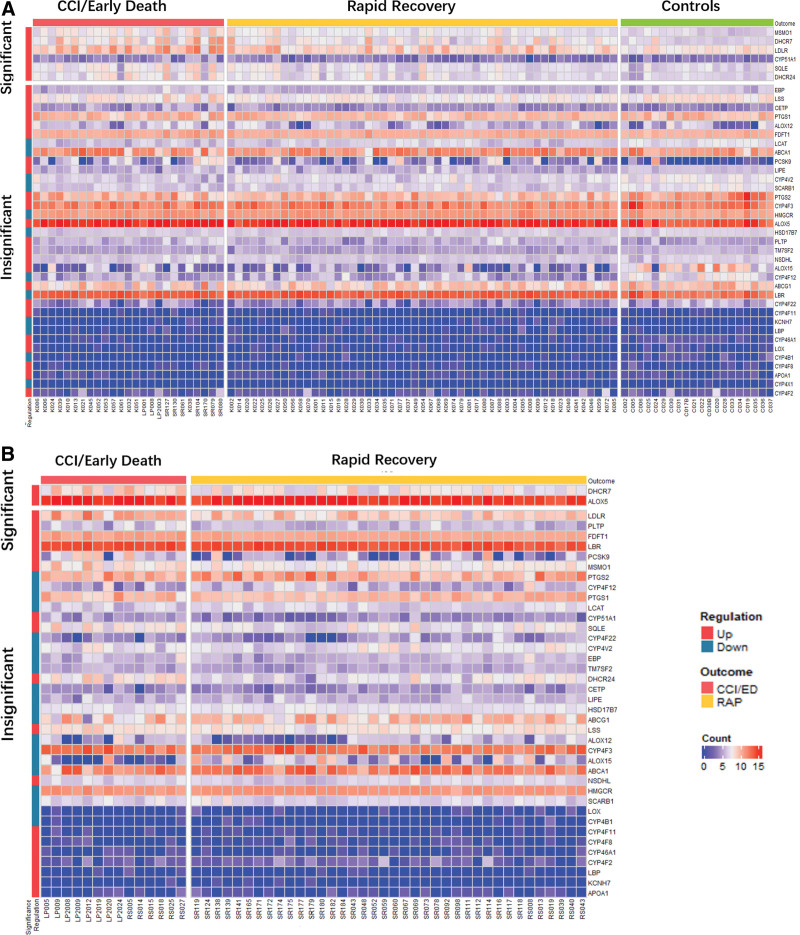
Figure 1 depicts the workflow for RNA-seq data analysis (Fig. 1A) and significantly differentially expressed genes for the derivation and validation cohorts (Fig. 1B) and by 90-day mortality (Fig. 1C). In the derivation cohort, 458 of 39,372 genes were differentially expressed by the patient outcome, including six of the 47 lipid metabolism genes of interest. In the validation cohort, 501 of 36,585 genes were identified as differentially expressed genes, including 2 lipid genes of interest. Of the 47 lipid metabolism genes of interest, there were 6 significant genes identified in the derivation cohort (CYP51A1, DHCR24, DHCR7, MSMO1, SQLE, and LDLR) and 2 genes identified in the validation cohort (DHCR7 and ALOX5). All of these genes were up-regulated in early death/CCI patients when compared with rapid recovery patients. Figure 2 displays heatmaps of differentially expressed genes for derivation and validation cohorts. Five of the significant derivation cohort genes encode enzymes that catalyze critical steps in the biosynthesis of cholesterol (CYP51A1, DHCR24, DHCR7, MSMO1, and SQLE). CYP51A1 is critical for cholesterol synthesis, steroid synthesis, and drug metabolism (41). LDLR encodes the LDL receptor which endocytoses LDL-C from circulation (18). Both significant genes from the validation cohort were up-regulated in CCI/early death patients compared with rapid recovery. ALOX5 is the critical enzyme for the generation of all leukotrienes, potent mediators of inflammation (42). The only gene identified to be significantly up-regulated in both cohorts was DHCR7. The log2fold change in DHCR7 expression between CCI/ED versus rapid recovery patients was 1.4020 in the derivation cohort, and 2.1563 in the validation cohort. All the differentially expressed genes for derivation and validation cohorts are presented in Supplemental Data File 1 (http://links.lww.com/CCX/B202).
Figure 1.
Transcriptomic data analysis methodology and results by outcome and mortality. A, Methodologic flow for transcriptomic data analysis for derivation and validation groups. B, Differentially expressed genes for derivation and validation groups by outcomes of chronic critical illness (CCI)/early death compared with rapid recovery. C, Differentially expressed genes for derivation and validation groups by 90-day mortality. ED = early death, RAP = rapid recovery.
Figure 2.
Heatmaps of differentially expressed genes for derivation (A) and validation (B) groups by outcomes of chronic critical illness (CCI)/early death compared with rapid recovery.
We performed a differential expression analysis by 90-day mortality. None of the lipid metabolism genes of interest were detected in the derivation cohort. However, DHCR7 and PLTP were detected and up-regulated in the validation cohort (Fig. 1). PLTP encodes a protein that is important for cholesterol and lipopolysaccharide clearance, and transfers phospholipids from triglyceride-rich lipoproteins. It also helps to regulate HDL size and is involved in cholesterol and lipopolysaccharide clearance (23).
We next examined gene expression in sepsis patients and healthy controls by RT-qPCR. Based on availability of total RNA, we picked 10 CCI, 12 early death, 12 rapid recovery patients, and 11 healthy controls for RT-qPCR. Demographics of patients included in RT-qPCR are presented in Supplemental Digital Content, Table 4 (http://links.lww.com/CCX/B201). Five of the six genes (LDLR, DHCR24, DHCR7, MSMO1, and SQLE) identified in the RNA-seq analysis were significantly up-regulated in comparison to controls, whereas CYP51A1 was not (Supplemental Digital Content, Fig. 1, http://links.lww.com/CCX/B201).
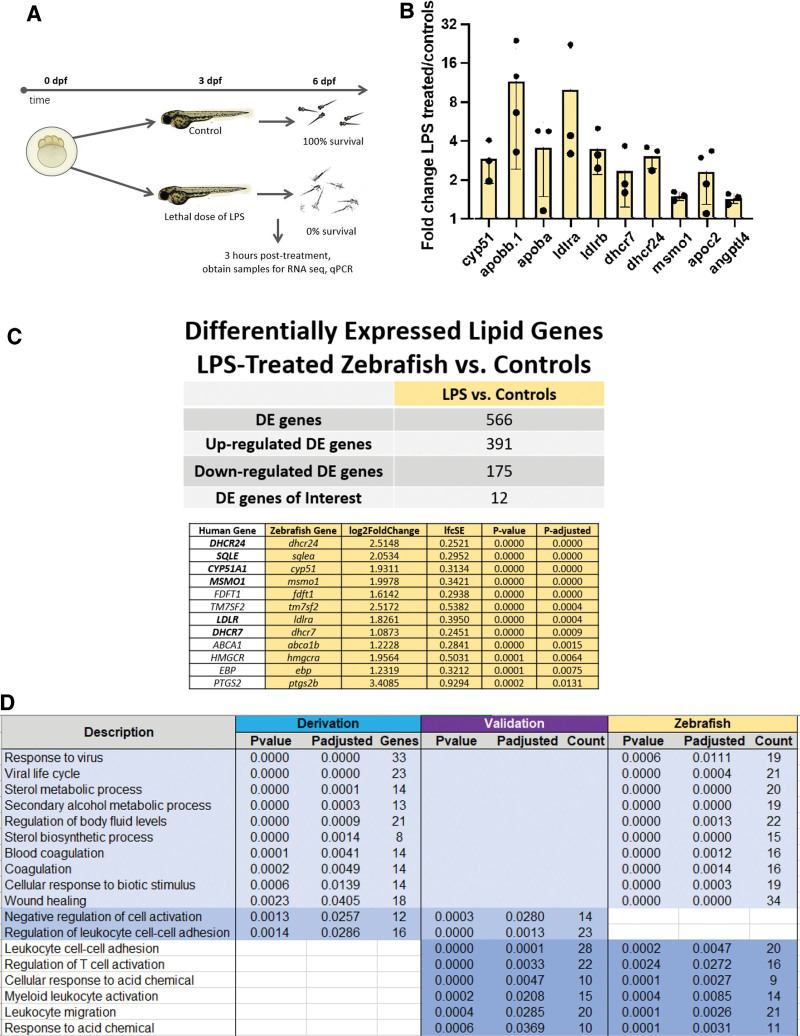
Workflow for zebrafish experiments with lipopolysaccharide versus controls is depicted in Figure 3A. RT-qPCR of cholesterol-related genes showed up-regulation of genes for the LDL receptor (ldlra, ldlrb), dhcr7, dhcr24, msmo1, and cyp51 in lipopolysaccharide-treated zebrafish compared with controls (Fig. 3B). Differential expression analysis of RNA-seq data from three lipopolysaccharide-treated zebrafish and three controls identified 12 lipid metabolism genes that were up-regulated in lipopolysaccharide-treated zebrafish compared with controls (Fig. 3C). Notably, six of the genes (dhcr7, dhcr24, sqlea, cyp51, msmo1, and ldlra) were also up-regulated in CCI/early death sepsis patients in the derivation cohort, as was dhcr7 in the validation cohort. Overlap of significantly differentially expressed genes between derivation, validation, and zebrafish groups is depicted in Figure 3D. Gene primers for zebrafish experiments are noted in Supplemental Data File 2 (http://links.lww.com/CCX/B203).
Figure 3.
Drug testing in zebrafish model of endotexemia. A, Methodology for zebrafish experiments. Zebrafish are treated with a lethal dose of lipopolysaccharide or maintained in control embryo medium at 3 (days postfertilization) dpf and examined for survival at 4, 5, and 6 dpf. B, Reverse transcriptase polymerase chain reaction (RT-qPCR) of cholesterol related genes from lipopolysaccharide-treated fish versus controls 3 hours after treatment at 3 dpf. Data represented as fold change lipopolysaccharide/controls. A value of 1 would signify no change, value greater than 1 is up-regulation in lipopolysaccharide treated versus controls, and value less than 1 is down-regulation. Individual dots represent separate experiments. The graph bars represent mean and sd. C, Differential expression analysis of RNA sequencing (RNA-seq) data from three lipopolysaccharide-treated zebrafish and three controls identified 12 lipid metabolism genes that were up-regulated in lipopolysaccharide-treated zebrafish compared with controls. D, Overlap of significantly differentially expressed genes between derivation, validation, and zebrafish groups.
We tested several cholesterol metabolism drugs in our zebrafish model including AY9944 (Dhcr7 inhibitor), triparanol (Dhcr24 inhibitor), atorvastatin (HMG-CoA reductase inhibitor), torcetrapib (Cetp inhibitor), and ezetimibe (cholesterol absorption inhibitor). Results of all zebrafish drug experiments are displayed in Supplemental Figure 2 (http://links.lww.com/CCX/B201). Varying concentrations of each drug were administered at 3 dpf (days postfertilization) with or without a dose of lipopolysaccharide that caused complete lethality by 4 dpf. For AY9944 (Dhcr7 inhibitor), 10–20 µM of AY9944 alone showed no effects on survival. When administered with lipopolysaccharide, the 10 µM dose led to partial protection against mortality, whereas 20 µM resulted in 100% survival up to 6 dpf. None of the other drugs tested protected against lipopolysaccharide death.
DISCUSSION
In this study, we performed an unbiased differential expression analysis of leukocyte gene expression RNA-seq data from diverse, prospective cohorts of sepsis patients. We further investigated 47 lipid metabolism genes to delineate lipid metabolic changes in sepsis patients by outcome and identified DHCR7 to be significantly and consistently up-regulated for patients with CCI/early death and in the 90-day mortality group when compared with healthy controls and rapid recovery patients. DHCR7 encodes an enzyme that removes the double bond in the B ring of sterols and catalyzes the conversion of 7-dehydroxycholesterol (7DHC) to cholesterol (43). 7DHC is also a precursor to vitamin D, catalyzed by DHCR7 (43). In a parallel set of RNA-seq studies conducted in a zebrafish endotoxemia model, we observed that dhcr7 was significantly up-regulated in samples from zebrafish that received lethal doses of lipopolysaccharide when compared with controls. Furthermore, pharmacologic blockade of Dhcr7 resulted in complete rescue from death. These results are consistent with dhcr7 having a potential mechanistic link to endotoxic death in a zebrafish endotoxemia model.
DHCR7 is a critical gene involved in cholesterol biosynthesis, immune regulation, and metabolism. Patients with loss of function mutations in DHCR7 develop Smith-Lemli-Optiz syndrome, which results in branchial and cardiac defects, electrolyte abnormalities (hypocalcemia, hyponatremia, and hyperkalemia), and extremely low cholesterol levels (< 38.7 mg/dL) associated with necrotizing enterocolitis, recurrent infections, sepsis-like episodes, and death in several patients (44). In a recent study, the genetic association of variants in the DHCR7 gene (and other genes for vitamin D metabolism) with subsequent bacterial pneumonia was studied (45). They found that genetic variants of CYP2R1 but not DHCR7, GEMIN2, or HAL were associated with increased risk of bacterial pneumonia.
Recently, the potential mechanistic role of DHCR7 in combatting systemic infections has been studied. Xiao and colleagues showed that DHCR7 inhibition or genetic ablation enhanced both in vivo and in vitro macrophage-mediated anti-viral function (46). They demonstrated that two DHCR7 inhibitors (AY9944 and tamoxifen) led to increased clearance of vesicular stomatitis virus (VSV) and Zika virus. AY9944 administered to virus-infected (VSV or murine cytomegalovirus) macrophages led to enhanced Ifnb production in control macrophages but failed to enhance Ifnb production in DHCR7-deficient macrophages. The treatment of macrophages with tamoxifen also resulted in enhanced Ifnb expression upon treatment with a TLR3 agonist or VSV. Tamoxifen has also been shown to enhance neutrophil-mediated phagocytosis and extracellular trap formation to clear bacteria and has been proposed as a potential agent for combatting multi-drug resistant gram-negative infections (47, 48).
We discovered a number of genes involved in the cholesterol synthesis pathway to be up-regulated in sepsis patients when compared with healthy controls. While this could be a general response to reduced LDL-C and HDL-C levels in sepsis, the expression of some of these genes discriminated sepsis patients with CCI/early death outcomes from those in the rapid recovery and control groups, suggesting potential bedside prognostic utility. Our mortality analysis also revealed some additional insights. The up-regulation of DHCR7 and PLTP for 90-day mortality emphasizes the important role that DHCR7 (and PLTP) may play in death from sepsis. In addition to regulating HDL size and facilitating cholesterol and lipopolysaccharide clearance, PLTP is critical to the immunomodulatory action of HDL and is a key factor in maintaining plasma sphingosine-1-phosphate levels (S1P) (22). S1P, which is primarily carried on HDL in association with apolipoprotein M, has antiapoptotic and chemotactic effects and levels decline in sepsis. Declining S1P levels have a strong inverse relationship with organ failure (49).
This study had several limitations. First, this was a small prospective study of gene expression from a single center. Findings from this analysis should be confirmed in a larger and multi-center study. However, to increase the generalizability of our results, we used a diverse cohort of patients (gender and race) and derived and validated our results in two separate cohorts. Our initial RNA-seq analysis involved single-end sequencing, whereas the validation involved paired-end sequencing. This difference was due to technical advances in the Department of Pathology that sequenced our samples but should not affect interpretation of our results. Though our lipopolysaccharide zebrafish model of endotoxemia is a sterile model, we were able to recapitulate several aspects of human sepsis, namely mortality and similar differential expression patterns for the lipid metabolism genes of interest. Finally, being an observational study, there is no way to infer causality between observed gene expression differences and outcomes.
CONCLUSIONS
In conclusion, this study identified DHCR7 up-regulation as potentially influencing poor outcomes after sepsis (CCI/early death) in humans. Our robust findings in human sepsis, confirmed in a validation cohort as well as with RT-qPCR analysis, were then recapitulated in a zebrafish endotoxemia model with similar differential expression of DHCR7 in lipopolysaccharide-treated zebrafish. Blockade of Dhcr7 led to complete rescue of lipopolysaccharide-treated zebrafish from death and may lead to therapeutic opportunities and drug repurposing for sepsis. These findings should be validated in larger, multi-center studies.
Supplementary Material
Footnotes
The authors have filed a provisional patent for the idea of using DHCR7 blockade for the treatment of sepsis. This work was supported by the National Institutes of Health grant R01GM133815 (to F.W.G.), National Institutes of Health grant R35HL150784 (to J.A.S.), National Institutes of Health grant P01AI042288 (to T.M.B.), and National Institutes of Health grant K12HL133304 (to V.J.). J.A.S. is the Henry and Mala Dorfman Family Professor of Pediatric Hematology/Oncology. The remaining authors have not disclosed any potential conflicts of interest.
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
REFERENCES
- 1.Rudd KE, Johnson SC, Agesa KM, et al. : Global, regional, and national sepsis incidence and mortality, 1990–2017: Analysis for the Global Burden of Disease Study. Lancet 2020; 395:200–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaieski DF, Edwards JM, Kallan MJ, et al. : Benchmarking the incidence and mortality of severe sepsis in the united states. Crit Care Med 2013; 41:1167–1174 [DOI] [PubMed] [Google Scholar]
- 3.Singer M, Deutschman CS, Seymour CW, et al. : The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016; 315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar G, Kumar N, Taneja A, et al. ; Milwaukee Initiative in Critical Care Outcomes Research (MICCOR) Group of Investigators: Nationwide trends of severe sepsis in the 21st century (2000–2007). Chest 2011; 140:1223–1231 [DOI] [PubMed] [Google Scholar]
- 5.Guirgis FW, Brakenridge S, Sutchu S, et al. : The long-term burden of severe sepsis and septic shock: Sepsis recidivism and organ dysfunction. J Trauma Acute Care Surg 2016; 81:525–532 [DOI] [PubMed] [Google Scholar]
- 6.Brakenridge SC, Efron PA, Cox MC, et al. : Current epidemiology of surgical sepsis: Discordance between inpatient mortality and 1-year outcomes. Ann Surg 2019; 270:502–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gardner AK, Ghita GL, Wang Z, et al. : The development of chronic critical illness determines physical function, quality of life, and long-term survival among early survivors of sepsis in surgical ICUs. Crit Care Med 2019; 47:566–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mira JC, Gentile LF, Mathias BJ, et al. : Sepsis pathophysiology, chronic critical illness, and persistent inflammation-immunosuppression and catabolism syndrome. Crit Care Med 2017; 45:253–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guirgis FW, Dodani S, Leeuwenburgh C, et al. : HDL inflammatory index correlates with and predicts severity of organ failure in patients with sepsis and septic shock. PLoS One 2018; 13:e0203813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morin EE, Guo L, Schwendeman A, et al. : HDL in sepsis - risk factor and therapeutic approach. Front Pharmacol 2015; 6:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catapano AL, Pirillo A, Bonacina F, et al. : HDL in innate and adaptive immunity. Cardiovasc Res 2014; 103:372–383 [DOI] [PubMed] [Google Scholar]
- 12.Khovidhunkit W, Kim M-S, Memon RA, et al. : Effects of infection and inflammation on lipid and lipoprotein metabolism: mechanisms and consequences to the host. J Lipid Res 2004; 45:1169–1196 [DOI] [PubMed] [Google Scholar]
- 13.İnal V, Yamanel L, Taşkin G, et al. : Paraoxonase 1 activity and survival in sepsis patients. Balkan Med J 2015; 32:183–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liao XL, Lou B, Ma J, et al. : Neutrophils activation can be diminished by apolipoprotein A-I. Life Sci 2005; 77:325–335 [DOI] [PubMed] [Google Scholar]
- 15.Murphy AJ, Woollard KJ, Hoang A, et al. : High-density lipoprotein reduces the human monocyte inflammatory response. Arterioscler Thromb Vasc Biol 2008; 28:2071–2077 [DOI] [PubMed] [Google Scholar]
- 16.Murphy AJ, Woollard KJ, Suhartoyo A, et al. : Neutrophil activation is attenuated by high-density lipoprotein and apolipoprotein A-I in in vitro and in vivo models of inflammation. Arterioscler Thromb Vasc Biol 2011; 31:1333–1341 [DOI] [PubMed] [Google Scholar]
- 17.De Nardo D, Labzin LI, Kono H, et al. : High-density lipoprotein mediates anti-inflammatory reprogramming of macrophages via the transcriptional regulator ATF3. Nat Immunol 2014; 15:152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Topchiy E, Cirstea M, Kong HJ, et al. : Lipopolysaccharide is cleared from the circulation by hepatocytes via the low density lipoprotein receptor. PLoS One 2016; 11:e0155030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walley KR: Role of lipoproteins and proprotein convertase subtilisin/kexin type 9 in endotoxin clearance in sepsis. Curr Opin Crit Care 2016; 22:464–469 [DOI] [PubMed] [Google Scholar]
- 20.Walley KR, Boyd JH, Kong HJ, et al. : Low low-density lipoprotein levels are associated with, but do not causally contribute to, increased mortality in sepsis. Crit Care Med 2019; 47:463–466 [DOI] [PubMed] [Google Scholar]
- 21.Walley KR, Francis GA, Opal SM, et al. : The central role of proprotein convertase subtilisin/kexin type 9 in septic pathogen lipid transport and clearance. Am J Respir Crit Care Med 2015; 192:1275–1286 [DOI] [PubMed] [Google Scholar]
- 22.Barker G, Winer JR, Guirgis FW, et al. : HDL and persistent inflammation immunosuppression and catabolism syndrome. Curr Opin Lipidol 2021; 32:315–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barker G, Leeuwenburgh C, Brusko T, et al. : Lipid and lipoprotein dysregulation in sepsis: Clinical and mechanistic insights into chronic critical illness. J Clin Med 2021; 10:1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trinder M, Walley KR, Boyd JH, et al. : Causal inference for genetically determined levels of high-density lipoprotein cholesterol and risk of infectious disease. Arterioscler Thromb Vasc Biol 2020; 40:267–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trinder M, Genga KR, Kong HJ, et al. : Cholesteryl ester transfer protein influences high-density lipoprotein levels and survival in sepsis. Am J Respir Crit Care Med 2019; 199:854–862 [DOI] [PubMed] [Google Scholar]
- 26.Xie Y, Meijer AH, Schaaf MJM: Modeling inflammation in zebrafish for the development of anti-inflammatory drugs. Front Cell Dev Biol 2021; 8:620984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang L, Liu C, Miller YI: Zebrafish models of dyslipidemia: Relevance to atherosclerosis and angiogenesis. Transl Res 2014; 163:99–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ka J, Jin SW: Zebrafish as an emerging model for dyslipidemia and associated diseases. J Lipid Atheroscler 2021; 10:42–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Elm E, Altman DG, Egger M, et al. ; STROBE Initiative: The strengthening the reporting of observational studies in epidemiology (STROBE)statement: Guidelines for reporting observational studies. J Clin Epidemiol 2008; 61:344–349 [DOI] [PubMed] [Google Scholar]
- 30.Percie du Sert N, Ahluwalia A, Alam S, et al. : Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol 2020; 18:e3000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guirgis FW, Black LP, Henson M, et al. : A hypolipoprotein sepsis phenotype indicates reduced lipoprotein antioxidant capacity, increased endothelial dysfunction and organ failure, and worse clinical outcomes. Crit Care 2021; 25:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guirgis FW, Black LP, Rosenthal MD, et al. : LIPid Intensive Drug Therapy for Sepsis Pilot (LIPIDS-P): Phase I/II clinical trial protocol of lipid emulsion therapy for stabilizing cholesterol levels in sepsis and septic shock. BMJ Open 2019; 9:e029348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris PA, Taylor R, Thielke R, et al. : Research Electronic Data Capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 2009; 42:377–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harris PA, Taylor R, Minor BL, et al. ; REDCap Consortium: The REDCap consortium: Building an international community of software platform partners. J Biomed Inform 2019; 95:103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loftus TJ, Mira JC, Ozrazgat-Baslanti T, et al. : Sepsis and critical illness research center investigators: Protocols and standard operating procedures for a prospective cohort study of sepsis in critically ill surgical patients. BMJ Open 2017; 7:e015136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Friedewald WT, Levy RI, Fredrickson DS: Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972; 18:499–502. Available at: http://www.ncbi.nlm.nih.gov/pubmed/4337382. Accessed September 18, 2014 [PubMed] [Google Scholar]
- 37.Berman HB:Bartlett’s test for homogeneity of variance. Available at: https://stattrek.com/anova/homogeneity/bartletts-test. Accessed July 25, 2022.
- 38.Dobin A, Davis CA, Schlesinger F, et al. : STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013; 29:15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liao Y, Smyth GK, Shi W: featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014; 30:923–930 [DOI] [PubMed] [Google Scholar]
- 40.Love MI, Huber W, Anders S: Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kaluzhskiy L, Ershov P, Yablokov E, et al. : Human lanosterol 14-alpha demethylase (CYP51A1) is a putative target for natural flavonoid luteolin 7,3’-disulfate. Molecules 2021; 26:2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun QY, Zhou HH, Mao XY: Emerging roles of 5-lipoxygenase phosphorylation in inflammation and cell death. Oxid Med Cell Longev 2019; 2019:2749173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luu W, Hart-Smith G, Sharpe LJ, et al. : The terminal enzymes of cholesterol synthesis, DHCR24 and DHCR7, interact physically and functionally. J Lipid Res 2015; 56:888–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Donoghue SE, Pitt JJ, Boneh A, et al. : Smith-Lemli-Opitz syndrome: Clinical and biochemical correlates. J Pediatr Endocrinol Metab 2018; 31:451–459 [DOI] [PubMed] [Google Scholar]
- 45.Çolak Y, Nordestgaard BG, Afzal S: Low vitamin D and risk of bacterial pneumonias: Mendelian randomisation studies in two population-based cohorts. Thorax 2021; 76:468–478 [DOI] [PubMed] [Google Scholar]
- 46.Xiao J, Li W, Zheng X, et al. : Targeting 7-dehydrocholesterol reductase integrates cholesterol metabolism and IRF3 activation to eliminate infection. Immunity 2020; 52:109–122.e6 [DOI] [PubMed] [Google Scholar]
- 47.Corriden R, Hollands A, Olson J, et al. : Tamoxifen augments the innate immune function of neutrophils through modulation of intracellular ceramide. Nat Commun 8369; 6:8369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watashi K, Inoue D, Hijikata M, et al. : Anti-hepatitis C virus activity of tamoxifen reveals the functional association of estrogen receptor with viral RNA polymerase NS5B. J Biol Chem 3276; 282:5–32772 [DOI] [PubMed] [Google Scholar]
- 49.Winkler MS, Ma¨rtz KB, Nierhaus A, et al. : Loss of sphingosine 1-phosphate (S1P) in septic shock is predominantly caused by decreased levels of high density lipoproteins (HDL). J Intensive Care 2019; 7:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.