Abstract
Psoriasis is a common chronic inflammatory skin disease with no cure. It is driven by the interleukin (IL)-23/IL-17A axis and type 17 T helper cells; however, recently, group 3 innate lymphoid cells (ILC3s) have also been implicated. Despite being the focus of much research, factors regulating the activity of ILC3s remain incompletely understood. Immune regulatory pathways are particularly important at barrier sites, such as the skin, gut, and lungs, which are exposed to environmental substances and microbes. CD200R1 is an immune regulatory cell surface receptor that inhibits proinflammatory cytokine production in myeloid cells. CD200R1 is also highly expressed on ILCs, where its function remains largely unexplored. We previously observed reduced CD200R1 signaling in psoriasis-affected skin, suggesting that dysregulation may promote disease. Here, we show that contrary to this, psoriasis models are less severe in CD200R1-deficient mice due to reduced IL-17 production. Here, we uncover a key cell-intrinsic role for CD200R1 in promoting IL-23-driven IL-17A production by ILC3s by promoting signal transducer and activator of transcription 3 activation. Therefore, contrary to its inhibitory role in myeloid cells, CD200R1 is required on ILC3 to promote IL-23-stimulated signal transducer and activator of transcription 3 activation, triggering optimal IL-17 production.
Introduction
Barrier sites such as the skin, are enriched for immune cells that can rapidly respond to stimulation by producing large quantities of proinflammatory cytokines. Innate lymphoid cells (ILCs) are one such cell type, with group 3 ILCs (ILC3s) responding to interleukin (IL)-23 and IL-1β by producing IL-17A and IL-22.1 These cytokines cause changes in epidermal cells, including hyperproliferation, dysregulated maturation, and epidermal production of cytokines, chemokines, and antimicrobial peptides, which are critical for protection against extracellular bacterial and fungal infection.2, 3 However, the IL-23/IL-17 axis can also drive psoriasis, a common, incurable chronic inflammatory skin disease, as demonstrated by the efficacy of biologics targeting this pathway.4 ILC3s are also implicated in psoriasis because they are a potent source of IL-17 and are increased in lesions.5, 6 Therefore, it is crucial to understand how ILC3 activity is regulated to prevent inappropriate activation.
Immunosuppressive pathways are key to maintaining immune homeostasis and are particularly important in barrier tissues, where there is constant exposure to environmental stimuli and commensal microbes. A key immunosuppressive pathway in myeloid cells is CD200R1 signaling, which has been shown to suppress proinflammatory cytokine production, reducing immune responses against self-antigens,7, 8, 9, 10 allergens,11 infectious agents,12 and cancer.13 CD200R1 is a cell surface receptor, which on engagement with its ligand, CD200, transmits a signaling cascade inhibiting mitogen-activated protein kinases and suppressing myeloid cell activation. CD200R1 is expressed on many immune cell types, and in addition to myeloid cells, is particularly highly expressed on ILC14, 15, 16, 17; however, its function on these cells remains underexplored.
We have previously shown that the ligand CD200 is reduced in nonlesional psoriatic skin17 and we therefore hypothesized that in healthy skin, CD200R1 signaling prevents inappropriate inflammation; however, in psoriasis, this is reduced, contributing to psoriasis susceptibility. To examine this hypothesis, here, we compare the severity of inflammation in psoriasis models in wild-type (WT) and CD200R1-deficient mice. This shows that, surprisingly, global CD200R1 deficiency reduces the severity of psoriasis-like skin inflammation due to a reduced IL-17A production. CD200R1 is highly expressed by ILCs and here, we show that CD200R1-deficient ILC3s are less able to produce IL-17A. This is associated with a vastly different transcriptomic profile, despite the surface phenotype being largely unchanged. We also demonstrate that the requirement for CD200R1 in ILC3 is cell-intrinsic. To understand why CD200R1KO ILC3s are unable to produce IL-17A optimally, signaling downstream of IL-23 stimulation was investigated, demonstrating that CD200R1 is required for optimal signal transducer and activator of transcription 3 (STAT3) activation in response to IL-23. Therefore, we describe a critical role for CD200R1 in promoting IL-17 production by ILC3 by enhancing IL-23-driven STAT3 activation.
Results
Global CD200R1 deficiency impairs interleukin-17 production in psoriasis models
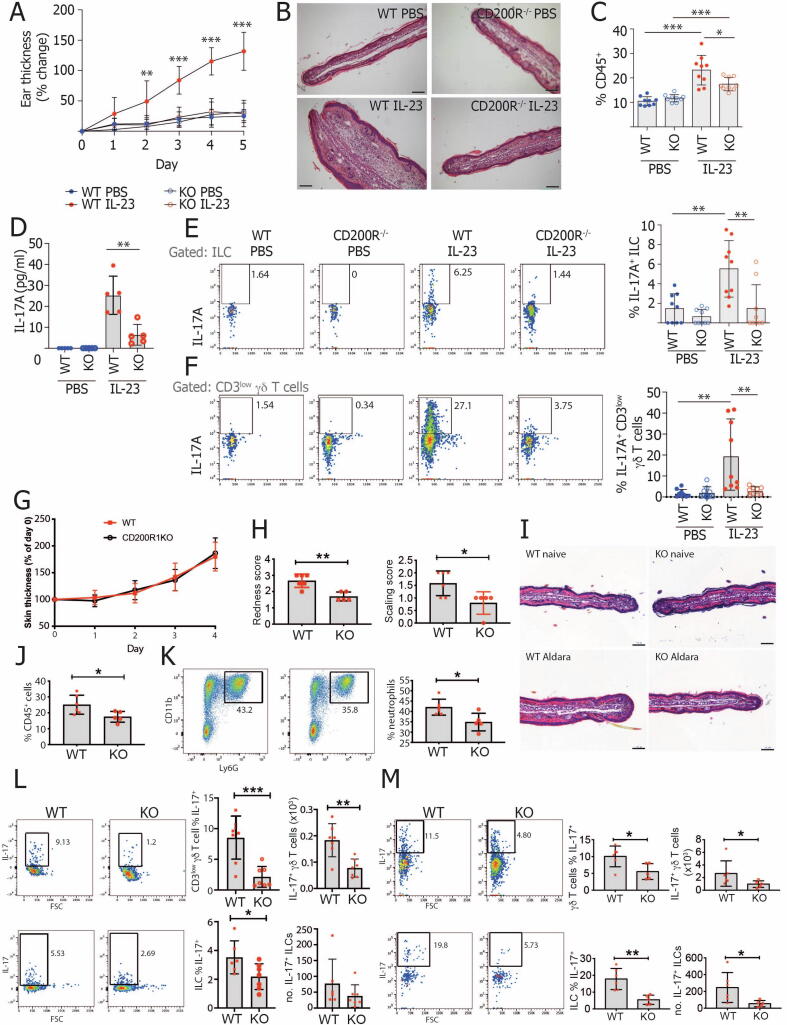
As we17 and others18 have demonstrated reduced CD200 levels in samples from patients with psoriasis and CD200R1 signaling has been shown to dampen immune responses,7, 8, 9, 10, 11, 12, 13 we hypothesized that the absence of CD200R1 would enhance inflammation in psoriasis models. Contrary to this, global CD200R1 deficiency protected mice from skin thickening in a model induced by repeated intradermal injection with IL-2319 (Figs. 1A and 1B). Total immune cells (CD45+) were increased in IL-23 treated WT mouse skin, but this increase was inhibited in CD200R1KO mice (Fig. 1C). To determine how CD200R1 promotes inflammation, skin IL-17A levels were measured because this is a critical cytokine driving inflammation. Enzyme-linked immunosorbent assay (ELISA) and flow cytometry demonstrated reduced IL-17A overall in the skin and reduced production by both ILCs and CD3low (dermal) γδ T cells in CD200R1KO mice (Figs. 1D–F and gating shown in Fig. 2C).
Fig. 1.
CD200R1 promotes IL-17A production in murine models of psoriasis. A-F, WT and CD200R1KO (KO) Ears were intradermally injected with 1µg IL-23, or phosphate-buffered saline daily for 5 consecutive days. A, Skin thickening measured using a digital micrometer. B, Hematoxylin & eosin stained skin sections. C, Proportion of live, single cells in skin that are CD45+. D, IL-17A level in media from overnight cultured skin cells. E, Proportion of skin ILCs (CD45+ Lin- CD3- TCRβ- γδTCR- Thy1+ CD127+) producing IL-17A. F, Proportion of skin CD3low γδ T cells producing IL-17A. G-M, Inflammation was induced by topically treating WT and CD200R1KO (KO) littermate ear skin with Aldara cream daily. G, Skin thickening measured using a digital micrometer. H, Skin redness and scaling scores. I, Hematoxylin and eosin-stained skin sections. J, Proportion of live, single cells in skin that are CD45+. K, Proportion of skin CD45+ cells that are neutrophils (CD45+ CD11b+ Ly6G+). L, Proportion and number of skin CD3low γδ T cells or ILCs producing IL-17A. M, Proportion and number of lymph node γδ T cells or ILCs producing IL-17A. Histology scale bars show 100 µm. Data shown are pooled from two independent experiments except for (B) and (I) showing representative histology, (D) showing representative enzyme-linked immunosorbent assay, (H, J, K and M) showing representative data from one experiment. A, C-F were analyzed by two-way analysis of variance followed by a Bonferroni post hoc test. H, J-M were analyzed by Student’s t test. A, C, E, F n = 9, D n = 5. G, H, J-M n = 5-7. IL = interleukin; ILC = innate lymphoid cell; KO = knockout; PBS = phosphate-buffered saline; WT = wild-type.
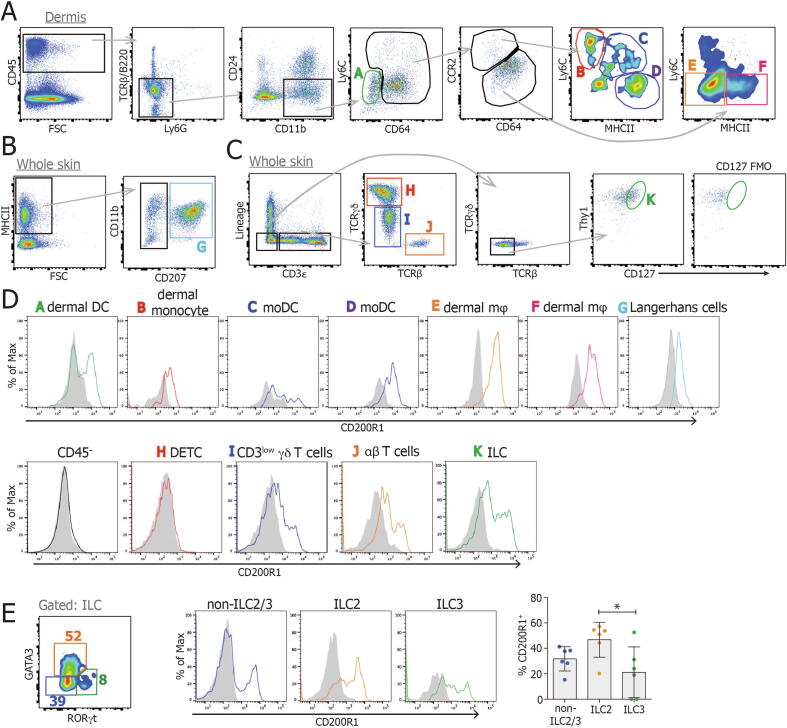
Fig. 2.
CD200R1 is expressed by skin ILCs. Dorsal skin cells were isolated and analyzed by flow cytometry using fluorescence minus one controls to set the gates. A, Dermal live CD45+ single cells were gated into various monocyte, dendritic cell and macrophage populations. B, Whole skin live CD45+ single cells were gated for Langerhans cells. C, Whole skin live CD45+ single cells were gated for T-cell subsets and ILCs. D, CD200R1 expression on each cell subset. E, Skin ILCs (CD45+ Lin- CD3- TCRβ- γδ TCR- Thy1+ CD127+) were gated for ILC2 (GATA3+), ILC3 (RORγt+) and non-ILC2/3 (GATA3- RORγt-), and CD200R1 expression was analyzed within each subset. Filled histograms show CD200R1KO data, lines show wild-type data. All data are representative of two replicate experiments except (E) showing data from two experiments which was analyzed by one-way analysis of variance. FSC = fetal calf serum; ILC = innate lymphoid cell; KO = knockout; moDC = monocyte-derived dendritic cells; TCR = T-cell receptor.
To investigate this further, a more complex psoriasis model was used, in which Aldara cream, (containing the toll-like receptor 7-agonist, imiquimod, and inflammasome activating isosteric acid) induces inflammation.20 After 4 days of treatment, skin thickening was similar in WT and CD200R1KO mice, but redness and scaling were slightly reduced in the absence of CD200R1 (Figs. 1G–I). The absence of CD200R1 reduced the accumulation of immune cells, in particular neutrophils, within the skin (Figs. 1J and 1K) and reduced the production of IL-17A by ILCs and γδ T cells within the skin and lymph node (LN) (Figs. 1L and 1M).
IL-17 is critical in combating fungal infections; therefore, to examine if CD200R1KO mice also have altered fungal immune responses in the skin, a Candida albicans infection model was performed, which showed CD200R1KO mice produce less IL-17 in response to fungal skin infection (Supplementary Fig. S1).
CD200R1 is highly expressed by ILCs
Given the reduction in IL-17A production in psoriasis-like skin inflammation models, the expression of CD200R1 on skin cells was examined. As anticipated from the literature, CD200R1 was expressed on skin macrophage and dendritic cell subsets (gated as described previously21) but was absent from nonimmune cells and dendritic epidermal T cells (Figs. 2A–D). CD200R1 was also expressed on a proportion of dermal γδ T cells and on a larger proportion of skin ILCs (Fig. 2D), as previously described.17 On splitting the ILCs into subsets based on their expression of lineage defining transcription factors, CD200R1 was shown to be expressed most highly on ILC2 but was also expressed on ILC3s and GATA3- RORγt- ILCs (which contain ILC1, as well as ILCs, which have lost expression of lineage defining transcription factors22) in the skin (Fig. 2E). Although dermal γδ T cells are thought to be the dominant drivers of inflammation in mouse models through their production of IL-17A, ILCs also produce IL-17A in these models and have been shown to be important contributors to inflammation.23 Given this, the relatively low level of expression of CD200R1 on dermal γδ T cells, the higher expression on ILCs, and the evidence that ILC3 contribute to human psoriasis,5, 6 the effect of CD200R1 on ILCs was examined.
CD200R1 promotes interleukin-17A production by ILCs
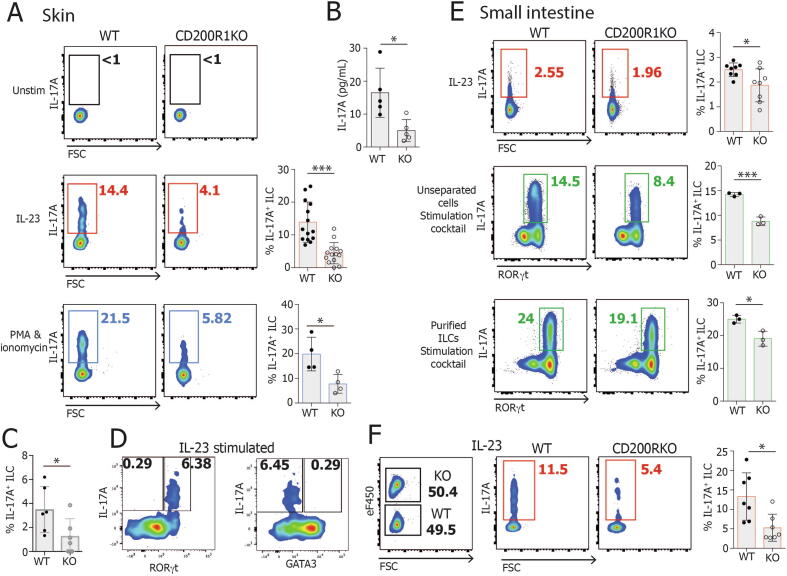
To determine if CD200R1 regulates IL-17 production by ILCs in a more direct manner, outside of the context of skin inflammation, dorsal skin cells were isolated and stimulated with IL-23 or phorbol 12-myristate 13-acetate (PMA) and ionomycin, demonstrating that CD200R1-deficient ILCs are less able to produce IL-17A than WT, regardless of the stimulatory agents used (Figs. 3A–C). IL-22 production was also examined and may be reduced in CD200R1KO ILCs, but this was not found to be statistically significantly different, potentially due to the relatively low levels of production (Supplementary Fig. S2A–B). The levels of IL-22 were reduced in the culture supernatant from CD200R1KO skin cells relative to WT (Supplementary Fig. S2B), but it is unclear if this is due to a reduction in IL-22 production by ILCs or another cell type present. To determine the ILC subtype producing IL-17A, IL-17A production was examined in RORγt+ and RORγt- cells; this showed that it is the RORγt+ ILCs, which are responsible for producing IL-17A in response to IL-23 stimulation (Fig. 3D). Similarly, to determine if the IL-17A producing cells could be inflammatory ILC2, which co-express GATA3 and RORγt, IL-17A production was examined on GATA3+ and GATA3- ILC populations. This showed that the IL-17A-producing cells are predominantly GATA3- RORγt+ (Fig. 3D) and are therefore ILC3 rather than inflammatory ILC2.
Fig. 3.
CD200R1 is required for efficient IL-17A production by ILCs. Dorsal skin or intestinal lamina propria cells from WT or CD200R1KO (KO) mice were stimulated in culture and IL-17A production by ILC was measured by flow cytometry. A, IL-17 production by dorsal skin ILCs (CD45+ Lin- CD3- TCRβ- γδ TCR- Thy1+ CD127+) stimulated with IL-23, or with PMA and ionomycin for 4 hours. B, IL-17A levels in cell supernatant from IL-23-stimulated skin cells. C, IL-17 production by dorsal skin ILCs from littermates in response to IL-23. D, IL-17A production in RORγt+ and RORγt- ILCs, and in GATA3+ and GATA3- ILCs in response to IL-23 stimulation. E, IL-17 production by intestinal lamina propria cells, or flow cytometric sorted intestinal lamina propria ILCs (CD45+ Lin- CD3- TCRβ- γδ TCR- Thy1+ CD127+) stimulated with IL-23 alone or PMA, Ionomycin, IL-23, IL-6, IL-2 and IL-1β for 5 hours. F, CD200R1KO dorsal skin cells were labeled with a fluorescent dye before being co-cultured in a 1:1 ratio with unlabeled WT cells. IL-17A production was analyzed in co-cultured cells stimulated with IL-23 overnight. A (PMA/Ionomycin stim) (n = 4), B (n = 5), E (stimulation cocktail) (n = 3) data shown are from one representative experiment. C (n = 6), E (IL-23 stim) (n = 8-9), F (n = 7) data from two independent experiments. A (IL-23 stim) (n = 14) data shown are from three independent experiments. Data were analyzed by Student’s t test. FSC = fetal calf serum; IL = interleukin; ILC = innate lymphoid cell; KO = knockout; TCR = T-cell receptor; WT = wild-type.
Similar to the skin, gut ILCs from CD200R1-deficient mice were less able to produce IL-17A after stimulation with IL-23, or with PMA, ionomycin, and a cocktail of cytokines, regardless of whether they are cultured unseparated or cultured after flow cytometric sorting to obtain a purified ILC population (Fig. 3-E). This reduction in IL-17A production by CD200R1deficient ILCs was found to be independent of secreted inhibitory factors in the CD200R1-deficient cell cultures because the co-culture of WT and CD200R1-deficient cells maintained the impaired IL-17A production by CD200R1KO ILCs (Fig. 3F). There was also no increase in known ILC3 inhibitory cytokines, IL-10, interferon (IFN)-γ, and IL-25 in the CD200R1-deficient cell cultures relative to WT cultures (Supplementary Fig. S2C), suggesting that CD200R1 directly affects IL-17A production rather than modulating the activity of an intermediary cell type in this in vitro model.
CD200R1 does not affect ILC3 subsets
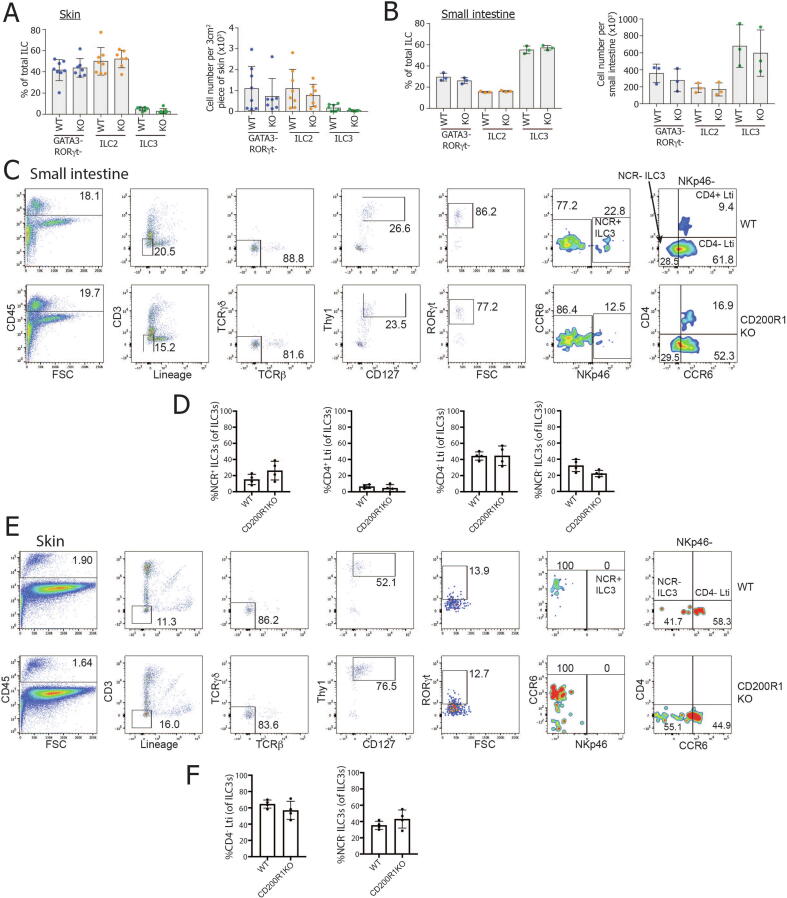
We hypothesized that the reduced IL-17 production seen in CD200R1KO ILCs may be due to either a reduction in ILC3s numbers or the lack of an ILC3 subset. To test this hypothesis, firstly, the proportion and numbers of ILC2, ILC3, and ILC lacking GATA3 and RORγt22 were examined in the skin and small intestinal lamina propria. This showed that there was no reduction in ILC3 in CD200R1KO, which could account for the reduced IL-17 production (Figs. 4A–B). ILC3 are heterogeneous, particularly in the small intestine, where four main subsets of ILC3s are seen.24 Natural cytotoxicity receptor (NCR+) (NKp46+) ILC3s are specialized in interacting with myeloid and stromal cells, CD4+ and CD4- lymphoid tissue inducer cell-like (CCR6+) ILC3s have roles in modulating innate and adaptive immune responses, and NCR- ILC3s are precursors of the NCR+ ILC3 subset. All of these subsets were observed in CD200R1KO small intestine at similar frequencies to WT mice (Figs. 4C–D), showing that the reduced IL-17 production in CD200R1KO ILCs is not due to a missing major ILC3 subset. Less is known about the subsets of ILC3 present in the skin, but applying similar gating as that used for the small intestine allowed the presence of CD4- lymphoid tissue inducer cell-like ILC3s and NCR- ILC3s to be observed. Similar to the small intestine, CD200R1 deficiency did not affect these skin ILC3 populations (Figs. 4E–F).
Fig. 4.
CD200R1KO mice do not lack any major subsets of ILC3s. A, The proportion and number of WT and CD200R1KO (KO) skin ILCs (CD45+ Lin- CD3- TCRβ- γδTCR- Thy1+ CD127+) within each subset (ILC2, GATA3+; ILC3 RORγt+; non-ILC2/3, GATA3- RORγt-). B, The proportion and number of small intestinal lamina propria ILCs within each subset. C, The proportion of ILC3s in each subset in WT and CD200R1KO small intestinal lamina propria. E and F, The proportion of ILC3s in each subset in WT and CD200R1KO skin. A data shown from two independent experiments (n = 7-9). B-F data are representative of two independent experiments, B n = 3, D and F n = 4. FSC = fetal calf serum; IL = interleukin; ILC = innate lymphoid cell; KO = knockout; NCR = natural cytotoxicity receptor; TCR = T cell receptor; WT = wild-type.
CD200R1-deficient ILC3s are transcriptomically very different from WT
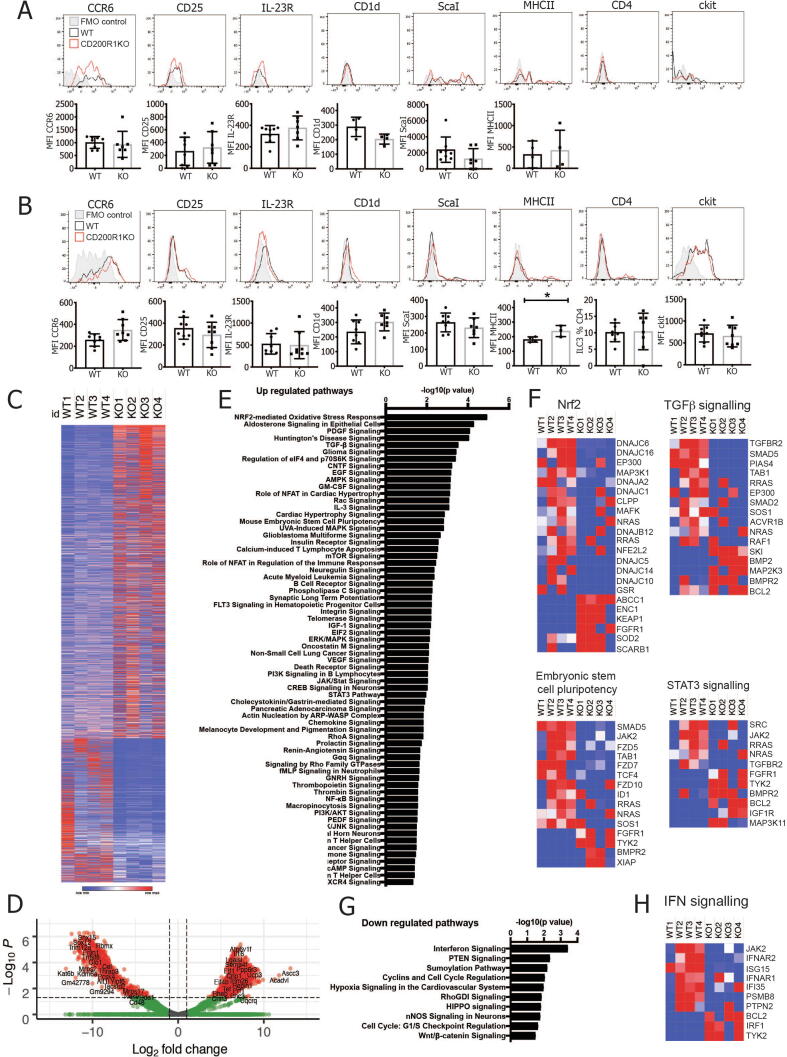
To determine why ILC3s from CD200R1-deficient mice are less able to produce IL-17A, the cell surface phenotype was analyzed by flow cytometry. Skin ILC3s showed undetectable CD4, IL-23R, and ckit and had only low levels of CD25 and CD1d expression. They expressed CCR6, Sca-1, and major histocompatibility complex class II, but CD200R1 deficiency does not appear to affect the expression of any of these markers (Fig. 5A). In the gut, the expression of all of the markers was observed; however, again, CD200R1 deficiency did not affect the levels of these markers (Fig. 5B), demonstrating that ILC3 from CD200R1-deficient mice have a similar cell surface phenotype to WT. IL-23R was expressed at an extremely low level due to a presumed combination of low expression level and the lack of appropriate reagents,25 making interpretation of the effect of CD200R1 on IL-23R expression problematic. Gut ILC3 are highly heterogeneous; so, there may be differences in the expression of some of these markers on a specific ILC3 subset. However, because the main ILC3 subsets are in equal proportions in CD200R1KO and WT mice (Figs. 4B–D), this seems unlikely.
Fig. 5.
CD200R1KO ILC3s are deficient at activating STAT3. WT and CD200R1KO (KO) ILC3s (CD45+ Lin- CD3- TCRβ- γδTCR- Thy1+ CD127+ RORγt+) cell surface marker expression in A, dorsal skin; B, small intestinal lamina propria. C, RNAseq analysis of flow cytometrically sorted ILC3s from WT (RORγt eGFP) and CD200R1KO (CD200R1KO RORγt eGFP) lamina propria. Log2fold change values are shown for all differentially regulated genes. D, Volcano plot of the RNAseq data. E, Pathways predicted to be upregulated in KO ILC3 by pathway analysis of the RNAseq data. F, Heat maps showing z scores for genes in pathways predicted to be enriched in KO ILC3. G, Pathways predicted to be reduced in KO ILC3 by pathway analysis of the RNAseq data. H, Heat maps showing z scores for genes in pathways predicted to be downregulated in KO ILC3. A, B. data are from two independent experiments n = 4-8. Data were analyzed by Student’s t test. GFP = green fluorescent protein; IFN = interferon; IL = interleukin; ILC = innate lymphoid cell; KO = knockout; RNAseq = RNA sequencing; STAT3 = signal transducer and activator of transcription 3; TCR = T-cell receptor; TGF-β = transforming growth factor-β; WT = wild-type.
To determine how CD200R1 affects ILC3 function, ILC3s from WT (RORc eGFP) and CD200R1-deficient (CD200R1KO RORc eGFP) mice were sorted from small intestine (total ILC3s isolated, gating, and purity shown in Supplementary Fig. S3) and RNA sequencing (RNASeq) was performed. Small intestinal cells were used to provide a large enough cell population to be isolated. ILC3s from CD200R1KO mice were found to have 3285 differentially regulated genes, with 2252 of those upregulated, and 1033 downregulated relative to WT (Fig. 5C). Despite there being more upregulated than downregulated genes in the ILC3s from CD200R1-deficient mice, there are less highly upregulated genes (with a log2 fold change of > 10) than there are highly downregulated genes (with a log2 fold change < -10) (Fig. 5D). Pathway analysis revealed many changes in ILC3s from CD200R1-deficient mice, but the most highly enriched pathway was the nuclear factor-erythroid 2-related factor 2 (Nrf2)-mediated oxidative stress response (Figs. 5E–F), which is known to protect against oxidative stress and reduce reactive oxygen species production but also inhibits type 17 T helper cell differentiation, STAT3 activation, and innate immune cell activation.26, 27 Indeed, other pathways enriched in CD200R1KO ILC3s included STAT3 signaling, as well as others of potential interest, including transforming growth factor-β signaling and embryonic stem cell pluripotency (Figs. 5E–F). A small number of pathways were downregulated in ILC3 from CD200R1KO mice, the most significantly altered being interferon signaling (Figs. 5G–H). This may reflect an inability to signal in response to interferons or a reduction in interferons in CD200R1KO mice. Overall, these data suggest that ILC3s from CD200R1-deficient mice are transcriptomically very different from WT and may have changes in cytokine signaling pathways; however, because these data are from global CD200R1-deficient mice, they do not distinguish between a role for CD200R1 on the ILC3s themselves versus CD200R1, affecting ILC3s through another cell type.
CD200R1 plays a cell-intrinsic role in promoting interleukin-17A production by ILC3
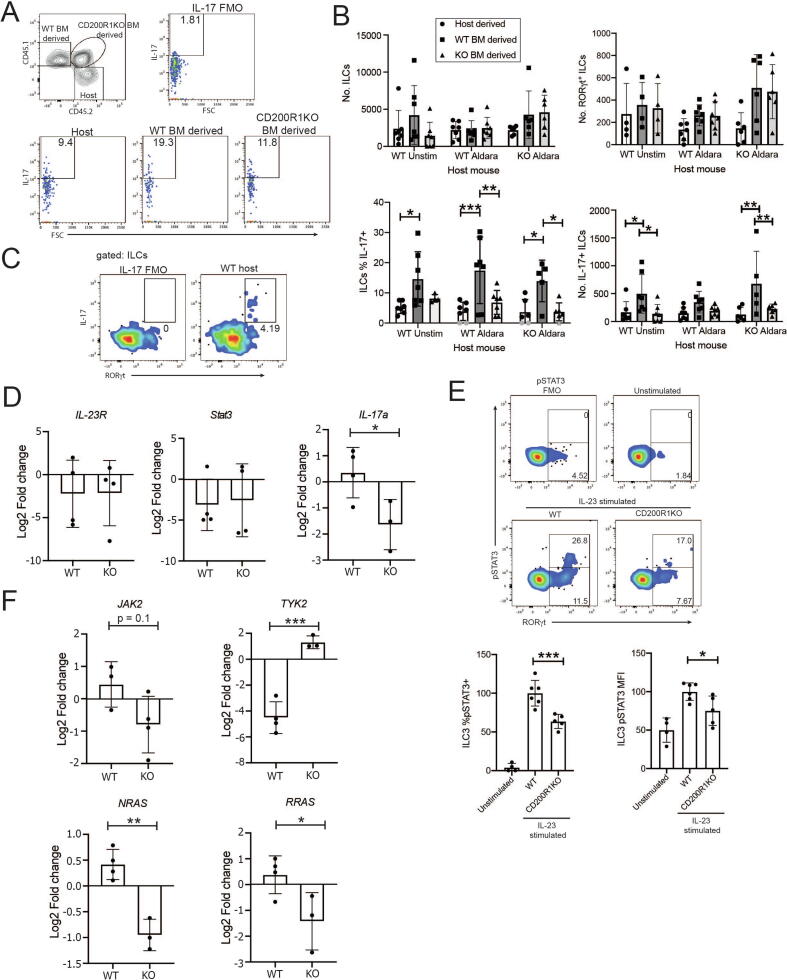
In Fig. 3E, we showed that sorted CD200R1KO gut ILC3 are deficient in IL-17A production, and in Fig. 3F, we showed that when CD200R1KO skin cells were co-cultured with WT skin cells, the CD200R1KO ILCs were deficient in IL-17A production, ruling out the effects of CD200R1 deficiency on other cell types in the cultures. This suggests that the effects of CD200R1 on IL-17A production by ILC may be cell-intrinsic, but these data do not exclude effects of CD200R1 on non-ILC influencing ILC activity long term. These findings only rule out the effect of CD200R1 being mediated by non-ILCs within the time frame of the experiment (24 hours). Therefore, to determine if CD200R1 affects ILC3 activity in a cell-intrinsic or extrinsic manner, chimeric mice were generated with a mixture of WT and CD200R1KO bone marrow, and skin inflammation was induced. This revealed that CD200R1KO ILCs are less able to produce IL-17A than WT ILCs in the skin (Figs. 6A–C). The overall number of ILCs in skin is unaffected by the genotype of the host mice or the genotype of the bone marrow, from which the ILCs are derived. However, the number of IL-17A-producing ILCs derived from CD200R1KO bone marrow is reduced relative to those derived from WT bone marrow in untreated WT and Aldara-treated CD200R1KO hosts (Fig. 6C). Together, this shows that CD200R1 promotes IL-17 production by skin ILC3s in a cell-intrinsic manner.
Fig. 6.
CD200R1 is required in a cell-intrinsic manner to promote IL-17A production by ILC3. A, Representative plots showing IL-17 production by ILC in mixed bone marrow (WT and CD200R1KO) chimeric mice which were stimulated in culture with PMA and ionomycin. B-C, IL-17 production by skin ILC in mixed bone marrow (WT and CD200R1KO) chimeric mice. B, numbers and proportion of ILC and ILC3s producing IL-17A in skin. In the bottom left bar chart, the background FMO value (1.81) was subtracted from each data point. Gray data points show data less than 2x the background (FMO) which were set to zero. C, Flow cytometry showing the ILC subset producing IL-17. D, Il-23r, Stat3 and Il-17a expression in gut WT or CD200R1KO ILC3 measured by RNAseq. E, Representative pSTAT3 staining in inguinal, axillary and brachial lymph node ILCs either unstimulated, IL-23-stimulated WT ILCs or IL-23-stimulated CD200R1KO ILCs relative to the pSTAT3 FMO control. Gated on ILCs. Lower numbers show the % of ILCs which are RORγt+. Upper numbers show the proportion of ILC3s staining for pSTAT3. Bar charts show the normalized % of ILC3s which are pSTAT3+ and the median fluorescence intensity for pSTAT3 in ILC3s. F, Expression of components of the IL-23R signaling pathway in gut WT or CD200R1KO ILC3 measured by RNAseq. B, E data are from two independent experiments. B n = 4-7, E n = 4-6. B, C data were analyzed by two-way analysis of variance followed by a Bonferroni post hoc test. D, F data were analyzed by Student’s t test. E data were analyzed by one-way analysis of variance followed by a Tukey’s post hoc test. BM = bone marrow; FMO = fluorescence minus one; IL = interleukin; ILC = innate lymphoid cell; KO = knockout; RNAseq = RNA sequencing; pSTAT3 = phosphorylation of signal transducer and activator of transcription 3; WT = wild-type.
CD200R1 promotes IL-17 production by ILC3 through enhancing STAT3 activation in response to IL-23
Because CD200R1KO ILC3s are less able to produce IL-17A in response to IL-23 and detecting IL-23R using antibodies is problematic,25 we investigated the Il-23r transcript level in our RNAseq data. This showed that CD200R1KO ILC3 do not have reduced Il-23r transcript levels (Fig. 6D). STAT3 is a crucial mediator of IL-23 signaling, and the STAT3 signaling pathway was found to be enriched in CD200R1KO ILC3 (Figs. 5E–F). This enrichment in signaling components may suggest there is an enhanced response to IL-23, or alternatively, it may reflect the cell compensating for a deficiency in signaling. The RNAseq data were again examined to determine if the Stat3 expression was affected by CD200R1. Stat3 levels were also found to be independent of CD200R1, but in contrast, the CD200R1-deficient ILC3s do have reduced Il-17a transcript levels as expected (Fig. 6D). Because the IL-23R levels appear to be unchanged in the absence of CD200R1 (Fig. 6D), we investigated signaling downstream of IL-23R by examining the phosphorylation of STAT3 (pSTAT3). This demonstrated that the proportion of pSTAT3-positive cells and the level of pSTAT3 is reduced in CD200R1-deficient ILC3s (Fig. 6E), showing that signaling in response to IL-23 is impaired in the absence of CD200R1. The RNAseq data suggest that STAT3 signaling may be affected by CD200R1 (Figs. 5E–F), which coincides with the reduction in pSTAT3 levels in stimulated ILC3s (Fig. 6E). To understand this further, the expression of the components of the IL-23R signaling pathway were determined in the RNAseq data. On engagement of IL-23R, JAK2 and TYK2 become activated and phosphorylate STAT3, allowing its dimerization and translocation to the nucleus, where it can modulate transcription of target genes.28 Jak2 expression was not affected by CD200R1; however, Tyk2, which is also activated rapidly in response to IL-23R engagement, was increased in CD200R1KO ILC3s (Fig. 6F). TYK2 has been shown to negatively regulate STAT3 activation,29 perhaps accounting for the reduced STAT3 phosphorylation observed in these cells. NRAS and RRAS mRNA levels were also reduced in CD200R1KO ILC3s, again showing that IL-23R signaling is reduced in the absence of CD200R1 (Fig. 6F) and providing an explanation for the reduced IL-17A production in the absence of CD200R1.
Discussion
Here, we show that CD200R1 promotes IL-17A production by ILCs in a cell-intrinsic manner and through the promotion of STAT3 phosphorylation. This is surprising because the CD200:CD200R1 pathway has been shown to inhibit proinflammatory cytokine production by myeloid cells30, 31 and suppress a variety of immune responses.7, 8, 9, 10, 11, 12, 13 However, most studies examining the role of CD200:CD200R1 signaling used exogenous ligand or blocked CD200R1 using antibodies. Therefore, the role of this pathway in ubiquitous knockout animals lacking CD200R1 in all cells has not previously been fully addressed.
We recently examined psoriasis-like skin inflammation in mice treated with a CD200R1 blocking antibody and found that neutrophil accumulation was increased, demonstrating that CD200R1 limits neutrophil recruitment.17 Here, we did not observe effects on neutrophil recruitment in the IL-23 injection model (data not shown) and only saw a slight decrease in neutrophils in the Aldara cream-induced psoriasis model. However, neutrophil recruitment effects may be masked in CD200R1-deficient mice due to compensation between the increase in neutrophil recruitment and the reduction in IL-17A production. Indeed, the skin thickening in response to psoriasis-like inflammation was not changed in CD200R1-deficient mice despite the reduction in IL-17A, which suggests that there may be an increase in an alternative inflammatory response in these mice. The fact that we did not observe a decrease in IL-17A production when CD200R1 was blocked in an in vitro ILC3 activation model using adult skin cells17 may suggest that CD200R1 promotes IL-17A production through the effects on the development or differentiation of ILC3s, or CD200R1 may be required over a longer period to affect IL-17A production.
Other work has shown that mice genetically lacking the ligand, CD200, have reduced suppression, including influenza and mouse coronavirus infections,12, 32, 33 in an experimental meningococcal septicemia model34 and in experimental autoimmune encephalitis and collagen-induced arthritis models.35 This suggests that endogenous CD200 does indeed inhibit diverse immune responses, which is opposite to the effects that we describe here for its receptor, CD200R1. However, recent papers suggest that the CD200:CD200R1 signaling pathway may not act as a straightforward ligand-receptor pair. The effect of exogenous CD200 is not mirrored by the effect of agonistic antibodies against CD200R1,36 suggesting that either the agonistic antibodies do not activate CD200R1 in the same way as CD200 or that CD200 has effects independent from CD200R1. Although there exists CD200R-like proteins, which appear to have activating functions and could account for differential effects of CD200, these do not appear to be capable of CD200 binding37, 38 and so probably do not account for these effects. Similarly, viruses are known to express CD200 homologues, which are proposed to allow viral evasion of the immune response39; however, it has been shown that some of these effects are independent of CD200R1.40 This again suggests that CD200 has effects independent of CD200R1. Conversely, in the gut, other ligands for CD200R1 have been identified,41 and it is not yet clear if these ligands play similar or distinct roles to CD200. In support of our finding here that CD200R1 can promote inflammation, a recent report suggests that CD200R1 switches from being suppressive to being stimulatory in the presence of IFN-α.42 Therefore, it is clear that we do not yet fully understand the different roles played by CD200 versus CD200R1 and how these effects are mediated. Gaining a better understanding of this pathway is crucial, particularly as therapeutics are being developed to target this pathway for the treatment of cancers.43
Although we have shown that CD200R1 promotes IL-17A production by ILC3s in a cell-intrinsic manner and that CD200R1 promotes STAT3 activation downstream of IL-23 stimulation, we have not shown that the reduced STAT3 activation in ILC3s in CD200R1KO mice is due to the lack of CD200R1 on the ILC3s themselves. Therefore, we cannot rule out the possibility that the effects of CD200R1 on STAT3 activation are mediated by another cell type and CD200R1 on ILC3s affects IL-17A production through another mechanism. One interesting finding here is that CD200R1 promotes ILC3 IL-17A production in response to PMA/ionomycin stimulation, which bypasses the requirement for STAT3 activation. Although we do not yet understand the mechanism by which CD200R1 acts, given the large transcriptomic differences, it seems likely that CD200R1 deficiency dysregulates the signaling pathway components more widely than just the STAT3 signaling pathway.
In Fig. 3, we show 20% of WT ILCs produce IL-17A in response to PMA/ionomycin stimulation. In the bone marrow chimeras, a similar proportion (15-20%) of the WT bone marrow-derived cells produce IL-17A, whereas lower proportions of the surviving host ILCs and the CD200R1KO bone marrow-derived ILCs produce IL-17A, indicating reduced activity. The CD200R1KO bone marrow-derived ILCs are less able to produce IL-17A due to their deficiency in CD200R1 because this is the only difference between these cells and the WT bone marrow-derived ILCs. The residual host ILC’s reduced ability to produce IL-17A was unexpected but may be due to radiation effects or the age of the mice (at analysis, they were 20-25 weeks old). In contrast to the idea that this is age-related, transcripts associated with IL-17 signaling have been shown to be enriched with age in ILCs.44 High-dose localized irradiation has been shown to induce IL-17 production by γδ T cells and ILCs45 rather than inhibit IL-17A production; however, the differences in the radiation dose and body site targeted may explain this difference. In the case of targeted radiation, ILCs could be recruited from other skin or body sites; in contrast, here, where whole body irradiation was used, ILCs were depleted globally, thus host ILCs cannot be recruited from other tissue sites. Therefore, it seems likely that whole body irradiation is responsible for the reduced ability of host ILC to produce IL-17A either by directly affecting ILC subsets or activity, or indirectly through effects on other cell types.
Here, the IL-17 production capacity of ILC was investigated in LNs, in addition to the skin. Clearly, the skin is the crucial tissue to investigate when studying skin inflammation models, but it has the disadvantage of requiring lengthy enzymatic digestion procedures to allow the isolation of cells, which can impact on the cell activity and phenotype. Conversely, isolating cells from LNs is much more straightforward; however, the contribution of lymph node derived IL-17 to skin inflammation is not well understood. Evidence shows that there is very little trafficking of ILCs from skin to the LN either in other inflammatory skin models or under homeostatic conditions.50 However, the Aldara model of psoriasis does involve systemic inflammation, including the induction of IL-17 in secondary lymphoid organs.20 Also, other inflammatory skin models have increased ILC numbers in skin draining lymph nodes due to increased proliferation and recruitment from the circulation.50 Therefore, an increase in IL-17A production in lymph nodes is likely to reflect ongoing inflammation systemically and probably also cutaneously.
In conclusion, here, we show a crucial novel role for CD200R1 in promoting IL-17A production by ILC3s showing that CD200:CD200R1 effects are more complex than just inhibiting immune responses.
Materials and methods
Mice
All animal experiments were locally ethically approved and were performed in accordance with the UK Home Office Animals (Scientific Procedures) Act 1986. C57BL/6 mice were obtained from Charles River Laboratories, Wilmington, MA, USA. CD200R1KO mice46 (provided by Prof. Tracy Hussell), RORc eGFP47 (provided by Prof. Gerard Eberl), CD45.148 (B.6SJL-Ptprca Pep3b/BoyJ–Ly5.1 mice, provided by Prof. Andrew MacDonald), CD200R1KO CD45.1, and CD200R1KO RORc eGFP mice (generated by crossing CD200R1KO with CD45.1 or RORc eGFP mice, respectively) were bred and maintained in specific pathogen-free conditions in house. Mice were 7-14 weeks old at the start of procedures, unless otherwise stated.
Skin inflammation models
The mice were anesthetized and ear thickness was measured daily using a digital micrometer (Mitutoyo UK, Andover, UK). Redness and scaling were scored daily. For the IL-23 intradermal injection model, the ears were injected intradermally with 1 μg of IL-23 (eBioscience, Waltham, MA, USA) or phosphate-buffered saline (PBS) daily for 5 consecutive days.19 For the Aldara cream model,20 the ears were treated topically with 20 mg of Aldara cream (Meda Pharmaceuticals, Bishops Stortford, UK) (containing imiquimod and isosteric acid) daily for 4 days.
Hematoxylin and eosin staining
Ear tissue was fixed in 10% neutral buffered formalin (Sigma Aldrich, Gillingham, UK), then embedded in paraffin and cut to 5 μm. Hematoxylin and eosin staining was carried out using an automated Shandon Varistain V24-4 (Waltham, MA, USA). Images were acquired using a 3D HISTECH Pannoramic-250 microscope slide-scanner (3D HISTECH, Budapest, Hungary). Snapshots were taken with Case Viewer software (3D HISTECH, Budapest, Hungary).
Cell isolation
Skin and LN
The ears were split in half and floated on 0.8% w/v trypsin (Sigma Aldrich, Gillingham, UK) at 37°C for 30 minutes; then, the epidermis was removed and both epidermis and dermis were chopped and digested in 0.1 mg/ml (0.5 Wunch units/ml) Liberase TM (Roche, Basel, Switzerland) at 37°C for 1 hour. Subcutaneous fat was removed from dorsal skin before floating on trypsin, as above. Tissue was chopped and digested with 1 mg/ml Dispase II (Roche, Basel, Switzerland) at 37°C for 1 hour. Digested skin tissue suspensions or inguinal, brachial, and axillary lymph nodes were passed through 70-µm cell strainers (Starstedt, Numbrecht, Germany) and the cells were counted.
Small intestinal lamina propria cells
The cells were isolated, as detailed previously.49 Briefly, the small intestines were excised and Peyer’s patches were removed. Intestines were sliced open, washed with PBS, cut into segments, and shaken in Roswell Park Memorial Institute 1640 Media (RPMI) with 3% fetal calf serum, 20 mM HEPES, and 1% penicillin streptomycin (Sigma Aldrich, Gillingham, UK). The tissue was transferred to prewarmed RPMI supplemented with 3% fetal calf serum, 20 mM HEPES, 1% penicillin streptomycin, 2 mM ethylenediaminetetraacetic acid (Fischer Scientific), and 14.5 mg/ml dithiothreitol (Sigma Aldrich, Gillingham, UK) and incubated at 37°C for 20 minutes with agitation. The tissue was shaken three times in prewarmed RPMI supplemented with 20 mM HEPES, 1% penicillin streptomycin, and 2 mM ethylenediaminetetraacetic acid and then was washed with PBS. Tissues were minced, then digested with 0.1 mg/ml Liberase Thermolysin Low (TL) (Roche, Basel, Switzerland) and 0.5 mg/ml DNaseI (Sigma Aldrich, Gillingham, UK) in RPMI for 30 minutes at 37°C. Cell suspensions were passed through 70-μm and 40-μm cell strainers (Starstedt, Numbrecht, Germany), then washed in complete RPMI [RPMI supplemented with 10% heat inactivated FBS, 1% penicillin streptomycin solution, 2 mM L-glutamine, 1 mM sodium pyruvate, 20 mM HEPES, 1X nonessential amino acid solution, 25 nM 2-mercaptoethanol (Sigma Aldrich, Gillingham, UK)].
Bone marrow cells
Fibulas and tibias were removed, and bone marrow cells were isolated using a needle and syringe and PBS. Red blood cells were lysed using ammonium-chloride-potassium lysis buffer (Lonza, Basel, Switzerland), and cells were washed and counted.
Flow cytometric analysis
Cells were incubated with 0.5 µg/ml anti-CD16/32 (2.4G2, BD Bioscience, Franklin Lakes, NJ, USA) and near infrared dead cell stain (Invitrogen, Waltham, MA, USA) before staining with fluorescently labeled antibodies. Cells were fixed with Foxp3/transcription factor buffer staining set (eBioscience, Waltham, MA, USA) between 30 minutes and 16 hours at 4°C.
For cytokine analysis, 10 mM of Brefeldin A was added to cell cultures for 4 hours before staining for the cell surface markers, as described previously. After overnight fixation, cells were permeabilized with Foxp3/transcription factor buffer staining set (eBioscience, Waltham, MA, USA) and were stained with intracellular markers or cytokines. Cells were analyzed on a BD Fortessa or LSR II flow cytometer. Data were analyzed using FlowJo (TreeStar, Ashland, OR, USA). Antibodies are detailed in Supplement Table S1.
For pSTAT3 staining, inguinal, axillary, and brachial LN cells were stained for surface markers, then stimulated with 100 ng/ml of IL-23 (Biolegend, San Diego, CA, USA) for 15 minutes. Cells were fixed with Phosflow Fix Buffer I (BD Biosciences, Franklin Lakes, NJ, USA)) at 37°C for 10 minutes before permeabilization at 4°C in Phosflow Perm Buffer III (BD Biosciences, Franklin Lakes, NJ, USA) for 30 minutes, staining with phycoerythrin-conjugated pSTAT3 (pY705) (BD Bioscience clone 4/P-STAT3) at room temperature for 30 minutes. Data were normalized to the mean stimulated WT data.
ILCs were gated as live, single cell, CD45+, Lin- (Ter119, F4/80, CD11b, CD11c, FcεRIα, Gr1, CD19), CD3-, T-cell receptor (TCR)β-, TCRγδ-, CD90.2+, CD127+.
In vitro innate lymphoid cell activation assay
Mouse dorsal skin cells were stimulated in culture with 40 ng/ml IL-23 (Biolegend, San Diego, CA, USA) alone or in combination with 40 ng/ml of IL-1β (eBioscience, Waltham, MA, USA) overnight. Supernatants were retained for cytokine analysis and cells were cultured with 10 mM Brefeldin A (Sigma Aldrich, Gillingham, UK) for 4-5 hours before staining for flow cytometric analysis. Alternatively, cells were stimulated with 50 ng/ml PMA and 500 ng/ml ionomycin (Sigma Aldrich, Gillingham, UK) and 10 mM Brefeldin A for 4 hours before staining for flow cytometric analysis. Similarly, small intestinal lamina propria cells were stimulated with 20 ng/ml IL- 23, IL-6, IL-2 (Biolegend, San Diego, CA, USA), and IL-1β in the presence of 10 μM Brefeldin A for 2 hours before PMA (50 ng/ml) and ionomycin (500 ng/ml) was added for a further 3 hours.
For co-culture of WT and CD200R1KO cells, dorsal skin cells were isolated and either the WT or CD200R1KO cells were labeled with 10 μM eF450-conjugated cell proliferation dye (eBioscience, Waltham, MA, USA) for 10 minutes in the dark at 37°C, then were washed and co-cultured with the unlabeled cells.
Enzyme-linked immunosorbent assay
Levels of IL-17A, IL-10, IFN-γ, and IL-25 were measured using Ready-SET-Go! enzyme-linked immunosorbent assay kits (eBioscience, Waltham, MA, USA), following the manufacturer’s instructions. Data plotted are the mean of at least two technical replicates for each individual sample.
Flow cytometric cell sorting
Cells were isolated from the small intestinal lamina propria and labeled with fluorescent antibodies as detailed previously before sorting on a BD FACSAria (BD Biosciences, Franklin Lakes, NJ, USA) Cell Sorter to a typical purity of > 95%.
RNA sequencing
ILC3s [live, single cell, CD45+, Lin- (Ter119, F4/80, CD11b, CD11c, FcεRIα, Gr1, CD19), CD3-, TCRβ--, TCRγδ-, CD90.2+, CD127+, GFP+] were sorted from RORc eGFP (WT) and CD200R1KO RORc eGFP (KO) small intestinal lamina propria, keeping cells from each mouse separate. RNA was isolated using the RNeasy Micro kit (QIAGEN, Hilden, Germany), following the manufacturer’s instructions. Contaminating genomic DNA was removed using gDNA eliminator columns. The quality and integrity of the RNA samples were assessed using a 2200 TapeStation (Agilent Technologies, Cheadle, UK), and libraries were generated using the TruSeq® stranded mRNA assay (Illumina, Inc., California, USA), according to the manufacturer’s protocol before sequencing with an Illumina HiSeq 4000 instrument. The output data were demultiplexed (allowing one mismatch) and BCL-to-Fastq conversion was performed using Illumina’s bcl2fastq software, version 2.17.1.14. Reads were quality trimmed using Trimmomatic_0.36 (PMID: 24695404), then mapped against the reference mouse genome, version mm10/GRCm38 using STAR_2.4.2 (PMID: 23104886). Counts per gene were calculated with HTSeq_0.6.1 (PMID:25260700) using annotation from GENCODE M12 (http://www.gencodegenes.org/). Normalization and differential expression were calculated with DESeq2_1.12.3 (PMID:25516281) and edgeR_3.12.1 (PMID:19910308). Ingenuity Pathways Analysis (QIAGEN, Hilden, Germany) was used to determine differentially regulated pathways.
Generating bone marrow chimeric mice
Male WT (C57BL/6) and CD200R1KO mice (both CD45.2) were exposed to a split dose of 10.5-11 Gy irradiation. Bone marrow was isolated from male WT (CD45.1 or CD45.1 × C57BL/6 F1) and CD200R1KO (either CD45.1 or CD45.1 × CD200R1KO F1) mice and donor T cells were removed using CD90.2 microbeads and LS MACS columns (Miltenyi Biotech, Bergisch Gladbach, Germany). Cells were counted and mixed at a 1 : 1 ratio before being adaptively transferred intravenously into the irradiated hosts. A total of 10-12 weeks after reconstitution, skin inflammation was induced by topical Aldara cream application daily (as described previously) for 6 days. A day after the final treatment, the mice were euthanized, and the cells analyzed by flow cytometry after stimulation with PMA and ionomycin to ensure cytokine production was detectable.
Statistics
Data were analyzed for normality using the Shapiro-Wilk test. When comparing two independent groups, a Student’s t test (for normally distributed data) was used. For three or more groups, data were analyzed by one-way analysis of variance (as normally distributed), or to determine the effects of two independent variables on multiple groups, a two-way analysis of variance was used, followed by a Bonferroni post hoc test. All statistical tests were performed using Prism Software (v8.4.2, GraphPad Software Inc., La Jolla, CA USA). A p value < 0.05 was considered significant. Error bars show standard deviation.
All experiments were performed at least twice (except for the RNAseq analysis), with at least three independent samples per group. Each data point represents an individual animal.
Author contributions
HL, SJ, LB, JC, MP, and AS performed experiments. HL, AO, SJ, and AS analyzed the data. AS designed the project and wrote the manuscript.
Declarations of competing interest
The authors have no competing interests to declare.
Funding
This work was funded by a precompetitive, open innovation award to the Manchester Collaborative Centre for Inflammation Research at the University of Manchester, AstraZeneca, and GSK and a Wellcome Trust and Royal Society, Sir Henry Dale Fellowship to AS (109375/Z/15/Z).
Acknowledgments
This research was funded by a precompetitive, open innovation award to the Manchester Collaborative Centre for Inflammation Research by the University of Manchester, AstraZeneca, and GSK and a Wellcome Trust and Royal Society, Sir Henry Dale Fellowship to AS (109375/Z/15/Z). The University of Manchester Bioimaging Facility microscopes used in this study were purchased with grants from the Biotechnology and Biological Sciences Research Council, Wellcome, and the University of Manchester Strategic Fund. The authors acknowledge assistance from Peter Walker, Roger Meadows, Gareth Howell, and Leo Zeef and the use of the University of Manchester Histology, Flow Cytometry, Genomic Technologies, Bioinformatics, and Biological Services facilities. The authors also acknowledge technical help from Peter Warn and Andrew Sharp on skin infection models and Tovah Shaw on intestinal cell isolation. The authors acknowledge intellectual expertise on group 3 ILCs from Matthew Hepworth and on CD200R1 from Tracy Hussell.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mucimm.2023.01.001.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The datasets generated during this study are available from the corresponding author on reasonable request.
REFERENCES
- 1.Cua D.J., Tato C.M. Innate IL-17-producing cells: the sentinels of the immune system. Nat. Rev. Immunol. 2010;10:479–489. doi: 10.1038/nri2800. [DOI] [PubMed] [Google Scholar]
- 2.Iwakura Y., Ishigame H., Saijo S., Nakae S. Functional specialization of interleukin-17 family members. Immunity. 2011;34:149–162. doi: 10.1016/j.immuni.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 3.Nograles K.E., et al. Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br. J. Dermatol. 2008;159:1092–1102. doi: 10.1111/j.1365-2133.2008.08769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ghoreschi K., Balato A., Enerbäck C., Sabat R. Therapeutics targeting the IL-23 and IL-17 pathway in psoriasis. Lancet. 2021;397:754–766. doi: 10.1016/S0140-6736(21)00184-7. [DOI] [PubMed] [Google Scholar]
- 5.Teunissen M.B.M., et al. Composition of innate lymphoid cell subsets in the human skin: enrichment of NCR(+) ILC3 in lesional skin and blood of psoriasis patients. J. Invest. Dermatol. 2014;134:2351–2360. doi: 10.1038/jid.2014.146. [DOI] [PubMed] [Google Scholar]
- 6.Villanova F., et al. Characterization of innate lymphoid cells in human skin and blood demonstrates increase of NKp44+ ILC3 in psoriasis. J. Invest. Dermatol. 2014;134:984–991. doi: 10.1038/jid.2013.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenblum M.D., Yancey K.B., Olasz E.B., Truitt R.L. CD200, a “no danger” signal for hair follicles. J. Dermatol. Sci. 2006;41:165–174. doi: 10.1016/j.jdermsci.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y., et al. CD200R1 agonist attenuates mechanisms of chronic disease in a murine model of multiple sclerosis. J. Neurosci. 2010;30:2025–2038. doi: 10.1523/JNEUROSCI.4272-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorczynski R.M., Chen Z., Lee L., Yu K., Hu J. Anti-CD200R ameliorates collagen-induced arthritis in mice. Clin. Immunol. 2002;104:256–264. doi: 10.1006/clim.2002.5232. [DOI] [PubMed] [Google Scholar]
- 10.Gorczynski R.M., Chen Z., Yu K., Hu J. CD200 immunoadhesin suppresses collagen-induced arthritis in mice. Clin. Immunol. 2001;101:328–334. doi: 10.1006/clim.2001.5117. [DOI] [PubMed] [Google Scholar]
- 11.Rosenblum M.D., et al. CD200 is a novel p53-target gene involved in apoptosis-associated immune tolerance. Blood. 2004;103:2691–2698. doi: 10.1182/blood-2003-09-3184. [DOI] [PubMed] [Google Scholar]
- 12.Snelgrove R.J., et al. A critical function for CD200 in lung immune homeostasis and the severity of influenza infection. Nat. Immunol. 2008;9:1074–1083. doi: 10.1038/ni.1637. [DOI] [PubMed] [Google Scholar]
- 13.Kretz-Rommel A., et al. CD200 expression on tumor cells suppresses antitumor immunity: new approaches to cancer immunotherapy. J. Immunol. 2007;178:5595–5605. doi: 10.4049/jimmunol.178.9.5595. [DOI] [PubMed] [Google Scholar]
- 14.Blom L.H., et al. The immunoglobulin superfamily member CD200R identifies cells involved in type 2 immune responses. Allergy. 2017;72:1081–1090. doi: 10.1111/all.13129. [DOI] [PubMed] [Google Scholar]
- 15.Weizman O.E., et al. ILC1 confer early host protection at initial sites of viral infection. Cell. 2017;171:795–808.e12. doi: 10.1016/j.cell.2017.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagasawa M., et al. KLRG1 and NKp46 discriminate subpopulations of human CD117(+)CRTH2(-) ILCs biased toward ILC2 or ILC3. J Exp Med. 2019;216:1762–1776. doi: 10.1084/jem.20190490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linley H., Jaigirdar S., Mohamed K., Griffiths C.E.M., Saunders A. Reduced cutaneous CD200:CD200R1 signaling in psoriasis enhances neutrophil recruitment to skin. Immun. Inflamm. Dis. 2022;10:e648. doi: 10.1002/iid3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li B., et al. Transcriptome analysis of psoriasis in a large case-control sample: RNA-seq provides insights into disease mechanisms. J. Invest. Dermatol. 2014;134:1828–1838. doi: 10.1038/jid.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chan J.R., et al. IL-23 stimulates epidermal hyperplasia via TNF and IL-20R2-dependent mechanisms with implications for psoriasis pathogenesis. J. Exp. Med. 2006;203:2577–2587. doi: 10.1084/jem.20060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Fits L., et al. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J. Immunol. 2009;182:5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 21.Tamoutounour S., et al. Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity. 2013;39:925–938. doi: 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 22.Dutton E.E., et al. Characterisation of innate lymphoid cell populations at different sites in mice with defective T cell immunity. Wellcome Open Res. 2017;2:117. doi: 10.12688/wellcomeopenres.13199.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pantelyushin S., et al. Rorγt+ innate lymphocytes and γδ T cells initiate psoriasiform plaque formation in mice. J. Clin. Invest. 2012;122:2252–2256. doi: 10.1172/JCI61862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valle-Noguera A., Gómez-Sánchez M.J., Girard-Madoux M.J.H., Cruz-Adalia A. Optimized protocol for characterization of mouse gut innate lymphoid cells. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.563414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Awasthi A., et al. Cutting edge: IL-23 receptor gfp reporter mice reveal distinct populations of IL-17-producing cells. J. Immunol. 2009;182:5904–5908. doi: 10.4049/jimmunol.0900732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao M., et al. Nuclear factor erythroid 2-related factor 2 deficiency exacerbates lupus nephritis in B6/lpr mice by regulating Th17 cell function. Sci. Rep. 2016;6:38619. doi: 10.1038/srep38619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thimmulappa R.K., et al. Nrf2 is a critical regulator of the innate immune response and survival during experimental sepsis. J. Clin. Invest. 2006;116:984–995. doi: 10.1172/JCI25790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pastor-Fernández G., Mariblanca I.R., Navarro M.N. Decoding IL-23 signaling cascade for new therapeutic opportunities. Cells. 2020;9:2044. doi: 10.3390/cells9092044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mori R., et al. TYK2-induced phosphorylation of Y640 suppresses STAT3 transcriptional activity. Sci. Rep. 2017;7:15919. doi: 10.1038/s41598-017-15912-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cherwinski H.M., et al. The CD200 receptor is a novel and potent regulator of murine and human mast cell function. J. Immunol. 2005;174:1348–1356. doi: 10.4049/jimmunol.174.3.1348. [DOI] [PubMed] [Google Scholar]
- 31.Jenmalm M.C., Cherwinski H., Bowman E.P., Phillips J.H., Sedgwick J.D. Regulation of myeloid cell function through the CD200 receptor. J. Immunol. 2006;176:191–199. doi: 10.4049/jimmunol.176.1.191. [DOI] [PubMed] [Google Scholar]
- 32.Karnam G., et al. CD200 receptor controls sex-specific TLR7 responses to viral infection. PLoS Pathog. 2012;8:e1002710. doi: 10.1371/journal.ppat.1002710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rygiel T.P., et al. Lack of CD200 enhances pathological T cell responses during influenza infection. J. Immunol. 2009;183:1990–1996. doi: 10.4049/jimmunol.0900252. [DOI] [PubMed] [Google Scholar]
- 34.Mukhopadhyay S., et al. Immune inhibitory ligand CD200 induction by TLRs and NLRs limits macrophage activation to protect the host from meningococcal septicemia. Cell Host Microbe. 2010;8:236–247. doi: 10.1016/j.chom.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 35.Hoek R.M., et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200) Science. 2000;290:1768–1771. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- 36.Pilch Z., et al. The pro-tumor effect of CD200 expression is not mimicked by agonistic CD200R antibodies. PLoS One. 2019;14:e0210796. doi: 10.1371/journal.pone.0210796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatherley D., Cherwinski H.M., Moshref M., Barclay A.N. Recombinant CD200 protein does not bind activating proteins closely related to CD200 receptor. J. Immunol. 2005;175:2469–2474. doi: 10.4049/jimmunol.175.4.2469. [DOI] [PubMed] [Google Scholar]
- 38.Wright G.J., et al. Characterization of the CD200 receptor family in mice and humans and their interactions with CD200. J. Immunol. 2003;171:3034–3046. doi: 10.4049/jimmunol.171.6.3034. [DOI] [PubMed] [Google Scholar]
- 39.Vaine C.A., Soberman R.J. The CD200-CD200R1 inhibitory signaling pathway: immune regulation and host-pathogen interactions. Adv. Immunol. 2014;121:191–211. doi: 10.1016/B978-0-12-800100-4.00005-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akkaya M., Kwong L.S., Akkaya E., Hatherley D., Barclay A.N. Rabbit CD200R binds host CD200 but not CD200-like proteins from poxviruses. Virology. 2016;488:1–8. doi: 10.1016/j.virol.2015.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kojima T., et al. Novel CD200 homologues iSEC1 and iSEC2 are gastrointestinal secretory cell-specific ligands of inhibitory receptor CD200R. Sci. Rep. 2016;6:36457. doi: 10.1038/srep36457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Vlist M., et al. Signaling by the inhibitory receptor CD200R is rewired by type I interferon. Sci. Signal. 2021;14:eabb4324. doi: 10.1126/scisignal.abb4324. [DOI] [PubMed] [Google Scholar]
- 43.Mahadevan D., et al. Phase I study of samalizumab in chronic lymphocytic leukemia and multiple myeloma: blockade of the immune checkpoint CD200. J. Immunother. Cancer. 2019;7:227. doi: 10.1186/s40425-019-0710-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Solá, P. et al. Local IL-17 orchestrates skin aging. bioRxivhttps://www.biorxiv.org/content/10.1101/2022.01.31.478459v1.full (2022) [Date accessed: 14 July 2022].
- 45.Liao W., Hei T.K., Cheng S.K. Radiation-induced dermatitis is mediated by IL17-expressing γδ T cells. Radiat. Res. 2017;187:454–464. doi: 10.1667/RR007CC.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boudakov I., et al. Mice lacking CD200R1 show absence of suppression of lipopolysaccharide-induced tumor necrosis factor-alpha and mixed leukocyte culture responses by CD200. Transplantation. 2007;84:251–257. doi: 10.1097/01.tp.0000269795.04592.cc. [DOI] [PubMed] [Google Scholar]
- 47.Lochner M., et al. In vivo equilibrium of proinflammatory IL-17+ and regulatory IL-10+ Foxp3+ RORgamma t+ T cells. J. Exp. Med. 2008;205:1381–1393. doi: 10.1084/jem.20080034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shen F.W., et al. Cloning of Ly-5 cDNA. Proc. Natl Acad. Sci. U. S. A. 1985;82:7360–7363. doi: 10.1073/pnas.82.21.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw T.N., et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. J. Exp. Med. 2018;215:1507–1518. doi: 10.1084/jem.20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dutton E.E., et al. Peripheral lymph nodes contain migratory and resident innate lymphoid cell populations. Sci Immunol. 2019;4:eaau8082. doi: 10.1126/sciimmunol.aau8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during this study are available from the corresponding author on reasonable request.