SUMMARY
During mitosis, chromosomes assemble kinetochores in order to dynamically couple with spindle microtubules 1,2. Kinetochores also function as signaling hubs directing mitotic progression by recruiting and controlling the fate of the Anaphase Promoting Complex/Cyclosome (APC/C) activator CDC-20 3-5. Kinetochores either incorporate CDC-20 into checkpoint complexes that inhibit the APC/C or dephosphorylate CDC-20, which allows it to interact with and activate the APC/C 4,6. The importance of these two CDC-20 fates likely depends on biological context. In human somatic cells the major mechanism controlling mitotic progression is the spindle checkpoint. By contrast, progression through mitosis during the cell cycles of early embryos is largely checkpoint-independent 7-10. Here, we first show that CDC-20 phosphoregulation controls mitotic duration in the C. elegans embryo and defines a checkpoint-independent temporal mitotic optimum for robust embryogenesis. CDC-20 phosphoregulation occurs at kinetochores and in the cytosol; at kinetochores, flux of CDC-20 for local dephosphorylation requires an ABBA motif on BUB-1 that directly interfaces with the structured WD40 domain of CDC-20 6,11-13. We next show that a conserved “STP” motif in BUB-1 that docks the mitotic kinase PLK-1 14 is necessary for CDC-20 kinetochore recruitment and timely mitotic progression. The kinase activity of PLK-1 is required for CDC-20 to localize to kinetochores and phosphorylates the CDC-20-binding ABBA motif of BUB-1 to promote BUB–1-CDC-20 interaction and mitotic progression. Thus, the BUB-1-bound pool of PLK-1 ensures timely mitosis during embryonic cell cycles by promoting CDC-20 recruitment to the vicinity of kinetochore-localized phosphatase activity.
eTOC BLURB
Houston et al. show that an optimal mitotic duration set independently of the spindle checkpoint by phosphoregulation of CDC-20 ensures robust C. elegans embryogenesis. They also show that CDC-20 flux through kinetochores, which is important for its activation, requires BUB-1-bound PLK-1, whose kinase activity enhances CDC-20 binding to BUB-1.
RESULTS & DISCUSSION
In the early C. elegans embryo at 20°C all 12 chromosomes align on the spindle and initiate anaphase ~180-200 seconds after nuclear envelope breakdown (NEBD). Eliminating spindle checkpoint signaling does not alter this timing 6,15, indicating that control of mitotic duration is achieved through a checkpoint-independent mechanism.
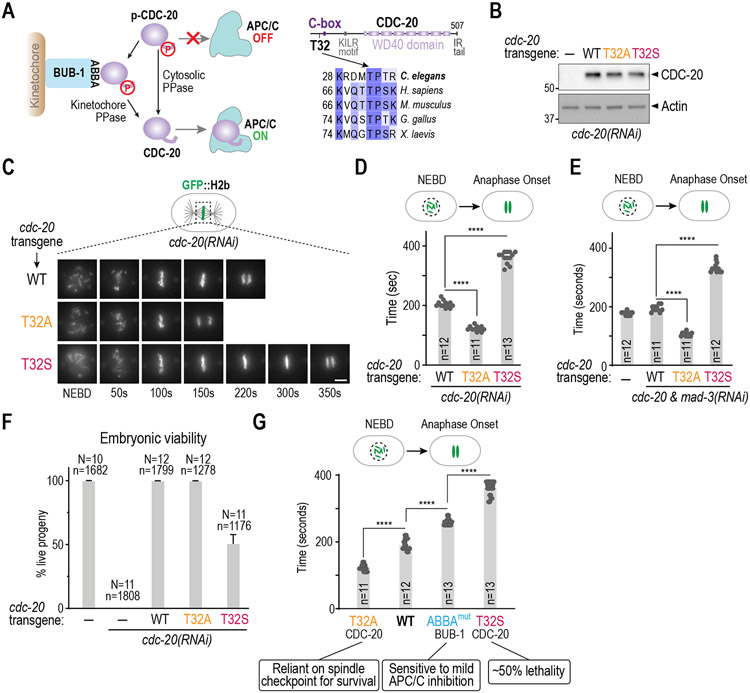
In prior work, we found that phosphoregulation of the APC/C activator CDC-20 is an important mechanism controlling mitotic duration in the early C. elegans embryo 6. Phosphorylation of Cdc20 by Cdk1/2 on sites adjacent to the C-box, a critical APC/C binding motif in the N-terminus, prevents it from binding and activating the APC/C 16-19. Dephosphorylation of Cdc20 by protein phosphatases at the kinetochore and in the cytosol reverses this inhibitory phosphorylation, promoting its binding to and activation of the APC/C (Figure 1A; 6,20-23). In C. elegans, kinetochore flux of CDC-20 requires its binding to the conserved ABBA motif of BUB-1, which positions CDC-20 in the vicinity of kinetochore-bound protein phosphatase 1 (PP1)6. Preventing phosphorylation of 3 Cdk sites in the CDC-20 N-terminus significantly shortens the NEBD-to-anaphase interval to ~120 seconds, indicating premature APC/C activation, and bypasses the contribution of kinetochore phosphatase activity to control of mitotic duration 6. The most functionally significant of the 3 sites is Thr32, which is positioned adjacent to the critical APC/C-binding C-box (Figure 1A-D; 6). To determine whether persistent CDC-20 phosphorylation has the opposite effect, we had engineered conventional phosphomimetic mutations. These mutations exhibited phenotypes associated with loss of CDC-20-activated APC/C function, including compromised osmotic integrity, that precluded analysis of mitosis in one-cell embryos 6. We therefore took a different approach based on conservation of a threonine as the critical target site and the preference of protein phosphatases for phospho-threonine over phospho-serine as a substrate 24,25. To make CDC-20 more persistently phosphorylated, we mutated Thr32 to serine. This subtle alteration significantly increased mitotic duration, with NEBD to anaphase onset taking on average ~360s, compared to ~200s in wild-type embryos (Figure 1B-D), indicating delayed APC/C activation. In T32S CDC-20 embryos, chromosome alignment was not delayed and no chromosome missegregation was observed (Figure 1C, Figure S1A). In addition, a GFP fusion of T32S CDC-20 localized normally to kinetochores (Figure S1B), and phenotypes caused by loss of APC/C function, such as meiotic arrest, osmotic integrity defects, and polarity defects in one-cell embryos 26,27 were not observed. Thus, T32S CDC-20 specifically slows the rate of APC/C activation, and thus delay anaphase onset, but does not perturb other mitotic processes. Notably, the effects of T32A and T32S CDC-20 mutants on mitotic duration were independent of the spindle checkpoint, as the same effects were observed in the absence of MAD-3, a critical checkpoint effector 3,5(Figure 1E; Figure S1C). These results establish that phosphoregulation of CDC-20 is a spindle checkpoint-independent mechanism controlling APC/C activation, and hence mitotic duration, during embryonic divisions.
Figure 1. CDC-20 phosphoregulation controls early embryonic mitotic duration independently of the spindle checkpoint.
(A) (left) Schematic of APC/C control via CDC-20 phosphoregulation; (right) Sequence alignments highlighting the conserved Thr32 Cdk phosphosite. (B) Immunoblot of lysates from worms with the indicated RNAi-resistant cdc-20 mutant transgenes after depletion of endogenous CDC-20; actin serves as a loading control. (C) Stills from timelapse movies imaging chromosomes in embryos from the indicated conditions. Scale bar is 5 μm. (D) & (E) Quantification of the NEBD-anaphase onset interval for the indicated conditions. Bar height is the mean value. n is the number of embryos quantified. (F) Embryonic viability for the indicated conditions. N is number of worms whose progeny were scored and n is the number of embryos; mean and 95% confidence interval are plotted. (G) Relationship between mitotic duration and embryonic phenotypes. Phenotype annotations for T32A CDC-20 and ABBAmut BUB-1 are from previously published work 6. Data for CDC-20 T32A and T32S are the same as Figure 1D. All p-values are from unpaired t-tests; ****:p<0.0001. See Figure S1 for supporting information.
We next assessed the effect of perturbing mitotic duration by altering CDC-20 phosphorylation on embryonic viability. T32A CDC-20, with accelerated anaphase onset, did not result in lethality but T32S CDC-20, with significantly delayed anaphase onset compared to wild-type embryos, exhibited ~50% lethality (Figure 1F). Thus, on its own and somewhat surprisingly, shorter mitotic duration is better tolerated than longer mitotic duration during embryonic development. T32S CDC-20 embryos arrested either in late stages of embryonic development or as dead, unhatched larvae (Figure S1D). When considered together, T32A CDC-20, T32S CDC-20 and the BUB-1 ABBA motif mutant that prevents CDC-20 kinetochore localization, along with the wildtype, represent a well-ordered series for assessing the effect of mitotic duration on embryogenesis (Figure 1G). T32A CDC-20, while viable on its own, exhibits strong synthetic lethality with loss of MAD-3, indicating that accelerating anaphase onset makes embryos reliant on the spindle checkpoint to prevent chromosome segregation errors and promote normal embryonic development6 (Figure 1G). By contrast, T32S CDC-20 exhibits significant lethality on its own (Figure 1G). On the other hand, ABBAMut BUB-1, which has delayed anaphase onset but a shorter mitotic duration than T32S CDC-20, is viable but sensitized to mild APC/C inhibition6 (Figure 1G). These results suggest that the mitotic duration observed in wild-type embryos, which is defined by dynamic changes in CDC-20 phosphorylation, represents an optimum needed to support robust embryonic development. In addition, there appears to be a mitotic duration length threshold, beyond which embryonic development is severely affected.
Understanding how mitotic duration is optimized to promote robust embryonic development requires elucidation of the mechanisms controlling the rate of APC/C activation through control of CDC-20 phosphorylation. In prior work we showed that CDC-20, but not the core APC/C, rapidly fluxes through kinetochores6. Preventing CDC-20 kinetochore localization by mutating BUB-1’s ABBA motif or preventing CDC-20 dephosphorylation at kinetochores by removing PP1 bound to KNL-1, both extended the NEBD to anaphase onset interval 6,28. Eliminating CDC-20 N-terminal phosphorylation suppressed the extension of mitotic duration observed in these mutants, leading to a model in which CDC-20 recruitment to kinetochores promotes its dephosphorylation by PP16. As noted above (Figure 1G), loss of CDC-20 phosphoregulation at kinetochores leads to embryos that are viable but highly sensitized to mild APC/C inhibition. Thus, kinetochores contribute to defining the optimum mitotic duration for robust embryogenesis by promoting CDC-20 dephosphorylation.
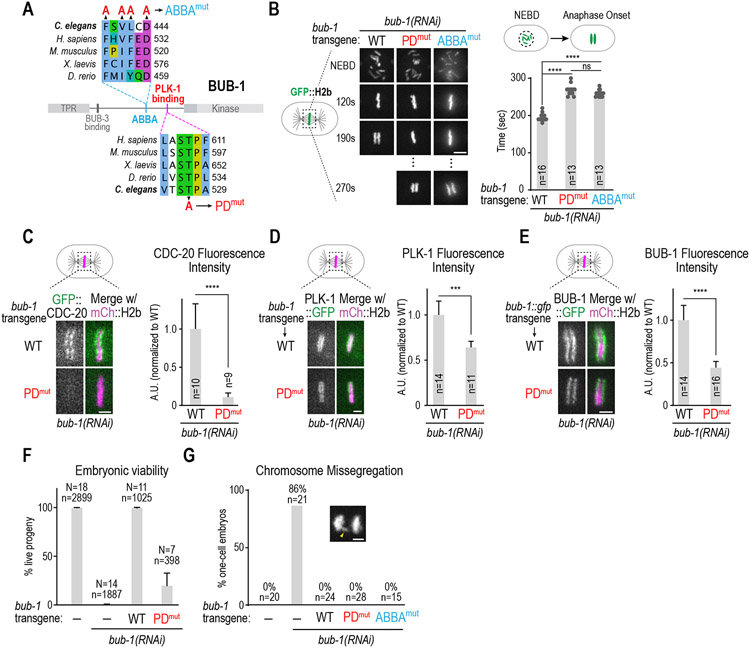
The key binding site for CDC-20 at the kinetochore is the conserved ABBA motif in BUB-1, which interfaces with the structured WD40 domain of CDC-20 (Figure 2A)6,11-13. in the process of analyzing other conserved regions of BUB-1, we found a conserved “STP” PLK-1 binding motif located ~90 aa C-terminal to the ABBA motif (Figure 2A 14,29,30) that is also required for CDC-20 to localize to kinetochores. In vitro binding assays have validated the “STP” motif in C. elegans BUB-1 as interacting with the Polo box domain of PLK-1 in a Thr527 phosphorylation-dependent manner31. Mutation of the PLK-1 docking motif of BUB-1 (T527A or PDmut) increased the NEBD to anaphase onset interval to the same extent as selective mutation of the BUB-1 ABBA motif (Figure 2B, Figure S2A). Consistent with this, CDC-20 was undetectable at kinetochores in PDmut BUB-1 (Figure 2C). Moreover, PDmut BUB-1 was deficient for spindle checkpoint signaling (Figure S2B), which requires CDC-20 incorporation into mitotic checkpoint complexes at unattached kinetochores 32,33. PDmut BUB-1 reduced but did not eliminate PLK-1 kinetochore localization, indicating the presence of an additional kinetochore-localized pool of PLK-1 31,34 (Figure 2D). PDmut BUB-1::GFP, which was expressed similarly to WT BUB-1::GFP, localized to kinetochores with a maximal level that was ~50% of WT BUB-1::GFP (Figure 2E; Figure S2A); this partial reduction is potentially due to reduced phosphorylation of KNL-1, the phosphoregulated kinetochore receptor of BUB-1 35,36. However, the reduction of kinetochore-localized PDmut BUB-1 is unlikely to account for the loss of kinetochore-localized CDC-20, as reducing BUB-1 levels to a similar extent as is observed in PDmut BUB-1 through engineered reduction of BUB-1 binding sites in KNL-1 35,37 did not eliminate CDC-20 kinetochore localization (Figure S2C). These data indicate that the PLK-1 docking site on BUB-1, while contributing to kinetochore pools of PLK-1 and BUB-1, is essential for CDC-20 kinetochore recruitment.
Figure 2. The PLK-1-docking site in BUB-1 is required for timely mitosis and CDC-20 kinetochore recruitment.
(A) Sequence alignments of the CDC-20 binding ABBA motif and the PLK-1 docking motif in BUB-1. Mutations engineered to disrupt CDC-20 binding (ABBAmut) and PLK-1 interaction (PDmut) are indicated. (B) (left) Images from timelapse movies of chromosomes for the indicated conditions. WT and PDmut BUB-1 were imaged using spinning disk confocal imaging while ABBAmut BUB-1 was imaged on a widefield microscope. (right) NEBD-anaphase onset interval for the indicated conditions. ABBAmut BUB-1 data are the same as in Figure 1G. Scale bar is 2 μm. (C-E) Representative images (left) and quantification of chromosomal fluorescence intensity at metaphase (right) in WT vs PDmut BUB-1 for the indicated proteins. n is the number of embryos quantified. Error bars are the 95% confidence interval. Scale bar is 2 μm. (F) Embryonic viability analysis for the indicated conditions. N is number of worms analyzed; n is total number of progeny scored. Error bars are the 95% confidence interval. (G) Frequency of anaphase chromosome missegregation scored from timelapse movies for the indicated conditions. Inset shows an example of lagging anaphase chromatin (arrowhead). n is the number of embryos quantified. Scale bar is 2 μm. p-values are from unpaired t-tests; ***:p<0.001, ****:p<0.0001. See Figure S2 for supporting information.
ABBAmut BUB-1 on its own does not exhibit significant embryonic lethality but is highly sensitive to mild APC/C inhibition 6. By contrast, PDmut BUB-1 exhibited significant embryonic lethality on its own (Figure 2F). BUB-1 inhibition leads to penetrant embryonic lethality and visible chromosome missegregation characterized by anaphase bridges 15,37; however, chromosome missegregation was not observed for either PDmut or ABBAmut BUB-1 in one-cell embryos (Figure 2G), indicating that missegregation was unlikely to cause the lethality observed with PDmut BUB-1. These data suggest that lethality observed in PDmut BUB-1 may reflect an as-yet-unknown role for BUB-1-bound PLK-1 during embryonic development.
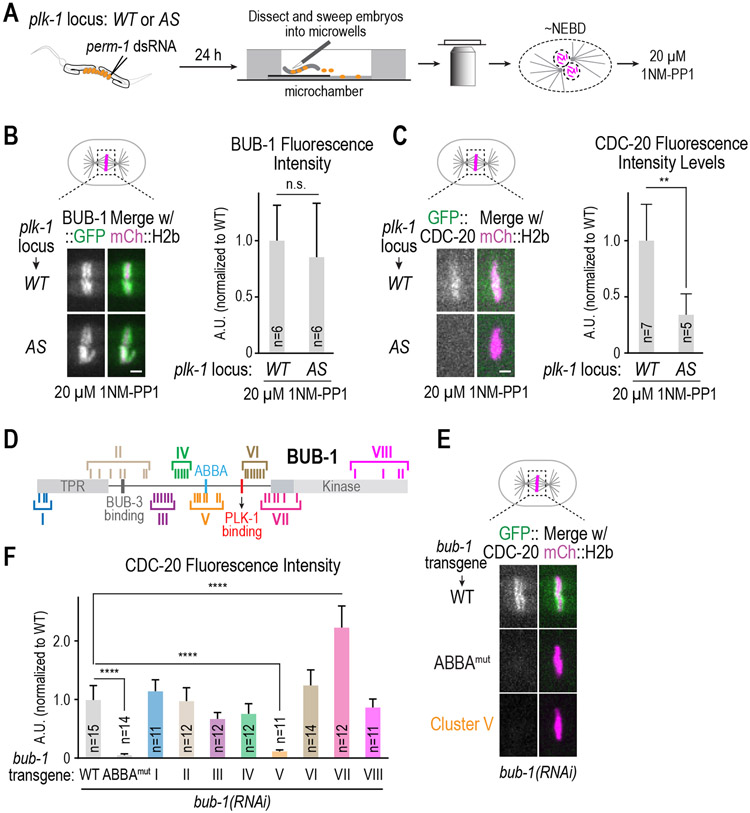
The results above suggests that PLK-1 docked onto BUB-1 is important to recruit CDC-20 to kinetochores. We next sought to address if PLK-1 kinase activity is important for this function. As PLK-1 is required for meiotic progression and mitotic entry 38, mutants that eliminate its kinase activity cannot be analyzed. We therefore used an analog-sensitive (AS) allele of PLK-1 engineered at the endogenous locus using genome-editing 39. To temporally control activity inhibition, we employed a method to permeabilize embryos 40 and added the inhibitor specifically targeting analog-sensitive PLK-1 to permeabilized embryos immobilized in microwells around the time of NEBD (Figure 3A); timed inhibition was essential as earlier addition of the inhibitor significantly delays/prevents NEBD and mitotic entry. We conducted this analysis in strains expressing in situ tagged BUB-1::GFP or transgene-encoded GFP::CDC-20. In the presence of wildtype PLK-1, following inhibitor addition kinetochore localization of both BUB-1 and CDC-20 was observed (Figure 3B,C). By contrast, following inhibitor addition in PLK-1AS, BUB-1 was still kinetochore-localized but CDC-20 kinetochore localization was significantly diminished (Figure 3B,C; Figure S2D). Without inhibitor addition, CDC-20 localized similarly in the presence of both wildtype and PLK-1AS (Figure S2E). As the inhibitor was added around NEBD, the localization of BUB-1 at kinetochores is likely due to PLK-1 phosphorylation of KNL-1 prior to inhibitor addition. Thus, PLK-1 kinase activity is important for CDC-20 kinetochore recruitment.
Figure 3. Role of PLK-1 kinase activity in CDC-20 kinetochore recruitment and identification of putative PLK-1 target sites.
(A) Schematic of assay used to permeabilize embryos and temporally control PLK-1 inhibition using an analog-sensitive (AS) allele engineered at the endogenous plk-1 locus. (B & C) Representative images (left) and quantification of chromosomal fluorescence intensity (right) for the indicated conditions. n is the number of embryos quantified. Scale bars, 2 μm. p-values are from unpaired t-tests; n.s.: not significant; **: p<0.01.(D) Schematic of BUB-1 highlighting 45 putative PLK-1 target sites and 8 clustered alanine mutants designed to assess effects on CDC-20 kinetochore recruitment. (E) Quantification of GFP::CDC-20 localization for the indicated conditions. n is the number of embryos quantified. p-values are from unpaired t-tests; ****:p<0.0001. (F) Representative images of CDC-20 localization in the indicated conditions. Scale bar, 2 μm. See Table S1 and Figures S2 and S3 for supporting information.
The primary binding site for CDC-20 at kinetochores is BUB-1’s ABBA motif, which is ~90 aa N-terminal to the PLK-1 docking site in BUB-1. Thus, an attractive model is that tethered PLK- 1 phosphorylates residues in BUB-1 to enhance the ABBA-dependent interaction with CDC-20. To gain insight into the mechanism by which PLK-1 is promoting CDC-20 kinetochore recruitment, we employed an unbiased in vivo mutagenesis screen to identify functionally relevant PLK-1 target sites in BUB-1, using an approach that previously yielded PLK-1 target sites important for cytokinesis and spindle assembly 39,41. We defined 45 putative PLK-1 sites in BUB-1 using phosphorylation consensus motifs and conservation criteria (Figure S3A, Table S1). We mutated these 45 sites in 8 clusters (Figure 3D; Figure S3B), integrated RNAi-resistant untagged BUB-1 transgenes encoding the 8 cluster mutants, and crossed the transgenes into a strain expressing GFP::CDC-20. Following endogenous BUB-1 depletion, CDC-20 localization was analyzed by live imaging. This screen revealed that the Cluster V mutant, in which 7 putative PLK-1 sites were mutated to alanine, has significantly reduced CDC-20 kinetochore localization and delayed anaphase onset (Figure 3E,F; Figure S3C). By contrast, kinetochore recruitment of BUB-3, which depends on BUB-1 15,37, was unaffected by Cluster V mutation (Figure S3D). In addition, the Cluster V mutant, as well as all other cluster mutants, rescued the embryonic lethality caused by BUB-1 depletion (Figure S3E). Thus, the Cluster V mutant does not affect BUB-1 kinetochore localization or its essential function(s) in embryo viability. Interestingly, the Cluster VII mutant, which alters a number of residues in the N-terminal extension involved in Bub1 kinase activation 42, was associated with elevated CDC-20 kinetochore localization and delayed anaphase onset (Figure 3F; Figure S3C,F), the reasons for which are currently unclear.
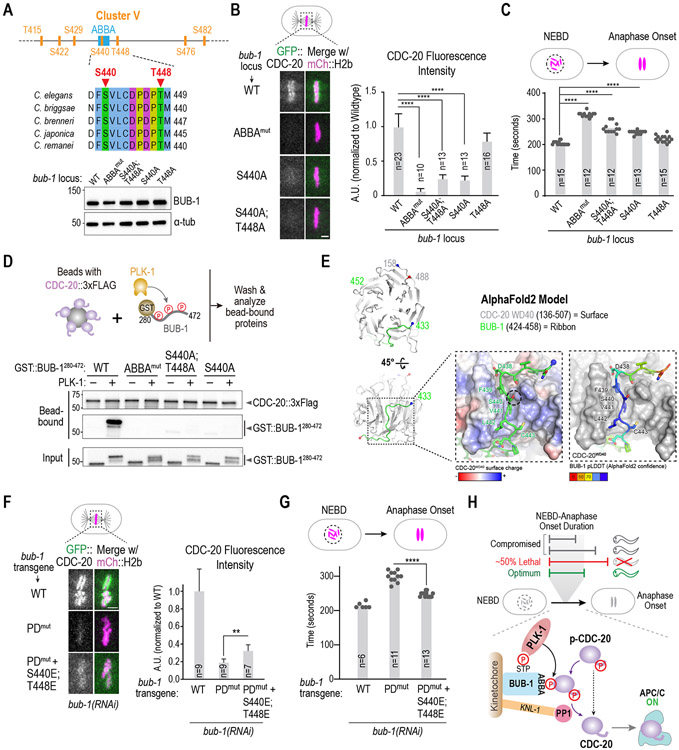
We next focused on which of the 7 mutated PLK-1 sites in Cluster V are critical for CDC-20 kinetochore recruitment. As S440 and T448 are conserved across Caenorhabditis species (Figure 4A), we mutated them to alanine, individually and in combination, at the endogenous bub-1 locus in a strain expressing GFP::CDC-20 and mCherry::H2b; all 3 homozygous mutants were viable and had normal BUB-1 levels (Figure 4A). Mutation of S440, which is within the ABBA motif, caused a major reduction in CDC-20 kinetochore recruitment, whereas mutation of T448 had a mild effect (Figure 4B). Of the other sites, S422, S476 and S482 are not conserved across Caenorhabditis species, and, while T415 is conserved, its mutation had no effect on CDC-20 kinetochore localization (not shown). Neither S440A nor S440A;T448A mutants of BUB-1 reduced CDC-20 kinetochore localization to the same extent as the ABBA mutant (Figure 4B). This observation is consistent with the idea that the ABBA motif of BUB-1 has intrinsic affinity for CDC-20 that is enhanced by PLK-1 phosphorylation.
Figure 4. Mechanism by which PLK-1 phosphorylation promotes CDC-20 kinetochore recruitment.
(A) (top) Details on sites mutated in Cluster V, which is centered around the ABBA motif. (bottom) Immunoblot of BUB-1 following genome-editing of the endogenous bub-1 locus to introduce the indicated mutations. The genome editing was conducted in a strain with transgene-encoded GFP::CDC-20 and mCherry::H2b. (B) Analysis of CDC-20 localization in the indicated conditions. n is the number of embryos quantified. Scale bar, 2 μm. (C) Quantification of NEBD-anaphase onset interval for the indicated conditions. n is the number of embryos quantified. Error bars are the 95% confidence interval. (D) (top) Schematic of assay used to assess interaction of an ABBA motif-containing BUB-1 fragment with full-length CDC-20; constitutively-active purified PLK-1 was used to phosphorylate the BUB-1 fragment. (bottom) Results of binding assay conducted with wildtype or indicated specific mutant GST::BUB-1 fragments. Top two immunoblots show proteins bound to CDC-20-coated beads; bottom immunoblot is of the input GST::BUB-1 fragments phosphorylated by PLK-1. Results shown are representative of two independent experiments. (E) AlphaFold2 model of C. elegans BUB-1 ABBA motif and C. elegans CDC-20. Two views show the overall position of the BUB-1 ABBA motif (green) docked against the WD40 beta-propeller domain (grey) in CDC-20. The start and ends of the shown chains are represented with blue and red colored spheres, respectively. The enlarged, space-filled zoom views show: overall surface charge on ABBA binding site on CDC-20 WD40 (left); confidence score of BUB-1 ABBA motif residues in this binding site (right). The Ser440 residue is outlined by a black dotted circle. (F) Analysis of CDC-20 localization in the indicated conditions, with quantification on the right. n is the number of embryos quantified. Scale bar, 2 μm. (G) Quantification of NEBD-anaphase onset interval for the indicated conditions. n is the number of embryos quantified. Error bars are the 95% confidence interval. (H) Model summarizing key findings of the study. All p-values are from unpaired t-tests; **:p<0.01; ****: p<0.0001. See Figure S4 for supporting information.
In agreement with the effect on CDC-20 kinetochore localization, the NEBD-Anaphase onset interval was significantly increased for S440A BUB-1, but the magnitude of the increase was less than for ABBAmut or PDmut BUB-1 (Figure 4C). The T448A mutant on its own did not significantly extend mitotic duration but S440A;T448A was slightly elevated relative to S440A alone (Figure 4C). As noted above, S440A and the S440A;T448A BUB-1 have a lesser effect on CDC-20 kinetochore localization than ABBAmut BUB-1 (Figure 4B). These observations are consistent with S440 being a major target site of BUB-1-bound PLK-1 whose phosphorylation enhances intrinsic affinity of the BUB-1 ABBA motif for CDC-20.
To test if phosphorylation at sites identified by the in vivo screen directly enhances affinity of BUB-1 for CDC-20, we expressed full-length C. elegans CDC-20 in a human cell protein expression system, bound it to beads, and combined the CDC-20-coated beads with a GST-fused BUB-1 fragment (aa 280-472) that contains the ABBA motif (Figure 4D). Without PLK-1 phosphorylation, the ABBA-containing BUB-1 fragment interacted weakly with CDC-20. By contrast, incubation of the BUB-1 fragment with PLK-1 (constitutively active T194D C. elegans PLK-1) in the presence of ATP strongly stimulated binding to CDC-20 (Figure 4D). Robust PLK- 1 phosphorylation was evident in the retarded gel mobility of the GST-BUB-1280-472 fragment (Figure 4D). Importantly, mutating residues in the ABBA motif that are essential for interaction with the WD40 domain of CDC-20 eliminated the phosphorylation-stimulated interaction, even though significant phosphorylation of the ABBAmut BUB-1 fragment was observed (Figure 4D). We next conducted the same analysis with GST::BUB-1 fragments with alanine mutations of S440 or S440;T448 and found that preventing phosphorylation at these sites greatly reduced the PLK-1 stimulated interaction between BUB-1 and CDC-20 (Figure 4D). Analysis of the input indicated that the S440A and the S440A;T448A mutant BUB-1 fragments were phosphorylated, as evidenced by their retarded gel mobility (Figure 4D); the difference in mobility between the mutant and WT GST::BUB-1 fragments is suggestive of phosphorylation of the mutated sites by PLK-1. Mass spectrometry of PLK-1 phosphorylated WT BUB-1 fragment confirmed phosphorylation of T448; unfortunately, despite multiple attempts with different protocols, the ABBA motif peptide including S440 was never detected (not shown). Nonetheless, the balance of in vitro and in vivo evidence strongly supports the conclusion that phosphorylation of S440 by PLK-1 significantly enhances the affinity of BUB-1 for CDC-20.
An Alphafold2 model of a C. elegans CDC-20–BUB-1 complex revealed that the BUB-1 ABBA motif residues interact with the β-propeller WD40 domain of CDC-20 in the inter-blade groove region (Figure 4E; Figure S4A;43); structural superposition indicated that the modeled binding of C. elegans BUB-1 ABBA motif to C. elegans CDC-20 is equivalent to the structurally characterized interaction of S. cerevisiae Acm1’s ABBA motif with the WD40 domain of the Cdc20 ortholog Cdh1 (Figure S4B;12). Notably, S440 of BUB-1 is positioned in the vicinity of a basic region on C. elegans CDC-20 (specifically R234), potentially suggesting favorable interactions in its phosphorylated state.
The biochemical data and Alphafold2 model suggest that electrostatic interactions mediated by phosphorylation contribute to the BUB-1 ABBA – CDC-20 interaction. A prediction of this model is that mimicking phosphorylation at the key sites should bypass the requirement for PLK-1 docking on BUB-1. To test this idea, we engineered phosphomimetic residues in S440 and T448 and combined them with the BUB-1 PLK-1-docking site mutation. The phosphomimetic residues on their own did not significantly affect CDC-20 localization or mitotic duration (Figure S4C-E). However, when S440E;T448E mutations were combined with PDmut, the phosphomimetic residues significantly restored CDC-20 localization and partially suppressed the delay in anaphase onset (Figure 4F,G); we note that, as PDmut reduces BUB-1 kinetochore levels by ~50% (Figure 2E), at most ~50% rescue would be expected. These data further strengthen the conclusion that BUB-1-tethered PLK-1 phosphorylates BUB-1 to enhance its affinity for CDC-20 (Figure 4H).
In summary, the results reported here highlight the importance of CDC-20 phosphoregulation in defining an optimum mitotic duration that supports robust C. elegans embryogenesis (Figure 4H). This mechanism represents a checkpoint-independent means of controlling mitotic duration and may also operate in other contexts. Our findings are most consistent with the biochemical event dictating the length of mitosis being the rate of CDC-20 dephosphorylation, both at the kinetochore and likely also in the cytosol (Figure 1A; Figure 4H). With respect to the kinetochore-based arm of CDC-20 dephosphorylation, BUB-1–bound PLK-1 plays a central role by phosphorylating the BUB-1 ABBA motif to promote its binding to CDC-20. ABBA motifs are present in multiple Cdc20-interacting proteins, including cyclin A, the Bub1-related protein BubR1, and Acm1 11,13,44. While a serine residue analogous to S440 is not present in vertebrate Bub1 ABBA motifs (Figure 2A), a serine is present at this location in the ABBA motifs of BubR1 and cyclin A 13. Whether phosphorylation of this serine or in residues adjacent to the ABBA motif enhances the affinity of Cdc20-interacting proteins in other species, analogous to what we report here for BUB-1 in C. elegans, will be important to address in the future. Finally, somewhat unexpectedly, prolonging mitosis by delaying anaphase onset has a more negative consequence on embryogenesis than accelerating anaphase onset. Addressing why embryogenesis is highly sensitive to prolonged mitoses will be important to address in future work.
STAR METHODS:
Resource Availability:
Lead Contact
Further information and requests for reagents or resources should be directed to the lead contact, Arshad Desai (abdesai@ucsd.edu).
Materials Availability
All unique and stable resources generated by this study are available from the Lead Contact without restriction.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to re-analyze the data reported in this paper are available from the lead contact upon request.
Experimental Model and Subject Details:
C. elegans strains (Key Resources Table) were maintained at 16-20°C using standard nematode growth medium plates seeded with OP-50 E. coli. Freestyle™ 293-F cells (ThermoFisher) were cultured at 37 °C and 8% CO2 with shaking at 125 rpm in flasks containing Freestyle™ 293 Expression Medium (ThermoFisher).
Key Resources Table:
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti- GST | Dammerman et al.51 | OD140 |
| Rabbit anti- BUB-1 (aa 287-665) | Oegema et al.52 | OD31-B |
| Rabbit anti- CDC-20 (aa 1-160) | Kim and Lara-Gonzalez et al.6 | OD220 |
| Mouse anti- α-tubulin (DM1α) | Sigma | Cat# T9026, RRID: AB_477593 |
| Mouse anti-actin (C4) | Millipore | Cat# MAB1501 RRID: AB_2223041 |
| Mouse anti-FLAG (M2) | Sigma | Cat# F1804 RRID: AB_439685 |
| Goat Anti-Rabbit IgG, HRP conjugated | Jackson Immunoresearch | Cat# 111-035-003 RRID: AB_231357 |
| Donkey Anti-Mouse IgG, HRP conjugated | Jackson Immunoresearch | Cat# 715-035-150 RRID: AB_234070 |
| Goat Anti-Mouse Light Chain Specific, HRP conjugated | Jackson Immunoresearch | Cat# 115-035-174 RRID: AB_2338512 |
| Donkey Anti-Rabbit IgG, HRP conjugated | Jackson Immunoresearch | Cat# 711-035-152 RRID: AB_10015282 |
| Donkey Anti-Mouse IgG, AP conjugated | Jackson Immunoresearch | Cat# 715-055-150 RRID: AB_234077 |
| Bacterial and virus strains | ||
| E. coli Strain: OP50-1 | Caenorhabditis Genetics Center (CGC) | N/A |
| Chemicals and Recombinant Proteins | ||
| Cas9-NLS | Macrolab, UC Berkeley QB3 | N/A |
| 1NM-PP1 | Cayman Chemical | Cat# 13330 |
| FM4-64 | Invitrogen | Cat# T13320 |
| Experimental models: Cell Lines | ||
| Freestyle™ 293-F | ThermoFisher | Cat# R79007, RRID: CVCL_D603 |
| Experimental models: Organisms/strains | ||
| C. elegans: Strain wild type N2 (ancestral) | CGC | N2 |
| C. elegans: Strain EG6429 ttTi5605 II; unc-119(ed3) III | Laboratory of Erik Jorgensen | EG6429 |
| C. elegans: Strain EG6701 ttTi4348 I; unc-119(ed3) III; oxEx1580 | Laboratory of Erik Jorgensen | EG6701 |
| C. elegans: Strain OD56 unc-119(ed3) III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | McNally et al.53 | OD56 |
| C. elegans: Strain OD991 unc-119(ed3)III;ltSi268[pOD/pTK013; Pbub-1::Bub1 reencoded; cb-unc-119(+)]II | Kim et al.37 | OD991 |
| C. elegans: Strain OD1702 unc-119(ed3)?III; ltSi560 [oxTi365; oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, tbg-1::gfp::tbb-2_3’UTR; cb-unc-119(+)]V | Kim et al.37 | OD1702 |
| C. elegans: Strain OD2003 unc-119(ed3)? mat-3(or344)III; ltSi560 [oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, tbg-1::gfp::tbb-2_3’UTR; cb-unc-119(+)]V | Kim and Lara-Gonzalez et al.6 | OD2003 |
| C. elegans: Strain OD2024 ltSi268[pOD1951/pTK013; Pbub-1::Bub1 reencoded; cb-unc-119(+)]II; unc-119(ed3)III?;ltSi560[oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, GFP::tbg-1::tbb-2_3’UTR; cb-unc-119(+)]V | Kim and Lara-Gonzalez et al.6 | OD2024 |
| C. elegans: Strain OD2498 ltSi251[pOD1940/pTK002; Pbub-1::GFP-Bub1 reencoded; cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | Kim et al.37 | OD2498 |
| C. elegans: Strain OD2548 ltSi817[pPLG048; Pfzy-1::fzy-1 reencoded::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)III | Kim and Lara-Gonzalez et al.6 | OD2548 |
| C. elegans: Strain OD2591 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)III?;; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | Kim and Lara-Gonzalez et al.6 | OD2591 |
| C. elegans: Strain OD2721 unc-119(ed3)III;ltSi1346[pOD3109/pTK070;Pbub-1::BUB-1 T527A reencoded; cb-unc-119(+)]II | This Study | OD2721 |
| C. elegans: Strain OD2794 ltSi1346[pTK070;Pbub-1::BUB-1 T527A reencoded; cb-unc-119(+)]II;unc-119(ed3)?III; ltSi560 [oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, tbg-1::gfp::tbb-2_3’UTR; cb-unc-119(+)]V | This Study | OD2794 |
| C. elegans: Strain OD2798 ltSi268[pOD1951/pTK013; Pbub-1::Bub1 reencoded; cb-unc-119(+)]II; unc-119(ed3)? mat-3(or344) III;ltSi560[oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, GFP::tbg-1::tbb-2_3’UTR; cb-unc-119(+)]V | This Study | OD2798 |
| C. elegans: Strain OD2921 san-1(lt42[gfp::tev::loxP::3xFlag::san-1])I; unc-119(ed3)?III; ltIs37[pAA64; pie-1/mCHERRY::his-58; unc-119 (+)]IV | Lara-Gonzalez et al.33 | OD2921 |
| C. elegans: Strain OD2941 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I;ltSi1[pOD809/pJE110; Pknl-1::KNL-1reencoded::RFP; cb-unc-119(+)]II;unc-119(ed3)III?;ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | Kim and Lara-Gonzalez et al.6 | OD2941 |
| C. elegans: Strain OD3032 ltSi817[pPLG048; Pfzy-1::fzy-1 reencoded::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)?III; ltSi560 [oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, tbg-1::gfp::tbb-2_3’UTR; cb-unc-119(+)]V | Kim and Lara-Gonzalez et al.6 | OD3032 |
| C. elegans: Strain OD3087 ltSi1001[pPLG069; Pfzy-1::fzy-1 reencoded T32A::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)III | Kim and Lara-Gonzalez et al.6 | OD3087 |
| C. elegans: Strain OD3090 ltSi1001[pPLG069; Pfzy-1::fzy-1 reencoded T32A::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)?III ltSi560 [oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, tbg-1::gfp::tbb-2_3’UTR; cb-unc-119(+)]V | Kim and Lara-Gonzalez et al.6 | OD3090 |
| C. elegans: Strain OD3849 ltSi1380[pTK102;Pbub-1::GFP::BUB-1 T527A reencoded; cb-unc-119(+)]II;unc-119(ed3)?III;ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD3849 |
| C. elegans: Strain OD4706 ltSi1518[pPLG331; Pfzy-1::fzy-1 reencoded T32S::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)III | This Study | OD4706 |
| C. elegans: Strain OD4793 ltSi1518[pPLG331; Pfzy-1::fzy-1 reencoded T32S::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)?III ltSi560 [oxTi365; oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, tbg-1::gfp::tbb-2_3’UTR; cb-unc-119(+)]V | This Study | OD4793 |
| C. elegans: Strain OD4800 bub-1(lt185[bub-1 F439A, V441A, L442A, D444A]I; unc-119(ed3)?III; ltSi560 [oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, tbg-1::gfp::tbb-2_3’UTR; cb-unc-119(+)]V | This Study | OD4800 |
| C. elegans: Strain OD4877 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1535[pOD2674/pTK050; Pbub-1::Bub1 ABBA mut 1 (F439A,V442A,L443A,D445A); cb-unc-119(+)]II; unc-119(ed3)?III; ltIs37[pAA64; pie-1/mCHERRY::his-58; unc-119 (+)]IV | This Study | OD4877 |
| C. elegans: Strain OD4963 ltSi1346[pOD3109/pTK070;Pbub-1::BUB-1 T527A reencoded; cb-unc-119(+)]II; plk-1[lt16[plk-1::GFP]::loxp::cb-unc-119(+)::loxp], unc-119(ed3)?III ;ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD4963 |
| C. elegans: Strain OD4964 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1346[pOD3109/pTK070;Pbub-1::BUB-1 T527A reencoded; cb-unc-119(+)]II; unc-119(ed3)III?;; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD4964 |
| C. elegans: Strain OD4965 ltSi268[pOD/pTK013; Pbub-1::Bub1 reencoded; cb-unc-119(+)]II; plk-1[lt16[plk-1::GFP]::loxp::cb-unc-119(+)::loxp], unc-119(ed3)?III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD4965 |
| C. elegans: Strain OD4966 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)III;ltSi268[pOD/pTK013; Pbub-1::Bub1 reencoded; cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD4966 |
| C. elegans: Strain OD4983 ltSi1346[pTK070;Pbub-1::BUB-1 T527A reencoded; cb-unc-119(+)]II; unc-119(ed3)?, mat-3(or344)III; ltSi560 [oxTi365; pPLG014; Pmex-5::GFP::his-11::tbb-2_3’UTR, tbg-1::gfp::tbb-2_3’UTR; cb-unc-119(+)]V | This Study | OD4983 |
| C. elegans: Strain OD5035 bub-1(lt82 [bub-1::gfp])I; unc-119(ed3)? III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5035 |
| C. elegans: Strain OD5051 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)III?;; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV; plk-1((lt106[plk-1 C52V] lt108[plk-1 L115G])III | This Study | OD5051 |
| C. elegans: Strain OD5067 bub-1(lt82 [bub-1::gfp])I; plk-1((lt106[plk-1 C52V] lt108[plk-1 L115G])III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)]IV; unc-119(ed3)?III; | This Study | OD5067 |
| C. elegans: Strain OD5136 ltSi1624[pOD3969/pJMH59; Pbub-1 reencoded cluster I (S2A, S16A, S22A); cb-unc-119(+)]II | This Study | OD5136 |
| C. elegans: Strain OD5155 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1624[pOD3969/pJMH59; Pbub-1 reencoded cluster I (S2A, S16A, S22A); cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV; | This Study | OD5155 |
| C. elegans: Strain OD5175 ltSi1642[pOD3971/pJMH61; Pbub-1 reencoded cluster III (T327A, S329A, T332A, S336A, S341A, S343A); cb-unc-119(+)]II; unc-119(ed3)? III; | This Study | OD5175 |
| C. elegans: Strain OD5179 ltSi1643[pOD3976/pJMH66; Pbub-1 reencoded cluster VIII (S840A, S915A, S955A, S967A); cb-unc-119(+)]II | This Study | OD5179 |
| C. elegans: Strain OD5192 ltSi1645[pOD3973/pJMH63; Pbub-1 reencoded cluster V (T415A, S422A, S429A, S440A, T448A, S476A, S482A); cb-unc-119(+)]II | This Study | OD5192 |
| C. elegans: Strain OD5200 ltSi1649[pOD3972/pJMH62; Pbub-1 reencoded cluster IV (S368A, T370A, S372A, S376A, T377A); cb-unc-119(+)]II; unc-119(ed3)? III | This Study | OD5200 |
| C. elegans: Strain OD5204 ltSi1653[pOD3974/pJMH64; Pbub-1 reencoded cluster VI (S548A, T549A, T564A, S573A, S583A, S588A, S593A); cb-unc-119(+)]II; unc-119(ed3)? III | This Study | OD5204 |
| C. elegans: Strain OD5207 ltSi1656[pOD3975/pJMH65; Pbub-1 reencoded cluster VII (S599A, S612A, S627A, T630A, T652A, S686A); cb-unc-119(+)]II | This Study | OD5207 |
| C. elegans: Strain OD5214 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1642[pOD3971/pJMH61; Pbub-1 reencoded cluster III (T327A, S329A, T332A, S336A, S341A, S343A); cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5214 |
| C. elegans: Strain OD5215 ltSi1660[pOD3970/pJMH60; Pbub-1 reencoded cluster II (T135A, T163A, S193A, S220A, T232A, S283A, T289A); cb-unc-119(+)]II | This Study | OD5215 |
| C. elegans: Strain OD5225 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1660[pOD3970/pJMH60; Pbub-1 reencoded cluster II (T135A, T163A, S193A, S220A, T232A, S283A, T289A); cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5225 |
| C. elegans: Strain OD5226 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3’UTR; cb-unc-119(+)]I; ltSi1649[pOD3972/pJMH62; Pbub-1 reencoded cluster IV (S368A, T370A, S372A, S376A, T377A); cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5226 |
| C. elegans: Strain OD5227 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1645[pOD3973/pJMH63; Pbub-1 reencoded cluster V (T415A, S422A, S429A, S440A, T448A, S476A, S482A); cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5227 |
| C. elegans: Strain OD5228 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1653[pOD3974/pJMH64; Pbub-1 reencoded cluster VI (S548A, T549A, T564A, S573A, S583A, S588A, S593A); cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5228 |
| C. elegans: Strain OD5229 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1656[pOD3975/pJMH65; Pbub-1 reencoded cluster VII (S599A, S612A, S627A, T630A, T652A, S686A); cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5229 |
| C. elegans: Strain OD5230 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1643[pOD3976/pJMH66; Pbub-1 reencoded cluster VIII (S840A, S915A, S955A, S967A); cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5230 |
| C. elegans: Strain OD5288 ltSi1340[pTK077;Pbub-3::BUB-3 reencoded::GFP; cb-unc-119(+)]I; ltSi268[pOD/pTK013; Pbub-1::Bub1 reencoded; cb-unc-119(+)]II; unc-119(ed3)? III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5288 |
| C. elegans: Strain OD5289 ltSi1340[pTK077;Pbub-3::BUB-3 reencoded::GFP; cb-unc-119(+)]I; ltSi1645[pOD3973/pJMH63; Pbub-1 reencoded cluster V (T415A, S422A, S429A, S440A, T448A, S476A, S482A); cb-unc-119(+)]II; unc-119(ed3)? III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5289 |
| C. elegans: Strain OD5290 ltSi1340[pTK077;Pbub-3::BUB-3 reencoded::GFP; cb-unc-119(+)]I; ltSi1535[pOD2674/pTK050; Pbub-1::Bub1 ABBA mut 1 (F439A,V442A,L443A,D445A); cb-unc-119(+)]II; unc-119(ed3)? III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5290 |
| C. elegans: Strain OD5306 bub-1(lt219[bub-1 S440A, T448A]I; ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)?III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)]IV; | This Study | OD5306 |
| C. elegans: Strain OD5307 bub-1(lt185[bub-1 F439A, V441A, L442A, D444A]I; ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)?III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)]IV; | This Study | OD5307 |
| C. elegans: Strain OD5319 bub-1(lt220[bub-1 S440A])I; ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)?III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)]IV; | This Study | OD5319 |
| C. elegans: Strain OD5327 bub-1(lt225[bub-1 S440E, T448E])I; ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)?III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)]IV; | This Study | OD5327 |
| C. elegans: Strain OD5349 bub-1(lt231[bub-1 T448A])I;ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; unc-119(ed3)?III; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)]IV; | This Study | OD5349 |
| C. elegans: Strain OD5438 ltSi1786[pOD5259/pJMH80; Pbub-1:bub-1 reencoded S440E, T448E, T527A::bub-1 3’UTR; cb-unc-119(+)]II | This Study | OD5438 |
| C. elegans: Strain OD5455 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi1786[pOD5259/pJMH80; Pbub-1:bub-1 reencoded S440E, T448E, T527A::bub-1 3'UTR; cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5455 |
| C. elegans: Strain OD5456 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3'UTR; cb-unc-119(+)]I; ltSi23[pOD1011/pJE161; Pknl-1::KNL-1reencoded(Mutant D291-505)::RFP; cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5456 |
| C. elegans: Strain OD5457 ltSi814[pPLG047; Pfzy-1::gfp::fzy-1::fzy-1 3’UTR; cb-unc-119(+)]I; ltSi44[pOD1039/pJE170; Pknl-1::KNL-1reencoded(Mutant D85-505)::RFP; cb-unc-119(+)]II; unc-119(ed3)III?; ltIs37 [pAA64; pie-1/mCHERRY::his-58; unc-119 (+)] IV | This Study | OD5457 |
| C. elegans: Strain PLG19: ccpSi10[pOD5264/pJMH85; Pfzy-1::gfp::fzy-1 T32S::fzy-1 3'UTR; cb-unc-119(+)]II; unc-119(ed3)III | This Study | PLG19 |
| Oligonucleotides | ||
| Primer 1 for synthesis of dsRNA targeting bub-1 (R06C7.8): AATTAACCCTCACTAAAGGCCTCATTGAACTTGGAAACC; Template: N2 Genomic DNA | Moyle et al.35 | N/A |
| Primer 2 for synthesis of dsRNA targeting bub-1 (R06C7.8): AATACGACTCACTATAGGGATCCGAATTGGCACATAA; Template: N2 Genomic DNA | Moyle et al.35 | N/A |
| Primer 1 for synthesis of dsRNA targeting fzy-1 (cdc-20) (ZK177.6) TAATACGACTCACTATAGGCAGCTTCAATTGCATGGAGA; Template: N2 Genomic DNA | Kim and Lara-Gonzalez et al.6 | N/A |
| Primer 2 for synthesis of dsRNA targeting fzy-1 (cdc-20) (ZK177.6): AATTAACCCTCACTAAAGGTGTTCTCACCTTGAATTTCGTG; Template: N2 Genomic DNA | Kim and Lara-Gonzalez et al.6 | N/A |
| Primer 1 for synthesis of dsRNA targeting san-1 (mad-3) (ZC328.4): TAATACGACTCACTATAGGCGAAGAACTTCAAAACCTGGA; Template: N2 Genomic DNA | Essex et al.15 | N/A |
| Primer 2 for synthesis of dsRNA targeting san-1 (mad-3) (ZC328.4): AATTAACCCTCACTAAAGGTTTGTCGGTCCAGATCCTTC; Template: N2 Genomic DNA | Essex et al.15 | N/A |
| Primer 1 for synthesis of dsRNA targeting zyg-1 (F59E12.2): AATTAACCCTCACTAAAGGTGGACGGAAATTCAAACGAT; Template: N2 cDNA | Essex et al.15 | N/A |
| Primer 2 for synthesis of dsRNA targeting zyg-1 (F59E12.2): TAATACGACTCACTATAGGAACGAAATTCCCTTGAGCTG; Template: N2 cDNA | Essex et al.15 | N/A |
| Primer 1 for synthesis of dsRNA targeting perm-1 (T01H3.4): TAATACGACTCACTATAGGAATTTTCTAGGTCGTCAATCTTCA; Template: N2 cDNA | Carvalho et al.40 | N/A |
| Primer 2 for synthesis of dsRNA targeting perm-1 (T01H3.4): AATTAACCCTCACTAAAGGCGAAAACGCGATCATTTTTA; Template: N2 cDNA | Carvalho et al.40 | N/A |
| crRNA sequence for bub-1 (R06C7.8) ABBA motif: CGGAUCAGGAUCACAGAGCA | Lara-Gonzalez et al.33 | N/A |
| crRNA sequence for dpy-10 (T14B4.7): GCUACCAUAGGCACCACGAG | Paix et al.45 | N/A |
| Repair template for bub-1 (R06C7.8) S440A/T448A: TTAGCTACTACGCAGATAGTTACTGACTTTGCAGTGCTCTGTGATCCTGATCCGGCAATGACCATTACACAGGAGCGTCCGAAAAAA | This Study | N/A |
| Repair template for bub-1 (R06C7.8) S440A: TTAGCTACTACGCAGATAGTTACTGACTTTGCAGTGCTCTGTGATCCTGATCCGGCAATGACCATTACACAGGAGCGTCCGAAAAAA | This Study | N/A |
| Repair template for bub-1 (R06C7.8) T448A: AGCTACTACGCAGATAGTTACTGACTTTTCTGTGCTCTGTGATCCTGATCCGGCAATGACCATTACACAGGAGCGTCCGAAAAAA | This Study | N/A |
| Repair template for bub-1 (R06C7.8) ABBAMut (F439A, V441A, L442A, D444A): CTGTCTTCGTTCCAGAAAGATCTTTAGCTACTACGCAGATAGTTACTGACgcTTCtGcGgcCTGTGcTCCTGATCCGACAATGACCATTACACAGGAGCGTCCGAAAAAAGTGTCGAA | Lara-Gonzalez et al.33 | N/A |
| Repair template for bub-1 (R06C7.8) S440E/T448E: TTAGCTACTACGCAGATAGTTACTGACTTTgaaGTGCTCTGTGATCCTGATCCGgaaATGACCATTACACAGGAGCGTCCGAAAAAA | This study | N/A |
| Recombinant DNA | ||
| Plasmid for CDC-20 expression in mammalian cells: pOD4316/pPLG352 [pCDNA 3.1(+) FZY-1::3xFLAG (C. elegans CDC-20)] | Lara-Gonzalez et al. 33 | pOD4316 |
| Plasmid for GST tagged BUB-1 fragment expression in E. coli: pOD4314/pPLG350 [pGEX-6P-1 GST::BUB-1 (280-472)] | Lara-Gonzalez et al.33 | pOD4314 |
| Plasmid for GST tagged BUB-1 ABBAmut fragment expression in E. coli: pOD4374/pPLG410 [pGEX-6P-1 GST::BUB-1 (280-472) ABBAMut] | Lara-Gonzalez et al.33 | pOD4374 |
| Plasmid for GST tagged BUB-1 S440A/T448A fragment expression in E. coli: pOD5239/pJMH75 [pGEX-6P-1 GST::BUB-1 (280-472) S440A/T448A] | This Study | pOD5239 |
| Plasmid for GST tagged BUB-1 S440A fragment expression in E. coli: pOD5236/pJMH72 [pGEX-6P-1 GST::BUB-1 (280-472) S440A] | This Study | pOD5236 |
| Software and algorithms | ||
| Fiji | Schindelin et al.50 | SCR_002285 |
| Prism | GraphPad | SCR_002798 |
| Adobe Illustrator | Adobe | SCR_010279 |
| Pymol | DeLano et al.49 | SCR_000305 |
| GPS Polo 1.0 | Xue et al.46 | SCR_016374 |
Method Details
Worm Strain Construction:
RNAi-resistant bub-1 and cdc-20 transgenes were previously described 6,35. Transgenes were cloned into pCFJ151 or pCFJ352 and injected into strains EG6429 or EG6701, respectively, along with constructs encoding for the Mos transposase and a mix of four negative selection markers against extrachromosomal plasmid arrays. One week after injection, integrants were selected on the basis of their rescuing of the unc phenotype of the parental strains and their lack of negative selection markers, and further confirmed by PCR.
RNAi:
dsRNAs were synthesized in vitro. Briefly, target sequences were amplified by PCR using oligos containing either T3 or T7 promoters at their 5’ end. Complementary RNAs were synthesized from the PCR templates using T3 or T7 RNA polymerases (MEGAscript T3 or T7 kit, ThermoFisher). Single-stranded RNA products were then purified using MEGAclear kit (ThermoFisher) and annealed by incubation at 68°C for 10 minutes followed by 37°C for 30 minutes. Resulting dsRNA were measured by A260nm using a nano-drop spectrophotometer.
For RNAi based depletions, L4 stage worms were injected with 0.8-1 mg/mL of dsRNA and incubated at 20°C for 36-48 hours before imaging-based experiments. For embryonic viability assays, L4 stage worms were injected with dsRNA and recovered at 20°C for 24 hours. Injected worms were singled and allowed to lay progeny for 24 hours at 20°C. Parental worms were then removed and after further 24 hr, progeny was scored as either viable or dead.
CRISPR-Cas9:
CRISPR-Cas9 Ribonucleoproteins (RNPs) were used for in situ mutagenesis 45. Target sequences were identified using CHOPCHOP (https://chopchop.cbu.uib.no/). Purified Cas9 protein was purchased from MacroLab. Repair oligos were ordered from Eton Biosciences as HPLC-purified products. TracrRNA and crRNAs were ordered from IDT.
A co-CRISPR strategy was used to facilitate the identification of edits. For this, a guide RNA mix was created using 100 μM tracrRNA, 25 μM dpy-10 crRNA, and 75 μM targeting crRNA. RNAs were annealed by incubation at 95 °C for 5 minutes, followed by hybridization at 22 °C for 5 minutes. Then, 2 μL of the RNA mix were mixed with 5 μL of 40 μM Cas9 protein and incubated at 22 °C for 5 minutes to generate RNP complexes. A repair oligo was added to the mix to a final concentration of 5.4 μM and the resulting mix was injected in the germline of adult worms. After 4-5 days, between 24-30 F1 progeny from plates with high proportion of dpy-10 mutant worms (“jackpot” broods) were singled out, and edits were screened through PCR-based amplification of the allele, followed by sanger sequencing. Edited strains were outcrossed 1 time to the parental strain to remove the dpy-10 mutated allele.
Fluorescence imaging of C. elegans embryos:
Gravid adult hermaphrodite worms were dissected in M9 buffer, early embryos were transferred onto a 2% agarose pad using a mouth pipet, covered with a 22x22 mm coverslip, and imaged at 20°C. For kinetochore localization assays, embryos were imaged on an Andor Revolution confocal system (Andor) coupled to a CSU-10 spinning disk confocal scanner (Yokogawa) and an electron multiplication back-thinned charge-coupled device camera (iXon, Andor), using either a 60x or 100x 1.4 NA Plan Apochromat objective; or using an inverted Axio Observer Z1 (Zeiss) microscope with a CSU-X1 spinning disk head (Yokogawa) and a 100x 1.3 NA Plan Apochromat Lens (Zeiss). A 6x2 μm z-stack was collected every 10 or 20 seconds.
For mitotic timing experiments, timelapse imaging of one cell embryos expressing GFP::H2b or mCherry::H2b was performed on a widefield deconvolution microscope (DeltaVision) connected to a charge-coupled device camera (pco.edge 5.5 sCMOS; PCO) and a 60x 1.42NA PlanApo N objective (Olympus). A 5x2 μm z-stack was collected every 10 or 20 seconds.
For imaging of GFP::MAD-3 in oocytes, a 10x1 μm z-stack was taken on a widefield deconvolution microscope (DeltaVision) connected to a charge-coupled device camera (pco.edge 5.5 sCMOS; PCO) and a 60x 1.42NA PlanApo N objective (Olympus).
PLK-1 inhibition assays:
The PLK-1 analog-sensitive (AS) allele (C52V and L115G) was previously described 39. For PLK-1 inhibition experiments, L4 stage worms were injected with perm-1 dsRNA to prevent formation of the drug-impermeable eggshell and incubated at 20 °C for 16-18 hours before imaging. Worms were dissected in custom made imaging chamber slides and embryos deposited into built-in wells. Embryos were monitored by DIC imaging, and 1NM-PP1 was added to the well to a final concentration of 20 μM after NEBD. Embryos were imaged every 20 seconds using 8x2 μm z stacks using DIC, RFP, and GFP channels. Embryo permeability was assessed at each imaging session by addition of lipophilic dye FM4-64, which binds to the plasma membrane of permeabilized but not impermeable embryos 39.
Protein Expression and Purification:
GST tagged BUB-1 (280-472) wildtype and mutant constructs were cloned into pGEX-6P-1 vectors and transformed into Bl21(DE3)pLys E. coli cells. Cultures were grown at an OD600 of 0.4-6 and induced overnight at 20°C with 0.4 mM IPTG. The next morning, cells were collected by centrifugation and re-suspended with lysis buffer (25 mM HEPES pH 7.5, 300 mM NaCl, 1 mM MgCl2, 0.1% Triton X-100, 10 mM βME), supplemented with 1 mM PMSF, complete EDTA protease inhibitors (Roche) and 1 mg/mL Lysozyme. Cells were lysed by sonication, and lysates were cleared by ultracentrifugation for 30 minutes at 40,000 rpm at 4°C in a 45Ti rotor. GST-tagged proteins were purified from the cleared lysates using glutathione-sepharose 4B beads (Cytiva), and incubated with end-over-end rotation for 2 hours at 4°C. Beads were washed 3 times with lysis buffer and eluted with 10 mM reduced glutathione in 50 mM Tris-HCl pH 8.0; 150 mM NaCl. Fractions containing the most protein were combined and dialyzed against storage buffer (25 mM HEPES pH 7.5, 200 mM NaCl, 1 mM MgCl2, 0.1% Triton X-100, 10 mM βME, 5% glycerol). Proteins were aliquoted, snap frozen in liquid nitrogen and stored at −80°C.
In vitro binding assays:
A construct encoding for CDC-20::3xFLAG under a CMV promoter was transfected into Freestyle 293-F cells (Thermo Fisher Scientific). 48 hours later, cells were harvested and washed with PBS before re-suspending in lysis buffer (20 mM Tris-Cl pH 7.5, 50 mM NaCl, 5 mM EGTA, 2 mM MgCl2, 0.5% Triton X-100, 1 mM DTT), supplemented with 1x complete Protease inhibitor cocktail (Millipore). Cells were sonicated for 6 minutes in an ice-cold water bath, then centrifuged for 15 minutes at 15,000xg at 4°C. Whole-cell lysates were then incubated with Anti-FLAG beads (Sigma) for 2 hours at 4°C and beads were washed four times with lysis buffer.
Meanwhile, 2 μM GST::BUB-1 (280-472) was phosphorylated with 500 nM constitutively active PLK-1 T194D (gift from Jeffrey Woodruff, UT Southwestern) in kinase buffer (20 mM Tris-Cl pH 7.5, 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, 0.2 mM ATP), for 1 hour at 23°C. Phosphorylated GST::BUB-1 was then directly added to CDC-20::3xFLAG bound to beads to a final GST::BUB-1 concentration of 100 nM. Binding was performed for 2 hours at 4°C, after which beads were washed with lysis buffer 4 times. Proteins were eluted from the beads by resuspending in SDS sample buffer before analysis by immunoblotting.
Preparation of worm lysates:
L4-stage worms were injected with dsRNA at a concentration of 1 μg/μL and incubated at 20°C for 36-48 hours. 60 worms per condition were washed with M9 plus 0.1% Tween 20, resuspended in SDS-sample buffer and lysed by sonication followed by boiling.
Immunoblotting:
For binding assays, samples were run on 4-15% gradient Mini-Protean gels (BioRad) and transferred to PVDF membranes using a TransBlot Turbo system (BioRad). Membranes were blocked for 1 hour in TBS-Tween 0.05% (TBS-T) with 5% skim milk and antibodies were incubated in either the same buffer or in 5% BSA. Membranes were developed using Super Signal West Femto Max Sensitivity Substrate (ThermoFisher) before imaging on ChemiDoc system (BioRad). For RNAi blots, the equivalent of 5-10 adult worms was loaded into 4-12% NuPAGE Bis-Tris gels (Invitrogen). Proteins were transferred overnight to nitrocellulose membranes, after which membranes were blocked and antibodies incubated in TBS-T with 5% milk. Membranes were developed using Western Bright Sirius substrate (Advansta) before imaging on ChemiDoc System (BioRad).
Screen for Putative PLK-1 sites:
Putative PLK-1 phosphorylation sites in C. elegans BUB-1 were identified using GPS Polo 1.0 46, with filter set to “all”. Sites were prioritized by comparing conservation with other closely related Caenorhabditis species, matching consensus polo motif [N/D/E/Y, X, S/T, F], and a Polo GPS score above 1.5. Sequences of Caenorhabditis bub-1 genes were aligned using MAFFT. The full list of sites is available in Supplemental Table 1.
AlphaFold Structure Prediction:
Structure prediction for CDC-20 (residues range: 136-507) and BUB-1 (residues range: 424-458) ABBA motif interaction was performed using AlphaFold2 ColabFold notebook 47,48. Structural analysis and depiction were carried out in PyMol 49.
Imaging analysis and quantifications:
Microscope images were processed using Fiji/ImageJ 50. NEBD was defined as the time point where free nuclear histone signal dissipates into the cytosol and spindle forces start pushing on chromosomes. Anaphase was the first time point where the separation of sister chromatids was evident. DNA decondensation was the time point where chromosome signals become dimmer and their area starts expanding.
Kinetochore intensities were measured by drawing a circle around the chromosome area (mCherry::H2b) and transferred to the GFP channel. After recording integrated intensity, the circle was expanded by 3 pixels to calculate local background signal.
For PLK-1 inhibition assays, kinetochore signals were quantified 60 seconds before anaphase onset. A 10 pixel wide linescan was drawn across the chromosomes and signal intensities were quantified as described before 35. Imaging depth variations in signal intensities were corrected by dividing by the mCherry::H2b signal.
For quantification of GFP::MAD-3 in germline oocytes, a box was drawn around the oocyte, and the integrated density was recorded for each frame in a 10 μm z-stack. Background signal was measured for an identical sized box outside the worm and subtracted from the germline signal. A worm strain without GFP::MAD-3 was imaged to estimate autofluorescence (Fig S1C).
Quantification and Statistical Analysis:
All graphs shown in the manuscript were created and analyzed in Prism (Graphpad). P values were calculated in Prism software (Graphpad) using unpaired t tests. P values are displayed as follows: ns = p>0.05; * = p<0.05; ** = p<0.01; *** = p<0.001; **** = p < 0.0001. Error bars display the 95% confidence interval.
Supplementary Material
HIGHLIGHTS.
An optimal mitotic duration is important for robust C. elegans embryogenesis
Activation of CDC-20 at kinetochores controls embryonic mitotic duration
Kinetochore recruitment of CDC-20 by BUB-1 requires BUB-1–bound PLK-1
Phosphorylation of BUB-1 by PLK-1 enhances binding and recruitment of CDC-20
ACKNOWLEDGMENTS
We thank members of the Oegema and Desai labs for their support, Federico Pelisch for sharing unpublished work and helpful discussions, Jeffrey Woodruff for the gift of constitutively active PLK-1, Aleesa Schlientz for comments on the manuscript, and Becky Green for input on the model figure. This work was supported by a grant from the NIH to A.D. (R01 GM074215), and a grant from the NIH to K.D.C. (R35 GM144121). T.K. was supported by a grant from the National Research Foundation of Korea (No. NRF-2020RC1C1008596). J. H. was supported by an NSF Graduate Research Fellowship (grant #1650112). A.D. and K.O. acknowledge salary support from the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing financial interests.
REFERENCES:
- 1.Musacchio A, and Desai A (2017). A Molecular View of Kinetochore Assembly and Function. Biology (Basel) 6. 10.3390/biology6010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheeseman IM (2014). The kinetochore. Cold Spring Harb Perspect Biol 6, a015826. 10.1101/cshperspect.a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lara-Gonzalez P, Pines J, and Desai A (2021). Spindle assembly checkpoint activation and silencing at kinetochores. Semin Cell Dev Biol 117, 86–98. 10.1016/j.semcdb.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lara-Gonzalez P, Kim T, and Desai A (2017). Taming the Beast: Control of APC/C(Cdc20)-Dependent Destruction. Cold Spring Harb Symp Quant Biol 82, 111–121. 10.1101/sqb.2017.82.033712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Musacchio A (2015). The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr Biol 25, R1002–1018. 10.1016/j.cub.2015.08.051. [DOI] [PubMed] [Google Scholar]
- 6.Kim T, Lara-Gonzalez P, Prevo B, Meitinger F, Cheerambathur DK, Oegema K, and Desai A (2017). Kinetochores accelerate or delay APC/C activation by directing Cdc20 to opposing fates. Genes Dev 31, 1089–1094. 10.1101/gad.302067.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clute P, and Masui Y (1995). Regulation of the appearance of division asynchrony and microtubule-dependent chromosome cycles in Xenopus laevis embryos. Dev Biol 171, 273–285. 10.1006/dbio.1995.1280. [DOI] [PubMed] [Google Scholar]
- 8.Duro J, and Nilsson J (2021). SAC during early cell divisions: Sacrificing fidelity over timely division, regulated differently across organisms: Chromosome alignment and segregation are left unsupervised from the onset of development until checkpoint activity is acquired, varying from species to species. Bioessays 43, e2000174. 10.1002/bies.202000174. [DOI] [PubMed] [Google Scholar]
- 9.Gerhart J, Wu M, and Kirschner M (1984). Cell cycle dynamics of an M-phase-specific cytoplasmic factor in Xenopus laevis oocytes and eggs. J Cell Biol 98, 1247–1255. 10.1083/jcb.98.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang M, Kothari P, and Lampson MA (2015). Spindle assembly checkpoint acquisition at the mid-blastula transition. PLoS One 10, e0119285. 10.1371/journal.pone.0119285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diaz-Martinez LA, Tian W, Li B, Warrington R, Jia L, Brautigam CA, Luo X, and Yu H (2015). The Cdc20-binding Phe box of the spindle checkpoint protein BubR1 maintains the mitotic checkpoint complex during mitosis. J Biol Chem 290, 2431–2443. 10.1074/jbc.M114.616490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.He J, Chao WC, Zhang Z, Yang J, Cronin N, and Barford D (2013). Insights into degron recognition by APC/C coactivators from the structure of an Acm1-Cdh1 complex. Mol Cell 50, 649–660. 10.1016/j.molcel.2013.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Fiore B, Davey NE, Hagting A, Izawa D, Mansfeld J, Gibson TJ, and Pines J (2015). The ABBA motif binds APC/C activators and is shared by APC/C substrates and regulators. Dev Cell 32, 358–372. 10.1016/j.devcel.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi W, Tang Z, and Yu H (2006). Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol Biol Cell 17, 3705–3716. 10.1091/mbc.e06-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Essex A, Dammermann A, Lewellyn L, Oegema K, and Desai A (2009). Systematic analysis in Caenorhabditis elegans reveals that the spindle checkpoint is composed of two largely independent branches. Mol Biol Cell 20, 1252–1267. 10.1091/mbc.E08-10-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alfieri C, Zhang S, and Barford D (2017). Visualizing the complex functions and mechanisms of the anaphase promoting complex/cyclosome (APC/C). Open Biol 7. 10.1098/rsob.170204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kramer ER, Scheuringer N, Podtelejnikov V, Mann M, and Peters JM (2000). Mitotic regulation of the APC activator proteins CDC20 and CDH1. Molecular Biology of the Cell 11, 1555–1569. DOI 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yudkovsky Y, Shteinberg M, Listovsky T, Brandeis M, and Hershko A (2000). Phosphorylation of Cdc20/Fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem Bioph Res Co 271, 299–304. DOI 10.1006/bbrc.2000.2622. [DOI] [PubMed] [Google Scholar]
- 19.Labit H, Fujimitsu K, Bayin NS, Takaki T, Gannon J, and Yamano H (2012). Dephosphorylation of Cdc20 is required for its C-box-dependent activation of the APC/C. Embo Journal 31, 3351–3362. 10.1038/emboj.2012.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bancroft J, Holder J, Geraghty Z, Alfonso-Perez T, Murphy D, Barr FA, and Gruneberg U (2020). PP1 promotes cyclin B destruction and the metaphase-anaphase transition by dephosphorylating CDC20. Mol Biol Cell 31, 2315–2330. 10.1091/mbc.E20-04-0252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hein JB, Garvanska DH, Nasa I, Kettenbach AN, and Nilsson J (2021). Coupling of Cdc20 inhibition and activation by BubR1. J Cell Biol 220. 10.1083/jcb.202012081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hein JB, and Nilsson J (2016). Interphase APC/C-Cdc20 inhibition by cyclin A2-Cdk2 ensures efficient mitotic entry. Nat Commun 7, 10975. 10.1038/ncomms10975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevah-Sitry D, Miniowitz-Shemtov S, Teichner A, Kaisari S, and Hershko A (2022). Role of phosphorylation of Cdc20 in the regulation of the action of APC/C in mitosis. Proc Natl Acad Sci U S A 119, e2210367119. 10.1073/pnas.2210367119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hein JB, Hertz EPT, Garvanska DH, Kruse T, and Nilsson J (2017). Distinct kinetics of serine and threonine dephosphorylation are essential for mitosis. Nat Cell Biol 19, 1433–1440. 10.1038/ncb3634. [DOI] [PubMed] [Google Scholar]
- 25.Cundell MJ, Hutter LH, Nunes Bastos R, Poser E, Holder J, Mohammed S, Novak B, and Barr FA (2016). A PP2A-B55 recognition signal controls substrate dephosphorylation kinetics during mitotic exit. J Cell Biol 214, 539–554. 10.1083/jcb.201606033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rappleye CA, Tagawa A, Lyczak R, Bowerman B, and Aroian RV (2002). The anaphase-promoting complex and separin are required for embryonic anterior-posterior axis formation. Dev Cell 2, 195–206. 10.1016/s1534-5807(02)00114-4. [DOI] [PubMed] [Google Scholar]
- 27.Golden A, Sadler PL, Wallenfang MR, Schumacher JM, Hamill DR, Bates G, Bowerman B, Seydoux G, and Shakes DC (2000). Metaphase to anaphase (mat) transition-defective mutants in Caenorhabditis elegans. J Cell Biol 151, 1469–1482. 10.1083/jcb.151.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Espeut J, Cheerambathur DK, Krenning L, Oegema K, and Desai A (2012). Microtubule binding by KNL-1 contributes to spindle checkpoint silencing at the kinetochore. Journal of Cell Biology 196, 469–482. 10.1083/jcb.201111107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh P, Pesenti ME, Maffini S, Carmignani S, Hedtfeld M, Petrovic A, Srinivasamani A, Bange T, and Musacchio A (2021). BUB1 and CENP-U, Primed by CDK1, Are the Main PLK1 Kinetochore Receptors in Mitosis. Mol Cell 81, 67–87 e69. 10.1016/j.molcel.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nguyen AL, Fadel MD, and Cheeseman IM (2021). Differential requirements for the CENP-O complex reveal parallel PLK1 kinetochore recruitment pathways. Mol Biol Cell 32, 712–721. 10.1091/mbc.E20-11-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor SJP, Bel Borja L, Soubigou F, Cheerambathur D, and Pelisch F (2022). BUB-1 and CENP-C recruit PLK-1 to Control Chromosome Alignment and Segregation During Meiosis I in C. elegans Oocytes BioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Piano V, Alex A, Stege P, Maffini S, Stoppiello GA, Veld PJHI, Vetter IR, and Musacchio A (2021). CDC20 assists its catalytic incorporation in the mitotic checkpoint complex. Science 371, 67–71. 10.1126/science.abc1152. [DOI] [PubMed] [Google Scholar]
- 33.Lara-Gonzalez P, Kim T, Oegema K, Corbett K, and Desai A (2021). A tripartite mechanism catalyzes Mad2-Cdc20 assembly at unattached kinetochores. Science 371, 64-+. 10.1126/science.abc1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lera RF, Potts GK, Suzuki A, Johnson JM, Salmon ED, Coon JJ, and Burkard ME (2016). Decoding Polo-like kinase 1 signaling along the kinetochore-centromere axis. Nat Chem Biol 12, 411–418. 10.1038/nchembio.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moyle MW, Kim T, Hattersley N, Espeut J, Cheerambathur DK, Oegema K, and Desai A (2014). A Bub1-Mad1 interaction targets the Mad1-Mad2 complex to unattached kinetochores to initiate the spindle checkpoint. J Cell Biol 204, 647–657. 10.1083/jcb.201311015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Espeut J, Lara-Gonzalez P, Sassine M, Shiau AK, Desai A, and Abrieu A (2015). Natural Loss of Mps1 Kinase in Nematodes Uncovers a Role for Polo-like Kinase 1 in Spindle Checkpoint Initiation. Cell Rep 12, 58–65. 10.1016/j.celrep.2015.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim T, Moyle MW, Lara-Gonzalez P, De Groot C, Oegema K, and Desai A (2015). Kinetochore-localized BUB-1/BUB-3 complex promotes anaphase onset in C. elegans. J Cell Biol 209, 507–517. 10.1083/jcb.201412035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chase D, Serafinas C, Ashcroft N, Kosinski M, Longo D, Ferris DK, and Golden A (2000). The polo-like kinase PLK-1 is required for nuclear envelope breakdown and the completion of meiosis in Caenorhabditis elegans. Genesis 26, 26–41. . [DOI] [PubMed] [Google Scholar]
- 39.Gomez-Cavazos JS, Lee KY, Lara-Gonzalez P, Li Y, Desai A, Shiau AK, and Oegema K (2020). A Non-canonical BRCT-Phosphopeptide Recognition Mechanism Underlies RhoA Activation in Cytokinesis. Curr Biol 30, 3101–3115 e3111. 10.1016/j.cub.2020.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carvalho A, Olson SK, Gutierrez E, Zhang K, Noble LB, Zanin E, Desai A, Groisman A, and Oegema K (2011). Acute drug treatment in the early C. elegans embryo. PLoS One 6, e24656. 10.1371/journal.pone.0024656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohta M, Zhao Z, Wu D, Wang S, Harrison JL, Gomez-Cavazos JS, Desai A, and Oegema KF (2021). Polo-like kinase 1 independently controls microtubule-nucleating capacity and size of the centrosome. J Cell Biol 220. 10.1083/jcb.202009083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang JS, Yang MJ, Li B, Qi W, Zhang C, Shokat KM, Tomchick DR, Machius M, and Yu HT (2008). Structure and Substrate Recruitment of the Human Spindle Checkpoint Kinase Bub1. Molecular Cell 32, 394–405. 10.1016/j.molcel.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans R, O’Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, et al. (2022). Protein complex prediction with AlphaFold-Multimer. BioRxiv. 10.1101/2021.10.04.463034 [DOI] [Google Scholar]
- 44.Burton JL, Xiong Y, and Solomon MJ (2011). Mechanisms of pseudosubstrate inhibition of the anaphase promoting complex by Acm1. EMBO J 30, 1818–1829. 10.1038/emboj.2011.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paix A, Folkmann A, and Seydoux G (2017). Precision genome editing using CRISPR-Cas9 and linear repair templates in C. elegans. Methods 121–122, 86–93. 10.1016/j.ymeth.2017.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, and Yao X (2005). GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res 33, W184–187. 10.1093/nar/gki393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, and Steinegger M (2022). ColabFold: making protein folding accessible to all. Nat Methods 19, 679–682. 10.1038/s41592-022-01488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeLano WL (2002). PyMOL: An Open-Source Molecular Graphics Tool. CCP4 Newsletter on Protein Crystallography 40, 44–53. [Google Scholar]
- 50.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dammermann A, Maddox PS, Desai A, and Oegema K (2008). SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J Cell Biol 180, 771–785. 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oegema K, Desai A, Rybina S, Kirkham M, and Hyman AA (2001). Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 153, 1209–1226. 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McNally K, Audhya A, Oegema K, and McNally FJ (2006). Katanin controls mitotic and meiotic spindle length. J Cell Biol 175, 881–891. 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to re-analyze the data reported in this paper are available from the lead contact upon request.