Abstract
In principle, the steady-state concentrations of biomolecules in complex systems can be far from the thermodynamic equilibrium concentrations of individual processes. This means that, in addition to thermodynamics, reaction kinetics may play an important role. This view is not fully reflected in combinatorial studies in biochemistry that focus on the selection of stably interacting molecules reflected by high equilibrium constants. For kinetically controlled processes in vivo, forward or backward reaction rates are critical but not necessarily an equilibrium state. Here we have studied the control of antisense RNA-mediated gene suppression in human cells on a general basis and in a way that excludes individual structure-specific influences. The complete antisense sequence space against the chloramphenicol acetyltransferase gene (cat) was generated and a kinetic selection technique was established to enrich for fast annealing antisense species. Selected sub-populations showed successively faster annealing which was related to increased inhibition of cat gene expression in HeLa cells, providing strong evidence for the view that the suppression of gene expression by antisense RNA is controlled kinetically regardless of specific RNA structures.
INTRODUCTION
In living cells most metabolites and gene products are thought to exist in a steady state which seems to be crucial for their biological functions. Their steady-state concentrations are the result of a variety of anabolic and catabolic processes and can be far from the thermodynamic equilibrium concentrations. Thus, the action of a given compound in living cells might be controlled thermodynamically and/or kinetically. However, this situation is not always adequately reflected by biochemical studies in which SELEX procedures and phage display techniques are used to identify high affinity interactions, represented by the equilibrium constant Keq, between nucleic acids or peptides and other types of biologically relevant molecules such as cofactors (1,2), amino acids (3) or proteins (4). Equilibrium selection is based on the assumption of a thermodynamic reaction control. However, looking at kinetically controlled processes in living cells would require a kinetic selection procedure in order to identify biologically active molecules. Evidence for kinetic control in vivo has been reported for important biological processes such as mitochondrial ATP synthesis (5), antibody–antigen interactions (6), 5S RNA gene transcription (7) and individual cases of antisense RNA-mediated gene regulation in bacteria (8–11) and mammalian cells (12). So far, SELEX-like kinetic in vitro selection techniques have not been developed, though some kinetic selection pressure could have occurred in conventional SELEX by increasing the stringency of selection via shortened incubation times or decreased concentrations (13).
Here we have investigated the control of antisense RNA-mediated gene suppression in mammalian cells on a more general level using a kinetic in vitro selection technique and by including a complete space of consecutive antisense sequences. In the case of antisense RNA-mediated regulation of gene expression, RNA structures crucially influence the efficiency of RNA–RNA double-strand formation in vitro (8,14,15) as well as effectiveness in cells (16,17). In specific cases, some light has been shed on the complex relationship between RNA structure, the kinetics of RNA–RNA annealing in vitro and efficacy in living cells (for reviews see refs 8,12). However, to exclude specific structural influences and to directly relate annealing kinetics and efficacy to each other one has to average over other individual properties of antisense RNA that affect inhibition. In this work we have used the complete antisense RNA sequence space directed against the chloramphenicol acetyltransferase (cat) mRNA and performed selection such that after several cycles of kinetic selection for fast RNA–RNA annealing the number of active species was still large enough to average over their individual characteristics, including their individual structures. Successively faster RNA–RNA annealing in vitro was related to stepwise increased inhibition of cat gene expression in human cells, providing experimental evidence for kinetic control of the action of antisense RNA in mammalian cells.
MATERIALS AND METHODS
Generation of a complete cat-directed antisense sequence space
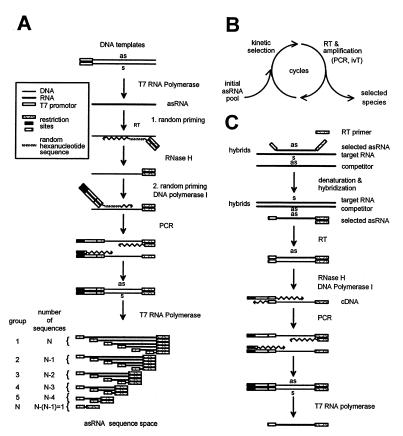
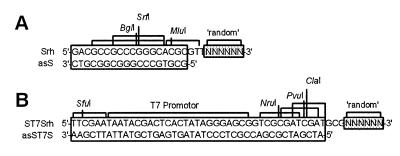
Generation of the complete space of consecutive cat-directed antisense sequences is based on two subsequent steps of random priming followed by primer extension (Fig. 1). It starts with in vitro transcription of RNA corresponding to the sequence of interest. First, random priming and cDNA synthesis were performed using an antisense RNA complementary to the full-length cat target mRNA (Fig. 1A). Aliquots of 100 pmol of this RNA of L nucleotides length (here 850 nt) were denatured at 75°C for 5 min, placed on ice and 4096/L × 100 pmol (here 482 pmol) of the first random primer (Srh) were added. This primer consisted of a random hexanucleotide priming site at its 3′-end and recognition sites for restriction endonucleases (Fig. 2A). The template:primer ratio was chosen such that statistically one primer annealed with one RNA molecule to yield successively shortened 5′-ends of the first cDNA strand. The ratio 4096/L takes into consideration the fact that for target sequences shorter than 4096 nt statistically only a part of the theoretically 4096 random hexanucleotide sequences will find a complementary sequence on the target molecule. Reverse transcription was performed for 2 h at 37°C in a volume of 60 µl using SuperScript Plus RNase H– reverse transcriptase (SSRT; Gibco BRL, Karlsruhe, Germany). Non-primed RNA was degraded by treatment with RNases T1 and VI for 30 min prior to elimination of unbound first random primer with PrimeErase Quick Push Columns (Stratagene, La Jolla, CA). The RNA/DNA hybrids were then treated with RNase H (Boehringer, Mannheim, Germany) for 30 min at 37°C. Second strand cDNA synthesis was performed with DNA polymerase I in the presence of 50 pmol of the second random primer (ST7Srh) to yield successively 5′ shortened second cDNA strands. The primer was annealed for 30 min at 16°C and the reaction was then incubated for 2 h at 37°C. The second random primer included a random hexanucleotide priming site, the T7 promoter and restriction sites (Fig. 2B). To avoid priming via other parts than the random hexanucleotide priming sites, both random primers were prehybridised before use with the antisense primers asS and asST7S, respectively (Fig. 2). These antisense primers were complementary to the random primer sequences except for the random hexanucleotide stretch and the three intermediate spacer nucleotides. The second cDNA strands were amplified by PCR to yield a pool of templates for the generation of variant antisense RNAs by in vitro transcription. Prior to all enzymatic reactions nucleic acids were extracted by phenol and precipitated with ethanol.
Figure 1.
Schematic depiction of a kinetic in vitro selection procedure for the identification of fast annealing antisense RNA species. (A) Schematic depiction of the generation of DNA templates for transcription of a complete antisense RNA sequence space via (i) in vitro transcription of the relevant sequence, (ii) two subsequent steps of random priming, (iii) amplification of templates and (iv) in vitro transcription of antisense RNA species. This method leads to N groups of antisense molecules consisting of sequences with identical 3′-ends with N equal to the length of the target RNA in nucleotides. Each of these two groups contain together L + 1 different sequences yielding a total sequence diversity of L/2(L + 1). (B) Kinetic selection cycle. (C) Amplification step of selected species.
Figure 2.
Specific design of the random primers used: (A) first random primer; (B) second random primer (see text and Fig. 1). Both random primers were prehybridised with antisense primers to favour priming by the random hexanucleotide sites.
Selection of fast hybridising antisense RNA
In principle, the kinetic selection step was performed as described previously (16). A [32P]uridine-labelled population of antisense sequences was synthesised by in vitro transcription using the cDNA library generated as described above. Fifty nanograms of this antisense sequence pool were incubated with a 10-fold molar excess of cat sense RNA in a hybridisation buffer containing 100 mM NaCl, 20 mM Tris–HCl, pH 7.4, and 10 mM MgCl2 at 37°C. The concentration of the target RNA was chosen such that ~5% of the antisense species annealed within the first 3 min (in the first round of selection). The incubation time was 2 min in the second and third rounds and 1 min in the fourth round of selection. After 1–3 min the hybridisation reaction was stopped by adding 10 vol of stop buffer (50 mM Tris–HCl, pH 8, 25 mM EDTA, 0.5% SDS, 7 M urea, bromophenol blue and xylene cyanol) precooled on ice. Single-stranded antisense RNA and target RNA/antisense RNA duplexes were seperated by native agarose gel electrophoresis, the duplex-containing gel slices were excised, the duplexes were isolated and the selected antisense species were amplified as described below.
Amplification of selected species
Hybrids of selected antisense RNA and target RNA were heat denatured (97°C for 3 min) in the presence of a 10-fold molar excess of full-length antisense cat RNA over the target RNA strands. Selected species were reverse transcribed (Superscript reverse transcriptase; Gibco BRL) and cDNA was amplified by PCR prior to RNA in vitro transcription.
Determination of annealing rate constants for complementary RNA
Observed association rate constants (kobs) were measured as described in detail (15). Briefly, radioactively labelled antisense RNA (2.5 nM final concentration) was incubated with the 850 nt long cat target RNA at 25 or 100 nM final concentration in hybridisation buffer (see above) at 37°C. Aliquots were withdrawn at different time points, transferred into 10 vol of precooled stop buffer (see above) and analysed by native polyacrylamide gel electrophoresis. Gels were dried, exposed to X-ray film and band intensities were determined using a phosphorimager (Molecular Dynamics, Sunnyvale, CA). Second order association rate constants were calculated as described (15).
Inhibition of cat gene expression in human cells
Aliquots of 300 ng/µl pool RNA and 30 ng/µl cat expression plasmid pRC-CMV-CAT (18) were co-microinjected into the nuclei of 300 HeLa cells. Expression of the cat gene was monitored 6 h post-injection.
RESULTS AND DISCUSSION
Generation of the complete antisense sequence space
To generate the complete antisense RNA sequence space against cat, DNA templates for in vitro transcription of antisense RNA were produced from a cat-directed full-length antisense RNA via two steps of randomly primed cDNA synthesis (Fig. 1A). The maximal theoretical diversity D of such created pools is given by the length L of the RNA target sequence according to the equation D = L/2 × (L + 1), which relates to ~3.6 × 105 species for the 850 nt long cat target sequence used here. Antisense species vary in length between only a few nucleotides and the full-length antisense RNA. In the subsequent selection cycles, however, antisense sequences shorter than ~14 nt will not be included for technical reasons (data not shown). This method allows the generation of genotypically defined sequence spaces on the level of RNA, cDNA and genomic sequences according to the sequence input.
In vitro selection for fast RNA–RNA annealing
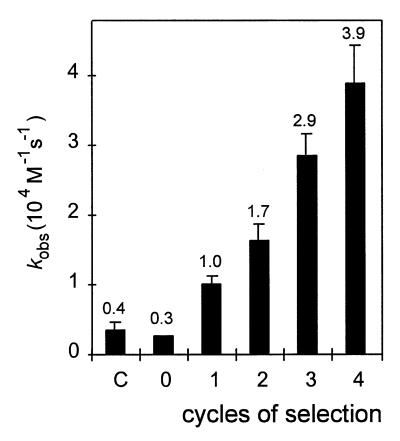
The initial pool of antisense species was incubated together with cat target RNA. Aliquots of the annealing reaction were fractionated into fast and slow hybridising species by gel electrophoretic separation of the single- and double-stranded fractions at early time points during the annealing reaction as described previously (16,19). The fast hybridising antisense molecules were contained in the double-stranded fraction. They were used as templates for cDNA synthesis, amplified by PCR and transcribed in vitro to yield a pool of faster annealing RNA species (Fig. 1B and C). Successive cycles of kinetic selection yielded progressively faster annealing subpopulations of antisense species with ~13-fold increased mean annealing rates after four cylces (Fig. 3).
Figure 3.
Observed mean annealing rate constants of pools of cat-directed antisense RNA species as a function of the number of cycles of selection and amplification. The numbers represent mean values of at least three independent measurements. The error bars represent the standard deviation. C, not selected control, four times reverse transcribed and PCR amplified; 0, initial pool; 1–4, one to four times selected species.
After the first round of selection and amplification the composition of the pool RNA shifts towards longer chain lengths due to the loss of RNA species smaller than ~50–60 nt (Fig. 4). The explanation for this phenomenon is technical in nature. The initial pool contains a large number of RNA species that cannot hybridise with the target RNA. One has to consider that the length of primer sequences of these species is 43 nt and that a consecutive stretch of ~14 nt is necessary for annealing under the experimental conditions used here. Thus, species of 50–60 nt in length would not be expected to anneal to the target even if they contain short antisense portions. Most of these species are apparently lost after the first selection cycle. The size fractions in each lane of the autoradiograph shown in Figure 4, including the prominent one above the 43 nt size markers, were quantified by phosphorimager analysis. When standardised to the total amount of RNA in each lane, the percentage of the 43–55 nt fraction decreased successively from ~42% in the starting pool (lane 0 in Fig. 4) to levels of 28 (lane 1), 19 (lane 2), 21 (lane 3) and 17% (lane 4). It should also be noted that the stringency of selection was stepwise increased by shortening the incubation time of the annealing reaction from 3 min in the first cycle to 1 min in the fourth cycle, which might have influenced the composition of the selected pools of antisense RNA, particularly the fourth one.
Figure 4.
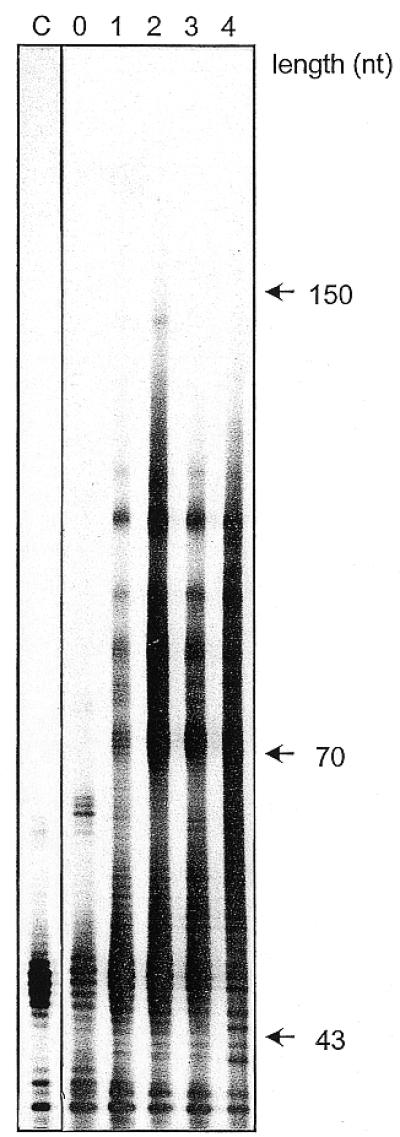
Chain length composition of the antisense RNA pools as a function of the number of cycles of selection and amplification. C, not selected control, four times reverse transcribed and PCR amplified; 0, initial pool; 1–4, one to four times selected species. Antisense RNA samples were analysed by polyacrylamide gel electrophoresis under denaturing conditions. Autoradiography of a 10% polyacrylamide gel.
The increased mean annealing rate of the fourth pool versus the first selected pool is not related to the increase in the mean length of the selected antisense species (Fig. 4), which is compatible with kinetic control but not thermodynamic control of antisense RNA-mediated gene suppression in this study. Further, this is reminiscent of the earlier finding that RNA–RNA double strands longer than ~30–50 bp do not show a regular relationship between annealing kinetics and duplex stability (16,19).
Fast RNA–RNA annealing is related to increased inhibition of gene expression
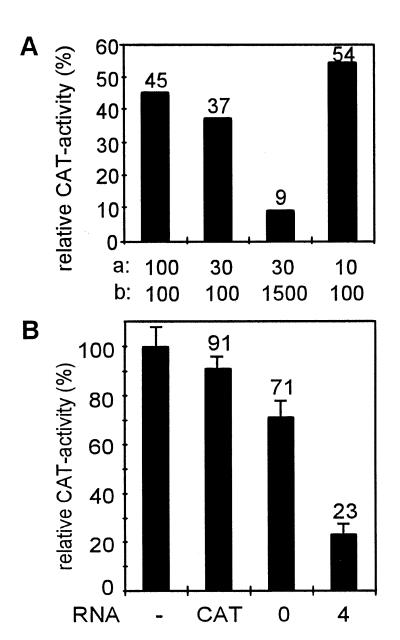
To analyse whether the in vitro annealing of antisense RNA is related to inhibition of gene expression, the initial pool and the four times selected RNA populations were tested for their ability to inhibit cat gene expression in HeLa cells. Pools of antisense RNA were microinjected into HeLa cells together with a cat expression plasmid (pRC-CMV-cat; 18) and cat expression was monitored 6 h post-injection because at this time point inhibition was maximal (data not shown). First, we determined the influence of the concentration of cat-directed antisense RNA and the cat expression plasmid on apparent gene expression (Fig. 5A). For the following experiments concentrations of 30 ng/µl cat expression plasmid and 300 ng/µl antisense RNA allowed an appropriate range to measure and compare cat expression. Strongest inhibition (23% CAT activity) was seen with the four times selected pool, which was significantly increased versus the initial pool (71% CAT activity; Fig. 5B). This experiment shows that fast RNA–RNA annealing in vitro is related to increased inhibition of cat expression. A slight reduction in cat expression (91% CAT activity) was observed with a cat sense control RNA (Fig. 5B). This reduction was significant and might be a sequence-specific or non-sequence-specific non-antisense effect.
Figure 5.
Inhibition of cat gene expression in HeLa cells by pools of cat-directed antisense RNA. (A) Influence of the amount of antisense RNA and cat expression plasmid on the extent of inhibition. The numbers in line a represent the cat plasmid concentration in ng/µl and in line b the antisense RNA concentrations in ng/µl. (B) Expression of cat in the presence of cat sense RNA (CAT) as a control, the initial pool (0) or the four times selected pool (4). Activity of CAT after microinjection of the cat expression vector alone (lane –) served as a standard (100% cat expression). The numbers represent mean values of at least three independent measurements with standard deviations (see error bars) below 10%.
The observed correlation between association kinetics in vitro and inhibition of cat expression in HeLa cells is of non-linear nature and maximal levels of inhibition approximate but do not reach complete inhibition under the experimental conditions used here, though stronger inhibition was measured at a higher excess of antisense RNA over the cat target gene (Fig. 5A). A similar observation was made with HIV-1-directed antisense RNA (16,17). A non-linear correlation in this work could indicate that in addition to the annealing kinetics, other parameters characteristic of antisense RNA species are suboptimal and could limit antisense effects under conditions under which annealing rates contribute at their maximal levels to the overall extent of inhibition. In the experimental approach used here, however, individual RNA structures cannot affect such parameters, which could include intracellular stability and localisation as well as binding of cellular factors.
Theoretical considerations of kinetic control
In the case of cooperative processes such as the formation of long chain double-stranded nucleic acids, values of the free reaction enthalpy (ΔGreact) can be substantially increased, i.e. the thermodynamic equilibrium is very much on the side of folded proteins or nucleic acid double strands. The structure-independent relationship between RNA–RNA annealing kinetics in vitro and the extent of inhibition in human cells described here provides strong evidence for kinetic control of antisense RNA-mediated suppression of gene expression. Regarding the endogenous action of the artificial cat-directed antisense RNA, kinetic control implies that the annealing rate is important but not the equilibrium state or the reverse reaction, which is usually very slow and negligible, as in the case of duplex formation between long chain complementary nucleic acids (>30 bp). For example, a 56 bp RNA double strand was stable in vitro at physiological ionic strength and temperature and simple dissociation of this duplex could not be measured (20). Thus, in view of the extremely slow dissociation and the high Keq values for long chain duplex RNA one could theoretically hypothesise that kass is critical for RNA–RNA annealing. In the case of non-cooperative processes, however, the kinetics of binding might be of increasing importance, once a minimum affinity is reached. For example, the natural maturation of antibodies seems to be thermodynamically controlled in the early phase of selection and at least partly kinetically controlled in the late selection phase (6).
Kinetic control of antisense RNA-mediated inhibition is plausible for several reasons. Firstly, for most low and high molecular weight metabolites in the living cell many reactions compete which each other. Long chain antisense RNA may either fulfil its regulatory role and bind to its target RNA or, alternatively, may be degraded or converted into a form that cannot hybridise or be transported to cellular loci at which the target will not be encountered. If gene expression is to be suppressed, then the antisense RNA has to bind faster to the target RNA than undergo these alternative and presumably irreversible processes. Secondly, regulation could be an important option: it seems to be of advantage in living cells if antisense RNA-mediated inhibition can be controlled, e.g. by altering the kinetics of the association step via cellular facilitators of RNA–RNA annealing or by modulation of RNA structure, including a ‘chaperone-like’ activity. Conversely, controlling the dissociation of long and stable RNA double strands seems to be unfavourable since melting of duplex RNA would require energy-consuming activities.
In vitro selection with genotypically defined sequence spaces
Compared to pools used in common SELEX, pools created as described here are substantially reduced in sequence diversity but, depending on specific purposes, can comprise the complete space of molecules that are active in the chosen selection step. In the case of an antisense RNA sequence space as the starting pool, in principle all antisense species share the property of annealing with their target in the selection step. Thus, an improvement in the phenotypic property of antisense species (here annealing kinetics) should be observable from the beginning of selection, whereas the extent of increased mean annealing kinetics is expected to be relatively small and presumably limited to maximal kass values of <107 to 107 M–1 s–1. This is in contrast to conventional SELEX with pools of completely random sequences. Considering the average annealing rate constant of the initial antisense pool to be 3 × 103 M–1 s–1 and further considering that association rate constants kass of long chain heteropolymeric complementary RNAs reach maximal levels of ~2 × 106 M–1 s–1 (14,21), the improvement in the selection parameter is only ~1000-fold. In contrast, in equilibrium selection (SELEX) starting with random sequences the dynamic range of the parameter selected for (Keq) is substantially greater. It should be noted that related approaches focusing on limited sequence spaces of relevant species have been undertaken previously (22,23).
Perspectives
In the kinetic selection described here one might expect that more rounds of selection for fast binding finally result in only a few antisense species. Theoretically, one should be able to identify the fastest annealing one. This, however, is not necessarily achievable and is not indicated by this work with cat-derived sequences: the differences between antisense species with respect to annealing kinetics are very small and may be extinguished by differing efficiencies in the reverse transcription or PCR steps. Further, it seems to be reasonable to assume that without specific pairing mechanisms which require highly specifc RNA structures, the same or very similar levels of RNA–RNA annealing can be achieved by a large number of the structurally different antisense RNA species included in this study since the cat sequences do not seem to have anything to do with antisense control. Thus, at maximal levels of annealing in this study, a pool could still be obtained containing a large number of suboptimally annealing antisense RNA species which did not differ enough with respect to kass to be resolved in the selection step. This work implies that kinetically controlled processes require a kinetic selection procedure in order to identify the most active molecules. In the future design of antisense and also other drugs, kinetic selection and evolution might help to explore new classes of therapeutic molecules that have been neglected by conventional SELEX procedures so far.
The purpose of using kinetic selection here was to study the relationship between annealing kinetics and biological effectiveness. If, however, the aim was to search for effective antisense species one should also consider less expensive and less time-consuming theoretical computer-based methods (17,24–26). Future studies to improve antisense RNA should include properties such as intracellular stability, subcellular localisation or binding of cellular factors that may affect the effectiveness of antisense RNA. As for RNA–RNA annealing, RNA structure also seems to be crucial for these properties and it will be interesting to see whether RNA structures can be found that allow an improvement in several or all parameters critical for the effectiveness of antisense RNA in vivo.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Roger Fischer for help with automatic microinjection. This work was funded by the Deutsche Forschungsgemeinschaft grant no. Sc14/1-3.
REFERENCES
- 1.Sassanfar M. and Szostak,J.W. (1993) Nature, 364, 550–553. [DOI] [PubMed] [Google Scholar]
- 2.Lorsch J.R. and Szostak,J.W. (1994) Biochemistry, 33, 973–982. [DOI] [PubMed] [Google Scholar]
- 3.Famulok M. and Szostak,J.W. (1992) J. Am. Soc., 114, 3990–3991. [Google Scholar]
- 4.Tuerk C. and Gold,L. (1990) Science, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 5.LaNoue K.F., Jeffries,F.M.H. and Radda,G.K. (1986) Biochemistry, 25, 7667–7675. [DOI] [PubMed] [Google Scholar]
- 6.Foote J. and Milstein,C. (1991) Nature, 352, 530–532. [DOI] [PubMed] [Google Scholar]
- 7.Seidel C.W. and Lawrence,J.P. (1992) J. Mol. Biol., 227, 1009–1018. [DOI] [PubMed] [Google Scholar]
- 8.Wagner E.G.H. and Simons,R.W. (1994) Annu. Rev. Microbiol., 48, 713–742. [DOI] [PubMed] [Google Scholar]
- 9.Case C.C., Roels,S.M., Jensen,P.D., Lee,J., Kleckner,N. and Simons,R.W. (1989) EMBO J., 8, 4297–4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hjalt T. and Wagner,G. (1992) Nucleic Acids Res., 20, 6723–6732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jain C. (1995) J. Mol. Biol., 246, 585–594. [DOI] [PubMed] [Google Scholar]
- 12.Sczakiel G. (1997) Antisense Nucleic Acid Drug Dev., 7, 439–444. [DOI] [PubMed] [Google Scholar]
- 13.Lorsch J.R. and Szostak,J.W. (1994) Nature, 371, 31–36. [DOI] [PubMed] [Google Scholar]
- 14.Persson C., Wagner,E.G. and Nordström,K. (1990) EMBO J., 9, 3767–3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckardt S., Romby,P. and Sczakiel,G. (1997) Biochemistry, 36, 12711–12721. [DOI] [PubMed] [Google Scholar]
- 16.Rittner K., Burmester,C. and Sczakiel,G. (1993) Nucleic Acids Res., 21, 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patzel V. and Sczakiel,G. (1998) Nature Biotechnol., 16, 64–68. [DOI] [PubMed] [Google Scholar]
- 18.Rittner K. (1993) PhD thesis, University Heidelberg, Heidelberg, Germany.
- 19.Kronenwett R., Haas,R. and Sczakiel,G. (1996) J. Mol. Biol., 259, 632–644. [DOI] [PubMed] [Google Scholar]
- 20.Homann M., Nedbal,W. and Sczakiel,G. (1996) Nucleic Acids Res., 24, 4395–4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walter N.G. and Burke,J.M. (1997) RNA, 3, 392–404. [PMC free article] [PubMed] [Google Scholar]
- 22.Dobbelstein M. and Shenk,T. (1995) J. Virol., 69, 8027–8034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singer B.S., Shtatland,T., Brown,D. and Gold,L. (1997) Nucleic Acids Res., 25, 781–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sczakiel G., Homann,M. and Rittner,K. (1993) Antisense Res. Dev., 3, 45–52. [DOI] [PubMed] [Google Scholar]
- 25.James W. and Cowe,E. (1997) Methods Mol. Biol., 74, 17–26. [DOI] [PubMed] [Google Scholar]
- 26.Denman R.B. (1993) Biotechniques, 15, 1090–1095. [PubMed] [Google Scholar]