Abstract
The nature of nuclear structures that are required to confer transcriptional regulation by distal enhancers is unknown. We show that long-range enhancer-dependent β-globin transcription is achieved in vitro upon addition of the DNA architectural protein HMG I/Y to affinity-enriched holo RNA polymerase II complexes. In this system, HMG I/Y represses promoter activity in the absence of an associated enhancer and strongly activates transcription in the presence of a distal enhancer. Importantly, nucleosome formation is neither necessary for long-range enhancer regulation in vitro nor sufficient without the addition of HMG I/Y. Thus, the modulation of DNA structure by HMG I/Y is a critical regulator of long-range enhancer function on both DNA and chromatin-assembled genes. Electron microscopic analysis reveals that HMG I/Y binds cooperatively to preferred DNA sites to generate distinct looped structures in the presence or absence of the β-globin enhancer. The formation of DNA topologies that enable distal enhancers to strongly regulate gene expression is an intrinsic property of HMG I/Y and naked DNA.
INTRODUCTION
Long-range transcriptional regulation is a process of fundamental importance in the control of many complex gene loci and mediates the phenomenon of promoter switching within multigene families. Other genetic control elements, such as insulators, silencers and locus control regions (LCRs) can also regulate gene expression over considerable distances (1). Nevertheless, little is known about the mechanisms by which enhancers and other genetic elements target specific promoters and regulate their activity at long range. Specific models to address these questions generally invoke direct contact between enhancer- and promoter-bound factors by DNA looping or indirect interactions between promoters and enhancers by protein tracking along the DNA (2,3). The contribution of DNA topology to the process of promoter–enhancer communication may also be important (4).
Previous studies have demonstrated that chromatin structure serves to package genes into an enhancer-responsive configuration. Indeed, long-range (>1 kb) enhancer-dependent transcription can be achieved in cell-free systems that use chromatin-assembled genes (5,6). Unexpectedly, however, we found that distal (2 kb) enhancer regulation of chick β-globin or human T cell receptor α (TCR α) chain genes is also observed in the absence of a separate chromatin assembly step when plasmids are incubated with high levels of tissue-specific protein extracts from the appropriate cell type (7,8). In these systems, enhancers were shown to increase the number of active promoters rather than the rate of transcription. Moreover, promoter derepression and DNA topology were found to play critical roles in this process. Our conclusions agree with in vivo analyses which support the ‘binary model’ of enhancer function in which activation of gene expression occurs by derepression of a given gene in an increased number of actively expressing cells rather than by an enhanced rate or level of transcription (9–11). In addition, in vitro studies using the TCR α gene demonstrated that binding of the DNA architectural protein HMG I/Y is essential for long-range enhancer-dependent transcription. HMG I/Y is known to regulate transcription and other nuclear processes through the formation of higher order protein–DNA complexes (12).
In our earlier studies using tissue-specific transcription extracts, the role of nucleosome formation in enhancer function could not be rigorously addressed (7,8). These extracts generally contain a significant amount of histones and chromatin assembly components and can generate nucleosomal structures on plasmid templates. Indeed, electron microscopic analysis of DNA templates incubated with HeLa protein extracts, prepared in a manner similar to our tissue-specific extracts, revealed a canonical bead-like nucleosomal structure (13). In addition, these extracts contain high levels of nuclease activity and the majority of plasmid DNA is degraded during the transcription reaction. Therefore, to examine the role of chromatin structure and to delineate the mechanism by which distal enhancers regulate their target promoters, we have used a well-defined system composed of affinity-purified holo RNA polymerase II and HMG I/Y. We have focused on the well-characterized chick β-globin gene, which is expressed at a precise stage in erythroid development by the generation of an active chromatin structure and the recruitment of a distal 3′-enhancer (14–16). This enhancer regulates both the adult β- and embryonic ɛ-globin promoters in a temporal manner and functions as a LCR to establish integration site-independent erythroid-specific gene expression (17). The β/ɛ enhancer has been shown to be completely functional in vitro using both the individual β-globin gene and cosmids containing the entire 40 kb β-globin chromosomal locus when assembled into synthetic nuclei (5) or transcribed as DNA by erythroid transcription extracts (8).
Here we show that long-range enhancer-dependent transcription in vitro minimally requires the DNA architectural protein HMG I/Y and affinity-enriched holo RNA polymerase II. In this system, HMG I/Y has the dual function of repressing promoter activity in the absence of the enhancer and activating transcription when the enhancer is present. Nucleosome formation is not needed to achieve distal enhancer regulation and is not sufficient for enhancer function without the addition of HMG I/Y. Significantly, HMG I/Y is required for enhancer activity on both DNA and chromatin-assembled genes. Finally, electron microscopy and DNA footprinting analyses reveal that HMG I/Y binds cooperatively to discrete regions of β-globin gene-containing plasmids and then self-associates to form DNA loops. The enhancer-responsive DNA structure results, in a large part, from preferred binding of HMG I/Y within the β-globin enhancer and distinct DNA loop structures are observed in the presence and absence of the enhancer. Thus, the DNA architectural protein HMG I/Y regulates β-globin transcription in a positive and negative manner that is dependent upon a 3′-distal enhancer irrespective of the chromatin status of the gene. In addition to its role in short-range transcriptional control, a general property of HMG I/Y may be to generate long-range enhancer dependence on a variety of genes by recognizing specific DNA structures rather than sequences.
MATERIALS AND METHODS
Preparation of plasmids, recombinant proteins and extracts for transcription and chromatin assembly
The β-globin gene plasmids pUC18ABC/Δ1 and a derivative deleted of the β/ɛ enhancer, pUC18AB/ΔC, have been described (8,18). All plasmids were prepared by the methodology of Triton X-100 lysis followed by CsCl centrifugation as described previously (7). Recombinant HMG I protein from the pET7C clone (a gift from Dr Ray Reeves) was expressed and purified by established procedures (19). Recombinant GST–TFIIS (a gift from Jack Greenblatt) was purified using GST–Sepharose according to the manufacturer’s protocol (Pharmacia, Uppsala, Sweden). HeLa cells were purchased from Cellex Bioscience, Inc. (Minneapolis, MS) and whole cell extracts were prepared as described (7). The chromatin assembly extract, S-190, was prepared from 6 h Drosophila embryos as described (20).
Western blot analyses
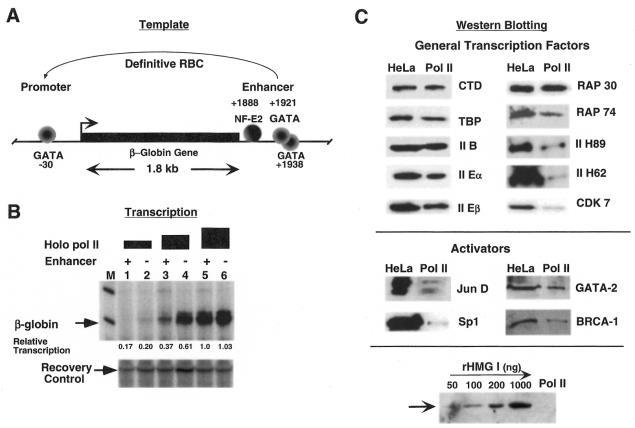
For general transcription factors and activators, 10 µl of HeLa whole cell extract or 3 µl of holo RNA polymerase II were analyzed by either 10% (low molecular weight proteins) or 5% (high molecular weight proteins) SDS–PAGE. Recombinant HMG I (50–1000 ng) was used as a positive control and calibration standard and was electrophoresed by 15% SDS–PAGE. Standard procedures were employed for the separation and transfer of proteins to nitrocellulose membranes. Western blots were performed with the appropriate antibody as indicated. The polyclonal antibodies against various general transcription factors (GTFs), RNA polymerase II CTD, Sp1, GATA-2 and HMG I (Santa Cruz Biotechnology Inc., Santa Cruz, CA) were used at 1000-fold dilution. The polyclonal antibody against JunD (Pharmingen, San Diego, CA) was used at 500-fold dilution. The ECL western blot detection system (Amersham, Uppsala, Sweden) was used in all analyses.
Preparation of human RNA polymerase II holoenzyme
Affinity purification of RNA polymerase II holoenzyme was carried out essentially as described (21). Briefly, the affinity column was prepared by immobilizing the recombinant GST–TFIIS fusion protein on glutathione–Sepharose 4B beads at a ligand concentration of 4 mg/ml. Approximately 20 ml of HeLa whole cell extract was loaded onto a 2.0 ml GST–TFIIS affinity column in a buffer containing 50 mM NaCl, 10 mM HEPES pH 7.9, 0.1 mM EDTA, 0.1 mM DTT and 10% glycerol (0.05 M buffer A). The column was washed with 10 bed vol of 0.05 M buffer A. The bound proteins were eluted as 500 µl fractions with 8 ml of 0.3 M buffer A. Each fraction was analyzed by western blotting for the presence of various GTFs and the RNA polymerase II subunit RPB1. Peak fractions were pooled and dialyzed against buffer A. The dialyzed protein fraction was concentrated ~10-fold by Centricon-10 concentrators (Amicon, Beverly, MA) before use in transcription assays.
In vitro transcription
Reactions containing 0.1 µg supercoiled β-globin plasmid DNA were incubated with 3 µl of holo RNAP II in a buffer containing 20 mM HEPES pH 7.9, 60 mM KCl, 5 mM MgCl2, 2% glycerol, 1 mM DTT, 20 U RNasin and 0.6 mM rNTPs in a final volume of 25 µl at 30°C for 60 min unless otherwise noted. Reactions were stopped with a buffer containing 20 µg carrier yeast tRNA, 20 µg proteinase K, 0.1% SDS and 10 mM EDTA. A radiolabeled DNA fragment (200 bp) was included as a recovery control at this stage. RNA transcripts were then purified by proteinase K digestion, phenol–chloroform extraction and ethanol precipitation. Samples were analyzed by AMV reverse transcriptase-mediated primer extension using a β-globin-specific 32P-labeled oligonucleotide primer before electrophoretic analysis on an 8% polyacrylamide/8 M urea/TBE gel. ImageQuant (Molecular Dynamics) was used to quantify the relative amounts of transcription reaction products after autoradiography. Complementation experiments involved preincubation of supercoiled DNA templates with FPLC-purified HMG I recombinant protein at 30°C in 20 mM HEPES, pH 7.9, 5 mM MgCl2, 50 mM KCl, 5 mM DTT and 200 µg/ml BSA in 10 µl of reaction volume followed by transcription with holo RNAP II as described above. At the maximal protein:DNA ratio, HMG I was present in ~200-fold molar excess. DNA topology remained unchanged after incubation with holo RNAP II and HMG I/Y during the transcription reaction as assayed by agarose gel electrophoresis (data not shown).
Supercoiled β-globin gene plasmids were preincubated with either buffer (a ‘no protein’ control for chromatin-induced repression) or HeLa nuclear extract (a source of holo RNA polymerase II) in the presence or absence of rHMG I and assembled into chromatin by incubating with a Drosophila S-190 extract and core histones for 4 h at 27°C. After chromatin assembly, an aliquot was removed for micrococcal nuclease analysis to assess the extent of nucleosomal assembly. Transcription of chromatin-assembled templates was carried out after incubation with HeLa nuclear extract and NTPs at 30°C for 60 min (22). Transcription reactions were stopped and analyzed as above.
DNase I footprinting
Supercoiled β-globin plasmids (0.2 µg) were incubated at 30°C for 15 min with holo RNAP II (6 µl) or recombinant HMG I under transcription buffer conditions as described above except in the absence of NTPs. DNase I-treated samples were phenol–chloroform extracted and ethanol precipitated. End-labeled primer (0.5 pmol) was annealed to the alkali-denatured template by heating at 65°C for 30 min in Vent buffer (40 mM NaCl, 10 mM Tris pH 8.9, 5 mM MgSO4). Annealed primers were extended with 0.4 U Vent DNA polymerase (New England Biolabs, Beverly, MA) and 0.5 mM dNTPs at 76°C for 10 min. The products were ethanol precipitated and analyzed on a 6% polyacrylamide/8 M urea/TBE sequencing gel.
Electron microscopy
Recombinant HMG I–DNA complexes were formed in a buffer containing 20 mM HEPES pH 7.4, 1 mM DTT and either 60 (for surface spreading) or 100 mM KCl (for direct visualization). Reactions included 50 or 100 ng linear DNA and 0–60 ng HMG I as indicated in the text. Incubations were carried out for 10 min at 30°C and the samples fixed with glutaraldehyde (0.6%) at room temperature for 5 min. The samples were filtered through 2 ml columns of Bio-Gel A-5m (Bio-Rad, Hercules) equilibrated with 10 mM Tris–HCl pH 7.6, 0.1 mM EDTA. For drop surface spreading, ammonium acetate was added to 0.25 M to the filtered sample and cytochrome c added to 4 µg/ml. A 50 µl drop was placed on a parafilm sheet for 1 min and the DNA picked up with a parlodion-covered grid. The sample was stained with uranyl acetate and dehydrated in 80% ethanol. The grids were rotary shadow cast with platinum:palladiun as described (23). For direct visualization, the DNA templates were linearized with Acc65I and ScaI and end-labeled as described (24). The reactions were carried out under the above conditions in the presence of 5 ng streptavidin and filtered through Bio-Gel A-5m. The filtered samples were mixed with a buffer containing spermidine, adsorbed to glow charged thin carbon foils, dehydrated through a water–ethanol series and rotary shadow cast with tungsten as described (25). Samples were visualized in a Philips 400 instrument. Micrographs were scanned from negatives using a Nikon LS 4500 multiformat film scanner and the contrast optimized and panels arranged using Adobe Photoshop. Morphometry measurements were conducted using a Summagraphics digitizer coupled to a Macintosh computer programmed with software developed by J. D. Griffith.
Measurement of the mass of the HMG I particles on DNA was according to the procedure described (26). In brief, HMG I–DNA complexes were fixed with 0.6% glutaraldehyde for 5 min at room temperature and isolated by gel filtration. Just prior to adsorption to the electron microscopic supports, streptavidin (Gibco BRL, Rockville, MD), which had been fixed in the same manner, was added to a concentration of 1 µg/ml and the mixture processed for electron microscopy. Micrographs were taken of HMG I–DNA complexes with at least five streptavidin particles lying nearby. Using a COHU CCD camera attached to a Macintosh computer and NIH IMAGE software, the mean projected area of the streptavidin particles was determined and this value compared to the projected area of the HMG I–DNA particle. Calculation of the HMG particle mass based on the 68 kDa mass of streptavidin was then determined.
RESULTS
Long-range enhancer-regulated transcription in vitro requires holo RNA polymerase II and HMG I/Y
Our previous analyses of long-range enhancer function in tissue-specific transcription extracts yielded important mechanistic information that was consistent between two distinct genes, β-globin and TCR α (7,8). However, a more refined system was needed to address detailed questions about long-range regulation because crude extracts degrade DNA rapidly and are capable of some nucleosome assembly. In particular, we wished to distinguish between the role of chromatin and the effects of HMG I/Y on DNA topology in generating enhancer-dependent gene regulation. We also wished to test current models of long-range promoter control by determining whether DNA loop formation occurs under enhancer-dependent transcription conditions and whether enhancers regulate promoters by activation or derepression or both. For this purpose, we devised conditions in which preparations of RNA polymerase II holoenzyme that contained all of the initiation factors required for basal promoter function could be complemented with HMG I/Y and various transcription factors, as needed, to reconstitute long-range enhancer-dependent transcription. The chick β-globin gene template used in these studies is diagrammed in Figure 1A. The promoter contains a non-canonical TATA box (–30 GATA), which interacts with the erythroid transcription factor GATA-1 (18), and is up-regulated by several nearby proteins, such as NF-E4, a member of the Sp1 family (27). The enhancer is located ~2 kb from the promoter and is regulated predominantly by two adjacently bound GATA-1 proteins and the erythroid heterodimer NF-E2, which is a member of the AP1 family (17).
Figure 1.
Transcription of β-globin genes by the holo RNA polymerase II complex. (A) Diagram of the chick β-globin gene and protein complexes on the promoter and 3′-enhancer regions. (B) In vitro transcription of the enhancer-containing (lanes 1, 3 and 5) and enhancerless (lanes 2, 4 and 6) β-globin gene plasmids using increasing amounts of affinity-enriched holo RNAP II: lanes 1 and 2, 1 µl; lanes 3 and 4, 2 µl; lanes 5 and 6, 3 µl. M indicates molecular weight markers and the lower panel shows a recovery control. Numbers below each lane indicate the relative transcription (in arbitrary units) measured by phosophorimager analysis of the gel. (C) Western blot analyses of holo RNAP II (3 µl) and HeLa whole cell extracts (10 µl ) using antibodies to various general transcription factors and activators. Not all of the general transcription factors are present in stoichiometric amounts in the holo RNA pol II preparations, as has been shown previously (21). Recombinant HMG I was detected in the concentration range 50–1000 ng protein and used as a positive control. HMG I is not a component of the holo RNA polymerase II preparation.
We fractionated ‘holo RNAP II’ from HeLa cells using TFIIS affinity chromatography, as described by Greenblatt and colleagues (21). This holocomplex contains TFIIB, IID (TBP + TAFIIs), IIE, IIF and IIH and is responsive to GAL4–VP16 and GAL4–Sp1 activators. As shown in Figure 1B, titration of holo RNAP II prepared in this manner transcribes β-globin templates in the presence or absence of the enhancer with similar efficiency at the highest enzyme level (lanes 5 and 6) but prefers the enhancerless template at lower enzyme levels (lanes 3 and 4), possibly due to non-specific competitive inhibition by the enhancer sequences. Thus, holo RNAP II is capable of forming a functional initiation complex on the β-globin –30 GATA box and activating basal transcription independent of the 3′-distal enhancer. A western blot analysis of holo RNAP II reveals that, in addition to the basal initiation factors, our preparation also contains the transcriptional activators AP1/junD, Sp1 and GATA-2, but no detectable HMG I/Y (Fig. 1C).
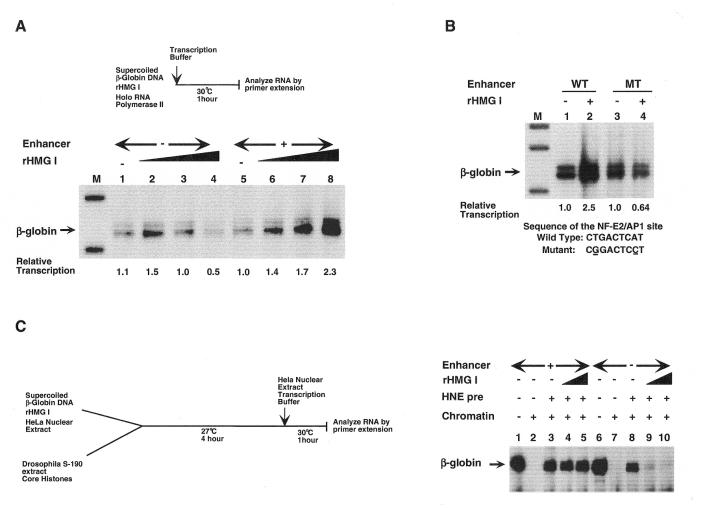
Our intention was to next add erythroid promoter- and enhancer-binding proteins, such as NF-E4, NF-E2 and GATA-1, to attempt to reconstruct enhancer-dependent β-globin transcription. Surprisingly, addition of the architectural protein HMG I/Y (recombinant form = rHMG I) to the holo RNAP II reactions was sufficient to generate enhancer regulation (Fig. 2A). This was achieved by the ability of rHMG I to selectively repress the β-globin promoter in the absence of the enhancer (compare lanes 1 and 4) and activate the promoter in the presence of the enhancer (compare lanes 5 and 8). Under basal transcription conditions, addition of HMG I increases promoter activity 2.3-fold in the presence of the enhancer (lanes 5 and 8) and almost 5-fold over the level obtained in the absence of the enhancer (lanes 4 and 8). Control experiments show that HMG I does not repress transcription of the enhancerless minimal adenovirus major late promoter or a minimal α-globin promoter, even over a wide range of protein concentrations (data not shown). This indicates that HMG I is not functioning as a general repressor but is selectively inactivating the β-globin promoter unless the enhancer is present to counteract this repression, as observed with TCR α genes in T-cell extracts (7).
Figure 2.
Long-range enhancer-dependent transcription of β-globin genes by holo RNA polymerase II is conferred by HMG I/Y. (A) rHMG I selectively represses transcription of enhancerless β-globin genes and activates transcription in the presence of the enhancer. Transcription of the enhancerless (lanes 1–4) and enhancer-containing (lanes 5–8) β-globin genes with 3 µl of holo RNAP II and increasing amounts of rHMG I: lanes 1 and 5, 0 ng; lanes 2 and 6, 15 ng; lanes 3 and 7, 30 ng; lanes 4 and 8, 60 ng. Numbers below each lane indicate the relative transcription (in arbitrary units) measured by phosophorimager analysis of the gel. (B) Mutation of the AP1/NF-E2 binding site within the β-globin gene enhancer abolishes enhancer-dependent activation by rHMG I. Transcription of β-globin genes containing the wild-type enhancer (lanes 1 and 2) or mutated enhancer (lanes 3 and 4) by 3 µl of holo RNAP II in the absence of rHMG I (lanes 1 and 3) or in the presence of 60 ng rHMG I (lanes 2 and 4). Numbers below each lane indicate the relative transcription (in arbitrary units) measured by phosophorimager analysis of the gel. (C) Enhancer-dependent transcription of chromatin-assembled β-globin genes requires HMG I/Y. Enhancer-containing (lanes 1–5) and enhancerless (lanes 6–10) β-globin gene plasmids were either mock-assembled (lanes 1 and 6) or chromatin-assembled using Drosophila S-190 extract (lanes 2–5 and 7–10). In some cases, HeLa nuclear extract (a source of holo RNA polymerase II) alone (lanes 3 and 8) or in combination with rHMG I at 30 (lanes 4 and 9) or 60 ng (lanes 5 and 10) was bound to the DNA templates prior to chromatin assembly for 10 min at 30°C. After assembly, reactions were transcribed and processed.
It is interesting that enhancer-dependent β-globin expression is achieved in this in vitro system in the absence of any erythroid transcription factors. One explanation is that our preparation of the HeLa holo RNAP II contains components (GATA-2 and AP1/junD) that are members of the GATA-1 and NF-E2 families which are able to interact with consensus binding sites within the β-globin enhancer. GATA-2 is a ubiquitous member of the GATA family that can substitute for GATA-1 to activate expression of globin genes from GATA sites in transfected non-erythroid cells (28,29). JunD is a member of the AP1 family, to which NF-E2 also belongs. Thus, it is possible that these factors might function in this system as do their erythroid-specific counterparts, GATA-1 and NF-E2, to form an active β-globin enhancer in the absence of normal tissue-specific constraints.
As further proof that the β-globin enhancer is functioning at long range (~2 kb) in this simplified in vitro system and requires rHMG I to mediate this process, DNA templates containing a linker-scanning mutation in the critical NF-E2/AP1-binding site within the enhancer were examined. This mutation abolishes NF-E2/AP1 interaction and reduces β-globin transcription 20-fold when measured by transient expression in erythroid cells (30). In our in vitro system the NF-E2/AP1-binding site mutation reduces transcription almost 5-fold in the presence of HMG I (Fig. 2B, compare lanes 2 and 4) but has no effect in the absence of this architectural protein (lanes 1 and 3). Thus, HMG I addition is required to observe transcriptional down-regulation by the NF-E2/AP1 mutation within the enhancer. This presumably occurs by the ability of HMG I/Y to generate a DNA structure that enables the β-globin promoter to interact with or be responsive to the distal enhancer. This effect is specific to HMG I since in vitro studies with the enhancer-dependent TCR α gene indicate that HMG I cannot be replaced by other proteins known to distort DNA structure, such as histone H1, core histones, YY1 or HMG1 (7). Although the fold activation and enhancer effect is much larger using erythroid transcription extracts, and comparable to that observed in transfection assays, this simplified system consisting of holo RNAP II and rHMG I recapitulates the essential requirements for long-range enhancer regulation. This is similar to the observation that formation of an initiation complex by general basal factors occurs on all RNA polymerase II genes and is the critical first step in promoter activation. Regulation of the initiation complex (or promoter–enhancer function) by tissue-specific activators, chromatin structure, etc. represent subsequent layers of transcriptional control.
Long-range enhancer-dependent transcription on chromatin-assembled genes requires HMG I/Y
The simplified system described above demonstrates that enhancer-dependent transcription of β-globin gene plasmids can occur in vitro in the absence of a canonical nucleosomal structure. We wished to examine whether enhancer regulation could still be achieved under these conditions if the DNA template was assembled into chromatin. This is particularly important because all genes function within a chromatin environment in their native state and long-range enhancers must counteract chromatin-mediated repression in vivo. β-Globin gene plasmids, containing and lacking the 3′ β/ɛ enhancer, were assembled into chromatin using Drosophila embryonic S-190 extracts under enhancer-responsive conditions. As shown in Figure 2C, assembly of β-globin gene plasmids into chromatin results in transcriptional repression in the presence or absence of the distal enhancer (compare lanes 1 with 2 and 6 with 7). Chromatin-mediated repression of both the enhancer-containing and enhancer-deleted β-globin gene plasmids could be relieved if, prior to chromatin assembly, DNA templates were preincubated with a HeLa nuclear extract, used as a source of holo RNA polymerase II, to generate promoter accessibility (compare lanes 2 with 3 and 7 with 8). Interestingly, if β-globin gene plasmids were preincubated with both rHMG I and HeLa nuclear extract, only enhancer-containing plasmids were transcribed (compare lanes 4 and 5 with 9 and 10). Promoters lacking the 3′ β/ɛ enhancer failed to counteract repression by chromatin and HMG I/Y even in the presence of HeLa nuclear extract, as observed on naked DNA templates (Fig. 2A). Significantly higher levels of transcription were seen in these experiments (Fig. 2C) as compared to those shown in Figure 2A and B. This is because transcription of chromatin templates was carried out using HeLa nuclear extracts whereas transcription of DNA templates was by holo RNA polymerase II alone. Based upon these results, we conclude that the DNA architectural factor HMG I/Y and holo RNA polymerase II are sufficient to confer long-range enhancer-dependent transcription in vitro regardless of the chromatin status of the gene.
Interaction of holo RNA polymerase II complex and HMG I/Y with the β-globin promoter and enhancer
To determine whether holo RNAP II could interact with the β-globin promoter or enhancer, DNase I footprinting on supercoiled β-globin gene plasmids was performed. As shown in Figure 3A, holo RNAP II binds strongly to the –30 GATA box and sequences downstream to +1 within the β-globin promoter. Interactions were also detected near binding sites for three activator proteins, NF-E4 (–50), β-CTF (–70) and β-AP2 (–90). Interestingly, purified Sp1 binds with high affinity to these regions and can presumably substitute for some of the erythroid activators. Thus, Sp1 or other components within the holo RNAP II complex may be interacting with the β-globin upstream promoter sequences. An analysis of the β-globin enhancer using supercoiled templates revealed that sequences in both the GATA-1 and NF-E2 consensus binding sites displayed enhanced DNase cleavage or protection in the presence of holo RNAP II (Fig. 3B). This suggests that components within this complex, such as GATA-2 and AP1, may interact with these critical regions to generate a functional enhancer in the presence of rHMG I. This is supported by the decreased transcription observed with β-globin genes that contain mutations in the NF-E2 enhancer site (Fig. 2B). Surprisingly, holo RNAP II binding to either the promoter or enhancer was unaffected by HMG I and holo RNAP II–promoter interactions were identical in the presence or absence of the enhancer (Fig. 3C and data not shown). In summary, these experiments indicate that the preparation of holo RNAP II used in our reactions contains components that interact with critical regions of both the β-globin promoter and enhancer, consistent with the transcriptional regulation that we observe in vitro. A similar footprinting analysis was conducted with rHMG I. In this case no binding was observed within the promoter from –100 to +120 (data not shown), which suggests that the ability of HMG I to repress enhancerless β-globin genes is not due to localized steric hindrance of holo RNAP II but possibly due to long-range inhibition. In contrast, multiple regions of HMG I interaction were detected in the enhancer which may be correlated with the activation of this control element (Fig. 4A). Note that HMG I/Y binding overlaps the critical GATA-1 binding sites in the β-globin enhancer region (Fig. 4B). Previous gel mobility shift experiments have shown that in the fetal γ-globin promoter both GATA-1 and HMG I/Y can bind to the same site simultaneously and synergistically activate transcription (31).
Figure 3.
Interaction of holo RNA polymerase II with the β-globin promoter and enhancer regions. (A) DNase I footprinting of holo RNAP II within the β-globin promoter region using supercoiled plasmid DNA. Lane 1, control, no protein on β-globin (+enhancer); lane 2, holo RNAP II on β-globin (+enhancer); lane 3, holo RNAP II on β-globin (–enhancer). Lanes A, C, G and T contain β-globin promoter DNA sequencing ladders. Bars to the right of the autoradiogram represent holo RNAP II sites of interaction. (B) DNase I footprinting of holo RNA polymerase II within the β-globin enhancer region using supercoiled plasmid DNA. Lane 1, DNase I digestion following 15 min incubation of holo RNAP II on β-globin (+enhancer); lane 2, DNase I digestion following 2 min incubation; lane 3, no protein control. Bars to the right of the autoradiogram represent holo RNAP II sites of interaction. (C) DNase I footprinting of holo RNA polymerase II within the β-globin promoter region using supercoiled plasmid DNA in the presence of 60 ng rHMG I. Lane 1, control, no protein on β-globin (+enhancer); lanes 2–4, DNase I digestion following 15 min incubation of holo RNAP II on β-globin (+enhancer) in the absence (lane 2) or presence of rHMG I; lane 3 (+enhancer) and lane 4 (–enhancer). Bars to the right of the autoradiogram represent holo RNAP II sites of interaction.
Figure 4.
Interaction of HMG I/Y with the β-globin enhancer. (A) DNase I footprinting of rHMG I within the β-globin enhancer region using supercoiled plasmid DNA. Lane 1, no protein on β-globin (+enhancer) plasmid; lanes 2 and 3, 30 and 60 ng rHMG I, respectively. M indicates DNA size markers. Bars to the right of the panel indicate regions protected by rHMG I. (B) Summary of transcription factors and HMG I/Y binding sites [from (A) and data not shown] within the β-globin enhancer region.
Electron microscopic analysis of HMG I/Y-associated β-globin DNA
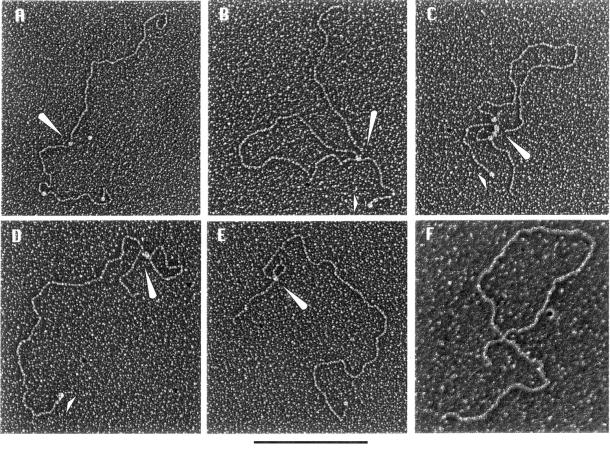
To gain insight into how promoter–enhancer communication occurs at a distance, we used electron microscopy to examine rHMG I binding to β-globin plasmids in the presence or absence of the distal enhancer. Linear DNA templates labeled at one end with biotin were incubated with HMG I, followed by addition of streptavidin to mark the location of the biotin, and prepared for electron microscopy. Maintaining the protein:DNA ratio identical to that in the in vitro transcription reactions, HMG I in the concentration range 0–30 ng/50 ng DNA was incubated with β-globin plasmids either containing or lacking the enhancer. At low HMG I concentrations the majority of the DNA molecules were linear and contained HMG I particles scattered along their length. Some molecules, however, contained loops with a HMG I particle at the base of the loop. As the concentration of HMG I was increased (corresponding to lanes 3 and 7 in Fig. 2A), a larger number of the enhancer-containing and enhancer-deleted DNAs showed one or more loops in addition to scattered HMG I particles. At high concentrations of HMG I (corresponding to lanes 4 and 8 in Fig. 2A) the majority of the molecules were multi-looped. A sampling of the types of molecules seen at an intermediate HMG I concentration (15 ng/50 ng DNA) is shown in Figure 5A–E. The size of the loops generated by HMG I was measured with reference to a streptavidin–biotin-labeled DNA end. In the case of enhancer-minus templates, the size of a single loop observed at intermediate HMG I concentrations varied from 1.0 to 2.3 kb. However, in the case of enhancer-plus templates, single loops of predominantly 4.0–4.5 kb were observed under identical conditions. Interestingly, the enhancer response required 100- to 200-fold molar excess of protein as compared to DNA. At this high ratio, one would anticipate that HMG I would bind all over the plasmid, particularly considering the low sequence specificity of HMG I/Y binding. On the other hand, our observation of loop structures of specific size under these conditions suggests that binding of HMG I to the β-globin gene is not random but rather a property of the DNA structure of the sequence being recognized. At high concentrations of HMG I, at which maximal enhancer activity was observed, most templates were in multi-looped structures generally consisting of a large loop and one or two small loops at the base of the large loop (as shown in Fig. 5C). It is possible that clustering of loops at the junction region may allow distal transcription factor binding sites to be juxtaposed. However, since it is difficult to trace the path of DNA entering and exiting the junction region any conclusion regarding the specific location of enhancer or promoter regions on these loops cannot be made.
Figure 5.
HMG I/Y forms DNA loops by cooperative binding and self-association. Electron micrographs of HMG I-mediated loops in streptavidin–biotin end-labeled β-globin DNA. (A–E) A sampling of the types of molecules seen at HMG I concentration of 15 ng for 50 ng DNA. Enhancer-containing β-globin DNA–HMG I complexes are shown in (A)–(C) and enhancerless β-globin DNA–HMG I complexes are shown in (D) and (E). (F) Demonstration of a protein-stabilized large loop in enhancer-containing β-globin DNA using the classic surface method employing a denatured film of cytochrome c protein. Arrows in (A)–(E) represent rHMG I particles bound to DNA and the short arrow in (D) represents the streptavidin–biotin tag. The size bar indicates 1 kb.
We further established that the loops observed during electron microscopy are mediated by HMG I using the classic surface spreading method employing a denatured film of cytochrome c protein. Here the surface tension forces spread the DNA out such that loops generated by DNA crossing over itself are rarely seen in DNAs of a few kilobases unless a loop has been formed by protein binding (Fig. 5F). This makes it possible to unambiguously score loops within large numbers of DNA molecules and to determine the sizes of the loops. Since supercoiled DNA is already ‘looped’ it was not possible to determine whether HMG I generated de novo loops or stabilized existing ones. However, multi-looped structures (with HMG I at the base of the loop) were also generated on supercoiled plasmids and this appeared to be an order of magnitude more efficient than when linear templates were used (data not shown).
Previous studies with the β-interferon enhanceosome demonstrated that HMG I molecules can bind cooperatively through protein–DNA and protein–protein interactions to synergistically activate transcription (32). In our studies the electron micrographs revealed a spectrum of rHMG I particle sizes bound along the DNA from only slightly larger than the metal shadowing grain (arrow, Fig. 5B) to very distinct particles at the loop junctions (Fig. 5B–E), some of which were larger than the 68 kDa streptavidin tag (arrow, Fig. 5D). To provide a more quantitative estimate of their size, the projected areas of HMG I particles in the micrographs were compared to that of streptavidin molecules bound to DNA and also included in the electron microscopic background as an internal size marker. This provides a means of correcting for metal coating and determining the mass of HMG I bound to DNA (26). The distribution of particle masses, shown in Figure 6, indicates that HMG I can bind to DNA as spherical, oligomeric particles predominantly in the size range spanning dimers to tetramers (monomers would be too small to be detected by metal shadow casting). The greater mass of the HMG I particles at DNA loop junctions, particularly in multi-looped molecules, suggests that cooperative binding of HMG I is involved in loop formation. Of note, most of the linear, non-looped structures showed very few distinct bound HMG I particles, suggesting that once several HMG I proteins assemble on DNA, they self-associate to form loops.
Figure 6.
HMG I/Y particle mass. Mass analysis of rHMG I complexes formed on DNA. Complexes of HMG I were formed on linear plasmids containing the β-globin gene enhancer and promoter regions as described in Materials and Methods. The mass of these molecules was calculated based on area measurements using streptavidin (68 kDa) as the standard.
DISCUSSION
The in vitro transcription and electron microscopy studies presented here demonstrate that enhancer-dependent transcription at a distance can be achieved in the absence of chromatin and requires only the DNA architectural factor HMG I/Y. Long-range enhancer-dependent transcription by HMG I/Y occurs through two separate actions: (i) down-regulation of promoter activity in the absence of an enhancer; (ii) strong activation of transcription from templates that contain an enhancer. The mechanistic basis for the down-regulation of promoter activity by HMG I/Y in the absence of an associated enhancer is not clear. However, a simplistic steric hindrance of the basal machinery by HMG I/Y can be ruled out based upon DNase I footprint analysis (Fig. 3C). On the other hand, transcriptional activation by the β-globin enhancer upon HMG I/Y addition correlates with the DNA structures observed in electron micrographs (Fig. 5). We propose that HMG I/Y-generated DNA structures could provide a framework within which enhancer–promoter interaction is stabilized and highly efficient. Importantly, the DNA topology established by HMG I/Y confers long-range enhancer regulation on both DNA and chromatin templates. This indicates that although nucleosome formation is not necessary for distal enhancer function in vitro, HMG I/Y can still generate an enhancer-responsive DNA structure within a chromatin context. Thus, a specific DNA topology that is stabilized by HMG I/Y presumably constitutes one of the initial events in the multiple steps required for long-range enhancer-dependent transcriptional regulation.
HMG I/Y has been reported previously to regulate expression of a variety of genes in either a positive or negative manner (12). For example, HMG I/Y is an activator of the β-interferon and interleukin-2 receptor genes and a repressor of the interleukin-4 and ɛ immunoglobulin genes. In these cases, localized bending of DNA by HMG I/Y is proposed to facilitate assembly of multi-component enhancer-binding complexes which control gene activity (33). In this regard, HMG I/Y has been shown to interact directly with DNA-bound transcription factors, including NF-κB, ATF-2/c-jun and SRF, to enhance their DNA-binding affinity and transactivation potential (33,34). Similarly, HMG I/Y can interfere with the DNA-binding activity of other factors to down-regulate transcription (35).
Our results indicate a distinct mechanism of regulation that controls long-range enhancer–promoter communication. In particular, we find that HMG I/Y acts indirectly through altering DNA structure and topology to regulate the β-globin gene in a positive or negative manner, depending upon the presence of a distal enhancer element. Moreover, these studies demonstrate that the essence of this regulation can be observed even in the absence of tissue-specific factors that bind directly to the β-globin enhancer and promoter regions. It is likely that the tissue- and stage-specific activators of β-globin genes function within this structural context and, together with the specific chromatin structure of the β-globin locus, generate the refined developmental control observed for this gene family.
Several sequence-specific transcription factors, such as Sp1 and SpGCF1, have been shown to multimerize and bring distal DNA regions into close proximity through looping (36–38). Indeed, GATA-1 can self-associate and potentially generate DNA loops between distal sites (39). However, we have consistently observed in our in vitro functional studies that natural promoter and enhancer complexes themselves are insufficient to support long-range enhancer-dependent transcription. The absolute requirement for HMG I/Y to activate transcription in vitro may reflect the need to create a specific DNA topology in which direct interaction between promoter and enhancer complexes can occur if their affinity is too weak to support a stable and/or functional association.
The observation that HMG I/Y can recognize a wide range of genetic elements with little sequence homology, including distinct enhancers of the β-interferon (40), TCR α (7) and β-globin genes, is not surprising. Rather, it is consistent with its role as an architectural factor to recognize specific DNA structures and create specialized DNA topologies within which proximal and distal control regions can regulate transcription. Such a system provides the flexibility for a single protein to mediate transcriptional control of a variety of genes. Our analyses show specificity for HMG I/Y to generate long-range enhancer regulation. However, it is likely that other DNA architectural factors may also serve a similar function on genes which contain appropriate recognition structures or sequences.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Jack Greenblatt for advice on the initial purification of holo RNA polymerase II and Katherine Jones for thoughtful comments on the manuscript. We also acknowledge excellent technical support from Thomas Wu and Aziz Ander. This project was funded by grants from the National Institutes of Health to B.M.E. (GM 38760) and to J.D.G. (GM 31819). R.B. is the recipient of an Arthritis Foundation Investigator Award and S.M. was supported by an NCI training grant (T32 CA 09156).
REFERENCES
- 1.Blackwood E.M. and Kadonaga,J.T. (1998) Science, 281, 61–63. [Google Scholar]
- 2.Ptashne M. (1986) Nature, 322, 697–701. [DOI] [PubMed] [Google Scholar]
- 3.Wang J.C. and Giaever,G.N. (1988) Science, 240, 300–304. [DOI] [PubMed] [Google Scholar]
- 4.Rippe K., von Hippel,P.H. and Langowski,J. (1995) Trends Biochem. Sci., 20, 500–506. [DOI] [PubMed] [Google Scholar]
- 5.Barton M.C. and Emerson,B.M. (1994) Genes Dev., 8, 2453–2465. [DOI] [PubMed] [Google Scholar]
- 6.Laybourn P.J. and Kadonaga,J.T. (1992) Science, 257, 1682–1685. [DOI] [PubMed] [Google Scholar]
- 7.Bagga R. and Emerson,B.M. (1997) Genes Dev., 11, 629–639. [DOI] [PubMed] [Google Scholar]
- 8.Barton M.C., Madani,N. and Emerson,B.M. (1997) Proc. Natl Acad. Sci. USA, 94, 7257–7262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majumder S., Miranda,M. and DePamphilis,M.L. (1993) EMBO J., 12, 1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters M.C., Fiering,S., Eidemiller,J., Magis,W., Groudine,M. and Martin,D.I. (1995) Proc. Natl Acad. Sci. USA, 92, 7125–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walters M.C., Magis,W., Fiering,S., Eidemiller,J., Scalzo,D., Groudine,M. and Martin,D.I. (1996) Genes Dev., 10, 185–195. [DOI] [PubMed] [Google Scholar]
- 12.Bustin M. and Reeves,R. (1996) Prog. Nucleic Acid Res. Mol. Biol., 54, 35–100. [DOI] [PubMed] [Google Scholar]
- 13.Sinha S.N., Hellwig,R.J., Allison,D.P. and Niyogi,S.K. (1982) Nucleic Acids Res., 10, 5533–5552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Choi O.R. and Engel,J.D. (1986) Nature, 323, 731–734. [DOI] [PubMed] [Google Scholar]
- 15.Hesse J.E., Nickol,J.M., Lieber,M.R. and Felsenfeld,G. (1986) Proc. Natl Acad. Sci. USA, 83, 4312–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGhee J.D., Wood,W.I., Dolan,M., Engel,J.D. and Felsenfeld,G. (1981) Cell, 27, 45–55. [DOI] [PubMed] [Google Scholar]
- 17.Reitman M. and Felsenfeld,G. (1988) Proc. Natl Acad. Sci. USA, 85, 6267–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fong T.C. and Emerson,B.M. (1992) Genes Dev., 6, 521–532. [DOI] [PubMed] [Google Scholar]
- 19.Nissen M.S., Langan,T.A. and Reeves,R. (1991) J. Biol. Chem., 266, 19945–19952. [PubMed] [Google Scholar]
- 20.Kamakaka R.T., Bulger,M. and Kadonaga,J.T. (1993) Genes Dev., 7, 1779–1795. [DOI] [PubMed] [Google Scholar]
- 21.Pan G., Aso,T. and Greenblatt,J. (1997) J. Biol. Chem., 272, 24563–24571. [DOI] [PubMed] [Google Scholar]
- 22.Armstrong J.A., Bieker,J.J. and Emerson,B.M. (1998) Cell, 95, 93–104. [DOI] [PubMed] [Google Scholar]
- 23.Thresher R. and Griffith,J. (1992) Methods Enzymol., 211, 481–490. [DOI] [PubMed] [Google Scholar]
- 24.Bortner C. and Griffith,J. (1990) J. Mol. Biol., 215, 623–634. [DOI] [PubMed] [Google Scholar]
- 25.Griffith J.D. and Christiansen,G. (1978) Annu. Rev. Biophys. Bioeng., 7, 19–35. [DOI] [PubMed] [Google Scholar]
- 26.Griffith J.D., Makhov,A., Zawel,L. and Reinberg,D. (1995) J. Mol. Biol., 246, 576–584. [DOI] [PubMed] [Google Scholar]
- 27.Gallarda J.L., Foley,K.P., Yang,Z.Y. and Engel,J.D. (1989) Genes Dev., 3, 1845–1859. [DOI] [PubMed] [Google Scholar]
- 28.Orkin S.H. (1992) Blood, 80, 575–581. [PubMed] [Google Scholar]
- 29.Yamamoto M., Ko,L.J., Leonard,M.W., Beug,H., Orkin,S.H. and Engel,J.D. (1990) Genes Dev., 4, 1650–1662. [DOI] [PubMed] [Google Scholar]
- 30.Reitman M., Lee,E., Westphal,H. and Felsenfeld,G. (1990) Nature, 348, 749–752. [DOI] [PubMed] [Google Scholar]
- 31.Magis W. and Martin,D.I. (1995) Biochem. Biophys. Res. Commun., 214, 927–933. [DOI] [PubMed] [Google Scholar]
- 32.Yie J., Liang,S., Merika,M. and Thanos,D. (1997) Mol. Cell. Biol., 17, 3649–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du W., Thanos,D. and Maniatis,T. (1993) Cell, 74, 887–898. [DOI] [PubMed] [Google Scholar]
- 34.Chin M.T., Pellacani,A., Wang,H., Lin,S.S., Jain,M.K., Perrella,M.A. and Lee,M.E. (1998) J. Biol. Chem., 273, 9755–9760. [DOI] [PubMed] [Google Scholar]
- 35.Klein-Hessling S., Schneider,G., Heinfling,A., Chuvpilo,S. and Serfling,E. (1996) Proc. Natl Acad. Sci. USA, 93, 15311–15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mastrangelo I.A., Courey,A.J., Wall,J.S., Jackson,S.P. and Hough,P.V. (1991) Proc. Natl Acad. Sci. USA, 88, 5670–5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su W., Jackson,S., Tjian,R. and Echols,H. (1991) Genes Dev., 5, 820–826. [DOI] [PubMed] [Google Scholar]
- 38.Zeller R.W., Griffith,J.D., Moore,J.G., Kirchhamer,C.V., Britten,R.J. and Davidson,E.H. (1995) Proc. Natl Acad. Sci. USA, 92, 2989–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Crossley M., Merika,M. and Orkin,S.H. (1995) Mol. Cell. Biol., 15, 2448–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thanos D. and Maniatis,T. (1995) Cell, 83, 1091–1100. [DOI] [PubMed] [Google Scholar]