Abstract
Non-long terminal repeat (LTR) retrotransposons or LINEs transpose by reverse transcription of an RNA intermediate and are thought to use the 3′ hydroxyl of a chromosomal cleavage to initiate synthesis of the first strand of the cDNA. Many of them terminate in a poly(dA) sequence at the 3′ end of the coding strand although some, like the I factor of Drosophila melanogaster, have 3′ ends formed by repeats of the trinucleotide TAA. We report results showing that I factor transcripts end a few nucleotides downstream of the TAA repeats and that these extra nucleotides are not integrated into chromosomal DNA during retrotransposition. We also show that the TAA repeats are not required for transposition and that I elements containing mutations affecting the TAA sequences generate transposed copies ending with tandem repeats of various types. Our results suggest that during integration the 3′ end of the I factor RNA template can pair with nucleotides at the target site and that tandem duplications are generated by the reverse transcriptase of the I factor in a manner that is reminiscent of the activity of the reverse transcriptases of telomerases. Reverse transcriptases of other non-LTR retrotransposons may function in a similar way.
INTRODUCTION
The mechanism by which the RNA transposition intermediates of non-long terminal repeat (LTR) retrotransposons or LINEs are reverse transcribed is still poorly understood. Non-LTR retrotransposons contain dA-rich sequences at the 3′ end of their coding strand which, in many cases, is poly(dA). They usually have two open reading frames (ORFs), the second of which encodes nuclease (1,2) and reverse transcriptase (3,4) activities. Most non-LTR retrotransposons are dispersed in the genome but some insert at specific sites. The latter include R1 and R2 which insert at specific sites in rRNA genes of insects (5), and HeT-A and TART in Drosophila melanogaster (6,7) which insert at chromosome ends and compensate for the absence of standard telomeric repeats.
Reverse transcription of the RNA transposition intermediate of non-LTR retrotransposons is thought to occur at the site of integration (reviewed in 8). According to current models, the endonuclease encoded by the element makes staggered single strand breaks at the target site and the reverse transcriptase uses the 3′ hydroxyl at one of the nicks to prime synthesis of the first DNA strand using the RNA transposition intermediate as template. Some steps of this process have been demonstrated in vitro using the site-specific R2Bm element (9–12). R2Bm is a non-LTR retrotransposon of Bombyx mori with a single ORF which encodes both reverse transcriptase and endonuclease activities. Reverse transcription of R2Bm initiates precisely at the 3′ end of the element provided that the RNA template contains additional nucleotides corresponding to DNA downstream of the site of insertion (12,13). The additional nucleotides at the 3′ end of the RNA presumably facilitate alignment of the RNA with the site of insertion during reverse transcription.
Very little is known about the sequences required at the 3′ ends of non-LTR retrotransposons that are not sequence specific to allow initiation of reverse transcription. L1Hs elements that occur in the human genome contain a putative polyadenylation signal near their 3′ ends and their transcripts contain poly(A) tails that appear to be at least partially reverse transcribed during transposition (14–16). This is not true for all non-LTR retrotransposons, however, since some have other A-rich sequences at their 3′ ends like the TAA repeats of the I factor of D.melanogaster (17), Dong of B.mori (18), LOA of Drosophila silvestris (19) and Q of Anopheles gambiae (20).
I factors are non-LTR retrotransposons that are not site specific and are present in some strains of D.melanogaster known as inducer (reviewed in 21,22). Strains devoid of functional I factors are called reactive. I factors are stable in inducer strains, but transpose at high frequencies in the germ line of the females, known as SF females, resulting from crosses between reactive females and inducer males (23). They have 4–8 repeats of the trinucleotide TAA at the 3′ end of the coding strand which contains two ORFs (17,24) (Fig. 1A). ORF2 putatively encodes nuclease, reverse transcriptase and RNase H activities. A 5.4 kb long RNA containing the complete sequence of the element, including UAA repeats, which is thought to be the transposition intermediate, is synthesized in the ovaries of SF females (25).
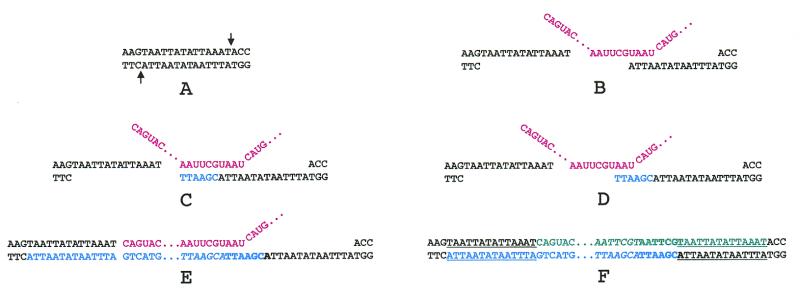
Figure 1.
Structures of the I factor, of the 3′ ends of transcripts of a wild-type I factor, and of its transposed copies. (A) Structure of the I factor. The symbols are as follows: ORF1 and ORF2, the two open reading frames; NB, zinc finger motifs; N, endonuclease domain; RT, reverse transcriptase domain; RNASE H, RNase H domain. The TAA repeats at the 3′ end are shown. Arrowheads indicate the positions and orientations of the oligonucleotides used in PCR and sequencing experiments. (B) Sequences of the 3′ ends of the wild-type I factor I954Y and of the two types of RNAs produced by this element. (C) Sequences of both ends of some transposed copies of the single I954Y I factor. The figure shows examples of the various types of transposed copies containing the same number of TAA repeats as I954Y (It1), and less (It2) or more (It3) TAA repeats than I954Y. Sequences corresponding to the I elements are shown in blue and flanking sequences in black. Target site duplications are underlined. Four out of the 12 insertions analyzed were at sites containing the sequence TAA which could be either part of the target site duplication or one of the TAA repeats at the end of the I factor (like It2).
In this paper we show that the transposition intermediates of a wild-type I factor extend a few nucleotides beyond the TAA repeats and that these extra nucleotides are not normally integrated into chromosomal DNA during retrotransposition. We have constructed I elements containing mutations affecting the TAA repeats and shown that they are not required for transposition. Transposed copies of these mutated elements have tandem repeats of various types at their 3′ ends. These might be produced by an activity of the reverse transcriptase of the I factor related to that of the reverse transcriptases of telomerases.
MATERIALS AND METHODS
Construction of the mutated I factors
pI97 was constructed by PCR amplification of pI954 (26) using primers 6851 (Fig. 1A: 5′-CTGCAGTCCATGGTACCAATCTATTAAC-3′) and 97 in the flanking DNA (5′-GGAATTCTTTTTTTTTTTTTGATAGATAGAATAGTTTAC-3′) and ligating the KpnI–EcoRI digested PCR products into the corresponding restriction sites of pI954. For pI99, primer 97 was replaced by 99 also in the flanking DNA (5′-GGAATTCTGATAGATAGAATAGTTTAC-3′). pI954Y results from the insertion of the yellow gene (27) into the XhoI site of pI954 (28).
Establishment of Drosophila transgenic lines
P-mediated transformation of the reactive strain JA was performed according to Spradling and Rubin (29) using the helper plasmid phs70Δ2-3wc. Flies containing transgenes I97 and I99 were selected on a medium containing neomycin (1 mg/ml) at 23°C. Males transformed with pI954Y were [y+] and had a single copy of the full-length I factor. In order to maintain a single copy of I954Y in the genome, transgenic males were crossed with JA reactive females at each generation (28). I factor transposition only occurs in the female germ-line. The structure of all transgenes was checked by Southern blot analyses.
Cloning transposed copies of the I factor
Inverse PCR experiments were carried out as follows. One microgram of genomic DNA was digested with 4–12 U of NdeII or BstYI overnight at the appropriate temperature. After phenol extraction, the DNA fragments were circularized with 400 U of T4 DNA ligase (New England Biolabs, Montigny, France) overnight at room temperature. After DNA precipitation and PCR amplifications with oligonucleotides 3 and 7 (Fig. 1A: 5′-ACCCTCTAGACCTTCTTAGC-3′ and 5′-TCGCAAGGTCGGCTTTAAGG-3′, respectively) or oligonucleotides 3 and 5 (Fig. 1A: 5′-GAGAGGCACGACTTATCTCTTCGG-3′), the PCR products were cloned with the Stratagene PCR Script Amp SK (+) cloning kit. Sequencing reactions were performed using the fidelity DNA sequencing system from Oncor (Appligene, Illkirch, France).
RLM-3′-RACE
The 3′ end of I factor RNA was analyzed by RNA ligase mediated RACE experiments with the following modifications of the method previously described (30). Ten micrograms of total RNA extracted with the RNAble (Eurobio, Les Ulis, France) from 50 flies were incubated for 15 min at room temperature with 10 U of DNase I (Gibco BRL, Cergy, France). Eighty picomoles of the anchor oligonucleotide (5′-CAACGGTGGTATATCCAGTGATTTT-3′) were ligated overnight to the RNA with 120 U of RNA ligase (Gibco BRL). The reverse transcription reaction was primed using 40 pmol of anchor primer (5′-AAAATCACTGGATATACCACCGTTG-3′) and incubated for 1 h at 37°C. Then nested PCR was carried out first using oligonucleotide 6851 (Fig. 1A) and the anchor primer and then second with oligonucleotides 6851, 3 or 1 (Fig. 1A: 5′-GTACATAACAAGCCAGCAATTAG-3′) and the anchor primer. PCR products were cloned as described previously. Colony blot screening was performed using a PCR fragment of the last 160 nt of the I factor and labeled with the Megaprime kit (Amersham, Les Ulis, France). Positive clones were sequenced (see above).
RESULTS
Transcripts of the I factor terminate within downstream flanking sequences
The results of previous experiments indicate that full-length I factor transcripts are not polyadenylated (25). We used RLM-3′-RACE to determine precisely the structure of the 3′ end of transcripts from full-length I factors. This was done using RNA extracted from SF females produced by crossing females of the reactive strain JA with males of strain 21.1 (28) which contains a single functional I factor, I954Y, with 5 TAA repeats at its 3′ end (Fig. 1B). We observed two types of transcript (Fig. 1B). Both were obtained in two independent experiments. Up to four nucleotides downstream of I954Y were present at the 3′ end of the RNAs, suggesting that transcription terminates a few nucleotides downstream of the TAA repeats.
In order to determine whether or not these additional nucleotides are copied by reverse transcriptase during I factor retrotransposition we sequenced the ends of transposed copies from the single element I954Y contained in the 21.1 transgenic line. These were cloned from DNA extracted from 50 males coming directly from SF females produced by crossing 21.1 males and JA reactive females in such a way that it is very likely that each transposed copy resulted from a single transposition event that occurred in the germ line of the SF females. They were cloned by inverse PCR using BstY1 digested DNA and primers 3 and 5, and sequenced using oligonucleotides 1 and 5 as primers (see Fig. 1A and Materials and Methods).
The sequences of both ends of 14 transposed copies were determined. Two of them were not flanked by target site duplications suggesting that they have been involved in chromosomal rearrangements that frequently occur in the SF female germ line (31). Target site duplications delimit the ends of the other 12 transposed I factors. Figure 1C summarizes the data. The 3′ ends of these transposed copies can be assigned to the last nucleotide preceding the target site duplication. None of the insertions contain the extra nucleotides present downstream of the UAA repeats in the I factor transcripts, indicating that these nucleotides are not integrated into chromosomal DNA during retrotransposition.
Transposed copies of I954Y terminate with 3, 4, 5 or 6 TAA repeats (see Fig. 1C) demonstrating that the number of repeats can vary during retrotransposition.
The TAA repeats at the 3′ end of I elements are not required for transposition
In order to investigate whether the terminal TAA repeats are essential for I factor transposition we constructed two elements with altered 3′ ends. I97 has the TAA repeats replaced by 13 deoxyadenosines (Fig. 2A) while I99 has these repeats deleted entirely (Fig. 3A). Transgenic lines containing these elements were established using the reactive stock JA for transgenesis, and were called 97-52 for I97, and 99-18 and 99-59 for I99. They were maintained by crosses en masse, allowing transposition to occur provided the mutated I elements retained the ability to transpose. After several generations, all lines behaved as typical inducer stocks (data not shown), indicating that elements I97 and I99 can transpose and that the transgenic stocks contain multiple copies resulting from retrotransposition of these mutated I elements.
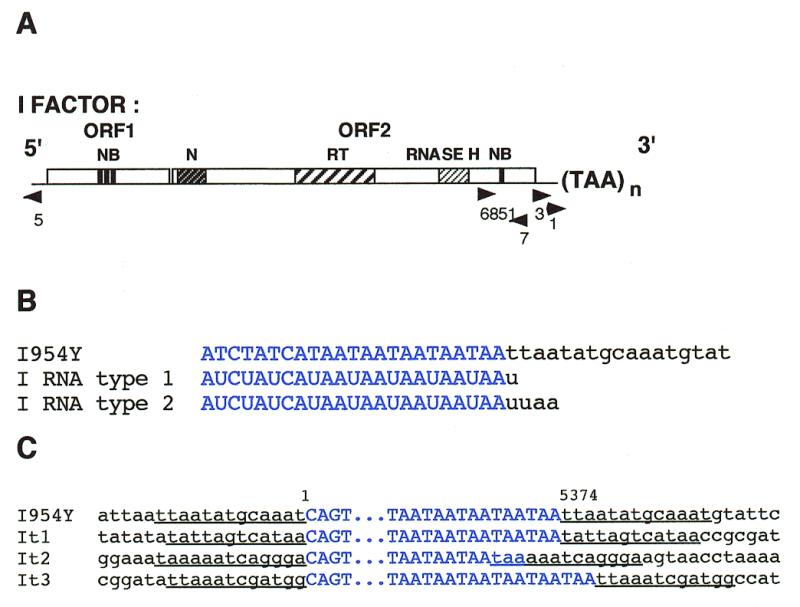
Figure 2.
Structure of the I97 mutated I factor containing an oligo(dA) and of the ends of I elements resulting from its transposition. (A) Structure of I97 showing the oligo(dA) instead of TAA repeats. The abbreviations are the same as in Figure 1A. (B) Examples of transposed copies of I97. The sequence of the I factor is that of the element inserted in the wIR3 mutation (21). I97 is the parental I element containing 13 deoxyadenosines, and IpA are transposed copies. The numbers correspond to the positions of the nucleotides in the I factor sequence (17). I element sequences are in blue, the poly(dA) sequences at the 3′ ends of the progenitor element and of its transposed copies are in red and flanking sequences are in black. Target sites duplications are underlined.
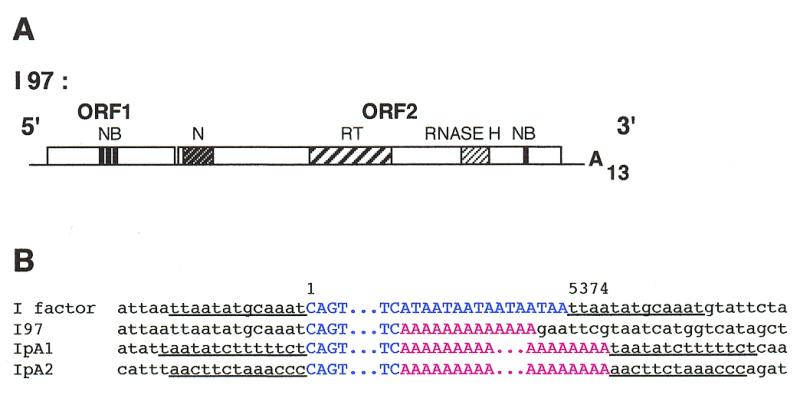
Figure 3.
Structure of the I99 mutated I factor devoid of TAA repeats and of an oligo(dA), and of the ends of I elements resulting from its transposition. (A) Structure of I99. The abbreviations are the same as in Figure 1A. (B and C) Examples of transposed copies of I99. The sequence of the I factor is that of the element inserted in the wIR3 mutation (21), I99 is the parental I element devoid of TAA repeats, IpdA are transposed copies of I99 that terminate by a poly(dA) sequence (B), and Ifs are transposed copies of I99 terminating with tandem duplications (C). Six IpdA and seven Ifs insertions were recovered. The numbers correspond to the positions of nucleotides in the I factor sequence (17). I element sequences are in blue and flanking sequences in black. Letters in green correspond to sequences directly flanking the 3′ end of the progenitor element I99 and to identical sequences at the 3′ end of its transposed copies. Letters in red correspond to poly(dA) sequences at the end of IpdA elements. The sequence TCG shown in violet in Ifs elements corresponds to three nucleotides that were not present in the DNA flanking I99 nor in the target sites of its transposed copies. Letters in italic in (C) indicate the duplication of the sequence AATTCGT at the end of Ifs elements. Target site duplications are underlined.
The reverse transcriptase of the I factor might generate repeats at the 3′ end of the element
We have determined the structure of the 3′ ends of transposed copies of I97. DNA was extracted from 50 flies of stock 97-52 and digested with NdeII. The ends of transposed copies of I97, named IpA elements, were cloned by inverse PCR experiments using oligonucleotides 3 and 7, and sequenced using primer 1 (Fig. 1A). The sequences of the 3′ end of 12 of these transposed copies were determined and found to have poly(dA) sequences which are much longer than that of I97, varying in size from 25 to more than 100 deoxyadenosines. The data are summarized in Figure 2B. None of the IpA elements that were sequenced are flanked by sequences identical to the DNA adjacent to I97 confirming that they were generated by transposition. These data demonstrate that an I element terminated by a poly(dA) sequence can transpose and hence that the TAA repeats are not essential for transposition.
In order to determine the structure of the sequence into which IpA elements had inserted we sequenced both ends of two further insertions (called IpA1 and IpA2) using the same method as that described above, except that NdeII was replaced by BstYI. Figure 2B shows that they are flanked by target site duplications, indicating that they are the results of normal retrotransposition. The duplication flanking IpA1 is 14 nt long while that flanking IpA2 is either 11 or 13 nt long depending on whether the first two deoxyadenosines at the 3′ junction belong to the target site or to the poly(dA) sequence. The poly(dA) sequences of IpA elements were presumably created during the retrotransposition process since they are not part of the target site duplications.
The ends of transposed copies of I99 in strains 99-18 and 99-59 were similarly analyzed. We sequenced the 3′ ends of 13 transposed copies. The data are summarized in Figure 3B and C. Surprisingly, retrotransposition of I99 generated two types of elements with different 3′ ends.
Elements of the first type (called IpdA) terminate with poly(dA) sequences (Fig. 3B). They begin after the deoxyguanosine which is the first nucleotide flanking I99. We sequenced both ends of three transposed copies of this category. Figure 3B shows that they are flanked by target site duplications that are not themselves composed solely of deoxyadenosines, suggesting again that the poly(dA) sequences were added during transposition. These elements have a few deoxyadenosines that could belong to either the poly(dA) sequence or the target site.
Three extra nucleotides are present between the 5′ end of IpdA3 and the target site duplication (Fig. 3B). These are not present at the 5′ junction of the donor element I99, suggesting that they are untemplated nucleotides that were added during the retrotransposition process, possibly during priming of the second strand as previously observed (32). We have made similar observations for a few other transposed I elements that we have analyzed in this study.
Transposed copies of the second type (called Ifs elements) are shown in Figure 3C. They have no poly(dA) sequences but have at their 3′ ends 11 nt that were adjacent to the progenitor element I99. Sequences downstream of these nucleotides are different from each other and from those flanking I99 indicating that each element had inserted at a different site. We sequenced both ends of four of these elements, Ifs1 and Ifs2 from transgenic line 99-18, and Ifs3 and Ifs4 from 99-59. Each insertion is flanked by a target site duplication that is 12 to 14 or 15 nt long depending on whether the deoxyguanosine present at the 3′ junction of Ifs1 is the last nucleotide of the transposed copy or the first of the target site (Fig. 3C). This indicates that Ifs elements were generated by transposition and that these transposed copies of I99 have taken with them some 3′ flanking sequences. All Ifs elements are flanked at the 3′ end by a tandem duplication of the sequence AATTCGT that was present only once in the DNA flanking the progenitor element I99.
DISCUSSION
Initiation of reverse transcription of the site-specific element R2Bm in vitro requires 250 nt of the 3′ untranslated region (UTR) (11) suggesting that the transposition complex interacts with this sequence. Since we have found that I elements with different 3′ ends can efficiently transpose, the 3′ terminal TAA repeats of wild-type I factors cannot be essential for recognition by the I factor reverse transcriptase. Any sequence or structure that it does recognize may lie in the 3′ UTR since functional I factors from D.melanogaster and Drosophila teissieri show very strong similarities in this region (24).
Synthesis of the first DNA strand of non-LTR retrotransposons is thought to be initiated at the 3′ end of the RNA transposition intermediate. We have shown here that the transposition intermediate of the I factor contains a few nucleotides flanking the element at its 3′ end and that these extra nucleotides are not integrated during retrotransposition in vivo. This suggests that reverse transcription does not initiate at the last nucleotide of the RNA template but a few nucleotides upstream, precisely at the 3′ end of the I element. Generation of precise junctions between the cDNA of the site-specific R2Bm retrotransposon and the insertion site requires RNA templates containing nucleotides from the DNA flanking the donor element (12,13). The presence of flanking sequences in the RNA might increase the precision of reverse transcription primed by a 3′ OH at the site of integration.
Termination of transcription and initiation of reverse transcription seem to be different for I factors and human L1 elements (L1Hs). Although the I factor does not contain any polyadenylation signal, its transcripts end just a few nucleotides downstream of the element indicating that it has strong signals for the end of transcription. The polyadenylation signal near the 3′ end of L1Hs appears to be very weak and, in a system to study transposition in cultured cells, transposed copies of this element contain long sequences that flanked the progenitor elements at their 3′ ends (33,34). These data indicate that reverse transcription of wild-type I factors initiates in the UAA repeats at the very end of the RNA transposition intermediate whereas that of L1Hs may initiate in flanking sequences at the end of the transcripts far from the end of L1Hs sequences.
Although I factors do not insert in a sequence-specific manner, association of DNA at the target site and nucleotides at the 3′ end of their transposition intermediates may nevertheless facilitate initiation of reverse transcription. Wild-type I factors insert into AT-rich regions and their targets often begin with the trinucleotide TAA (21) while most I elements terminating with a poly(dA) sequence have inserted into target sites beginning with two or more deoxyadenosines (Figs 2B and 3B). Similar observations have been reported for the human L1 elements and the Alu, B1, B2 and ID elements all which end with poly(dA) (2,35). Association of the transposition intermediate and target DNA may stabilize the reverse transcription complex promoting the initiation of the synthesis of the first cDNA strand. With this in mind it may be possible to modify the sequence at the 3′ end of an I factor to target it to particular chromosomal sites.
The transposed copies of I99, the modified I factor with no TAA repeats, are particularly informative. These are of two types (Fig. 3B and C): those followed by a poly(dA) sequence (IpdA insertions) and those followed by the sequence GAATTCGTAATTCG (Ifs insertions). The target site duplications flanking insertions of each type start with a few nucleotides related to those which lie immediately downstream of I99, a short run of deoxyadenosines in the case of IpdA insertions and TA or TAA in the case of Ifs insertions. This may reflect base pairing of the target site with RNA transposition intermediates which extend beyond the end of the I factor. Furthermore, the 3′ end of each Ifs insertion contains a tandem duplication of the sequence AATTCGT.
A mechanism by which these tandem repeats could be generated is shown in Figure 4. The RNA transposition intermediate, which extends beyond the end of the I factor, pairs with a few nucleotides of chromosomal DNA after cleavage of the target site by the endonuclease encoded by ORF2 (Fig. 4A and B). This allows reverse transcriptase to initiate DNA synthesis using as primer the 3′ OH at the end of the strand of the chromosomal DNA (Fig. 4C). After polymerization of a few nucleotides the RNA template dissociates and reanneals to a short complementary sequence in the newly synthesized DNA (Fig. 4D). Continued reverse transcription generates a tandem duplication in the flanking DNA before entering the body of the I factor (Fig. 4E). Once synthesis of the first strand of I factor DNA is complete, the RNase H encoded by the element degrades the RNA template in the DNA/RNA hybrid molecule. Then reverse transcriptase can synthesize the second DNA strand using the 3′ OH at the other end of the target site as primer and the newly synthesized DNA as template. This process produces a transposed copy of the I factor with a short repeated sequence at its right hand end and flanked by a target site duplication (Fig. 4F).
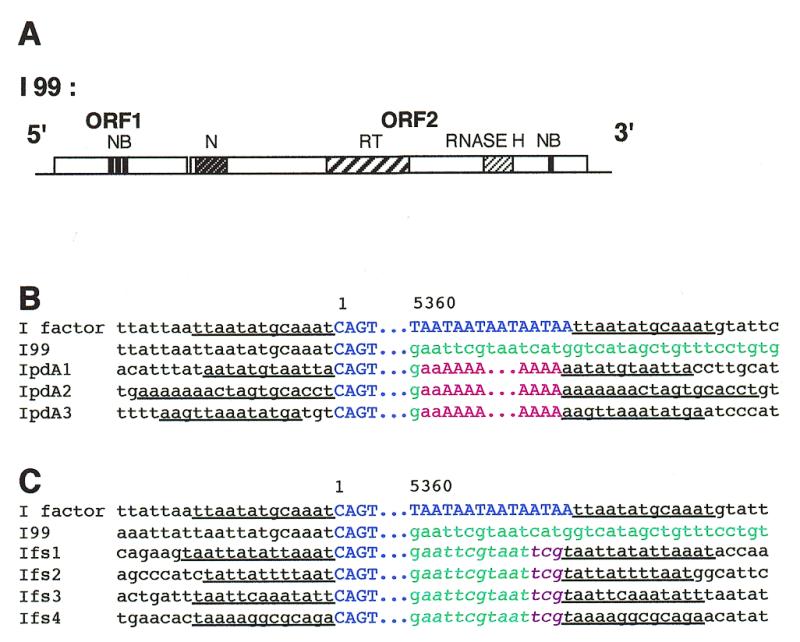
Figure 4.
A model for integration of the I factor generating a tandem repeat at the 3′ end of the element (see text). (A) The I factor encoded endonuclease cleaves chromosomal DNA at the target for integration (arrows). (B) The RNA transposition intermediate anneals to the target DNA. (C) Reverse transcription initiates using the chromosomal DNA as primer and RNA as template. (D) The RNA template dissociates from the chromosomal DNA and anneals to the newly synthesized sequence. (E) Reverse transcription extends into the I factor to complete first DNA strand synthesis. (F) After degradation of the RNA by RNase H, second strand synthesis is completed to generate a tandem duplication and the target site duplication. The target DNA is indicated in black, the RNA transposition intermediate in red, the first cDNA strand in blue and the second cDNA strand in green. The two copies of the tandem repeat generated at the 3′ end of the I factor are shown in italic and bold. The target site duplication is underlined.
Figure 4 illustrates this process for Ifs1 but IpdA insertions could be generated in a similar manner with adjacent poly(dA) sequences formed by a series of disassociation and reannealing events which may be favored by a homopolymeric sequence. The poly(dA) sequences following IpdA insertions are unlikely to have resulted from polyadenylation of RNA transposition intermediates since the potential polyadenylation signal nearest to the 3′ end of the I factor is 73 bp upstream. This process can account for variation in the number of TAA repeats at the 3′ end of wild-type elements since elements of this type generally insert at sites containing a TAA sequence (21). A similar mechanism can also explain the presence of more complex repeats at the 3′ end of some other non-LTR retrotransposons as previously suggested (36). Although non-LTR retrotransposons that end with a poly(dA) sequence, like mammalian L1 elements, have potential polyadenylation signals near their 3′ ends, the possibility that a poly(A) sequence in an RNA transposition intermediate is amplified during transposition by the process described above cannot be ruled out.
Our results suggest that the first steps of DNA synthesis by reverse transcriptases of non-LTR retrotransposons might be similar to the generation of telomeric repeats. The reverse transcriptase component of telomerase repeatedly copies a short sequence of the RNA component of this enzyme using as primer the 3′ OH of the extending chromosome end to which the telomerase RNA can associate by a few base pairs (reviewed in 37–39). The telomeres of D.melanogaster chromosomes do not comprise short tandem repeats but copies of the two non-LTR retrotransposons HeT-A (6) and TART (7) which insert preferentially at chromosome ends. Telomerase may have evolved from a primitive nucleic acid replication system with the reverse transcriptase component subsequently taking on a life of its own as a non-LTR retrotransposon. The ancestors of Diptera may then have lost telomerase by mutation and recruited the related activities of TART and HeT-A to preserve the ends of their chromosomes. It is also interesting that reverse transcription of non-LTR retrotransposons shows similarities to telomere addition to fragmented chromosomes in Euplotes crassus. Ciliates undergo chromosome fragmentation and telomere addition when the genome of the micronucleus is converted to the transcriptionally-active macronucleus. Fragmentation occurs at specific sites that are devoid of telomeric repeats and is followed by telomere addition by telomerase in a way that might be related to reverse transcription and integration of non-LTR retrotransposons (40), providing an additional similarity between these retroelements and telomeres.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Isabelle Busseau for her help at various stages of this work. This work has been supported by the CNRS and by grants from the Association pour la Recherche sur le Cancer (ARC) and The Wellcome Trust (036248/Z/92/Z).
REFERENCES
- 1.Feng Q., Moran,J.V., Kazazian,H.H. and Boeke,J.D. (1996) Cell, 87, 905–916. [DOI] [PubMed] [Google Scholar]
- 2.Feng Q., Scumann,G. and Boeke,J.D. (1998) Proc. Natl Acad. Sci. USA, 95, 2083–2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gabriel A. and Boeke,J.D. (1991) Proc. Natl Acad. Sci. USA, 88, 9794–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mathias M.L., Scott,A.F., Kazazian,H.H., Boeke,J.D. and Gabriel,A. (1991) Science, 254, 1808–1810. [DOI] [PubMed] [Google Scholar]
- 5.Jakubczak J.L., Burke,W.D. and Eickbush,T.H. (1991) Proc. Natl Acad. Sci. USA, 88, 3295–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biessman H., Valgeirsdottir,K., Lofsky,A., Chin,C., Ginther,B., Levis,R.W. and Pardue,M.-L. (1992) Mol. Cell. Biol., 12, 3910–3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levis R.W., Ganesan,R., Houtchens,K., Tolar,L.A. and Sheen,F. (1993) Cell, 75, 1083–1093. [DOI] [PubMed] [Google Scholar]
- 8.Finnegan D.J. (1997) Curr. Biol., 7, 245–248. [DOI] [PubMed] [Google Scholar]
- 9.Xiong Y. and Eickbush,T.H. (1988) Cell, 55, 235–246. [DOI] [PubMed] [Google Scholar]
- 10.Luan D.D., Korman,M.H., Jakubczak,J.L. and Eickbush,T.H. (1993) Cell, 72, 595–605. [DOI] [PubMed] [Google Scholar]
- 11.Luan D.D. and Eickbush,T.H. (1995) Mol. Cell. Biol., 15, 3882–3891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luan D.D. and Eickbush,T.H. (1996) Mol. Cell. Biol., 16, 4726–4734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eickbush D.G., Luan,D.D. and Eickbush,T.H. (2000) Mol. Cell. Biol., 20, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fanning T. (1983) Nucleic Acids Res., 11, 5073–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voliva C.F., Martin,S., Hutchison,III,C.A. and Edgell,M.H. (1984) J. Mol. Biol., 178, 795–813. [DOI] [PubMed] [Google Scholar]
- 16.Skowronsky J., Fanning,T.G. and Singer,M.F. (1988) Mol. Cell. Biol., 8, 1385–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fawcett D.H., Lister,C.K., Kellet,E. and Finnegan,D.J. (1986) Cell, 47, 1007–1015. [DOI] [PubMed] [Google Scholar]
- 18.Xiong Y. and Eickbush,T.H. (1993) Nucleic Acids Res., 21, 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felger I. and Hunt,J.A. (1992) Genetica, 85, 119–130. [DOI] [PubMed] [Google Scholar]
- 20.Besansky N.J., Bedell,J.A. and Mukabayire,O. (1994) Insect Mol. Biol., 3, 49–56. [DOI] [PubMed] [Google Scholar]
- 21.Bucheton A. (1990) Trends Genet., 6, 16–21. [DOI] [PubMed] [Google Scholar]
- 22.Busseau I., Chaboissier,M.-C., Pélisson,A. and Bucheton,A. (1994) Genetica, 93, 101–116. [DOI] [PubMed] [Google Scholar]
- 23.Picard G. (1976) Genetics, 83, 107–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abad P., Vaury,C., Pelisson,A., Chaboissier,M.-C., Busseau,I. and Bucheton,A. (1989) Proc. Natl Acad. Sci. USA, 86, 8887–8891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chaboissier M.-C., Busseau,I., Prosser,J., Finnegan,D.J. and Bucheton,A. (1990) EMBO J., 9, 3557–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pritchard M.A., Dura,J.-M., Pelisson,A., Bucheton,A. and Finnegan,D.J. (1998) Mol. Gen. Genet., 214, 533–540. [DOI] [PubMed] [Google Scholar]
- 27.Geyer P.K. and Corces,V.G. (1987) Genes Dev., 1, 996–1004. [DOI] [PubMed] [Google Scholar]
- 28.Chaboissier M.-C., Bucheton,A. and Finnegan,D.J. (1998) Proc. Natl Acad. Sci. USA, 95, 11781–11785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Spradling A.C. and Rubin,G.M. (1982) Science, 218, 348–353. [DOI] [PubMed] [Google Scholar]
- 30.Huang L., Zhu,Y. and Anders,D.G. (1996) J. Virol., 70, 5272–5281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Busseau I., Pélisson,A. and Bucheton,A. (1989) Mol. Gen. Genet., 218, 222–228. [DOI] [PubMed] [Google Scholar]
- 32.Jensen S. and Heidmann,T. (1991) EMBO J., 10, 1927–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran J.V., Holmes,S.E., Naas,T.P., DeBerardinis,R.J., Boeke,J.D. and Kazazian,H.H. (1996) Cell, 87, 917–927. [DOI] [PubMed] [Google Scholar]
- 34.Moran J.V., DeBerardinis,R.J. and Kazazian,H.H. (1998) Science, 283, 1530–1534. [Google Scholar]
- 35.Jurka J. (1997) Proc. Natl Acad. Sci. USA, 94, 1872–1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogiwara I., Miya,M., Oshima,K. and Okada,M. (1999) Mol. Biol. Evol., 16, 1238–1250. [DOI] [PubMed] [Google Scholar]
- 37.Blackburn E.H. (1992) Annu. Rev. Biochem., 61, 113–129. [DOI] [PubMed] [Google Scholar]
- 38.Zakian V.A. (1995) Science, 270, 1601–1607. [DOI] [PubMed] [Google Scholar]
- 39.Prescott J.C. and Blackburn,E.H. (1999) Curr. Opin. Genet. Dev., 9, 368–373. [DOI] [PubMed] [Google Scholar]
- 40.Klobutcher L.A. (1999) Mol. Cell, 4, 695–704. [DOI] [PubMed] [Google Scholar]