Abstract
This is the first report of a complete mitochondrial genome sequence from a photosynthetic member of the stramenopiles, the chrysophyte alga Chrysodidymus synuroideus. The circular-mapping mitochondrial DNA (mtDNA) of 34 119 bp contains 58 densely packed genes (all without introns) and five unique open reading frames (ORFs). Protein genes code for components of respiratory chain complexes, ATP synthase and the mitoribosome, as well as one product of unknown function, encoded in many other protist mtDNAs (YMF16). In addition to small and large subunit ribosomal RNAs, 23 tRNAs are mtDNA-encoded, permitting translation of all codons present in protein-coding genes except ACN (Thr) and CGN (Arg). The missing tRNAs are assumed to be imported from the cytosol. Comparison of the C.synuroideus mtDNA with that of other stramenopiles allowed us to draw conclusions about mitochondrial genome organization, expression and evolution. First, we provide evidence that mitochondrial ORFs code for highly derived, unrecognizable versions of ribosomal or respiratory genes otherwise ‘missing’ in a particular mtDNA. Secondly, the observed constraints in mitochondrial genome rearrangements suggest operon-based, co-ordinated expression of genes functioning in common biological processes. Finally, stramenopile mtDNAs reveal an unexpectedly low variability in genome size and gene complement, testifying to substantial differences in the tempo of mtDNA evolution between major eukaryotic lineages.
INTRODUCTION
Mitochondrial genome analysis has been recognized as a valuable tool for resolving evolutionary relationships among the various eukaryotic lineages. While about 150 complete mitochondrial DNA (mtDNA) sequences have been published over the last 20 years, these data still cover only a small fraction of the major extant eukaryotic groups. The vast majority of mtDNAs sequenced are from animals (about 95), approximately 15 are from fungi and two are from plants, whereas protists, which represent by far the largest and most diverse eukaryotic assemblage, are conspicuously under-represented with a total of only about 25 genomes sequenced. For numerous protist divisions, not even a single representative mtDNA has been completely sequenced at the present time (1).
This study focuses on a large group of flagellated protists that are characterized by particular ‘hair’-covered flagella, and are therefore named ‘stramenopiles’ (‘straw-haired’). At the level of cellular ultrastructure, this group is defined by tubulocristate mitochondria and tripartite flagellar hairs. Also termed heterokonts, they consist of nearly 20 divisions, including photosynthetic organisms (‘chromophytes’) such as chrysophyte (golden), phaeophyte (brown), diatom, raphidophyte and xanthophyte algae, as well as non-photosynthetic taxa such as bicosoecids, labyrinthulomycetes and oomycetes (2). The latter two groups had originally been included in the true fungi, due to their fungal-like morphology and absorptive nutrition. However, cytoskeletal ultrastructure (2) and molecular data (3–6) unambiguously classify oomycetes and labyrinthulomycetes as members of heterokont protists. Certain stramenopile species are well studied owing to their contribution to overall biomass (diatoms) and polysaccharide production (brown algae), others for being major infectious agents of agricultural importance, e.g., the potato late blight pathogen Phytophthora infestans.
Stramenopile morphology ranges from giant kelps to nearly bacteria-sized golden algae, from myceliar oomycetes to yeast-like thraustochytrids and from amoeboid chlamydomyxetes to silicious shell-forming diatoms. Reproductive strategies found in this group (including sexual and asexual propagation) are similarly heterogeneous, as is their geographic distribution, spanning numerous freshwater, marine and terrestrial habitats in virtually all climate regions (for an overview, see 7). Such a spectacular range of organismal variety raises the question whether stramenopiles are an evolutionarily ancient group or rather more rapidly diversifying than other eukaryotic lineages. However, available nuclear rRNA sequence data provide few clues as to the order in which the major stramenopile divisions have emerged in evolutionary time (8), or the specific relationship of stramenopiles to other tubulocristate protists such as dinoflagellates, ciliates and apicomplexans (5). Especially controversial is the question whether or not stramenopiles were originally photosynthetic, i.e., whether heterotrophic lineages such as oomycetes, bicosoecids and labyrinthulomycetes lost their chloroplasts during evolution or never had any. In this regard, genomic data such as complete mitochondrial or chloroplast DNA sequences from stramenopiles are key to a better understanding of the evolutionary history of these highly diverse creatures.
The Organelle Genome Megasequencing Program (OGMP) has been investigating mtDNAs from several stramenopiles and presents here the first detailed description of a complete mitochondrial genome from a photosynthetic member of this assemblage, Chrysodidymus synuroideus. This golden freshwater alga was formally described in 1962 (9), but an in-depth light and electron-microscopical characterization was only published 30 years later (10). Chrysodidymus synuroideus is a unicellular organism with cells 5–10 µm in length that congregates as two-cell ‘colonies’ resembling a stretched-out pair of sausages. Two flagella of unequal length emerge at the anterior end, the longer of which carries tripartite tubular hairs; both flagella and hairs are covered with siliceous scales. The body also is covered by scales that closely resemble those of the better known genus Mallomonas (solitary cells) and especially Synura, which are colonial organisms. Scale morphology and kinetid architecture are among the features that place Chrysodidymus in the same family (Synuraceae, elevated by some authors to order Synurales and/or class Synurophyceae) as Mallomonas and Synura. No member of the Synuraceae is known to be mixotrophic (to prey on micro-organisms). Images of C.synuroideus can be inspected at the Protist Image Database (PID) at http://megasun.bch.umontreal.ca/protists/protists.html
We describe here the gene content and genome organization of C.synuroideus mtDNA, and compare these features with those from other, photosynthetic and non-photosynthetic, stramenopiles. On the basis of this comparison, we discuss mitochondrial genome diversity and evolution within this eukaryotic group. Phylogenetic issues will be addressed in detail in a subsequent paper together with the release of further stramenopile mtDNA sequences.
MATERIALS AND METHODS
The complete mtDNA sequence of C.synuroideus has been deposited in GenBank (accession number AF222718).
Culture of C.synuroideus and mtDNA isolation
Chrysodidymus synuroideus Prowse was originally isolated from Jyme Lake, a sphagnum bog, located at Kemp Biological Station, Vilas Co., WI, USA (10). This culture has been deposited with the UTEX Algal Culture Collection, Austin, TX, USA (LB 2713). Uni-algal cultures were established (11), and log phase cells (5.9 × 104 cells/ml) from a 10 l culture were used to extract total DNA using a protocol designed by Chesnick and Cattolico (12). Mitochondrial, chloroplast and nuclear DNA were separated through CsCl–bisbenzimide isopycnic centrifugation, by which mtDNA forms the most A+T-rich band.
Cloning and DNA sequencing
Mitochondrial DNA was physically sheared by nebulization (13), and a size fraction of 500–3000 bp was recovered after agarose gel electrophoresis. The DNA was incubated with a mixture of T7 DNA polymerase and Escherichia coli DNA polymerase I (the Klenow fragment) to generate blunt ends, and then cloned into the SmaI site of the phagemid pFBS (B.F.Lang, unpublished results), a shortened derivative of pBluescript II KS+ (Stratagene, La Jolla, CA). Recombinant plasmids containing mtDNA inserts were identified by colony hybridization using mtDNA as a probe. Clones contained in this random library encompassed the entire C.synuroideus mitochondrial genome.
DNA sequencing and data analysis
DNA sequencing was performed by the dideoxy chain termination method (14), using single-stranded DNA as a template and [35S]dATP as a label. Acrylamide gels, dried onto the glass plate (15), were autoradiographed, and sequences were entered manually into computer files. In addition, automated sequencing was performed on a Li-Cor 4000L apparatus, using an end-labeled primer and a cycle-sequencing protocol (Amersham, Piscataway, NJ).
Sequence readings were assembled using the XBAP package (16) and the consensus sequence was integrated, together with feature annotations, in the masterfile format (http://megasun.bch.umontreal.ca/ogmp/ogmpid.html ). The FASTA program (17) was employed for searches in local databases and the BLAST network service (18) for similarity searches in GenBank at the National Center for Biotechnology Information. Custom-made batch utilities were used for submitting queries and browsing the output (BBLAST, TBOB, BFASTA and FOB). Multiple protein alignments were performed with the CLUSTAL W program (19), integrated into the GDE package (Genetic Data Environment; 20). A number of additional programs, including multiple sequence file manipulation, pre-processing, conversion and batch utilities for XBAP, FASTA and GDE, as well as a masterfile maintenance suite have been developed by the OGMP. These utilities are described in more detail and are available through the OGMP website at http://megasun.bch.umontreal.ca/ogmp/ogmpid.html
RESULTS
Physical properties, gene content and architecture of C.synuroideus mtDNA
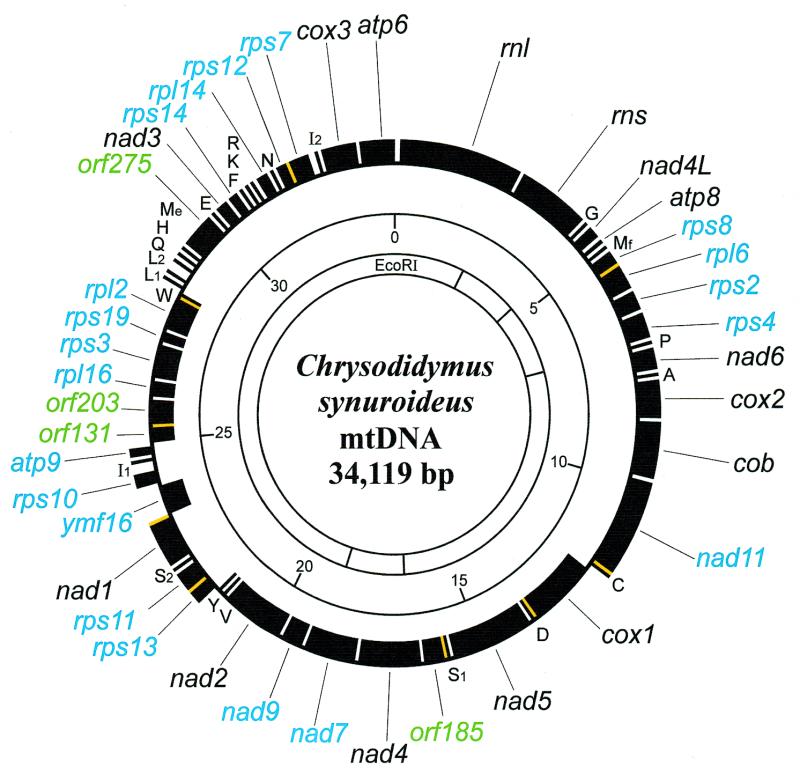
Figure 1 depicts the physical and gene map of the circular 34 119 bp C.synuroideus mtDNA. Its overall A+T content (75.9%) and its higher A+T content in coding regions (87.5%) compared to intergenic regions (75.2%) is typical for mtDNAs. Genes are densely packed, making up 94.3% of the entire sequence, leaving intergenic regions between 1 and 207 nt, with a majority being 1–30 nt long and only five exceeding 100 nt. Genome compactness is further evidenced by the overlap of nine pairs of adjacent genes by 2–71 nt (average 30 nt). Genes are encoded on both DNA strands, and the gene arrangement suggests two major transcription units, each covering about one-half of the genome. A conspicuous stem-loop structure has been detected between trnW(cca) and trnL(uag) that might serve as a control element of a bi-directional promoter. In the mtDNA of the red alga, Chondrus crispus, a hairpin of comparable size has been demonstrated to play a role in transcription initiation (21), however, no significant sequence similarity between the C.synuroideus and C.crispus stem–loops is discernable.
Figure 1.
Gene and physical map of the C.synuroideus mitochondrial genome. Black blocks represent genes and ORFs which are transcribed clockwise (outside of circle) or counter-clockwise (inside of circle). Names of tRNA genes are indicated by the amino acid (one-letter code) they specify. Mf, Me, initiator and elongator trnM(cau); I1, trnI(uau); I2, trnI(gau); L1, trnL(cag); L2, trnL(aag); L3, trnL(caa); L4, trnL(cua); R1, trnR(acg); R2, trnR(ccu); R3, trnL(ucu); S1, trnS(gcu); S2, trnS(gga). Color coding of gene names discriminates genes typically found in mtDNAs of animals and fungi (black), protists (blue) and hypothetical protein-coding genes (ORFs) (green). The second circle shows the size scale, and the inner ring the EcoRI restriction fragments.
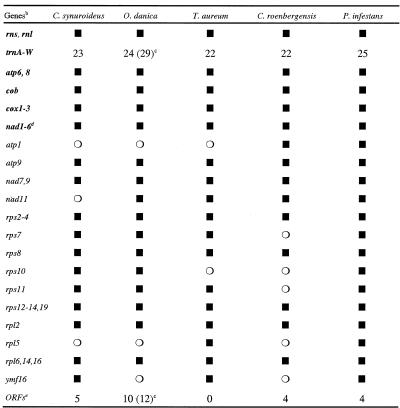
Table 1 lists the 63 genes (all without introns) and open reading frames (ORFs) residing in C.synuroideus mtDNA. Protein genes code mostly for components involved in respiration and oxidative phosphorylation. These are subunits of NADH:ubiquinone oxidoreductase (respiratory complex I), ubiquinone:cytochrome c oxidoreductase (complex III), cytochrome oxidase (complex IV) and ATP-synthase (complex V). In addition, genes for 15 protein components of the small and large subunits of the mitoribosome are found in this mtDNA (rps2–4, 7, 8, 10–14, 19 and rpl2, 6, 14, 16), as well as a gene (ymf16) encoding a conserved protein of as yet unassigned function.
Table 1. Gene content in stramenopile mtDNAsa.
aFor species abbreviations, see legend to Figure 2. Filled squares indicate presence, open circle absence of gene. Gene maps of T.aureum and O.danica mtDNAs are deposited at http://megasun.bch.umontreal.ca/ogmp/projects/individual.html
bGenes common to animal and fungal mtDNAs are shown in bold.
cNumber in brackets includes duplicated genes copies.
dIncludes nad1-4, nad4L and nad5-6.
eORFs larger than 60 amino acids, except orf32 (32 residues) in P.infestans mtDNA.
All the above genes could be readily identified at the sequence level, since no extraordinary deviations in length or in otherwise conserved domains are apparent. Only the identification of orf150 as ribosomal protein gene rps2 proved difficult. Support for this assignment is provided by the fact that orf150 is embedded in a cluster of ribosomal protein genes (rps8–rpl6–orf150–rps4), in the identical relative position as rps2 in the mtDNAs of P.infestans (22–24) and the bicosoecid Cafeteria roenbergensis (25,26). In addition, ORFs reminiscent of, and positionally equivalent to, the latter rps2 genes were detected in two other stramenopile mtDNAs, namely the chrysophyte Ochromonas danica and the labyrinthulomycete Thraustochytrium aureum (G.Burger, B.F.Lang and M.W.Gray, unpublished results). We performed multiple protein alignments including the mitochondrially-encoded counterparts of five Rps2 from stramenopiles and from other protists with little derived mitochondrial gene sequences, as well as selected chloroplast and bacterial Rps2 proteins. The alignment in Figure 2 shows the C-terminal half of the proteins (corresponding to residues 165–222 in the E.coli protein) that is best conserved across all taxa. Within stramenopiles, the putative S2 homologs appear to deviate progressively, starting from P.infestans and C.roenbergensis to O.danica, T.aureum and C.synuroideus, with the counterparts of the latter two bearing only barely discernible vestiges of their bacterial ancestor. With the identification of C.synuroideus orf150 as rps2, the total number of conserved genes in this mitochondrial genome totals 58. In addition, this mtDNA harbors a total of five hypothetical proteins (ORFs) of 60 residues or more that have no counterparts in other genomes.
Figure 2.
Multiple alignment of organellar and bacterial S2 proteins. Protein sequences were deduced from rps2 nucleotide sequences, and aligned using Clustal W (19), followed by manual adjustments. bc, cp, mt, bacterial, chloroplast or mitochondrial gene. GenBank accession numbers of protein sequences are indicated in brackets. E.coli, Escherichia coli (γ-proteobacterium; P02351); C.burn., Coxiella burnetii (α-proteobacterium; AAD33342); R.prow., Rickettsia prowasekii (α-proteobacterium; Q9ZE61); E.grac., Euglena gracilis (euglenoid; P30389); R.amer., Reclinomonas americana (jakobid; AAD11918); J.libe., Jakoba libera (jakobid; G.Burger, M.W.Gray and B.F.Lang, unpublished results); M.poly., Marchantia polymorpha (embryophyte; P26864); R.sali., Rhodomonas salina (cryptophyte; G.Burger, M.W.Gray and B.F.Lang, unpublished results); T.aest., Triticum aestivum (embryophyte; CAA74226); P.infe., Phytophthora infestans (oomycete; NP_041459); C.roen., Cafeteria roenbergensis (bicosoecid; AAF05802); O.dani., Ochromonas danica (chrysophyte); T.aure., Thraustochytrium aureum (labyrinthulomycete; G.Burger, M.W.Gray, B.F.Lang, unpublished results); C.synu., C. synuroideus (chrysophyte, this report). Unpublished sequences are available at http://megasun.bch.umontreal.ca/ogmp/projects/projects.html ). In columns with five or more identical residues, amino acids have been highlighted by colors that have PAM250 values ≥0 with regard to the most abundant amino acid of the corresponding column. Numbers to the left of the protein sequences indicate the position where the listed sequences start. Numbers in brackets indicate the number of residues not shown in the alignment. Dashes indicate alignment gaps and asterisks the C-terminus of the proteins.
The standard genetic code is used in the translation of C.synuroideus mitochondrial-encoded proteins. Table 2 shows that TAA is preferred over the TAG stop codon, whereas TGA codons do not occur at all. Codon frequency is biased by a very rare usage of CTC and CTG (Leu), and a complete lack of CGG (Arg) codons. We scrutinized the highly divergent rps2 sequence for potentially abnormal codon usage which might indicate that it is a pseudogene, with an active copy residing in the nucleus. However, the codon frequency of rps2 is not significantly different from that of all other protein-coding genes nor is that of the five ORFs (Table 2). Therefore, we assume that all assigned genes, as well as ORFs in this genome, are translated.
Table 2. Codon frequency in genes and ORFs of C.synuroideus mtDNAa.
| AAb | Codon | Genes | ORFs | AA | Codon | Genes | ORFs | AA | Codon | Genes | ORFs | AA | Codon | Genes | ORFs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | % | % | % | % | % | % | % | ||||||||
| F | TTT | 95 | 94 | S | TCT | 37 | 37 | Y | TAT | 89 | 90 | C | TGT | 89 | 70 |
| F | TTC | 5 | 6 | S | TCC | 3 | 8 | Y | TAC | 11 | 10 | C | TGC | 11 | 30 |
| L | TTA | 78 | 71 | S | TCA | 29 | 25 | * | TAA | 94 | 75 | * | TGA | – | – |
| L | TTG | 8 | 10 | S | TCG | 6 | 2 | * | TAG | 6 | 25 | W | TGG | 100 | 100 |
| L | CTT | 9 | 11 | P | CCT | 38 | 58 | H | CAT | 90 | 71 | R | CGT | 35 | 19 |
| L | CTC | 0 | – | P | CCC | 7 | 16 | H | CAC | 10 | 29 | R | CGC | 5 | 6 |
| L | CTA | 5 | 7 | P | CCA | 47 | 16 | Q | CAA | 96 | 88 | R | CGA | 15 | 0 |
| L | CTG | 0 | 1 | P | CCG | 9 | 11 | Q | CAG | 4 | 12 | R | CGG | – | 0 |
| I | ATT | 59 | 44 | T | ACT | 45 | 50 | N | AAT | 88 | 88 | S | AGT | 20 | 25 |
| I | ATC | 5 | 6 | T | ACC | 8 | 7 | N | AAC | 12 | 12 | S | AGC | 4 | 4 |
| I | ATA | 35 | 49 | T | ACA | 42 | 40 | K | AAA | 91 | 88 | R | AGA | 41 | 62 |
| M | ATG | 100 | 100 | T | ACG | 5 | 3 | K | AAG | 9 | 12 | R | AGG | 4 | 12 |
| V | GTT | 56 | 47 | A | GCT | 48 | 60 | D | GAT | 92 | 88 | G | GGT | 57 | 36 |
| V | GTC | 4 | – | A | GCC | 6 | – | D | GAC | 8 | 12 | G | GGC | 3 | – |
| V | GTA | 35 | 44 | A | GCA | 35 | 33 | E | GAA | 88 | 81 | G | GGA | 31 | 64 |
| V | GTG | 4 | 9 | A | GCG | 11 | 7 | E | GAG | 12 | 19 | G | GGG | 10 | – |
aZero, frequency ≤0.5 and >0%; minus, not a single occurrence of the corresponding codon in the particular reading frames. The root square deviation (RMSD) and correlation index (R) of the codon frequencies of individual protein-coding genes and ORFs was compared with that of all assigned protein-coding genes taken together (excluding rps2). In the case of assigned genes, RMDS and R values vary between 0.326/0.616 (atp9) and 0.063/0.982 (nad1). Codon frequenceis of rps2 and the ORFs are not significantly different from those assigned genes, because their values fall within the range observed for assigned genes, namely 0.206/0.840 (rps2), 0.230/0.790 (orf131), 0.275/0.716 (orf185), 0.195/0.831 (orf203) and 0.146/0.902 (orf275).
bAA, cognate amino acid in one-letter code; asterisk, stop codon.
In addition to these 38 protein-coding genes and ORFs, a total of 25 RNA genes are present in C.synuroideus mtDNA, coding for 23 tRNAs and the small and large subunit (SSU, LSU) rRNAs. The two rRNAs display conventional eubacteria-like secondary structures with predicted sizes of 2586 and 1579 nt, the LSU rRNA being significantly shorter than the E.coli counterpart (2904 nt). A 5S rRNA gene was not detected. Mitochondrially-encoded tRNAs display standard cloverleaf secondary structures and permit translation of nearly all codons found in protein-coding mitochondrial genes. As in a number of other protists, two isoleucine-specifying tRNA genes are present, trnI(gau) and trnI(cau). ATA (Ile) codons are assumed to be recognized by the gene product of trnI(cau), whose C in the anticodon is post-translationally modified to lysidine which pairs with A but not with G (27,28). Transfer RNAs absent from C.synuroideus mtDNA are ones recognizing ACN (Thr) and CGN (Arg) codons, which occur abundantly in protein genes of this genome. Import of cytoplasmic tRNAs is believed to compensate for the missing mitochondrial tRNA genes. Finally, a conspicuous tRNA-like sequence is present, downstream of rps10 and overlapping its C-terminus by 10 nt. Despite a typical amino-acyl stem and T arm, the lack of universally conserved nucleotides and of canonical secondary structure folding capacity in the D and anticodon stem–loops suggest that this sequence element is not a functional tRNA gene. It might be a remnant of a previously active tRNA gene that is now functionally replaced by a nuclear-encoded counterpart, but no specific features are discernable in this tRNA-like sequence that would be characteristic of the two genes missing from the mtDNA, i.e., trnR(ucg) or trnT(ugu). Alternatively, the tRNA-like sequence might be a vestige of an intra-molecular recombination process, since breakpoints involved in major genome rearrangements within mitochondrial genomes often coincide with tRNA genes (G.Burger, B.F.Lang and M.W.Gray, unpublished results).
Mitochondrial gene complement in stramenopiles
Table 1 compares the gene content of mtDNAs from five stramenopiles: the photosynthetic C.synuroideus described here, another golden alga, O.danica, and three non-photosynthetic taxa, the labyrinthulomycete T.aureus (for references, see Table legend), the bicosoecid C.roenbergensis (25,26) and the oomycete P.infestans (22–24). All these stramenopile mtDNAs code for LSU and SSU rRNAs and 22–25 tRNAs, and share a common set of 27 protein-coding genes that include, in addition to the fungal/animal repertoire, genes typical for protists, i.e., atp9, nad7, 9, 11; rps3, 4, 12–14, 19 and rpl2, 6, 14, 16. These genomes differ with respect to the absence/presence of seven genes, namely, atp1, rps7, 10, 11, rpl2, 5 and ymf16. Most similar in mitochondrial gene content are the two chrysophyte algae, which differ in the presence of two genes only: nad11 is absent from C.synuroideus but present in O.danica, and vice versa in the case of ymf16. The absence of rpl5 and atp1 from both the C.synuroideus and O.danica genomes suggests that these genes were lost in a recent, shared chrysophyte ancestor. Differences in gene numbers between the other three stramenopile mtDNAs vary from three to five, with Phytophthora containing the largest mitochondrial gene set (22–24). Finally, it should be mentioned that the gene types retained or lost in stramenopile mtDNAs corroborate the notion of an overall trend in the order in which genes were lost from mitochondria (or migrated to the nucleus), a trend that appears to be common to all eukaryotic lineages, as we have discussed in a recent review (1).
DISCUSSION
Mitochondrial DNAs of protists typically contain 0–15 ORFs, five on average, which have no counterparts in any other organism [see the Organelle Genome Database (GOBASE) at http://megasun.bch.umontreal.ca/gobase/ ; 29]. Our working hypothesis is that these ORFs may be highly divergent and, therefore, unrecognizable versions of ‘missing’ respiratory or ribosomal protein genes (30). This hypothesis draws support from the comparative data on stramenopile mtDNAs reported here. The stramenopile mitochondrial rps2 gene is an excellent illustration of gradual deterioration of sequence conservation across this lineage, to a degree where the most derived members would not have been recognized without knowledge of the intermediate ones.
From the complete sequence data of the photosynthetic stramenopile C.synuroideus (this report), in combination with data from four other taxa of this group (see Results), typical characteristics of the stramenopile mitochondrial genome begin to emerge. These mitochondrial DNAs range between 34 and 43 kb in size and harbor 29–34 protein-coding genes, as many as 28 of which are common to all members of this assemblage (Table 1). Of similar low variability is the number of genes coding for tRNA species in these mtDNAs, ranging from 23 to 25. Interestingly, the sets of mitochondrial-encoded tRNA genes from all taxa studied lack one specifying ACN (Thr), which must have been lost from an ancestral mtDNA soon after the emergence of the stramenopile lineage. In fact, trnT is the most frequently absent tRNA gene among protist mtDNAs (26).
The above observations reveal a narrow range of variation in genome size and number of mitochondrially-encoded genes within known stramenopiles, although the extensive variety in morphology, ecology and physiology within this assemblage may have suggested otherwise. A similarly narrow range of mtDNA variation was observed previously within rhodophyte protists (31). This situation contrasts sharply with the extraordinary mtDNA diversity in the chlorophyte lineage (32) and exemplifies large differences in the tempo of mtDNA evolution between major protist phyla.
Although introns are absent from the mtDNA of all five of the stramenopiles compared in this study (26), the brown alga Pylaiella littoralis, also a stramenopile, was reported to harbor group II introns in the cox1 and rnl genes (33; the complete mtDNA sequence is not available for this species). We believe, however, that introns were not ancestrally present in stramenopile mitochondrial genomes. The fact that the rnl introns of P.littoralis are highly similar to cyanobacterial introns suggests that phaeophytes only recently acquired these introns via horizontal transfer. An even more recent acquisition of the same intron type has been suggested for mtDNA of the red alga Porphyra purpurea (31), indicating that this cyanobacteria-like intron is exceptionally active in lateral propagation.
Mitochondrial genomes, and those from protists in particular, exhibit a number of characters that testify to the bacterial ancestry of this organelle. Relics of the bacterial str, S10, spc and α operons, which mainly comprise ribosomal proteins, have been detected in numerous protist and plant mtDNAs, with the most highly conserved operons in the jakobid flagellate Reclinomonas americana (34). Chrysodidymus synuroideus mtDNA (Fig. 1) comprises four arrays in which 14 of its 15 ribosomal protein genes are grouped together. Notably, only part of these arrays exhibit the ancestral, bacteria-like gene order. In the cluster rps8–rpl6–rps2–rps4, which we discovered in all five stramenopiles compared in this report, only the first gene pair reflects a bacterial arrangement (spc operon), whereas addition of rps2 and rps4 to this cluster must have arisen secondarily and specifically in stramenopiles. A second riboprotein gene array in C.synuroideus mtDNA that has most probably been assembled secondarily is rps14–rpl14–rps12–rps7. In eubacterial genomes, only the last two genes are found adjacent (str operon), whereas the other two are non-adjacent members of the spc operon. Most likely true relics of the original, bacterial gene arrangement are the two arrays rpl2–rps19–rps3–rpl16 and rps13–rps11, which correspond to the S10 and α operons in E.coli, and occur in mtDNAs of several protists, namely in the jakobid R.americana (34), the rhizopod amoeba Acanthamoeba castellanii (35), the green alga Nephroselmis olivacea (32) and in a primitive plant, the liverwort Marchantia polymorpha (36).
Conservation of mitochondrial gene arrays that are organized like ancestral, bacterial operons strongly suggests that expression of these genes is temporally and/or stoichiometrically co-ordinated by a mechanism involving co-transcription/co-translation, known from bacterial systems. An even stronger indication that this mode of concerted regulation is indeed active in certain mitochondria is the finding of secondary clustering of groups of mitochondrial genes functioning in a common biological process (the ‘guilt by association’ principle; 37,38). Among the organisms featuring secondary clustering of mitochondrial riboprotein genes, C.synuroideus exhibits two of the most evident examples, as outlined above (see also ref. 39 for data on the green alga Prototheca wickerhamii).
Acknowledgments
ACKNOWLEDGEMENTS
We thank M. W. Gray (Dalhousie University, Halifax, NS, Canada) for critical comments on the manuscript, C. O’Kelly (Bigelow Laboratory for Ocean Science, West Boothbay Harbor, ME, USA) for helpful information concerning organismal aspects of this work, L. Forget for excellent technical assistance in library construction and sequencing and L. E. Graham (University of Wisconsin, Madison, WI, USA) for providing organismal images of C.synuroideus to the Protist Image Database. This investigation was supported by the Medical Research Council, Canada, grant SP-14226, salary support by the Canadian Institute for Advanced Research (CIAR) to B.F.L. and G.B., a generous academic equipment grant by Sun microsystems (Palo Alto, CA, USA) and the donation of an automatic sequencer by Li-Cor (Lincoln, NE, USA).
DDBJ/EMBL/GenBank accession no. AF222718
REFERENCES
- 1.Lang B.F., Gray,M.W. and Burger,G. (1999) Annu. Rev. Genet., 33, 351–397. [DOI] [PubMed] [Google Scholar]
- 2.Patterson D.J. (1989) In Green,J.C., Leadbeater,B.S.C. and Diver,W.L. (eds), Chromophyte Algae: Problems and Perspectives. Clarendon Press, Oxford, UK, pp. 357–379.
- 3.Gunderson J.H., Elwood,H., Ingold,A., Kindle,K. and Sogin,M.L. (1987) Proc. Natl Acad. Sci. USA, 84, 5823–5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ariztia E.V., Andersen,R.A. and Sogin,M.L. (1991) J. Phycol., 27, 428–436. [Google Scholar]
- 5.Leipe D.D., Wainright,P.O., Gunderson,J.H., Porter,D., Patterson,D.J., Valois,F., Himmelreich,S. and Sogin,M.L. (1994) Phycologia, 33, 369–377. [Google Scholar]
- 6.Saunders G.W., Potter,D., Paskind,P. and Andersen,A. (1995) Proc. Natl Acad. Sci. USA, 92, 244–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marquis L., Corliss,J.O., Melkonian,M. and Chapman,D.J. (eds) (1990) Handbook of Protoctista. Jones and Bartlett, Boston, MA, pp. 454–466.
- 8.Andersen R.A., Potter,D., Bidigare,R.R., Latasa,M., Rowan,K. and O’Kelly,C.J. (1998) J. Phycol., 34, 286–298. [Google Scholar]
- 9.Prowse G.A. (1962) Gardens Bull. (Singapore), 19, 105–146. [Google Scholar]
- 10.Graham L.E., Graham,J.M. and Wujek,D.E. (1993) J. Phycol., 29, 330–341. [Google Scholar]
- 11.Chesnick J.M., Tuxbury,K., Coleman,A., Burger,G. and Lang,B.F. (1996) J. Phycol., 32, 452–456. [Google Scholar]
- 12.Chesnick J.M. and Cattolico,R.A. (1993) Methods Enzymol., 224, 168–176. [DOI] [PubMed] [Google Scholar]
- 13.Okpodu C.M., Robertson,D., Boss,W.F., Togasaki,R.K. and Surzycki,S.J. (1994) Biotechniques, 16, 154–159. [PubMed] [Google Scholar]
- 14.Sanger F., Nicklen,S. and Coulson,A.R. (1977) Proc. Natl Acad. Sci. USA, 74, 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lang B.F. and Burger,G. (1990) Anal. Biochem., 188, 176–180. [DOI] [PubMed] [Google Scholar]
- 16.Dear S. and Staden,R. (1991) Nucleic Acids Res., 19, 3907–3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pearson W.R. (1990) Methods Enzymol., 183, 63–98. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 19.Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith S.W., Overbeek,R., Woese,C.R., Gilbert,W. and Gillevet,P.M. (1994) Comput. Appl. Biosci., 10, 671–675. [DOI] [PubMed] [Google Scholar]
- 21.Richard O., Bonnard,G., Grienenberger,J.M., Kloareg,B. and Boyen,C. (1998) J. Mol. Biol., 283, 549–557. [DOI] [PubMed] [Google Scholar]
- 22.Lang B.F. (2000) GenBank accession no. U17009.
- 23.Lang B.F. and Forget,L. (1993) In O’Brien,S.J. (ed), Genetic Maps. Locus Map of Complex Genomes. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 3.133–3.135.
- 24.Paquin B., Laforest,M.J., Forget,L., Roewer,I., Wang,Z., Longcore,J. and Lang,B.F. (1997) Curr. Genet., 31, 380–395. [DOI] [PubMed] [Google Scholar]
- 25.Burger G. (1999) GenBank accession no. AF193903.
- 26.Gray M.W., Lang,B.F., Cedergren,R., Golding,G.B., Lemieux,C., Sankoff,D., Turmel,M., Brossard,N., Delage,E., Littlejohn,T.G., Plante,I., Rioux,P., Saint-Louis,D., Zhu,Y. and Burger,G. (1998) Nucleic Acids Res., 26, 865–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muramatsu T., Nishikawa,K., Nemoto,F., Kuchino,Y., Nishimura,S., Miyazawa,T. and Yokoyama,S. (1988) Nature, 336, 179–181. [DOI] [PubMed] [Google Scholar]
- 28.Weber F., Dietrich,A., Weil,J.-H. and Marechal-Drouard,L. (1990) Nucleic Acids Res., 18, 5027–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korab-Laskowska M., Rioux,P., Brossard,N., Littlejohn,T.G., Gray,M.W., Lang,B.F. and Burger,G. (1998) Nucleic Acids Res., 26, 138–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burger G., Zhu,Y., Littlejohn,T.G., Greenwood,S.J., Schnare,M.N., Lang,B.F. and Gray,M.W. (2000) J. Mol. Biol., 297, 365–380. [DOI] [PubMed] [Google Scholar]
- 31.Burger G., Saint-Louis,D., Gray,M.W. and Lang,B.F. (1999) Plant Cell, 11, 1675–1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turmel M., Lemieux,C., Burger,G., Lang,B.F., Otis,C., Plante,I. and Gray,M.W. (1999) Plant Cell, 11, 1717–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontaine J.M., Rousvoal,S., Leblanc,C., Kloareg,B. and Loiseaux-de Goër,S. (1995) J. Mol. Biol., 251, 378–389. [DOI] [PubMed] [Google Scholar]
- 34.Lang B.F., Burger,G., O’Kelly,C.J., Cedergren,R., Golding,G.B., Lemieux,C., Sankoff,D., Turmel,M. and Gray,M.W. (1997) Nature, 387, 493–497. [DOI] [PubMed] [Google Scholar]
- 35.Burger G., Plante,I., Lonergan,K.M. and Gray M.W. (1995) J. Mol. Biol., 245, 522–537. [DOI] [PubMed] [Google Scholar]
- 36.Oda K., Yamato,K., Ohta,E., Nakamura,Y., Takemura,M., Nozato,N., Akashi,K., Kanegae,T., Ogura,Y., Kohchi,T. and Ohyama,K. (1992) J. Mol. Biol., 223, 1–7. [DOI] [PubMed] [Google Scholar]
- 37.Dandekar T., Snel,B., Huynen,M. and Bork,P. (1998) Trends Biochem. Sci., 23, 324–328. [DOI] [PubMed] [Google Scholar]
- 38.Niehrs C. and Pollet,N. (1999) Nature, 402, 483–487. [DOI] [PubMed] [Google Scholar]
- 39.Wolff G., Plante,I., Lang,B.F., Kück,U. and Burger,G. (1994) J. Mol. Biol., 237, 75–86. [DOI] [PubMed] [Google Scholar]