Abstract
Non-homologous DNA end joining (NHEJ) is considered the major pathway of double-strand break (DSB) repair in mammalian cells and depends, among other things, on the DNA end-binding Ku70/80 heterodimer. To investigate the function of Ku in NHEJ we have compared the ability of cell-free extracts from wild-type CHO-K1 cells, Ku80-deficient xrs6 cells and Ku80-cDNA-complemented xrs6 cells (xrs6-Ku80) to rejoin different types of DSB in vitro. While the two Ku80-proficient extracts were highly efficient and accurate in rejoining all types of DNA ends, the xrs6 extract displayed strongly decreased NHEJ efficiency and accuracy. The lack of accuracy is most evident in non-homologous terminus configurations containing 3′-overhangs that abut a 5′-overhang or blunt end. While the sequences of the 3′-overhangs are mostly preserved by fill-in DNA synthesis in the Ku80-proficient extracts, they are always completely lost in the xrs6 extract so that, instead, small deletions displaying microhomology patches at their breakpoints arise. In summary, our results are consistent with previous results from Ku-deficient yeast strains and indicate that Ku may serve as an alignment factor that not only increases NHEJ efficiency but also accuracy. Furthermore, a secondary NHEJ activity is present in the absence of Ku which is error-prone and possibly assisted by base pairing interactions.
INTRODUCTION
Double-strand breaks (DSB), probably the most disruptive type of lesion in DNA, may arise spontaneously in the cell or after exposure to DNA-damaging agents, such as ionizing radiation (IR). If left unrepaired, DSB leads to broken chromosomes and cell death; if repaired improperly, mutations, chromosome rearrangements and oncogenic transformation may arise. Repair of DSB is achieved by at least three different mechanisms: (i) homologous recombination repair (HRR), a highly accurate process that usually restores the precise DNA sequence at the break; (ii) single-strand annealing (SSA), a process that leads to the formation of deletions; or (iii) non-homologous DNA end joining (NHEJ) that joins two broken ends directly. While HRR and SSA involve the members of the Rad52 epistasis group and strictly require extensive sequence homology, the Rad52-independent NHEJ process can dispense with sequence homology (1–4). Although increasing evidence indicates that vertebrate cells are also quite proficient at repairing DSB by HRR (5) during S and G2, NHEJ is still considered the major pathway of DSB repair.
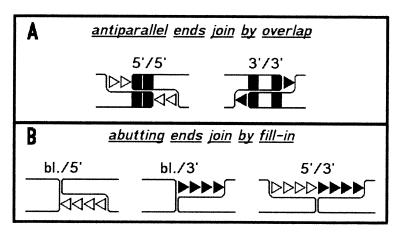
First reports on NHEJ date back to the early eighties (6–8). In the following years, the mechanisms of NHEJ were studied in detail in mammalian cells (9–12), extracts from Xenopus eggs (13,14) and later in yeast (15–17). NHEJ includes (i) the simple ligation of complementary (cohesive) protruding single strands (PSS) or blunt (bl.) ends which leads to the precise restoration of the broken sequence and (ii) the joining of non-complementary (non-homologous) DNA termini. Studies in Xenopus egg extracts showed that NHEJ is a highly accurate process that tends to preserve the sequences of interacting non-homologous DNA ends by generating two major types of products (overlap and fill-in). The type of product formed depends on the structure of ends being joined; overlap junctions typically arise during the joining of DNA ends containing anti-parallel PSS (5′/5′ and 3′/3′) while fill-in junctions are formed between abutting DNA ends (bl./5′, bl./3′ and 5′/3′). In the first case, the ends form by pairing of single fortuitously complementary bases, incompletely matched overlaps whose structure determines the patterns of subsequent repair reactions (Fig. 1; 18,19). In the second case, the sequences of participating 5′- and/or 3′-PSS are fully preserved by fill-in DNA synthesis in a process in which the ends are transiently held together by non-covalent interactions while the 3′-hydroxyl group of the 5′-PSS or blunt end is used as a primer to direct repair synthesis of the 3′-PSS (Fig. 1B; 14). Based on these data, we postulated in 1990 that an alignment factor may function to maintain the two ends in alignment to facilitate their biochemical reconfiguration into a ligatable structure (14). Similar joining events have been observed in vivo and in vitro in mammalian cells (10,11,20,21) indicating that the mechanisms found in the Xenopus system also apply for mammalian systems. The identity of the putative alignment factor, however, remained unknown.
Figure 1.
Major pathways of accurate NHEJ as observed in vitro in Xenopus eggs (14,19) and mammalian cells (12,21). (A) DNA ends with anti-parallel 5′- or 3′-PSS form short mismatched overlaps at positions of complementary base pairs (black boxes) which determine the patterns of DNA fill-in synthesis (open and black triangles; 18,19). (B) Sequences of 5′- and 3′-PSS in abutting terminus configurations are preserved by fill-in synthesis (14). While fill-in of 5′-PSS (open triangles) can be primed at the recessed 3′-OH group of the same end, fill-in of 3′-PSS (black triangles) can be primed only at the 3′-OH of the abutting terminus which may be a blunt (bl.) end or a 5′-PSS.
Analysis of hamster cell lines hypersensitive to IR and defective in DSB repair led to the identification of four complementation groups involved in NHEJ (22,23). The corresponding genes (XRCC4–7) encode the XRCC4 protein, an essential co-factor of DNA ligase IV (24–26), and the three components of the DNA-dependent protein kinase (DNA-PK) represented by the 86 and 70 kDa subunits of the heterodimeric Ku complex (Ku70/80) and the catalytic subunit of the protein kinase (DNA-PKCS), respectively (27–29). Due to its property of binding to a variety of end structures, including blunt ends and 5′- and 3′-PSS (for review see 30), the highly abundant Ku70/80 heterodimer is a particularly attractive candidate for the putative alignment factor (23,31). This assumption is supported by the analysis of IR-sensitive hamster cells bearing mutations in the XRCC5 gene (encodes Ku80) such as the xrs6 cell line (27,32) which is devoid of Ku end-binding activity and does not express detectable levels of the Ku70 subunit, whose stability requires the presence of Ku80 (33,34). These mutants are defective in general DSB repair and the joining of broken DNA ends created during V(D)J recombination (27,35,36). In accordance with a function of Ku in alignment, Saccharomyces cerevisiae rad52 mutant strains defective in the yeast Ku70/80 homolog not only display strongly decreased efficiency of NHEJ but also decreased accuracy of NHEJ as expressed by elevated levels of deletions (37,38). This led to the suggestion that Ku might be involved in protecting DNA ends from degradation thereby enhancing the accuracy of NHEJ. However, in hamster cells, this issue is still controversial (39–41).
To investigate whether the Ku heterodimer is able to exert alignment function and enhance the accuracy of NHEJ, we have examined the ability of cell-free extracts prepared from the Ku80-deficient xrs6 cell line (27,32) to join complementary and non-complementary DNA ends using a plasmid rejoining assay employed previously to study NHEJ in Xenopus egg extracts (13,14). As controls, cell-free extracts from the parental wild-type chinese hamster ovary (CHO)-K1 cell line and a Ku80-cDNA-complemented xrs6 cell line (xrs6-Ku80; 42) were used. We show that NHEJ activity is strongly decreased in the xrs6 extracts. Furthermore, the accuracy of NHEJ, as expressed by the formation of precise ‘overlap’ and ‘fill-in’ junctions (Fig. 1), is also significantly decreased in the xrs6 extracts. This becomes most evident in terminus configurations containing 3′-PSS. Our results indicate that Ku80, in conjunction with Ku70, may indeed function as the previously postulated alignment factor (14).
MATERIALS AND METHODS
Materials
All three cell lines (CHO-K1, xrs6 and xrs6-Ku80) used in this study were purchased from the European Collection of Cell Culture (Wiltshire, UK), cell culture media and all additives from Biochrom (Berlin, Germany). The V3 (43) and XR-1 (44) mutant CHO cell lines used in one control experiment were generous gifts from E. Dikomey (University of Hamburg, Germany) and S. Critchlow and S. P. Jackson (Cambridge University, UK), respectively. Enzymes and reagents for immunoblotting [nitro blue tetrazolium chloride (NBT)/5-bromo-4-chloro-3-indoyl-phosphate (BCIP)] were obtained from Roche Diagnostics (Mannheim, Germany), proteinase K from Sigma (Munich, Germany), N-ethyl-N-nitroso urea (ENU) from Serva (Heidelberg, Germany), kits for gel extraction of DNA and plasmid minipreparation from Qiagen (Hilden, Germany), [α-32P]dCTP (3.000 Ci/mM), random priming kit and T7-dideoxy sequencing kit from Amersham/Pharmacia Biotech (Eschborn, Germany), microdialysis filters from Millipore. Polyclonal anti-mouse-Ku86 and -Ku70 antisera were purchased from Santa Cruz Biotech (Heidelberg, Germany) and the secondary alkaline phosphatase-coupled antibody from Jackson Immuno Research Laboratories (Baltimore, MD).
Cell culture
The xrs6 cell line, originally derived from the CHO-K1 line on the basis of sensitivity to IR (32), is deficient in XRCC5 and fails to express functional Ku80 protein because of a splice site mutation that results in a 13 nt insertion and a frame shift (42). Control cell lines included the parental wild-type CHO-K1 cell line and the xrs6-Ku80 cell line, which is derived from xrs6 by stable transfection of the hamster wild-type XRCC5 cDNA (42). All three cell lines were grown at 37°C in a humidified 5% CO2 atmosphere in Ham’s F-12 medium enriched with 10% fetal calf serum, 2 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin.
Cell-free extracts
For all experiments described in this study, whole cell extracts were used which were prepared exactly as described previously (21). Approximately 5 × 108 cells of each cell line were used in each preparation to yield 0.5–1 ml of extract with a protein concentration ranging between 6 and 10 mg/ml. For most reproducible conditions, extracts from all three cell lines were prepared on the same day using the same freshly made solutions. Stored in 50 µl aliquots in liquid nitrogen, extracts remained active for 6–12 months. Prior to use in NHEJ reactions, the extract volume needed was dialyzed against freshly prepared M-buffer [50 mM 3-(n-morpholino)-2-hydroxy-propane sulfonic acid (MOPSO)-NaOH pH 7.5, 40 mM KCl, 10 mM MgCl2, 5 mM 2-mercaptoethanol] on microdialysis filters (0.025 µm) for 30 min at 4°C (45).
DNA substrates
The three substrates for cohesive and blunt end ligation were derived from the pSP65 (3 kb; Promega, Mannheim, Germany) by linearization with a single restriction enzyme (RE) (Bam, 5′-PSS; Pst, 3′-PSS; Sma, blunt ends). The eight substrates for NHEJ were derived from a 4.2 kb modified pSP65 containing a 1.2 kb λ-DNA insert between the restriction sites used for substrate preparation (pSP65/λ; 19). Generation of 3 kb linear plasmid substrates containing two non-homologous ends was controlled by quantitative excision of the 1.2 kb λ-insert. Each substrate was named after the pair of RE used in its preparation [(i) antiparallel ends: Bam/Asp and Bam/Sal, 5′/5′; BstX/BstX and Kpn/Pst, 3′/3′; (ii) abutting ends: Sma/Sal, bl./5′; Sma/Pst, bl./3′; Bam/Pst and Sac/Sal, 5′/3′ and 3′/5′, respectively]. Although obtained from cleavage with BstXI (CCAN5/NTGG) only, the BstX/BstX substrate contains non-complementary termini, due to the presence of different linkers upstream (5′-CCACTAAG/GTGG) and downstream (5′-CCACTTTG/GTGG) of the λ-insert. All substrates were gel purified.
Assay for NHEJ and analysis of products
In the K1-positive control, ligation and NHEJ was found to occur efficiently within 6 h at 25°C in the presence of 1–2 ng DNA/µl and 4–10 µg protein/µl. In standard reactions, 10 ng of each substrate were incubated for the times indicated at 25°C in a total volume of 10 µl containing 6–8 µg/µl of extract protein (always adjusted to the extract with the lowest protein concentration) in M-buffer supplemented with 1 mM ATP pH 7.5, 200 µM dNTPs (50 µM each) and 50 ng/µl bovine serum albumin (BSA). After adjustment to 20 mM Tris–HCl pH 7.5, 10 mM EDTA, 1% SDS, reactions were stopped by incubation at 65°C for 5 min. Samples were digested for 30 min at 37°C with 2 mg/ml proteinase K and further purified by phenol extraction and ethanol precipitation. Equivalents of 2 ng substrate DNA were electrophoresed in 1% agarose gels containing 1 µg/ml ethidium bromide (EthBr) to separate open circle (oc) from covalently closed circle (ccc) products and visualized by in situ gel-hybridization (14) using a pSP65-specific probe labeled with [α-32P]dCTP by random priming. For quantification of reaction products in gels, a phosphorimaging facility (Fuji, Düsseldorf, Germany) was used. Circular joined products were cloned by transformation of 4 ng equivalents of substrate DNA of each NHEJ sample in Escherichia coli strain DH5α. Cloned NHEJ products were purified by miniscale extraction and subjected to restriction analysis (where applicable) or T7-dideoxy sequencing.
Assay for preferential repair synthesis as a control of extract quality
For preparation of the ethylated plasmid substrate to measure excision repair synthesis, a 6 mg/ml solution of ENU in 2-(n-morpholino)-ethane sulfonic acid (MES) buffer (1 mM MES–HCl pH 6.0, 60 mM NaCl, 0.5 mM EDTA), diluted from 100 mg/ml stock in dimethyl sulfoxide (DMSO), was mixed with an equal volume of 100 mg/ml pSP65 in alkylation buffer [50 mM 3-(n-morpholino)-propane sulfonic acid (MOPS)-NaOH pH 7.4, 60 mM NaCl, 0.5 mM EDTA]. The resulting ENU concentration of 3 mg/ml generates about four to five 6O-ethyl-guanines per plasmid molecule and a spectrum of other alkylation adducts (46). After incubation at 37°C for 60 min, pSP65 DNA was purified by three washes with 2 ml TE in centricon tubes (10 kDa MW cut off, Amicon, Witten, Germany). To measure preferential repair synthesis, 50 ng of ethylated pSP65 (3 kb) together with 50 ng of untreated pSP65/λ (4.2 kb) were incubated for the times indicated at 25°C in a total volume of 10 µl containing 36 µg of whole cell extract protein in M-buffer supplemented with 2 µCi [α-32P]dCTP, 0.8 µM dCTP, 50 µM each dATP, dTTP, dGTP, 1 mM ATP, pH 7.5, and 50 ng/µl BSA. After adjustment to 10 mM Tris–HCl pH 7.5, 10 mM EDTA and 0.5% SDS, reactions were terminated by incubation at 65°C. Samples were purified as described for NHEJ assays and split into two equal aliquots, one of which was cleaved with EcoRI while the other remained untreated. Differential 32P incorporation was analyzed by separation of total EcoRI-digested and untreated samples in 1% agarose gels containing 1 µg/ml EthBr and subsequent autoradiography. Thereafter, gels were hybridized to a pSP65-specific 32P-labeled probe as described for NHEJ-assays to control the DNA content per lane.
Western blots and gel shift assays
For immunoblotting, Ku-specific antisera were used (1:100 dilution) and visualized with the NBT/BCIP reagents as specified in the supplier’s manual. DNA end binding activity of Ku was assayed with the help of gel shift experiments essentially as described previously (47,48); 1 ng of a 5′-terminally phosphate-labeled double-stranded 69mer oligonucleotide was incubated in a total volume of 5 µl in M-buffer with varying amounts (0.5–15 µg) of K1- and xrs6-Ku80 extract protein in the presence or absence of 250 ng circular plasmid competitor for 15 min at room temperature. Samples were separated by 4.5% native PAGE.
RESULTS
General assay for ligation and NHEJ in vitro and control of extract quality
Plasmids linearized with a single RE represent homologous substrates with cohesive 5′- or 3′-PSS or blunt ends to measure the efficiency of ligation. Non-homologous substrates, generated by cleavage with two different RE, contain non-complementary DNA ends and are used to measure the efficiency of NHEJ. Extract-mediated end joining converts both substrate types into monomeric oc and ccc and various linear multimers which are separated in agarose gels. Isolation of single NHEJ events for analysis of junctional sequences is achieved by cloning of the circular products in E.coli. Using a total of three homologous and eight non-homologous substrates, we have examined whole cell extracts from the Ku80-deficient xrs6-cell line (32), the wild-type CHO-K1 (K1) parent and the hamster Ku80-cDNA-complemented xrs6 cell line (xrs6-Ku80; 42) to investigate the role of Ku in NHEJ.
Since xrs6 cells in vivo are defective in DSB repair (27), cell-free extracts from these cells should display decreased NHEJ efficiency when compared to extracts from CHO-K1 or xrs6-Ku80. To verify that observed differences in NHEJ are not caused by fluctuations in extract quality, we have compared the quality of different extract batches in an independent assay that measures excision-repair-mediated DNA synthesis on a plasmid containing alkylation damage. For this, a supercoiled ENU-treated plasmid (3 kb) was incubated together with a larger native control plasmid (4.2 kb). Excision of the ethyl-adducts from the ENU-treated plasmid leaves single-stranded gaps that are filled by repair DNA synthesis as detected by incorporation of radiolabeled dNTPs. Time-dependent incorporation of radiolabel occurs preferentially in the 3 kb ENU-treated plasmid while the 4.2 kb native control plasmid is labeled to a significantly lower extent (Fig. 2, top). The controls at the bottom show that all lanes contain the same amounts of DNA to verify that preferential labeling of the 3 kb plasmid is not due to higher amounts of input substrate. This experiment shows that all three extracts are capable of performing preferential repair synthesis on the damaged plasmid. The fact that the efficiency of repair synthesis is nearly the same in all three extracts demonstrates that the quality of the extract preparations is comparable and that observed differences in NHEJ can indeed be ascribed to the different properties of the three cell lines. All experiments described below were performed with extracts whose quality had been checked in this assay.
Figure 2.
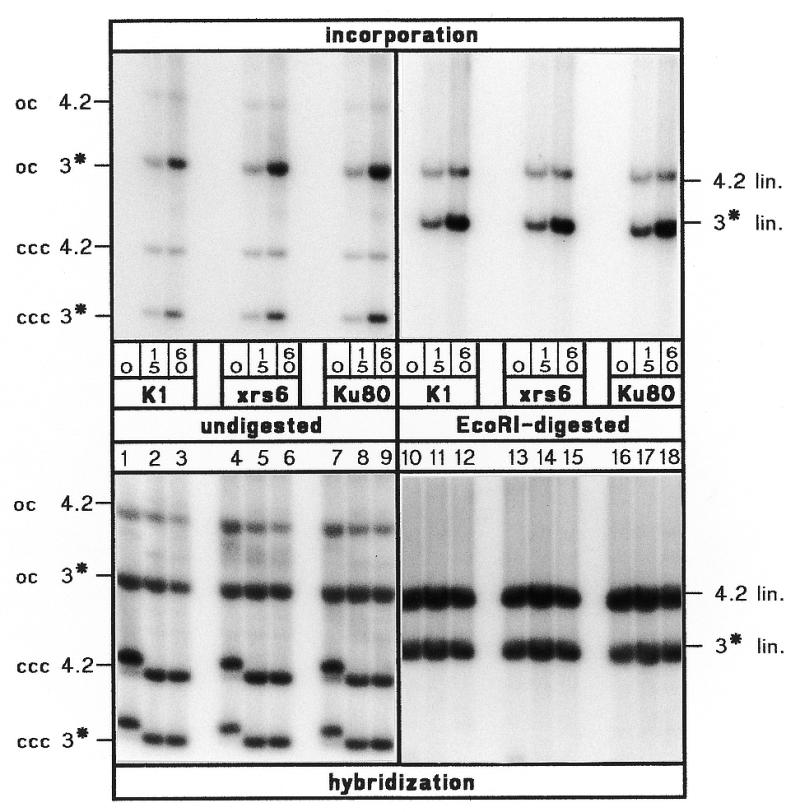
Control of extract quality. Top, kinetics (times in min below each lane) at 25°C of incorporation of [α-32P]dCTP in supercoiled 3 kb ENU-treated (asterisk) and 4.2 kb native plasmid substrates in extracts from CHO-K1 (K1), xrs6 and xrs6-Ku80 (Ku80) cells. Lanes 1–9, ccc and oc forms of the plasmid substrates. Lanes 10–18, the same samples after linearization with EcoRI show preferential accumulation of radiolabel in the 3 kb band. Bottom, same lanes as at top but after hybridization to a 32P-labeled plasmid probe for control of DNA content. Shift of ccc bands at time 0 (lanes 1, 4 and 7) results from different levels of positive supercoil induced by EthBr in the gel which is caused by residual negative supercoil (relaxed upon extract treatment) plasmids purified from E.coli.
Efficiency of ligation and NHEJ is decreased in xrs6 extracts
A representative example of the different joining properties of the three extracts is shown in Figure 3. As reflected by the high levels of circle formation, most efficient ligation of cohesive 5′- (Bam) and 3′-PSS (Pst), and blunt ends (Sma), and NHEJ of non-homologous 5′/5′-PSS (Bam/Asp) is observed in the K1 extract. By contrast, circle formation is strongly decreased in xrs6 indicating that the absence of Ku affects the joining of all substrate types, however, to varying degrees depending on the type of ends being joined. Generally, the strongest impairment is observed for Bam (13.8% of wild-type), Sma (14%) and Bam/Asp (15.7%) while Pst is much less affected (46%) (Table 1). In the Ku80-complemented xrs6-Ku80 extracts, the inhibition of ligation and NHEJ is reverted confirming that the observed defect in xrs6 is indeed caused by the absence of Ku80. Still, ccc product formation in xrs6-Ku80 reaches on average only 50% of the wild-type level (Table 1). Possible reasons for this difference are discussed below. Thus we may summarize that the efficiency of ligation and NHEJ is decreased by about a factor of 5 in xrs6 and a factor of 2 in xrs6-Ku80. Furthermore, the average efficiency of cohesive and blunt end ligation is about twice as high as the efficiency of NHEJ in all three extracts reflecting the higher complexity of the NHEJ reaction.
Figure 3.
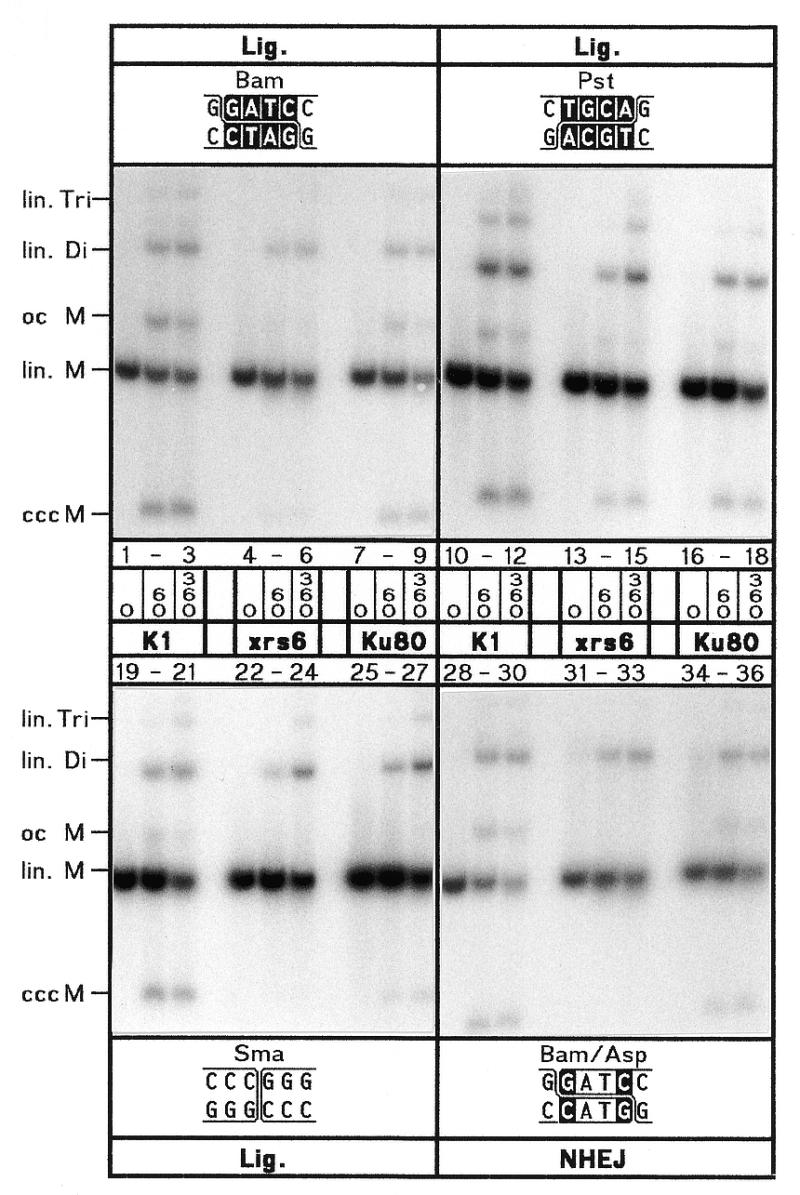
Efficiency of NHEJ in the different extracts. Kinetics (times in min) at 25°C of ligation (Lig.) of cohesive 5′- (Bam) and 3′-PSS (Pst), and blunt ends (Sma) and NHEJ of non-complementary 5′/5′-PSS (Bam/Asp) in extracts from CHO-K1 (K1), xrs6 and xrs6-Ku80 (Ku80) cells. Terminus configurations are shown at the top of each panel with base matches as white letters. Accurate NHEJ of Bam/Asp is known to involve formation of a mismatched overlap whose nicks are closed by ligation (19). Reaction products as marked on the left side: lin., linear; oc, open circle; ccc, covalently closed circle; M, monomer, 3 kb; Di, dimer, 6 kb; Tri, trimer, 9 kb.
Table 1. Efficiency of ligation and NHEJ in the different extracts.
| Terminus | CHO-K1 | xrs6 | xrs6-Ku80 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| configurations | % ccc | % Di | % ccc | % wt | % Di | %wt | % ccc | % wt | % Di | % wt |
| Bam (5′-coh.) | 21.7 | 7.9 | 3.0 | 13.8 | 5.6 | 70.9 | 12.3 | 56.8 | 5.7 | 72.2 |
| Pst (3′-coh.) | 12.6 | 11.3 | 5.8 | 46.0 | 10.6 | 93.8 | 7.2 | 57.2 | 8.7 | 77.0 |
| Sma (bl.) | 11.4 | 9.3 | 1.6 | 14.0 | 9.6 | 103.2 | 3.4 | 29.9 | 10.3 | 110.8 |
| Average lig. | 15.2 | 9.5 | 3.5 | 22.8 | 8.6 | 90.5 | 7.6 | 50.0 | 8.2 | 86.8 |
| Bam/Asp (5′/5′) | 8.3 | 12.1 | 1.3 | 15.7 | 12.1 | 100.0 | 4.3 | 51.8 | 9.6 | 79.3 |
| Bam/Sal (5′/5′) | 9.7 | 11.4 | 1.7 | 17.5 | 11.0 | 96.5 | 2.5 | 25.8 | 10.3 | 90.3 |
| BstX/BstX (3′/3′) | 8.3 | 11.2 | 1.2 | 14.5 | 10.1 | 90.2 | 2.9 | 35.0 | 9.5 | 84.8 |
| Kpn/Pst (3′/3′) | 4.8 | 11.4 | 1.5 | 31.3 | 12.0 | 105.3 | 1.7 | 35.5 | 12.7 | 111.4 |
| Sma/Sal (bl./5′) | 8.5 | 11.2 | 1.6 | 18.8 | 11.9 | 106.3 | 2.9 | 34.1 | 12.8 | 114.3 |
| Sma/Pst (bl./3′) | 2.4 | 9.6 | 1.1 | 45.9 | 7.1 | 74.0 | 3.8 | 158.7 | 13.5 | 140.6 |
| Bam/Pst (5′/3′) | 9.8 | 10.9 | 1.5 | 15.3 | 9.0 | 82.6 | 5.9 | 60.2 | 13.8 | 126.6 |
| Sac/Sal (3′/5′) | 4.1 | 13.2 | 1.0 | 24.4 | 6.5 | 49.2 | 5.3 | 130.0 | 12.1 | 91.7 |
| Average NHEJ | 7.0 | 11.4 | 1.4 | 20.0 | 10.0 | 87.8 | 3.7 | 52.9 | 11.8 | 103.5 |
The efficiency of formation of ccc monomers (ccc) and linear dimers (Di) from the three homologous (ligation, lig.) and eight non-homologous substrates (NHEJ) after 6 h at 25°C was measured by phosphorimager analysis of gels as radioactivity per product band. Corrected for background and product size (dimers are twice as large as circles) and expressed as percentage of total radioactivity per lane (% ccc, % Di) to compensate for the differences in the amount of substrate input, these values are set in relation to the corresponding values obtained in the CHO-K1 wild-type extract (% wt) which is considered to be 100%.
In contrast to circle formation, the formation of linear multimers is much less strongly affected in xrs6 (dimers, average 90% of wild-type; Table 1). This unexpected result could be explained in two ways: either the formation of linear multimers is not or is less dependent on Ku than the formation of circles or a secondary NHEJ mechanism exists that is mainly active in the absence of Ku and preferentially promotes the formation of linear dimers. The first possibility appears unlikely because in another cell-free system, the formation of linear multimers was shown to be dependent on Ku (49). On the other hand, a study performed in Xenopus egg extracts showed that the formation of linear multimers is even increased upon immuno-depletion of Ku (50). A detailed interpretation of these seemingly conflicting results together with our own data is presented in the Discussion.
Accuracy of DNA end joining is decreased in xrs6 extracts
To investigate the hypothesis that Ku enhances the accuracy of DNA end joining (37,38,41) it is important to define the term ‘accuracy’. While it is obvious that ‘accurate ligation’ of complementary ends restores the original restriction site used to create the DSB, the definition of ‘accurate NHEJ’ is not self-evident because the joining of non-complementary ends necessarily causes a change in the original sequence. Still, general rules for NHEJ were established because extracts from Xenopus eggs (14,18,19) and mammalian cells (21) generate highly reproducible spectra of junctions by two main NHEJ pathways, designated as ‘overlap’ and ‘fill-in’ mechanisms with the former joining anti-parallel and the latter abutting ends (Fig. 1). In the following, all junctions formed by the ‘overlap’ or ‘fill-in’ pathway are therefore defined as ‘accurate’.
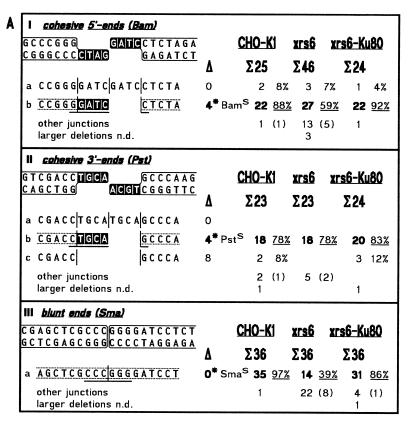
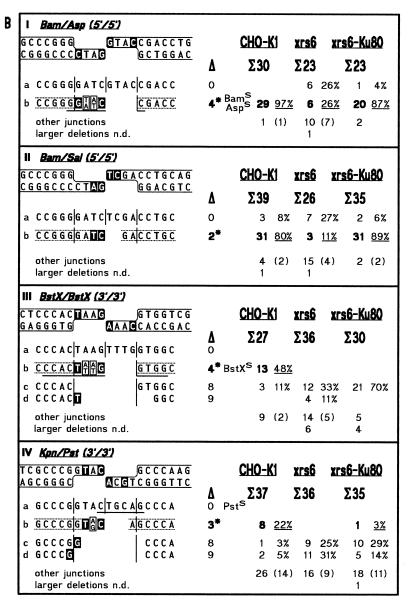
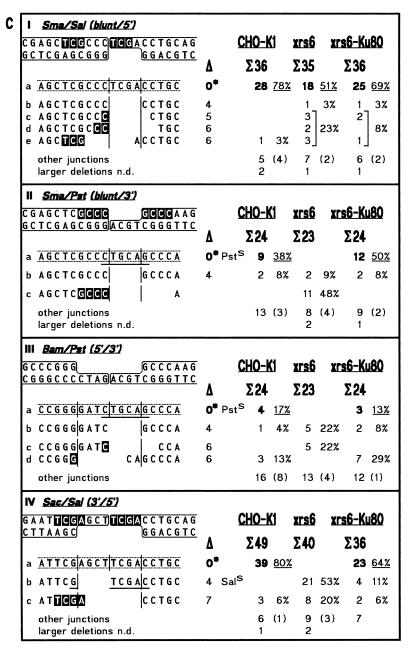
In this study, we have analyzed the sequences of 1024 cloned junctions obtained by end joining of 11 DNA substrates in the three extracts. The 273 junctions derived from ligation of cohesive 5′- (Bam), 3′- (Pst) and blunt ends (Sma) are shown in Figure 4A. NHEJ junctions are subdivided in 377 junctions derived from substrates containing anti-parallel ends (5′/5′, Bam/Asp, Bam/Sal; 3′/3′, BstX/BstX, Kpn/Pst) which are joined by ‘overlap’ formation (Fig. 4B) and 374 junctions derived from substrates containing abutting ends (bl./5′, Sma/Sal; bl./3′, Sma/Pst; 5′/3′, Bam/Pst; 3′/5′, Sac/Sal) which are joined by ‘fill-in’ (Fig. 4C). To facilitate direct comparison, the accuracy values for ligation and NHEJ are summarized in Table 2.
Figure 4.
Spectra of junctions generated by DNA end joining in extracts from CHO-K1, xrs6 and xrs6-Ku80 cells. (A) Ligation of cohesive and blunt ends; (B) NHEJ of anti-parallel ends by overlap formation; (C) NHEJ of abutting ends by fill-in. Terminus configurations including flanking double-strand sequences are shown at the top of each panel with complementary bases used in junction formation highlighted by white letters. Junctions are listed below as top strand sequences with vertical lines marking breakpoint (middle) and blunt positions (left and right). Note that only the main junctions are shown while less frequent events are summarized under ‘other junctions’, which comprise all three possible types of interactions (between two PSS, one PSS with the flanking duplex of the other terminus and two flanking duplexes). ‘Larger deletions’ designate junctions not determined (n.d.) due to failure of primer annealing during sequencing. Sequences between broken lines highlight ‘accurate’ junctions of which the ones formed by overlap from Bam/Asp, BstX/BstX and Kpn/Pst contain mismatches that yield two different sequences (boxes with two bases) after segregation in E.coli (18,19). Total numbers of bases deleted from each junction are listed below the open triangles with asterisks marking the ‘accurate’ junction. Underlining indicates a restored restriction site used for quicker analysis (e.g. BamS). Microhomology patches (white letters) found at breakpoints are ascribed arbitrarily to the left end although the actual origin of the affected bases cannot be determined unambiguously. Frequencies of junctions obtained in the different extracts are shown on the right. Σ indicates total numbers of junctions analyzed, bold numbers and underlined percentages frequencies of ‘accurate’ junctions, numbers in brackets the fraction of ‘other junctions’ displaying microhomologies of 1–3 bp at their breakpoints.
Table 2. Fidelity of ligation and NHEJ.
| Terminus | CHO-K1 | xrs6 | xrs6-Ku80 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| configurations | Σ jcts | Σ acc. | % acc. | Σ jcts | Σ acc. | % acc. | Σ jcts | Σ acc. | % acc. |
| Ligation cohesive/blunt | 84 | 75 | 89 | 105 | 59 | 56 | 84 | 73 | 87 |
| 5′/5′ | 25 | 22 | 88 | 46 | 27 | 59 | 24 | 22 | 92 |
| 3′/3′ | 23 | 18 | 78 | 23 | 18 | 78 | 24 | 20 | 83 |
| bl./bl. | 36 | 35 | 97 | 36 | 14 | 39 | 36 | 31 | 86 |
| NHEJ antiparallel ends | 133 | 81 | 61 | 121 | 9 | 7 | 123 | 50 | 41 |
| 5′/5′ | 69 | 60 | 87 | 49 | 9 | 18 | 58 | 49 | 84 |
| 3′/3′ | 64 | 21 | 33 | 72 | 0 | 0 | 65 | 1 | 2 |
| NHEJ abutting ends | 133 | 80 | 60 | 121 | 18 | 15 | 120 | 63 | 53 |
| bl./5′ | 36 | 28 | 78 | 35 | 18 | 51 | 36 | 25 | 69 |
| bl./3′ | 24 | 9 | 37 | 23 | 0 | 0 | 24 | 12 | 50 |
| 5′/3′ | 73 | 43 | 59 | 63 | 0 | 0 | 60 | 26 | 43 |
| Total | 350 | 236 | 67 | 347 | 86 | 25 | 327 | 186 | 57 |
The ‘accuracy’ of ligation and NHEJ was derived from a total of 1024 junctional sequences and determined individually for each extract and each group of terminus configurations (underlining marks groups comprising two different substrates) in absolute numbers of total junctions (Σ jcts) and ‘accurate’ junctions (Σ acc.), and the corresponding percentages (% acc.).
Spectra of junctions formed by ligation of cohesive and blunt ends
The spectra of ligation junctions (Fig. 4A) show that the accuracy of 5′-PSS and blunt end ligation is significantly decreased in xrs6 and restored to wild-type levels in xrs6-Ku80 (Table 2). Interestingly, the accuracy of 3′-PSS ligation remains unchanged in xrs6 indicating that the dependence of ligation on Ku varies with the type of ends being joined with blunt ends (39%) < cohesive 5′-PSS (59%) < 3′-PSS (78%, no difference to wild-type; note that wild-type accuracy is slightly reduced with respect to the other two substrates). In a first approximation, ligation accuracy appears to be related to ligation efficiency because blunt ends and 5′-PSS yielded much lower ccc product levels (∼14% of wild-type) than 3′-PSS (46%; Table 1). This may be due to the substrate specificity of ligase IV which, together with the XRCC4 protein, is known to be involved in NHEJ in vivo (51,52).
Spectra of junctions formed by NHEJ of anti-parallel ends
Substantial fractions of the junctional spectra formed in the K1 extract display the typical features of ‘overlap’ and ‘fill-in’ junctions and are therefore defined as ‘accurate’ [Fig. 4B, (b) junctions; Fig. 4C, (a) junctions]. This result confirms that the rules for ‘accurate’ NHEJ, originally established for the Xenopus system, are generally applicable.
The ‘accuracy’ of anti-parallel 5′-PSS joining by ‘overlap’ formation is highest in both K1 (87%) and xrs6-Ku80 (84%) (Fig. 4B, Ib and IIb). In xrs6, however, the fraction of these ‘accurate’ junctions is much smaller (18%) so that accuracy is decreased by about a factor of 5 (Table 2). Instead, NHEJ in xrs6 has shifted from ‘overlap’ formation to another pathway (Ia, 26%; IIa, 27%) in which both 5′-PSS are blunted by fill-in followed by blunt end ligation. In some cases (not shown explicitly in Figure 4 but summarized in the fraction of ‘other junctions’), the 5′-PSS have lost some nucleotides while the rest is conserved by fill-in and blunt end ligation (I, 4%; II, 23%), a phenomenon also observed with ligation of cohesive Bam 5′-PSS (Fig. 4A, I, 14%). This indicates that, in the absence of Ku, ‘overlap’ formation is competed by fill-in of 5′-PSS to yield blunt ends, which are subsequently ligated.
The ‘accuracy’ of anti-parallel 3′-PSS joining is generally strongly decreased in K1 (33%, Table 2; Fig. 4B, IIIb and IVb), but in xrs6, not a single junction fulfills the criteria of ‘accurate’ ‘overlap’ formation (0%). Instead, most junctions have formed by complete loss of both 3′-PSS followed by blunt end ligation (Fig. 4B, IIIc and d, IVc and d). Since blunt end ligation was found to be inefficient and inaccurate in xrs6 (see Table 1 and Fig. 4A, III) it cannot be ruled out that the single base matches present at the junctional breakpoints of IIIc, and IVc and d (Fig. 4B) stimulate ligation by stabilizing the joining intermediate.
Strikingly, the accuracy of anti-parallel 3′-PSS joining is not restored to wild-type levels in xrs6-Ku80 (2%) so that the spectra of 3′/3′ junctions resemble more closely the ones found in xrs6 which strictly contrasts the results obtained from antiparallel 5′-PSS joining. Together with the fact that the overall joining efficiencies are decreased in xrs6-Ku80 (50% wild-type, Table 1) this indicates that the features of xrs6-Ku80 are not identical to those of the CHO-K1 parent so that the complemented cell line takes an intermediate position between the mutant and the wild-type. To investigate the reason for the observed differences between K1 and xrs6-Ku80, we performed western blots to detect potential differences in the amount of Ku in the two extracts, and gel shift experiments to assay for the potential differences in DNA end binding activity of Ku (47,48). While no differences in Ku expression levels were observed in western blots, a minimal reduction of DNA end binding activity (~1.5-fold) was detected in the xrs6-Ku80 extract (not shown). Whether this difference is sufficient to account for the observed differences in general NHEJ efficiency and accuracy of anti-parallel 3′-PSS joining is unknown but may indicate that Ku is not able to develop its activities to the full in xrs6-Ku80. This could be due to the fact that the xrs6-Ku80 cell line is complemented with the Ku80 cDNA but not the genomic sequence which may contain additional elements important for the regulation and/or post-translational modification of Ku80.
In an attempt to achieve complete restoration of the accuracy of antiparallel 3′-PSS joining we performed mixing experiments between the xrs6 extract and extracts from two other mutant CHO cell lines, V3 and XR-1, which have normal levels of Ku but are defective in DNA-PKCS and XRCC4, respectively (24,43,44,53). Under these conditions, all proteins are derived from their genomic sequences. Both the V3 as well as the XR-1 extract alone exhibit not only strongly decreased efficiency (not shown) but also accuracy of NHEJ as shown in Table 3 for BstX/BstX (compare Fig. 4B, III). The decrease in accuracy is less pronounced in V3 than in XR-1 indicating that DNA-PKCS is less important for NHEJ accuracy than XRCC4. Remarkably, mixing of both mutant extracts with the xrs6 extract results in restoration of the accuracy of BstX rejoining to wild-type levels. This indicates that the lack of accuracy of 3′-PSS joining in the xrs6-Ku80 cell line could be due to slightly different interactions between the proteins required for the trimming of 3′-PSS and the Ku protein expressed from its cDNA with respect to the one expressed from its genomic sequence.
Table 3. Restoration of accuracy in mixed extracts.
| K1 | xrs6 | Xrs6-Ku80 | V3 | XR-1 | xrs6 × V3 | xrs6 × XR-1 |
|---|---|---|---|---|---|---|
| 13 of 27 | 0 of 36 | 0 of 30 | 8 of 24 | 1 of 21 | 14 of 24 | 12 of 24 |
| 48% | 0% | 0% | 33% | 5% | 58% | 50% |
Given are the fractions of clones and corresponding percentages of ‘accurate’ joining of anti-parallel 3′-PSS (BstX/BstX; see Fig. 4B, III) in the wild-type K1 extract, the complemented xrs6-Ku80 extract, the mutant xrs6, V3 and XR-1 extracts and mixtures of xrs6 with V3 (xrs6 × V3) and xrs6 with XR-1 extract (xrs6 × XR-1), respectively.
Spectra of junctions formed by NHEJ of abutting ends
As expected, the joining of abutting terminus configurations in K1 is achieved mainly by the ‘accurate’ ‘fill-in’ pathway which conserves the entire sequences of both 5′- and 3′-PSS by fill-in DNA synthesis (Fig. 4C). The accuracy of this pathway varies with the type of ends being joined with Sma/Sal (I) and Sac/Sal (IV) displaying the highest accuracy (78 and 80%, respectively) followed by Sma/Pst (II) with intermediate (38%) and Bam/Pst (III) with lowest accuracy (17%). This shows that NHEJ in K1 is able to fill in 3′-PSS by DNA synthesis that is primed at the partner terminus, a feature an alignment factor is supposed to have (14). Although slightly reduced, accuracy patterns obtained in xrs6-Ku80 resemble those of K1, reflecting the same substrate specificity (Fig. 4C and Table 2).
The difference in accuracy of the two 5′/3′-terminus configurations Sac/Sal and Bam/Pst observed in K1 (80 versus 17%) and xrs6-Ku80 (64 versus 13%) is striking and not easily explained but suggests that, apart from the structural configuration of the ends, the sequences of involved overhangs may also be important. In this context, it may be of relevance that an A is filled in at the ultimate position of the 5′-PSS in Sac/Sal. As many DNA polymerases are capable of adding single nucleotides to blunt ends, preferentially A or a nucleotide identical to the last one filled in (54,55), it is possible that untemplated addition of a second A to the 3′-OH of the blunted 5′-PSS facilitates pairing with the ultimate T of the partner 3′-PSS which would enhance fill-in of the 3′-PSS and thus result in the high accuracy achieved with this substrate (56,57). In Bam/Pst, by contrast, a C is filled in the ultimate position of the 5′-PSS and a T would have to be added in order to facilitate pairing, a configuration expected to reduce the accuracy of this joining reaction.
With the exception of Sma/Sal, the spectra of junctions produced in xrs6 are completely different from the ones produced in K1 and xrs6-Ku80. Joining of Sma/Sal yields 51% of ‘accurate’ junctions (Fig. 4C, I) which indicates that xrs6 has a strong capability of filling 5′-PSS. The fact that the accuracy of Sma/Sal joining is even higher than the one obtained for blunt end ligation (39%, Fig. 4A, III) indicates the possibility that the polymerase responsible for 5′-PSS fill-in might be able to support subsequent blunt end ligation by transiently stabilizing the joining intermediate. In spite of this strong polymerase activity in xrs6 which also conserves the 5′-PSS sequences of the other abutting terminus configurations (Fig. 4C, III and IV), not a single ‘accurate’ junction is formed from substrates containing 3′-PSS (Table 2). Instead, all 3′-PSS sequences are lost entirely leading to blunt end ligation between the trimmed 3′-PSS and the partner blunt end (Fig. 4C, IIb) or the filled 5′-PSS (IIIb and IVb). The fact that the ability to conserve entire 3′-PSS sequences is restored to almost wild-type levels in xrs6-Ku80 demonstrates that the ‘fill-in’ joining pathway is strictly dependent on Ku.
Besides blunt end joining, another type of NHEJ that creates small deletions with microhomologies at the breakpoints is preferentially realized in xrs6 (Fig. 4C, IIc, 48% and IVc, 20%), while in K1 and xrs6-Ku80, the same homology patches are not used at all (IIc) or with significantly lower frequency (IVc; both 6%). This suggests that NHEJ in the absence of Ku is assisted by base pairing interactions which is confirmed by results obtained in vivo in mammalian cells (41) and yeast (37,38). The fact that the use of terminus-flanking microhomology patches leads to the generation of small deletions is consistent with the idea that the fidelity of NHEJ is reduced in Ku-deficient cells (37–39).
Stability of DNA ends is decreased in xrs6
To investigate whether the accuracy of end joining is related to the stability of DNA ends, we have calculated the total number of each type of end (5′-PSS, 3′-PSS and blunt) from the 1024 junctions analyzed, taking into account that each junction originates from the interaction of two ends (Table 4). In K1- and xrs6-Ku80, more 5′-PSS (79%; 76%) and blunt ends (81%; 75%) participate in the formation of ‘accurate’ junctions than 3′-PSS (49%; 31%). This bias is even more pronounced in xrs6 (31 and 35% versus 13%), and remarkably, the 13% of ‘accurate’ junctions stem exclusively from accurate ligation of cohesive 3′-PSS (78% of all ligation junctions are ‘accurate’) while not a single 3′-PSS participated in the formation of an ‘accurate’ NHEJ junction.
Table 4. Stability of ends.
| Terminus | 5′-PSS | 3′-PSS | Blunt ends | ||||||
|---|---|---|---|---|---|---|---|---|---|
| configurations | CHO | xrs6 | Ku80 | CHO | xrs6 | Ku80 | CHO | xrs6 | Ku80 |
| Σ ends | 297 | 288 | 260 | 271 | 276 | 262 | 132 | 130 | 132 |
| Σ ends in acc. jcts | 235 | 36 | 153 | 130 | 36 | 80 | 107 | 46 | 99 |
| 79% | 31% | 76% | 49% | 13% | 31% | 81% | 35% | 75% | |
| lig. | 50 | 92 | 48 | 46 | 46 | 48 | 72 | 72 | 72 |
| acc. lig. | 88% | 59% | 92% | 78% | 78% | 83% | 97% | 39% | 86% |
| NHEJ | 247 | 196 | 212 | 225 | 230 | 214 | 60 | 58 | 60 |
| acc. NHEJ | 77% | 18% | 72% | 42% | 0% | 19% | 62% | 31% | 62% |
| Σ ends in inacc. jcts | 62 | 198 | 63 | 141 | 240 | 182 | 25 | 84 | 33 |
| 21% | 69% | 24% | 51% | 87% | 69% | 17% | 65% | 25% | |
| Loss of term. nucl. | |||||||||
| –0 | 22/7% | 92/32% | 28/11% | 10/4% | 0 | 6/2% | 18/14% | 45/35% | 19/14% |
| –1 | 6/2% | 12/4% | 9/3% | 13/5% | 0 | 7/3% | – | – | – |
| –2 | 8/3% | 17/6% | 3/1% | 24/9% | 1*/0.4% | 19/7% | – | – | – |
| –3 | 7/2% | 11/4% | 5/2% | 19/7% | 23*/8% | 24/9% | – | – | – |
| –4 | 8/3% | 15/5% | 11/4% | 30/11% | 84/30% | 74/28% | – | – | – |
| Beyond blunt | 11/4% | 51/18% | 7/3% | 45/16% | 132/48% | 53/20% | 7/5% | 39/30% | 14/11% |
The total number of ends (Σ), calculated individually for each terminal structure (5′-PSS, 3′-PSS and blunt), was derived from the total of 1024 junctions taking into account that two ends have participated in the formation of each junction. Given are the absolute numbers of ends (Σ) involved in ligation (lig.) and NHEJ and the distribution of the corresponding ‘accurate’ junctions (acc. jcts, acc. lig., acc. NHEJ). The fraction of ‘inaccurate’ junctions (inacc. jcts) comprises the numbers of ends involved in both ‘inaccurate’ ligation and NHEJ junctions. For the evaluation of the stability of a particular type of end only ‘inaccurate’ junctions are considered. Stability is reflected by the absolute number and percentages of intact ends (–0). In addition, the values for loss of 1, 2, 3 and 4 nt (coincides with the blunt end position) from 5′- and 3′-PSS are given [asterisks in the xrs6/3′-PSS column indicate loss from 3′-PSS derived from junctions with a 3 bp (1 case) and 1 bp microhomology (23 cases) at the breakpoint]. Deletions affecting the flanking duplex are summarized in the ‘beyond blunt’ fraction.
The stability of DNA ends in xrs6 was further analyzed in the fraction of ‘inaccurate’ junctions (Table 4) where end stability is expressed as percentage of ends that remained intact (–0 nt; for 5′- and 3′-PSS this means sequence conservation by fill-in DNA synthesis), or have lost 1–4 nt, or have suffered from deletions reaching into the flanking duplex region (‘beyond blunt’). In xrs6, the fraction of junctions in which at least one end is degraded ‘beyond blunt’ is strongly elevated (64 versus 18% and 22% in K1 and xrs6-Ku80, respectively). Although the extent of duplex degradation is generally only modest (xrs6: 20%, –1 nt; 32%, –2 to 14 nt; 12%, larger. K1: 7%, –1 nt; 7%, –2 to 9 nt; 4%, larger. xrs6-Ku80: 9%, –1 nt; 6%, –2 to 8 nt; 8%, larger) opposed to in vivo studies (39,41), this result indicates that degradation of DNA ends is minimized in the presence of Ku.
Furthermore, we found that blunt ends are the most stable ends in K1 and xrs6-Ku80 (–0, both 14%), followed by 5′-PSS (7%; 11%) and finally 3′-PSS (4%; 2%). In xrs6, blunt ends and 5′-PSS exhibit similar stability (35 and 32%) indicating that there is a strong tendency for fill-in of 5′-PSS to generate blunt ends that may subsequently undergo blunt end ligation. By contrast, the stability of 3′-PSS in xrs6 is zero with most ends being degraded to the blunt end position (–4 nt, 30%) or beyond (48%). The few cases in which 3 nt (–1, 0.4%) and 1 nt (–3, 8%), respectively, of the 3′-PSS sequences are conserved are without exception derived from junctions containing a microhomology of 3 or 1 bp, respectively, at their breakpoints. This indicates that the conservation of parts of 3′-PSS sequences can be realized only in junctions that provide base match possibilities between the 3′-PSS and the partner terminus. In ‘genuine’ blunt end junctions (no homology at breakpoint), however, 3′-PSS are always degraded to the blunt position or beyond.
In conclusion, our results show that xrs6 cells completely lack the ability to perform DNA fill-in synthesis on 3′-PSS and, furthermore, are hardly able to form ‘overlap’ junctions. This indicates that Ku, on the one hand, facilitates the fill-in of 3′-PSS in abutting terminus configurations, the crucial step in the ‘fill-in’ joining pathway (14), and, on the other hand, enhances the formation of mismatched overlaps between anti-parallel PSS, the crucial step in the ‘overlap’ pathway (18,19). Together these data provide strong evidence that Ku acts as an alignment factor in NHEJ.
DISCUSSION
Using DSB repair competent and deficient CHO cell lines, we have established an in vitro system that facilitates studying the functions of DSB repair proteins, such as the Ku70/80 heterodimer described here, in the mammalian NHEJ process. Our data provide direct evidence that the Ku70/80 heterodimer is required for efficient and accurate joining of complementary and non-complementary ends.
In vivo studies in Ku70/80-deficient yeast strains (37,38) indicated that Ku might be of crucial importance for the efficiency and fidelity of NHEJ, presumably by protecting DNA ends from degradation and thus enhancing the accuracy of NHEJ. Data obtained from transfection of linear DNA substrates in the Ku80-deficient xrs6 cell line, however, remained controversial with regard to this aspect because some authors observed decreased levels of efficiency and increased levels of end degradation (39) while others did not and therefore suggested that Ku is not required for efficient and ‘accurate’ NHEJ (40,41). The cause of these different results is probably the transfection procedure itself, because terminal degradation of the substrate molecules is not necessarily a result of the NHEJ reaction but may occur in the nucleus and/or during passage of the substrate through the cytoplasm. Thus, only a fraction of the substrate molecules undergoing NHEJ would maintain their original terminal structures, which in turn would significantly influence the spectra of NHEJ products observed. This problem is minimized in vitro because joining-active extract components gain access to the substrates directly upon incubation so that possibly observed terminal degradation is indeed the result of the NHEJ process itself.
In summary we have shown that the efficiency and accuracy of ligation as well as NHEJ is significantly decreased in the absence of Ku and that accuracy is related to the type of ends being joined as reflected by the differential stability of the different DNA ends (blunt ≈ 5′-PSS >> 3′-PSS) in xrs6. In this context, it is remarkable that ligation of cohesive 3′-PSS is very efficient and accurate (3′-PSS > 5′-PSS > blunt ends) in the absence of Ku while NHEJ of non-homologous terminus configurations containing 3′-PSS is not (accuracy of zero in all cases: 3′/3′ = bl./3′ = 5′/3′ < 5′/5′ < bl./5′). This apparent paradox may be explained by the properties of the proteins involved in ligation and NHEJ. Ligation is expected to require only XRCC4 and DNA ligase IV which is known to be essential for NHEJ in vivo (51,52). In another in vitro system containing ligase IV, highest ligation efficiency was observed for 3′-PSS > 5′-PSS > blunt ends (49) which is consistent with our results. A substrate preference of ligase IV for 3′-PSS would explain why ligation of cohesive 3′-PSS can occur efficiently and accurately in xrs6 and is not significantly stimulated by Ku in K1- and xrs6-Ku80. On the other hand, the efficiency and accuracy of cohesive 5′-PSS and blunt end ligation is considerably increased in K1- and xrs6-Ku80, which indicates that Ku may enhance the efficiency of DNA ligase IV on less preferred DNA ends and is consistent with results from in vitro studies using purified Ku and ligase IV (58).
In contrast to ligation, NHEJ is expected to involve other factors in addition to XRCC4 and ligase IV, such as DNA polymerase(s) and exonuclease(s). As postulated in the ‘fill-in’ joining pathway, ‘accurate’ NHEJ of 3′-PSS in abutting terminus configurations occurs by DNA fill-in synthesis primed at the 3′-OH group of the partner terminus (14). This pathway is entirely blocked in xrs6, since not a single ‘accurate’ junction is formed from terminus configurations containing 3′-PSS. This result strongly suggests that Ku may act as an alignment factor by transiently holding abutting ends together and thus allowing the polymerase to prime DNA synthesis at the partner terminus and to bridge the gap. Previously, DNA polymerases were suggested to take the part of an alignment factor due to their ability to perform DNA synthesis on discontinuous templates (56,57,59). This possibility, however, appears to be unlikely because DNA polymerase β, which was implicated in NHEJ recently (60), is not able to perform this type of reaction under physiological conditions (61). Furthermore, xrs6 cells which are expected to contain similar DNA polymerase levels as K1 cells display efficient polymerase activity on 5′-PSS but are unable to fill 3′-PSS. This suggests that DNA polymerases alone are not able to exert alignment function.
In addition to facilitating discontinuous DNA synthesis in abutting terminus configurations, Ku may also stabilize short mismatched overlaps formed between insufficiently stable single complementary base pairs of anti-parallel PSS and thus permits fill-in synthesis, nick ligation and other repair reactions to occur on these intermediates. Together these results show that Ku fulfills all previously proposed criteria of an alignment factor in NHEJ (14,19). The previous finding of elevated frequency of imprecise NHEJ in cells with mutated Ku (37–39) has led to the argument that Ku protects DNA ends from degradation. However, in the light of Ku acting as an alignment factor, we rather support the model of Ramsden and Gellert (58) which suggests that processing of ends and end joining are competitive processes, and Ku acts to increase the frequency of accurate end joining by stabilizing crucial intermediates thus increasing the rate of accurate NHEJ.
As seen by the strong tendency in xrs6 for fill-in of 5′-PSS to generate blunt ends and degradation of 3′-PSS to the blunt position, blunt end joining is one of two alternative NHEJ pathways in the absence of Ku. The other Ku-independent pathway involves the formation of short deletions at sites of microhomology. This becomes most evident in the fractions of junctions displaying a 4 bp microhomology at their breakpoints (see Fig. 4C, II and IV) which is significantly elevated in xrs6 (6.3 versus 0.3% in K1 and 0.9% in xrs6-Ku80). The underlying reaction is reminiscent of mechanisms designated as direct-repeat end joining (62,63) or microhomology-driven SSA (45,64,65) both of which use small homology patches such as direct repeats. Basically, such a mechanism can proceed by exonuclease-mediated trimming or helicase-mediated duplex unwinding of the ends to generate extended 3′-PSS that can undergo either SSA at the exposed microhomology patches or duplex invasion to create a D-loop. In this context, it is interesting to note that increased fractions of deletions with breakpoint homologies have been found in vivo in xrs6 cells (41) and Ku-deficient yeast strains (37,38), and in vitro in Ku-devoid protein fractions partially purified from Xenopus egg extracts (45) and calf thymus (66). Together with the data presented here, this suggests that an error-prone pathway that creates deletions is dominant in the absence of Ku.
In this context, it is interesting that the absence of Ku appears to affect mainly the formation of circles while the formation of linear multimers is much less strongly influenced. This intriguing phenomenon could be in part explained by the fact that Ku promotes DNA looping in vitro by self-association which would lead to the preferential formation of circles (67). On the other hand, Baumann and West showed that the formation of linear multimers is also strictly dependent on Ku in extracts derived from human lymphoblastoid cell lines (49). Interestingly, these authors neither obtained any circles in their extracts nor did they retain multimer formation activity upon addition of several inhibitors of NHEJ. These results indicate a crucial difference between their extracts and ours, which is due to the different preparation procedures rather than the different cell lines used.
In the light of Labhart’s results showing that Xenopus egg extracts retain a strong multimer formation activity even after immuno-depletion of Ku while circle formation is almost completely abolished (50)—which is fully consistent with our own results obtained in xrs6—it is likely that dimer formation in the absence of Ku is the result of a secondary Ku-independent NHEJ activity. This NHEJ activity, which appears to be absent in the extracts of Baumann and West (49), is not only error-prone but also promotes preferentially the formation of linear multimers. In the presence of Ku, both circles and multimers would be formed at about similar levels by accurate NHEJ. In addition to that, part of the multimer fraction could arise by the secondary error-prone NHEJ pathway. In the absence of Ku, this pathway can become fully active and promote the formation of linear multimers (but no circles) so that the wrong impression could arise as if Ku-dependent NHEJ formed only circles.
Why does the secondary Ku-independent NHEJ pathway form mainly linear multimers and hardly any circles? Interestingly, the linear dimers formed in the Ku-depleted Xenopus egg extract are mainly arranged in head-to-head (H/H) and tail-to-tail (T/T) but rarely in head-to-tail (H/T) orientation, although random joining of linear molecules is expected to create twice as many H/T as either H/H or T/T molecules (50). This is reminiscent of a mechanism observed previously in human cell extracts and purified protein fractions thereof (65,68,69). Since the fraction of multimers formed in our extracts are composed mainly of H/H and T/T products whose junctions did not arise by ligation of the corresponding complementary ends (E.Feldmann, W.Goedecke and P.Pfeiffer, unpublished result), microhomology-driven SSA (45,64,65) appears to be suited best to account for multimer formation in our extracts. As pointed out previously by Young et al. (65), the palindromic nature of the plasmid polylinker used for substrate preparation exhibits more extended regions of complete homology in H/H and T/T orientation than in H/T orientation, which leads to pairing in the region of homology and loss of a certain number of nucleotides. A prediction of this model is that microhomology-driven SSA of substrate molecules in H/T orientation would be less efficient, and the precise number of base pairs lost from the junction would be dependent on the position of small fortuitous patches of homology. This is consistent with the elevated fractions in xrs6 of junctions (all of which are H/T because they are derived from cloned circles) containing 4 bp microhomologies at their breakpoints and would explain why, in the absence of Ku, the fraction of H/T products (circles) decreases while the fraction of multimers (mainly H/H and T/T) remains largely unchanged. Further experiments are required to investigate this interesting phenomenon.
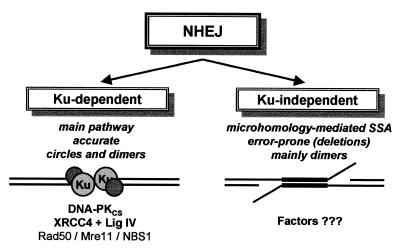
In conclusion, the results presented here suggest that Ku enhances the efficiency of ‘accurate’ NHEJ by functioning as an alignment factor. In the absence of Ku, NHEJ is not completely disabled but considerably less efficient and error-prone in that secondary mechanisms based on blunt end joining and microhomology-driven SSA are used preferentially (Fig. 5). Further studies in cell-free extracts from other DSB repair deficient cell lines will help to elucidate the functions of the other NHEJ proteins and identify the factors involved in Ku-independent NHEJ.
Figure 5.
Features of the Ku-dependent and -independent pathway of NHEJ. In the Ku-dependent pathway, the Ku-heterodimers (grey circles) bind to the ends of a broken DNA duplex (black lines) and serve as alignment factors to mediate, together with DNA-PKCS, XRCC4 and ligase IV, accurate NHEJ that forms both circles and linear dimers. The Rad50/Mre11/NBS1 nuclease complex (in yeast NBS1 is Xrs2) is probably also involved in this pathway (4). In the error-prone Ku-independent pathway, microhomology patches (black boxes) are used for SSA that forms mainly dimers containing deletions. The factors involved in this pathway are presently unknown but the Rad52 protein, one of the key players in homologous recombination, might be a possible candidate because it binds, like Ku, to DNA ends and promotes SSA (3).
Acknowledgments
ACKNOWLEDGEMENTS
We thank Drs M. F. Rajewsky, J. Thomale, C. Schunck and the members of AGI for stimulating discussions and laboratory space. This work was funded by a grant (98.053.1/2) to P.P. by the Wilhelm Sander Stiftung für Krebsforschung. P.P. is holder of a fellowship of the Heisenberg program of the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Shinohara A. and Ogawa,T. (1995) Trends Biochem. Sci., 20, 387–391. [DOI] [PubMed] [Google Scholar]
- 2.Kanaar R., Hoeijmakers,J.H.J. and van Gent,D.C. (1998) Trends Cell Biol., 8, 483–489. [DOI] [PubMed] [Google Scholar]
- 3.Haber J.E. (1999) Nature, 398, 665–667. [DOI] [PubMed] [Google Scholar]
- 4.Critchlow S.E. and Jackson,S.P. (1998) Trends Biochem. Sci., 23, 394–398. [DOI] [PubMed] [Google Scholar]
- 5.Thacker J. (1999) Biochimie, 81, 77–85. [DOI] [PubMed] [Google Scholar]
- 6.Pellicer A., Robins,D., Wold,B., Sweet,R., Jackson,J., Lowy,I., Roberts,J.M., Sim,G.K. and Silverstein,S. (1980) Science, 209, 1414–1422. [DOI] [PubMed] [Google Scholar]
- 7.Perucho M., Hanahan,D. and Wigler,M. (1980) Cell, 22, 309–317. [DOI] [PubMed] [Google Scholar]
- 8.Orr-Weaver T.L. and Szostak,J.W. (1983) Microbiol. Mol. Biol. Rev., 63, 349–404. [Google Scholar]
- 9.Wilson J.H., Berget,P.B. and Pipas,J.M. (1982) Mol. Cell. Biol., 2, 1258–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roth D.B, Porter,T.N. and Wilson,J.H. (1985) Mol. Cell. Biol., 5, 2599–2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roth D.B. and Wilson,J.H. (1986) Mol. Cell. Biol., 6, 4295–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roth D.B. and Wilson,J.H. (1988) In Kucherlapati,R. and Smith,G. (eds), Genetic Recombination. Am. Soc. Microbiol., Washington, DC, pp. 621–653.
- 13.Pfeiffer P. and Vielmetter,W. (1988) Nucleic Acids Res., 16, 907–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thode S., Schäfer,A., Pfeiffer,P. and Vielmetter,W. (1990) Cell, 60, 921–928. [DOI] [PubMed] [Google Scholar]
- 15.Goedecke W., Pfeiffer,P. and Vielmetter,W. (1994) Nucleic Acids Res., 22, 2094–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kramer K.M., Brock,J.A., Bloom,K., Moore,J.K. and Haber,J.E. (1994) Mol. Cell. Biol., 14, 1293–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mézard C. and Nicolas,A. (1994) Mol. Cell. Biol., 14, 1278–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pfeiffer P., Thode,S., Hancke,J., Keohavong,P. and Thilly,W.G. (1994) Mutagenesis, 9, 527–535. [DOI] [PubMed] [Google Scholar]
- 19.Pfeiffer P., Thode,S., Hancke,J. and Vielmetter,W. (1994) Mol. Cell. Biol., 14, 888–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bøe S.-O., Sodroski,J., Helland,D.E. and Farnet,C.M. (1995) Biochem. Biophys. Res. Commun., 215, 987–993. [DOI] [PubMed] [Google Scholar]
- 21.Daza P., Reichenberger,S., Göttlich,B., Hagmann,M., Feldmann,E. and Pfeiffer,P. (1996) Biol. Chem., 377, 775–786. [DOI] [PubMed] [Google Scholar]
- 22.Jeggo P.A. (1990) Mutat. Res., 239, 1–16. [DOI] [PubMed] [Google Scholar]
- 23.Jackson S.P. and Jeggo,P.A. (1995) Trends Biol. Sci., 20, 412–415. [DOI] [PubMed] [Google Scholar]
- 24.Li Z., Otevrel,T., Gao,Y., Cheng,H.-L., Seed,B., Stamato,T.D., Taccioli,G.E. and Alt,F.W. (1996) Cell, 83, 1079–1089. [DOI] [PubMed] [Google Scholar]
- 25.Critchlow S.E., Bowater,R. and Jackson,S.P. (1997) Curr. Biol., 7, 588–598. [DOI] [PubMed] [Google Scholar]
- 26.Grawunder U., Wilm,M., Wu,X., Kulesza,P., Wilson,T.E., Mann,M. and Lieber,M.R. (1997) Nature, 388, 492–495. [DOI] [PubMed] [Google Scholar]
- 27.Taccioli G.E., Gottlieb,T.M., Blunt,T., Priestly,A., Demengeot,J., Mizuta,R., Lehmann,A.R., Alt,F.W., Jackson,S.P. and Jeggo,P.A. (1994) Science, 265, 1442–1445. [DOI] [PubMed] [Google Scholar]
- 28.Blunt T., Finnie,N.J., Taccioli,G.E., Smith,G.C., Demengeot,J., Gottlieb,T.M., Mizuta,R., Varghese,A.J., Alt,F.W., Jeggo,P.A. and Jackson,S.P. (1995) Cell, 80, 813–823. [DOI] [PubMed] [Google Scholar]
- 29.Gu Y., Jin,S., Gao,Y., Weaver,D.T. and Alt,F.W. (1997) Proc. Natl Acad. Sci. USA, 94, 8076–8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Featherstone C. and Jackson,S.P. (1999) Mutat. Res., 434, 3–15. [DOI] [PubMed] [Google Scholar]
- 31.Roth D.B., Lindahl,T. and Gellert,M. (1995) Curr. Biol., 5, 496–499. [DOI] [PubMed] [Google Scholar]
- 32.Jeggo P.A. and Kemp,L.M. (1983) Mutat. Res., 112, 313–327. [DOI] [PubMed] [Google Scholar]
- 33.Chen F., Peterson,S.R., Story,M.D. and Chen,D.J. (1996) Mutat. Res., 362, 9–19. [DOI] [PubMed] [Google Scholar]
- 34.Errami A., Smider,V., Rathmell,W.K., He,D.M., Hendrickson,E.A., Zdzienicka,M.Z. and Chu,G. (1996) Mol. Cell. Biol., 16, 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pergola F., Zdzienicka,M.Z. and Lieber,M.R. (1993) Mol. Cell. Biol., 13, 3464–3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taccioli G.E., Rathburn,G., Oltz,E., Stamato,T., Jeggo,P.A. and Alt,F.W. (1993) Science, 260, 207–210. [DOI] [PubMed] [Google Scholar]
- 37.Boulton S.J. and Jackson,S.P. (1996) EMBO J., 15, 5093–5103. [PMC free article] [PubMed] [Google Scholar]
- 38.Boulton S.J. and Jackson,S.P. (1996) Nucleic Acids Res., 24, 4639–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang F. and Jasin,M. (1996) J. Biol. Chem., 271, 14405–14411. [DOI] [PubMed] [Google Scholar]
- 40.Han J.-O., Stehen,S.B. and Roth,D.B. (1997) Mol. Cell. Biol., 17, 2226–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kabotyanski E.B., Gomelsky,L., Han,J.-O., Stamato,T.D. and Roth,D.B. (1998) Nucleic Acids Res., 26, 5333–5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singleton B.K., Priestley,A., Steingrimsdottir,H., Gell,D., Blunt,T., Jackson,S.P., Lehman,A.R. and Jeggo,P.A. (1997) Mol. Cell. Biol., 17, 1264–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whitmore G.F., Varghese,A.J. and Gulyas,S. (1989) Int. J. Radiat. Biol., 56, 657–665. [DOI] [PubMed] [Google Scholar]
- 44.Stamato T.D., Weinstein,R., Giaccia,A. and Mackenzie,L. (1983) Somat. Cell Mol. Genet., 9, 165–173. [Google Scholar]
- 45.Göttlich B., Reichenberger,S., Feldmann,E. and Pfeiffer,P. (1998) Eur. J. Biochem., 258, 387–395. [DOI] [PubMed] [Google Scholar]
- 46.Engelbergs J. (1998) PhD Thesis. University of Essen, Essen, Germany.
- 47.Getts R.C. and Stamato,T.D. (1994) J. Biol. Chem., 269, 15981–15984. [PubMed] [Google Scholar]
- 48.Rathmell W.K. and Chu,G. (1994) Mol. Cell. Biol., 14, 4741–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baumann P. and West,S.C. (1998) Proc. Natl Acad. Sci. USA, 95, 14066–14070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Labhart P. (1999) Mol. Cell. Biol., 19, 2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson T.E., Grawunder,U. and Lieber,M.R. (1997) Nature, 388, 495–498. [DOI] [PubMed] [Google Scholar]
- 52.Frank K.M., Sekiguchi,J.M., Seidl,K., Swat,W., Cheng,H.-L., Davidson,L., Kangaloo,L. and Alt,F.W. (1998) Nature, 396, 173–177. [DOI] [PubMed] [Google Scholar]
- 53.Peterson S.R., Kurimasa,A., Oshimura,M., Dynan,W.S., Bradbury,E.M. and Chen,D.J. (1995) Proc. Natl Acad. Sci. USA, 92, 3171–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark J.M. (1988) Nucleic Acids Res., 16, 9677–9688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Clark J.M. (1991) Gene, 104, 75–80. [DOI] [PubMed] [Google Scholar]
- 56.King J.S., Fairley,C.F. and Morgan,W.F. (1994) J. Biol. Chem., 269, 13061–13064. [PubMed] [Google Scholar]
- 57.King J.S., Fairley,C.F. and Morgan,W.F. (1996) J. Biol. Chem., 271, 20450–20457. [DOI] [PubMed] [Google Scholar]
- 58.Ramsden D.A. and Gellert,M. (1998) EMBO J., 17, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Islas A.L., Fairley,C.F. and Morgan,W.F. (1998) Nucleic Acids Res., 26, 3729–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wilson T.E. and Lieber,M.R. (1999) J. Biol. Chem., 274, 23599–23609. [DOI] [PubMed] [Google Scholar]
- 61.Reichenberger S. and Pfeiffer,P. (1998) Eur. J. Biochem., 251, 81–90. [DOI] [PubMed] [Google Scholar]
- 62.Thacker J., Chalk,J., Ganesh,A. and North,P. (1992) Nucleic Acids Res., 20, 6183–6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thacker J. (1999) Acad. Sci. Paris, 322, 103–108. [DOI] [PubMed] [Google Scholar]
- 64.Lehman C.W., Trautman,J.K. and Carroll,D. (1994) Nucleic Acids Res., 22, 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicolás A.L., Munz,P.L. and Young,C.S.H. (1995) Nucleic Acids Res., 23, 1036–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mason R.M., Thacker,J. and Fairman,M.P. (1996) Nucleic Acids Res., 24, 4946–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cary R.B., Peterson,S.R., Wang,J., Bear,D.G., Bradbury,E.M. and Chen,D.J. (1997) Proc. Natl Acad. Sci. USA, 94, 4267–4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Derbyshire M.K., Epstein,L.H., Young,C.S.H., Munz,P.L. and Fishel,R. (1994) Mol. Cell. Biol., 14, 156–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nicolas A.L. and Young,C.S.H. (1994) Mol. Cell. Biol., 14, 170–180. [DOI] [PMC free article] [PubMed] [Google Scholar]