Abstract
Tetramethylammonium (TMA) chloride, dimethyl sulfoxide and formamide are known to increase, under certain conditions, the specificity and efficiency of the polymerase chain reaction (PCR). We compared the ability of several TMA derivatives and some other reagents to increase the specificity of PCR and to improve the yield of amplification. A novel combination of the enhancer TMA and oxalate as anion is demonstrated to be a powerful enhancer of PCR. Addition of 2 mM TMA oxalate to the PCR mixture decreases the formation of non-specific DNA fragments and increases the yield of specific PCR products.
INTRODUCTION
The polymerase chain reaction (PCR) is a versatile method for rapid amplification of selected DNA segments. The numerous protocols designed for different PCR applications almost always require an optimization step when put into practice (1). In some cases, especially when complex cDNA or genomic DNA is used as the PCR template and the calculated annealing temperature is relatively low, multiple non-specific bands cannot be eliminated although all experimental parameters are seemingly optimized. It has been reported that some reagents like tetramethylammonium (TMA) chloride (2–4), dimethyl sulfoxide (5–9), formamide (10), ammonium sulfate (9,11), betaine (8,12,13), acetamide (14) and non-ionic detergents (15) are capable of improving the efficacy and specificity of PCR. These reagents are often used as components of the commercial optimization and enhancer kits for PCR. In attempting to amplify fragments of mouse and rat Thy-1 gene by PCR we found that commercially available TMA derivatives as well as some other compounds were unable to suppress non-specific PCR products or decreased the yield of specific fragments to an unacceptable level. To overcome these inadequacies, we tested various TMA derivatives and several other reagents, finding that the nature of the counter ion has a decisive role in PCR performance.
MATERIALS AND METHODS
PCR additives
TMA chloride, TMA hydrogen sulfate, ammonium chloride, benzyldimethylhexadecylammonium chloride, betaine monohydrate and dimethyl sulfoxide were obtained from Fluka Chemie AG (Buchs, Switzerland). Hexadecyltrimethylammonium (HTA) bromide was obtained from Sigma Chemical Co. (St Louis, MO), and formamide was from USB Corp. (Cleveland, OH). TMA oxalate and TMA acetate were prepared by neutralizing TMA hydroxide (Fluka) with oxalic acid and acetic acid, respectively. HTA oxalate was prepared by neutralizing HTA hydroxide (Fluka) with oxalic acid. The pH of all reagents used as additives was adjusted to 8.0–8.6.
PCR and its evaluation
PCR was carried out with 300 ng of genomic DNA template, isolated from rat or mouse tail as described by Birren et al. (16), 0.5 µM of each primer (Generi Biotech, s.r.o., Hradec Králové, Czech Republic, see below), 0.2 mM dNTPs (Pharmacia LKB Biotechnology AB, Uppsala, Sweden), 10 mM Tris–HCl (pH 8.8), 50 mM KCl, 0.1% Triton X-100, 1.5 mM MgCl2 and 2.5 U Taq DNA polymerase (Promega Corp., Madison, WI) in a final volume of 50 µl. The primers used in initial experiments recognized the rat (primer 1: ATG AAC CCA GTC ATC AGC A; primer 2: ATA GTT TTA TTG GAG CTT GT) or mouse (primer 3: ATG AAC CCA GCC ATC AGC G; primer 4: GGG TAA GGA CCT TGA TAT AGG; primer 5: TCT CGG GCG CGA ATC CCA TG; primer 6: CAC AGA GAA ATG AAG TCC AG) Thy-1 gene sequences. Annealing temperature for each primer was calculated from melting temperature (Tm) as described by Dieffenbach and Dveksler (1). PCR was carried for 30 cycles with 15 s denaturation at 94°C, 15 s annealing at 49, 56 and 58°C with primers 1 + 2, 3 + 4 and 5 + 6, respectively, followed by 30 s extension at 72°C. Cycling was started by 1 min denaturation at 94°C and terminated by 1 min incubation at 72°C. Most of the experiments were performed using thermocycler PTC-200 (MJ Research, Inc., Watertown, MA). Optimal annealing temperature in the presence and absence of TMA oxalate was evaluated using RoboCycler Gradient 96 (Stratagene, La Jolla, CA). The intensity of the ethidium bromide stained bands was determined by densitometric analysis of the gels using a gel documentation system and GelBase/Gelblot software (UVP, Cambridge, UK). The ability of the tested reagents to increase the efficiency of amplification was defined as a ratio of the densitometric value of the specific DNA band determined after PCR in the presence of a given concentration of the additive and in its absence. Efficiency of PCR without any additives was taken as 1.0. The specificity of amplification was calculated as a ratio of the densitometric value of the specific band and that of all bands amplified by PCR. By definition, maximal value of specificity, in the absence of non-specific bands, equals 1.0. The concentrations of the additives that inhibited amplification of the specific bands by 90% were also calculated from dose–response curves.
RESULTS
Preliminary experiments with combination of primers 1 and 2, recognizing the rat Thy-1 gene sequence, indicated that the specific fragment of 1026 bp and several other ‘non-specific’ fragments were amplified. These ‘non-specific’ fragments were not eliminated when the annealing temperature, concentration of Mg2+ ions and concentration of DNA polymerases were optimalized (not shown). To determine whether the specificity and/or efficiency could be improved, various additives (see Table 1) were added to the PCR mixture at a final concentration between 0.01 mM and 2 M, and the PCR products were analyzed after electrophoretic fractionation on agarose gels and staining with ethidium bromide. An example of the analysis of one of the compounds tested, TMA oxalate, is shown in Figure 1A. In the absence of the compound (lane 1) the expected DNA fragment was generated by PCR together with several others. With increasing concentrations of the reagent there was an increase in the amount of the specific product and a decrease in non-specific bands. At 2 mM TMA oxalate only the specific band was observed. The intensity of the ethidium bromide stained bands was determined by densitometric analysis as described in the Materials and Methods. The data presented in Figure 1B show the ability of the tested reagent to improve PCR efficiency and specificity (for definitions see Materials and Methods), calculated from a gel sample reproduced in Figure 1A. In a control PCR (without any additive) the specificity was 0.2 and efficiency was, by definition, 1.0. Maximum specificity (1.0) and efficiency (2.2) were observed at 2 mM TMA oxalate. At a concentration of 9 mM TMA oxalate, the amplification of the specific band was inhibited by 90%. Similar analyses were performed for all compounds tested and summary data are presented in Table 1. These data indicate that maximal efficiency values were also markedly increased when TMA chloride (5 mM) and formamide (0.5 M) were used. Only a slight increase in efficiency was found in samples supplemented with TMA hydrogen sulfate or HTA bromide. All other additives, including DMSO, did not improve PCR efficiency (maximal efficiency ≤1.1). The specificity was increased from 0.2 to 0.5 when TMA chloride (20 mM) or HTA bromide (0.5 mM) were used. Dimethyl sulfoxide (1.4 M) and formamide (1 M) increased the specificity to 0.6 and 0.8, respectively. Only specific bands (specificity 1.0) were observed in reaction mixtures supplemented with TMA oxalate (at a concentration of 2 mM) and TMA hydrogen sulfate (50 mM).
Table 1. Effect of the tested additives on efficiency, specificity and inhibition of PCR.
| Tested additives | Maximal efficiencya | Maximal specificitya | 90% inhibitiona |
|---|---|---|---|
| TMA chloride | 1.9(5 mM) | 0.5(20 mM) | (35 mM) |
| TMA oxalate | 2.2(2 mM) | 1.0(2 mM) | (9 mM) |
| TMA acetate | 1.0[<10 mM] | 0.4(20 mM) | (40 mM) |
| TMA hydrogen sulfate | 1.2(0.5 mM) | 1.0(50 mM) | (70 mM) |
| Ammonium chloride | 1.0[<20 mM] | 0.2[<20 mM] | (50 mM) |
| Benzyldimethylhexadecylammonium chloride | 1.1(0.2 mM) | 0.4(1 mM) | (1.0 mM) |
| HTA bromide | 1.2(0.05 mM) | 0.5(0.5 mM) | (0.8 mM) |
| HTA oxalate | 1.0[<0.2 mM] | 0.2[<0.2 mM] | (0.5 mM) |
| Betaine monohydrate | 1.1(100 mM) | 0.4(750 mM) | (900 mM) |
| Dimethyl sulfoxide | 1.0[<1.4 M] | 0.6(1.4 M) | (1.6 M) |
| Formamide | 1.4(0.5 M) | 0.8(1 M) | (2.0 M) |
| PCR without any tested additives /control value/ | /1.0/ | /0.2/ | – |
aNumbers in parentheses indicate concentrations of the additives at which maximal efficiency, maximal specificity or 90% inhibition of PCR were attained. Numbers in brackets indicate a range of non-inhibitory concentrations.
Figure 1.
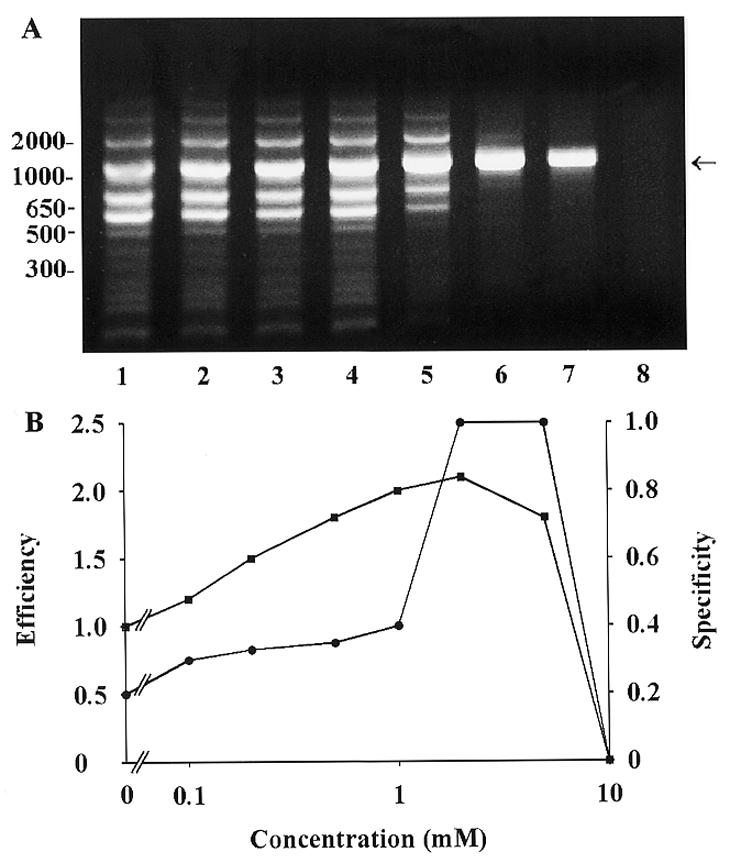
Analysis of PCR products formed in the absence or presence of different concentrations of TMA oxalate. (A) Ethidium bromide stained DNA fragments generated by PCR, using PTC 200 thermocycler, and fractionated by electrophoresis in 1% agarose. Lane 1, negative control, without TMA oxalate; lanes 2–8, TMA oxalate at final concentrations in mM: 0.1 (lane 2), 0.2 (lane 3), 0.5 (lane 4), 1 (lane 5), 2 (lane 6), 5 (lane 7) and 10 (lane 8). The arrow shows specific PCR product, 1026 bp fragment of mouse Thy-1 gene. Positions of DNA markers in bp are indicated on the left. (B) Efficiency (squares) and specificity (circles), determined by densitometric analysis of the data in (A), as a function of the concentration of TMA oxalate.
The tested reagents also differed in their ability to inhibit PCR. Data presented in Table 1 indicate that HTA bromide and HTA oxalate inhibited PCR at relatively low concentrations; 90% inhibition at 0.8 and 0.5 mM, respectively. Others showed medium inhibitory effect (TMA derivatives) or weak inhibition (betaine monohydrate, dimethyl sulfoxide, formamide). Interestingly, in many cases the concentration that gave the highest specificity also significantly inhibited the yield of the specific product.
To verify that TMA oxalate can improve the efficiency and/or specificity of PCR we also used as a template mouse genomic DNA and two other sets of primers (primers 3 + 4 and 5 + 6) recognizing the mouse Thy-1 gene. Using these primers, DNA fragments of 813 and 549 bp were amplified. Importantly, the yield of these fragments was increased ∼2-fold when PCR was supplemented with 2 mM TMA oxalate (data not shown). An increase in specificity and/or efficiency was also observed with several other sets of primers using mouse or rat genomic DNA as a template in PCRs which did not give satisfying results under routinely used conditions. By means of RoboCycler gradient 96 we evaluated the optimal annealing temperature for amplification of rat Thy-1 DNA fragment using primers 1 and 2 in the presence or absence of 2 mM TMA oxalate. Our data indicated that the optimal annealing temperature was 49°C and did not change after an addition of 2 mM TMA oxalate.
DISCUSSION
Data presented in this study indicate that a novel combination of TMA enhancer with oxalate anion increases the yield and specificity of PCR using genomic DNA as a template and primers which did not give satisfying results under otherwise optimalized conditions. Different concentrations of TMA chloride have been previously used to improve PCR. Thus, 60 mM TMA chloride was found to be optimal for amplification of a gene fragment coding for a surface protein of Paramecium primaurelia (2), whereas 10–100 µM TMA chloride was used for amplification of cDNA for tumor necrosis factor-β and interleukin-1α (3). Even lower concentrations of TMA chloride (5 µM) were recommended for routine use in PCR identification of Ehrlichia species (4). We have found that TMA chloride enhanced amplification of the Thy-1 gene fragment. However, maximal efficiency and specificity was observed, respectively, at 5 and 20 mM of TMA chloride. Using TMA oxalate, maximum increase in efficiency and specificity was observed at the same concentration of 2 mM. Optimal annealing temperature was not significantly different in the presence or absence of 2 mM TMA oxalate, thus corroborating the previous finding that optimal annealing temperature is not changed in the presence of 60 mM TMA chloride (2).
Several lines of evidence indicated that TMA oxalate surpassed TMA chloride and other reagents as a PCR enhancer. First, addition of TMA oxalate maximally increased the yield of PCR as inferred from dose–response curves. Secondly, at 2 mM TMA oxalate, only specific PCR products were observed. Although TMA hydrogen sulfate and some other reagents also remarkably increased the PCR specificity, maximal effect was always accompanied by decreased efficiency. TMA oxalate was the only reagent in which maximal specificity and maximal efficiency were observed at the same concentration. Thirdly, most of the reagents gave maximum specificity at concentrations which were close (≤2-fold difference) to the concentrations inhibiting the yield of PCR by 90%. In contrast, TMA oxalate showed a 90% inhibition at concentrations 4.5-fold higher than those required for maximum specificity.
The molecular mechanism of the enhancing effect of TMA oxalate on PCR is unknown. Previous data indicated that at 3 M TMA chloride the thermal stability of an AT base pair was identical to that of a GC base pair (17,18). The slight increase of the AT base pair stability at 2 mM TMA oxalate could explain the observed enhancing effect of this additive, especially when AT enriched primers are used. Alternatively, TMA oxalate could slightly inhibit the strand annealing through a decreased GC base pair stability. Partial inhibition of strand annealing could lead to a more efficient binding of primers to the templates and possibly also to a more efficient amplification, not hindered by partially hybridized DNA templates. However, our finding that the optimal annealing temperature is similar in control and TMA–oxalate supplemented samples suggest that changes in base stability caused by 2 mM TMA oxalate are likely to be relatively small. Another possibility is that low concentrations of TMA oxalate reduce the secondary structure of oligonucleotide primers that could, consequently, bind more efficiently to the DNA template. Nor can a direct effect of TMA oxalate on the catalytic properties of DNA polymerase be excluded.
In conclusion, our data indicate that an addition of TMA oxalate to PCR increases the specificity and efficiency of low performance PCR when complex genomic DNA is used as a template. The best results are obtained when PCR mixture is supplemented with 2 mM TMA oxalate.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Hana Mrázová for skilled technical assistance. The work was supported by grant FB-C2/24/99 from Ministry of Industry and Trade of the Czech Republic and by grants 312-98-K205, 310/00/0205 and 204/00/0204 from the Grant Agency of the Czech Republic. The research of P.D. was supported in part by an International Research Scholar’s award from Howard Hughes Medical Institute.
REFERENCES
- 1.Dieffenbach C.W. and Dveksler,G.S. (1995) PCR Primer: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY.
- 2.Chevet E., Lemaître,G. and Katinka,M.D. (1995) Nucleic Acids Res., 23, 3343–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung T., Mak,K. and Fong,K. (1990) Nucleic Acids Res., 18, 4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warner C.K. and Dawson,J.E. (1996) In Persing,D.H. (ed.), PCR Protocols for Emerging Infectious Diseases. ASM Press, Washington DC.
- 5.Winship P.R. (1989) Nucleic Acids Res., 17, 1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bookstein R., Lai,C.-C., To,H. and Lee,W.-H. (1990) Nucleic Acids Res., 18, 1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sidhu M.K., Liao,M.-J. and Rashidbaigi,A. (1996) Biotechniques, 21, 44–47. [DOI] [PubMed] [Google Scholar]
- 8.Baskaran N., Kandpal,R.P., Bhargava,A.K., Glynn,M.W., Bale,A. and Weissman,S.M. (1996) Genome Res., 6, 633–638. [DOI] [PubMed] [Google Scholar]
- 9.Watanabe M., Abe,K., Aoki,M., Kameya,T., Itoyama,Y., Shoji,M., Ikeda,M., Iizuka,T. and Hirai,S. (1996) Neurol. Res., 18, 16–18. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar G., Kapelner,S. and Sommer,S.S. (1990) Nucleic Acids Res., 18, 7465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olive D.M., Simsek,M. and Al-Mufti,S. (1989) J. Clin. Microbiol., 27, 1238–1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissensteiner T. and Lanchbury,J.S. (1996) Biotechniques, 21, 1102–1108. [DOI] [PubMed] [Google Scholar]
- 13.Hengen P.N. (1997) Trends Biochem. Sci., 22, 225–226. [DOI] [PubMed] [Google Scholar]
- 14.Reysenbach A.L., Giver,L.J., Wickham,G.S. and Pace,N.R. (1992) Appl. Environ. Microbiol., 58, 3417–3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bachmann B., Lüke,W. and Hunsmann,G. (1990) Nucleic Acids Res., 18, 1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Birren B., Green,E.D., Klapholz,S., Myers,R.M. and Roskams,J. (1997) Genome Analysis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, NY.
- 17.Melchior W.B. and Von Hippel,P.H. (1973) Proc. Natl Acad. Sci. USA, 70, 298–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs K.A., Rudersdorf,R., Neill,S.D., Dougherty,J.P., Brown,E.L. and Fritsch,E.F. (1988) Nucleic Acids Res., 16, 4637–4650. [DOI] [PMC free article] [PubMed] [Google Scholar]


