Abstract
Oxygen supplementation is life saving for premature infants and for COVID-19 patients but can induce long-term pulmonary injury by triggering inflammation, with xenobiotic-metabolizing CYP enzymes playing a critical role. Murine studies showed that CYP1B1 enhances, while CYP1A1 and CYP1A2 protect from, hyperoxic lung injury. In this study we tested the hypothesis that Cyp1b1-null mice would revert hyperoxia-induced transcriptomic changes observed in WT mice at the transcript and pathway level. Wild type (WT) C57BL/6J and Cyp1b1-null mice aged 8–10 weeks were maintained in room air (21% O2) or exposed to hyperoxia (>95% O2) for 48h. Transcriptomic profiling was conducted using the Illumina microarray platform. Hyperoxia exposure led to robust changes in gene expression and in the same direction in WT, Cyp1a1-, Cyp1a2-, and Cyp1b1-null mice, but to different extents for each mouse genotype. At the transcriptome level, all Cyp1-null murine models reversed hyperoxia effects. Gene Set Enrichment Analysis identified 118 hyperoxia-affected pathways mitigated only in Cyp1b1-null mice, including lipid, glutamate, and amino acid metabolism. Cell cycle genes Cdkn1a and Ccnd1 were induced by hyperoxia in both WT and Cyp1b1-null mice but mitigated in Cyp1b1-null O2 compared to WT O2 mice. Hyperoxia gene signatures associated positively with bronchopulmonary dysplasia (BPD), which occurs in premature infants (with supplemental oxygen being one of the risk factors), but only in the Cyp1b1-null mice did the gene profile after hyperoxia exposure show a partial rescue of BPD-associated transcriptome. Our study suggests that CYP1B1 plays a pro-oxidant role in hyperoxia-induced lung injury.
Keywords: Hyperoxia, Cytochrome P450, CYP1B1, Bronchopulmonary dysplasia, Lung, Oxidative stress
Graphical abstract
1. Introduction
Oxygen supplementation is a life-saving treatment used for patients with a multitude of diseases [1]. However, excess oxygen supplementation not only causes acute, but also chronic, lung damage [2]. Hyperoxic lung injury occurs when excess oxygen triggers inflammatory processes resulting in damage to pulmonary tissues [3]. The importance of understanding the underlying mechanisms in hyperoxia-mediated lung injury has increased tremendously in the past few years as more patients have required oxygen supplementation for the treatment of acute respiratory distress syndrome (ARDS) following COVID-19 infections [4]. Another group at risk of hyperoxic lung injury due to supplemental oxygen requirements are premature infants, who could develop bronchopulmonary dysplasia (BPD) [5], the most common morbidity associated with preterm birth.
Hyperoxic lung injury caused by supplemental oxygen may necessitate even more oxygen, further contributing to the lung injury. Thus, the treatment for the problem enhances the problem, creating a vicious cycle of injury. The optimal oxygenation level that may satisfy the patient's oxygen requirements while minimizing the pulmonary tissue damage is challenging to implement clinically [6,7]. To improve management of patients requiring supplemental oxygen, a better understanding of the underlying mechanisms through which hyperoxia perpetuates lung injury is needed. Altering or mitigating the adverse effects of these mechanisms may provide us with a solution in which patients may be given the required oxygen supplementation without the risk of hyperoxic lung injury.
One such mechanism shown to play a critical role in hyperoxic lung injury involves the xenobiotic-metabolizing cytochrome 450 (CYP) enzymes [8]. CYP enzymes have been studied extensively for their contribution in drug and xenobiotic metabolism. This family of enzymes is found throughout the body, most significantly in the liver and lungs [9]. These phase-I oxidative enzymes may contribute to oxidative stress due to production of reactive oxygen species (ROS), resulting in lipid peroxidation and DNA damage that may perpetuate hyperoxic lung injury [8]. Interestingly, CYP enzymes also play roles in ROS detoxification [10].
Three commonly studied CYP enzymes have been linked to hyperoxic lung injury, specifically the CYP1A1, CYP1A2, and CYP1B1 enzymes [3,8,11]. While CYP1A1 and 1B1 are constitutively present in lung, CYP1A2 is predominantly expressed in liver. Murine studies have shown that the CYP1B1 enzyme may contribute to hyperoxic lung injury as Cyp1b1-null mice had reduced lung injury compared to WT mice upon exposure to hyperoxia [10]. In contrast, studies have shown that Cyp1a1-null and Cyp1a2-null mice experience more significant lung injury compared to WT mice in hyperoxic environments [11]. Therefore, the CYP1A1 and CYP1A2 enzymes attenuate, while CYP1B1 potentiates, hyperoxic lung injury [10].
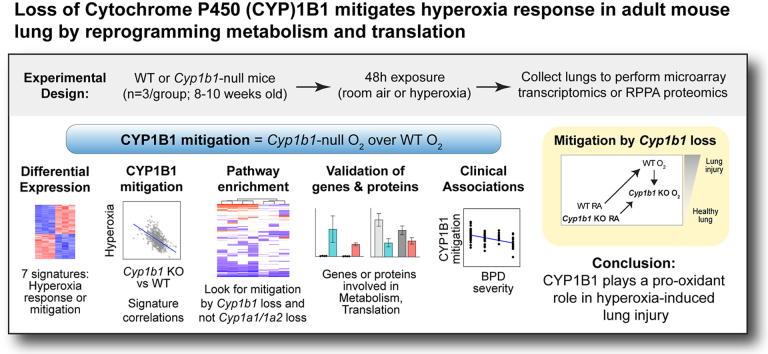
Although prior studies established the relationship between these enzymes and lung injury, the underlying mechanisms remain unclear. Because of the various interconnected pathways that these CYP enzymes influence, these mechanisms are difficult to untangle. We recently reported transcriptomic and proteomic profiling of lung samples of WT, Cyp1a1-null, and Cyp1a2-null mice exposed to hyperoxia [11]; after hyperoxic injury, DNA repair was induced in WT but suppressed in both mutants, while early estrogen response was suppressed in WT but induced in both mutants; apoptosis was suppressed in WT and Cyp1a1-null but induced in Cyp1a2-null mice. To analyze the mechanisms by which CYP1B1 contributes to hyperoxic lung injury, we exposed WT and Cyp1b1-null mice to hyperoxia (95% O2) treatment for 48 h and subjected the lung tissues to transcriptomics microarray and Reverse Phase Protein Array (RPPA) analysis. We sought to identify key pathways altered by hyperoxia in Cyp1b1-null but not in Cyp1a1-or Cyp1a2-null mice as well as to examine which of the lung hyperoxia signatures associated with various pathologic lung conditions: BPD, lung cancer, acute respiratory distress syndrome (ARDS), and idiopathic pulmonary fibrosis (IPF). Fig. 1 demonstrates the overarching analytical approach deployed in this research. As the phenotype observed in Cyp1b1-null mice has been opposite to that in mice lacking Cyp1a1 or Cyp1a2, we speculated that CYP1B1 plays a pro-oxidant role in oxygen injury. Thus, we hypothesize that unique molecular pathways in Cyp1b1-null mice under hyperoxic conditions mitigate lung injury.
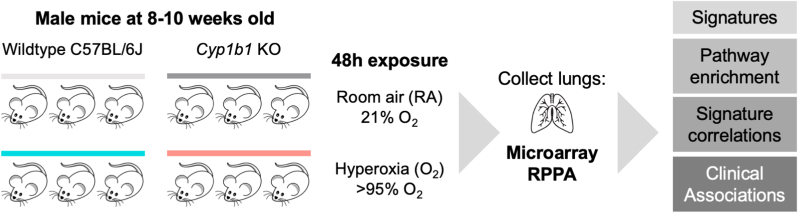
Fig. 1.
Experimental design. Wildtype (WT) C57BL/6J mice and Cyp1b1−/− (Cyp1b1-null) mice were exposed for 48 h to either room air (RA) or hyperoxia (O2). Lungs were collected and assayed for gene expression using microarray or for protein expression using Reverse Phase Protein Array (RPPA) platforms. We evaluated the transcriptional phenotype of Cyp1b1-null mice using gene signatures, enriched pathways, gene signature correlations, and clinical associations with potential lung disease endpoints.
2. Materials and methods
2.1. Animals
Male mice were used for all studies. C57BL/6J wild type (WT) mice were purchased from Charles River Laboratories (Wilmington, Delaware). Cyp1a1-null and Cyp1a2-null mice, on a C57BL/6J background, have been previously described [11]. Cyp1b1-null mice, on a C57BL/6J background, were generated by Dr. Frank J. Gonzalez and have been previously described [12]. The Institutional Animal Care and Use Committee of Baylor College of Medicine approved the study (Protocol No: AN-907), which was performed in accordance with federal guidelines for the humane care and use of laboratory animals. Animals were maintained at the Feigin Center animal facility at Baylor College of Medicine (Houston, Texas). Animals were kept under a 12-h day/night light cycle and were provided purified tap water and food (Purina Rodent Lab Chow 5001 from Purina Mills, Inc., Richmond, Indiana) ad libitum. Mice used in these experiments were between 8- and 10-weeks old.
2.2. Hyperoxia exposure and tissue harvesting
Male mice were randomly selected for each treatment group and were maintained in room air (21% O2) as a control group or exposed to hyperoxia (>95% O2) using pure O2 at 5 L/min for 48h, in a sealed Plexiglass chamber. The oxygen concentration in the plexiglass chamber was monitored by an oxygen analyzer (Getronics, Kenilworth, New Jersey). Following hyperoxia exposure, animals were anesthetized with sodium pentobarbital (200 mg/kg i.p.) and euthanized by exsanguination while under deep anesthetization. Lung tissues were collected and snap frozen at −80 °C for future analysis.
2.3. RNA isolation
RNA extraction and quality control were performed as described previously, with minor modifications [11]. Three biological replicates per experimental group were used for these analyses. Total RNA from frozen lung samples was isolated using the miRNeasy kit as per the manufacturer's standard protocols (Qiagen, Valencia, CA, USA). Sample RNA concentration was measured using a Nanodrop-8000 (Thermo Scientific, Wilmington, DE, USA) and quality checks were done using the NanoDrop spectrophotometer and the Agilent Bioanalyzer. The 260/280 and 260/230 ratios needed to be greater than 1.8 to meet RNA quality control. Additionally, RNA samples needed an RNA Integrity Number (RIN) value between 7 and 10.
2.4. Microarray transcriptomics analysis
RNA profiling using Illumina Gene Expression MouseWG-6 v2.0 Expression BeadChip Kit was performed at the Laboratory for Translational Genomics at Baylor College of Medicine as previously described [11]. Preprocessing of the microarray data, including quality control, background adjustment, and variance stabilization, was performed using the Lumi package [13] implemented in the R statistical system. A transcript was considered ‘detected’ at a p-value cutoff of <0.01. Significantly changed probes were determined using the limma package [14] as implemented in the R statistical system, with significance achieved for fold change exceeding 1.5x and FDR-adjusted p-value<0.05. Differentially expressed probes were depicted as heatmaps using the heatmap package implemented in the R statistical system. Enriched pathways were determined using the Gene Set Enrichment Analysis (GSEA) method [15], with significance achieved at FDR<0.25. Selected genes were validated by qRT-PCR, as previously described [11].
2.5. Reverse phase protein array (RPPA)
Frozen whole lung tissues were homogenized with a T25 Ultra-Turrax (Janke & Kunkel) at 8000 rpm in lysis buffer (1% Triton X-100, 50 mM HEPES pH 7.4, 150 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 100 mM NaF, 10 mM Na pyrophosphate, 1 mM Na3VO4, 10% glycerol, containing freshly added protease and phosphatase inhibitors from Roche Applied Science Cat. # 05056489001 and 04906837001, respectively). The volume of lysis buffer was 1 ml per 40 mg of tissue. After centrifugation to remove debris, the supernatant was collected, protein concentration was measured, and the lysates were diluted to 1.5 mg/ml using lysis buffer. Three parts lysate were added to one part 4x SDS sample buffer (40% Glycerol, 8% SDS, 0.25 M Tris.HCL pH 6.8, 10% ß-mercaptoethanol), boiled for 5 min, and stored at −80 °C. Reverse phase protein array analysis (RPPA) was performed in the Functional Proteomics RPPA core facility at the MD Anderson Cancer Center as previously described [11]. In summary, tissue lysates were serially diluted by increasing two-fold ratios for 5 consecutive dilutions (ranging from undiluted to 1:16 dilution) and arrayed on nitrocellulose coated slides in an 11 × 11 format. Samples were assayed with antibodies by tyramide-based signal amplification approach and visualized using DAB colorimetric reaction. Slides were scanned on a flatbed scanner to generate 16-bit tiff image. Spots from tiff images were identified and the density was quantified by Array-Pro Analyzer. The protein expression was normalized by integrating all dilution curves for all samples and all antibodies, using the Supercurve algorithm [16].
2.6. Analysis of human cohorts using murine gene signatures
As previously reported [17], we computed activity scores for murine gene signatures in human cohorts using summed z-scores. Briefly, in a human cohort each gene was transformed to z-scores. For a murine gene signature and for every specimen, z-scores of increased genes were added, and z-scores of decreased genes were subtracted, computing a summed z-score. Human orthologs of mouse genes were determined using the BioMart repository [18]. We determined correlations of activity scores in human cohorts using the Python scientific library, with significance achieved at p < 0.05. For human cohorts with control and case specimens, we used a numerical encoding of the control/case, then performed Pearson correlation analysis between signature scores and numerically encoded clinical variables; significance was achieved for p < 0.05. Signature correlation results were plotted using GraphPad Prism.
For gene signature correlation, the following lung cohorts were downloaded and used: lung explant controls GSE151052, TCGA Lung Adenocarcinoma [19], TCGA Lung squamous carcinoma [20], and normal GTEx lungs [21]. Bronchopulmonary dysplasia (BPD) blood transcriptomics from newborns were downloaded using the dataset GSE32472 [22]. ARDS patients’ blood transcriptomics were downloaded using the dataset GSE32707 [23]. Idiopathic pulmonary fibrosis (IPF) transcriptomics datasets used included GSE93606 [24], GSE53845 [25], GSE35145 [26], GSE24206 [27], and GSE110147 [28].
2.7. Statistical analysis
Statistical significance was achieved using a parametric two-tailed unequal variance Student's t-test, with significance being achieved at p < 0.05.
3. Results
3.1. Disruption of Cyp1b1 mitigated the murine response to hyperoxia
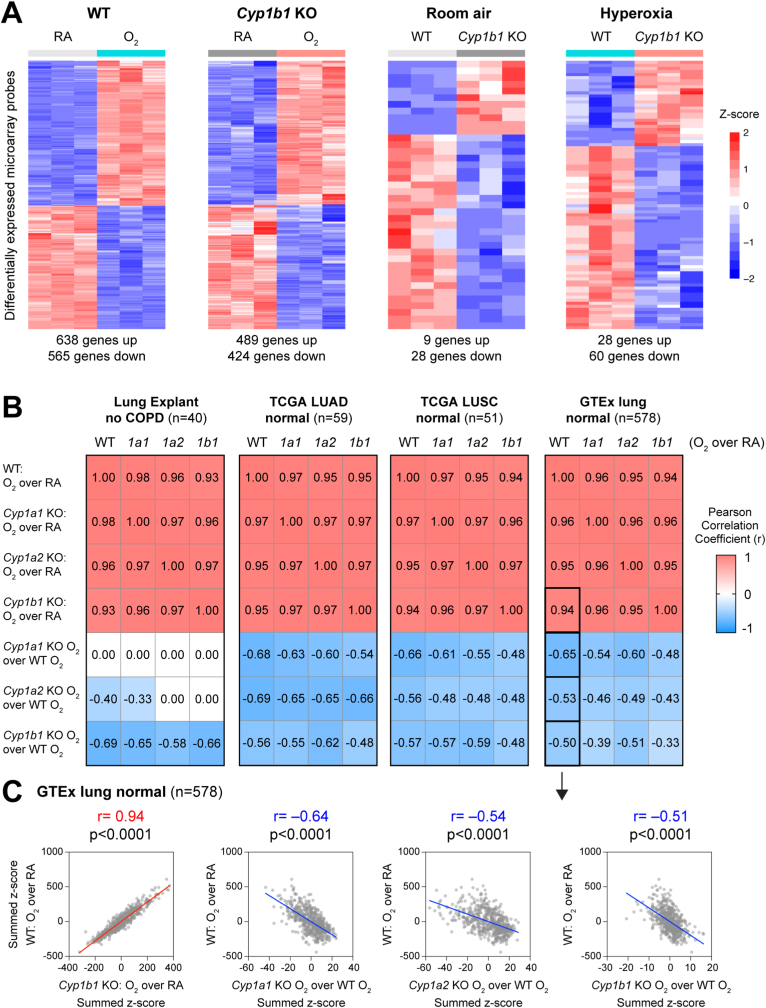
Using transcriptome microarray analysis, we determined the hyperoxia exposure response of WT and Cyp1b1-null mice (Fig. 2A). Whereas a robust hyperoxia response was observed in both WT (with 638 genes increased and 565 genes decreased) and Cyp1b1-null mice (with 489 induced and 424 decreased genes), minimal differences were detected between WT and Cyp1b1-null mouse lungs under either normoxia (9 induced and 28 decreased genes) or hyperoxia (28 induced and 60 decreased genes) conditions. In a previous publication, our group reported signatures of hyperoxia exposure for other murine models genetically modified to delete Cytochrome P450 genes, specifically Cyp1a1-null and Cyp1a2-null mice [11]. To compare the relationship between gene signatures, while accounting for unclear gene-gene relationships, we used the analytical tool of Pearson Correlation based on gene activity scores over transcriptome profiles of lungs from human cohorts, a computationally established methodology [17,29,30]. Specifically, we computed gene signature summed z-scores independently over several transcriptomic cohorts of normal human lungs, including explants, normal adjacent tissues compiled by The Cancer Genome Atlas (TCGA) for lung adenocarcinoma (LUAD) [19], lung squamous carcinoma (LUSC) [20], and also lungs compiled by the Genotype-Tissue Expression (GTEx) project [21] (Fig. 2B). Interestingly, we determined that hyperoxia transcriptomic responses of WT, Cyp1a1-null, Cyp1a2-null, and Cyp1b1-null mice were highly correlated, with a Pearson Correlation Coefficient in the range 0.93–0.98 (p < 0.05). Strikingly, signatures comparing the hyperoxia exposure in genetically modified mice Cyp1a1-null, Cyp1a2-null, and Cyp1b1-null with the hyperoxia exposure in WT mice revealed a negative correlation in multiple cohorts with the canonical (WT) hyperoxia response (p < 0.05) (Fig. 2B). For each of the CYP mutant mice, we observed negative correlations over the GTEx human lungs between the WT hyperoxia response and the mutant hyperoxia to WT hyperoxia signatures, ranging from −0.50 to −0.65 (p < 0.05) (Fig. 2C), suggesting that loss of these CYP enzymes might reverse some of the effects of hyperoxia through commonly regulated genes.
Fig. 2.
Cyp1b1-null mice exhibited a robust hyperoxia exposure response and potentially mitigated the wildtype response. A. Heatmaps of differentially expressed microarray probes, assessing hyperoxia response in WT or Cyp1b1-null mice (left) or differences between WT and Cyp1b1-null mice under either normoxia (RA) or hyperoxia (O2) conditions (right). The number of differentially expressed genes is listed below each heatmap (fold change exceeding 1.5X, FDR<0.05). B. Correlation of hyperoxia or mitigation gene signatures (as summed z-scores) using genes expressed in four cohorts of normal human lungs (GSE151052, TCGA LUAD, TCGA LUSC, GTEx lung). Pearson Correlation Coefficient was used, with significance achieved at p < 0.05. Black boxes indicate the four correlations shown as scatterplots in Panel C. C. Scatterplots indicating the correlation of the hyperoxia transcriptomic response in WT versus the hyperoxia response in the Cyp1b1-null mouse model, or versus the mitigation of the hyperoxia response by genetically modified mouse models Cyp1a1-null, Cyp1a2-null, or Cyp1b1-null. Axes show summed z-scores for the indicated comparison, with each point representing a patient sample from the GTEx human lung cohort.
3.2. Hyperoxic damage is attenuated in Cyp1b1-null mice via a coordinated metabolic program
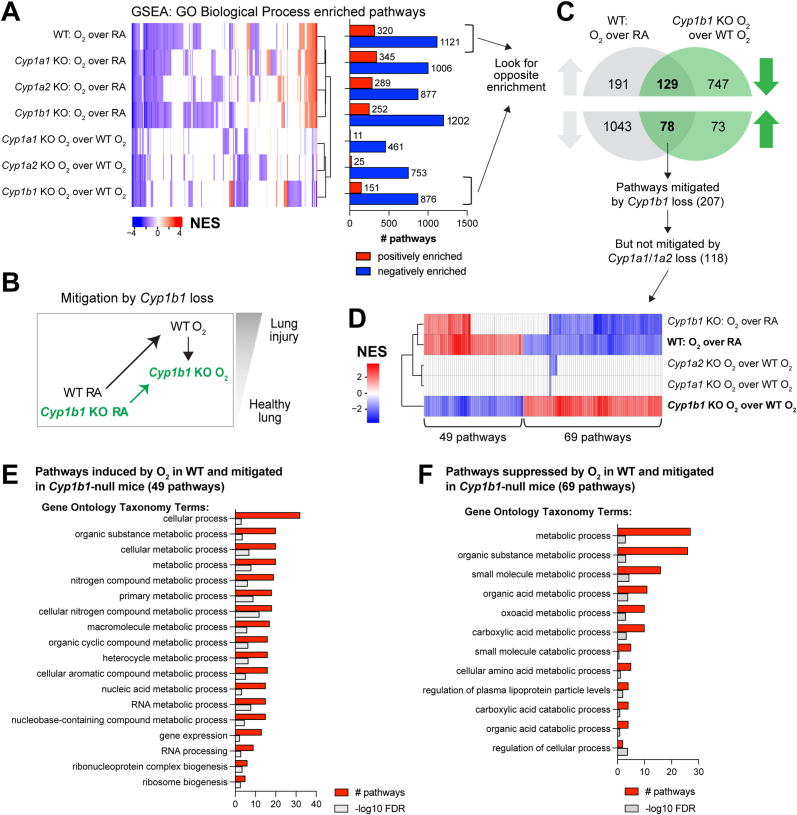
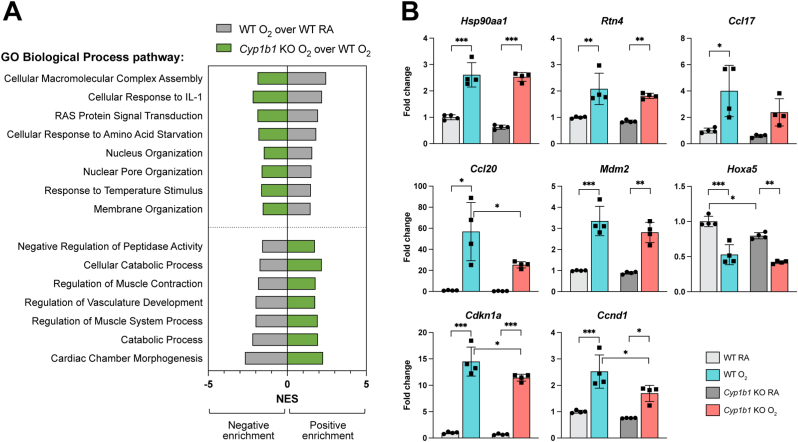
We have shown that hyperoxia exposure led to correlated transcriptomic responses in WT mice and in null models for Cyp1a1, Cyp1a2, and Cyp1b1 (Fig. 2B). A similar observation was substantiated by enriched pathway profiles by GSEA using the Gene Ontology Biological Processes (GOBP) pathway compendium (Fig. 3A, Supplemental Table 1). Enriched pathways showed a robust hyperoxia response, similar across the WT and the three mouse models Cyp1a1-null, Cyp1a2-null, Cyp1b1-null. However, when evaluating GSEA pathways enriched in the comparisons between the hyperoxia exposure in genetically modified mouse models versus the hyperoxia exposure in WT mice, Cyp1a-null and Cyp1a2-null associated pathways clustered together, consistent with the previously reported phenotypes [11], and there were clear clusters of either up- or down-regulated pathway specific to the Cyp1b1-null mice (Fig. 3B, D). Based on the reported phenotypes (e.g. Cyp1a1-null and Cyp1a2-null mice showed a more severe phenotype in terms of oxygen injury compared to WT, whereas Cyp1b1-null mice showed a less severe phenotype), on the clustering patterns observed based on signature correlations (Fig. 2B and C), and on the enriched GSEA pathways (Fig. 3A), we proposed a potential mechanism explaining the protective effect observed in the Cyp1b1-null model. Specifically, the phenotype of Cyp1b1-null is likely explained by pathways that are enriched in both WT mice and Cyp1b1-null mice after hyperoxia exposure but are partially mitigated in hyperoxia-exposed Cyp1b1-null mice compared to hyperoxia-exposed WT mice (Fig. 3B). These pathways are highlighted in Fig. 3C. Analysis of the GSEA enriched pathways revealed 207 pathway candidates possibly explaining the protective effect of Cyp1b1 loss (Fig. 3C), with 129 increased and 78 decreased pathways after hyperoxia exposure. However, since the Cyp1b1-null phenotype is opposite to that of the Cyp1a1-null and Cyp1a2-null mice, we further excluded the pathways that are also mitigated in either of the Cyp1a1-null or Cyp1a2-null mice upon exposure to hyperoxia, resulting in a refined collection of 118 pathways (Fig. 3C and D). Overall, we determined 69 positively enriched and 49 negatively enriched pathways after hyperoxia exposure that show potential mitigation only in the Cyp1b1-null mice (Fig. 3D). Based on the GOBP taxonomy, we carried out enrichment of taxonomy terms separately in the mitigated pathway based on their direction in the WT mice hyperoxia response (Fig. 3E and F, Supp Figs. 1A and 1B) (significance at FDR<0.25). Mitigated pathways both induced and decreased by hyperoxia enriched for organic substance metabolic processes, but induced pathways showed a preference for nitrogen metabolism, whereas suppressed pathways showed a preference for catabolic or carboxylic acid metabolism.
Fig. 3.
Use of enriched pathways to dissect a transcriptional program specific to the Cyp1b1-null mice mitigating the effects of hyperoxia exposure in WT mice. A. Heatmap with unbiased clustering of enriched pathways by Gene Set Enrichment Analysis (GSEA) from comparisons of hyperoxia response and potential mitigation of hyperoxia effects in mouse models Cyp1a1-, Cyp1a2-, Cyp1b1-null (KO). Number of positively and negatively enriched pathways are shown in bar graph. B. Hypothesized hyperoxia mitigation model in Cyp1b1-null mice. C. Venn diagram to identify candidate pathways to explain the mitigation of hyperoxia effects in Cyp1b1-null mice, where pathway enrichment is oppositely enriched. D. Pathways mitigated in the Cyp1b1-null mice but not in Cyp1a1-null or Cyp1a2-null mice. E. Top gene ontology enriched terms in the 49 hyperoxia-induced pathway candidates for hyperoxia effects mitigated in the Cyp1b1-null mice. F. Top gene ontology enriched terms in the 69 hyperoxia-suppressed pathway candidates for hyperoxia effects mitigated in the Cyp1b1-null mice.
Based on the candidate pathways involved in mitigation of the hyperoxia exposure effects in Cyp1b1-null mice, we conducted validation of gene expression changes using qRT-PCR analysis (Fig. 4A). We chose genes involved either in pathways induced by hyperoxia, such as nucleus or membrane organization, or suppressed by hyperoxia, such as development and catabolic processes (Fig. 4B). We expanded our selection to genes changed in hyperoxia in WT and Cypb1-null mice by a fold change of least 1.25x, FDR<0.05. Genes such as 90 kDa heat shock protein (Hspn90aa1), Reticulon 4 (Rtn4), C–C motif chemokine ligand 17 (Ccl17), and C–C motif chemokine ligand 20 (Ccl20) were induced by hyperoxia in both WT and Cyp1b1-null mice (Fig. 4B); these genes span hyperoxia-induced pathways including cellular macromolecular complex assembly, response to temperature stimulus, and cellular response to interleukin. Mouse double minute 2 (Mdm2) was induced by hyperoxia in both WT and Cyp1b1-null mice and is part of several hyperoxia-mediated suppressed pathways, including cardiac chamber morphogenesis and catabolic processes. Homeobox A5 (Hoxa5) was suppressed by hyperoxia in both WT and Cyp1b1-null mice and is involved in the hyperoxia-suppressed pathway of regulation of vasculature development. Interestingly, cyclin dependent kinase inhibitor 1A (Cdkn1a) was induced by hyperoxia in both WT and Cyp1b1-null mice but mitigated in Cyp1b1-null O2 mice compared to WT O2 mice and is a part of hyperoxia-induced pathways (response to temperature stimulus, RAS protein signal transduction, and cellular response to amino acid starvation). The cell cycle pathway was induced by hyperoxia in WT mice and suppressed in all mutant murine models; hence, it was not selected as a Cyp1b1-specific mitigation pathway. However, we noticed that Hoxa5, Mdm2, and Cdkn1a were all cell cycle-specific genes mitigated at the transcriptional level, based on microarray data, only in the Cyp1b1-null mouse model. We selected another cell cycle gene, Cyclin D1 (Ccnd1), mitigated based on the microarray data only in the Cyp1b1-null mice and demonstrated that it was induced by hyperoxia in both WT and Cyp1b1-null mice but mitigated in Cyp1b1-null O2 mice compared to WT O2 mice (Fig. 4B).
Fig. 4.
Gene-level evaluation of hyperoxia exposure responses mitigated in Cyp1b1-null mice. A. Selected pathways enriched by hyperoxia in WT (grey bars) and mitigated in Cyp1b1-null mice (green bars). B. qRT-PCR to assess mRNA expression of genes involved in a typical hyperoxia response and found in the enriched pathway genesets. Fold change relative to WT normoxia (RA) for each treatment is shown, with asterisks indicating significance (*p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.0001). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
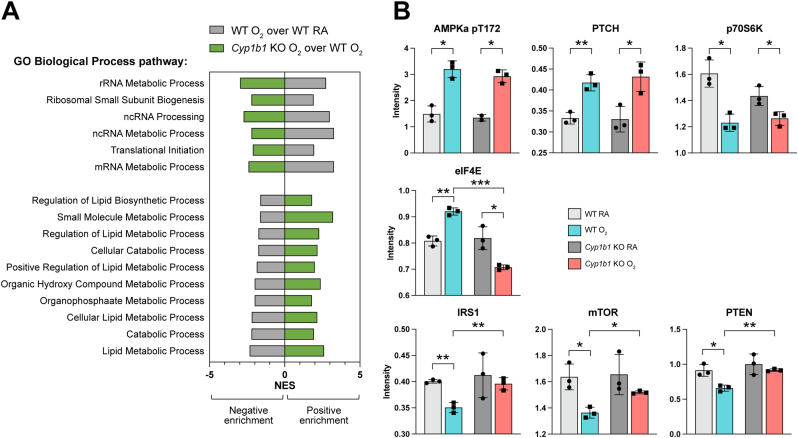
Using a reverse phase protein array (RPPA) platform, we assessed proteins associated with the 118 mitigated pathways (Fig. 5A). We determined proteins similarly increased by hyperoxia exposure in both WT and Cyp1b1-null mice, specifically phospho-AMPK-alpha (T172) and protein patched homolog 1 (PTCH), and similarly decreased by hyperoxia exposure p70S6K (Fig. 5B). Interestingly, we also identified proteins affected by hyperoxia in WT mice but mitigated between Cyp1b1-null O2 mice and WT O2 mice: eukaryotic translation initiating factor 4E (eIF4E) was induced by hyperoxia in WT then decreased in Cyp1b1-null O2 mice, whereas insulin receptor substrate 1 (IRS1), mammalian target of rapamycin (mTOR), and phosphatase and tensin homolog (PTEN) were decreased by hyperoxia in WT mice but induced in Cyp1b1-null O2 mice (Fig. 5B). Overall, the proteomic validation data indicates a strong association with mitigation of metabolic processes in Cyp1b1-null O2 mice, in particular catabolism and lipid metabolism (Fig. 5A).
Fig. 5.
Protein-level evaluation of hyperoxia exposure responses mitigated in Cyp1b1-null mice. A. Selected pathways enriched by hyperoxia in WT (grey bars) and mitigated in Cyp1b1-null mice (green bars). B. Reverse Phase Protein Array (RPPA) to assess levels of proteins affected by hyperoxia and found in the enriched pathway genesets. Protein intensity values for each treatment are shown, with asterisks indicating significance (*p < 0.05, **p < 0.005, ***p < 0.0005). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.3. Hyperoxic responses in WT and cytochrome P450 mutant mice associated with adult and pediatric lung diseases
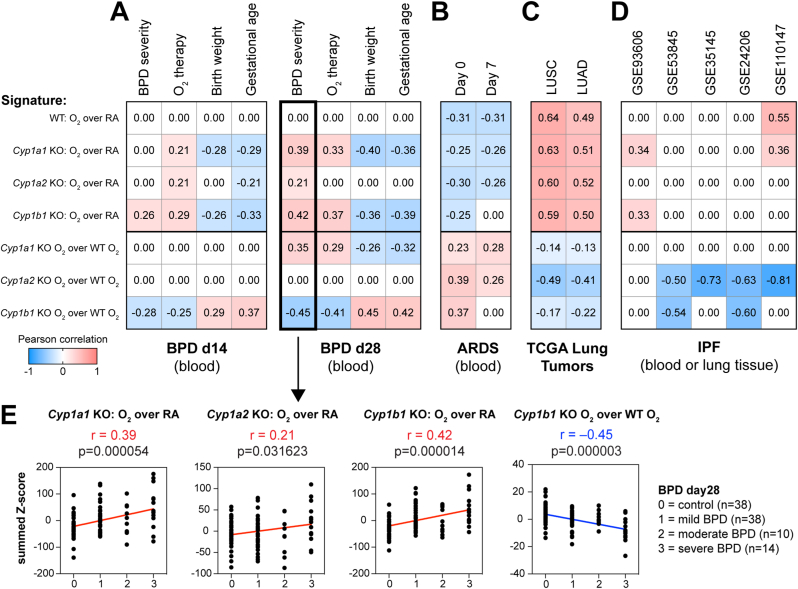
As demonstrated numerous times in cancer biology, gene signatures obtained from in vitro or in vivo experiments can be utilized to mine public disease datasets and interrogate quantitative associations with disease traits, such as cancer vs control or cancer grade/stage [29,31,32]. Recently, our group demonstrated that such approaches are effective outside of the cancer realm, integrating murine hyperoxia gene signatures with BPD blood gene expression profiles [17]. Blood transcriptomics from a human newborn cohort collected at PND14 and PND28 [22] indicated that signatures of hyperoxia exposure in murine lung of either WT or Cyp1a1-null, Cyp1a2-null, or Cyp1b1-null models associated positively with severity of BPD or the need for O2 supplementation and, conversely, associated negatively with birth weight or gestational age (Fig. 6A). Strikingly, the hyperoxia response of Cyp1b1-null mice, but not Cyp1a1-null or Cyp1a2-null mice, relative to the WT hyperoxia response associated with reduced risk of BPD or oxygen supplementation in d14 BPD samples; the Cyp1a1-null hyperoxia treatment positively associated with BPD risk in d28 BPD samples, indicating a further exacerbation of the hyperoxia deleterious effects (Fig. 6A, E). Using an adult human ARDS blood transcriptome cohort GSE32707 [23], hyperoxia exposure signatures in WT or the three mutant mice and hyperoxia mitigation signatures in all three mutant mice showed opposite correlations with risk of progression to ARDS, but did not distinguish between Cyp1a1-null, Cyp1a2-null, or Cyp1b1-null (Fig. 6B). Using lung transcriptomics data from The Cancer Genome Atlas, hyperoxia exposure signatures associated with cancer risk in both lung adenocarcinoma (LUAD) [19] and lung squamous carcinoma (LUSC) [20], whereas mutant hyperoxia mitigation signatures showed association with reduced cancer risk for all three Cyp1a, Cyp1a2, and Cyp1b1 genes (Fig. 6C). Analysis of both blood and lung transcriptomic cohorts for Idiopathic Pulmonary Fibrosis (IPF) patients showed that whereas hyperoxia exposure associated with IPF risk, hyperoxia mitigation signatures in either Cyp1a2-null or Cyp1b1-null were associated with reduced IPF risk (Fig. 6D, Supplemental Fig. 2).
Fig. 6.
Clinical associations of disease progression with hyperoxia exposure responses in a wide range of lung disease models were reversed in the Cyp1b1-null hyperoxia mitigation signature. Publicly available transcriptomics data from blood or lung tissues were used to perform clinical associations between hyperoxia signatures or hyperoxia mitigation signatures (summed z-scores) and disease progression. Data sets used were (A) Bronchopulmonary dysplasia (BPD) blood transcriptomics from newborns at d14 or d28 (GSE32472), (B) ARDS patient blood samples at d0 and d7 (GSE32707), (C) adenocarcinoma (LUAD) or squamous (LUSC) lung tumor samples from TCGA, and (D) Idiopathic pulmonary fibrosis (IPF) transcriptomics from blood (GSE93606) or lung tissues (GSE53845, GSE35145, GSE24206, and GSE110147). E. Detailed scatterplots from panel A (black boxes) show the association of hyperoxia exposure signatures in Cyp1a1-null, Cyp1a2-null, or Cyp1b1-null mice or the hyperoxia response mitigation signature of Cyp1b1-null mice with severity of BPD at d28. Associations were evaluated using the parametric Pearson correlation, with the Pearson correlation coefficient (r) and p-values indicated.
4. Discussion
Several murine studies have demonstrated the protective effects of CYP1B1 enzymes against hyperoxic lung injury [8,33]. The aim of this study was to analyze transcriptomic responses unique to Cyp1b1-null mice exposed to hyperoxia that may provide insight into the underlying injury mechanisms contributed by CYP1B1 enzymes in hyperoxic environments. In order to parse through these mechanisms, a systematic approach was developed to analyze differences in pathways, gene expression, and protein levels amongst WT, Cyp1a1-, Cyp1a2-, and Cyp1b1-null mice. Integration with human blood or lung transcriptomics enabled the analysis of our murine hyperoxia gene signatures with those identified in BPD.
The phenotype previously reported by our group is that Cyp1b1-null mice exposed to hyperoxia exhibit milder effects compared to WT mice exposed to hyperoxia [10]. To understand that difference, we evaluated the very same comparison at gene signature and enriched pathway level and compared it with the hyperoxia response of WT mice (Fig. 2A). However, following approaches commonly used to tease apart the role of different genotypes in hyperoxia exposure [11,34], we have also analyzed neonatal hyperoxic lung injury compare the hyperoxic response of WT mice to the hyperoxic response of the mutant mice (Fig. 2B and C). Hyperoxia exposure led to changes in gene expression in the same direction but at different extents in WT, Cyp1a1-, Cyp1a2-, and Cyp1b1-null mice. To delineate the potential mechanism by which CYP1B1 enzymes contribute to hyperoxic lung injury, enriched pathways due to transcriptomic responses for WT, Cyp1a1-null, Cyp1a2-null, and Cyp1b1-null mice exposed to both normoxic and hyperoxic environments were analyzed and compared. First, enriched pathways common to both WT and Cyp1b1-null mice were removed from consideration. Second, because CYP1A enzymes have shown protective, rather than contributory, effects on hyperoxic lung injury, enriched pathways common to Cyp1a1-null, Cyp1a2-null, and Cyp1b1-null mice were also removed. Out of 207 pathways attenuated in Cyp1b1-null mice exposed to hyperoxia, 39 were attenuated in the Cyp1a1-null and Cyp1a2-null mice as well after hyperoxia exposure; those pathways included six cell cycle related pathways and four apoptotic pathways. However, simple mitigation of a pathway or collection of related pathways is not sufficient to mitigate the hyperoxia phenotypes. Following this analysis, 118 pathways were determined to be either uniquely up- or down-regulated in Cyp1b1-null mice exposed to hyperoxia. Of the 118 pathways identified, 69 decreased and 49 induced pathways by hyperoxia exposure in WT mice were mitigated in Cyp1b1-null mice. The induced pathways included nitrogen and folic acid metabolism while decreased pathways demonstrated carboxylic acid, lipid, fatty acid, glutamate, and amino acid metabolism. These unique pathways may give insight into the potential mechanisms by which CYP1B1 enzymes promote hyperoxic lung injury. Accumulation of intracellular nitrogen is toxic to cells [35]. Therefore, mitigation of hyperoxic induction of nitrogen metabolism in Cyp1b1-null mice may reduce the accumulation of nitrogen and prevent resulting cellular damage. Promotion of nitrogen accumulation may be one possible mechanism in which CYP1B1 enzymes contribute to hyperoxic lung injury. Conversely, hyperoxia resulted in a decrease in carboxylic acid metabolism in WT mice, which is essential in many phase-I reactions for xenobiotic and endogenous molecular metabolism [36]. Cyp1b1-null mice restored carboxylic acid metabolism, indicating that the presence of CYP1B1 enzymes in WT mice may be interfering with carboxylic acid metabolism in the setting of hyperoxia. Therefore, decreased carboxylic acid metabolism may offer another potential mechanism in which CYP1B1 enzymes contribute to lung injury under hyperoxic conditions.
Differences in gene expression between Cyp1b1-null and WT O2 mice were also analyzed to determine other possible mechanisms. Some gene expression alterations caused by hyperoxia were common to both Cyp1b1-null and WT mice, such as the induction of Mdm2 and the suppression of Hoxa5. Hyperoxia-driven Cdkn1a and Ccnd1 gene induction was mitigated in Cyp1b1-null mice compared to WT mice. Lack of Ckdn1a expression has been shown to worsen lung injury secondary to increased cellular apoptosis [37]. Ccnd1 may have a similar mechanism of action, as it serves as a regulator of CDK kinases, impacting cell cycle progression [38]. Decreased expression of these cell cycle regulators in Cyp1b1-null mice may prevent regeneration of damaged lung tissue in the setting of hyperoxic injury in comparison to WT mice.
The elongation factor eIF4E is a crucial component of translation initiation in the process of protein synthesis [39]. Our analysis found that in a hyperoxic environment, eIF4E protein expression was induced in WT mice but decreased in Cyp1b1-null mice. Inhibition of this protein has become a key mechanistic target in cancer drug development [39]. Because eIF4E inhibition may prevent cancer development and progression, Cyp1b1-null mice may have some level of protection from uncontrolled cellular proliferation due to the decreased eIF4E levels in hyperoxic exposure [40,41]. Cyp1b1-null mice also maintained higher expression of the oncogenic protein mTOR and the tumor suppressor protein PTEN after hyperoxia exposure than WT. Because of the contradictory mechanisms of these two proteins, further analysis would need to be conducted to determine the net effect the increased levels have on hyperoxic lung injury in Cyp1b1-null mice [[42], [43], [44]].
For further analysis, the gene signatures from these murine models were integrated with human blood or lung transcriptome of pulmonary diseases. The hyperoxic gene signatures of all the murine models, WT, Cyp1a1-null, Cyp1a2-null, and Cyp1b1-null, associated positively with BPD and the need for O2 supplementation and negatively with birth weight and gestational age. This result indicates that exposure to hyperoxia is a determining factor for BPD, regardless of the presence of CYP1 enzymes. However, an important finding showed that the hyperoxic signature of Cyp1b1-null mice had a reduced risk of BPD and oxygen supplementation, while Cyp1a1-null and Cyp1a2-null mice hyperoxic signatures did not demonstrate this association. Hyperoxia response gene signatures in WT, Cyp1a1-null, Cyp1a2-null, and Cyp1b1-null mice correlated strongly with transcriptome dysregulation seen in other lung diseases, such as ARDS, lung cancer or IPF, and conversely hyperoxia gene expression for each mouse mutant compared to WT associated with lower disease risk; however, there was no risk reduction specific to Cyp1b1-null mice as seen for BPD. These results further validate the hypothesis that CYP1B1 enzymes contribute to hyperoxic lung injury.
5. Conclusion
In conclusion, our analysis provides insight into possible mechanisms by which CYP1B1 enzymes contribute to hyperoxic lung injury. Based on these results, CYP1B1 enzymes may lead to nitrogen accumulation and interfere with carboxylic acid metabolism in hyperoxic environments, which can be toxic to cells. Furthermore, CYP1B1 enzymes may play a role in the induction of Cdkna1 and Ccnd1 transcription under hyperoxic conditions, allowing for continued proliferation of damaged cells and further contributing to lung injury. Similarly, induction of the transcription initiating protein eIF4E may also allow for continued proliferation in this setting. More studies are required to analyze these identified pathways unique to the Cyp1b1-null model in order to further understand the mechanistic interactions of these pathways as well as to identify potential targets for therapy to treat or prevent pulmonary disease development in hyperoxic settings.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
SLG, MJR, TG, and CC were partially funded by The Cancer Prevention and Research Institute of Texas (CPRIT) RP170005 and RP210227, NIH/NCI P30 shared resource grant CA125123, NIH/NIEHS grants P30 ES030285, and NIMHD P50MD015496. KL was supported by R01HL144775 and R01HL146395. BM was supported by 1P42 ES0327725, R01HL129794, 1R01ES029382, 1R21ES032739.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2023.102790.
Contributor Information
Cristian Coarfa, Email: coarfa@bcm.edu.
Bhagavatula Moorthy, Email: bmoorthy@bcm.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
Supplemental figure 1Categorization of enriched pathways mitigated in the hyperoxia exposed Cyp1b1-null mice compared to hyperoxia exposed WT mice. Enriched pathways (rows) from Fig. 3 were categorized by Gene Ontology taxonomy terms (columns). Forty-nine pathways were induced (A) and sixty-nine pathways were suppressed (B) in WT mice by hyperoxia exposure and mitigated in Cyp1b1-null O2 mice. Blue squares indicate inclusion of an enriched pathway under the parent taxonomy term, while some enriched pathways did not belong to the most represented taxonomy terms (grey squares).
Supplemental figure 2Clinical associations of disease progression with hyperoxia exposure responses in Idiopathic Pulmonary Fibrosis (IPF) were reversed in the Cyp1b1-null hyperoxia mitigation signature. A. Signature correlation analysis indicated that WT hyperoxia response associated with progression to IPF using transcriptomics from blood (GSE93606) or lung tissues (GSE53845, GSE35145, GSE24206, and GSE110147). However, hyperoxia mitigation signatures of Cyp1b1-null or Cyp1a2-null mice showed anti-correlation with progression to IPF. B. Detailed scatterplots from panel A (black boxes) show the association of hyperoxia exposure signatures in Cyp1a1-null, Cyp1a2-null, or Cyp1b1-null mice or the hyperoxia response mitigation signature of Cyp1b1-null mice with severity of IPF. Associations were evaluated using the parametric Pearson correlation, with the Pearson correlation coefficient (r) and p-values indicated.
Data availability
Data will be made available on request.
References
- 1.Duke T., Graham S.M., Cherian M.N., et al. Oxygen is an essential medicine: a call for international action. Int. J. Tubercul. Lung Dis. 2010;14(11):1362–1368. [PMC free article] [PubMed] [Google Scholar]
- 2.Kallet R.H., Matthay M.A. Hyperoxic acute lung injury. Respir. Care. 2013;58(1):123–141. doi: 10.4187/respcare.01963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stading R., Couroucli X., Lingappan K., et al. The role of cytochrome P450 (CYP) enzymes in hyperoxic lung injury. Expet Opin. Drug Metabol. Toxicol. 2021;17(2):171–178. doi: 10.1080/17425255.2021.1853705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiersinga W.J., Rhodes A., Cheng A.C., et al. Pathophysiology, transmission, diagnosis, and treatment of coronavirus disease 2019 (COVID-19): a review. JAMA. 2020;324(8):782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- 5.Couroucli X.I., Liang Y.H., Jiang W., et al. Prenatal administration of the cytochrome P4501A inducer, Beta-naphthoflavone (BNF), attenuates hyperoxic lung injury in newborn mice: implications for bronchopulmonary dysplasia (BPD) in premature infants. Toxicol. Appl. Pharmacol. 2011;256(2):83–94. doi: 10.1016/j.taap.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brower R.G., Lanken P.N., MacIntyre N., et al. Higher versus lower positive end-expiratory pressures in patients with the acute respiratory distress syndrome. N. Engl. J. Med. 2004;351(4):327–336. doi: 10.1056/NEJMoa032193. [DOI] [PubMed] [Google Scholar]
- 7.Investigators I.-R., the A. New Zealand Intensive Care Society Clinical Trials G, et al. Conservative Oxygen Therapy during Mechanical Ventilation in the ICU. N. Engl. J. Med. 2020;382(11):989–998. doi: 10.1056/NEJMoa1903297. [DOI] [PubMed] [Google Scholar]
- 8.Veith A., Moorthy B. Role of cytochrome P450s in the generation and metabolism of reactive oxygen species. Curr Opin Toxicol. 2018;7:44–51. doi: 10.1016/j.cotox.2017.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zanger U.M., Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 2013;138(1):103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 10.Veith A.C., Bou Aram B., Jiang W., et al. Mice lacking the cytochrome P450 1B1 gene are less susceptible to hyperoxic lung injury than wild type. Toxicol. Sci. 2018;165(2):462–474. doi: 10.1093/toxsci/kfy154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lingappan K., Maity S., Jiang W., et al. Role of cytochrome P450 (CYP)1A in hyperoxic lung injury: analysis of the transcriptome and proteome. Sci. Rep. 2017;7(1):642. doi: 10.1038/s41598-017-00516-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buters J.T., Sakai S., Richter T., et al. Cytochrome P450 CYP1B1 determines susceptibility to 7, 12-dimethylbenz[a]anthracene-induced lymphomas. Proc. Natl. Acad. Sci. U. S. A. 1999;96(5):1977–1982. doi: 10.1073/pnas.96.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du P., Kibbe W.A., Lin S.M. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24(13):1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- 14.Ritchie M.E., Phipson B., Wu D., et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ju Z., Liu W., Roebuck P.L., et al. Development of a robust classifier for quality control of reverse-phase protein arrays. Bioinformatics. 2015;31(6):912–918. doi: 10.1093/bioinformatics/btu736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grimm S.L., Dong X., Zhang Y., et al. Effect of sex chromosomes versus hormones in neonatal lung injury. JCI Insight. 2021;6(13) doi: 10.1172/jci.insight.146863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Durinck S., Moreau Y., Kasprzyk A., et al. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21(16):3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 19.Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–525. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Consortium G.T., Laboratory D.A., Coordinating Center -Analysis Working G., et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pietrzyk J.J., Kwinta P., Wollen E.J., et al. Gene expression profiling in preterm infants: new aspects of bronchopulmonary dysplasia development. PLoS One. 2013;8(10) doi: 10.1371/journal.pone.0078585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dolinay T., Kim Y.S., Howrylak J., et al. Inflammasome-regulated cytokines are critical mediators of acute lung injury. Am. J. Respir. Crit. Care Med. 2012;185(11):1225–1234. doi: 10.1164/rccm.201201-0003OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Molyneaux P.L., Willis-Owen S.A.G., Cox M.J., et al. Host-microbial interactions in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2017;195(12):1640–1650. doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DePianto D.J., Chandriani S., Abbas A.R., et al. Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax. 2015;70(1):48–56. doi: 10.1136/thoraxjnl-2013-204596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanders Y.Y., Ambalavanan N., Halloran B., et al. Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2012;186(6):525–535. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meltzer E.B., Barry W.T., D'Amico T.A., et al. Bayesian probit regression model for the diagnosis of pulmonary fibrosis: proof-of-principle. BMC Med. Genom. 2011;4:70. doi: 10.1186/1755-8794-4-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cecchini M.J., Hosein K., Howlett C.J., et al. Comprehensive gene expression profiling identifies distinct and overlapping transcriptional profiles in non-specific interstitial pneumonia and idiopathic pulmonary fibrosis. Respir. Res. 2018;19(1):153. doi: 10.1186/s12931-018-0857-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geng C., Rajapakshe K., Shah S.S., et al. Androgen receptor is the key transcriptional mediator of the tumor suppressor SPOP in prostate cancer. Cancer Res. 2014;74(19):5631–5643. doi: 10.1158/0008-5472.CAN-14-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor B.S., Schultz N., Hieronymus H., et al. Integrative genomic profiling of human prostate cancer. Cancer Cell. 2010;18(1):11–22. doi: 10.1016/j.ccr.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geng C., Kaochar S., Li M., et al. SPOP regulates prostate epithelial cell proliferation and promotes ubiquitination and turnover of c-MYC oncoprotein. Oncogene. 2017;36(33):4767–4777. doi: 10.1038/onc.2017.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abbas H.A., Bui N.H.B., Rajapakshe K., et al. Distinct TP63 isoform-driven transcriptional signatures predict tumor progression and clinical outcomes. Cancer Res. 2018;78(2):451–462. doi: 10.1158/0008-5472.CAN-17-1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dinu D., Chu C., Veith A., et al. Mechanistic role of cytochrome P450 (CYP)1B1 in oxygen-mediated toxicity in pulmonary cells: a novel target for prevention of hyperoxic lung injury. Biochem. Biophys. Res. Commun. 2016;476(4):346–351. doi: 10.1016/j.bbrc.2016.05.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coarfa C., Zhang Y., Maity S., et al. Sexual dimorphism of the pulmonary transcriptome in neonatal hyperoxic lung injury: identification of angiogenesis as a key pathway. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;313(6):L991–L1005. doi: 10.1152/ajplung.00230.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zou D., Li J., Fan Q., et al. Reactive oxygen and nitrogen species induce cell apoptosis via a mitochondria-dependent pathway in hyperoxia lung injury. J. Cell. Biochem. 2019;120(4):4837–4850. doi: 10.1002/jcb.27382. [DOI] [PubMed] [Google Scholar]
- 36.Skonberg C., Olsen J., Madsen K.G., et al. Metabolic activation of carboxylic acids. Expet Opin. Drug Metabol. Toxicol. 2008;4(4):425–438. doi: 10.1517/17425255.4.4.425. [DOI] [PubMed] [Google Scholar]
- 37.Blazquez-Prieto J., Huidobro C., Lopez-Alonso I., et al. Activation of p21 limits acute lung injury and induces early senescence after acid aspiration and mechanical ventilation. Transl. Res. 2021;233:104–116. doi: 10.1016/j.trsl.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J.W., Rhee C.K., Kim T.J., et al. Effect of pravastatin on bleomycin-induced acute lung injury and pulmonary fibrosis. Clin. Exp. Pharmacol. Physiol. 2010;37(11):1055–1063. doi: 10.1111/j.1440-1681.2010.05431.x. [DOI] [PubMed] [Google Scholar]
- 39.Romagnoli A., D'Agostino M., Ardiccioni C., et al. Control of the eIF4E activity: structural insights and pharmacological implications. Cell. Mol. Life Sci. 2021;78(21–22):6869–6885. doi: 10.1007/s00018-021-03938-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao J., Teng L., Yang S., et al. MNK as a potential pharmacological target for suppressing LPS-induced acute lung injury in mice. Biochem. Pharmacol. 2021;186 doi: 10.1016/j.bcp.2021.114499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li X., Shan C., Wu Z., et al. Emodin alleviated pulmonary inflammation in rats with LPS-induced acute lung injury through inhibiting the mTOR/HIF-1alpha/VEGF signaling pathway. Inflamm. Res. 2020;69(4):365–373. doi: 10.1007/s00011-020-01331-3. [DOI] [PubMed] [Google Scholar]
- 42.Populo H., Lopes J.M., Soares P. The mTOR signalling pathway in human cancer. Int. J. Mol. Sci. 2012;13(2):1886–1918. doi: 10.3390/ijms13021886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hua H., Kong Q., Zhang H., et al. Targeting mTOR for cancer therapy. J. Hematol. Oncol. 2019;12(1):71. doi: 10.1186/s13045-019-0754-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen C.Y., Chen J., He L., et al. PTEN: tumor suppressor and metabolic regulator. Front. Endocrinol. 2018;9:338. doi: 10.3389/fendo.2018.00338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figure 1Categorization of enriched pathways mitigated in the hyperoxia exposed Cyp1b1-null mice compared to hyperoxia exposed WT mice. Enriched pathways (rows) from Fig. 3 were categorized by Gene Ontology taxonomy terms (columns). Forty-nine pathways were induced (A) and sixty-nine pathways were suppressed (B) in WT mice by hyperoxia exposure and mitigated in Cyp1b1-null O2 mice. Blue squares indicate inclusion of an enriched pathway under the parent taxonomy term, while some enriched pathways did not belong to the most represented taxonomy terms (grey squares).
Supplemental figure 2Clinical associations of disease progression with hyperoxia exposure responses in Idiopathic Pulmonary Fibrosis (IPF) were reversed in the Cyp1b1-null hyperoxia mitigation signature. A. Signature correlation analysis indicated that WT hyperoxia response associated with progression to IPF using transcriptomics from blood (GSE93606) or lung tissues (GSE53845, GSE35145, GSE24206, and GSE110147). However, hyperoxia mitigation signatures of Cyp1b1-null or Cyp1a2-null mice showed anti-correlation with progression to IPF. B. Detailed scatterplots from panel A (black boxes) show the association of hyperoxia exposure signatures in Cyp1a1-null, Cyp1a2-null, or Cyp1b1-null mice or the hyperoxia response mitigation signature of Cyp1b1-null mice with severity of IPF. Associations were evaluated using the parametric Pearson correlation, with the Pearson correlation coefficient (r) and p-values indicated.
Data Availability Statement
Data will be made available on request.