Abstract
Host cell factor (HCF, C1, VCAF or CFF) is a cellular protein that is required for transcription activation of herpes simplex virus (HSV) immediate-early (IE) genes by the virion protein VP16. The biological function of HCF remains unclear. Recently we identified a cellular transcription activator, Luman. As with VP16, the transactivation function of Luman is also regulated by HCF. Here we report a second human protein, Zhangfei (ZF) that interacts with HCF in a fashion similar to Luman and VP16. Although ZF shares no significant sequence homology with Luman, the two proteins have some structural similarities. These include: a basic domain–leucine zipper (bZIP) region, an acidic activation domain and a consensus HCF-binding motif. Unlike Luman, or most other bZIP proteins, ZF by itself did not appear to bind consensus bZIP-binding sites. It was also unable to activate promoters containing these response elements. Although in transient expression assays ectopically expressed ZF was unable to block transactivation by VP16 of a HSV IE promoter, ZF could prevent the expression of several HSV proteins in cells infected with the virus. The ability of ZF to block the synthesis of the HSV IE protein ICP0 relied on its binding to HCF, since a mutant of ZF that was unable to bind HCF was also unable to prevent viral IE protein expression.
INTRODUCTION
Viral transcription factors are often used as paradigms for the study of eukaryotic transcription regulation. Among these, the herpes simplex virus (HSV) protein VP16 is the most studied model for the combinatorial control of gene expression by selective protein–protein interactions (1,2). VP16 (also known as Vmw65 or αTIF) is a structural protein and is brought into cells upon infection as a component of the infecting HSV virion. In the infected cell VP16 initiates the viral replicative process by activating expression of the immediate-early (IE or α) class of viral genes. These genes code for regulatory proteins that subsequently promote expression of the delayed- early (E or β) and late (L or γ) temporal classes of HSV genes. The transcription activation of IE genes by VP16 requires the involvement of at least two cellular proteins, the POU domain protein Oct-1 (3–8) and a host cell factor, HCF (also called C1, VCAF or CFF) (9–12). Unlike most other transcription activators, VP16 does not bind to DNA directly, but is recruited by Oct-1 to TAATGARAT motifs (R is a purine) present in all IE gene promoters. Upon infection of permissive cells, VP16 first forms a complex with HCF. This association subsequently promotes the interaction of this complex with Oct-1, which is bound to the TAATGARAT motifs (3,5–7,13,14).
The HCF gene encodes a large 2035 amino acid protein. However, HCF mainly exists in human cells as a family of polypeptides, which are the products of proteolytic cleavage of the primary HCF protein. The resulting N- and C-terminal fragments of HCF remain held together by non-covalent bonds (15–17). Despite its known accessory role in VP16-activated transcription, the cellular function of HCF in host cells remains undetermined. The finding of homologs in species as diverse as humans, insects and nematodes (10,18–22) suggests that HCF may play a critical role in host cells that is conserved in evolution. HCF has recently been implicated in regulation of the cell cycle (23). In the hamster cell line tsBN67 a single point mutation in the HCF gene, Pro→Ser at position 134 (P134S), confers a temperature-sensitive phenotype on the protein. At the non-permissive temperature the growth of tsBN67 cells is arrested at the G0/G1 decision point of the cell cycle. While the mutation has no apparent effect on the stability and post-translational processing of HCF, it prevents VP16 from binding HCF and subsequently abolishes transcription activation by VP16 (23,24).
Recently, we (25–27) and others (28) identified a human basic domain–leucine zipper (bZIP) protein, Luman/LZIP, that interacts with HCF. Luman is a transcription factor of the CREB/ATF gene family with a potent acidic activation domain (27). Luman and VP16 compete with each other for the binding of HCF in vitro (25) and their mechanism for binding to HCF appears to be the same. Luman and its homologs in mice, LZIP (29), and in Drosophila, dCREB-A/BBF-2 (30,31), all have a conserved HCF-binding motif, (D/E)HXY(S/A). This motif is also found in VP16 as well as in VP16 homologs in other herpesviruses (25,27). Studies have shown that conserved residues in this motif in VP16 (32–36), Luman and dCREB-A/BBF-2 (27,28) are critical for their binding to HCF. The P134S mutation in HCF also disrupts its association with Luman and dCREB-A/BBF-2, as it does with VP16 (27,28). These data strongly suggest that VP16 mimics Luman in its interaction with HCF. The advantage of this mimicry to HSV is not fully understood, although recent studies have suggested a role for HCF in the establishment of latency and reactivation of HSV (26,37).
Here we report the identification of yet another human protein that interacts with HCF. This protein, Zhangfei (ZF), is also a bZIP transcription factor but it did not appear to bind to consensus bZIP sites such as the cAMP response element (CRE), the CCAAT/enhancer-binding protein (C/EBP) binding site and the AP-1 site. While in transient expression assays ZF was unable to prevent the activation by VP16 of a reporter gene linked to a HSV IE promoter, it did block the expression of HSV proteins of the IE, E and L temporal classes in cells infected with the virus. The ability of ZF to prevent the synthesis of IE protein ICP0 depended on binding HCF, since a mutant of ZF that could not bind HCF was unable to block ICP0 expression.
MATERIALS AND METHODS
Yeast two-hybrid screening and cloning of ZF cDNA
The nucleotide sequence of ZF cDNA is deposited in GenBank (accession no. AF039942). The ZF cDNA clone resulted from the same yeast two-hybrid screening previously reported for the Luman clone (25). Briefly, a bait plasmid pGBThcfNC that contains the functional HCF coding sequence was used to screen a human HeLa cDNA library (MatchMaker cDNA Library; Clontech) according to the manufacturer’s instructions. An estimated 5 × 106 independent colonies were screened. The phenotype of the positive colonies was confirmed by a separate β-galactosidase colony lift assay. The cDNA library plasmids were isolated by a leu phenotype rescue strategy.
Plasmids and mutagenesis
The plasmid pGEX-KG for constructing glutathione S-transferase (GST) fusion proteins was a gift from Gerry Weinmaster (University of California, Los Angeles, CA). Plasmids pM1, for constructing GAL4 fusion proteins, and pG5EC, a plasmid containing the coding sequences of the gene for chloramphenicol acetyltransferase (CAT) linked to five GAL4-binding motifs in its promoter region, were obtained from Ivan Sadowski (University of British Columbia, Vancouver, Canada). All the HCF plasmids used in this study contain the functional version of HCF, HCF(NC) (19,25). The P143S mutation of HCF, HCF(P134S), was made by introducing a C→T transition using a PCR strategy, as described previously (27). The construction of plasmids for expressing HCF and HCF(P134S) and their GAL fusion proteins in mammalian cells has been described elsewhere (25,27).
Plasmid pGAL-ZF was constructed by subcloning the EcoRI–XhoI cDNA fragment containing the entire predicted coding sequence of ZF into pM1 between EcoRI and SalI sites. To construct pcZF for expressing ZF protein in mammalian cells, the same cDNA fragment was subcloned into pcDNA3 (Invitrogen) between the same restriction sites. To construct pGEX-ZF, an oligonucleotide linker (5′-GATCCAATGGAGAATTCCTGACGGATATCGGGCCCT and 5′-CTAGAGGGCCCGATATCCTGCAGGAAATTCTCCATTG) was inserted between the BamHI and XbaI sites of pGEX-KG to realign the reading frame at the EcoRI site. The EcoRI–XhoI cDNA fragment of ZF was subsequently cloned into the new pGEX vector between the same restriction sites.
Deletion mutants of ZF were generated by PCR. These mutants included ZF1-30 (5′-GGAATTCATGGAGGAGGAGGCGATCGC and 5′-CCGCTCGAGGTCCAGGAGATCCGCCAGTT), ZF1-60 (5′- GGAATTCATGGAGGAGGAGGCGATCGC and 5′-CCGCTCGAGCAGG CCGCCGCTATCCGAGC), ZF31-272 (5′-GGAATTCATGCCCAGGCAACC- GGACTGGCA and 5′-CCGCTCGAGCTACATTTTAAGA- GAAGACG), ZF61-272 (5′-GGAATTCATGTGGAGAGGGGACGATGAC and 5′-CCGCTCGAGCTACATTTTAAGAGAAGACG) and ZF75-272 (5′-GGAATTCATGCAGCGCTTCTCTGACCT and 5′-CCGCTCGAGCTACATTTTAAGAGAAGACG), where the numbers following ZF are amino acid positions in the presumed ZF protein and the oligonucleotides in parentheses are the primers used in the specific PCR. All left primers had an attached EcoRI site in front of an ATG, while right primers had a XhoI site to facilitate subsequent cloning. All PCR were performed on a MJR PTC-200 thermal cycler. A typical 50 µl reaction mixture consisted of 1 ng of pcZF plasmid DNA, 0.2 mM each of dATP, dTTP, dGTP and dCTP, 0.2 µM primers, 1× KlenTaq buffer and 1× KlenTaq polymerase mixture (Clontech). The PCR program began with 1 min denaturation at 94°C, followed by 25 cycles of 30 s denaturation at 94°C, 30 s annealing at 56°C and 4 min extension at 72°C, and ended with a final 8 min extension at 72°C. The PCR reactions were subjected to electrophoresis in a 0.8 or 2% agarose gel. The PCR bands were excised and DNA was eluted, purified and digested with EcoRI and XhoI. These fragments were cloned between the same sites in pcDNA3 and pGEX or between the EcoRI and SalI sites in pM1. The nucleotide sequences of all the deletion mutants were confirmed by DNA sequencing.
Amino acid substitutions in ZF were generated by site-directed mutagenesis as described elsewhere (25,27,38). These mutants were then cloned into the same expression vectors, pcDNA3, pM1 and pGEX, as the wild-type ZF. The construction of Luman and VP16 plasmids have been described previously (25,38,39).
Cell culture, transfections and CAT assays
The growth conditions of COS7 and Vero cells and the method of transfection have been described previously (25,38,39). CAT expression was measured using a CAT enzyme-linked immunosorbent assay (ELISA) kit (Boehringer Mannheim) according to the instructions provided by the manufacturer. Each transfection was repeated at least once and the mean and standard error are shown in the figures.
Purification of GST fusion proteins, in vitro transcription and translation, electrophoretic mobility assays (EMSA) and GST pull-down
The GST fusion proteins were produced and purified using glutathione–Sepharose beads (Pharmacia) from Escherichia coli strain BL21(DE3) (Novogen) (25). A rabbit reticulocyte in vitro transcription–translation system (TnT; Promega) was used to produce 35S-labeled proteins according to the manufacturer’s protocol. GST pull-down assays were performed as described previously (25). To ensure that each GST pull-down reaction contained the same amount of GST fusion protein, the concentration of each sample of glutathione beads with fusion proteins was adjusted so that when examined by SDS–PAGE each reaction contained the same intensity of Coomassie blue stained protein band.
Antibodies and immunofluorescent microscopy
The antibody against ZF was raised in rabbits immunized with purified GST–ZF fusion protein as previously described (40). The monoclonal antibodies against HSV gB and ICP5 were a gift from Dr Lenore Pereira, (University of California, San Francisco, CA), monoclonal antibodies against ICP0 were purchased from the Rumbaugh-Goodwin Institute and a hybridoma cell line (58-S) secreting antibodies against ICP4 was obtained from the American Type Tissue Culture Collection.
Cells were grown on uncoated coverslips and transfected with pcZF by the calcium phosphate precipitation method (41). Two days post-transfection, cells were washed once in phosphate-buffered saline and fixed in cold methanol for 30 min at room temperature. Cells were stained for immunofluorescence using a technique described previously (26). To determine the effects of ZF on HSV protein synthesis, cells transfected with a plasmid expressing ZF were infected, 20 h following transfection, with HSV-1 strain KOS at a multiplicity of infection (m.o.i.) of 10 p.f.u./cell. Six or 12 h later cells were fixed and stained for immunofluorescence using a rabbit antibody against ZF and a monoclonal antibody against a HSV protein. Alexa546-labeled goat anti-rabbit and Alexa488-labeled goat anti-mouse antibodies (Molecular Probes Inc.) were used as secondary antibodies. The slides were observed using a Zeiss Axioskop microscope equipped for epifluorescence and the appropriate filters. To determine the number of ZF-expressing cells that also expressed HSV proteins, the first 100 cells seen on a slide that stained for ZF (Alexa546) were examined for HSV protein staining (Alexa488). Any cell having detectable levels of Alexa488 fluorescence was considered as expressing HSV protein. Figures for this report were prepared using Adobe Illustrator 7.0 and Adobe Photoshop 4.0 (Adobe Systems Inc.).
RNA analysis
Multiple-tissue northern blots (Clontech) containing poly(A)+ RNA from adult and fetal human tissues were hybridized with ZF and actin cDNA according to the manufacturer’s instructions. The ZF probe was prepared by EcoRI and XhoI excision from plasmid pcZF. Both ZF and actin probes were labeled by the random labeling method (42).
RESULTS
ZF encodes a bZIP protein which is localized on chromosome 11q14
The human protein HCF is largely defined by its involvement in VP16-induced transcription complex formation. To understand the cellular function of HCF and to identify its cellular ligands, we used HCF as bait and screened a HeLa cDNA library using the yeast two-hybrid strategy. Out of an estimated 5 × 106 colonies screened, two positive cDNA clones were identified. The first clone that was confirmed and characterized was Luman (25–27). Here we present the results obtained with the other clone, Zhangfei (named after a legendary Chinese warrior who was contemporary with Luman, ~220 AD).
The isolated cDNA clone contained a 2577 bp insert. A BLAST search (43) against the human EST (expressed sequence tag) database of GenBank recovered an additional 375 nt sequence at the 5′-end. Attempts to amplify additional sequences at the 5′-end of the cDNA by RACE–PCR did not yield further upstream sequences from two HeLa cDNA libraries, including one library specifically designed for 5′-RACE (Marathon-Ready HeLa cDNA; Clontech). The final 2952 nt cDNA sequence contained only one open reading frame (ORF) larger than 200 nt, starting from nt 458 (Fig. 1). This ORF was 819 nt long and was preceded by a Kozak-like sequence. Upstream from the ORF there were multiple stop codons in all three reading phases. Sequence analysis (Fig. 1) showed that ZF had a basic domain and a leucine zipper of six heptad leucine repeats separated by a conserved six amino acid spacer. A search of GenBank did not locate significant matches outside the bZIP region. Beyond the bZIP region there was no significant homology between ZF and Luman.
Figure 1.
The nucleotide sequence of the ZF ORF and the known 5′-UTR. The identified cDNA contains only one ORF larger than 200 nt, with multiple upstream stop codons (lower case and bold) in all three reading frames. The ZF protein encoded by this ORF is 272 amino acids long with a predicted molecular mass of 29 kDa. This protein contains a bZIP region (in brackets) with six perfect leucine heptad repeats (bold and underlined). A conserved six amino acid spacer (dotted underlined) separating the basic domain and leucine zipper is also present. In the N-terminal region is an acidic domain (underlined), rich in negatively charged amino acids. Close to the C-terminal end it has a HCF-binding motif (boxed). The 1676 bp 3′-UTR containing multiple polyadenylation sites is not shown.
PCR-based sequence tagged sites (STSs) have been used as landmarks for construction of various types of genomic maps, including radiation hybridization maps of the human genome. By using Electronic PCR, a web-based STS match program (http://www.ncbi.nlm.nih.gov/STS/ ), ZF was matched to two physically mapped STSs, WI-30362 and WIAF-2104-STS on chromosome 11. Searching GeneMap’98 (http://www.ncbi.nlm.nih.gov/genemap98/ ) revealed that the physical positions of these two STSs are found to be at practically the same location, or 298.59 and 299.04 cR3000 on chromosome 11. The gene for ZF was therefore on the long arm of chromosome 11 between markers D11S1354 and D11S1311, which corresponds to the q14.1 region on the cytogenetic ideogram of GeneMap’98.
ZF has an acidic activation domain
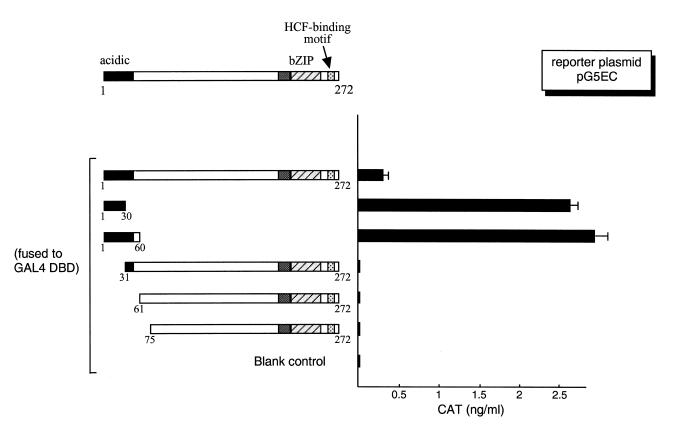
A charge distribution analysis of the presumed ZF ORF showed that it has a highly negatively charged N-terminus. Of the first 30 amino acids, 11 are negatively charged Asp or Glu residues, with no positively charged residues (Fig. 1). To test whether the acidic domain in ZF is a transcription activation domain, as are the ones in Luman and VP16, a series of deletion mutants of ZF (Fig. 2, left) were made and fused to the GAL4 DNA-binding domain (DBD). These plasmids were co-transfected into COS7 cells with a CAT reporter plasmid, pG5EC, in which CAT expression is under the control of GAL4 upstream activation sequences (UAS). CAT ELISA assays were performed to quantify the strength of activation by these constructs. Results from these transient transfection experiments (Fig. 2, right) showed that the acidic region is an activation domain and that the first 30 amino acids are sufficient to activate transcription. We also found that deletion of the first 30 amino acids disrupted transcription activation. Interestingly, in the form of GAL fusion protein, full-length ZF appeared to be a poor activator when compared to ZF (1–30). Removal of the sequence downstream from the activation domain region apparently increased the strength of activation (Fig. 2, right). Similar results were also observed with VP16 (44).
Figure 2.
The N-terminal region of ZF, rich in negatively charged amino acids, is an activation domain. On the left is the schematic representation of the structure of the ZF protein. The numbers indicate the positions of the amino acid. ZF and its deletion mutants were fused to the GAL4 DNA-binding domain. The same amount (0.5 µg) of each plasmid was introduced into COS7 cells along with the reporter plasmid pG5EC (0.5 µg), which has five copies of the GAL4 UAS in the promoter region linked to the CAT gene. The parental vector expressing only the GAL4 DBD, pM1, was used as the blank control. The CAT activity was measured by ELISA 48 h post-transfection.
ZF has a unique basic domain which does not appear to bind consensus bZIP sites
The bZIP gene family is usually considered to consist of three subfamilies: CRE-binding CREB/ATF proteins, C/EBP and AP-1(-like) proteins. The proteins within each of these families preferentially bind to their own consensus elements. To study the DNA-binding specificity of ZF, EMSA were carried out using purified GST-linked ZF proteins and double-stranded oligonucleotides containing consensus binding sequences (i.e. CRE, 5′-GCCGGTGACGTCATCGCAT, C/EBP, 5′-GGTATTGCGTAATTGATAT, and AP-1, 5′-ACCGGTGACTCAATGGCT) compiled from the binding site distribution matrix database TFMATRIX (45). Surprisingly, our EMSA results showed that ZF did not bind any of the three oligonucleotides, even when as much as 2 mg ZF protein was used (data not shown). In transient transfection assays, ZF could not activate transcription from promoters containing these binding sites either. Concurrent overexpression of HCF by co-transfecting cells with a plasmid expressing HCF did not increase transactivation by ZF (data not shown).
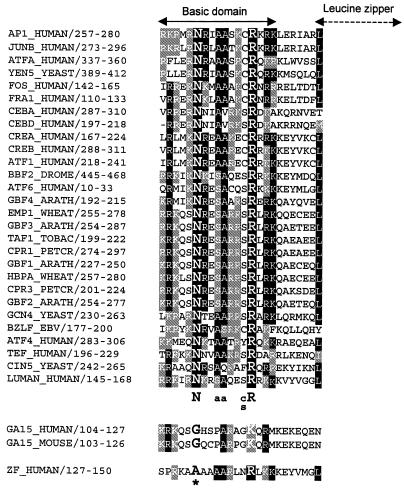
The unexpected EMSA results led us to re-examine the amino acid sequence of the ZF basic domain, which is believed to be directly responsible for DNA binding in all bZIP proteins (46,47). All basic domains of bZIP proteins are closely related: a sequence alignment of 30 representative bZIP proteins is shown in Figure 3. Among all bZIP proteins there is a consensus quintet sequence, NXXAAXX(C/S)R (X stands for any amino acid), which is critical for DNA binding (46–48). In the quintet sequence, the N and R residues are absolutely conserved (with GA15 the only known exception). The N residue in particular is believed to cause a ‘cap’ in the basic region that bends the protein to fit in the major groove of the DNA (46,48) or helps to induce a helical fork structure that positions the protein on the DNA (47,49). Mutation of the Asn residue results in a marked loss of DNA-binding activity (50). In an expanded amino acid sequence alignment (not shown) of 87 bZIP proteins retrieved from the SWISSPROT database, CHOP (also called GADD153 or DDIT3), encoded by locus GA15, is the only protein that does not have the N residue. Coincidentally, CHOP cannot bind DNA by itself (51–53), although it can bind to DNA by forming heterodimers with other bZIP proteins (54). An examination of the ZF sequence revealed that it also lacks the N residue in its basic domain (Fig. 3). Furthermore, the N-terminal portion of its basic region (before the conserved A136 and A137) bears little resemblance to other bZIP proteins. It has been shown that the residues N-terminal to the quintet sequence are also important for DNA binding (55–58). Therefore, it is likely that the conformational change caused by the lack of the Asn and/or other features in the N-terminal portion of the basic region impair the DNA-binding ability of ZF.
Figure 3.
A sequence alignment of basic regions of 30 representative bZIP proteins and ZF. The numbers following the names of the gene locus indicate the location of the segment in the encoded protein. All bZIP proteins, except GA15 (or CHOP, GADD153 or DDIT3) and ZF, share a consensus quintet sequence, NXXAAXX(C/S)R (X stands for any amino acid), which is essential for DNA binding. ZF lacks the absolutely conserved residue N, which is critical for protein conformation; the amino portion of the basic region bears little resemblance to other bZIP factors.
ZF associates with HCF through the binding motif DHDYA
Sequence alignment of ZF with VP16 and Luman as well as their homologs (Fig. 4A) showed that they share a conserved sequence motif, (D/E)HXY(S/A). Previous studies have shown that this motif is required for HCF binding in VP16 (32–36) and Luman (27,28). To compare ZF and Luman in their binding of HCF and to test whether this motif was also responsible for ZF binding to HCF, we changed the two conserved residues, H and Y, at positions 222 and 224, as well as the charged residue, D, at position 221 (Fig. 4A).
Figure 4.
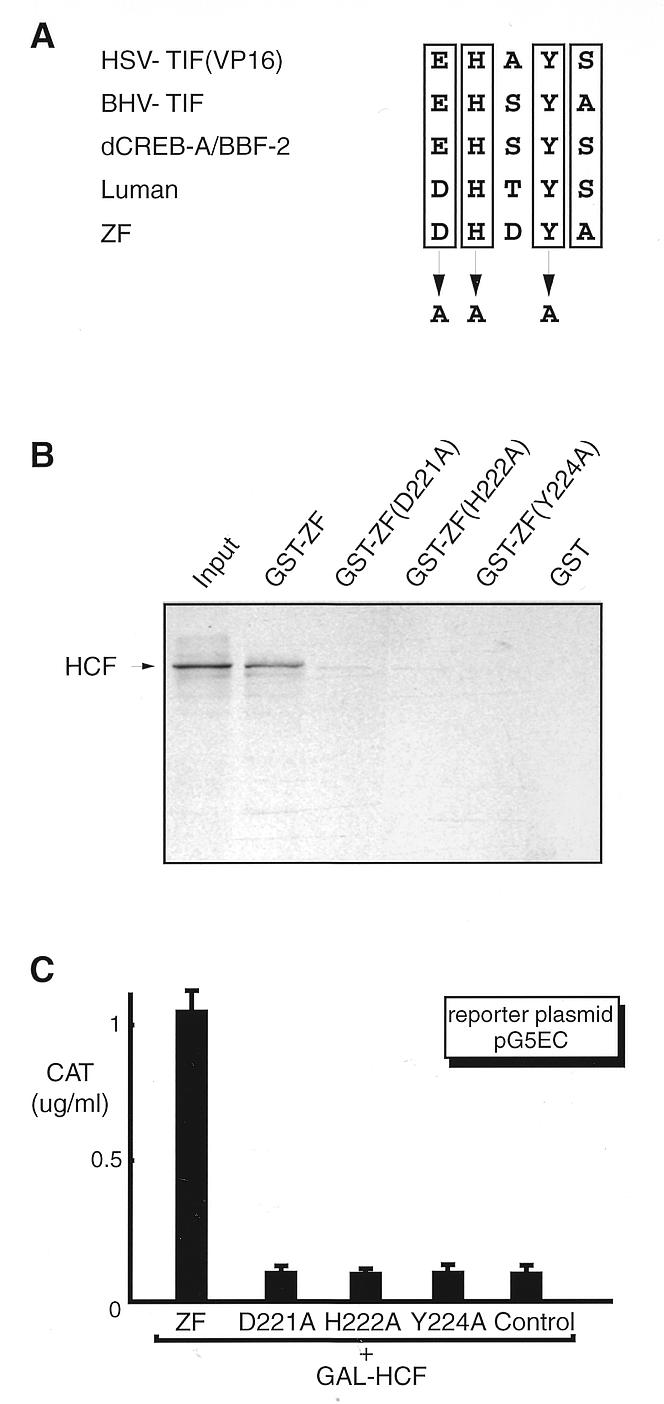
Mutagenesis study of the HCF-binding motif in ZF. (A) The consensus sequence, [D/E]HXY[S/A], found in VP16 and Luman as well as their homologs is proved to be a HCF-binding motif. To test whether this motif in ZF is also involved in HCF binding, we mutated two absolutely conserved residues, H222 and Y224, and the negatively charged residue D221 by site-directed mutagenesis. (B) GST pull-down assay. GST and all GST fusion proteins were produced in E.coli strain BL21(DE3), coupled to glutathione–Sepharose beads. After incubation with [35S]HCF, the beads were washed and analyzed by 10% SDS–PAGE. The lane labeled input represents one-tenth of the [35S]HCF incubation mixture. (C) Mammalian two-hybrid assay. Plasmids pcZF, pcZF(D221A), pcZF(H222A) and pcZF(Y224A) were introduced into COS7 cells separately with reporter pG5EC and the plasmid expressing GAL–HCF. An aliquot of 0.5 µg DNA was used for each plasmid. Plasmid pcDNA3 was used as a control. The CAT activity was measured by ELISA 48 h post-transfection.
A GST pull-down assay (Fig. 4B) and a mammalian two-hybrid assay (Fig. 4C) were performed to examine the effects of these mutations on the in vitro as well as in vivo binding of HCF by ZF. In GST pull-down assays, recombinant GST fusion proteins were produced in bacteria and coupled to glutathione–Sepharose beads. HCF was transcribed and translated in vitro in the presence of [35S]methionine. The radiolabeled HCF proteins were incubated with glutathione–Sepharose beads coupled with GST–ZF and mutants or GST alone. After extensive washing, proteins retained on the beads were eluted and separated by SDS–PAGE. We found that while wild-type ZF bound efficiently with HCF, none of the three ZF mutants did.
We also used a mammalian two-hybrid assay to test the in vivo association of ZF with HCF. As with the yeast two-hybrid system, a mammalian expression vector that produced the GAL4 DBD fusion protein, GAL–HCF, was introduced into COS7 cells with pcZF or plasmids specifying its mutants. Since ZF proteins have their own activation domain, a chimeric ZF protein with GAL4 activation domain was not needed. Plasmid pG5EC containing GAL4-binding motifs was used as the reporter. Like Luman, ZF interacted with HCF in COS7 cells, but ZF mutants failed to interact with HCF. Thus, all three mutations disrupted the association of ZF with HCF in both in vitro and in vivo assays.
Since our results indicated that, like VP16, ZF could also bind HCF, we tried to determine if ZF could compete with VP16 for binding of HCF, thereby reducing VP16 activation of IE promoters. However, in cells co-transfected with a plasmid specifying VP16 and up to a 20-fold excess of a plasmid specifying ZF, the ability of VP16 to activate a basal HSV IE promoter was unaffected (results not shown).
ZF associates with HCF but not its mutant HCF(P134S)
To compare ZF, Luman and VP16 in their binding of HCF, we examined the interaction of ZF with a particular mutant of HCF, P134S. The P134S mutation causes a temperature-sensitive phenotype in HCF and leads to growth arrest of cells at the G0/G1 transitional point of the cell cycle. Under these conditions, the association of HCF with VP16 (23,24) and Luman (27,28) and the transactivation function of these proteins is abolished. To test whether the HCF P134S mutation also affects ZF binding to HCF, we examined the binding of ZF to HCF and HCF(P134S) in a GST pull-down assay (Fig. 5A and B) as well as in a mammalian two-hybrid assay (Fig. 5C), as described above.
Figure 5.
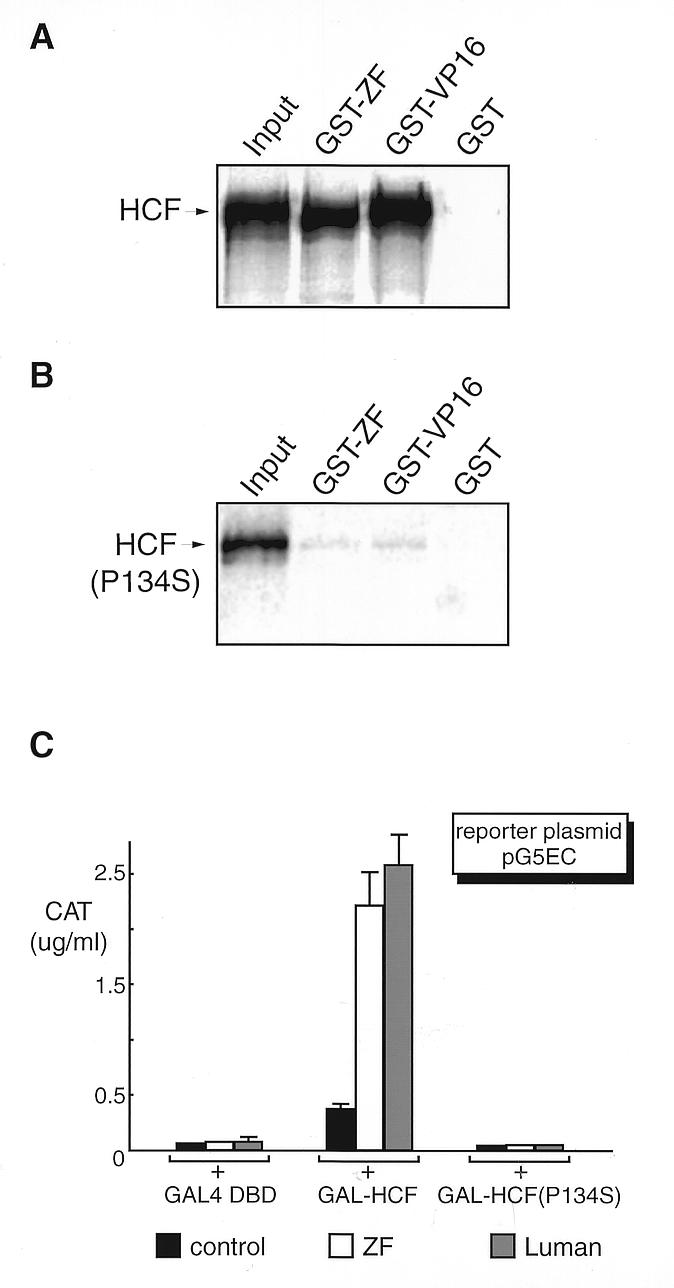
Like VP16, ZF binds to HCF but not to the mutant HCF(P134S), as determined by GST pull-down assay (A and B) and by mammalian two-hybrid assay (C). GST and GST fusion proteins of ZF and VP16 were produced in E.coli strain BL21(DE3), coupled to Sepharose beads. HCF was labeled with [35S]methionine by in vitro transcription and translation in a rabbit reticulocyte system (TnT; Promega). The protein-bound beads were incubated with an equivalent amount of 35S-labeled HCF (A) or HCF(P134S) (B). After extensive washing, proteins were eluted, fractionated by 10% SDS–PAGE and visualized by autoradiography. The first lane on the figure represents one-tenth of the input. Plasmids expressing GAL–HCF, GAL–HCF(P134) and GAL DBD alone (C) were co-transfected into COS7 cells with either pcZF, pcLuman or their parental vector pcDNA3 (0.5 µg each), with pG5EC as the reporter. The CAT activity was measured 48 h post-transfection.
We found that wild-type HCF bound efficiently to ZF and VP16 GST fusion proteins but not to GST itself (Fig. 5A). The HCF(P134S) mutant (Fig. 5B) was impaired in binding of ZF and VP16, as previously documented for VP16 (24) and Luman (27,28). The mammalian two-hybrid assay (Fig. 5C) also confirmed the same finding.
Effect of ZF on HSV expression
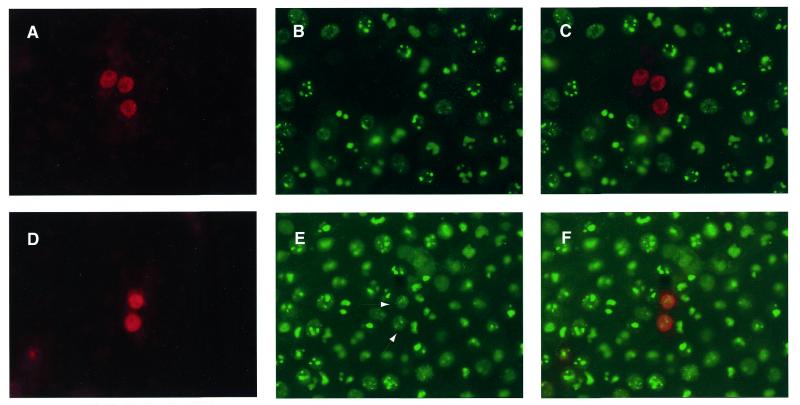
Recently we found that cells expressing Luman appear to be non-permissive for HSV replication (26). To determine if ZF would have a similar effect, cells transfected with ZF were infected with HSV. A m.o.i. of 10 p.f.u. was used to ensure infection of all cells. The cells were then fixed at 6 and 12 h after infection and stained simultaneously for ZF and one of several HSV proteins. These included the IE proteins ICP0 and ICP4, E proteins VP16 and gB and the L protein ICP5. Cells fixed 6 h after infection were stained for the IE proteins whereas cells fixed later were used to detect proteins of the E and L temporal classes. Very few cells that stained for ZF contained detectable levels of the HSV proteins. In one representative experiment, the percentages of ZF staining cells that also expressed the individual HSV proteins were: ICP0, 28%; ICP4, 5%; gB, 2%; VP16, 14%; ICP5, 10%. In contrast, almost all cells that did not contain ZF stained for the HSV proteins. To determine if the association of ZF with HCF was required for its suppression of HSV protein expression, we transfected cells with either ZF or the ZF(Y224A) mutant that showed reduced levels of HCF binding (Fig. 4B). The cells were then infected with HSV at a high m.o.i., fixed 6 h after infection and stained for ZF and ICP0. While ZF prevented infected cells from expressing ICP0 (only 24% of ZF expressing cells contained ICP0), the mutant did not efficiently block ICP0 synthesis. Eighty-three percent of cells expressing ZF Y224A contained ICP0. Figure 6A and B and the composite Figure 6C show three ZF-expressing cells that do not contain detectable levels of ICP0. Figure 6D and E and the composite Figure 6F show two cells expressing ZF Y224A that also stain for ICP0 (arrowheads in Fig. 6E).
Figure 6.
ZF prevents HSV ICP0 expression in virus-infected cells in a HCF-dependent manner. Vero cells were transfected with plasmids expressing either ZF (A–C) or ZF Y224A (D–F). The cells were then infected with HSV-1 at a m.o.i. of 10 p.f.u./cell. Six hours after infection the cells were fixed and stained simultaneously for both ZF (Alexa546, red) and ICP0 (Alexa488, green). The cells were then visualized in a fluorescent microscope using either a 546 (A and D) or 450–490 nm filter (B and E). The red and green images were combined (C and F) using Northern Eclipse 5.0 software. In (E) the arrowheads indicate ICP0 stained nuclei that also stain for ZF Y224A.
Northern blot analysis of ZF
To investigate the expression pattern of ZF, we performed northern blot analyses to determine the mRNA distribution of ZF in human tissues (Fig. 7). The poly(A)+ RNAs from eight different adult tissues (Fig. 7, left) and four fetal tissues (Fig. 7, right) were hybridized with radiolabeled ZF cDNA. The same blots were rehybridized with β-actin probe after stripping the previous probe and used as a loading reference. A 4.6 kb signal was detected in all tissues, although at different intensities. In adult tissues, ZF message was most abundant in heart, liver and skeletal muscle, moderately abundant in kidney and pancreas and was present at the lowest level (barely detectable) in lungs. In fetal tissues, however, the message was most abundant in kidney and only a very low amount was detected in heart, lung and liver.
Figure 7.
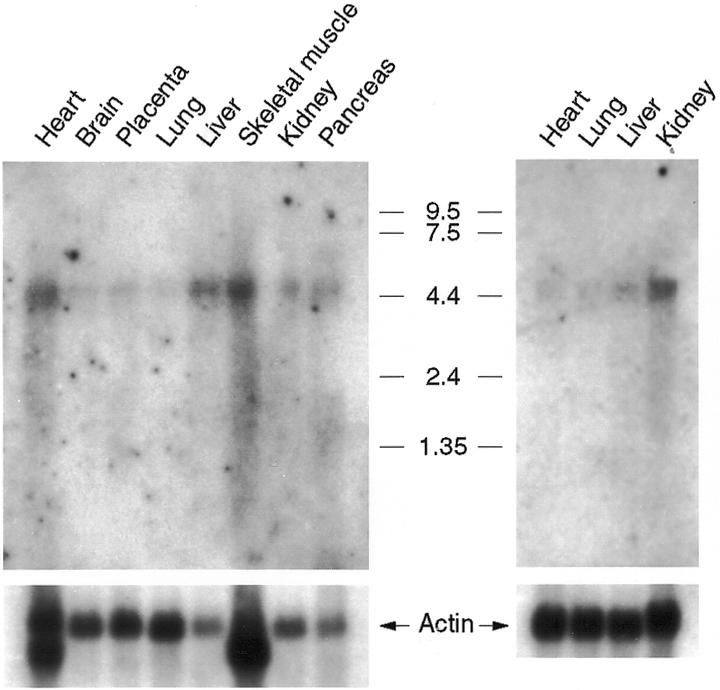
Northern blot analysis of ZF. MTN northern blots (Clontech) containing poly(A)+ mRNA from adult and fetal human tissues were hybridized with 32P-labeled full-length ZF cDNA. Molecular size standards (in kilobases) and tissues of origin are indicated. The same blots were hybridized with the β-actin probe and used as a loading reference. Note that the human β-actin cDNA probe cross-hybridizes to muscle-type actin in heart and skeletal muscle.
A western blot-based protein survey was also conducted in which protein samples (Protein Medley; Clontech) isolated from different human tissues (i.e. heart, brain, placenta, lung, liver, skeletal muscle, kidney and spleen) were used. However, no ZF protein was detected in any of the tissue samples (data not shown). We also found that the cell lines we tested for ZF, including HeLa, COS7 and 293, also did not have Z.
DISCUSSION
Recently we screened a HeLa cDNA library using the yeast two-hybrid strategy to identify cellular targets of HCF. Luman, the first clone that we isolated and characterized, is a bZIP protein that binds CRE and activates CRE-containing promoters (25,27). It also efficiently activates the promoters of genes coding for the HSV IE proteins ICP0 and ICP4 as well as the promoter for the HSV latency-related transcript (26). Here we report a second HCF-binding protein, ZF, that is also a bZIP protein.
Although ZF has no significant homology with Luman beyond the bZIP region, the two proteins have some structural and functional features in common: a bZIP region, an acidic activation domain and a consensus HCF-binding motif, (D/E)HXY(S/A). Our mutation analyses suggest that ZF, Luman and VP16 share the same mechanism of binding to HCF (27). All three proteins also have acidic transcription activation domains.
One unusual feature of ZF cDNA is its exceptionally long 5′- and 3′-untranslated regions (UTRs). In this cDNA clone the predicted ZF ORF is 819 nt long, followed by a 1676 nt 3′-UTR. The known sequence upstream from the ZF ORF is 457 nt; however, with a single message of 4.6 kb detected by northern blot (Fig. 7), it likely has 1.6 kb of additional 5′-sequence. Nevertheless, we think that the presumed 819 bp ZF ORF is the true and complete ORF. Our reasons include: (i) structurally, it encodes a protein that resembles the first identified HCF-binding protein, Luman; (ii) the sequence around the initiation codon, GAGATGG (Fig. 1), conforms well to the Kozak consensus sequence for mammalian protein biosynthesis, with a purine at position –3 and a G at position +4 (59); (iii) there are stop codons in all three reading frames upstream from the presumed initiation codon and multiple polyadenylation signals downstream from the ZF ORF; (iv) the nucleotide composition is different in the presumed coding and non-coding regions. The GC content of the ZF ORF is 64%; however, the known 457 nt 5′-UTR has a GC content of 71% and the 1676 nt 3′-UTR has a GC content of only 33%. More remarkably, the GC% of the 50 nt sequence immediately upstream of the presumed start codon ATG is 86%, while the GC content is only 64% in the first 50 nt of the ORF immediately after the start codon. The failure to recover further 5′-UTR sequence by RACE is likely because the full-length 4.6 kb transcript is not represented in the two HeLa cDNA libraries. The unusually high GC content at the 5′-end and a possible resulting secondary structure may have blocked the movement of DNA polymerase during first strand cDNA synthesis. Another possibility for the shorter transcript is alternative transcription splicing in HeLa cells. Besides ZF, exceptionally long 5′- and 3′-UTRs have been reported in a number of mammalian genes. These long UTRs are usually a means of additional translational regulation (60–65). Accordingly, unusually long 5′- and 3′-UTRs may imply unusual regulatory mechanisms of ZF expression.
We have recently shown that Luman not only interacts with HCF, but also requires this association for its transactivation of CRE-containing promoters (27). Mutations that block HCF binding also impair the ability of Luman to transactivate CRE promoters. For ZF however, it was rather difficult to assess the effect of HCF on transactivation or inhibition by ZF, due to lack of a reporter system. Although ZF could activate transcription from GAL4 UAS-containing promoters as a GAL4 DBD fusion protein, this activation was found to be independent of HCF binding, as is the case with Luman (27). GAL4 fusion proteins of the HCF binding mutants of ZF or just the first 30 amino acid activation domain could efficiently activate transcription from a GAL4 UAS promoter (Fig. 2; unpublished observation). Although the association with HCF may increase the binding of ZF to DNA, we did not observe any increase in transcription activation by ZF from promoters containing CRE, C/EBP or AP-1 sites in our transfection assays when additional HCF was provided (data not shown). Neither did we see an enhancement of DNA-binding ability of ZF by addition of purified recombinant HCF in EMSA.
In spite of these observations, there are indications that HCF binding can alter the ability of ZF to activate transcription. We found that the activation strength of ZF when tethered to GAL4 promoters through its interaction with GAL–HCF appeared much stronger than that of a direct fusion protein of ZF with the GAL4 DBD (compare the CAT activity of GAL–HCF/ZF in Fig. 5 with GAL–ZF in Fig. 2). Similar observations have been made with VP16 and Luman (27; unpublished data). We hypothesize that in the mammalian two-hybrid assay (Fig. 6) GAL–HCF provided a functionality that enhanced activation by ZF, in addition to merely recruiting ZF to the promoter. As with Luman (27), HCF may, at least under these conditions, either unmask the activation domain of ZF or synergistically activate transcription with ZF.
In contrast to Luman, which binds CRE and activates CRE-containing promoters (25,27), ZF did not appear to bind consensus bZIP sites by itself. A possible reason for the inability of ZF to recognize these promoters may be the absence of a critical Asn residue in its basic domain (46,48). An alternative explanation may be that the ZF protein in transfected cells has an aberrant secondary structure. While it is difficult to rule out this possibility, the protein does appear to have biological activity. It suppresses the expression of HSV proteins in cells infected with the virus in a HCF-dependent manner (Fig. 6). These results suggest that ZF may recognize cis-acting regulatory sequences in association with other DNA-binding proteins. To the authors’ knowledge, CHOP, the only other bZIP protein that does not have an Asn residue in its basic domain, does not bind DNA by itself either. Although CHOP can bind to DNA by heterodimerizing with other bZIP proteins (54), it can also function as a transcription inhibitor by preventing the DNA-binding activity of other bZIP proteins (51–53). We have not identified a DNA-binding partner for ZF. However, our preliminary experiments show that while ZF can form homodimers, it cannot associate with Luman (data not shown).
Like Luman, ectopically expressed ZF could block the expression of HSV proteins in cells infected with the virus. ZF prevented the expression of all three temporal classes of HSV proteins. At least for ICP0, binding of HCF by ZF appeared to be required for blocking protein synthesis. The mechanism by which ZF prevented HSV protein synthesis is not known. One possibility could be that it sequesters HCF, making it unavailable for VP16-activated IE expression. However, in transient expression assays ZF co-expressed with VP16 could not block VP16 activation of the ICP0 promoter. This suggests that the mechanism by which ZF influences HSV gene expression may be more complex than just sequestering of HCF.
The identification of a second human HCF-binding transcription activator suggests that HCF, as a co-activator or facilitator, may regulate the activity of several transcription factors. The alphaherpesviruses (HSV is a member of this subfamily) may have evolved to exploit this mechanism to modulate their complicated replicative cycles. We are currently trying to determine the biological roles of Luman and ZF in both normal and virus-infected animals and how HCF might affect them.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Ping Yang for technical assistance and Dr Stephen Walker for critical reading of an earlier draft of this manuscript. This work was funded by an operating grant to V.M. from the Natural Sciences and Engineering Research Council of Canada. R.L. was a post-doctoral fellow of the Saskatchewan Health Services Utilization and Research Commission.
REFERENCES
- 1.O’Hare P. (1993) Semin. Virol., 4, 145–155. [Google Scholar]
- 2.Thompson C. and McKnight,S. (1992) Trends Genet., 8, 232–236. [Google Scholar]
- 3.Kristie T.M. and Roizman,B. (1987) Proc. Natl Acad. Sci. USA, 84, 71–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsden H.S., Campbell,M.E., Haarr,L., Frame,M.C., Parris,D.S., Murphy,M., Hope,R.G., Muller,M.T. and Preston,C.M. (1987) J. Virol., 61, 2428–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKnight J.L., Kristie,T.M. and Roizman,B. (1987) Proc. Natl Acad. Sci. USA, 84, 7061–7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Hare P. and Goding,C.R. (1988) Cell, 52, 435–445. [DOI] [PubMed] [Google Scholar]
- 7.Stern S., Tanaka,M. and Herr,W. (1989) Nature, 341, 624–630. [DOI] [PubMed] [Google Scholar]
- 8.Roizman B. and Sears,A.E. (1995) In Field,B.N., Knipe,D.M. and Howley,P.M. (eds), Field’s Virology, 3rd Edn. Lippincott-Raven, Philadelphia, PA, Vol. 2, pp. 2231–2296.
- 9.Katan M., Haigh,A., Verrijzer,C.P., van der Vliet,P.C. and O’Hare,P. (1990) Nucleic Acids Res., 18, 6871–6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kristie T.M., LeBowitz,J.H. and Sharp,P.A. (1989) EMBO J., 8, 4229–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stern S. and Herr,W. (1991) Genes Dev., 5, 2555–2566. [DOI] [PubMed] [Google Scholar]
- 12.Xiao P. and Capone,J.P. (1990) Mol. Cell. Biol., 10, 4974–4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhys C.M., Ciufo,D.M., O’Neill,E.A., Kelly,T.J. and Hayward,G.S. (1989) J. Virol., 63, 2798–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Preston C.M., Frame,M.C. and Campbell,M.E. (1988) Cell, 52, 425–434. [DOI] [PubMed] [Google Scholar]
- 15.Kristie T.M., Pomerantz,J.L., Twomey,T.C., Parent,S.A. and Sharp,P.A. (1995) J. Biol. Chem., 270, 4387–4394. [DOI] [PubMed] [Google Scholar]
- 16.Wilson A.C., LaMarco,K., Peterson,M.G. and Herr,W. (1993) Cell, 74, 115–125. [DOI] [PubMed] [Google Scholar]
- 17.Wilson A.C., Peterson,M.G. and Herr,W. (1995) Genes Dev., 9, 2445–2458. [DOI] [PubMed] [Google Scholar]
- 18.Wilson A.C., Cleary,M.A., Lai,J.S., LaMarco,K., Peterson,M.G. and Herr,W. (1993) Cold Spring Harbor Symp. Quant. Biol., 58, 167–178. [DOI] [PubMed] [Google Scholar]
- 19.LaBoissière S., Walker,S. and O’Hare,P. (1997) Mol. Cell. Biol., 17, 7108–7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristie T.M. (1997) J. Biol. Chem., 272, 26749–26755. [DOI] [PubMed] [Google Scholar]
- 21.Johnson K.M., Mahajan,S.S. and Wilson,A.C. (1999) J. Virol., 73, 3930–3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu Y., Hengartner,M.O. and Herr,W. (1999) Mol. Cell. Biol., 19, 909–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goto H., Motomura,S., Wilson,A.C., Freiman,R.N., Nakabeppu,Y., Fukushima,K., Fujishima,M., Herr,W. and Nishimoto,T. (1997) Genes Dev., 11, 726–737. [DOI] [PubMed] [Google Scholar]
- 24.Wilson A.C., Freiman,R.N., Goto,H., Nishimoto,T. and Herr,W. (1997) Mol. Cell. Biol., 17, 6139–6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu R., Yang,P., O’Hare,P. and Misra,V. (1997) Mol. Cell. Biol., 17, 5117–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu R. and Misra,V. (2000) J. Virol., 74, 934–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu R., Yang,P., Padmakumar,S. and Misra,V. (1998) J. Virol., 72, 6291–6297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Freiman R.N. and Herr,W. (1997) Genes Dev., 11, 3122–3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burbelo P.D., Gabriel,G.C., Kibbey,M.C., Yamada,Y., Kleinman,H.K. and Weeks,B.S. (1994) Gene, 139, 241–245. [DOI] [PubMed] [Google Scholar]
- 30.Abel T., Bhatt,R. and Maniatis,T. (1992) Genes Dev., 6, 466–480. [DOI] [PubMed] [Google Scholar]
- 31.Smolik S.M., Rose,R.E. and Goodman,R.H. (1992) Mol. Cell. Biol., 12, 4123–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haigh A., Greaves,R. and O’Hare,P. (1990) Nature, 344, 257–259. [DOI] [PubMed] [Google Scholar]
- 33.Hayes S. and O’Hare,P. (1993) J. Virol., 67, 852–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lai J.S. and Herr,W. (1997) Mol. Cell. Biol., 17, 3937–3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simmen K.A., Newell,A., Robinson,M., Mills,J.S., Canning,G., Handa,R., Parkes,K., Borkakoti,N. and Jupp,R. (1997) J. Virol., 71, 3886–3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu T.J., Monokian,G., Mark,D.F. and Wobbe,C.R. (1994) Mol. Cell. Biol., 14, 3484–3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kristie T.M., Vogel,J.L. and Sears,A.E. (1999) Proc. Natl Acad. Sci. USA, 96, 1229–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Misra V., Walter,S., Yang,P., Hayes,S. and O’Hare,P. (1996) Mol. Cell. Biol., 16, 4404–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misra V., Walker,S., Hayes,S. and O’Hare,P. (1995) J. Virol., 69, 5209–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Misra V., Bratanich,A.C., Carpenter,D. and O’Hare,P. (1994) J. Virol., 68, 4898–4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen C. and Okayama,H. (1988) Biotechniques, 6, 632–638. [PubMed] [Google Scholar]
- 42.Feinberg A.P. and Vogelstein,B. (1983) Anal. Biochem., 132, 6–13. [DOI] [PubMed] [Google Scholar]
- 43.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sadowski I., Ma,J., Triezenberg,S. and Ptashne,M. (1988) Nature, 335, 563–564. [DOI] [PubMed] [Google Scholar]
- 45.Wingender E., Dietze,P., Karas,H. and Knuppel,R. (1996) Nucleic Acids Res., 24, 238–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vinson C.R., Sigler,P.B. and McKnight,S.L. (1989) Science, 246, 911–916. [DOI] [PubMed] [Google Scholar]
- 47.O’Neil K.T., Hoess,R.H. and DeGrado,W.F. (1990) Science, 249, 774–778. [DOI] [PubMed] [Google Scholar]
- 48.Kerppola T.K. and Curran,T. (1991) Science, 254, 1210–1214. [DOI] [PubMed] [Google Scholar]
- 49.Ellenberger T.E., Brandl,C.J., Struhl,K. and Harrison,S.C. (1992) Cell, 71, 1223–1237. [DOI] [PubMed] [Google Scholar]
- 50.Neuberg M., Schuermann,M. and Muller,R. (1991) Oncogene, 6, 1325–1333. [PubMed] [Google Scholar]
- 51.Fornace A.J. Jr, Nebert,D.W., Hollander,M.C., Luethy,J.D., Papathanasiou,M., Fargnoli,J. and Holbrook,N.J. (1989) Mol. Cell. Biol., 9, 4196–4203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Park J.S., Luethy,J.D., Wang,M.G., Fargnoli,J., Fornace,A.J.,Jr, McBride,O.W. and Holbrook,N.J. (1992) Gene, 116, 259–267. [DOI] [PubMed] [Google Scholar]
- 53.Ron D. and Habener,J.F. (1992) Genes Dev., 6, 439–453. [DOI] [PubMed] [Google Scholar]
- 54.Ubeda M., Wang,X.Z., Zinszner,H., Wu,I., Habener,J.F. and Ron,D. (1996) Mol. Cell. Biol., 16, 1479–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gentz R., Rauscher,F.J.d., Abate,C. and Curran,T. (1989) Science, 243, 1695–1699. [DOI] [PubMed] [Google Scholar]
- 56.Neuberg M., Schuermann,M., Hunter,J.B. and Muller,R. (1989) Nature, 338, 589–590. [DOI] [PubMed] [Google Scholar]
- 57.Landschulz W.H., Johnson,P.F. and McKnight,S.L. (1989) Science, 243, 1681–1688. [DOI] [PubMed] [Google Scholar]
- 58.Turner R. and Tjian,R. (1989) Science, 243, 1689–1694. [DOI] [PubMed] [Google Scholar]
- 59.Kozak M. (1997) EMBO J., 16, 2482–2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Faber P.W., van Rooij,H.C., van der Korput,H.A., Baarends,W.M., Brinkmann,A.O., Grootegoed,J.A. and Trapman,J. (1991) J. Biol. Chem., 266, 10743–10749. [PubMed] [Google Scholar]
- 61.Cohen T., Nahari,D., Cerem,L.W., Neufeld,G. and Levi,B.Z. (1996) J. Biol. Chem., 271, 736–741. [DOI] [PubMed] [Google Scholar]
- 62.Chuang T.H., Hamilton,R.T. and Nilsen Hamilton,M. (1995) Gene, 162, 303–308. [DOI] [PubMed] [Google Scholar]
- 63.Horiuchi T., Macon,K.J., Kidd,V.J. and Volanakis,J.E. (1990) J. Biol. Chem., 265, 6521–6524. [PubMed] [Google Scholar]
- 64.Imataka H., Nakayama,K., Yasumoto,K., Mizuno,A., Fujii Kuriyama,Y. and Hayami,M. (1994) J. Biol. Chem., 269, 20668–20673. [PubMed] [Google Scholar]
- 65.Keaveney M., Parker,M.G. and Gannon,F. (1993) J. Mol. Endocrinol., 10, 143–152. [DOI] [PubMed] [Google Scholar]