Abstract
Advances in kidney genomics in the last twenty years has opened the door for more precise diagnosis of kidney disease and identification of new and specific therapeutic agents. Despite these advances, an imbalance exists between low resource and affluent regions of the world. Individuals of European ancestry from US, UK, and Iceland, account for 16% of the world population, but represent >80% of all genome-wide association studies (GWAS). South Asia, South-East Asia, Latin America, and Africa together account for 57% of the world population but less than 5% of GWAS. Implications of this difference include limitations in new variant discovery, inaccurate interpretation of the effect of genetic variants in non-European populations, and unequal access to genomic testing and novel therapies in resource-poor regions. It also further introduces ethical, legal, and social pitfalls, and may ultimately propagate global health inequities. Ongoing efforts to reduce the imbalance in low-resource regions include funding and capacity building, population-based genome sequencing, population-based genome registries, and genetic research networks. More funding, training, and capacity building for infrastructure and expertise is needed in resource-poor regions. Focusing on this will ensure multiple fold returns on investments in genomic research and technology.
Keywords: South Asia, Southeast Asia, Latin America and Africa, genomic resources imbalance, APOL1, CKD, genetics, genetic testing
Introduction
Chronic kidney disease (CKD) is an important cause of morbidity and mortality worldwide1–3, and it is estimated that 3.7% of the world population are living with CKD1. CKD is a leading cause of non-communicable disease-mortality and the global burden due to CKD is increasing1–3. The increased incidence and prevalence is attributed to incomplete understanding of etiology and pathogenesis of most causes of CKD. The downstream consequences of this incomplete understanding of disease mechanisms is the lack of effective and robust strategies for disease prevention and treatment. This effect is even more pronounced in countries with limited resources, where the factors above are compounded by environmental pollution, high prevalence of infectious diseases, and limited resources for the treatment and prevention of CKD1–3. Thus, morbidity and mortality associated with CKD is more noticeable in resource poor countries compared with high-income countries. In addition, in developed countries where more resources are available, there is a glaring disparity in the prevalence and severity of CKD in different populations. A good example is the United States, where people of African ancestry are disproportionately affected by CKD and tend to have worse outcomes because of years of enshrined racism, inequity, and unequal resource allocation in the health care system.
The completion of the Human Genome Project (HGP) and subsequent development of efficient analytical pipelines for genomic data has had a positive impact in our understanding of the molecular pathogenesis of chronic kidney diseases4. There is now a rapid turn-around in making genetic diagnoses as well as in the identification of both new causes of monogenic kidney diseases and risk loci for complex or polygenic kidney diseases5–7. For example, in the last twenty years more than 650 single gene causes of different CKD were identified5–7. In addition, multiple disease risk loci were identified for the more common kidney diseases with complex inheritance patterns such as diabetic kidney disease (DKD), IgA nephropathy (IgAN), focal and segmental glomerulosclerosis (FSGS) and other types of nephrotic syndrome7. The payoff of these advances includes but are not limited to: 1) more precise clinical diagnosis, 2) better understanding of disease mechanisms, 3) improved understanding of biologic basis of health disparities, 4) identification of novel and specific diagnostic tools, 5) identification of disease pathogenesis driven new therapies, and 6) development of a robust approach to precision therapeutics.
Despite these obvious benefits of the genomic revolution, there are disparities and inequalities in the populations targeted for the generation of genomics and other omics data. For example, individuals of European ancestry account for approximately 16% of the World population, yet represent 80–96% of all published genome wide association studies (GWAS)8–12. In the Genome Aggregation Database (gnomAD), data are available on next generation sequencing (NGS) from 138,632 individuals comprising of 15,496 whole genome sequencing (WGS) and 123,136 whole exome sequencing (WES).13 European populations make up 55% of all the NGS data despite the fact that they constitute only 16% of the total world population (Figure 1). In contrast, only 8.7% of African NGS are represented in the gnomAD database even though they account for 15.4% of the total world population13. In addition, review of the 81 monogenic causes of nephrotic syndrome reported in the literature showed that race and ethnicity of the participants was reported in only 66 of the discovery cohort (Supplementary Table 1). Of these reports, the initial discoveries were made predominantly in people of European, Middle Eastern, and Turkish ancestries (Supplementary Table 1). Similar patterns are observed in kidney disease related genome wide association studies.
Figure 1: Publicly available next generation sequencing by ancestries.
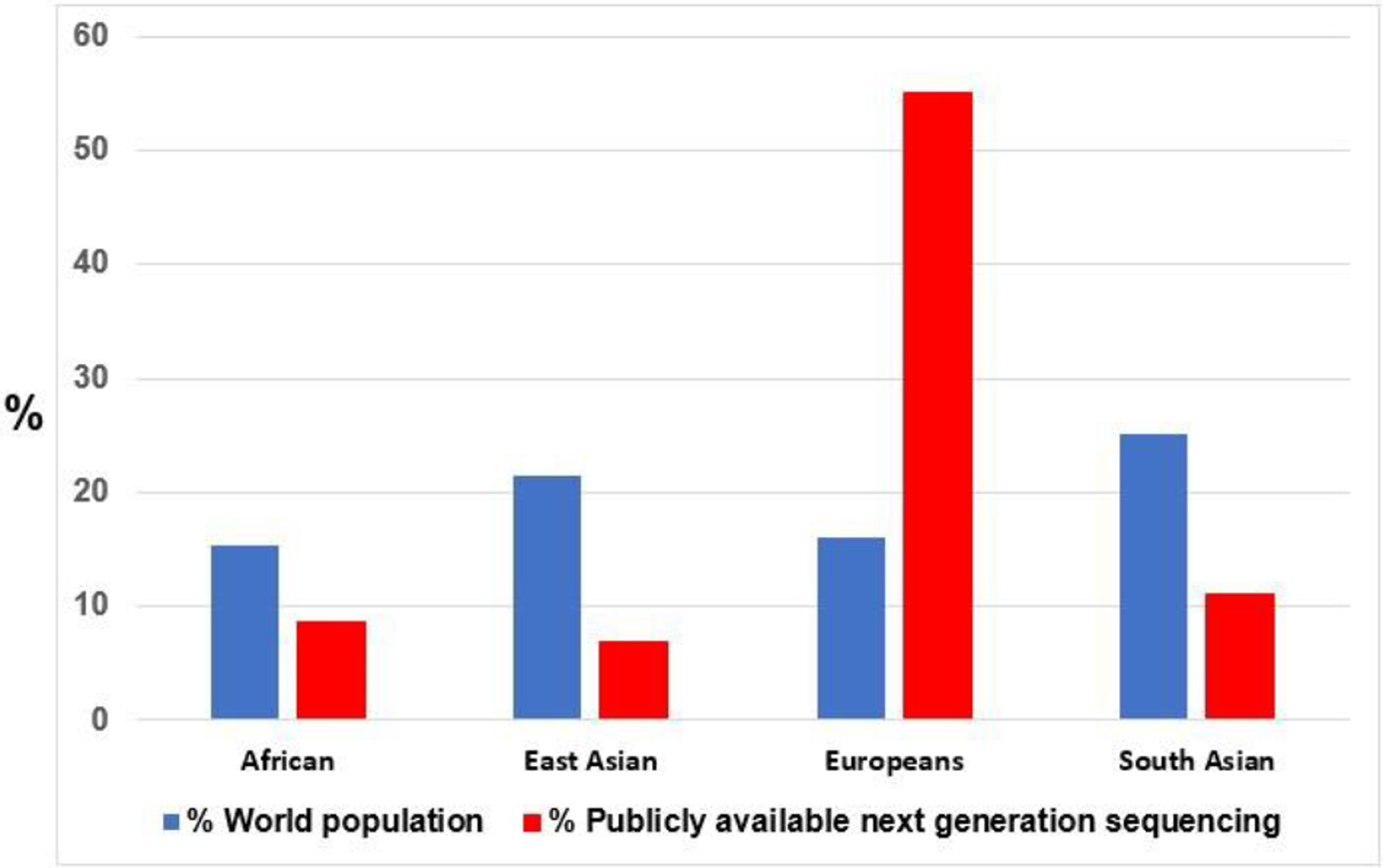
Publicly available next generation sequencing by African, East Asian, European and South Asian Ancestries (red bar). Blue bar represent the proportion of each ancestries as a percentage of the world population.
An appraisal of the National Human Genome Research Institute (NHGRI) catalogue of GWAS using the broad search term “kidney phenotype” yielded 65 studies (https://www.ebi.ac.uk/gwas/efotraits/EFO_0003086 Accessed 5/2/2022)14. The search showed disparity in race/population, geography, age, and phenotype studied (Supplementary Table 2). Based on the location of the senior authors, 37.5%, 37.5%, 25%, 0%, 0%, 0% and 0% of the studies were carried out in North America, Europe, East Asia, South Asia, South East Asia, Latin America, and Africa respectively (Figure 2). As expected, most of the participants in these studies are of European ancestry. The reasons for this imbalance includes lack of resources for conducting genomic studies in poor countries, bureaucratic bottleneck associated with collaborative genomic research between investigators in developed countries and resource limited countries, and the reluctance of investigators to study diverse populations because of challenges in dealing with genetic ancestral architecture. Irrespective of the reasons, the consequences of not studying diverse populations for genetic risk are numerous and include exaggeration of already existing inequities in access to high quality healthcare, incomplete understanding of genetic architecture of different diseases, missed opportunities to determine the effect of gene and environment interaction in phenotypic expression, and misuse of Eurocentric disease polygenic risk scores8–13. A good example is extrapolating the rare variant findings in European populations to a more diverse African population even though rare variants are generally population specific8–13. All these gaps have the potential to dilute the benefits of the genomic revolution especially in ethnic minorities and resource poor countries and further exacerbate the already existing disparity and inequity in health.
Figure 2: Proportion of continental population and kidney related GWAS studies in the NHGRI catalogue.
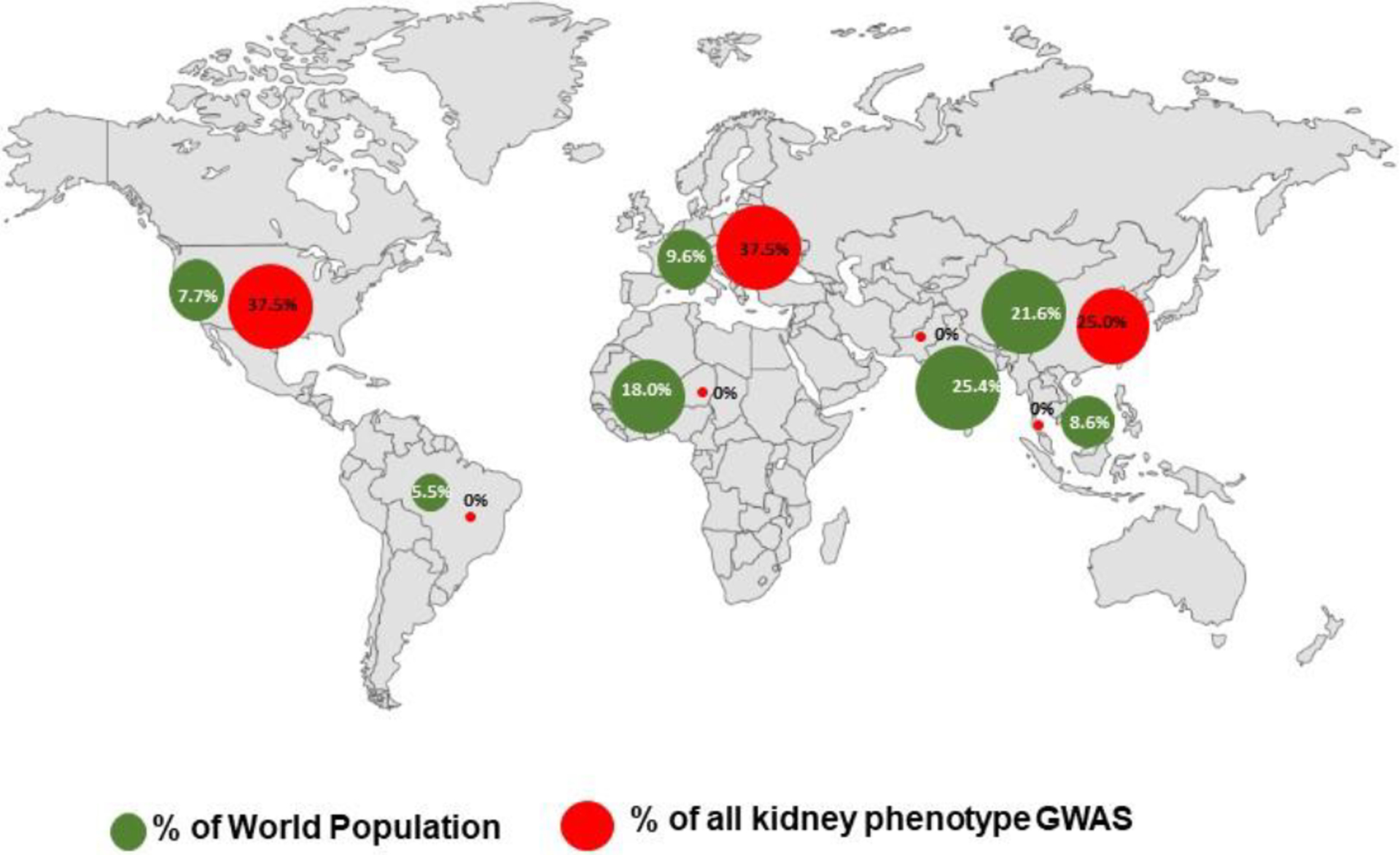
Figure 2 shows the proportion of the continental population and kidney related GWAS studies across Africa (0%), Southeast Asia (0%), Latin America (0%), South Asia (0%), Europe (37.5%), North America (37.5%), and East Asia (25%). Green dots represents percentage of World population, Red dots represent percentage of reported GWAS.
In this review, investigators with expertise in genomic medicine in resource poor regions will discuss the state of genomic data generation, examine the implications of genomic data imbalance, and provide recommendations for accelerating genomic research in low-income countries.
Genomic research in Africa
Africa has a population of about 1.4 billion people and 15.4% of the world’s population. It is made up of 54 countries and covers 20% of the Earth’s land area as the second largest continent. Chronic Kidney disease (CKD) is a public health crisis in Africa with an overall estimated prevalence of 15.8% (189 million people).15 Despite the increased risk and accelerated progression of CKD in individuals of recent African ancestry, only a minimal portion of individuals needing kidney replacement therapy in Africa can receive treatment. Delayed diagnosis, rudimentary facilities for the management of CKD, and exorbitant cost of treatment of CKD prevent the majority from accessing care. Similar to parts of Asia, Africans undergoing kidney replacement therapy have unsustainable out of pocket expenses and often sell assets to fund kidney transplantation or dialysis (Figure 3). The case in Figure 3 illustrates some of the major constraints associated with genetic testing and genomic research in Africans with CKD.
Figure 3:
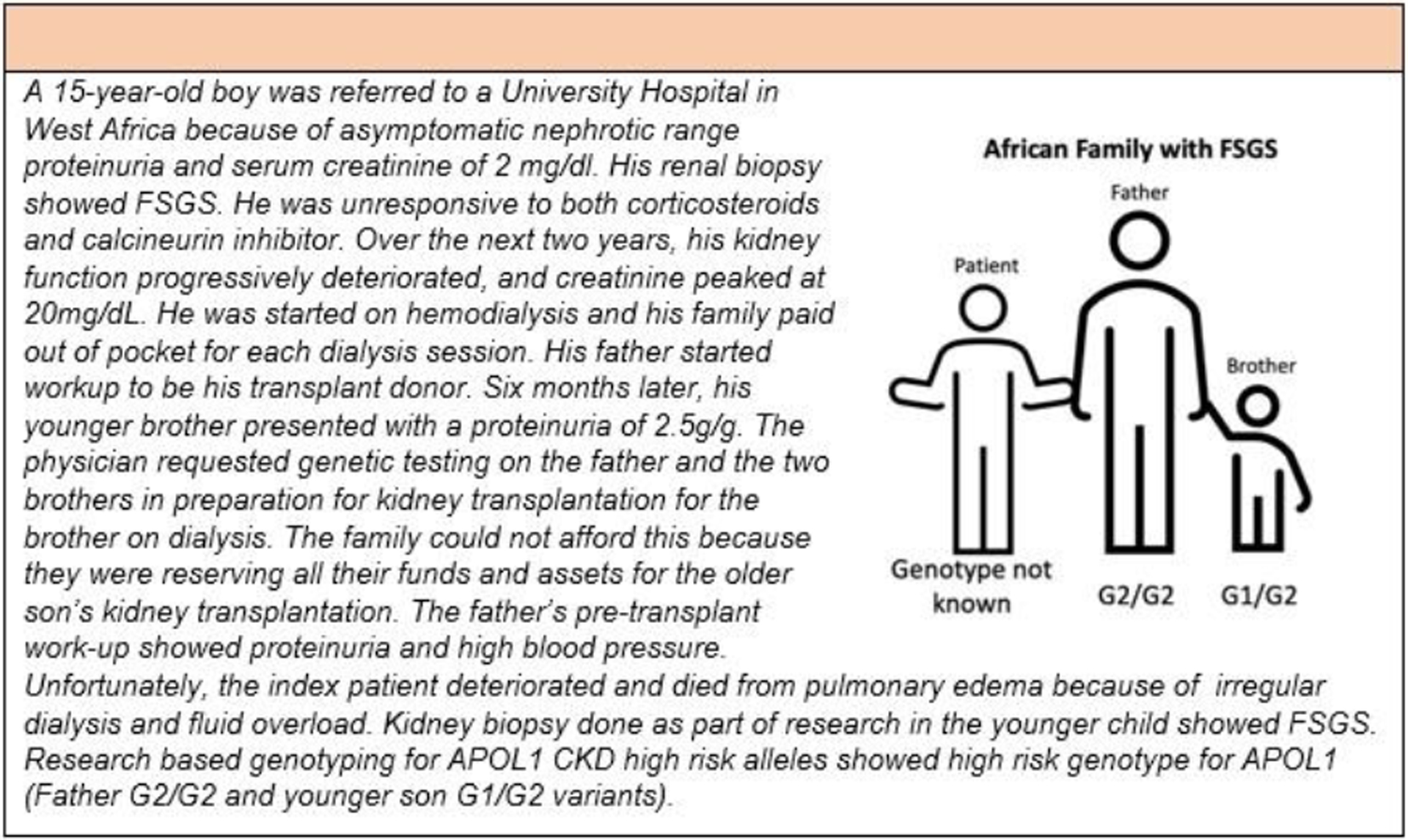
Case Study of genetic testing in an African with Focal Segmental Glomerulosclerosis.
To date, Africans are underrepresented in genome wide association studies (Figure 1). In our search of the English literature, we identified only one CKD GWAS study that was carried out in Africa (Table 1).16, 17 The pitfall of this imbalance is that the findings of >75% of all CKD GWAS studies cannot be extrapolated to Africans because of genetic diversity within Africa and difference in linkage disequilibrium between Africans and Europeans.18 This was clearly illustrated in the single GWAS of estimated glomerular filtration rate (eGFR) in a population in East Africa (n=3288) which reported a unique African specific variant at the glycine amidinotransferase (GATM), distinct from previously reported eGFR variants in European studies.16, 19–20 To the best of our knowledge, there are no published studies of large-scale next generation sequencing of CKD patients in Africa, some of the small scale studies using different genomic methods is shown in Supplementary Table 3.21 Most of the studies focused on replication of candidate genes rather than large scale discovery studies because of limited infrastructure and funding for research.21
Table 1:
Discovery studies of genetics of CKD from populations with ancestries in resource poor regions of the World
| Study | Population | Method | Phenotype | Loci/gene |
|---|---|---|---|---|
| Jarolim et al. Deletion in erythrocyte band 3 gene in malaria-resistant Southeast Asian ovalocytosis. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11022–6. | South East Asia Cases: 30 Controls: 30 |
Positional cloning | Distal RTA and ovalocytosis syndrome | SLC4A1 |
| Kopp et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008 Oct;40(10):1175–84. | African American Cases: 190 Controls: 222 |
MALD | FSGS ESKD |
MYH9 |
| Kao et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008 Oct;40(10):1185–92. | African American Cases: 1372 Controls: 806 |
MALD | Non-diabetic ESKD | MYH9 |
| Genovese et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010 Aug 13;329(5993):841–5. | African American Cases: 205 Controls: 180 |
Fine mapping of MYH9 locus | FSGS ESKD |
APOL1 |
| Woo et al. Parallel genotyping of 10,204 single nucleotide polymorphisms to screen for susceptible genes for IgA nephropathy. Ann Acad Med Singap. 2009 Oct;38(10):894–9. | South East Asian Cases: 28 Controls: 13 |
GWAS | IgA Nephropathy | GCM1 TNR TRDN |
| Nanayakkara et al. An integrative study of the genetic, social and environmental determinants of chronic kidney disease characterized by tubulointerstitial damages in the North Central Region of Sri Lanka. J Occup Health. 2014;56(1):28–38. | South Asian Cases: 311 Controls: 504 |
GWAS | CKD of unknown etiology | SLC13A3 |
| Gbadegesin et al. HLA-DQA1 and PLCG2 Are Candidate Risk Loci for Childhood-Onset Steroid-Sensitive Nephrotic Syndrome. J Am Soc Nephrol. 2015 Jul;26(7):1701–10 | South Asian Cases: 214 Controls: 149 |
Exome association study | Childhood steroid Sensitive nephrotic syndrome | HLA-DQA1 |
| Pungsrinont et al. Association between intelectin-1 variation and human kidney stone disease in northeastern Thai population. Urolithiasis. 2021 Dec;49(6):521–532 | South East Asian Cases: 105 Controls: 105 |
GWAS | Kidney stone | ITLN1 |
| Fatumo et al. Discovery and fine-mapping of kidney function loci in first genome-wide association study in Africans. Hum Mol Genet. 2021 Jul 28;30(16):1559–1568. | African Cases: 3,288 Controls: 8,224 |
GWAS | eGFR | GATM |
Table showing the major discovery studies of genetic studies from resource poor regions of the world. RTA: renal tubular acidosis, GWAS-genome wide association studies, APOL1-ApoliporotienL1, egfr-estimated glomerular filtration rare, FSGS- focal sclerosing glomerulosclerosis. IgA-Immunoglobulin
Historical and Evolutionary Evidence on the Importance of Advancing Genomic Research on the African Continent: APOL1 as a case study
APOL1 kidney risk variants arose in the African population less than 10,000 years ago. APOL1 variantsare absent in all but African ancestry genome and spread globally due to the slave trade from the16th and 19th centuries. The APOL1 gene risk variants for CKD are two risk haplotypes (G1 and G2) that arose in African populations presumably due to the protection from African sleeping sickness in individuals carrying these variants.22–23 Investigators proposed that the APOL1 risk variants arose after the “Out of Africa” expansion drove populations to other parts of the world, therefore APOL1 variants are predominantly observed in individuals of recent African ancestry.23–25 The risk variants are rare or non-existent in those without recent African ancestry (European ancestry) because the ancestors of modern Europeans migrated from Africa before the risk alleles originated. Circulating ApoL1 protects against Trypanosoma brucei and other Trypanosoma that infect animals.23 Humans are resistant to T.brucei because of the APOL1, because of this, a sub-species of the parasite emerged T. brucei rhodensiense which is resistant to APOL1 mediated lysis.26 However, ApoL1 protein modified by carriers with genetic risk of G1 and G2 are resistant to inactivation by T. brucei rhodensiense and having two copies of APOL1 risk variants (G1/G1, G1/G2, G2/G2) increases the risk of CKD in a recessive manner but the penetrance is incomplete.23, 26–27 In Africa having two copies of the APOL1 risk variant is protective against sleeping sickness in areas with high burden of sleeping sickness. However, in the US, and other areas that are non-endemic for trypanosomiasis the selective advantage is absent but the CKD consequences are present in African ancestry population.25 The prevalence of the high risk APOL1 genotype is about 13% in African Americans in the US, however the lifetime risk of CKD in patients with CKD is less than 25% suggesting the existence of additional CKD genetic risk factors, and possible genetic and environmental modifiers of the APOL1 high risk genotype.28–29 These findings demonstrate the importance of in-depth study of African genome and the urgent need for increase in pace of genomic research in Africa.
Genomic Research in Brazil and Latin America
Latin America is a geographic area where 8.4% of the world population live in 21 countries with different degrees of socioeconomic development. This region is also marked by social inequality which impacts negatively on health care systems and scientific research.30 The admixture population of Latin America started with Northeastern Asia people that arrived 15,000 to 18,000 years ago, followed by recent arrivals of Europeans and Africans 500 years ago, during and after the period of conquering and colonization. More recently, new wave of Europeans and populations from Asia represent the Latin America admixture population, finally resulting in the general Brazilian genetic ancestry of 68.8–79.5% of European, 10.3–29.3% of African, and 7.3–18.5% of Native American, these proportions vary between regions in Brazil.31–32
Population genomic data on diverse non-European and multi-ethnic admixture populations are scarce, and the inhabitants of Brazil, the largest country of Latin America, are not represented in the worldwide genomic data bank. In addition, there is no discovery GWAS or causal genes for kidney phenotypes from this region. Yet, specific data from this population is necessary for meaningful interpretation of existing genomic data sets. Another unique aspect of admixture in this region and most likely other regions worldwide is that phenotypes such as skin color and/or hair pigmentation does not correlate with genetic ancestry, therefore ethnicity determined by physical characteristics or self-declaration is usually misleading31–32. Therefore, GWAS and large-scale next generation sequencing in patients with CKD in Latin America populations is a unique opportunity to discover new loci related to ancestry-specific effects, and its interaction with known loci and the southern hemisphere’s unique environment. This specific information is also essential for drug discoveries and personalized use of existing therapeutic agents.7 Most of the genetic studies from this region are replication studies and results from these studies reinforce the need for concerted efforts for large-scale genomic studies (Supplementary Table 4).33–35
A recent example is the study of APOL1 high-risk genotype and steroid resistant nephrotic syndrome in Brazil35. In 318 pediatric patients with nephrotic syndrome followed in three tertiary pediatric nephrology centers in Brazil, APOL1 high risk genotype was found in 19 patients (6%), and it was associated with faster progression to end stage kidney disease in those with steroid resistant nephrotic syndrome and/or FSGS. Patients who harbor APOL1 high-risk alleles manifested nephrotic syndrome at older age, as observed in previous studies in African Americans, but unexpectedly, 9/19 (47%) of the cohort in Brazil self-declared as White.35 This study showed that the self-declaration of ethnicity and race in Brazil and possibly other countries in Latin America does not correlate with the ancestry loci driven risk, and corroborates the necessity for large scale genomic studies of CKD in this admixed population. There are also differences in the prevalence of APOL1 high-risk genotype among Latin American countries. For example, the prevalence of high-risk APOL1 genotype in Latinos living in the US varied between 0.1 to 1%, and it is associated with countries and regions of origin in Latin America.36
In addition, a study of pediatric patients with nephrotic syndrome who received kidney transplantation showed that only 8.4% of them have monogenic nephrotic syndrome (NS) compared with 10–30% reported from Europe suggesting that the genetic architecture of nephrotic syndrome in Brazil is likely to be different from published data from Europe and North America.37
Genomic studies in South Asia
South Asia, comprising eight nations, is home to one-fourth of the world’s population while occupying only 3.5% of its land area.38 Almost a quarter of the South Asian population falls below the international poverty line of $1.25/day.39 Morbidities related to infectious illnesses have declined with improvements in economy and levels of hygiene, leading to increased emphasis on non-communicable diseases. Hence, rare inherited diseases have emerged as a public health concern, particularly given high rates of consanguinity within certain populations in south Asia. Current expenditures on health per capita, however, remains low at 60.6 US dollars, and spending on research and development is merely 0.3–1.3% of the gross domestic product, making genetic testing virtually inaccessible.40–41 Within Asia, genetic ancestry is strongly correlated with both linguistic affiliations and geography similar to Africa.42 Consistent with social history, the extant populations inhabiting northern regions of south Asia show genetic similarities to the Indo-European populations of central Asia, while the population in the peninsular region and Sri Lanka appears to be derived from the early colonizers arriving from Africa along the southern exit route.43 Despite its genomic diversity, Asia is significantly under-represented in current genetic studies and reference genome databases. For example, in the catalogue of GWAS studies of different CKD, apart from the exome-wide association study in children with nephrotic syndrome and a study of CKD of undetermined origin (CKDu) from Sri Lanka, there is no other GWAS that focuses exclusively on South Asian population.44–45
As recent as 10 years ago, confirming genetic diagnosis for rare kidney diseases in most countries in South Asia required samples to be shipped to high-end research laboratories in developed nations. Costs of diagnostic testing in leading international laboratories are prohibitive at approximately 400–600 US dollars (2010) for inherited diseases such as primary hyperoxaluria and Alport disease. Occasionally, testing was facilitated free of cost by enterprising clinicians getting in touch with individual researchers, who performed testing either on humanitarian grounds, as personal favors, in lieu of participation in disease registries, and/or in the interest of furthering knowledge of disease pathogenesis. These options enabled etiological diagnosis in anecdotal cases, a few of which were published in the early 2000s (Supplementary Table 5).
Availability of next generation sequencing within the region, as part of funded research projects as well as through private laboratories, has catapulted exome sequencing into the center stage of diagnostic investigations. In most tertiary care centers across India, it is now possible to obtain clinical exome sequencing at a relatively affordable price ($200-$350) as part of work up for patients with suspected inherited nephropathies while whole exome sequencing costs 50% more. Thus, next-generation sequencing brought about a major turnaround in the access to genetic diagnosis.
On the research front, advances in sequencing techniques have clarified the genetic etiology of inherited kidney diseases in south Asia, which is often at variance with European studies. Research initiatives initiated locally to provide etiological diagnosis for specific syndromes through intramural grants at research institutes or government funding have enabled registries to explore the inherited basis of specific diseases or syndromes, including nephrotic syndrome, atypical hemolytic uremic syndrome, tubulopathies, cystic kidney diseases, and identification of founder mutations. For example, similar to Europeans, variations in NPHS1 are the leading cause of congenital nephrotic syndrome in India; however, certain mutations are very common and/or represent founder mutations, such that testing for just three variants in NPHS1 is likely to provide the diagnosis in half the cases of congenital nephrotic syndrome.46 Sri Lanka and Andhra Pradesh in India are described as hotspots of CKD of unknown origin (CKDu). While this ‘chronic interstitial nephritis in agricultural communities’ has been largely ascribed to exposure to agrochemicals/herbicides, one GWAS suggested a potential locus on chromosome 10 near the SLC13A3 gene for CKDu.45, 47 It should be noted that most of the genomic research activities in this region are limited to replication of findings from Europe and North America with few discovery studies in South Asians. A shift to well conducted discovery studies for monogenic diseases and identification of risk variants for different CKD will define better the genetic architecture of CKD In South Asia.
Genomic studies in South-East Asia
Southeast Asia consists of 11 countries, namely Brunei, East Timor, Indonesia, Singapore, Malaysia, Laos, Cambodia, Myanmar, Vietnam, Thailand, and Philippines. Covering about 3% of Earth’s total land area, it has a population of 669 million (~ 8.5% of the world’s population). CKD is a common non-communicable disease in the region with an estimated overall prevalence ranging from 8.6% in Indonesia and Vietnam to 33% in Singapore.48 The genetic underpinning of CKD is not well studied even though the Southeast Asian population has distinct genetic variations from the well-studied European ancestry population.49
Like the other regions, most of the studies on monogenic diseases or GWAS are replication studies of genes, variants, or disease risk loci found in other populations (Supplementary Table 6). However, these studies are important as they highlight the important differences. A good example is the recent study of genetics of glomerulopathies by the DragoN (Deciphering Diversities: Renal Asian Genetics Network) collaborative group.50 In this study, 183 probands with suspected genetic glomerulopathies were studied with gene panels for glomerulopathies and genetic diagnosis was made in 14% of the probands. In this study, >30% of monogenic glomerulopathies were due to variants in COL4A4 or COL4A5 genes.50 In addition, the prevalence of NPHS1 and NPHS2 mutations (common causes of monogenic NS in Europe) was low in this study.50–52 The diagnostic yield and the common genetic causes of glomerulopathies were different from what has been commonly reported from Europe and North America suggesting different genetic architecture of glomerulopathies.50 The reasons for this is unclear, but it may be due to substantial genetic admixture with the Malays and Chinese in South East Asia, and high infant mortality rates in early-onset nephrotic syndrome in low-resource settings. A monogenic disease discovery study reported from this region is the identification of mutations in SLC4A1 as a cause of distal renal tubular acidosis and ovalocytosis syndrome.53–54 This is a common monogenic disease found exclusively among populations in South East Asia, the red cell abnormality (ovalocytosis) is believed to have evolved as a protective mechanism against plasmodium falciparum malaria.54 These findings are clear illustrations of host genetic adaptation to malaria in Southeast Asia and further support the need for gene and environment interaction studies.
There are also few genetic association studies in patients with CKD from South East Asia.55–56 Despite the fact that IgA nephropathy is a common kidney disease in this region, most of the discovery studies were carried out in populations derived from East Asia and North America.57–59 The only discovery study in IgA nephropathy from this region was a Singapore study performed in 28 Chinese and 30 controls.55 While 42 genes were identified as possible susceptibility genes for IgA nephropathy, this study was grossly underpowered to imply any meaningful association or clinical significance. Another northeastern Thai study involving in 216 patients with nephrolithiasis and 216 controls identified the variant c.326T > A (rs2274907) in intelectin-1 (ITLN1) gene as a risk factor for nephrolithiasis. However, just like the IgA nephropathy report, this study was carried out in a small cohort and the significance is unclear.56 There is a clear need for large collaborative studies of different CKD phenotypes in order to gain insight into the genetic landscape of CKD, and potentially identify important novel and major disease risk variants, similar to how variants in APOL1 genes were discovered as major drivers of excess of CKD in people of African ancestry.23, 60–61
Implications of the imbalance in CKD genomic data in resource poor countries
The identification of variants in APOL1 high risk genotypes as the major genetic driver of CKD in people of African ancestry and subsequent discovery of compounds that can prevent or treat APOL1 associated nephropathy is a clear illustration of the need for equitable representation of different ancestral populations in genomic research.23 All the regions in this review are not well represented in publicly available genome database, yet they are likely to have distinct genetic variations different from currently available genomic data. In addition, most of the populations are not monolithic and are comprised of admixed populations that can enrich currently available genomic data and facilitate the discovery of novel diseases associated variations in the human genome. Some of the inherent problems associated with leaving these regions and other regions behind in the genomic revolution include:
Limitations to discovery of new variants in different populations: Genetic discovery relies on studies in the population of interest and needs to be of adequate power to detect an association, which in turn depends on effect size and the frequency of the variants. For example, variants that are common in individuals of African ancestry but rare in European populations cannot be discovered even with large scale studies in Europeans. This is clearly illustrated by the discovery of APOL1 CKD risk variants in Africans, and identification of causal genes for distal renal tubular acidosis and ovalocytosis syndrome in South East Asian population.23, 53 Furthermore, medications developed targeting molecular pathways may not be generalizable globally due to the regional differences in genetic architecture.
Danger of using polygenic risk scores derived from other populations: A polygenic risk score (PRS) estimates the genetic risk to a trait or disease by summing up the independent risk associated with a trait or disorder using the most informative GWAS data. There are limitations in the generalizability of polygenic risk scores derived from European ancestry to other populations. Polygenic risk scores derived from European samples predict individual risk better in individuals of European ancestry than non-Europeans. For example, the poor performance of PRS discovered in European population in individuals of African ancestry may arise from diverse population structure, differences in variant frequencies, and differences in linkage disequilibrium patterns between African and European populations.62–63 This was clearly illustrated by a recent study in which the investigators found that prediction accuracy of PRS for anthropometric measurement and blood panel in the UK Bio Bank using European derived statistics performed poorly in Africans.63 Indeed, if current polygenic risk scores continue to be used, health disparities may be exacerbated, especially in individuals from resource poor countries.
Unequal access to genomic testing and counseling services in resource poor regions: A downstream effect of the low-level of genomic research in these low resource regions is the paucity of genomic testing and counselling services. As seen in the case illustration from Africa (Figure 3), most people in these regions do not have ready access to clinical genetic testing, they do not have information about where they are available, and the tests are expensive and out of reach of many of them.
Ethical, legal, and social implications (ELSI) of the imbalance of genomic research: Several studies have demonstrated significant ethical and scientific challenges in using genetic data without adequate representation from other populations.9, 64 Return of results is crucial and with the paucity of genetic counsellors in low-resource regions of the world, return of genomic results to participants is challenging. We cannot achieve health equity until when access to genetic testing and counselling is fair and balanced across populations irrespective of economic status or geography. Cultural differences across populations should also be taken into consideration when sharing genomic results. Additionally, there is need for genomic knowledge to be globally applicable and free of bias during implementation.64
Strategies to improve genetic testing, research and clinical implications
To address this imbalance and to benefit maximally from the huge investments in genomic research so far, concerted efforts must be put in place to reduce the gap in genomic studies between resource endowed and resource poor regions. It is obvious that region-specific strategies are more likely to work. Two important points to highlight are that regardless of the source of the funding for genomic research, partnerships with local scientists, and community groups is crucial. Additionally, clarity on who has ownership of the data and data sovereignty needs to be well documented prior to the onset of the research. Below is a description of ongoing programs that are addressing this gap in different regions and proposed programs that may work across regions.
Africa
The Human Hereditary and Health in Africa Kidney Disease Research Network (H3A-KDRN) is a research funded by the National Institute of Health in the US, consisting of 16 academic medical centers in five African countries (Tanzania, Cameroon, Nigeria, Ghana, and South Africa).65 It is a part of the H3Africa Consortium funded by the NIH in the US and the UK Wellcome Trust.66 The goal of the H3Africa consortium is to identify the genetics underpinnings of both non-communicable and communicable diseases in Africa including CKD.67–68 The kidney disease cohort has recruited over 12,000 participants (cases with CKD and controls) and enrolled an additional 4000 participants with CKD in a prospective longitudinal cohort. The study has performed 750 renal biopsies and conducted GWAS in 3000 research participants.65 Adopting continent-wide efforts like the H3Africa framework in other low -resource settings may be crucial for playing catch-up in genomic research.
Latin America
The Brazilian public health system Sistema Unico de Saude (SUS) does not cover genetic testing and most Brazilians cannot afford out of pocket cost associated with genetic testing. In this context, The National Policy for the Comprehensive Care of Persons with Rare Diseases was launched in 2014 to regulate the integral care of people with genetic diseases and other rare conditions. With political and economic downturn, the availability of genetic testing through this program is still very limited.69 In 2021, the Agência Nacional de Saúde (ANS) in Brazil established the normative Resolution 465/21 that mandate genetic testing for hereditary cancers, neurologic, and syndromic or multisystemic diseases for private health sector that covers 25% of Brazilian population. This program will likely increase the number of Brazilian patients with access to genetic studies. Unfortunately, this program does not cover CKD, therefore advocacy efforts is ongoing to include CKD in this program.
At the population level, Brazil, the largest country in Latin America, is leading the effort to generate population based genomic data. The ABraOM (online Archive of Brazilian Mutations) is a Web-based public database that initially comprised exonic variants of 609 individuals, and more recently included genomic data of 1,171 unrelated individuals from the Saude, Bem-Estare Envelhecimento (SABE) study (Table 2). The SABE study is a study of health, well-being, and aging sponsored by the Pan American Health organization (PAHO), where seven Latin America countries including Brazil are participating. In Mexico, 300 non-related self-identified Mestizo individuals from seven states were sequenced for Ancestry analysis as part of the Mexican Genome Diversity Project (MGDP). The proportion of Native American ancestry was higher than in Brazil, varying from 36.2 to 66%, and was inversely proportional to the contribution of European ancestry, depending on the Mexico region. The African and East Asian ancestry contributions, in turn, are homogeneous among the seven states. Interestingly, a significant number of private haplotypes were found in the analyzed Mexican individuals. These initial findings pointed out the importance of increasing the number of Latino population sequencing to better understand their unique genomic variability and its potential implication in common complex disease in this specific population.70 In October 2021, the oriGen initiative was launched, this program is proposing to sequence the DNA of 100,000 Mexicans in order to create a genetic information database in collaboration with National Institute of Genomic Medicine, this study will be the largest study in Latin America when completed. (https://conecta.tec.mx/en/news/national/research/deciphering-mexican-genome-challenge-origen-project, accessed 06/09/2022).
Table 2:
Ongoing initiatives to bridge the genomic research gap between resources limited and resources endowed regions
| Initiative type | Name | Region/Country |
|---|---|---|
| International funding and capacity building | H3A-KDRN | Africa |
| Legislation | Normative Resolution 465/21 | Latin America/Brazil |
| Population based genome sequencing | ABraOM | Latin America/Brazil |
| oriGEN | Latin America/Mexico | |
| Genome Asia 100K Project | South Asia | |
| Registries and database | Genomics for Understanding Rare Diseases: India Alliance Network (GUaRDIAN) | South Asia |
| SAGE: Genetic variants from South Asian whole genomes and exomes | South Asia | |
| IGVdb, INDEX-db, IGDD, ClinIndb: DNA variations database of Indian population | South Asia/India | |
| Clinical genomic tools | GOMED | South Asia |
| Professional network | Brazilian Network for Pediatric Nephrotic Syndrome (REBRASNI) | Latin America/Brazil |
| DragoN network (Asia renal genetics network) | Southeast Asia | |
| NephQuest (Cohort study, bio repository and genetics of steroid resistant nephrotic syndrome) | South Asia | |
| Bio repository and registry for hemolytic uremic syndrome | South Asia | |
| Registry for renal tubular disorders | South Asia |
H3A-KDRN-Human Hereditary and Health in Kidney Disease Research Network AbraOM- Arquivo Brasileiro Online de Mutações (Online Archive of Brazilian Mutation) GoMED-Genomics and other omics tools for enabling medical decision
Research Institutes and professional societies are also collaborating to ensure universal availability of genetic testing, a recent initiative is that of the Brazilian Network for Pediatric Nephrotic Syndrome (REBRASNI) that was launched by three Brazilian public universities, the University of Sao Paulo, Federal University of Sao Paulo, and State University of Campinas. The goal of this program is to promote research in nephrotic syndrome for pediatric patients, generate resources for research from the government and private sector, and collaborate with other Investigators within and outside Brazil.71 A summary of some of these programs can be found in Table 2.
South Asia
A number of programs and initiatives sponsored by the government and large institutions are ongoing in this region to increase the pace of discoveries and address the imbalance (Table 2). The Genome Asia 100K Project was launched in 2016, the aim of this project is to sequence the genomes of 100,000 Asian individuals.72 Following the pilot phase of this project, 1739 genome sequences were published from samples across 64 Asian countries, the completion of the project is expected to profoundly change our understanding of the genomic diversity of south Asia.72 Establishment of registries and collaborative efforts among researchers in this region of the World is also addressing the problem associated with dearth of genomic data and understanding of the genetic basis of different CKD. A good example is the Genomics for Understanding Rare Diseases: India Alliance Network (GUaRDIAN), this program intends to provide genomic solutions for rare diseases in India.73 Resources available publicly through this program include the SAGE, a compendium of genetic variants integrating South Asian whole genomes and exomes,74 IGVdb, a DNA variation database of Indian population useful in understanding various aspects of human disease biology, including disease predisposition, adverse drug reaction and population migration.75 The INDEX-db is a database of genetic variations from the Indian population,76 the Indian Genetic Disease Database (IGDD),77 the India Allele Finder, a web-based annotation tool for identifying common alleles in next-generation sequencing data of Indian origin,78 and the ClinIndb, a catalog of the frequencies of rare and deleterious variants in the Indian population created using high-throughput genotyping and sequencing methods.79 Registries specific to kidney diseases were established and/or shared preliminary findings on inherited basis of disease, include those for childhood nephrotic syndrome (NephQuest), tubular diseases, hemolytic uremic syndrome and CAKUT (Supplementary Table 5).46
Concerted efforts to bridge the gaps in access to genetic diagnosis include partnerships between clinicians and researchers to set up a comprehensive rare disease care model in south India, and the creation of the Genomics and other omics technologies for Enabling Medical Decision (GOMED).80 This is a long-term outreach program of the Council of Scientific and Industrial Research (CSIR) Institute of Genomics and Integrative Biology, that will enable equitable access to all patients to state-of-the-art genetic testing.81
South-East Asia
Initiatives are also ongoing in this region Deciphering Diversities: Renal Asian Genetics Network (DragoN) is a South Asia and South East Asia initiative that will facilitate large-scale genomic studies in the region (Table 2). Precision Health Research, Singapore (PRECISE) will enable more genetic testing. Genetic tests remain unaffordable to the majority of the people in Singapore despite its high gross domestic product (GDP). In addition, the lack of genetic counsellors, low genetic literacy amongst nephrologists and the public, and the implications of a genetic diagnosis on existing insurance policies and future insurability are huge barriers in increasing the uptake of genetic testing in Singapore. These barriers are currently being addressed by a public clinical implementation project initiated by the Precision Health Research, Singapore (PRECISE) (https://www.npm.sg/).
Recommendations to bridge the genomic research gap
As genomic research continues to advance and new therapeutic targets emerge, it is crucial that infrastructure and funding for research expands in low-resource regions to ensure balance in genomic research globally. There is need for multiple stakeholder engagement and investment (government, research establishments, public and private sectors, international scientific organizations). Table 3 shows some recommendations to bridge the gap in genomic advances between low -resource and more affluent regions of the world.82
Table 3:
Recommendations to bridge the gaps in genomic research in resource poor regions82
| Increase representation of resource poor countries in genomic research | |
|---|---|
| Funders | Include resource poor countries in large funding announcements Focused studies engaging diverse populations in the regions Fund community engagement efforts and community based participatory research |
| Researchers | Shift in focus from replication to discovery studies Prioritize community engagement and community based participatory research Collaborations with researchers in the diaspora Include ethical, legal, and social implications in genomic research Focus on larger studies where complex traits can be investigated Design culturally sensitive studies |
| Regional government, public and private sectors | Increase funding for genomic research by home countries Provide infrastructure for genomic research and testing Consider public-private partnerships to advance genomic research Provide training opportunities Focus on large campaigns educating the population on genomic research Develop more tools to interrogate population specific genomes |
| Improve access to genomic services | |
| Funders | Requirement from funders to include diverse population in studies Funding and support for investigators in Western countries working in poor resource regions |
| Researchers | Ensure return of results and genetic counselling to participants Design implementation studies to integrate genomic research in clinical setting |
| Regional government, public and private sectors | Ensure that majority of the population have access to genomic services Build expertise and infrastructure for genetic counselling and return of results Fund training initiatives for specialists in genomic research Policies to include genetic testing in national and private health insurance schemes61 |
Conclusion
Despite several efforts across South Asia, South East Asia, Latin America and Africa to increase genomic research in CKD, there remains a paucity of studies focused on kidney disease from these regions. Without addressing genomic research globally, we cannot improve global health inequities in kidney disease. This is an important global issue, whichcalls for nephrologists to advocate on behalf of our patients. It is our social responsibility to address inequities. Funding agencies, federal governments, private partnerships, and researchers also have unique role to play in initiatives to correct the global imbalance in genomic research.
Supplementary Material
Financial Support:
TI is funded by the National Institute of Diabetes Digestive and Kidney Diseases (NIDDK) K23DK119542 and Boston Medical Center. KN is funded by the National Medical Research Council Singapore, Clinician Scientist Award, CSAINVmay009. RG is supported by National Institutes of Health/NIDDK grants 5R01DK098135 and 5R01DK094987, NIH/NICHD 1R21HD104176-01, and Duke Health Scholars award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest : The authors have no conflict of interest
References
- 1].Xie Y, Bowe B, Mokdad AH, Xian H, Yan Y, Li T, et al. Analysis of the Global Burden of Disease study highlights the global, regional, and national trends of chronic kidney disease epidemiology from 1990 to 2016. Kidney Int. 2018. Sep;94(3):567–581. [DOI] [PubMed] [Google Scholar]
- 2].GBD Chronic Kidney Disease Collaboration. Global, regional, and national burden of chronic kidney disease, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2020. Feb 29;395(10225):709–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3].Stanifer JW, Jing B, Tolan S, Helmke N, Mukerjee R, Naicker S, et al. The epidemiology of chronic kidney disease in sub-Saharan Africa: a systematic review and meta-analysis. Lancet Glob Health. 2014. Mar;2(3):e174–81. [DOI] [PubMed] [Google Scholar]
- 4].Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001. Feb 15;409(6822):860–921. doi: 10.1038/35057062. Erratum in: Nature 2001 Aug 2;412(6846):565. [DOI] [PubMed] [Google Scholar]
- 5].Hildebrandt F Genetic kidney diseases. Lancet. 2010. Apr 10;375(9722):1287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6].Groopman EE, Povysil G, Goldstein DB, Gharavi AG. Rare genetic causes of complex kidney and urological diseases. Nat Rev Nephrol. 2020. Nov;16(11):641–656. doi: 10.1038/s41581-020-0325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7].KDIGO Conference Participants. Genetics in chronic kidney disease: conclusions from a Kidney Disease: Improving Global Outcomes (KDIGO) Controversies Conference. Kidney Int. 2022. Apr 20:S0085–2538(22)00278–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8].Bustamante CD, Burchard EG, De la Vega FM. Genomics for the world. Nature. 2011. Jul 13;475(7355):163–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9].Martin AR, Kanai M, Kamatani Y, Okada Y, Neale BM, Daly MJ. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat Genet. 2019. Apr;51(4):584–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10].Borrell LN, Elhawary JR, Fuentes-Afflick E, Witonsky J, Bhakta N, Wu AHB, et al. Race and Genetic Ancestry in Medicine - A Time for Reckoning with Racism. N Engl J Med. 2021. Feb 4;384(5):474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11].Ruan Y, Lin YF, Feng YA, Chen CY, Lam M, Guo Z; Stanley Global Asia Initiatives, He L, Sawa A, Martin AR, Qin S, Huang H, Ge T. Improving polygenic prediction in ancestrally diverse populations. Nat Genet. 2022. May 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12].Fatumo S, Chikowore T, Choudhury A, Ayub M, Martin AR, Kuchenbaecker K. A roadmap to increase diversity in genomic studies. Nat Med. 2022. Feb;28(2):243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13].Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020. May;581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14]. [5/2/2022]. https://www.ebi.ac.uk/gwas/efotraits/EFO_0003086 Accessed.
- 15].Kaze AD, Ilori T, Jaar BG, Echouffo-Tcheugui JB: Burden of chronic kidney disease on the African continent: a systematic review and meta-analysis. BMC Nephrol, 19: 125, 2018. 10.1186/s12882-018-0930-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16].Fatumo S, Chikowore T, Kalyesubula R, Nsubuga RN, Asiki G, Nashiru O, et al. : Discovery and fine-mapping of kidney function loci in first genome-wide association study in Africans. Hum Mol Genet, 30: 1559–1568, 2021. 10.1093/hmg/ddab088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17].Mills MC, Rahal C: A scientometric review of genome-wide association studies. Communications biology, 2: 9, 2019. 10.1038/s42003-018-0261-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18].Mersha TB, Abebe T: Self-reported race/ethnicity in the age of genomic research: its potential impact on understanding health disparities. Human Genomics, 9, 2015 10.1186/s40246-014-0023-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19].Wuttke M, Li Y, Li M, Sieber KB, Feitosa MF, Gorski M, et al. : A catalog of genetic loci associated with kidney function from analyses of a million individuals. Nat Genet, 51: 957–972, 2019. 10.1038/s41588-019-0407-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20].Morris AP, Le TH, Wu H, Akbarov A, van der Most PJ, Hemani G, et al. : Trans-ethnic kidney function association study reveals putative causal genes and effects on kidney-specific disease aetiologies. Nat Commun, 10: 29, 2019. 10.1038/s41467-018-07867-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21].George C, Yako YY, Okpechi IG, Matsha TE, Kaze Folefack FJ, Kengne AP: An African perspective on the genetic risk of chronic kidney disease: a systematic review. BMC medical genetics, 19: 187, 2018. 10.1186/s12881-018-0702-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22].Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, et al. : Evolution of the primate trypanolytic factor APOL1. Proceedings of the National Academy of Sciences of the United States of America, 111: E2130–E2139, 2014. 10.1073/pnas.1400699111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23].Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, et al. : Association of Trypanolytic ApoL1 Variants with Kidney Disease in African Americans. Science, 329: 841–845, 2010. 10.1126/science.1193032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24].Freedman BI, Kopp JB, Langefeld CD, Genovese G, Friedman DJ, Nelson GW, et al. : The apolipoprotein L1 (APOL1) gene and nondiabetic nephropathy in African Americans. J Am Soc Nephrol, 21: 1422–1426, 2010. 10.1681/ASN.2010070730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25].Friedman DJ, Pollak MR: Genetics of kidney failure and the evolving story of APOL1. J Clin Invest, 121: 3367–3374, 2011. 10.1172/jci46263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26].Xong HV, Vanhamme L, Chamekh M, Chimfwembe CE, Van Den Abbeele J, Pays A, et al. : A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell, 95: 839–846, 1998. 10.1016/s0092-8674(00)81706-7 [DOI] [PubMed] [Google Scholar]
- 27].Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van den Abbeele J, et al. : Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature, 422: 83–87, 2003. 10.1038/nature01461 [DOI] [PubMed] [Google Scholar]
- 28].Kiberd BA, Clase CM: Cumulative risk for developing end-stage renal disease in the US population. J Am Soc Nephrol, 13: 1635–1644, 2002 [DOI] [PubMed] [Google Scholar]
- 29].Friedman DJ, Pollak MR: Apolipoprotein L1 and Kidney Disease in African Americans. Trends in Endocrinology and Metabolism, 27: 204–215, 2016. 10.1016/j.tem.2016.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30]. [5/20/2022]. https://www.statista.com/topics/3287/latin-america/#dossierKeyfigures Accessed.
- 31].Salzano FM, Sans M. Interethnic admixture and the evolution of Latin American populations. Genet Mol Biol. 2014. Mar;37(1 Suppl):151–70. doi: 10.1590/s1415-47572014000200003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32].Pena SD, Di Pietro G, Fuchshuber-Moraes M, Genro JP, Hutz MH, Kehdy Fde S, Kohlrausch F, et al. The genomic ancestry of individuals from different geographical regions of Brazil is more uniform than expected. PLoS One. 2011. Feb 16;6(2):e17063. doi: 10.1371/journal.pone.0017063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33].W10- Vajgel G, Lima SC, Santana DJS, Oliveira CBL, Costa DMN, Hicks PJ, et al. J Rheumatol. 2020. Aug 1;47(8):1209–1217. doi: 10.3899/jrheum.190684. Epub 2019 Nov 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34].Riella C, Siemens TA, Wang M, Campos RP, Moraes TP, Riella LV, et al. APOL1-Associated Kidney Disease in Brazil. Kidney Int Rep. 2019. Mar 20;4(7):923–929. doi: 10.1016/j.ekir.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35].Watanabe A, Guaragna MS, Belangero VMS, Casimiro FMS, Pesquero JB, de Santis Feltran L, et al. APOL1 in an ethnically diverse pediatric population with nephrotic syndrome: implications in focal segmental glomerulosclerosis and other diagnoses. Pediatr Nephrol. 2021. Aug;36(8):2327–2336. doi: 10.1007/s00467-021-04960-w. Epub 2021 Feb 14. [DOI] [PubMed] [Google Scholar]
- 36].Kramer HJ, Stilp AM, Laurie CC, Reiner AP, Lash J, Daviglus ML, Rosas SE, et al. African Ancestry-Specific Alleles and Kidney Disease Risk in Hispanics/Latinos. J Am Soc Nephrol. 2017. Mar;28(3):915–922. doi: 10.1681/ASN.2016030357. Epub 2016 Sep 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37].Feltran LS, Varela P, Silva ED, Veronez CL, Franco MC, Filho AP, et al. Targeted Next-Generation Sequencing in Brazilian Children With Nephrotic Syndrome Submitted to Renal Transplant. Transplantation. 2017. Dec;101(12):2905–2912. doi: 10.1097/TP.0000000000001846. [DOI] [PubMed] [Google Scholar]
- 38].Wikipedia contributors South Asia. Wikipedia, The Free Encyclopedia. May 25, 2022, 19:25 UTC. Available at: https://en.wikipedia.org/w/index.php?title=South_Asia&oldid=1089809764. Accessed May 29, 2022 [Google Scholar]
- 39].Poverty & Equity Data Portal”. Available at: povertydata.worldbank.org. Accessed May 29, 2022
- 40].World Health Organization Global Health Expenditure database. Available at apps.who.int/nha/database and Current health expenditure per capita (current US$) - South Asia | Data (worldbank.org); Accessed May 29, 2022
- 41].Wikipedia contributors. List of countries by research and development spending. Wikipedia, The Free Encyclopedia. May 31, 2022, 04:47 UTC. Available at: https://en.wikipedia.org/w/index.php?title=List_of_countries_by_research_and_development_spending&oldid=1090750504. Accessed June 1, 2022 [Google Scholar]
- 42].HUGO Pan-Asian SNP Consortium, Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, Calacal GC, et al. ; Indian Genome Variation Consortium. Mapping human genetic diversity in Asia. Science. 2009. Dec 11;326(5959):1541–5 [DOI] [PubMed] [Google Scholar]
- 43].Majumder PP. The human genetic history of South Asia. Curr Biol. 2010. Feb 23;20(4):R184–7 [DOI] [PubMed] [Google Scholar]
- 44].Gbadegesin RA, Adeyemo A, Webb NJ, Greenbaum LA, Abeyagunawardena A, Thalgahagoda S, et al. Mid-West Pediatric Nephrology Consortium. HLA-DQA1 and PLCG2 Are Candidate Risk Loci for Childhood-Onset Steroid-Sensitive Nephrotic Syndrome. J Am Soc Nephrol. 2015. Jul;26(7):1701–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45].Nanayakkara S, Senevirathna ST, Abeysekera T, Chandrajith R, Ratnatunga N, Gunarathne et al. An integrative study of the genetic, social and environmental determinants of chronic kidney disease characterized by tubulointerstitial damages in the North Central Region of Sri Lanka. J Occup Health. 2014;56(1):28–38. [DOI] [PubMed] [Google Scholar]
- 46].Joshi A, Sinha A, Sharma A, Shamim U, Uppilli B, Sharma P, et al. NephQuest Consortium. Next-generation sequencing for congenital nephrotic syndrome: A multi-center cross-sectional study from India. Indian Pediatr 2021; 58: 445–451 [PubMed] [Google Scholar]
- 47].Jayasumana C Chronic Interstitial Nephritis in Agricultural Communities (CINAC) in Sri Lanka. Semin Nephrol. 2019. May;39(3):278–283. [DOI] [PubMed] [Google Scholar]
- 48].Liyanage T, Toyama T, Hockham C, Ninomiya T, Perkovic V, Woodward M, Fukagawa M, et al. Prevalence of chronic kidney disease in Asia: a systematic review and analysis. BMJ Glob Health. 2022. Jan;7(1):e007525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49].HUGO Pan-Asian SNP Consortium, Abdulla MA, Ahmed I, Assawamakin A, Bhak J, Brahmachari SK, Calacal GC, et al. ; Indian Genome Variation Consortium. Mapping human genetic diversity in Asia. Science. 2009. Dec 11;326(5959):1541–5. [DOI] [PubMed] [Google Scholar]
- 50].Lu L, Yap YC, Nguyen DQ, Chan YH, Ng JL, Zhang YC, et al. Deciphering Diversities: Renal Asian Genetics Network (DRAGoN). Multicenter study on the genetics of glomerular diseases among southeast and south Asians: Deciphering Diversities - Renal Asian Genetics Network (DRAGoN). Clin Genet. 2022. May;101(5–6):541–551. [DOI] [PubMed] [Google Scholar]
- 51].Trautmann A, Lipska-Ziętkiewicz BS, Schaefer F. Exploring the Clinical and Genetic Spectrum of Steroid Resistant Nephrotic Syndrome: The PodoNet Registry. Front Pediatr. 2018. Jul 17;6:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52].Warejko JK, Tan W, Daga A, Schapiro D, Lawson JA, Shril S, et al. Whole Exome Sequencing of Patients with Steroid-Resistant Nephrotic Syndrome. Clin J Am Soc Nephrol. 2018. Jan 6;13(1):53–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53].Jarolim P, Palek J, Amato D, Hassan K, Sapak P, Nurse GT, et al. Deletion in erythrocyte band 3 gene in malaria-resistant Southeast Asian ovalocytosis. Proc Natl Acad Sci U S A. 1991. Dec 15;88(24):11022–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54].Wrong O, Bruce LJ, Unwin RJ, Toye AM, Tanner MJ. Band 3 mutations, distal renal tubular acidosis, and Southeast Asian ovalocytosis. Kidney Int. 2002. Jul;62(1):10–9. [DOI] [PubMed] [Google Scholar]
- 55].Woo KT, Lau YK, Wong KS, Zhao Y, Chan CM. Parallel genotyping of 10,204 single nucleotide polymorphisms to screen for susceptible genes for IgA nephropathy. Ann Acad Med Singap. 2009. Oct;38(10):894–9. [PubMed] [Google Scholar]
- 56].Pungsrinont T, Nettuwakul C, Sawasdee N, Rungroj N, Sritippayawan S, Yenchitsomanus PT. Association between intelectin-1 variation and human kidney stone disease in northeastern Thai population. Urolithiasis. 2021. Dec;49(6):521–532. [DOI] [PubMed] [Google Scholar]
- 57].Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet. 2014. Nov;46(11):1187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58].Li M, Foo JN, Wang JQ, Low HQ, Tang XQ, Toh KY, Yin PR, et al. Identification of new susceptibility loci for IgA nephropathy in Han Chinese. Nat Commun. 2015. Jun 1;6:7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59].Li M, Wang L, Shi DC, Foo JN, Zhong Z, Khor CC, et al. Genome-Wide Meta-Analysis Identifies Three Novel Susceptibility Loci and Reveals Ethnic Heterogeneity of Genetic Susceptibility for IgA Nephropathy. J Am Soc Nephrol. 2020. Dec;31(12):2949–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60].Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008. Oct;40(10):1175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61].Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008. Oct;40(10):1185–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62].Peterson RE, Kuchenbaecker K, Walters RK, Chen CY, Popejoy AB, Periyasamy S, et al. : Genome-wide Association Studies in Ancestrally Diverse Populations: Opportunities, Methods, Pitfalls, and Recommendations. Cell, 179: 589–603, 2019. 10.1016/j.cell.2019.08.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63].Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. : Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet, 100: 635–649, 2017. 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64].Jooma S, Hahn MJ, Hindorff LA, Bonham VL: Defining and Achieving Health Equity in Genomic Medicine. Ethn Dis, 29: 173–178, 2019. 10.18865/ed.29.S1.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65].Adu D, Ojo A: Overview of The Human Heredity and Health in Africa Kidney Disease Research Network (H3A-KDRN). Kidney360, 2: 129–133, 2021. 10.34067/kid.0002592020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66].Rotimi C, Abayomi A, Abimiku A, Adabayeri VM, Adebamowo C, Adebiyi E, et al. : Research capacity. Enabling the genomic revolution in Africa. Science, 344: 1346–1348, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67].Osafo C, Raji YR, Olanrewaju T, Mamven M, Arogundade F, Ajayi S, et al. : Genomic approaches to the burden of kidney disease in Sub-Saharan Africa: the Human Heredity and Health in Africa (H3Africa) Kidney Disease Research Network. Kid Intll, 90: 2–5, 2016. 10.1016/j.kint.2015.12.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68].Osafo C, Raji YR, Burke D, Tayo BO, Tiffin N, Moxey-Mims MM, et al. : Human Heredity and Health (H3) in Africa Kidney Disease Research Network: A Focus on Methods in Sub-Saharan Africa. Clin J Am Soc Nephrol 10: 2279–2287, 2015. 10.2215/CJN.11951214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69].Araújo Neto LA, Teixeira LA. New problems of a new health system: the creation of a national public policy of rare diseases care in Brazil (1990s-2010s). Salud Colect. 2020. Apr 5;16:e2210. English. doi: 10.18294/sc.2020.2210. [DOI] [PubMed] [Google Scholar]
- 70].Silva-Zolezzi I, Hidalgo-Miranda A, Estrada-Gil J, Fernandez-Lopez JC, Uribe-Figueroa L, Contreras A, et al. Analysis of genomic diversity in Mexican Mestizo populations to develop genomic medicine in Mexico. Proc Natl Acad Sci U S A. 2009. May 26;106(21):8611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71].Feltran LS, Watanabe A, Guaragna MS, Machado IC, Casimiro FMS, Neves PDMM, et al. Brazilian Network of Pediatric Nephrotic Syndrome (REBRASNI). Kidney Int Rep. 2019. Nov 21;5(3):358–362. doi: 10.1016/silvaj.ekir.2019.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72].GenomeAsia100K Consortium. The GenomeAsia 100K Project enables genetic discoveries across Asia. Nature 576, 106–111 (2019). 10.1038/s41586-019-1793-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73].GUaRDIAN Consortium, Sivasubbu S, Scaria V. Genomics of rare genetic diseases-experiences from India. Hum Genomics. 2019. Sep 25;14(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74].Hariprakash JM, Vellarikkal SK, Verma A, Ranawat AS, Jayarajan R, Ravi R, et al. SAGE: a comprehensive resource of genetic variants integrating South Asian whole genomes and exomes. Database (Oxford). 2018;2018:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75].Narang A, Das RR, Chaurasia A, Mukhopadhyay A, Mukerji M, Dash D. IGVBrowser--a genomic variation resource from diverse Indian populations. Database (Oxford). 2010;2010:baq022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76].Ahmed PH, Vidya V, More RP, Viswanath B, Jain S, Rao MS, et al. INDEX-db: The Indian Exome Reference Database (Phase I). J Comput Biol. 2019;26(3):225–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77].Pradhan S, Sengupta M, Dutta A, Bhattacharyya K, Bag SK, Dutta C, et al. Indian genetic disease database. Nucleic Acids Res. 2011;39(Database issue):D933–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78].Zhang JF, James F, Shukla A, Girisha KM, Paciorkowski AR. India Allele Finder: a web-based annotation tool for identifying common alleles in next-generation sequencing data of Indian origin. BMC Res Notes. 2017. Jun 27;10(1):233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79].Narang A, Uppilli B, Vivekanand A, Naushin S, Yadav A, Singhal K, et al. ; TRISUTRA Ayurgenomics Consortium, Prasher B, Sengupta S, Mukerji M, Faruq M. Frequency spectrum of rare and clinically relevant markers in multiethnic Indian populations (ClinIndb): A resource for genomic medicine in India. Hum Mutat. 2020. Nov;41(11):1833–1847 [DOI] [PubMed] [Google Scholar]
- 80].Nilakantam SR, Bhat D, Ravi MD, Dayananda CM, Basavanagowdappa H, Kumar KJ. Comprehensive Rare Disease Care model for screening and diagnosis of rare genetic diseases - an experience of private medical college and hospital, South India. Intractable Rare Dis Res. 2020. Aug;9(3):179–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81].Genomics and other omics technologies for Enabling Medical Decision. Available at Genomics and other omics tools for enabling Medical Decisions (GOMED) (igib.in). Accessed 1 June 2022 [Google Scholar]
- 82].Jooma S, Hahn MJ, Hindorff LA, Bonham VL. Defining and Achieving Health Equity in Genomic Medicine. Ethn Dis. 2019. Feb 21;29(Suppl 1):173–178. doi: 10.18865/ed.29.S1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


