Abstract
The biological properties of spherical nucleic acids (SNAs) are largely independent of nanoparticle core identity but significantly affected by oligonucleotide surface density. Additionally, the payload-to-carrier (i.e., DNA-to-nanoparticle) mass ratio of SNAs is inversely proportional to core size. While SNAs with many core types and sizes have been developed, all in vivo analyses of SNA behavior have been limited to cores >10 nm in diameter. However, “ultrasmall” nanoparticle constructs (<10 nm diameter) can exhibit increased payload-to-carrier ratios, reduced liver accumulation, renal clearance, and enhanced tumor infiltration. Therefore, we hypothesized that SNAs with ultrasmall cores exhibit SNA-like properties, but with in vivo behavior akin to traditional ultrasmall nanoparticles. To investigate, we compared the behavior of SNAs with 1.4-nm Au102 nanocluster cores (AuNC-SNAs) and SNAs with 10-nm gold nanoparticle cores (AuNP-SNAs). Significantly, AuNC-SNAs possess SNA-like properties (e.g., high cellular uptake, low cytotoxicity) but show distinct in vivo behavior. When intravenously injected in mice, AuNC-SNAs display prolonged blood circulation, lower liver accumulation, and higher tumor accumulation than AuNP-SNAs. Thus, SNA-like properties persist at the sub-10-nm length scale and oligonucleotide arrangement and surface density are responsible for the biological properties of SNAs. This work has implications for the design of new nanocarriers for therapeutic applications.
Keywords: Nanoparticle Biodistribution, Spherical Nucleic Acids, Ultrasmall Nanoparticles, Blood Circulation, Tumor Accumulation
Graphical Abstract
The biological behavior of spherical nucleic acids with ultrasmall 1.4-nm gold nanocluster cores was compared to conventionally sized spherical nucleic acids with 10-nm gold nanoparticle cores. Ultrasmall spherical nucleic acids have a higher payload-to-carrier ratio and more efficient cellular uptake than larger spherical nucleic acids, as well as prolonged blood circulation, lower liver accumulation, and higher tumor accumulation in vivo.
Introduction
Spherical nucleic acids (SNAs) are nanostructures consisting of densely packed, highly oriented oligonucleotides, typically arranged around a spherical nanoparticle core.1,2 This three-dimensional architecture imbues the constituent oligonucleotides with properties distinct from their linear analogues, including rapid cellular uptake in high quantity without the need for ancillary transfection agents,3,4 decreased nuclease susceptibility,5 and low cytotoxicity.6,7 Since their discovery, SNAs have made a significant impact in biomedicine,8,9 with established utility in disease detection,10,11 cancer immunotherapy,12–16 and gene regulation.17–20
Early work on the SNA platform was based on constructs synthesized from 13-nm gold nanoparticle cores (Au-SNAs).3,10,6 Because the oligonucleotides are covalently conjugated to the gold nanoparticle core, it is easy to both control the loading density and to calculate the drug-to-carrier (i.e., DNA-to-gold) ratios with high precision, allowing for a comprehensive understanding of the structure-function relationships of SNAs in biological settings.21 High surface loading of oligonucleotides increases the efficiency of SNA cellular uptake, a key consideration when designing systems for intracellular delivery.22 Furthermore, it has been shown that increasing the oligonucleotide surface density on the SNA not only protects the oligonucleotides from degradation but also reduces overall cellular toxicity without negatively affecting activity.23–25
Size is a key factor that influences nanoparticle retention within the body, as well as blood circulation half-life and tissue distribution following systemic administration.26 Generally, nanoparticles with hydrodynamic diameters greater than 10 nm exceed the renal filtration threshold and accumulate in the liver and spleen, where they are retained for prolonged periods.27–30 Conversely, nanoparticles that are smaller than 10 nm (“ultrasmall” nanoparticles) exhibit different behavior from larger nanoparticles.31 Ultrasmall nanoparticles are cleared through the kidneys – an elimination route that is faster than the hepatic accumulation and slow metabolism of larger nanoparticles.32–34,31 While this leads to lower accumulation in off-target organs (i.e., decreased off-target toxicity), it also decreases accumulation in the target organ (e.g., tumor).35 However, the fraction of nanoparticles that do reach the target site penetrate the tissue more efficiently.36 Moreover, cell entry by ultrasmall nanoparticles is highly efficient.37–39 Thus, for many nanoparticle systems, there is a tradeoff at the ultrasmall scale and it is important to consider if the benefits of faster elimination and lower off-target accumulation outweigh the drawbacks of decreased accumulation at the target site.
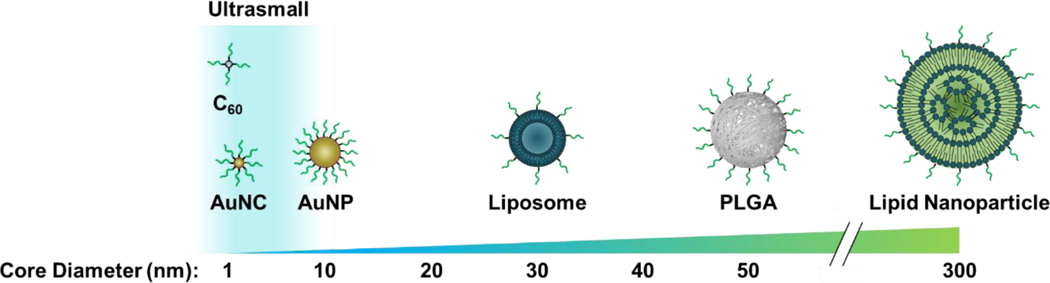
While SNAs have been synthesized from a variety of nanoparticle core types and sizes (Figure 1),1,40–44 ranging from molecularly pure 1-nm buckminsterfullerene C60 scaffolds41 to 300-nm lipid nanoparticles,44 all in vivo analyses have been limited to SNAs with cores exceeding 10 nm in diameter.18,43–46 Thus, little is known about the in vivo behavior of SNAs synthesized from ultrasmall nanoparticle cores. SNAs in this size regime, for example molecular SNAs synthesized from T8 polyoctahedral silsesquioxane (POSS) cores and buckminsterfullerene C60 cores, are capable of independently entering cells in vitro like their larger SNA counterparts; however, only molecular SNAs synthesized from the C60 core exhibit gene silencing activity, defining the lower limit of size and oligonucleotide loading density required for SNA-like behavior.41 While these molecular SNAs enter cells, their in vivo behavior has not been characterized.
Figure 1. SNA core materials and sizes.
Cores smaller than 10 nm exist within the ultrasmall size regime. Image is not drawn to scale.
To investigate the effect of an ultrasmall core on SNA in vivo properties, we synthesized SNAs from molecularly well-defined 1.4-nm gold nanocluster cores (AuNC-SNAs) and compared their biological behavior to conventional SNAs with 10-nm gold nanoparticle cores (AuNP-SNAs) (Figure S1). The gold nanocluster was chosen as the SNA core material for these analyses for several reasons. First, having both cores made of gold isolates our study to effects due to differences in size, rather than material. Second, while POSS and C60 have a finite and fixed number of attachment sites determined by their chemical structures, the loading density of oligonucleotides onto gold nanoclusters is only limited by the electrostatic and steric interactions from attaching thiolated oligonucleotides to the particle surface, thus overcoming a limitation of molecular SNAs and allowing for a higher surface loading density. Finally, gold can be sensitively traced in cells and tissues. We hypothesized that AuNC-SNAs would possess the attractive properties of traditional Au-SNAs (e.g., high cellular uptake, low cytotoxicity) and simultaneously mitigate their biological limitations (e.g., high liver accumulation, inability to be renally cleared, reduced targeting efficiency). Simultaneously, we hypothesized that moving to an ultrasmall size regime would maximize both the oligonucleotide surface density and drug-to-carrier ratio – two important parameters to consider when designing nanoscale therapeutics.22–25
Results and Discussion
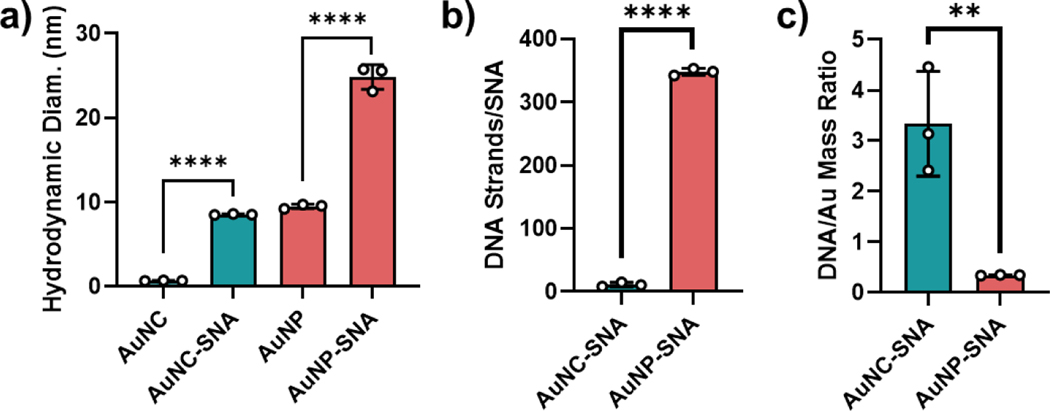
To synthesize AuNC-SNAs, Au102(p-mercaptobenzoic acid (p-MBA))44 nanoclusters were first prepared according to established protocols to serve as 1.4-nm AuNC cores (Figure S2).47 These materials were further functionalized with a 20-thymine thiolated DNA sequence to ensure that the effects observed in downstream studies were due to the overall construct, rather than the oligonucleotide sequence. Successful functionalization of the DNA to AuNCs to afford AuNC-SNAs was confirmed by the increase in hydrodynamic diameter measured by dynamic light scattering (DLS) (Figures 2a and S3). For comparison, larger AuNP-SNAs were synthesized from 10-nm gold nanoparticle cores according to literature protocols and their functionalization was confirmed by DLS.48,21
Figure 2. Characterization of AuNC-SNAs and AuNP-SNAs.
(a) DLS of bare AuNCs, AuNC-SNAs, bare AuNPs, and AuNP-SNAs. Error bars represent the standard deviation of 3 measurements. (b) DNA loading of AuNC-SNAs and AuNP-SNAs, reported on a per-particle basis. Error bars represent the standard deviation of 3 separately prepared batches of SNAs. (c) DNA-to-gold mass ratio of AuNC-SNAs and AuNP-SNAs. Error bars represent the standard deviation of 3 separately prepared batches of SNAs. Statistical analysis was performed using an unpaired t-test, where “**” represents a P value of <0.01 and “****” represents a P value of <0.0001.
The extent of DNA loading on both AuNC-SNAs and AuNP-SNAs was then quantified using OliGreen fluorescence assays. As expected, the absolute number of DNA strands per particle was higher for AuNP-SNAs (348 strands/particle) than AuNC-SNAs (11 strands/particle) due to the significantly larger surface area of the AuNP core (Figure 2b). However, the surface loading density of DNA on AuNC-SNAs was significantly higher as compared to AuNP-SNAs, along with an 11.5-fold higher DNA-to-gold mass ratio (3.21 vs. 0.28, Figure 2c). Thus, AuNC-SNAs have a higher payload-to-carrier ratio than larger AuNP-SNAs, such that each AuNC-SNA can deliver more DNA payload relative to gold material.
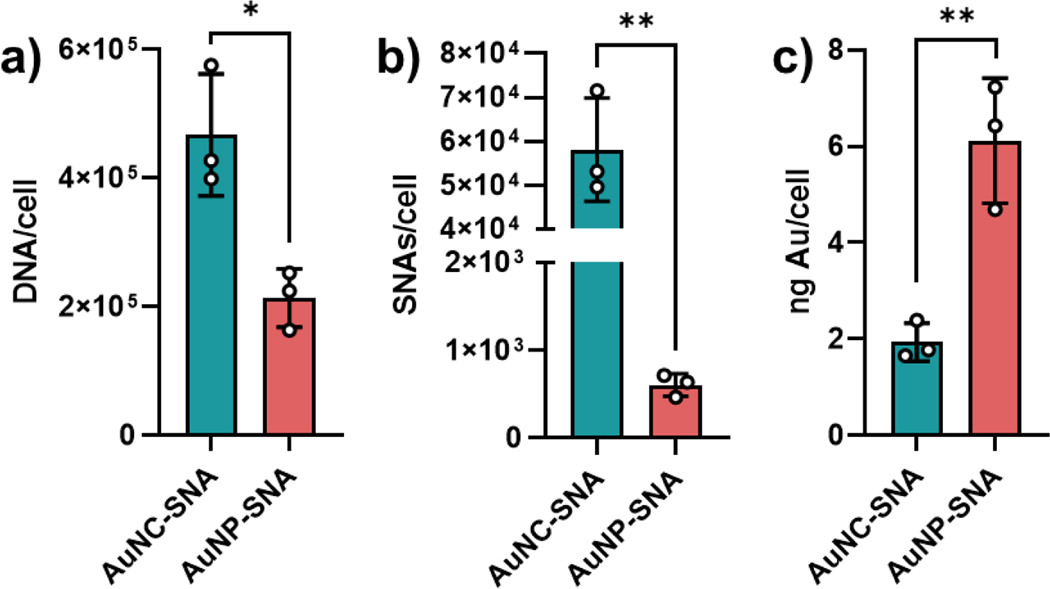
To determine the cellular uptake of AuNC-SNAs by cells, SK-OV-3 ovarian cancer cells were incubated with AuNC-SNAs at a concentration of 250 nM by DNA and compared to the uptake of AuNP-SNAs at the same DNA concentration. Cellular uptake was measured as a function of gold concentration using inductively coupled plasma mass spectrometry (ICP-MS), which was used to calculate both nanoparticle and DNA concentration. After 30 minutes of incubation, we observed that AuNC-SNAs delivered significantly more DNA (Figure 3a) and particles (Figure 3b) but significantly less gold (Figure 3c) per cell than their larger AuNP-SNA counterparts. Thus, AuNC-SNAs are more efficient at delivering DNA into cancer cells than AuNP-SNAs.
Figure 3. Cellular uptake of AuNC-SNAs and AuNP-SNAs.
(a) Cellular uptake of SNAs by DNA amount. (b) Cellular uptake of SNAs by SNA amount. (c) Cellular uptake of SNAs by gold amount. Error bars represent the standard deviation of 3 biological replicates. Statistical analysis was performed using an unpaired t-test, where “*” represents a P value of <0.05 and “**” represents a P value of <0.01.
It should be noted that two batches of cells were treated with AuNC-SNAs and AuNP-SNAs (at 250 nM with respect to DNA), respectively. Since the AuNC-SNAs have a lower DNA loading than AuNP-SNAs, a higher concentration of AuNC-SNAs (32 nM by SNA) than AuNP-SNAs (0.71 nM by SNA) is needed to achieve the 250 nM total DNA concentration. Thus, it may appear that the greater delivery of SNA particles by the AuNC-SNAs could be attributed to the fact that more AuNC-SNAs were introduced to the cells than AuNP-SNAs (45-fold difference). Cellular uptake of SNAs is directly proportional to treatment concentration up to a saturation point (at least 20 nM),22 so one would expect that a 45-fold increase in SNA concentration would result in an approximately 45-fold (if below the saturation concentration) or lower (if above the saturation concentration) increase in cellular uptake due to differences in SNA treatment concentration alone. However, AuNC-SNAs entered cells at a 96-fold greater particle number than AuNP-SNAs. Thus, the difference in SNA particle internalization cannot solely be attributed to the higher number of AuNC-SNAs treated to the cells compared to AuNP-SNAs, but also to more efficient uptake of AuNC-SNAs. AuNC-SNAs have a greater surface loading density than AuNP-SNAs, which has previously been observed to increase cellular uptake.22 In addition to superior cellular uptake, AuNC-SNAs matched the in vitro biological safety profile of AuNP-SNAs and exhibited similarly low cytotoxicity at treatment-relevant concentrations (Figure S4).17,18,20
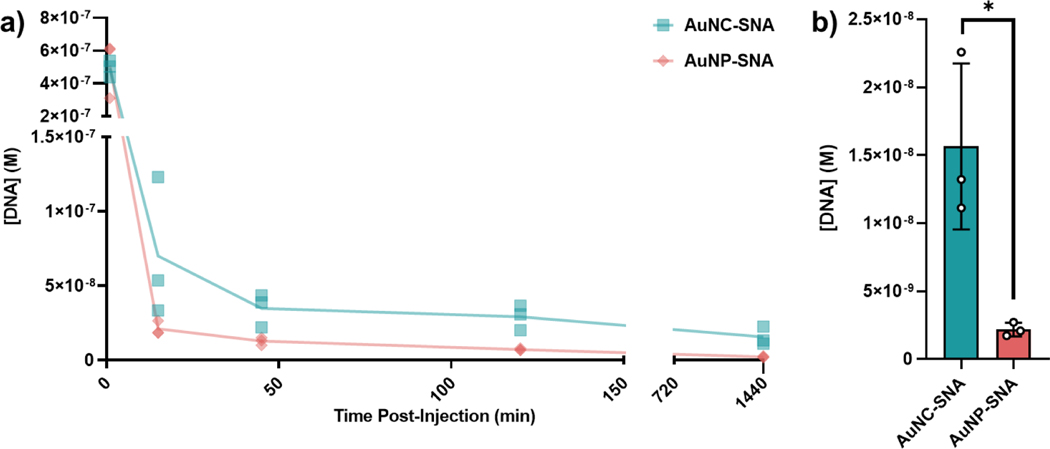
Next, we investigated the in vivo biological behavior of AuNC-SNAs as compared to AuNP-SNAs in mice bearing orthotopic 4T1 triple negative breast cancer (TNBC) tumors. Animals were intravenously (IV) administered a 3 nmol DNA equivalent of either AuNC-SNAs or AuNP-SNAs. At set timepoints, gold concentration in the blood was measured by ICP-MS and used to calculate the DNA blood concentration as a function of time (Figures 4a and S5). Remarkably, AuNC-SNAs remained in circulation for significantly prolonged periods relative to AuNP-SNAs, with detectable gold concentrations in the blood at 24 hours post-injection (Figure 4b). We hypothesized that a driver of the prolonged circulation of AuNC-SNAs was reduced liver sequestration. Additionally, prolonged circulation of AuNC-SNAs could lead to enhanced tumor accumulation. To investigate, we analyzed the biodistribution of both SNAs in these organs.
Figure 4. Blood circulation of AuNC-SNAs and AuNP-SNAs.
a) DNA blood concentration as a function of time post-IV injection when delivered as AuNC-SNAs (blue) or AuNP-SNAs (peach). Dots represent biological replicates. b) Final DNA blood concentration at 24 h post-injection. Error bars represent the standard deviation of 3 biological replicates. Statistical analysis was performed using an unpaired t-test, where “*” represents a P value of <0.05.
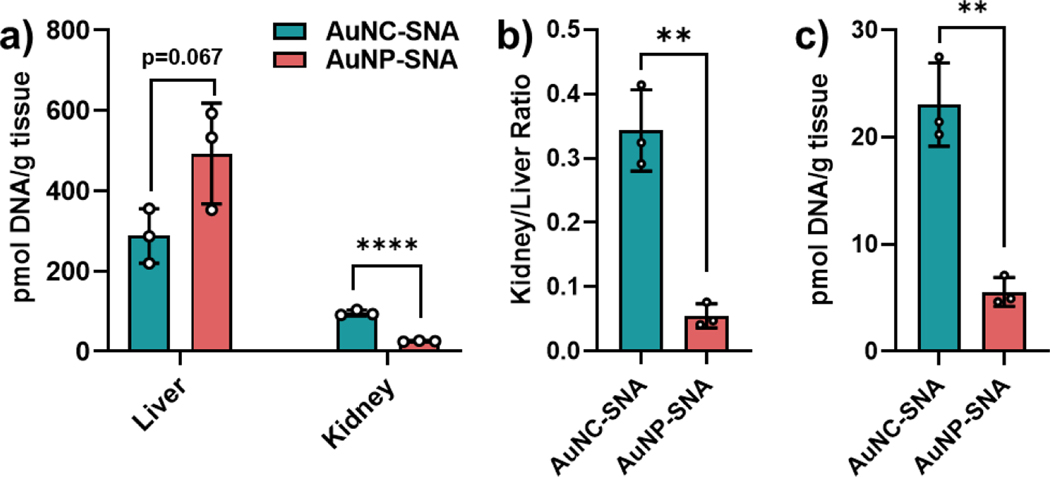
To assess the biodistribution of SNAs with either core size, mice bearing orthotopic 4T1 tumors were administered a 3 nmol DNA equivalent of either AuNC-SNAs or AuNP-SNAs via tail vein IV. At 24 h post-injection, animals were sacrificed and perfused, and tissues were collected and processed for ICP-MS analysis (Figure S6). As expected, both nanoparticle constructs accumulated in the liver and kidney, in agreement with conventional nanoparticle clearance pathways (Figure 5a).27–29 However, the AuNC-SNAs showed significantly lower liver accumulation coupled with increased kidney accumulation as compared to their larger AuNP-SNA counterparts, resulting in a higher kidney-to-liver ratio and revealing a distinct skew towards clearance by the kidney (Figure 5b). Moreover, AuNC-SNAs delivered a much higher payload to the site of interest (i.e., DNA to tumor) than their larger counterparts (Figure 5c). This is in contrast to what has been reported for other ultrasmall nanoparticles,35,38,49,50 and the greater tumor accumulation of AuNC-SNAs is likely due to a combination of prolonged blood circulation, greater tumor infiltration,36 and more efficient uptake into cancer cells. Finally, not only do AuNC-SNAs deliver more DNA payload to the tumor than their larger counterparts, but they also deliver significantly less gold to off-target organs (Figure S7), owing to their high DNA-to-gold ratios.
Figure 5. SNA accumulation in organs.
(a) DNA accumulation from AuNC-SNAs and AuNP-SNAs in the liver and kidney. (b) Kidney/liver ratio of DNA accumulation from each SNA. (c) Tumor accumulation of DNA from AuNC-SNAs and AuNP-SNAs. Error bars represent the standard deviation of 3 biological replicates. Statistical analysis was performed using an unpaired t-test, where “**” represents a P value of <0.01 and “****” represents a P value of <0.0001.
Conclusion
Through these studies, we found that SNA-like biological properties persist down to the sub-10-nanometer length scale in vivo, further indicating that the unique properties of SNAs arise from the highly oriented arrangement and surface density of the DNA, rather than the identity of the nanoscale core. Furthermore, AuNC-SNAs possess the favorable characteristics of ultrasmall nanoparticles without their pharmacokinetic drawbacks, providing important insight into the design of maximally effective nanoscale constructs for therapeutic applications. AuNC-SNAs were able to independently enter cells without the use of transfection agents, functioning similarly to larger 10-nm AuNP-SNAs. However, reducing the size of the core led to a significantly increased drug-to-carrier ratio in AuNC-SNAs and the delivery of higher quantities of DNA into cells than AuNP-SNAs. Moreover, AuNC-SNAs exhibited the same advantages of other ultrasmall nanoparticles (lower off-target organ accumulation, renal clearance) while subverting their disadvantages – instead, exhibiting longer circulation in blood and greater tumor accumulation. Thus, SNAs in the ultrasmall size regime have unusual properties that can be leveraged to develop next-generation nanoparticle therapeutics. These findings on the behavior of AuNC-SNAs can be used to inform the design of new types of SNAs and other nanostructures in the ultrasmall regime, such as DNA dendrons.51
Experimental Section
Oligonucleotide Synthesis.
DNA oligonucleotides (sequence: 5’ TTT-TTT-TTT-TTT-TTT-TTT-TT-(Spacer 18)2-SH 3’) were synthesized on a MerMade 12 system (LGC Biosearch Technologies) using 2’-O-triisopropylsilyloxymethyl-protected phosphoramidites (Glen Research). DNA oligonucleotides consisted of DNA base cyanoethyl phosphoramidites (Glen Research), two spacer-18 (18-O-dimethyoxytritylhexaethyleneglycol,1-[(2-cyanoethyl)-(N,N-diisopropyl)] phosphoramidites (Glen Research), and a thiol modification added by using 1-O-dimethoxytrityl-propyl-disulfide,1’-succinyl-lcaa-controlled pore glass beads. After synthesis, DNA oligonucleotides were deprotected following the manufacturer’s protocol (Glen Research). Deprotected DNA oligonucleotides were purified by high-performance liquid chromatography on a C18 column using 0.1 M triethylammonium acetate and acetonitrile as the solvents. The 5’-DMT group was removed from purified DNA oligonucleotides by treating with 20% acetic acid at room temperature for 1 h and extracting 3 times with ethyl acetate. The oligonucleotide solution was lyophilized and suspended in DNase/RNase-free water. Synthesis of the DNA oligonucleotide was confirmed using matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry.
Au102(p-MBA)44 Nanocluster Synthesis.
Au102(p-MBA)44 nanoclusters (gold nanoclusters) were synthesized following the method developed by Levi-Kalisman et al.52 Aqueous solutions of 28 mM HAuCl4·3H2O, 95 mM p-mercaptobenzoic acid, 300 mM NaOH, methanol, and water were combined into a final mixture of 11.89 mL 3 mM HAuCl4, 9 mM p-MBA, 47% (v/v) methanol and mixed in a round bottom flask for 1 h at room temperature. Next, 500 μL 150 mM NaBH4 was added, and the reaction was allowed to continue for 12 h. The product was precipitated by adding 2 M ammonium acetate to 80 mM in the mixture and centrifuging at 6000 rpm for 10 min. The precipitate was air-dried overnight, then re-dissolved in 200 μL water. To purify by fractional precipitation, methanol and 2 M ammonium acetate were added to the solution to a concentration of 0.12 M ammonium acetate, 60% methanol, and the solution was centrifugated at 6000 rpm for 10 min. The supernatant was collected and transferred to a centrifuge tube. Methanol and 2M ammonium acetate were added to a concentration of 0.12 M ammonium acetate, 80% methanol, and the solution was centrifuged at 6000 rpm for 10 min. The precipitate was lyophilized until dry, then re-dissolved in water.
AuNC-SNA Synthesis.
Gold nanoclusters were functionalized with thiolated DNA oligonucleotides via salt-aging. First, 62 nmol DNA was incubated in 100 mM dithiothereitol (DTT, pH 8) for 1 h to reduce the dithiol and purified using Nap-5 exclusion columns (Cytivia). The purified DNA was added to 2 mL 2.6 μM gold nanocluster suspension and incubated with shaking for 30 min at room temperature. Next, 0.05% Tween-20 was added to the solution, which was then vortexed thoroughly. The salt concentration was gradually increased to 0.5 M by adding NaCl, sonicating, and vortexing every 15 min while shaking, followed by an overnight incubation. Unattached oligonucleotides were removed by washing with 1% Tween-20 2 times, then 1× Dulbecco’s phosphate-buffered saline (DPBS) 3 times in Amicon Ultra 30K molecular weight cutoff spin filters (MilliporeSigma). SNAs were stored at 4 °C for up to 3 months.
AuNP-SNA Synthesis.
AuNPs (Ted Pella) 10 nm in diameter were functionalized with thiolated DNA oligonucleotides via salt-aging. First, 62 nmol DNA was incubated in 100 mM dithiothereitol (DTT, pH 8) for 1 h to reduce the disulfides and purified using Nap-5 exclusion columns (Cytivia). The purified DNA was added to 10 mL 7.7 nM 10-nm AuNP suspension and incubated with shaking for 30 min at room temperature. Then, 0.05% Tween-20 was added to the solution, which was then vortexed thoroughly. The salt concentration was gradually increased to 0.5 M by adding NaCl, sonicating, and vortexing every 15 min while shaking, followed by an overnight incubation. Unattached oligonucleotides were removed by washing with 1% Tween-20 2 times, then 1X DPBS 3 times in Amicon Ultra 50K molecular weight cutoff spin filters (MilliporeSigma). SNAs were stored at 4 °C for up to 3 months.
SNA Characterization.
SNA size was measured by diluting in 1 × DPBS and performing DLS using a Zetasizer (Malvern). SNA concentration was determined by measuring the absorbance spectra of the SNAs using a Cary-5000 spectrophotometer (Agilent). AuNC-SNA concentration was calculated using the absorbance at 510 nm and an extinction coefficient of 1.75 × 105 M−1 cm−1. AuNP-SNA concentration was calculated using the absorbance at 515 nm and an extinction coefficient of 1.01 × 108 M−1 cm−1. To measure DNA loading on SNAs, 2.5 nM SNAs were incubated with 20 nM potassium cyanide (KCN) at 50 °C until the gold dissolved. Next, 25 μL of this solution was added in triplicate to a 96-well plate. A standard curve of DNA in a solution identical to the SNA solution was prepared. Then, 75 μL water was added to all wells. For SNAs with unlabeled DNA, 100 μL Quanti-iT OliGreen solution (Invitrogen) was added to all wells, and samples were analyzed by measuring OliGreen fluorescence (λex = 480 nm) with a BioTek Cytation 5 imaging reader. The fluorescence of the SNA solution was compared to the standard curve to calculate the DNA concentration. DNA loading of SNAs was then calculated by taking the ratio of DNA concentration to gold core concentration. Surface loading density of DNA on SNAs was calculated by dividing the moles of DNA on each SNA by the surface area of the gold core. DNA-to-gold mass ratio was calculated by dividing the mass of DNA on each SNA by the mass of the gold core.
Cellular Uptake of SNAs.
SK-OV-3 cells were treated with 250 nM DNA equivalent of AuNC-SNAs and AuNP-SNAs in Opti-MEM for 30 min and 5 h. At the end of their treatment time, the cells were washed with 1× DPBS, 1:10 heparin solution in 1× DPBS, then 1× DPBS to remove any SNAs that remained outside the cells or dead cells. The cells were then trypsinized using 150 μL TrypLE Express (Thermo Fisher). 10 μL cells were stained with 10 μL Trypan Blue (Thermo Fisher) and their concentration was measured using an Invitrogen Countess II automated cell counter. Next, 120 μL cells were dissolved in 50 mL 2% HCl, 2% HNO3. The gold concentration in this solution was measured by performing ICP-MS. ICP-MS was performed on a computer-controlled (QTEGRA software) Thermo iCapQ ICP-MS (Thermo Fisher Scientific, Waltham, MA, USA) operating in STD mode and equipped with a ESI SC-2DX PrepFAST autosampler (Omaha, NE, USA). Internal standard was added inline using the prepFAST system and consisted of 1 ng/mL of a mixed element solution containing Bi, In, 6Li, Sc, Tb, Y (IV-ICPMS-71D from Inorganic Ventures). Online dilution was also carried out by the prepFAST system and used to generate a calibration curve consisting of 2, 4, 20, 100, and 200 ppb Au. Each sample was acquired using 1 survey run (10 sweeps) and 3 main (peak jumping) runs (40 sweeps). The isotopes selected for analysis were 197Au (for SNA quantification) and 89Y, 115In, 159Tb, and 209Bi (chosen as internal standards for data interpolation and machine stability). Instrument performance was optimized daily through autotuning followed by verification via a performance report (passing manufacturer specifications). The amount of DNA taken up into each cell was calculated by converting gold atom amount in each sample to gold nanoparticle mole amount (which is equivalent to SNA mole amount), converting moles of SNA to number of SNA particles, multiplying by the number of DNA strands per SNA, and dividing by the number of cells. The amount of SNAs taken up into each cell was calculated by converting gold atom amount in each sample to gold nanoparticle mole amount (which is equivalent to SNA mole amount), converting moles of SNA to number of SNA particles, and dividing by the number of cells. The amount of gold taken up into each cell was calculated by converting gold atom amount in each sample to gold mass and dividing by the number of cells.
Cytotoxicity of SNAs.
In 96-well cell culture dishes with 5,000 SK-OV-3 cells per well, SNAs in Opti-MEM were incubated with SK-OV-3 cells in triplicate for 48 h. The wells were washed with 1× DPBS 3 times. 50 μL 1× DPBS and 50 μL CellTiter-Glo 2.0 reagent (Promega) were added to the wells and fluorescence was measured using a BioTek Cytation 5 imaging reader. Cell viability was normalized to cells treated with Opti-MEM only.
In Vivo Analyses.
All animal procedures were approved by the Northwestern University IACUC. Female mice (Balb/C) were inoculated with 1 × 106 4T1 cells in the right inguinal mammary fat pad via subcutaneous injection. Both blood circulation and biodistribution experiments began at day 7 post-inoculation, when tumors were palpable (n = 3 per group). Animals were intravenously administered either AuNC-SNAs or AuNP-SNAs (3 nmol with respect to DNA) as a single 100 μL bolus. As a negative control, an additional set of animals were administered saline. To calculate the SNA concentration in blood, blood samples (n = 3 per time point) was collected in a heparinized tube via retro-orbital blood draw and stored on ice before digestion and analysis by ICP-MS (vide infra). For the biodistribution analysis at 24 h post-IV injection, animals (n = 3 per group) were humanely euthanized by cardiac perfusion while anesthetized. Tissues (tumor, liver, spleen, kidney, heart, lung) were harvested post-cardiac perfusion with 1X DPBS and stored on ice. For ICP-MS analysis, organs were weighed (wet mass) and lyophilized to facilitate digestion (vide infra).
SNA Content in Blood and Organs by ICP-MS.
Collected blood and organs were digested with 50% HCl, 50% HNO3 at 37 °C for three days. The solution was diluted to 2% HCl, 2% HNO3, and the gold concentration was measured by ICP-MS. ICP-MS was performed on a computer-controlled (QTEGRA software) Thermo iCapQ ICP-MS (Thermo Fisher Scientific, Waltham, MA, USA) operating in STD mode and equipped with a ESI SC-2DX PrepFAST autosampler (Omaha, NE, USA). Internal standard was added inline using the prepFAST system and consisted of 1 ng/mL of a mixed element solution containing Bi, In, 6Li, Sc, Tb, Y (IV-ICPMS-71D from Inorganic Ventures). Online dilution was also carried out by the prepFAST system and used to generate a calibration curve consisting of 2, 4, 20, 100, and 200 ppb Au. Each sample was acquired using 1 survey run (10 sweeps) and 3 main (peak jumping) runs (40 sweeps). The isotopes selected for analysis were 197Au (for SNA quantification) and 89Y, 115In, 159Tb, and 209Bi (chosen as internal standards for data interpolation and machine stability). Instrument performance was optimized daily through autotuning followed by verification via a performance report (passing manufacturer specifications). The amount of DNA in each sample was calculated by converting gold atom amount in each sample to gold nanoparticle mole amount (which is equivalent to SNA mole amount), converting moles of SNA to number of SNA particles, multiplying by the number of DNA strands per SNA, and dividing by the volume of blood or mass of tissue. The amount of gold in each sample was calculated by converting gold atom amount in each sample to gold mass and dividing by the volume of blood or mass of tissue.
Statistical Analyses.
All statistical analyses were performed using GraphPad Prism. For bar graphs, bars represent mean and error bars represent standard deviation. For line graphs, lines represent mean. For all experiments, n = 3 (replicates are described in figure captions). All P values were calculated using α = 0.05. For comparisons between two groups, means were compared using an unpaired, two-tailed t-test. P values were multiplicity adjusted to account for multiple comparisons. Cytotoxicity results were fit with a 3-parameter logistic curve using a least-squares fit.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under Awards R01CA208783, R01CA257926, and R01CA275430. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. C.E.C. was supported by a Postdoctoral Fellowship, PF-20–046-01 - LIB, from the American Cancer Society, as well as the Eden and Steven Romick Postdoctoral Fellowship through the American Committee for the Weizmann Institute of Science. M.K.V. thanks Jungsoo Park for providing the PLGA graphic in Figure 1. MALDI-TOF analysis of DNA was performed at the Integrated Molecular Structure Education and Research Center (IMSERC) mass spectrometry (MS) facility at Northwestern University, which has received support from the Soft and Hybrid Nanotechnology Experimental (SHyNE) Resource (NSF ECCS-2025633), the State of Illinois, and the International Institute for Nanotechnology (IIN). Gold analysis for cellular uptake, blood circulation, and biodistribution quantification was performed at the Northwestern University Quantitative Bioelement Imaging Center (QBIC) generously supported by the National Institutes of Health under grant S10OD020118.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest Disclosure: The authors declare no conflict of interest.
Contributor Information
Cassandra E. Callmann, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, Evanston, IL 60208, USA
Matthew K. Vasher, Department of Biomedical Engineering, International Institute for Nanotechnology, Northwestern University, Evanston, IL 60208, USA
Anindita Das, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, Evanston, IL 60208, USA.
Caroline D. Kusmierz, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, Evanston, IL 60208, USA
Chad A. Mirkin, Department of Chemistry, International Institute for Nanotechnology, Northwestern University, Evanston, IL 60208, USA Department of Biomedical Engineering, International Institute for Nanotechnology, Northwestern University, Evanston, IL 60208, USA.
Data Availability Statement:
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- (1).Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ A DNA-Based Method for Rationally Assembling Nanoparticles into Macroscopic Materials. Nature 1996, 382 (6592), 607. 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- (2).Cutler JI; Auyeung E; Mirkin CA Spherical Nucleic Acids. J. Am. Chem. Soc 2012, 134 (3), 1376–1391. 10.1021/ja209351u. [DOI] [PubMed] [Google Scholar]
- (3).Rosi NL; Giljohann DA; Thaxton CS; Lytton-Jean AKR; Han MS; Mirkin CA Oligonucleotide-Modified Gold Nanoparticles for Intracellular Gene Regulation. Science 2006, 312 (5776), 1027–1030. 10.1126/science.1125559. [DOI] [PubMed] [Google Scholar]
- (4).Choi CHJ; Hao L; Narayan SP; Auyeung E; Mirkin CA Mechanism for the Endocytosis of Spherical Nucleic Acid Nanoparticle Conjugates. Proc Natl Acad Sci U S A 2013, 110 (19), 7625–7630. 10.1073/pnas.1305804110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Seferos DS; Prigodich AE; Giljohann DA; Patel PC; Mirkin CA Polyvalent DNA Nanoparticle Conjugates Stabilize Nucleic Acids. Nano Lett. 2009, 9 (1), 308–311. 10.1021/nl802958f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Giljohann DA; Seferos DS; Prigodich AE; Patel PC; Mirkin CA Gene Regulation with Polyvalent siRNA-Nanoparticle Conjugates. J Am Chem Soc 2009, 131 (6), 2072–2073. 10.1021/ja808719p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Massich MD; Giljohann DA; Schmucker AL; Patel PC; Mirkin CA Cellular Response of Polyvalent Oligonucleotide−Gold Nanoparticle Conjugates. ACS Nano 2010, 4 (10), 5641–5646. 10.1021/nn102228s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Mahajan AS; Stegh AH Spherical Nucleic Acids as Precision Therapeutics for the Treatment of Cancer—From Bench to Bedside. Cancers 2022, 14 (7), 1615. 10.3390/cancers14071615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Huang Z; Callmann CE; Wang S; Vasher MK; Evangelopoulos M; Petrosko SH; Mirkin CA Rational Vaccinology: Harnessing Nanoscale Chemical Design for Cancer Immunotherapy. ACS Cent. Sci 2022, 8 (6), 692–704. 10.1021/acscentsci.2c00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Seferos DS; Giljohann DA; Hill HD; Prigodich AE; Mirkin CA Nano-Flares: Probes for Transfection and MRNA Detection in Living Cells. J. Am. Chem. Soc 2007, 129 (50), 15477–15479. 10.1021/ja0776529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Prigodich AE; Seferos DS; Massich MD; Giljohann DA; Lane BC; Mirkin CA Nano-Flares for MRNA Regulation and Detection. ACS Nano 2009, 3 (8), 2147–2152. 10.1021/nn9003814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Radovic-Moreno AF; Chernyak N; Mader CC; Nallagatla S; Kang RS; Hao L; Walker DA; Halo TL; Merkel TJ; Rische CH; Anantatmula S; Burkhart M; Mirkin CA; Gryaznov SM Immunomodulatory Spherical Nucleic Acids. PNAS 2015, 112 (13), 3892–3897. 10.1073/pnas.1502850112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Wang S; Qin L; Yamankurt G; Skakuj K; Huang Z; Chen P-C; Dominguez D; Lee A; Zhang B; Mirkin CA Rational Vaccinology with Spherical Nucleic Acids. Proceedings of the National Academy of Sciences 2019, 116 (21), 10473–10481. 10.1073/pnas.1902805116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Callmann CE; Cole LE; Kusmierz CD; Huang Z; Horiuchi D; Mirkin CA Tumor Cell Lysate-Loaded Immunostimulatory Spherical Nucleic Acids as Therapeutics for Triple-Negative Breast Cancer. Proceedings of the National Academy of Sciences 2020, 117 (30), 17543–17550. 10.1073/pnas.2005794117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Callmann CE; Kusmierz CD; Dittmar JW; Broger L; Mirkin CA Impact of Liposomal Spherical Nucleic Acid Structure on Immunotherapeutic Function. ACS Cent. Sci 2021, 7 (5), 892–899. 10.1021/acscentsci.1c00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Teplensky MH; Dittmar JW; Qin L; Wang S; Evangelopoulos M; Zhang B; Mirkin CA Spherical Nucleic Acid Vaccine Structure Markedly Influences Adaptive Immune Responses of Clinically Utilized Prostate Cancer Targets. Advanced Healthcare Materials 2021, 10 (22), 2101262. 10.1002/adhm.202101262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Zheng D; Giljohann DA; Chen DL; Massich MD; Wang X-Q; Iordanov H; Mirkin CA; Paller AS Topical Delivery of siRNA-Based Spherical Nucleic Acid Nanoparticle Conjugates for Gene Regulation. PNAS 2012, 109 (30), 11975–11980. 10.1073/pnas.1118425109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Jensen SA; Day ES; Ko CH; Hurley LA; Luciano JP; Kouri FM; Merkel TJ; Luthi AJ; Patel PC; Cutler JI; Daniel WL; Scott AW; Rotz MW; Meade TJ; Giljohann DA; Mirkin CA; Stegh AH Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Science Translational Medicine 2013, 5 (209), 209ra152–209ra152. 10.1126/scitranslmed.3006839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Randeria PS; Seeger MA; Wang X-Q; Wilson H; Shipp D; Mirkin CA; Paller AS siRNA-Based Spherical Nucleic Acids Reverse Impaired Wound Healing in Diabetic Mice by Ganglioside GM3 Synthase Knockdown. PNAS 2015, 112 (18), 5573–5578. 10.1073/pnas.1505951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Nemati H; Ghahramani M-H; Faridi-Majidi R; Izadi B; Bahrami G; Madani S-H; Tavoosidana G. Using siRNA-Based Spherical Nucleic Acid Nanoparticle Conjugates for Gene Regulation in Psoriasis. Journal of Controlled Release 2017, 268, 259–268. 10.1016/j.jconrel.2017.10.034. [DOI] [PubMed] [Google Scholar]
- (21).Hurst SJ; Lytton-Jean AKR; Mirkin CA Maximizing DNA Loading on a Range of Gold Nanoparticle Sizes. Anal. Chem 2006, 78 (24), 8313–8318. 10.1021/ac0613582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Giljohann DA; Seferos DS; Patel PC; Millstone JE; Rosi NL; Mirkin CA Oligonucleotide Loading Determines Cellular Uptake of DNA-Modified Gold Nanoparticles. Nano Lett. 2007, 7 (12), 3818–3821. 10.1021/nl072471q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Barnaby SN; Perelman GA; Kohlstedt KL; Chinen AB; Schatz GC; Mirkin CA Design Considerations for RNA Spherical Nucleic Acids (SNAs). Bioconjugate Chem. 2016, 27 (9), 2124–2131. 10.1021/acs.bioconjchem.6b00350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Yamankurt G; Stawicki RJ; Posadas DM; Nguyen JQ; Carthew RW; Mirkin CA The Effector Mechanism of siRNA Spherical Nucleic Acids. PNAS 2020, 117 (3), 1312–1320. 10.1073/pnas.1915907117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Vasher MK; Yamankurt G; Mirkin CA Hairpin-like siRNA-Based Spherical Nucleic Acids. J. Am. Chem. Soc 2022, 144 (7), 3174–3181. 10.1021/jacs.1c12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Hoshyar N; Gray S; Han H; Bao G. The Effect of Nanoparticle Size on in Vivo Pharmacokinetics and Cellular Interaction. Nanomedicine (Lond) 2016, 11 (6), 673–692. 10.2217/nnm.16.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Longmire M; Choyke PL; Kobayashi H. Clearance Properties of Nano-Sized Particles and Molecules as Imaging Agents: Considerations and Caveats. Nanomedicine 2008, 3 (5), 703–717. 10.2217/17435889.3.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Sonavane G; Tomoda K; Makino K. Biodistribution of Colloidal Gold Nanoparticles after Intravenous Administration: Effect of Particle Size. Colloids and Surfaces B: Biointerfaces 2008, 66 (2), 274–280. 10.1016/j.colsurfb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- (29).Hirn S; Semmler-Behnke M; Schleh C; Wenk A; Lipka J; Schäffler M; Takenaka S; Möller W; Schmid G; Simon U; Kreyling WG Particle Size-Dependent and Surface Charge-Dependent Biodistribution of Gold Nanoparticles after Intravenous Administration. European Journal of Pharmaceutics and Biopharmaceutics 2011, 77 (3), 407–416. 10.1016/j.ejpb.2010.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wang J; Liu G. Imaging Nano–Bio Interactions in the Kidney: Toward a Better Understanding of Nanoparticle Clearance. Angewandte Chemie International Edition 2018, 57 (12), 3008–3010. 10.1002/anie.201711705. [DOI] [PubMed] [Google Scholar]
- (31).Zhu GH; Azharuddin M; Islam R; Rahmoune H; Deb S; Kanji U; Das J; Osterrieth J; Aulakh P; Ibrahim-Hashi H; Manchanda R; Nilsson PH; Mollnes TE; Bhattacharyya M; Islam MM; Hinkula J; Slater NKH; Patra HK Innate Immune Invisible Ultrasmall Gold Nanoparticles—Framework for Synthesis and Evaluation. ACS Appl. Mater. Interfaces 2021, 13 (20), 23410–23422. 10.1021/acsami.1c02834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Zhao Y; Sultan D; Detering L; Luehmann H; Liu Y. Facile Synthesis, Pharmacokinetic and Systemic Clearance Evaluation, and Positron Emission Tomography Cancer Imaging of 64 Cu–Au Alloy Nanoclusters. Nanoscale 2014, 6 (22), 13501–13509. 10.1039/C4NR04569F. [DOI] [PubMed] [Google Scholar]
- (33).Park S; Aalipour A; Vermesh O; Yu JH; Gambhir SS Towards Clinically Translatable in Vivo Nanodiagnostics. Nat Rev Mater 2017, 2 (5), 1–20. 10.1038/natrevmats.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Yu M; Xu J; Zheng J. Renal Clearable Luminescent Gold Nanoparticles: From the Bench to the Clinic. Angewandte Chemie International Edition 2019, 58 (13), 4112–4128. 10.1002/anie.201807847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Kang H; Rho S; Stiles WR; Hu S; Baek Y; Hwang DW; Kashiwagi S; Kim MS; Choi HS Size-Dependent EPR Effect of Polymeric Nanoparticles on Tumor Targeting. Advanced Healthcare Materials 2020, 9 (1), 1901223. 10.1002/adhm.201901223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Huang K; Ma H; Liu J; Huo S; Kumar A; Wei T; Zhang X; Jin S; Gan Y; Wang PC; He S; Zhang X; Liang X-J Size-Dependent Localization and Penetration of Ultrasmall Gold Nanoparticles in Cancer Cells, Multicellular Spheroids, and Tumors in Vivo. ACS Nano 2012, 6 (5), 4483–4493. 10.1021/nn301282m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Huo S; Jin S; Ma X; Xue X; Yang K; Kumar A; Wang PC; Zhang J; Hu Z; Liang X-J Ultrasmall Gold Nanoparticles as Carriers for Nucleus-Based Gene Therapy Due to Size-Dependent Nuclear Entry. ACS Nano 2014, 8 (6), 5852–5862. 10.1021/nn5008572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Huo S; Chen S; Gong N; Liu J; Li X; Zhao Y; Liang X-J Ultrasmall Gold Nanoparticles Behavior in Vivo Modulated by Surface Polyethylene Glycol (PEG) Grafting. Bioconjugate Chem. 2017, 28 (1), 239–243. 10.1021/acs.bioconjchem.6b00488. [DOI] [PubMed] [Google Scholar]
- (39).Wu M; Guo H; Liu L; Liu Y; Xie L. Size-Dependent Cellular Uptake and Localization Profiles of Silver Nanoparticles. Int J Nanomedicine 2019, 14, 4247–4259. 10.2147/IJN.S201107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Banga RJ; Chernyak N; Narayan SP; Nguyen ST; Mirkin CA Liposomal Spherical Nucleic Acids. J Am Chem Soc 2014, 136 (28), 9866–9869. 10.1021/ja504845f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Li H; Zhang B; Lu X; Tan X; Jia F; Xiao Y; Cheng Z; Li Y; Silva DO; Schrekker HS; Zhang K; Mirkin CA Molecular Spherical Nucleic Acids. Proceedings of the National Academy of Sciences 2018, 115 (17), 4340–4344. 10.1073/pnas.1801836115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Zhu S; Xing H; Gordiichuk P; Park J; Mirkin CA PLGA Spherical Nucleic Acids. Advanced Materials 2018, 30 (22), 1707113. 10.1002/adma.201707113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Ferrer JR; Sinegra AJ; Ivancic D; Yeap XY; Qiu L; Wang J-J; Zhang ZJ; Wertheim JA; Mirkin CA Structure-Dependent Biodistribution of Liposomal Spherical Nucleic Acids. ACS Nano 2020, 14 (2), 1682–1693. 10.1021/acsnano.9b07254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Sinegra AJ; Evangelopoulos M; Park J; Huang Z; Mirkin CA Lipid Nanoparticle Spherical Nucleic Acids for Intracellular DNA and RNA Delivery. Nano Lett. 2021, 21 (15), 6584–6591. 10.1021/acs.nanolett.1c01973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Chinen AB; Guan CM; Ko CH; Mirkin CA The Impact of Protein Corona Formation on the Macrophage Cellular Uptake and Biodistribution of Spherical Nucleic Acids. Small 2017, 13 (16), 1603847. 10.1002/smll.201603847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Sita TL; Kouri FM; Hurley LA; Merkel TJ; Chalastanis A; May JL; Ghelfi ST; Cole LE; Cayton TC; Barnaby SN; Sprangers AJ; Savalia N; James CD; Lee A; Mirkin CA; Stegh AH Dual Bioluminescence and Near-Infrared Fluorescence Monitoring to Evaluate Spherical Nucleic Acid Nanoconjugate Activity in Vivo. Proceedings of the National Academy of Sciences 2017, 114 (16), 4129–4134. 10.1073/pnas.1702736114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Girard M; Wang S; Du JS; Das A; Huang Z; Dravid VP; Lee B; Mirkin CA; Cruz MO de la. Particle Analogs of Electrons in Colloidal Crystals. Science 2019, 364 (6446), 1174–1178. 10.1126/science.aaw8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Elghanian R; Storhoff JJ; Mucic RC; Letsinger RL; Mirkin CA Selective Colorimetric Detection of Polynucleotides Based on the Distance-Dependent Optical Properties of Gold Nanoparticles. Science 1997, 277 (5329), 1078–1081. 10.1126/science.277.5329.1078. [DOI] [PubMed] [Google Scholar]
- (49).Choi HS; Ipe BI; Misra P; Lee JH; Bawendi MG; Frangioni JV Tissue- and Organ-Selective Biodistribution of NIR Fluorescent Quantum Dots. Nano Lett. 2009, 9 (6), 2354–2359. 10.1021/nl900872r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Wilhelm S; Tavares AJ; Dai Q; Ohta S; Audet J; Dvorak HF; Chan WCW Analysis of Nanoparticle Delivery to Tumours. Nat Rev Mater 2016, 1 (5), 1–12. 10.1038/natrevmats.2016.14. [DOI] [Google Scholar]
- (51).Distler ME; Teplensky MH; Bujold KE; Kusmierz CD; Evangelopoulos M; Mirkin CA DNA Dendrons as Agents for Intracellular Delivery. J. Am. Chem. Soc 2021, 143 (34), 13513–13518. 10.1021/jacs.1c07240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Levi-Kalisman Y; Jadzinsky PD; Kalisman N; Tsunoyama H; Tsukuda T; Bushnell DA; Kornberg RD Synthesis and Characterization of Au102(p-MBA)44 Nanoparticles. J. Am. Chem. Soc 2011, 133 (9), 2976–2982. 10.1021/ja109131w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.