Abstract
Spatial transcriptomics and proteomics have independently transformed our understanding of complex biological processes; however, integration of these modalities is currently limited. To overcome this challenge, we developed Spatial PrOtein and Transcriptome Sequencing (SPOTS) for high-throughput integration of transcriptome and protein profiling within the spatial context. Applying SPOTS to spleen and breast cancer samples revealed that spatially-resolved multi-omic integration provides a comprehensive perspective on key biological processes in health and disease.
The crosstalk between cells in their native microenvironment is critical to our understanding of mechanisms regulating diverse aspects of biology in health and disease. Single-cell genomics approaches (e.g., single-cell RNA-seq) have substantially increased the throughput and granularity of our ability to study biological processes in tissues1,2. Nonetheless, the spatial context is lost during tissue dissociation required for single-cell experiments. In contrast, spatial transcriptomics (ST) enables in-depth molecular characterization of tissues in a spatially informed manner3. However, while being widely accessible and scalable, this method and similar recent advances, including High-Definition Spatial Transcriptomics (HDST)4, Slide-seqV25, and Seq-Scope6, are currently limited to unimodal transcriptomic characterization. On the other hand, several imaging-based methods have been developed that measure highly multiplexed protein expression in tissues by iterative antibody staining (4i)7, IBEX8, imaging mass spectrometry (MIBI)9, IMC10, MALDI-Imaging11 or using Antibody co-detection by indexing (CODEX)12. Others have computationally integrated CODEX data with CITE-seq on single cells (STvEA)13 to obtain spatial multi-omic data in silico.
Recent studies have begun to integrate protein and transcriptome information. For example, Jiang et al. have demonstrated the ability to measure a limited number of RNAs and proteins by converting RNA detection into an imaging mass spectrometry readout (PANINI)14. Alternatively, Liu et al. create spatially encoded gene and protein expression libraries using DNA-barcoded antibodies in a custom sophisticated microfluidics apparatus (DBiT-seq)15. However, these methods either rely on manual microarray design and automated robotic systems that are not readily accessible16 or show limited sensitivity for detecting the expected variability of proteins of interest17 and have not been demonstrated on highly fibrotic tissue such as solid tumors.
To address these challenges, we developed Spatial PrOtein and Transcriptome Sequencing (SPOTS), a novel multimodal approach that enables simultaneous recording of whole transcriptomes and a large panel of extracellular proteins in intact tissues (Fig. 1a; Supplementary Fig. 1; Supplementary Table 1). By leveraging the poly-A capture technology of the Visium slide18 (Supplementary Fig. 2a), SPOTS can simultaneously measure extracellular/intracellular protein levels via polyadenylated antibody-derived tag-conjugated (ADT-conjugated) antibodies, as in CITE-seq19 (Supplementary Fig. 2b), and mRNA expression while preserving tissue architecture (Supplementary Fig. 2c,d). We demonstrated that by supplementing Visium’s transcriptome profiling with >30 protein markers, SPOTS yields a more granular landscape of tissues, including cell types, biological processes, and phenotypes, in a highly reproducible manner.
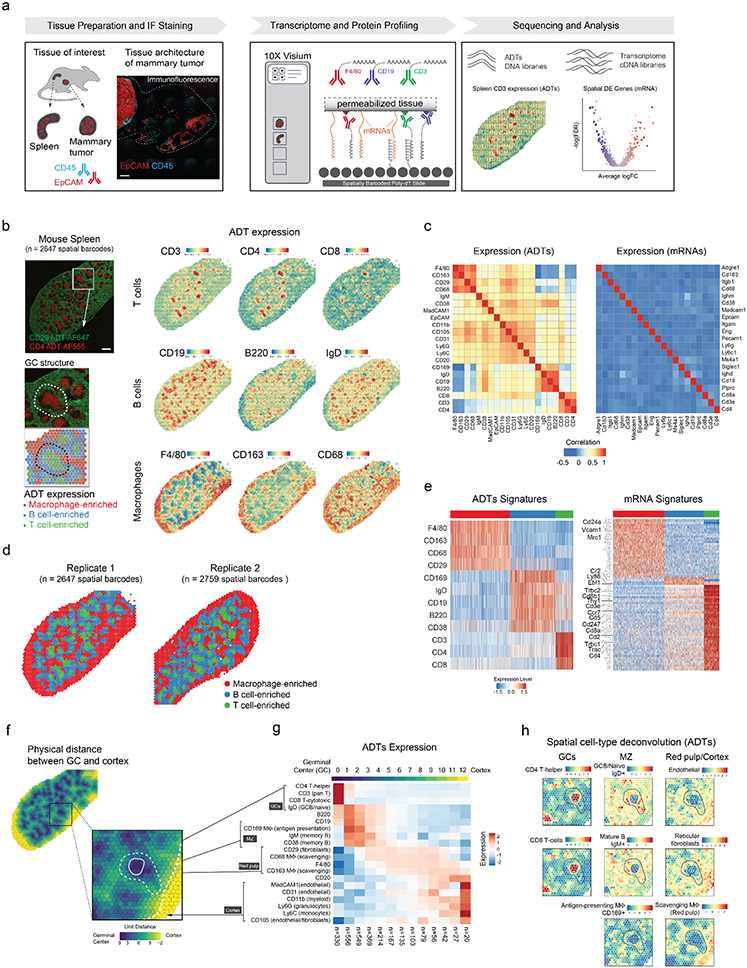
Figure 1: Spatial PrOtein and Transcriptome Sequencing (SPOTS).
(a) Overview of SPOTS workflow outline. Fresh tissue samples were collected and embedded in OCT. Tissue sections (10μm) were processed following SPOTS protocol (including staining with fluorescent and ADT antibodies, permeabilization, mRNA capture, reverse transcription, and second-strand synthesis; Methods) and sequenced for downstream analysis. See Supplementary Figure 1 for detailed SPOTS schematic. Scale bar 200μm.
(b) Normalized ADT levels of key surface markers for mouse spleen. Left panel: immunofluorescence (IF) staining for CD29 (green) and CD4 (red) with inset showing the germinal center (GC) architecture and its underlying spatial barcodes. Right panel: normalized ADT levels of marker genes for T cells (CD3, CD4, CD8), B cells (CD19, B220, IgD), and Macrophages (F4-80, CD163, CD68).
(c) Correlation between the 21 ADTs and their corresponding mRNA expression levels across two biological replicates (Supplementary Table 1).
(d) Spatial barcode clustering and major cell type enrichment (Macrophage, B cell, and T cell) based on ADTs of two biological replicates (Supplementary Table 2).
(e) ADT and mRNA signatures for each cluster of spatial barcodes. Left panel: heatmap showing expression levels (Z-score) of differentially expressed ADTs for each cluster. Right panel: heatmap showing expression levels (Z-score) of differentially expressed mRNAs in 3,000 downsampled spatial barcodes. Key marker genes are highlighted (Supplementary Table 3).
(f) Physical distance (color scale) from the center of GCs for each spatial barcode.
(g) Spatial gene expression patterns of ADTs in the spleen. ADT Expression pattern (Z-score) from GC to spleen cortex and their anatomical definition (black boxes). The top color bar represents the physical distance as in the right panel. The number of spatial barcodes in each bin is labeled at the bottom. MZ, marginal zone.
(h) Cell-type deconvolution based on ADTs expression (Z-score) of each spatial barcode overlaid onto the spleen tissue. Deconvolution values are provided in Supplementary Table 2.
First, we optimized the assay on splenic tissues as they contain diverse immune cell populations that are compartmentalized in well-defined structures of germinal centers (GCs), marginal zones (MZs), and red pulp/stroma regions20 (Fig. 1b). To achieve maximal retrieval of mRNAs and ADTs, we examined several methods of tissue fixation, antibody staining, and permeabilization conditions (Methods). The standard Visium workflow requires using methanol (MeOH) as a fixative which is optimal for mRNA capture. As MeOH fixation is less compatible with the staining of most antibody clones, we optimized the assay with paraformaldehyde (PFA) fixation, which is optimal for immunostaining (Supplementary Fig. 2e).
We initially observed low cDNA yields and poor antibody signal (Supplementary Fig. 2f). We hypothesized that the tissue permeabilization enzyme used as a part of the Visium Gene Expression kit (10x Genomics) workflow was not sufficient to release PFA-fixed mRNA and ADT molecules for capture on the slide. Therefore, we increased the permeabilization strength by using the tissue removal enzyme from the Visium Tissue Optimization kit (10x Genomics) in combination with 1% sodium dodecyl sulfate (SDS). These technical modifications increased the yields of both mRNA and ADT signals (Supplementary Fig. 2f). Finally, to evaluate ADT binding specificity, we immunostained spleens with dual-labeled ADT- and fluorophore-conjugated antibodies for CD4 and CD29 to highlight the CD4+ follicular T helper cells (Tfh) and CD29+ stromal cells20,21. We detected a significant ADT binding to slide surface areas without tissue (Supplementary Fig. 2g), which we hypothesized is due to the direct binding of the ADT’s poly-A tail to poly-dT oligos on the slide surface and not binding via cell epitopes. The non-specific ADT binding to spatial barcodes poses a major challenge as it hinders assay fidelity and reduces the signal-to-noise ratio. To circumvent this non-specific binding, we added poly-dT oligos into the staining buffer to inhibit ADT binding to tissue-free surface spatial barcodes, which ultimately eliminated the binding in regions without tissue, restored antibody specificity, and improved the signal-to-noise ratio (Supplementary Fig. 2h).
We next assessed SPOTS’ ability to capture cellular heterogeneity in the murine spleen using the ADT-conjugated antibodies. We designed a 21-plex panel that allows the identification of spleen constituents, including B cells (CD19, CD20, CD220, IgD, IgM, CD38), T cells (CD3, CD8, CD4), scavenging and antigen-presenting macrophages (CD169, F4/80, CD163, CD68, CD11b), granulocytes (Ly6G, Ly6C), endothelial cells (CD31, MadCAM1), and fibroblasts (CD29, CD105) (Supplementary Table 1, Supplementary Fig. 3a). Importantly, the spatial ADT expression patterns recapitulated the expected spleen structure22 in two biological replicates as revealed by IHC and dual-labeled CD4 and CD29 antibodies, showing CD4+ T cell-enriched GCs and CD29+ stroma-enriched red pulp/cortex (Fig. 1b and Supplementary Fig. 3b,c). Furthermore, we observed a high reproducibility (Pearson’s r = 0.998, p-value < 2.2e-16) in both the ADT-conjugated antibodies and mRNA expression across the two independent biological replicates, which enabled cell clustering without any explicit batch effect correction (Supplementary Fig. 4a).
Consistent with robust signal detection and a higher abundance of protein compared to mRNA, the protein co-expression patterns showed a stronger correlation structure than the corresponding mRNAs (Fig. 1c)23. For instance, ADT markers of T cells (CD3, CD4, CD8) were positively correlated with each other and negatively correlated with macrophage markers (F4/80, CD163, CD68). However, such covariations between key cell-type marker genes were lacking in mRNA (Fig. 1c), as reported in the original CITE-seq study, where ADTs are less prone to ‘dropout’ events compared to mRNAs19. Accordingly, we leveraged the protein modality (ADTs) to cluster the spatial barcodes into phenotypically distinct groups that were enriched in varying levels of macrophages, B cells, and T cells (Fig. 1d). In general, cell-type specific mRNAs, such as T-cell receptor-related genes (Cd3e, Cd4, Cd8a, Trac, Trbc1/2, Ccr7, Lat), were efficiently captured, as were their corresponding surface protein counterparts (CD3, CD4, CD8; Fig. 1e). Surface protein and mRNA abundance also showed a positive correlation (Pearson’s r = 0.53, p-value < 2.2e-16) in B cell-enriched regions in which key marker genes (IgD, IgM, CD19, CD38) were well expressed as both protein and mRNA (140±0.97 ADT UMIs, 19±0.15 mRNA UMIs; mean±s.e.m) (Supplementary Fig. 4b). However, such agreement declined in macrophage-enriched regions (Pearson’s r = 0.23, p-value < 2.2e-16) in which key marker genes (CD11b, CD163, F4/80, CD68) were more lowly expressed as mRNA (109±0.91 ADT UMIs, 0.36±0.01 mRNA UMIs; mean±s.e.m), as previously reported19,24.
Next, we incorporated spatial information into our analysis to identify spatial protein and gene expression patterns within the mouse spleen. First, we annotated GC-specific genes (n = 163 genes, GSE12366) and plotted ADTs expression as a function of physical distance from GC centers (Fig 1f-g and Supplementary Fig. 4c) along with ADT-based cell-type deconvolution analysis (Fig 1h and Supplementary Fig. 4d; Methods). Importantly, this analysis revealed that beyond the detection of various splenic cell types, SPOTS was able to recapitulate their function/activation states as a function of their spatial location. For example, the GC centers (regions 0; outlined) contained mostly CD4 T-helper, CD8 T cells, and IgD+ GC/naive B cells. However, the adjacent marginal zones (regions 2-4; dotted outline) contained IgM+ memory/mature B cells that were IgD- (as expected following a class-switching) and a specialized subset of antigen-presenting CD169+ macrophages that were shown to interact directly with B and CD4 helper cells through CD16925,26. In addition, in regions 5-10 (red pulp/stroma), we identified CD29+ fibroblasts and red pulp macrophages, which are characterized by the expression of scavenging and lysosomal markers CD163 and CD68 and are involved in the clearance of aged red blood cells from circulation27. Finally, and as predicted, the cortex areas (regions 11-12) were enriched with fibroblasts (CD105), blood vessels (CD31, MadCAM1), and other myeloid cells, such as macrophages, Ly6C+ monocytes, and Ly6G+ granulocytes (Supplementary Fig. 4d). Collectively, these analyses demonstrated that SPOTS was able to reconstruct the spatial distribution of multiple cell types in tissues with high resolution and phenotypic fidelity and provides granular information about their function and activation state in space.
Following the successful recapitulation of tissue structure and spatial signals, we examined whether SPOTS and Visium-alone capture comparable RNA signals. By analyzing spleen tissues, we noticed higher gene and UMI detection (Supplementary Fig. 5a) and less saturated gene expression libraries in Visium-only samples (Supplementary Fig. 5b). Nonetheless, these variations had a marginal effect on the overall assay performances, as plotting the detected ADT UMIs vs RNA UMIs in defined splenic regions demonstrates a strong positive correlation between the mRNA and protein signals across the major cell-types in spleens, as well as positive correlation between SPOTS- and Visum-detected genes (Supplementary Fig. 5c-d). Next, we then compared SPOTS’ performance with SM-Omics and Guilliams et al., two studies that integrate TotalSeq-A antibodies with ST technology16,17. While SPOTS and Guilliams’s methods rely on the Visium platform (55μm spatial barcodes, 100μm center to center distance) and template switched RT-PCR to amplify cDNA, SM-Omics relies on ST slide (100μm spatial barcodes, 200μm center to center distance) and in-vitro transcription to amplify cDNA. Through these comparative analyses, we demonstrated that SPOTS substantially improved the overall performances of these methodologies in every parameter tested, including signal capture (number of detected genes and ADTs), signal resolution, cell clustering, and discovery power in differential gene expression across tissue regions (Supplementary Fig. 6 and 7). These results highlight that by combining robust protein and mRNA modalities, SPOTS provided more detailed spatial molecular profiles of cellular phenotypes than either modality alone.
Unlike spleen tissues where immune cells are highly abundant and compartmentalized, the tumor microenvironment (TME) is highly variable and complex, leading to difficulties in characterizing less abundant cell types (e.g. T cells; Supplementary Fig. 8a) with current ST technologies28. Although several single-cell RNA-seq analyses of breast carcinomas have provided a granular characterization of the tumor-infiltrating leukocytes2,29,30, the precise location of these signatures and their physical relation to each other are incompletely resolved. Therefore, we applied SPOTS to murine breast tumors, utilizing the MMTV-PyMT transgenic mouse model31 (Fig. 2a). To adapt the method for highly fibrotic tissue such as solid tumors, we modified the SPOTS tissue permeabilization protocol, thereby increasing the generalizability of SPOTS (Fig. 2a and Supplementary Fig. 8b; Methods). To capture the full spectrum of the immune landscape in tumors, we designed a 32-plex ADT-conjugated antibody panel spanning major cell-lineages and activation states, including tumor epithelial cells (EpCAM, KIT), fibroblasts (PDGFRA, PDPN), myeloid cells (CD11b, CD11c, F4/80, MHC-II, Ly6G, Ly6C), lymphoid cells (NK1.1, B220, CD4, CD8), and endothelial cells (CD31, TIE-2) (Supplementary Table 1). First, we examined whether SPOTS and Visium-alone capture comparable RNA signals in breast cancer tissue. Unlike spleens, we found no significant difference in RNA capture when adding the ADT modality in comparison to Visium-alone, as both methods showed equivalent expression of housekeeping genes (Supplementary Fig. 9a,b) and detected a comparable number of UMIs and genes (Supplementary Fig. 9c). Moreover, by plotting the sequencing saturation curves, we found that gene expression libraries in SPOTS were less saturated, indicating that SPOTS also preserves RNA library complexity (Supplementary Fig. 9d).
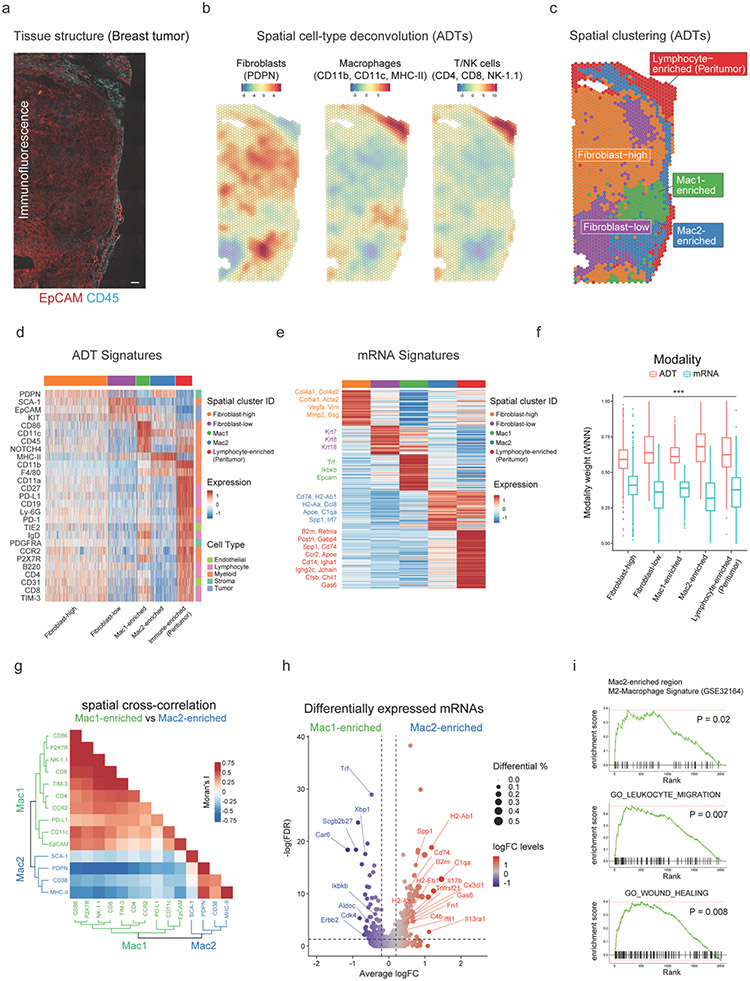
Figure 2: SPOTS reveals two spatially distinct macrophages in breast cancer TME.
(a) IF staining of EpCAM (red) and CD45 (cyan) in breast cancer tissue. Scale bar 200μm.
(b) Cell-type deconvolution based on ADTs (Z-score) of each spatial barcode overlaid onto the breast cancer tissue. Deconvolution values are provided in Supplementary Table 4.
(c) Spatial barcode clustering results with annotation of major cell type enrichment based on ADTs (Supplementary Table 4).
(d) Heatmap showing ADT signatures (Z-score) for each cluster (Supplementary Table 5).
(e) Heatmap showing the average expression levels (Z-score) of differentially expressed mRNAs for each cluster. Key marker genes are colored and highlighted. The complete list of differentially expressed mRNAs is listed in Supplementary Table 5.
(f) Boxplot showing relative contributions (WNN modality weights) to spatial clustering of mRNA and ADT modalities. Paired t-test, *p<0.05, **p<0.01, ***p<0.001, otherwise not significant (n.s).
(g) Heatmap showing the spatial cross-correlation (bivariate Moran’s I) of ADTs in Mac1- and Mac2-enriched regions with dendrograms showing hierarchical clustering of ADTs.
(h) Volcano plot showing log fold changes (logFC) of the top 5,000 most variable genes between Mac1- and Mac2-enriched spatial barcodes and their significance (y-axis; −log10 scale). Genes are dotted and colored by logFC levels (color scale). The size of the dot represents the difference in the fraction of detection between the two groups. Macrophage-related genes are annotated. P-values were determined by the Wilcoxon Rank Sum test. Vertical dotted lines represent ±0.2 logFC. Horizontal dotted lines represent false discovery rate (FDR) of 0.05 (−log10 scale). Raw and FDR corrected P-values and logFC values are listed in Supplementary Table 5.
(i) Gene Set Enrichment Analysis (GSEA) of Mac2-enriched spatial barcodes based on mRNA expression.
In order to characterize the complex interactions and the spatial gene expression heterogeneity within the TME, we first performed cell-type deconvolution32 and clustering33 analyses (Fig. 2b,c; Methods) and revealed several distinct tumor and peritumor regions enriched with varying levels of immune and stromal cells (Fig. 2d,e and Supplementary Fig. 10a). As observed in the splenic tissue, the correlation between an ADT and its corresponding mRNA was high when the gene was well expressed (Supplementary Fig. 10b). Conversely, given the dilution of RNA from multiple cells per spatial barcode, markers of underrepresented cells (either by cell number and/or RNA content) (Supplementary Fig. 10c,d) may lose correlation with their corresponding protein, as demonstrated by comparing the ADT- and mRNA-based captures of Epcam (tumor cells) and Ptprc (immune cells). While we observed a positive correlation with EpCAM immunofluorescence signals for both ADT (Pearson’s r = 0.29) and mRNA (Pearson’s r = 0.28) data, for less represented immune cell markers, the correlation of CD45 IF signals to mRNA was low (Pearson’s r = 0.05), but was effectively rescued by using the ADT modality (Pearson’s r = 0.22) (Supplementary Fig. 10e,f).
Indeed, by using a weighted nearest neighbor (WNN) based multimodal integration method33, we found that the ADT modality had a greater impact on clustering assignment compared to mRNA (Fig. 2f; 0.63 vs 0.37 WNN weights, 95%CI of mean differences 0.25-0.27, paired t-test p-value < 2.2e-16). These results further reinforced that by integrating both the ADT and mRNA modalities, SPOTS provided a more precise molecular characterization of immune cell populations in disorganized tissues, such as solid tumors.
We next examined two regions that were enriched with macrophage/myeloid markers (Mac1- and Mac2-enriched; Fig. 2c,d). Phenotypically, Mac1-enriched regions exhibited immunostimulatory ADTs of myeloid cells (CD86, CD11c) while Mac2-enriched regions displayed immunosuppressive ADTs (PD-L1; Fig. 2g). Structurally, the Mac2-enriched areas were in close proximity to a lymphocyte-enriched region (Fig. 2c) that was enriched with T cell exhaustion markers (PD-1, TIM-3), in-line with the well-documented immunosuppressive function of tumor-associated macrophages31. These results were further validated by differential mRNA expressions between Mac1- and Mac2-enriched spatial barcodes (Spp1, C1qa; Fig. 2h), and gene set enrichment analyses for M2-macrophages and wound healing signatures (Fig. 2i). These data suggest that M2-macrophages are forming an immunosuppressive barrier at tumor borders (Fig. 2c), leading to immune exhaustion and evasion, as previously shown in breast cancer30.
Here, we present SPOTS, a method that allows simultaneous measurement of a large panel of protein markers and whole transcriptomes in intact tissues. Direct comparison of SPOTS with Visium alone showed comparable RNA capture despite altering the fixation method and adding the protein modality in SPOTS (Supplementary Fig. 5 and 9). While it is likely that SPOTS may reduce RNA capture in some tissues, our results suggest that deeper sequencing can effectively compensate for this while still benefiting from a rich and informative protein modality. Further comparisons to current ST methodologies that integrate protein and transcriptome information16,17 have demonstrated that beyond technical improvements (55μm vs 100μm spatial barcode size, 100μm vs 200μm center to center distance), SPOTS substantially improves the overall performance, including signal capture (number of detected genes and ADTs), signal resolution, and cell clustering and enhances the discovery power in differential gene expression analysis across tissue regions (Supplementary Fig. 6 and 7). Therefore, we envision that SPOTS–a technique that makes use of the broadly available Visium technology and polyadenylated DNA-barcoded antibodies–may be readily integrated into emerging higher resolution ST technologies4,5, and further improve the molecular characterization of cell types and states in tissues. Future developments in SPOTS to enable profiling at single cell resolution (Visium HD), to capture somatic mutations in tumor cells34 or TCR sequences of tumor-infiltrating T cells35 would also enhance our understanding of cell-cell interactions in the immune microenvironment. Collectively, our data across normal spleen and breast tumors demonstrated that SPOTS enables both in-depth gene expression analysis and highly multiplexed immunophenotyping and is highly reproducible, simple to execute, and compatible with commercially available platforms. Broad adoption of SPOTS by the genomics community will pave the way towards a spatially-resolved multimodal map of tissue organization and function in health and disease.
Methods
SPOTS Protocol.
An extended protocol describing all recommended workflow, reagents, volumes, critical steps, and optional stopping points is provided as Supplementary Note Extended SPOTS protocol. A schematic of the workflow can also be found in Supplementary Fig. 1.
Mouse work.
Animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of the Research Animal Resource Center (RARC) at Weill Cornell Medicine (Protocol: 2016-0058). All mouse strains involved in this work including MMTV-PyMT (Stock No: 022974) and wild-type C57BL/6J (Stock No: 000664) were purchased from The Jackson Laboratory (USA). Females of the MMTV-PyMT model developed spontaneous tumors after ~110 days post-birth. To harvest fresh tumors, mice were euthanized, and mammary gland tumors were collected, washed three times in PBS-1% BSA, and embedded in OCT for storage at −80°C.
List of antibodies used for SPOTS.
The polyadenylated Biolegend TotalSeq™-A anti-mouse antibodies and clones used were: CD4 (Clone RM4-5); CD8a (Clone 53-6.7); CD366 (Tim-3) (Clone RMT3-23); CD279 (PD-1) (Clone RMP1-30); CD117 (c-kit) (Clone 2B8); Ly-6C (Clone HK1.4); CD11b (Clone M1/70); Ly-6G (Clone 1A8); CD19 (Clone 6D5); CD45 (Clone 30-F11); CD25 (Clone PC61); CD45R/B220 (Clone RA3-6B2); CD11c (Clone N418); F4/80 (Clone BM8); I-A/I-E (Clone M5/114.15.2); NK-1.1 (Clone PK136); Ly-6A/E (Sca-1) (Clone D7); CD3 (Clone 17A2); CD274 (B7-H1, PD-L1) (Clone MIH6); CD27 (Clone LG.3A10); CD20 (Clone SA275A11); CD86 (Clone GL-1); MadCAM1 (Clone MECA-367); CD163 (Clone S15049I); CD192 (CCR2; Clone SA203G11); CD169 (Siglec-1) (Clone 3D6.112); CD326 (Ep-CAM) (Clone G8.8); IgM (Clone RMM-1); CD38 (Clone 90); CD68 (Clone FA-11); CD29 (Clone HMβ1-1); IgD (Clone 11-26c.2a); CD140a (Clone APA5); CD11a (Clone M17/4); CD105 (Clone MJ7/18); P2X7R (Clone 1F11); CD1d (CD1.1, Ly-38) (Clone 1B1); Notch 4 (Clone HMN4-14); CD31 (Clone 390); CD202b (Tie-2, CD202) (Clone TEK4); Podoplanin (Clone 8.1.1). See Supplementary Fig. 11 for staining validations of selected clones and Supplementary Table 1 for a complete list of antibodies, clones, and barcodes used for SPOTS.
Fluorophore conjugation of ADT-conjugated antibodies.
Biolegend TotalSeq™-A anti-mouse antibodies CD4 (Clone RM4-5) and CD29 (Clone HMβ1-1) were conjugated with Alexa 555 or Alexa 647 with the Antibody Labeling Kit (ThermoFisherScientific) according to manufacturer instructions.
Pre-SPOTS quality control and optimization.
In advance of SPOTS, sections were obtained for RNA quality measure with the RNeasy Plus kit (Qiagen). Tissues with an RNA integrity number (RIN) of 7 or greater were used for future experiments. The optimal ADT concentrations were determined by titrations of fluorescently-tagged matched clones using immunofluorescence (IF). Appropriate permeabilization conditions for both the pre-staining permeabilization and the full tissue permeabilization were determined using the 10x Genomics Tissue Optimization kit (Supplementary Table 1).
Solutions for SPOTS.
See extended SPOTS protocol for a point-by-point description of the workflow (Supplementary Note, Supplementary Fig. 1). The staining and blocking solutions were adapted as previously described17. The following solutions were made immediately in advance of the experiment and were kept on ice. Each solution is given based on 1 fiduciary square/well on the Visium slide. Calculations should be made with a minimum 20% surplus to accommodate for dead volume. 2x Blocking buffer (400μl per sample): 120μL 20x SSC, 8μL 10% Tween 20, 160μL 10% BSA, 32μL 10μg/μl sheared salmon sperm, 40μL 20 U/μl RNAse inhibitor, 40μL water. Pre-staining permeabilization solution (75μL per sample): 46.875μL 375mM saponin (10X), (Thermo Fisher J63209.AK), 0.94μL RNAse inhibitor, 45.94μL 1X PBS. Antibody staining solution (50μl per sample): 25μL 2x blocking buffer, 10μL 100μM blocking oligos (dT25), fluorescent and TotalSeq™-A (Biolegend, USA) antibodies as titrated (Supplementary Table 1), antibodies and water together make up the remaining 15μL. Blocking solution (75μL per sample): 37.5μL 2x blocking buffer, 1.9μL 200 mM Ribonucleoside Vanadyl Complex (heated to 65°C shortly before use), 5μL Fc block (BioLegend, USA), 30.6μL water. Wash buffer (600μL per sample): 300μL 2X blocking buffer, 300μL water.
SPOTS.
See extended SPOTS protocol for a point-by-point description of the workflow (Supplementary Note, Supplementary Fig. 1). 10μm thick, OCT-embedded sections were deposited on the 10x Genomics Visium slides; slides were stored at −80°C for up to 1 week in advance of the experiment. Slides were moved on dry ice to the thermocycler and then incubated for 1 minute at 37°C on the Thermocycler Adaptor. The slides were then immersed in 1% PFA in PBS for 10 minutes at 25°C to fix the tissue. The slides were washed by dunking five times into a falcon tube with 3x SSC and then secured in the cassette. Each sample was blocked in 75μl of Blocking solution for 15 minutes at 4°C. Post-blocking, 75μl of the Pre-staining permeabilization solution was added to each sample at 4°C for 10 minutes. The solution was removed, and 50μl of Antibody staining solution was added to each sample. The slide was then incubated and protected from light for 90 minutes at 4°C with the antibodies. The samples were then washed with 150μl of Wash buffer four times for one minute each. A final pre-imaging wash was conducted by dunking the slide twenty times in a falcon tube with 3x SSC. The slide was imaged at 10X magnification to detect APC (CD45) and PE (EpCAM) fluorophores using the Axio Observer 7 Inverted Microscope (Zeiss Microscopy).
After imaging, the tissue was fully permeabilized for 9 minutes for spleen tissue and 27 minutes for breast cancer tissue using the Tissue Optimization Tissue Removal Enzyme (per sample: 3.5μL Tissue Removal Enzyme (10x Genomics, 3000387), 59.5μl 1x SSC, 7μl 10% SDS). Each well was then washed three times with 150μl of 0.1x SSC. The remainder of the experiment was conducted using the 10x Genomics Gene Expression protocol, starting with step 2.2 Reverse Transcription, including Visium Spatial Gene Expression Library Construction. The only deviations from the original protocol were the addition of additive primer to amplify ADT during Second Strand Synthesis (Step 3.0) and cDNA Amplification (Step 4.2) (8.8μl of 100μM and 4μl of 0.2μM additive primer were added to each reaction, respectively) (Supplementary Table 1) and retention of the supernatant in step 4.3 cDNA Cleanup - SPRIselect. Two sequential 1.9x SPRIselect cleanups (130μl and 95μl of SPRI for the first and second cleanups, respectively) were performed on said supernatant and the final product was eluted in 45μl of water. The ADT product from the supernatant was amplified and indexed using the SI-PCR and TruSeq Small RNA RPIx primers before Illumina sequencing (Supplementary Table 1). The master mix and thermocycler conditions for this reaction were as follows: 45μl ADT, 50μl of Amp Mix (2000047), 2.5μl of 20μM TruSeq Small RNA RPI index primer, 2.5μl of 20μM SI-PCR primer; 95°C for 3 minutes > (95°C for 20 seconds, 60°C for 30 seconds, 72°C for 20 seconds, 10 cycles) > 75°C for 5 minutes > 4°C hold. Approximately 50,000 and 8,500 reads were designated per spatial barcode for gene expression and ADT sequencing, respectively. Transcript (GEX) and antibody derived tag (ADT) libraries were sequenced on the Illumina Nextseq500 (spleen) or Novaseq6000 (mouse breast cancer).
PolyT blocking oligo length and wash temperature titration.
Dual-tagged (ADT+fluorophore) antibodies were incubated in antibody staining solution (see above) with different polyT blocking oligos for 30 minutes at 4°C to allow blocking oligos to bind to antibody-oligos. Next, the solutions were pipetted on top of different capture areas of two Visium slides. One slide was then incubated at 37°C and the other at 4°C for one hour. Subsequently, each capture area was washed 5x with 150μl 3x SSC at the respective temperature with buffers pre-headed to 37°C or pre-cooled to 4°C. The slide was disassembled from the slide cassette and a pre-imaging wash was conducted by dunking the slide twenty times in a falcon tube with 3x SSC at 25°C. The slide was imaged without a coverslip in a slide scanner (InnoScan 710, Innopsys) with the lowest intensity and gain settings.
Visium with fresh frozen tissues.
Fresh frozen mouse spleen or breast cancer tissues embedded in OCT were sectioned (10μm) onto Visium slides and processed as described in the Visium User Guide (10x Genomics). Mouse spleen was permeabilized for 25 minutes and breast tissue for 10 minutes. Approximately 50K reads per tissue-covered spot were designated to each sample when sequenced with NovaSeq 6000 S1 reagents.
Spleen data analysis.
The sequencing data and histology images were processed using SpaceRanger (v1.3.0 and pre-release v.2.0, 10x Genomics) with manual tissue alignment using Loupe Browser (v5.0, 10x Genomics) to obtain raw UMI count spot matrices for ADT and GEX. We analyzed the data using Seurat 4.0. Briefly, ADT data from the two spleen replicates were normalized using CLR normalization across each cell with Seurat’s NormalizeData function, and the transcriptomic data were normalized using log-transformed transcript per million (logTPM). We scaled and centered the normalized ADT data and then performed Principal Component Analysis (PCA) using Seurat’s RunPCA. We selected the first 10 PCs to build a shared nearest neighbor graph using Seurat’s FindNeighbors function and clustered the spatial barcodes using the FindClusters function with resolution=0.2. Upon clustering, we noticed a cluster of 15 spatial barcodes out of the 5,421 total barcodes that are sporadically spread and have a significantly lower number of ADTs compared to the rest (18 vs. 1,248 UMIs). We reasoned that these spatial barcodes may represent technical failures, and we removed them from downstream analysis and retained 5,406 spatial barcodes (2,647 from Replicate 1 and 2,759 from Replicate 2). There were 10 spatial barcodes near the edge of the slides that were excluded for visualization purposes. In total, we retained four clusters of spatial barcodes that were enriched in macrophages and B, T, and epithelial cells, and we excluded the epithelial cell-enriched cluster for visualization purposes (Fig. 1d). The clustering result and annotation are listed in Supplementary Table 2. To visualize the sparse mRNA data, we randomly sampled 30 barcodes for each cluster and created a downsample of 3,000 spatial barcodes.
Breast cancer data analysis.
Tumor tissues from the MMTV-PyMT mouse model were first aligned onto the Visium slides with Loupe Browser 5.0 (10x Genomics) using IF images as input. The resulting alignment JSON files were used as input for SpaceRanger software (v.1.3.0 10x Genomics), and the transcriptomic reads were mapped to a custom genome (mm10 with PyMT transgene). The ADT reads were mapped to the antibody barcode sequences (Supplementary Table 1) with the 4,992 whitelist tissue spatial barcodes provided by 10x Genomics as input using the Python package CITE-seq-Count v1.4.336. The count matrices of the transcriptome and ADT data were then normalized using log-transformed transcript per million (logTPM). For the Visium comparison experiment, HE images of the tumor tissues from four MMTV-PyMT mice were used as input for SpaceRanger software (v.1.3.0 10x Genomics) and the transcriptomic reads were mapped to a custom genome (mm10).
Saturation analysis.
The saturation curve was generated by cellranger count. The formula for calculating this metric is as follows: Sequencing Saturation=1-(n_deduped_reads/n_reads) where n_deduped_reads = Number of unique (valid cell-barcode, valid UMI, gene) combinations among confidently mapped reads, and n_reads = Total number of confidently mapped, valid cell-barcode, valid UMI reads.
Data visualization.
To better visualize the deconvolution results of the breast cancer SPOTS data, we utilized local Gi* statistics37, which is similar to local Moran’s I (spatial autocorrelation) and local Geary’s C statistics38, to create smooth expression profiles for visualization and diagnostics (Fig. 2b). The local Gi* statistics for each feature/gene was defined as , where xj is the expression value of a feature in cell j and wij is the spatial weight between cell i and j; the term is interpreted as weighted neighborhood or spatial lag. By definition, local Gi* statistics is a Z-score, and its P-value can be calculated using permutation-based methods38 or mathematical derivations39. Because Visium tissue barcodes are arranged on hexagonal lattices, we defined the weight matrix using hexagonal nearest neighbor distance (HNN)40, which reflects its true spatial orientation. We assigned the weights using HNN with a gaussian kernel of width 1 unit distance, and only neighbors within a 3 hexagonal distance were considered. We measured the physical distance from the center of GCs (defined by T cell-enriched regions) using HNN. The physical distance from the center of GCs for all the spatial barcodes are listed in Supplementary Table 2.
Cell type deconvolution.
We performed deconvolution of the breast cancer SPOTS data using SPOTlight v0.1.7 R package32 with default parameters, except we set min_cont=0 which did not remove any cells, and a recent unbiased scRNA-seq data of wild type MMTV-PyMT mouse model29 as reference. Briefly, the scRNA-seq data were downloaded from GSE158677 and analyzed using Seurat 4.033. Upon clustering, we identified six major cell types including cancer-associated fibroblasts (CAFs), Tumor, Endothelial, Myeloid, B, and T/NK cells. Differentially expressed genes were calculated by Seurat’s FindAllMarkers function with parameters min.pct=0.5 and logfc.threshold=1 and used for deconvolution with SPOTlight’s spotlight_deconvolution function. For the ADT deconvolution in breast cancer tissues (Fig. 2b), we first scaled the cell type ADTs (Tumor cells: EpCAM, KIT; CAFs: PDPN; Macrophages: CD11c, CD86, F4/80; B cells: CD19, B220; T/NK cells: CD4, CD8, NK-1.1) using cosine normalization41 and calculated the mean expression of their ADTs for each cell type. For the ADT deconvolution in spleens (Fig. 1h, Supplementary Fig. 4d), we first scaled the cell type ADTs for: CD4 T-helper (CD3, CD4), Cytotoxic T cells (CD8, CD3), GCB (B220, CD19, IgD), mature B cells (B220, CD19, IgM, CD38), antigen-presenting macrophages (CD169, F4/80), red-pulp/scavenging macrophages (F4/80, CD163, CD68), reticular fibroblast (CD29, CD105), endothelial cells (CD31, CD29, CD105), monocytes (Ly6C, CD11b), granulocytes (Ly6C, CD11b, Ly6G) using cosine normalization41 and calculated the mean expression of their ADTs for each cell type. The deconvoluted percentages were calculated as the fraction of the cell type ADT expressions over the total sum of all ADTs. The scaled deconvolution values were calculated using local Gi* statistics as described above. The IF staining intensity was quantified for each spatial barcode and normalized to the 0-1 range, and outlier values below 1st or above 99th percentiles were removed. The deconvolution values for each barcode are listed in Supplementary Table 2 for spleen and Supplementary Table 4 for breast cancer.
Integration.
In order to integrate the transcriptome and ADT data and assess the contributions of each modality (Fig. 2f), the WNN weights were calculated using Seurat’s FindMultiModalNeighbors function using the first 30 PCs of the transcriptome and first 10 PCs of ADTs. The WNN weights of mRNA and ADT modalities are listed in Supplementary Table 3.
Spatial cross-correlation analysis.
The spatial cross-correlation (bivariate Moran’s I) of each gene can be defined by either univariate or bivariate Moran’s I (Fig. 2g). In the univariate case, Moran’s I of any feature X was calculated as , where wij is a spatial weight matrix with wii = 0 and W = 𝛴wij, and N is the number of total spatial barcodes. The P-value of spatial autocorrelation (univariate Moran’s I) can be obtained by using mathematical derivations42. In the spatial cross-correlation (bivariate case)43, Moran’s I between feature X and Y was calculated as , and the P-value can be obtained by using mathematical derivations44.
Spatial clustering.
To identify spatially distinct cell populations in the breast cancer tumor microenvironment, we modified the current clustering scheme, which is solely based on expression or ADT data by incorporating the spatial information of each spatial barcode (Fig. 2c). We adopted a method that has been developed in the field of spatial statistics43, where multivariate spatial correlation (MSC) was defined as (Z is the z-score of the data matrix). This M matrix captures the spatial relationship between each gene across tissue spatial barcodes and is used for subsequent principal component analysis. This approach entails that the spatial correlation matrix (M) must be positive semidefinite, like the covariance matrix used in Principal Component Analysis (PCA), where the eigenvalues must be nonnegative. However, the M matrix is not positive semidefinite and can have negative eigenvalues as noted in the original publication. To resolve this, another spatial correlation45 matrix L with , such that , is positive semidefinite. By using this L matrix, we performed the singular value decomposition (SVD) in the same manner as PCA, and alternative sparse solutions can be obtained accordingly46. We termed this approach the Spatial Component Analysis (SCA) to imply its connection to canonical PCA. For clustering of the ADT data, we used the HNN distance weights as described in the Data visualization and the first 10 Spatial Components (SCs) as input to build the shared nearest neighbor graph and then clustered the spatial barcodes using the FindClusters function with resolution = 0.3. A more detailed analysis of SCA versus a canonical PCA for spatial data analysis can be found in the R package spots47 tutorial (https://stevexniu.github.io/spots). Clustering result and annotation are reported in Supplementary Table 3.
Differential expression analysis.
Differentially expressed (DE) genes for each cluster were calculated using Seurat’s FindAllMarkers function with default parameter or logfc.threshold = 0.1 for ADT data. For DE genes between Mac1- and Mac2-enriched regions, the top 5,000 variable genes with the highest dispersion values were selected using the FindVariableFeatures function (Fig. 2h). Gene Set Enrichment Analysis (GSEA; Fig. 2i) was performed using the fgsea v1.16 R package48 and GO term biological process gene sets (c5.bp.v7.1.symbols) with 10,000 permutations. M2 macrophage signatures genes were selected from GSE32164. The spleen germinal center signature genes were selected from GSE12366. The DE genes of spleen and breast cancer samples are listed in Supplementary Table 4 and Supplementary Table 5, respectively.
Comparison to SM-omics and Guilliams et al.
Data from SM-omics and Guilliams et al. were downloaded from the original publications16,17. We processed and normalized the data as previously described. The cell type and metadata were provided by the original publications.
Supplementary Material
Acknowledgments
We acknowledge J. Chew and Y. Yin from 10x Genomics Headquarters for critical discussions and K. Ganapathy for helping coordinate 10x Genomics and Weill Cornell interactions. X.N has the right to list himself first in author order on his CV. D.A.L. is supported by the Burroughs Wellcome Fund Career Award for Medical Scientists, Valle Scholar Award, Leukemia Lymphoma Scholar Award, the Sontag Foundation Distinguished Scientist Award, the Mark Foundation Emerging Leader Award, and the National Institutes of Health Director’s New Innovator Award (DP2-CA239065). This work was supported by the CEGS award (RM1 HG011014) and Emerson (NPT Charitable Grant).
Footnotes
Competing interests
M.S., P.M., and P.R. are current employees of 10x Genomics, Sweden. D.A.L. has served as a consultant for Abbvie, AstraZeneca, and Illumina, and is on the Scientific Advisory Board or equity holder of Mission Bio, Pangea, Alethiomics, and C2i Genomics; D.A.L. has received prior research funding from BMS, 10x Genomics, Ultima Genomics, and Illumina unrelated to the current manuscript. Other authors declare no competing interests.
Data and code availability
All raw data generated in this study have been deposited to GEO with the accession code GSE198353 and token kzknoeywtlahtgh. The R markdown files for SPOTS data analysis are included in the Supplementary Notes and also on Github (https://github.com/stevexniu/SPOTS-paper). High-resolution images of the tissues used in this study are available on the Figshare website (https://figshare.com/account/home#/projects/143019). To enable easy access to the SPOTS computational pipeline, the auxiliary R package “spots" can be found on the CRAN (https://CRAN.R-project.org/package=spots) and GitHub (https://github.com/stevexniu/spots) websites.
References
- 1.Tabula Muris Consortium et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azizi E et al. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell 174, 1293–1308.e36 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao T et al. Spatial genomics enables multi-modal study of clonal heterogeneity in tissues. Nature 601, 85–91 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vickovic S et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stickels RR et al. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat. Biotechnol 39, 313–319 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho C-S et al. Microscopic examination of spatial transcriptome using Seq-Scope. Cell 184, 3559–3572.e22 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gut G, Herrmann MD & Pelkmans L Multiplexed protein maps link subcellular organization to cellular states. Science 361, (2018). [DOI] [PubMed] [Google Scholar]
- 8.Radtke AJ et al. IBEX: A versatile multiplex optical imaging approach for deep phenotyping and spatial analysis of cells in complex tissues. Proc. Natl. Acad. Sci. U. S. A 117, 33455–33465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angelo M et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med 20, 436–442 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giesen C et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods 11, 417–422 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Walch A, Rauser S, Deininger S-O & Höfler H MALDI imaging mass spectrometry for direct tissue analysis: a new frontier for molecular histology. Histochem. Cell Biol 130, 421–434 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goltsev Y et al. Deep Profiling of Mouse Splenic Architecture with CODEX Multiplexed Imaging. Cell 174, 968–981.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Govek KW et al. Single-cell transcriptomic analysis of mIHC images via antigen mapping. Sci Adv 7, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang S et al. Combined protein and nucleic acid imaging reveals virus-dependent B cell and macrophage immunosuppression of tissue microenvironments. Immunity 55, 1118–1134.e8 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu Y et al. High-Spatial-Resolution Multi-Omics Sequencing via Deterministic Barcoding in Tissue. Cell 183, 1665–1681.e18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vickovic S et al. SM-Omics is an automated platform for high-throughput spatial multi-omics. Nat. Commun 13, 795 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guilliams M et al. Spatial proteogenomics reveals distinct and evolutionarily conserved hepatic macrophage niches. Cell 185, 379–396.e38 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ståhl PL et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016). [DOI] [PubMed] [Google Scholar]
- 19.Stoeckius M et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim HK & O’Neill HC Identification of Stromal Cells in Spleen Which Support Myelopoiesis. Front Cell Dev Biol 7, 1 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng H-W et al. Origin and differentiation trajectories of fibroblastic reticular cells in the splenic white pulp. Nat. Commun 10, 1739 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Béguelin W et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 23, 677–692 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Beyer A & Aebersold R On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell 165, 535–550 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Mimitou EP et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol 39, 1246–1258 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chávez-Galán L, Olleros ML, Vesin D & Garcia I Much More than M1 and M2 Macrophages, There are also CD169(+) and TCR(+) Macrophages. Front. Immunol 6, 263 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ravishankar B et al. Marginal zone CD169+ macrophages coordinate apoptotic cell-driven cellular recruitment and tolerance. Proc. Natl. Acad. Sci. U. S. A 111, 4215–4220 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagelkerke SQ et al. Red pulp macrophages in the human spleen are a distinct cell population with a unique expression of Fc-γ receptors. Blood Adv 2, 941–953 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Biancalani T et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valdés-Mora F et al. Single-cell transcriptomics reveals involution mimicry during the specification of the basal breast cancer subtype. Cell Rep. 35, 108945 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Keren L et al. A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell 174, 1373–1387.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Franklin RA et al. The cellular and molecular origin of tumor-associated macrophages. Science 344, 921–925 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elosua-Bayes M, Nieto P, Mereu E, Gut I & Heyn H SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 49, e50 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hao Y et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nam AS et al. Somatic mutations and cell identity linked by Genotyping of Transcriptomes. Nature 571, 355–360 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hudson WH & Sudmeier LJ Localization of T cell clonotypes using spatial transcriptomics. bioRxiv 2021.08.03.454999 (2021) doi: 10.1101/2021.08.03.454999. [DOI] [Google Scholar]
- 36.Roelli P, bbimber, Flynn B, santiagorevale & Gui G Hoohm/CITE-seq-Count: 1.4.2 (2019). doi: 10.5281/zenodo.2590196. [DOI] [Google Scholar]
- 37.Ord JK & Getis A Local spatial autocorrelation statistics: Distributional issues and an application. Geogr. Anal 27, 286–306 (2010). [Google Scholar]
- 38.Anselin L Local indicators of spatial association-LISA. Geogr. Anal 27, 93–115 (2010). [Google Scholar]
- 39.Getis A & Ord JK The analysis of spatial association by use of distance statistics. Geogr. Anal 24, 189–206 (2010). [Google Scholar]
- 40.Middleton L & Sivaswamy J Edge detection in a hexagonal-image processing framework. Image Vis. Comput 19, 1071–1081 (2001). [Google Scholar]
- 41.Haghverdi L, Lun ATL, Morgan MD & Marioni JC Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat. Biotechnol 36, 421–427 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moran PAP Notes on continuous stochastic phenomena. Biometrika 37, 17–23 (1950). [PubMed] [Google Scholar]
- 43.Wartenberg D Multivariate spatial correlation: A method for exploratory geographical analysis. Geogr. Anal 17, 263–283 (1985). [Google Scholar]
- 44.Czaplewski RL Expected Value and Variance of Moran’s Bivariate Spatial Autocorrelation Statistic for a Permutation Test. (U.S. Department of Agriculture, Forest Service, Rocky Mountain Forest and Range Experiment Station, 1993). [Google Scholar]
- 45.Lee S-I Developing a bivariate spatial association measure: An integration of Pearson’s r and Moran's I. J. Geogr. Syst 3, 369–385 (2001). [Google Scholar]
- 46.Robert Frost H Eigenvectors from Eigenvalues Sparse Principal Component Analysis. J. Comput. Graph. Stat 31, 486–501 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Niu X spots: Spatial Component Analysis. (2022). doi: 10.5281/zenodo.6918175. [DOI] [Google Scholar]
- 48.Korotkevich G, Sukhov V & Sergushichev A Fast gene set enrichment analysis. bioRxiv 060012 (2019) doi: 10.1101/060012. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.